SUMMARY
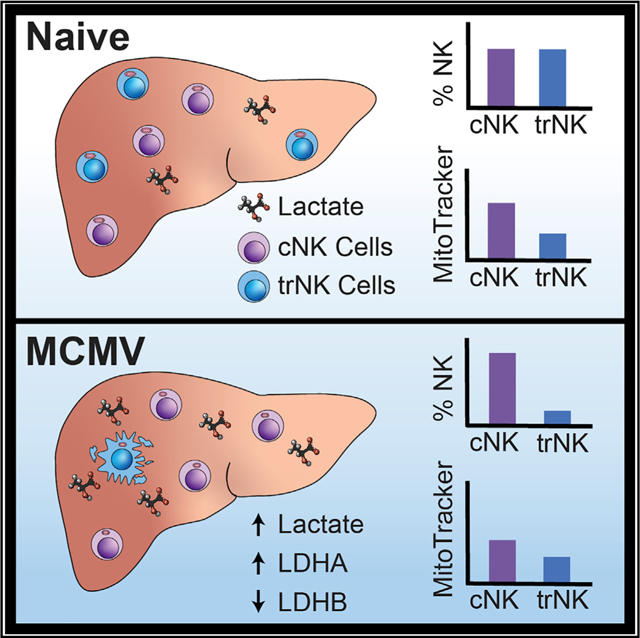
The liver harbors two main innate lymphoid cell (ILC) populations: conventional NK (cNK) cells and tissue-resident NK (trNK) cells. Using the MCMV model of infection, we find that, in contrast to liver cNK cells, trNK cells initially undergo a contraction phase followed by a recovery phase to homeostatic levels. The contraction is MCMV independent because a similar phenotype is observed following poly(I:C)/CpG or α-GalCer injection. The rapid contraction phase is due to apoptosis, whereas the recovery phase occurs via proliferation in situ. Interestingly, trNK cell apoptosis is not mediated by fratricide and not induced by liver lymphocytes or inflammatory cytokines. Instead, we find that trNK cell apoptosis is the consequence of an increased sensitivity to lactic acid. Mechanistic analysis indicates that trNK cell sensitivity to lactate is linked to impaired mitochondrial function. These findings underscore the distinctive properties of the liver-resident NK cell compartment.
In Brief
In this study, Dodard et al. evaluate the kinetics of the liver NK cell compartment in response to viral infection. They show that, in contrast to conventional NK cells, tissue-resident NK cells undergo apoptosis, which is due to a higher sensitivity to lactic acid and impaired mitochondrial function.
Graphical Abstract
INTRODUCTION
Natural killer (NK) cells are crucial members of the innate immune system and play key roles in early control of pathogens and tumor development by facilitating synergy between the innate and adaptive immune responses (Babić et al., 2011). NK cell subsets are often classified as members of the innate lymphoid cell (ILC) family, which is comprised of NK cells, ILC1, ILC2, ILC3, or LTi cells, depending on their transcription factor requirements and cytokine production (Vivier et al., 2018). NK cells require Tbx21 (T-bet) and Eomesodermin (Eomes); ILC1 and tissue-resident NK (trNK) cells require T-bet; ILC2 cells require GATA3; and ILC3 and LTi cells require RORγt (Colonna, 2018; Fang and Zhu, 2017; Ishizuka et al., 2016; Sonnenberg and Artis, 2015; Vivier et al., 2018). A role of ILCs has been shown in several biological settings, including tissue repair, inflammation, and immune defense (Artis and Spits, 2015; Cortez et al., 2015; McKenzie et al., 2014; Serafini et al., 2015). Within the past decade, extensive research has revealed the varied lineage, differentiation, and developmental pathways of ILCs. Often, these variations are dependent on the environmental cues of peripheral sites, the microbiome, and cytokine stimulation (Daussy et al., 2014; Gury-BenAri et al., 2016; Marçais et al., 2013; Peng et al., 2016).
Most of our knowledge regarding NK cells was derived from studies using human PBMCs and mouse splenic NK cells, which are referred to as circulating or conventional NK (cNK) cells (Sojka et al., 2014; Yokoyama et al., 2013). Recent studies have shown that ILC subsets can be distinguished based on transcription factor expression. cNK cells are positive for T-bet and Eomes, whereas trNK cells express T-bet but are negative for Eomes (Daussy et al., 2014; Sojka et al., 2014). trNK cells are non-circulating and, in addition to the liver, reside in peripheral organs such as the salivary glands, kidneys, and uterus (Erick and Brossay, 2016). Earlier studies found that a large portion of liver NK cells differed from the mature cNK cell phenotype because of their lack of DX5 expression and exclusive TRAIL expression (Crispe, 2009; Gordon et al., 2012; Kim et al., 2002; Takeda et al., 2005). However, the absence of DX5 was of limited use in identifying trNK cells because immature cNK cells also lack DX5 expression (Yokoyama et al., 2013). Because of these complications, studies performed on NK cells within the liver were not able to distinguish between cNK and trNK cells until recently (Peng et al., 2013; Weizman et al., 2017).
Regarding their function, hepatic trNK cells have been shown to produce interferon γ (IFN-γ) more rapidly than cNK cells during murine cytomegalovirus (MCMV) infection and to be beneficial during infection (Weizman et al., 2017). In contrast, they have been shown to negatively regulate the antiviral responses of hepatic T cells during lymphocytic choriomeningitis virus (LCMV) infection (Zhou et al., 2019). Previous work from our lab examined the immune response of the liver NK cell compartment following MCMV infection (Robbins et al., 2004). However, when these studies were performed, the markers to separate the heterogeneous ILC population were not available. In the present study, we revisit these studies and characterize the kinetics and immune response of each of the hepatic NK cell populations to viral challenge and other stimuli. Surprisingly, we found that, following a variety of biological stresses, trNK cells undergo rapid apoptosis, which is the consequence of an increased sensitivity to lactic acid and impaired mitochondrial function. This is followed by a recovery phase to homeostatic levels.
RESULTS
Hepatic cNK and trNK Cells Have Different Kinetic Responses after MCMV Infection
Our laboratory previously characterized the kinetics of the liver NK cell compartment in response to viral infection (Robbins et al., 2004). However, these studies were performed before the description of different populations of NK cells (Peng et al., 2013). Therefore, we sought to revisit these studies to investigate hepatic NK cell populations individually, using markers specific for cNK cells and trNK cells. Following cardiac puncture and liver perfusion, we found that hepatic cNK cell and trNK cell populations were equally represented (Figure 1A–C). The reported number and frequency of trNK cells in the liver have not been consistent across the literature, most likely because of the fact that liver perfusion and cardiac puncture are not always performed. Following MCMV infection, we found that liver cNK cells expand, with a peak in absolute numbers on day 7 post-infection, and then contract to steady-state levels on day 14 post-infection (Figures 1A–1C). This kinetic profile mostly parallels the splenic NK cell compartment during the immune response to MCMV infection (Robbins et al., 2004; Sun et al., 2009). In contrast, we unexpectedly found that trNK cells initially undergo a rapid contraction phase before returning to homeostatic cell numbers by day 7 and frequency by day 14 post-infection (Figures 1A–1C). Taken together, these data indicate that liver cNK and trNK cells have inverse kinetic profiles in response to MCMV infection.
Figure 1. Liver NK Cell Populations Have Distinct Kinetic Responses to MCMV Infection.
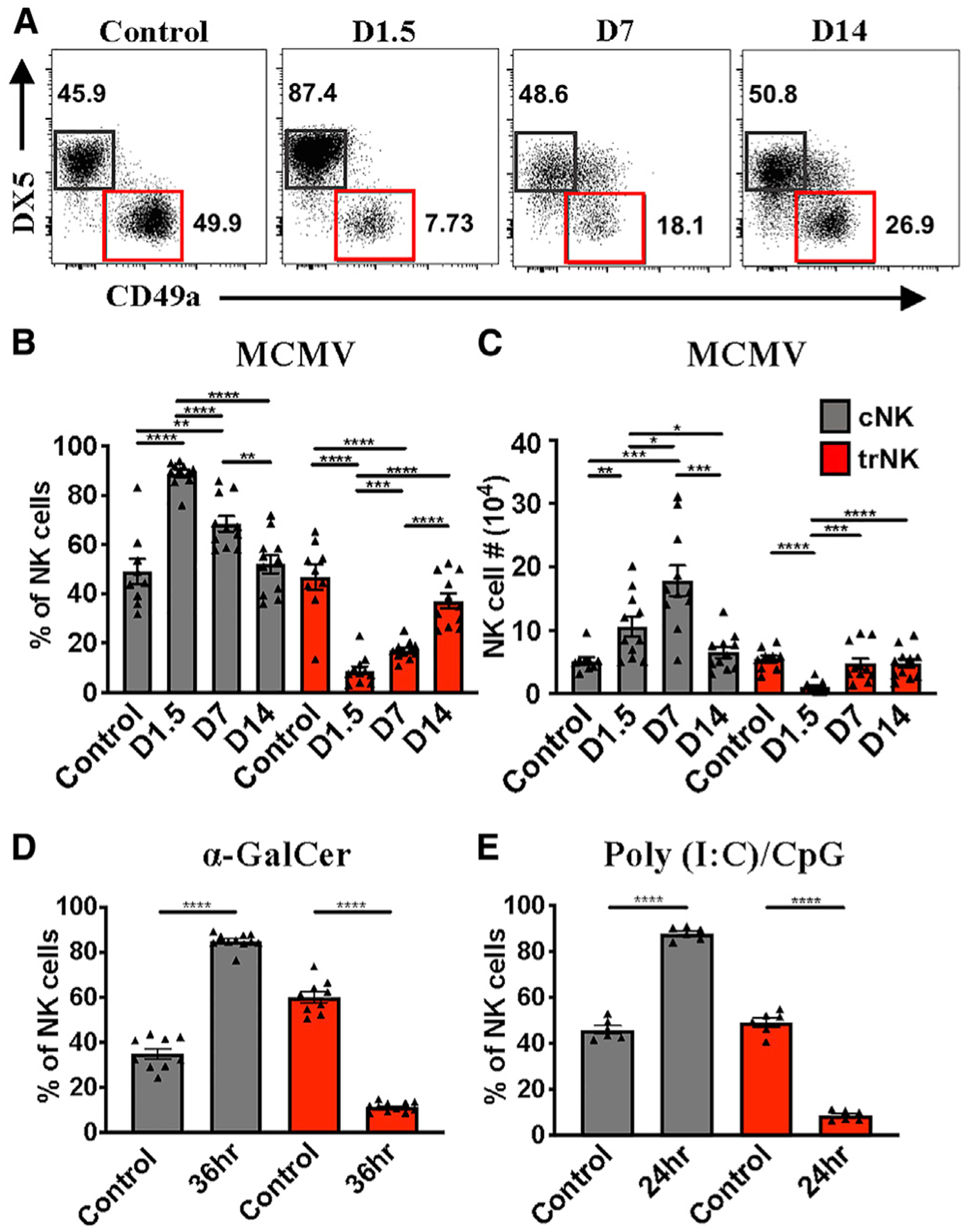
(A) Representative staining of liver cNK (gray) and trNK (red) cells at the indicated time points post-MCMV infection in C57BL/6 mice. Samples are first gated on TCRβ−CD3−NK1.1+ cells.
(B and C) Frequency (B) and absolute numbers (C) of liver cNK and trNK cell populations 0, 1.5, 7, or 14 days post-infection (n = 9–11).
(D) Frequency of liver cNK and trNK populations 36 h after α-GalCer treatment (n = 9–10).
(E) Frequency of liver cNK and trNK cell populations 24 h after poly(I:C)/CpG treatment (n = 6).
Data are representative of (A) or pooled from 2–3 experiments (B–E); error bars indicate SEM. *p < 0.05, **p < 0.01,***p < 0.001, and ****p < 0.0001.
The Hepatic trNK Compartment Cellularity Change Is Not Specific to MCMV Infection
Splenic NK cell expansion during MCMV has been well characterized and is dependent on increased sensitivity of Ly49H+ NK cells, primarily to interleukin-15 (IL-15), following Ly49H engagement with its MCMV-derived ligand m157 (Sun et al., 2009). Because trNK cells do not express Ly49H, we expected that they would behave like Ly49H− NK cells. Regarding the unexpected contraction phase, we hypothesized that it was induced directly by MCMV viral proteins or as an indirect consequence of the viral infection. To distinguish between these two possibilities, we utilized α-galactosylceramine (α-GalCer) and poly(I:C)/CpG to indirectly induce NK cell activation via inflammatory cytokines. Interestingly, trNK cells also contract rapidly in response to α-GalCer (Figures 1D and S1) and poly(I:C)/CpG injection (Figure 1E). From these data, we conclude that viral proteins are dispensable for liver trNK cell contraction.
Hepatic trNK Cells Proliferate and Home to the Liver
The early decrease in trNK cell number following activation could also potentially be due to their migration to other organs. However, we considered this unlikely because liver trNK cells do not circulate throughout the cardiovascular system like cNK cells (Björkström et al., 2016; Peng et al., 2013; Sojka et al., 2014). In support of these findings, we found that liver trNK cells predominantly homed to the liver following adoptive transfer of congenic liver trNK cells into Rag2−/−IL-2Rγ−/− mice (Figure 2A). When we examined other organs, such as the spleen and blood, few transferred cells were found (Figure 2A). We next investigated whether the cells that home to the liver proliferated. Using cell proliferation dye (CPD), we found that trNK cells proliferated, but not to the extent of their cNK cell counterparts (Figures 2B and 2C). Taken together, our data demonstrate that liver trNK cells home to, contract, and proliferate within the liver microenvironment.
Figure 2. Liver trNK Cells Home to/Proliferate within the Liver and Undergo Apoptosis after MCMV Infection.
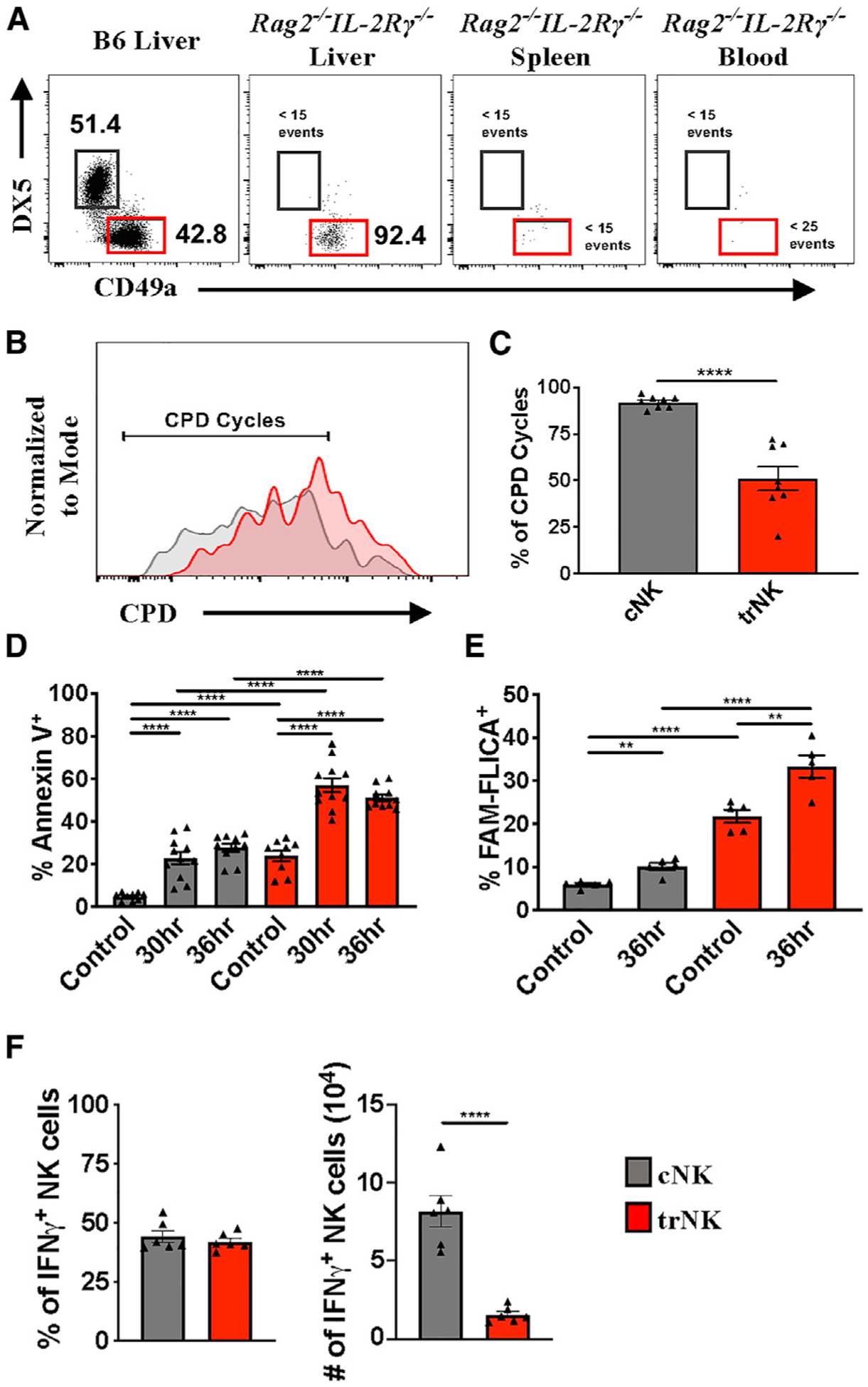
(A) Representative staining of liver trNK cells (red) 10 days post-transfer into Rag2−/−IL-2Rγ−/− mice.
(B) Representative proliferation of liver cNK (gray) and trNK cells (red) on day 4 post-transfer using cell proliferation dye (CPD).
(C) Frequency of liver cNK and trNK cells greater than indicated cycle on day 4 post-transfer (n = 8).
(D) Frequency of Annexin V+ liver cNK and trNK cell populations 30 and 36 h post-MCMV infection (n = 9–11).
(E) Frequency of FAM-FLICA+ liver cNK and trNK cell populations 36 h post-MCMV infection (n = 5).
(F) Frequency and numbers of liver cNK and trNK cells producing IFN-γ 36 h following MCMV infection (n = 6).
Data are representative of (A and B) or pooled from 2–3 experiments (C–F); error bars indicate SEM. *p < 0.05, **p < 0.01,***p < 0.001, and ****p < 0.0001.
Hepatic trNK Cells Undergo Apoptosis after MCMV Infection
Because we excluded migration to other tissues, we hypothesized that the activation-induced trNK cell disappearance was due to cell death. To investigate this possibility, we measured liver NK cell apoptosis following MCMV infection. We found that, in comparison with cNK cells, trNK cells preferentially undergo apoptosis 30 and 36 h post-MCMV infection, as indicated by Annexin V staining (Figure 2D). These findings were further confirmed using an assay for active caspases (FAM-FLICA) (Figure 2E). Similar results were obtained following α-GalCer treatment (Figure S1D). Altogether, these data demonstrate the distinctive sensitivity of the liver trNK cell compartment to activation and/or stress.
Liver trNK Cells Respond Early and Transiently within the Liver Microenvironment
Given the sensitivity of trNK cells to stimulation and their high rate of apoptosis, we reasoned that trNK cells may not contribute significantly to the antiviral immune response at early time points post-infection. However, it has been shown recently that trNK cells confer early host protection at initial sites of infection, but this was observed using a hydrodynamic injection method (Weizman et al., 2017). Using a traditional intraperitoneal (i.p.) MCMV infection model, we found that the frequency of IFN-γ+ trNK cells and cNK cells was comparable on day 1.5 after infection (Figure 2F). However, given the high rate of apoptosis within the trNK cell subset, IFN-γ+ cNK cells outnumbered IFN-γ trNK cells (Figure 2F). Similarly, the number of cNK cells producing IFN-γ was significantly higher 6 h after α-GalCer injection (data not shown). Taken together, these data indicate that the trNK effector response is rapid but less powerful than the cNK cell response at this time point.
Liver trNK Cell Disappearance Is Independent of IL-1, IL-12, IL-18, and IFN-γ
To gain insight into the mechanism leading to disappearance of the trNK cell compartment, we investigated the role of the inflammatory cytokines IL-12 and IL-18, which are produced at high levels during MCMV infection. To do this, we used mice conditionally deficient for IL-12Rβ2 and Myd88 in the NK lineage. Although beyond the scope of this paper, we did not find any obvious differences in NK cell development in these mice (Figures S2A and S2B and data not shown). NCR.Cre.IL-12Rβfl/fl, NCR.Cre.MyD88fl/fl, and corresponding heterozygous littermate controls were infected with MCMV for 36 h. Unexpectedly, we found that the trNK cell compartment contracted in frequency and number in all tested genotypes (Figures 3A, 3B, and S2C). Because the IL-1 cytokine family, which includes IL-18, signals through the Myd88 adaptor, these data indicate that IL-1, IL-12, and IL-18 are individually dispensable for the observed trNK cell phenotype. Notably, blocking IFN-γ also had no effect on trNK cell loss (Figure S2D).
Figure 3. Liver trNK Cell Disappearance Is Independent of IL-1, IL-12, IL-18, and Type 1 IFN Signaling.
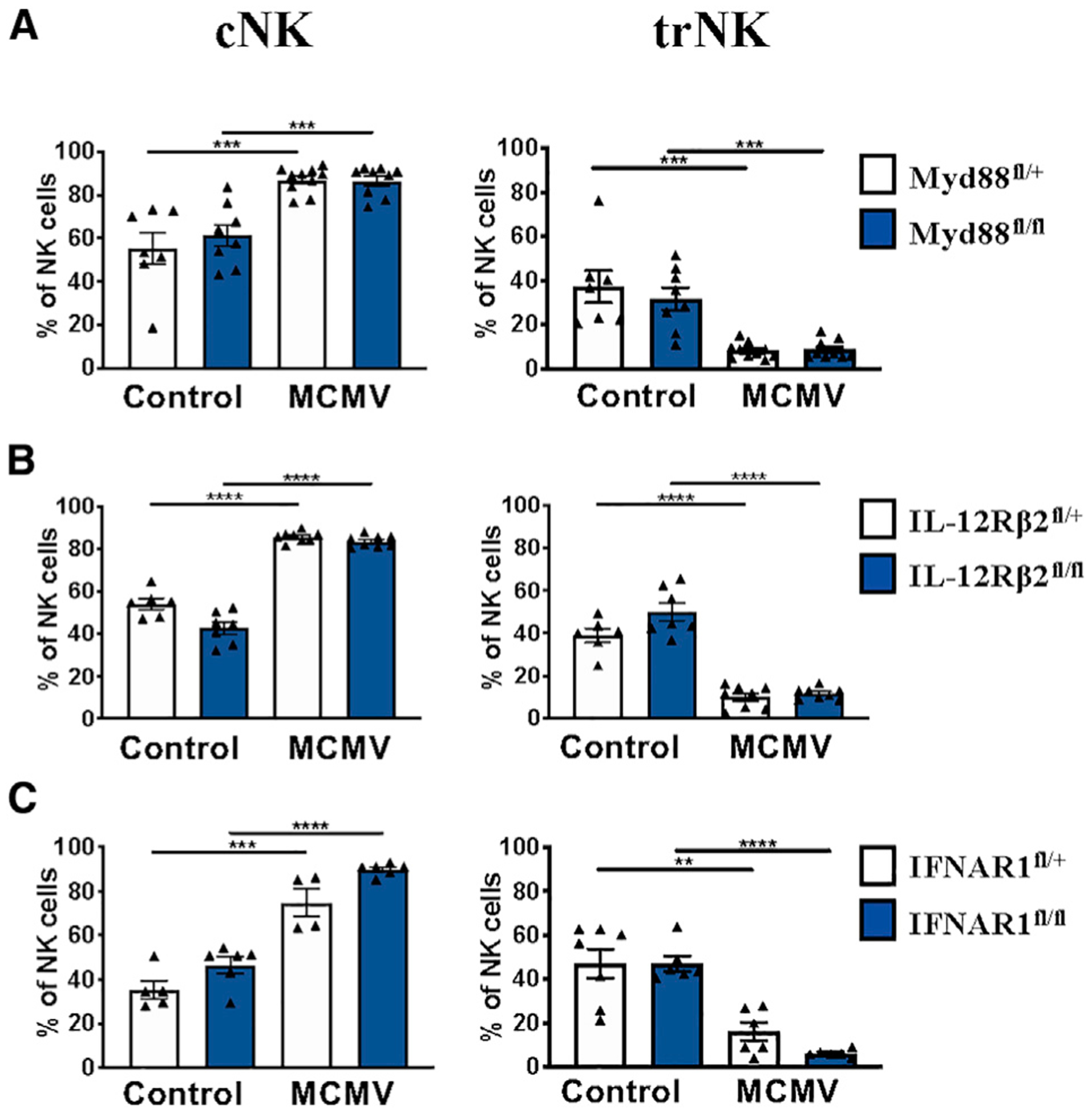
(A) Frequency of liver NK populations from MyD88fl/fl and MyD88fl/+ controls 36 h post-MCMV infection (n = 7–10).
(B) Frequency of liver NK cell populations from IL12Rβ2fl/fl and IL12Rβ2fl/+ controls 36 h post-MCMV infection (n = 6–8).
(C) Frequency of liver NK populations from IFNAR1fl/fl and IFNAR1fl/+ controls 36 h post-MCMV infection (n = 4–6).
Data are pooled from 2–3 experiments (A–C); error bars indicate SEM. *p <0.05, **p < 0.01,***p < 0.001, and ****p < 0.0001.
trNK Cell Apoptosis Is Not due to Killing by Other Lymphocytes
It has been shown that IFNAR-deficient NK cells exhibit more apoptosis compared with wild-type NK cells because of fratricide (Madera et al., 2016). We therefore hypothesized that trNK cells would be even more apoptotic in the absence of type 1 IFN signaling. Using mice that are conditionally deficient for the type I IFN receptor in the NK cell lineage, we found that the trNK compartment is equally reduced compared with littermate controls (Figure 3C), excluding a role of type I IFN. In addition, trNK cells unable to respond to type I IFN and IL-12 also contracted (Figure S2E).
We next investigated whether TRAIL was responsible for trNK cell apoptosis via fratricide. To this end, we took advantage of mice that are TRAIL deficient. We (Figures S2F and S2G) and others (Almeida et al., 2018; Sheppard et al., 2018; Turchinovich et al., 2018) found that, when Ncr1 is absent, trNK cells no longer express TRAIL. Using Ncr1gfp mice, in which an IRES-GFP replaces Ncr1, abolishing Ncr1 expression, we found that TRAIL-deficient trNK cells also underwent apoptosis (Figure S2H).
Liver trNK cell apoptosis could be mediated by other liver lymphocyte subsets. For instance, during LCMV infection, cNK cells have been shown to regulate the CD4+ T cell compartment (Waggoner et al., 2011). To explore the potential of trNK cell lysis mediated by adaptive lymphocytes within the liver microenvironment, Rag1−/− mice were infected with MCMV for 36 h. We found that the trNK cell kinetics in infected Rag1−/− mice were comparable with C57BL/6 mice, ruling out a role of T cells and B cells in trNK cell apoptosis (Figure S2I). We next investigated whether trNK cell lysis was mediated by cNK cells. We tested this indirectly, following adoptive transfer of trNK cells into Rag2−/−IL-2Rγ−/− mice. In the recipient mice, the only lymphocytes present were the transferred trNK cells. The recipient mice were allowed to reconstitute for 6 days before MCMV infection. On day 1.5 post-infection, we found that liver trNK cells also contracted, as indicated by total cell number (Figure S2J). Taken together, we can conclude that liver trNK cell disappearance is not caused by apoptosis induced by other lymphocytes.
trNK Apoptosis Is the Consequence of a Higher Sensitivity to Lactic Acid
Inflammation results in increased release of lactate (Haas et al., 2015). In humans, lactate is a metabolite of glycolysis, which can blunt tumor immunosurveillance by T and NK cells because of increased apoptosis. This cell death occurs as a result of impaired mitochondrial function (Harmon et al., 2019). Because MCMV infection leads to liver inflammation, we first measured lactate levels in the liver on day 1.5 after infection. We found that MCMV-induced liver inflammation resulted in an increased level of lactate (Figure 4A). We then measured liver levels of lactate dehydrogenase (LDH), which mediates bidirectional conversion of pyruvate and lactate, using qRT-PCR analysis of global liver LDH RNA transcripts. LDH is composed of two different subunits, LDHA and LDHB. LDHA catabolizes pyruvate to lactate, whereas LDHB converts lactate to pyruvate (Doherty and Cleveland, 2013). In agreement with the observed lactate increase, we found elevated LDHA levels and, conversely, reduced LDHB levels (Figure 4B). Interestingly, RNA sequencing (RNA-seq) analysis of sorted trNK cells shows that they appear to increase their endogenous LDHB after infection (Figure S3B), possibly to cope with the elevated lactate levels.
Figure 4. trNK Apoptosis Is the Consequence of Higher Sensitivity to Lactic Acid.
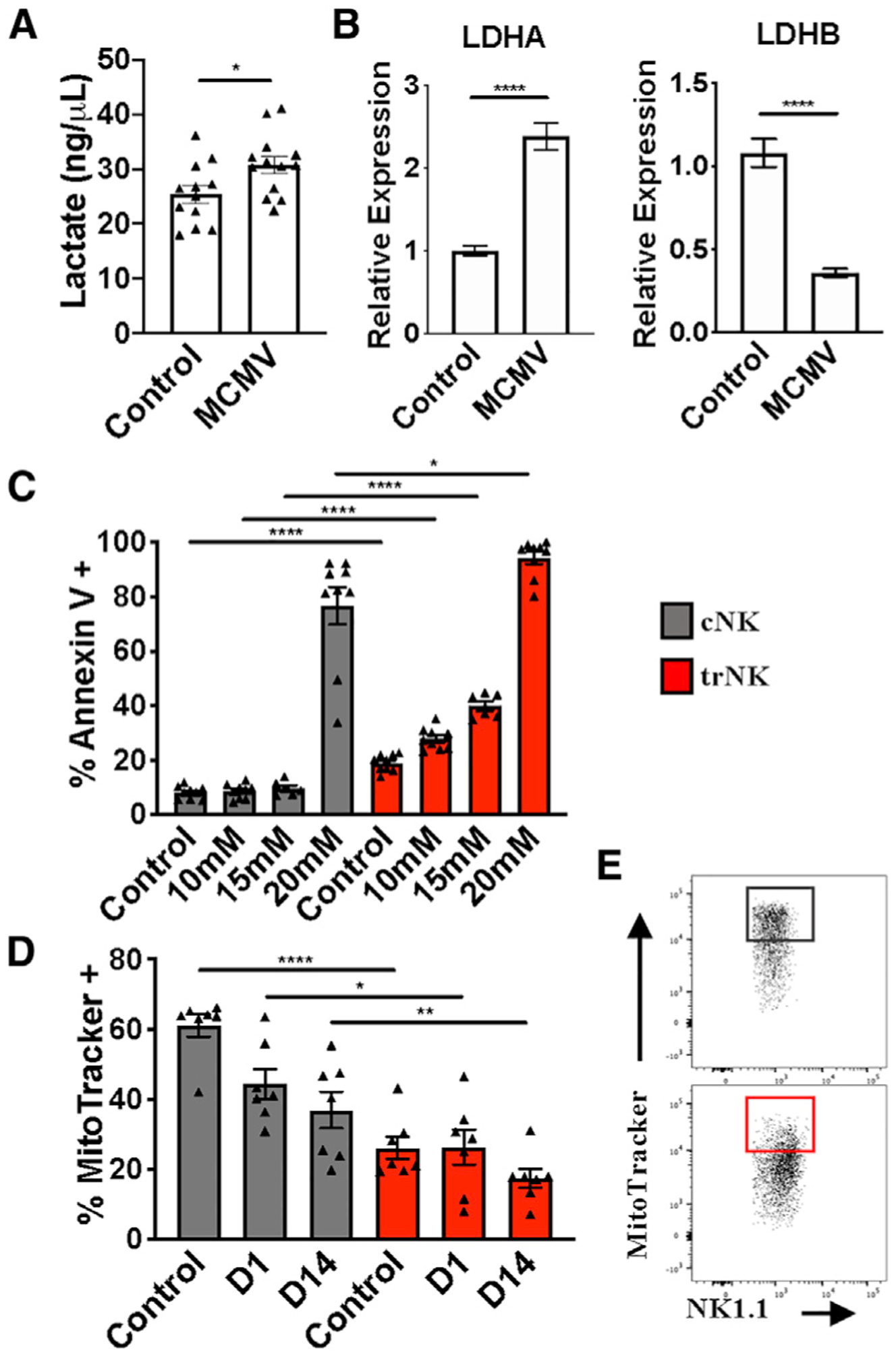
(A) Liver lactate levels 36 h post-MCMV infection (n = 12–13).
(B) LDH RNA transcript expression at steady state and on day 1.5 post-MCMV infection (representative of two experiments, n = 5).
(C) Frequency of Annexin V+ liver cNK (gray) and trNK (red) cells following 4-h incubation at the indicated concentrations of lactic acid (n = 9).
(D) Frequency of MitoTracker-positive liver cNK (gray) and trNK (red) cells at steady state and on days 1 and 14 post-MCMV infection (n = 7).
(E) Representative gating of MitoTracker+ frequencies on liver cNK (gray) and trNK (red) cells.
Data are representative of (E) or pooled from 2–3 experiments (A–D); error bars indicate SEM. *p < 0.05, **p < 0.01,***p < 0.001, and ****p < 0.0001.
We then compared the sensitivity of liver cNK cells and trNK cells with increased doses of lactate in vitro. We found that lactate preferentially induced apoptosis of trNK cells (Figure 4C). Importantly, acidification alone cannot account for the observed effect because a similar pH level attained with HCl (hydrochloric acid) addition did not increase apoptosis (Figure S3A). These data indicate that MCMV-induced lactate levels in the liver differentially affect trNK and cNK cell apoptosis. Interestingly, analysis of the RNA-seq data (Quatrini et al., 2018; Robinette et al., 2015), comparing liver trNK cells and cNK cells at steady state and following MCMV infection, reveals expression differences in mitochondrial function, apoptosis, and reactive oxygen species (ROS) production pathways, which are all linked to lactate metabolism (Figure S3B). Notably, mitochondrial carbonic anhydrase 5b (Car5b) is expressed approximately 16-fold more in liver cNK cells than trNK cells (Figures S3B and S3C), suggesting that mitochondrial CO2 conversion is impaired in trNK cells. In support of this hypothesis, using MitoTracker, we found that mitochondrial membrane potential is lower in liver trNK cells than in cNK cell counterparts at steady state and on day 1 post-infection. This trend is maintained even on day 14, when infection is cleared from the liver (Figures 4D and 4E). Overall, the data demonstrate that the trNK cell rapid contraction phase is linked to impaired mitochondrial function, which leads to a higher sensitivity to lactate.
DISCUSSION
Because of the relatively recent characterization of liver trNK cells, their exact roles have not been clearly defined. Some studies have shown evidence of a protective role during viral infection, whereas others have demonstrated a regulatory role of these cells (Li et al., 2017; Weizman et al., 2017; Zhou et al., 2019). Early reports have also shown that liver trNK cells develop a memory-like phenotype in response to skin contact hypersensitivity (O’Leary et al., 2006; Paust et al., 2010). Approximately 15 years ago, our lab characterized the liver NK cell kinetic response during MCMV infection (Robbins et al., 2004). Here we revisited these findings using markers that allowed us to distinguish trNK cells from cNK cells. We found an unexpected kinetic response of hepatic trNK cells following activation from viral challenge or stimulation. Liver trNK cells rapidly contract in number and frequency via apoptosis because of inflammatory stimuli. Because these cells rarely migrate to other tissues (Sojka et al., 2014; Figure 2A), it is likely that they are replenished from cells that survived in situ, which is supported by the active proliferation observed (Figures 2B and 2C). We initially thought that fratricide was the cause of the trNK cell contraction, as reported by others for cNK cells (Madera et al., 2016). However, using a variety of immunodeficient animals, we ruled out this pathway. We then reasoned that MCMV-induced inflammatory cytokines were specifically affecting liver trNK cells. Unexpectedly, contraction of the trNK cell compartment was also observed in mice that are conditionally deficient for the main inflammatory cytokine receptors (IL-12R, IFNAR, IL-18R, and IL-1R) in the NK cell lineage. Instead, our data imply that inflammation indirectly targets liver trNK cells through an increase in lactate concentration, caused by the higher sensitivity of trNK cells to lactate compared with their cNK counterparts. Our data also show that lactate accumulation may occur with many different insults to the liver, resulting in an early contraction phase of the trNK cell subset. Together with a recent study (Harmon et al., 2019), our work suggests that the increased susceptibility of trNK cells to lactate is due to their impaired mitochondrial function. In agreement with these findings, the RNA-seq data suggest that mitochondrial CO2 production is impaired in trNK cells, which would lead to a greater sensitivity to acidic changes. Importantly, Car5b expression is strongly reduced in trNK cells (Figure S3B). Mitochondrial carbonic anhydrases are required to convert CO2 to bicarbonate, which is necessary to provide resistance to pH changes in the acidic and basic directions (Shah et al., 2013). Understanding the role of mitochondrial carbonic anhydrases in trNK cells warrants further inquiry. Another important question is the lactate source. Although it is beyond the scope of this manuscript, it is clear that B cell and T cell lymphocyte lactate production is dispensable because trNK cells still undergo apoptosis in RAG-1-deficient mice. Notably, the lactate transporters MTC1 and MTC4 are poorly expressed in liver trNK cell and cNK cells, as reported by the Immunological Genome Project (http://www.immgen.org/), suggesting that these cells are not equipped to deal with lactate excess.
Regarding effector functions, it has been reported that trNK cells produce IFN-γ before cNK cells during MCMV infection and confer early host protection (Weizman et al., 2017). Our data confirm that trNK cell IFN-γ production precedes cNK cell IFN-γ production during MCMV infection. However, we found that the main IFN-γ producers are cNK cells. Differences observed with a previous study could be due to the route of infection: intraperitoneal (this study) versus hydrodynamic (Weizman et al., 2017). The high sensitivity of liver trNK cells to initial stimuli suggests that their activation is tightly regulated. Overactivation of the NK cell compartment may be beneficial in controlling pathogens and tumor development but may also induce hepatocellular damage and inhibit liver regeneration.
Collectively, the data presented here illustrate that hepatic trNK cells have distinctive properties and functions, which differ from the well-studied cNK cell population. Given the parallels between human and murine trNK cells, therapeutic agents capable of modulating the functions of liver trNK cells and reducing lactate levels during disease may help retain hepatic trNK cell function, affecting many different areas of disease research.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests for information and reagents can be obtained from the lead contact, Laurent Brossay (Laurent_brossay@brown.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
This study did not generate any unique datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6 and B6.SJL mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and Taconic Biosciences (Germantown, NY), respectively. Rag2−/−IL-2Rγ−/− mice were purchased from Taconic Biosciences and maintained in-house. Rag1−/−, B6;129P2(SJL)-Myd88tm1Defr/J (Myd88fl/fl), B6;129-NCR1tm10man/J (NCR1gfp), and B6(Cg)-Ifnar1tm1.1Ees/J (Ifnar1fl/fl) mice were purchased from The Jackson Laboratory. IL-12Rβ2fl/fl mice were generated at the Brown University Transgenic Facility. Both age and sex-matched mice (6 − 14 weeks) were used for this study. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Brown University, and were conducted in accordance with institutional guidelines for animal care. All mice were maintained at Brown University in AAALAC-accredited, pathogen-free facilities.
Generation of IL-12Rβ2fl/fl mice
IL-12Rβ2fl/fl mice were generated at the Brown University Transgenic Facility using a PG00231_Z_3_B02 plasmid construct purchased from the Knockout Mouse Project (KOMP) Repository. Following linearization of the plasmid with AsiS1 enzyme, the plasmid was electroporated in C57BL/6 embryonic stem cells (ES cells). ES cells were selected for homologous recombination by Southern blot and long-range PCR. Selected ES cells were microinjected into albino C57BL/6 blastocysts at the Brown University Mouse Transgenic and Gene Targeting Facility. Chimeric offspring were genotyped for the presence of the IL-12Rβ2 transgene by amplifying the neomycin probe. Founder mice were then crossed to ACTB:FLPe B6J mice (Jackson Labs, #005703) to delete the FRT-Neo-FRT cassette. Genotyping of IL-12Rβ2-floxed mice was performed using primers with amplicons of a nucleotide product from the wild-type allele and a nucleotide product from the targeted allele. Successful deletion of the neomycin cassette was also confirmed by PCR.
Mouse infection and treatments
Mice were infected intraperitoneally (i.p.) with 5 × 104 plaque forming units (PFU) of MCMV-RVG102. MCMV-RVG102 stocks were grown in vivo and isolated from salivary glands, as previously described (Anderson et al., 2019). Mice were treated i.p. with 2 μg α-Galactosylceramine (α-GalCer) or 100 μg of Poly (I:C) and 50 μg CpG oligodeoxynuleotides.
METHOD DETAILS
Isolation of murine lymphocytes
Mice were sacrificed by cervical dislocation and cardiac puncture following isoflurane treatment. Livers were perfused with 1% PBS-serum prior to removal. Hepatic lymphocytes were obtained by homogenizing with the E.01 program on a GentleMACS (Miltenyi Biotec), and filtered through nylon mesh. Samples were washed three times in 1% PBS-serum and overlayed on a two-step discontinuous Percoll gradient (GE Healthcare Bio-Sciences). Lymphocytes were harvested from the gradient interface and washed once in 1% PBS-serum.
Reagents, Abs, and flow cytometry analysis
Samples were resuspended in 1% PBS-serum and labeled with mAbs for 20 minutes on ice, in the dark. For intracellular staining of cytokines, cells were first surface stained, followed by fixation and permeabilization with cytofix/cytoperm and 1X PermWash (BD Biosciences). For intranuclear staining, cells were surface stained, then fixed and permeabilized using the FoxP3 transcription factor staining buffer set (Invitrogen). Apoptosis was evaluated by staining lymphocytes with Annexin V+ antibody in Annexin V binding buffer for 15 minutes at room temperature, in the dark. Additionally, apoptosis was evaluated via caspase activity using FAM-FLICA Poly Caspase Assay kit (ImmunoChemistry Technologies) according to the manufacturer’s protocol. For mitochondrial staining, lymphocytes were stained with MitoTracker Green (Invitrogen) for 30 minutes in accordance with manufacture recommendations. Events were collected on a FACSAria III (BD), and data were analyzed using FlowJo (FlowJo, LLC).
Adoptive transfer of NK cells
Under sterile conditions, NK cells were sorted from the liver of B6.SJL (CD45.1+) congenic mice. A FACSAria III cell sorter (BD) was used to purify hepatic cNK cells (NK1.1+ CD3− TCRb− DX5+ CD49a−) and trNK cells (NK1.1+ CD3− TCRb− DX5− CD49a+). Donor cells were intravenously injected into recipient Rag2 / IL-2Rγ−/− or C57BL/6 (CD45.2+) mice. Recipient mice were sacrificed for experiments at indicated days post-injection.
In vivo proliferation analysis
Under sterile conditions, hepatic lymphocytes from naive B6.SJL (CD45.1+) mice were labeled for 10 minutes at 37°C, in the dark, with 10 μM eFluor 450 Cell Proliferation Dye (eBioscience) in PBS. Cells were subsequently stained with specific mAbs and adoptively transferred into Rag2−/−IL-2Rγ−/− mice. Proliferation of specific NK cell subsets was analyzed in recipient livers on day 4 post-injection.
In vitro lactic acid incubation
Hepatic lymphocytes were incubated in RPMI media (GE LifeSciences) with indicated concentrations of L-Lactic Acid (Sigma-Aldrich) for 4 hours at 37°C.
Quantitative real-time PCR (qRT-PCR)
Mouse liver tissue was homogenized using Buffer RLT Plus (QIAGEN) according to manufacturer recommendations. Isolation of RNA was performed using the RNeasy Plus Mini Kit (QIAGEN). RNA was then quantified and analyzed for integrity using NanoDrop 2000/2000c (ThermoFisher). Complimentary DNA was synthesized using a iSCRIPT reverse transcription supermix (Bio-Rad) and amplified using iTaq Universal SYBR Green Supermix (Bio-Rad).
Lactate measurements
Livers were homogenized with the E.01 program on a GentleMACS (Miltenyi Biotec). Samples were spun down, and the supernatant was collected and filtered through Amicon Ultra – 0.5mL centrifugal filter units (Millipore-Sigma). Lactate measurements were acquired using a Lactate Assay kit (Sigma-Aldrich) and read using a Synergy HT (BioTek) plate reader.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were achieved with Prism Version 7.0 (GraphPad Software). Unpaired two-tailed Student t tests were used to compare cell populations from different mice. Differences considered significant when p < 0.05 (*), very significant when p < 0.01 (**), highly significant when p < 0.001 (***), and extremely significant when p < 0.0001 (****).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC anti-mouse CD49a | Thermo Fischer Scientific | Cat#: 17-0454-82; RRID: AB_469400 |
| APC anti-mouse CD11b | Thermo Fischer Scientific | Cat#: 17-1031-80; RRID: AB_1106993 |
| APC-eF780 anti-mouse CD45 | Thermo Fischer Scientific | Cat#: 47-0451-82; RRID: AB_1548781 |
| APC-eF780 anti-mouse CD45.1 APC-eF780 anti-mouse CD45.2 | Thermo Fischer Scientific Thermo Fischer Scientific | Cat#: 47-0453-82; RRID: AB_1582228 Cat#: 47-0454-82; RRID: AB_1272175 |
| BV510 anti-mouse TCRb | BioLegend | Cat#: 109234; RRID: AB_2562350 |
| BV570 anti-mouse CD45 | BioLegend | Cat#: 103136; RRID: AB_2562612 |
| BV605 anti-mouse CD3 | BioLegend | Cat#: 564009; RRID: AB_2732063 |
| BV711 anti-mouse CD49a | BioLegend | Cat#: 564863; RRID: AB_2738987 |
| BV785 anti-mouse NK1.1 | BioLegend | Cat#: 108749; RRID: AB_2564304 |
| eF450 anti-mouse CD3 | Thermo Fisher Scientific | Cat#: 48-0032-82; RRID: AB_1272193 |
| eF450 anti-mouse DX5 | Thermo Fisher Scientific | Cat#: 48-5971-82; RRID: AB_10671541 |
| eF450 anti-mouse IFN-γ | Thermo Fischer Scientific | Cat#: 48-7311-82; RRID: AB_1834366 |
| FITC anti-mouse Annexin V | BioLegend | Cat#: 640906 |
| FITC anti-mouse CD27 | Thermo Fischer Scientific | Cat#:11-0271-82; RRID: AB_465001 |
| FITC anti-mouse TCRβ | BioLegend | Cat#:109206; RRID: AB_313429 |
| PE anti-mouse CD49a | BioLegend | Cat#:142604; RRID: AB_10945158 |
| PE anti-mouse IFN-γ | Thermo Fischer Scientific | Cat#: 12-7311-82; RRID: AB_466193 |
| PE anti-mouse TRAIL | Thermo Fischer Scientific | Cat#: 12-5951-81; RRID: AB_466056 |
| PE-Cy7 anti-mouse IFN-γ | Thermo Fischer Scientific | Cat#: 25-7311-82; RRID: AB_469680 |
| PE-Cy7 anti-mouse KLRG1 | Thermo Fischer Scientific | Cat#: 25-5893-82; RRID: AB_1518768 |
| PerCP-Cy5.5 anti-mouse NK1.1 | BD Biosciences | Cat#: 45-5941-82; RRID: AB_914361 |
| PerCP-Cy5.5 anti-mouse TCRβ | BioLegend | Cat#: 109228; RRID: AB_1575173 |
| PerCP-eF710 anti-mouse/rat/human CD27 | Thermo Fischer Scientific | Cat#: 46-0271-80; RRID: AB_1834448 |
| Purified anti-mouse CD16/CD32 (Fc block, 2.4G2) | In-house produced | N/A |
| Purified anti-mouse IFN-γ | Bio X Cell | Cat#: BE0312; RRID: AB_2736992 |
| Purified anti-mouse IgG Isotype Control | Bio X Cell | Cat#: BE0091; RRID: AB_1107773 |
| Virus Strains | ||
| MCMV-RVG102 | In-house produced | Henry et al., 2000 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ammonium chloride | Fisher Scientific | Cat# A661-500 |
| Brilliant Stain Buffer | BD Biosciences | Cat#: 563794 |
| CPG ODN 1826 | InvivoGen | Cat#: tlrl-modn |
| Fixation and Permeabilization Solution | BD Biosciences | Cat#: 554722 |
| Fixation/Permeabilization Concentrate | eBioscience | Cat#: 00-5123-43 |
| Fixation/Permeabilization Diluent | eBioscience | Cat#: 00-5223-56 |
| Heparin sodium salt | Sigma-Aldrich | Cat#: H3393-500KU |
| Percoll | GE Healthcare | Cat#: 17-0891-01 |
| Permeabilization Buffer (10X) | eBioscience | Cat#: 00-8333-56 |
| Perm/Wash Buffer | BD Biosciences | Cat#: 554723 |
| Annexin V Binding Buffer (10X) | BD Biosciences | Cat#: 556454 |
| Polyinosinic-polycytidylic acid potassium salt (Poly(I:C)) | Sigma-Aldrich | Cat#: P9582-50MG |
| UltraComp eBeads, Compensation Beads | Invitrogen | Cat#: 01-2222-42 |
| L-(+)- Lactic acid solution | Millipore Sigma | Cat#: L1875-100ML |
| Critical Commercial Assays | ||
| RNeasy Plus Mini Kit (50) | QIAGEN | Cat#: 74134 |
| iScript Reverse Transcription Supermix | BIORAD | Cat#: 1708840 |
| Cell Proliferation Dye, eF450 | eBioscience | Cat#: 65-0842-85 |
| DNeasy Blood and Tissue Kit | QIAGEN | Cat#: 69506 |
| Lactate Assay Kit | Millipore Sigma | Cat#: MAK064-1KT |
| Amicon Ultra - 0.5mL centrifugal filter units | Millipore Sigma | Cat#: UFC501096 |
| Experimental Models: Mouse Strains | ||
| B6.SJL | Taconic | Cat#: 4007 |
| B6.SJL | Jackson | Cat#: 002014 |
| C57BL/6 | Jackson | Cat#: 000664 |
| C57BL/6 | Taconic | Cat#: B6 |
| Myd88fl/fl/ | Jackson | Cat#: 008888 |
| IL-12Rp2fl/fl | In House | N/A |
| IFNAR1fl/fl | Jackson | Cat#: 028256 |
| NCRgfp/gfp | Jackson | Cat#: 022739 |
| RAG1−/− | Jackson | Cat#: 002216 |
| Rag2−/−IL-2Rg−/− | Taconic | Cat#: 4111 |
| Oligonucleotides | ||
| Ldha Forward: 5’-TATCTTAATGAAGGACTTGGCGGA TGAG-3’ | IDT | Brand et al., 2016 |
| Ldha Reverse: 5’- GGAGTTCGCAGTTACACAGTAGTC-3’ |
IDT | Brand et al., 2016 |
| Ldhb Forward: 5’-TTGTGGCCGATAAAGATTACTCTG TGAC-3’ | IDT | Brand et al., 2016 |
|
Ldhb Reverse:5’- AGGAATGATGAACTTGAACACGTTGA −3’ |
IDT | Brand et al., 2016 |
| 18S Forward: 5’-ACCGATTGGATGGTTTAGTGAG-3’ | IDT | Brand et al., 2016 |
| 18S Reverse: 5 -CCTACGGAAACCTTGTTACGAC-3’ | IDT | Brand et al., 2016 |
| Software and Algorithms | ||
| CFX Maestro | Bio-Rad | N/A |
| FlowJo, v10 | FlowJo, LLC (Tree Star, Inc.) | https://www.flowjo.com |
| Prism 7.0 | GraphPad Software | https://www.graphpad.com |
| Other | ||
| BD FACSAria III | BD Biosciences | N/A |
| CFX384 Real-Time System | Bio-Rad | N/A |
| gentleMACS | Miltenyi Biotec | N/A |
| MACSQuant | Miltenyi Biotec | N/A |
| Synergy HT | BioTek | N/A |
| NanoDrop 2000/2000c | ThermoFisher | N/A |
Highlights.
Hepatic conventional NK and tissue-resident NK cells differ in kinetic response to MCMV
Hepatic trNK cells undergo rapid apoptosis during liver inflammation
trNK cell apoptosis is due to lactate sensitivity and impaired mitochondrial function
ACKNOWLEDGMENTS
We thank Kevin Carlson for cell sorting, Céline Fugère for i.v. injections, and Samantha Borys for illustration of the graphical abstract. We thank Dr. Courtney Anderson for scientific discussions and reading the manuscript. This work was supported by NIH research grants R01 AI46709 (to L.B.), R01 AI122217 (to L.B.), and F31 CA243305 (to A.T.). G.D. is supported by research supplement 3R01AI122217-S1 to promote diversity. The FACSAria was funded by NCCR equipment grant 1S10RR021051 (to L.B.) and upgraded to a FACSAria III by Provost’s equipment fund. E.V. is supported by funding from the European Research Council (ERC) under the European Union Horizon 2020 Research and Innovation Program (TILC, grant agreement 694502); Agence Nationale de la Recherche, including the PIONEER Project (ANR-17-RHUS-0007); Equipe Labellisée “La Ligue” (Ligue Nationale contre le Cancer); MSDAvenir, Innate Pharma; and institutional grants to the CIML (INSERM, CNRS, and Aix-Marseille University) and to Marseille Immunopole. S.U. is supported by funding from the ERC under the European Union Horizon 2020 Research and Innovation Program (TILC, grant agreement 648768), Agence Nationale de la Recherche (ANR-14-CE14-0009-01), and the ARC Foundation (PGA120140200817).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107855.
CONFLICTS OF INTEREST
E.V. is an employee of Innate-Pharma.
REFERENCES
- Almeida FF, Tognarelli S, Marçais A, Kueh AJ, Friede ME, Liao Y, Willis SN, Luong K, Faure F, Mercier FE, et al. (2018). A point mutation in the Ncr1 signal peptide impairs the development of innate lymphoid cell subsets. OncoImmunology 7, e1475875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CK, Reilly EC, Lee AY, and Brossay L (2019). Qa-1-Restricted CD8(+) T Cells Can Compensate for the Absence of Conventional T Cells during Viral Infection. Cell Rep. 27, 537–548.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D, and Spits H (2015). The biology of innate lymphoid cells. Nature 517, 293–301. [DOI] [PubMed] [Google Scholar]
- Babić M, Krmpotić A, and Jonjić S (2011). All is fair in virus-host interactions: NK cells and cytomegalovirus. Trends Mol. Med 17, 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkström NK, Ljunggren HG, and Michaëlsson J (2016). Emerging insights into natural killer cells in human peripheral tissues. Nat. Rev. Immunol 16, 310–320. [DOI] [PubMed] [Google Scholar]
- Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et al. (2016). LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell Metab 24, 657–671. [DOI] [PubMed] [Google Scholar]
- Colonna M (2018). Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity 48, 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez VS, Robinette ML, and Colonna M (2015). Innate lymphoid cells: new insights into function and development. Curr. Opin. Immunol 32, 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispe IN (2009). The liver as a lymphoid organ. Annu. Rev. Immunol 27, 147–163. [DOI] [PubMed] [Google Scholar]
- Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, et al. (2014). T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med 211, 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty JR, and Cleveland JL (2013). Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest 123, 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erick TK, and Brossay L (2016). Phenotype and functions of conventional and non-conventional NK cells. Curr. Opin. Immunol 38, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, and Zhu J (2017). Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J. Exp. Med 214, 1861–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, and Reiner SL (2012). The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, et al. (2016). The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell 166, 1231–1246.e13. [DOI] [PubMed] [Google Scholar]
- Haas R, Smith J, Rocher-Ros V, Nadkarni S, Montero-Melendez T, D’Acquisto F, Bland EJ, Bombardieri M, Pitzalis C, Perretti M, et al. (2015). Lactate Regulates Metabolic and Pro-inflammatory Circuits in Control of T Cell Migration and Effector Functions. PLoS Biol. 13, e1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon C, Robinson MW, Hand F, Almuaili D, Mentor K, Houlihan DD, Hoti E, Lynch L, Geoghegan J, and O’Farrelly C (2019). Lactate-Mediated Acidification of Tumor Microenvironment Induces Apoptosis of Liver-Resident NK Cells in Colorectal Liver Metastasis. Cancer Immunol. Res 7, 335–346. [DOI] [PubMed] [Google Scholar]
- Henry SC, Schmader K, Brown TT, Miller SE, Howell DN, Daley GG, and Hamilton JD (2000). Enhanced green fluorescent protein as a marker for localizing murine cytomegalovirus in acute and latent infection. J. Virol. Methods 89, 61–73. [DOI] [PubMed] [Google Scholar]
- Ishizuka IE, Constantinides MG, Gudjonson H, and Bendelac A (2016). The Innate Lymphoid Cell Precursor. Annu. Rev. Immunol 34, 299–316. [DOI] [PubMed] [Google Scholar]
- Kim S, Iizuka K, Kang HS, Dokun A, French AR, Greco S, and Yokoyama WM (2002). In vivo developmental stages in murine natural killer cell maturation. Nat. Immunol 3, 523–528. [DOI] [PubMed] [Google Scholar]
- Li T, Wang J, Wang Y, Chen Y, Wei H, Sun R, and Tian Z (2017). Respiratory Influenza Virus Infection Induces Memory-like Liver NK Cells in Mice. J. Immunol 198, 1242–1252. [DOI] [PubMed] [Google Scholar]
- Madera S, Rapp M, Firth MA, Beilke JN, Lanier LL, and Sun JC (2016). Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J. Exp. Med 213, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais A, Viel S, Grau M, Henry T, Marvel J, and Walzer T (2013). Regulation of mouse NK cell development and function by cytokines. Front. Immunol 4, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie ANJ, Spits H, and Eberl G (2014). Innate lymphoid cells in inflammation and immunity. Immunity 41, 366–374. [DOI] [PubMed] [Google Scholar]
- O’Leary JG, Goodarzi M, Drayton DL, and von Andrian UH (2006). T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol 7, 507–516. [DOI] [PubMed] [Google Scholar]
- Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, and von Andrian UH (2010). Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat. Immunol 11, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, and Tian Z (2013). Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest 123, 1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Wisse E, and Tian Z (2016). Liver natural killer cells: subsets and roles in liver immunity. Cell. Mol. Immunol 13, 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrini L, Wieduwild E, Escaliere B, Filtjens J, Chasson L, Laprie C, Vivier E, and Ugolini S (2018). Endogenous glucocorticoids control host resistance to viral infection through the tissue-specific regulation of PD-1 expression on NK cells. Nat. Immunol 19, 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins SH, Tessmer MS, Mikayama T, and Brossay L (2004). Expansion and contraction of the NK cell compartment in response to murine cytomegalovirus infection. J. Immunol 173, 259–266. [DOI] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, and Colonna M; Immunological Genome Consortium (2015). Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol 16, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini N, Vosshenrich CA, and Di Santo JP (2015). Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol 15, 415–428. [DOI] [PubMed] [Google Scholar]
- Shah GN, Rubbelke TS, Hendin J, Nguyen H, Waheed A, Shoemaker JD, and Sly WS (2013). Targeted mutagenesis of mitochondrial carbonic anhydrases VA and VB implicates both enzymes in ammonia detoxification and glucose metabolism. Proc. Natl. Acad. Sci. USA 110, 7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S, Schuster IS, Andoniou CE, Cocita C, Adejumo T, Kung SKP, Sun JC, Degli-Esposti MA, and Guerra N (2018). The Murine Natural Cytotoxic Receptor NKp46/NCR1 Controls TRAIL Protein Expression in NK Cells and ILC1s. Cell Rep 22, 3385–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, et al. (2014). Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3, e01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, and Artis D (2015). Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat. Med 21, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Beilke JN, and Lanier LL (2009). Adaptive immune features of natural killer cells. Nature 457, 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, Yagita H, Kinoshita K, Okumura K, and Smyth MJ (2005). TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood 105, 2082–2089. [DOI] [PubMed] [Google Scholar]
- Turchinovich G, Ganter S, Barenwaldt A, and Finke D (2018). NKp46 Calibrates Tumoricidal Potential of Type 1 Innate Lymphocytes by Regulating TRAIL Expression. J. Immunol 200, 3762–3768. [DOI] [PubMed] [Google Scholar]
- Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. (2018). Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066. [DOI] [PubMed] [Google Scholar]
- Waggoner SN, Cornberg M, Selin LK, and Welsh RM (2011). Natural killer cells act as rheostats modulating antiviral T cells. Nature 481, 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weizman OE, Adams NM, Schuster IS, Krishna C, Pritykin Y, Lau C, Degli-Esposti MA, Leslie CS, Sun JC, and O’Sullivan TE (2017). ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell 171, 795–808.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama WM, Sojka DK, Peng H, and Tian Z (2013). Tissue-resident natural killer cells. Cold Spring Harb. Symp. Quant. Biol 78, 149–156. [DOI] [PubMed] [Google Scholar]
- Zhou J, Peng H, Li K, Qu K, Wang B, Wu Y, Ye L, Dong Z, Wei H, Sun R, and Tian Z (2019). Liver-Resident NK Cells Control Antiviral Activity of Hepatic T Cells via the PD-1-PD-L1 Axis. Immunity 50, 403–417.e4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.


