Highlights
-
•
Radiomics studies in NSCLC suffer from a number of limitations.
-
•
No single radiomic signature has been translated into clinical use.
-
•
Identification of limitations can help future studies to expedite biomarker translation.
Abbreviations: AUC, area under the curve; CI, concordance index; HR, hazard ratio; ROI, region of interest; RQS, radiomics quality score; TRIPOD, Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis
Keywords: Radiomics, Imaging biomarkers, Lung cancer, Personalized medicine
Abstract
Radiomics has become a popular image analysis method in the last few years. Its key hypothesis is that medical images harbor biological, prognostic and predictive information that is not revealed upon visual inspection. In contrast to previous work with a priori defined imaging biomarkers, radiomics instead calculates image features at scale and uses statistical methods to identify those most strongly associated to outcome. This builds on years of research into computer aided diagnosis and pattern recognition. While the potential of radiomics to aid personalized medicine is widely recognized, several technical limitations exist which hinder biomarker translation. Aspects of the radiomic workflow lack repeatability or reproducibility under particular circumstances, which is a key requirement for the translation of imaging biomarkers into clinical practice. One of the most commonly studied uses of radiomics is for personalized medicine applications in Non-Small Cell Lung Cancer (NSCLC). In this review, we summarize reported methodological limitations in CT based radiomic analyses together with suggested solutions. We then evaluate the current NSCLC radiomics literature to assess the risk associated with accepting the published conclusions with respect to these limitations. We review different complementary scoring systems and initiatives that can be used to critically appraise data from radiomics studies. Wider awareness should improve the quality of ongoing and future radiomics studies and advance their potential as clinically relevant biomarkers for personalized medicine in patients with NSCLC.
1. Introduction
Lung cancer remains the leading cause of cancer-related mortality worldwide [1]. The 5 year survival for patients with non-small cell lung cancer (NSCLC), the most common form of the disease, is 10−20% [2,3]. Despite advances in treatment options in recent years, survival rates have changed little [3,4].
Given the patient variability and tumor heterogeneity of this cancer, personalizing treatment is key to improving survival beyond the current poor prognosis [5]. One requirement for successful delivery of personalized medicine is the identification and validation of biomarkers that can predict which patients will benefit from a given therapy. There is an unmet need for such biomarkers in lung cancer [6].
Medical imaging plays a key role in the diagnosis and treatment of lung cancer, making the use of image-based biomarkers to guide clinical decision-making attractive. Over the last several decades, a number of biomarkers derived from CT, PET and MRI that measure tumor size, shape and texture, or quantify aspects of the tumor microenvironment have been used in lung cancer studies for diagnosis, prediction, prognostication and response monitoring [[6], [7], [8]].
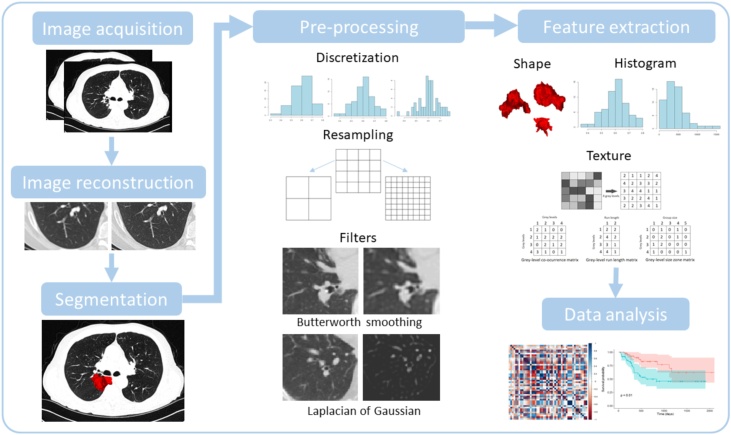
There is currently substantial interest in using computer algorithms to extend this approach to extract tens to thousands of image ‘features’ in an analysis pipeline strategy termed ‘radiomics’. Such methods test the hypothesis that medical images harbor data that will provide biomarkers for personalized medicine, but that the optimum biomarkers are not readily determined a priori [9]. Imaging biomarker studies postulate that medical images contain biological, prognostic and predictive information that is not apparent when clinicians view scans [10]. In radiomics, this information is extracted from digital images using computer algorithms to form ‘radiomic signatures’, a type of quantitative imaging biomarker formed by combining the radiomics features that have the strongest association to the measured outcome. The radiomics workflow consists of a series of steps [11]. summarized in Fig. 1. Proponents of radiomics hypothesize that these data-driven approaches will select the most statistically significant signature that relates to an outcome measure of interest. This approach is extremely popular, but to date the resultant imaging biomarkers have not been validated as useful tools for personalized medicine [12].
Fig. 1.
Visualization of the steps in the radiomics workflow. First, images are acquired and reconstructed. The region of interest is then segmented, from which features will be extracted. Next, pre-processing steps are performed to modify the images before feature extraction. Shape, first order (or histogram) and texture features are then extracted from the region of interest. Finally, data analysis steps attempt to find correlations between features and the specified outcome.
CT is the most commonly used modality worldwide for diagnosis, treatment planning, and follow-up in all stages of lung cancer, meaning that informative imaging biomarkers discovered from these data could be translated rapidly into clinical practice. In this review, we summarize the literature supporting use of CT radiomic biomarkers to guide decision-making in patients with NSCLC.
We appraise the published reports of CT radiomics biomarkers as predictive, prognostic or biologically informative tools and review literature highlighting methodological limitations. Our aims are to evaluate how robust the conclusions of these studies are and to assess how well the current standardization and reporting tools inform readers of the potential limitations when interpreting their results.
2. The potential of radiomics for personalized decision-making in NSCLC
A review of the literature found 43 CT image based studies that evaluated the prognostic or predictive role of radiomic signatures in patients with NSCLC (Table 1). Three of these studies, together with a further 21 we separately identified, evaluated the role of radiomic signatures in appraising aspects of tumor biology including genomic or pathologic biomarkers, signalling pathways, and disease classification in NSCLC (Table 2).
Table 1.
Radiomics studies in NSCLC, categorized into sections based on their investigated endpoint. The Data column specifies the total number of patients involved in the study, in brackets split by training and validation cohorts if applicable and specifying other cancer types of cohorts if applicable. Note: Studies marked with * are validation studies and their RQS score components refer to methodology based on the previous published data. This table has been simplified to clarify presentation – more details for each study are available in Supplementary Table 2.
| Reference | NSCLC stage | Data (training + validation) | Radiomic features in final model | Result |
|---|---|---|---|---|
| Overall survival | ||||
| Aerts et al. 2014 [13] | 1−3b | 647 pCT (422 + 225) |
Shape, first order and texture | CI = 0.65 |
| Van Timmeren et al. 2017 [14]* | 1−4 | 252 pCT and CBCT (102 + 56 + 94) |
Shape, first order and texture | CI = 0.69, 0.61, 0.59 (pCT) CI = 0.66,0.63,0.59 (CBCT) |
| Grossman et al. 2017 [15]* | 1−3 | 351 diagnostic CT (262 + 89) |
Shape, first order and texture | CI = 0.60 |
| Grossman et al. 2017 [15] | 1−3 | 351 diagnostic CT (262 + 89) |
Not specified | CI = 0.61 |
| Yu et al. 2017 [16] | 1 | 442 diagnostic CT (147 + 295) |
First order and texture | CI = 0.64 |
| Chaddad et al. 2017 [17] | 1−3b | 315 pCT | Shape and texture | Average AUC = 0.70−0.76 |
| Fave et al. 2017 [18] | 3 | 107 4DCT end of exhale, pCT and CBCT | Shape and texture | CI = 0.672 |
| Li et al. 2017 [19] | 1−2a | 59 follow up CT | Texture | AUC = 0.81 |
| Li et al. 2017 [20] | 1−2a | 92 4DCT Average-CT or 50 % phase-CT |
Shape and first order | AUC = 0.728 |
| Tang et al. 2018 [21] | 1−3 | 290 staging CT (114 + 176) |
Shape, first order and texture | CI = 0.72 |
| Bianconi et al. 2018 [22] | 1−3 | 203 pCT | Shape and texture | HR = 1.06−1.48 |
| De Jong et al. 2018 [23]* | 4 | 195 diagnostic CT | Shape, first order and texture | CI = 0.576 |
| Lee et al. 2018 [24] | 1−3 | 339 CT | Shape, first order and texture | CI = 0.772 |
| He et al. 2018 [25] | 1−3 | 186 CT (298 after oversampling (223 + 75)) |
Not specified | AUC = 0.9296 |
| Starkov et al. 2018 [26] | 1 | 116 pCT | Texture | High risk vs low risk median p-values = 0.04–0.07 |
| Yang et al. 2018 [27] | 1−4 | 371 CT (239 + 132) |
First order and texture | CI = 0.702 |
| Wang et al. 2019 [28] | 3 | 70 pre-treatment and 97 post-treatment CT from 118 patients | Texture | CI = 0.743 |
| Shi et al. 2019 [29] | 3 | 11 CBCT from 23 patients | First order | HR = 0.21 |
| Van Timmeren et al. 2019 [30] | 1−4 | 337 pCT and 2154 CBCTs from 337 patients (141 + 94 + 61 + 41) |
First order and texture | CI = 0.59, 0.54, 0.57 |
| Huang et al. 2019 [31] | 1−4 | 371 CT (254 + 63 + 54) |
Shape, first order and texture | CI = 0.621, 0.649 |
| Franceschini et al. 2019 [32] | 1−2 | 102 4DCT start of inspiration (70 + 32) |
Shape and texture | AUC = 0.85 |
| Local or metastatic recurrence | ||||
| Coroller et al. 2015 [33] | 2−3 | 182 pCT (98 + 84) |
First order and texture | CI = 0.6 |
| Mattonen et al. 2016 [34] | 1 | 45 follow-up CT | First order and texture | AUC = 0.85 |
| Huynh et al. 2016 [35] | 1−2 | 113 CT | First order and texture | Median CI = 0.67 |
| Huynh et al. 2017 [36] | 1−2a | 112 CT and AIP CT | Shape, first order and texture | AIP radiomics CI = 0.667 FB radiomics CI = 0.601 |
| Fave et al. 2017 [18] | 3 | 107 4DCT end of exhale, pCT and CBCT | Shape and texture | CI = 0.632, 0.558 (DM, LRR) |
| Li et al. 2017 [19] | 1−2a | 59 follow up CT | Texture | AUC = 0.80, 0.80 (RFS, LR-RFS) |
| Li et al. 2017 [20] | 1−2a | 92 4DCT Average-CT or 50 % phase-CT |
Shape | AUC = 0.747, 0.690 (RFS, LL-RFS) |
| Dou et al. 2018 [37] | 2−3 | 200 pCT (100 + 100) |
Texture | CI = 0.65 |
| Ferreira Junior et al. 2018 [38] | 1−4 | 68 CT (52 + 16) |
Shape and texture | AUC = 0.75, 0.71 (lymph node metastasis, DM) |
| Yang et al. 2018 [39] | 1−3 | 159CT (106 + 53) |
Shape, first order and texture | AUC = 0.856 |
| Zhong et al. 2018 [40] | 1−2 | 492 CT | First order and texture | AUC = 0.972 |
| Lafata et al. 2019 [41] | 1 | 70 CT | Texture | Maximum AUC = 0.72, 0.83, 0.60 (recurrence, LR, non-LR) |
| Akinci D’Antonoli et al. 2019 [42] | 1−2b | 124 CT | Shape, first order and texture | AUC 0.731, 0.750 (LR, DM) |
| He et al. 2019 [43] | Not specified | 717CT (423 + 294) |
First order and texture | CI = 0.734 |
| Xu et al. 2019 [44] | 3−4 | 132 CT (106 + 26) |
Texture | AUC = 0.642 |
| Franceschini et al. 2019 [32] | 1−2 | 102 4DCT start of inspiration (70 + 32) |
Shape, first order and texture | AUC = 0.73 |
| Ferreira-Junior et al. 2019 [45] | 1−4 | 85 CT | Shape, first order and texture | AUC = 0.92, 0.84 (DM, nodal metastasis) |
| Cong et al. 2019 [46] | 1a | 649 venous phase CT (455 + 194) |
Shape, first order and texture | AUC = 0.851 |
| Treatment response, disease-free or progression-free survival | ||||
| Coroller et al. 2016 [47] | 2−3 | 127 pCT | Shape, first order and texture | Median AUC = 0.65, 0.61 (GRD, pCR) |
| Huang et al. 2016 [48] | 1−2 | 282 CT (141 + 141) | First order and texture | HR = 2.09 |
| Song et al. 2016 [49] | 1−4 | 152 CT (80 + 72) |
Texture | HR = 2.35, 2.75 |
| Coroller et al. 2017 [50] | 2−3 | 85 pCT | Shape, first order and texture | Median AUC = 0.68, = 0.71 (pCR, GRD) |
| Tunali et al. 2019 [51] | 3b-4 | 228 CT | Texture | AUC = 0.804 |
| Franceschini et al. 2019 [32] | 1−2 | 102 4DCT start of inspiration (70 + 32) |
Texture | AUC = 0.88 |
| Lung toxicity | ||||
| Moran et al. 2017 [52] | 1 | 14 diagnostic CT | First order and texture | AUC = 0.689−0.750 |
| Krafft et al. 2018 [53] | Not specified | 192 50 % 4DCT phase | First order and texture | Average AUC = 0.68 |
| Staging | ||||
| Yuan et al. 2018 [54] | 1 | 327 CT | First order and texture | AUC = 0.938 |
| Yang et al. 2019 [55] | 1−3 | 256 CT | First order and texture | AUC = 0.93 |
Abbreviations: AUC, area under the curve; CBCT, cone-beam CT; CI, concordance index; DFS, disease free survival; DM, distant metastasis; GRD, gross residual disease; H&N, head and neck; HR, hazard ratio; LR, local relapse; LRR, local regional recurrence; LR-RFS, loco-regional recurrence-free survival; OS, overall survival; pCR, pathological complete response; pCT, radiotherapy planning CT scan; PFS, progression free survival; RFS, recurrence free survival.
Table 2.
Radiomics studies in NSCLC with an aspect of biology as the endpoint. The column labeled ‘Data’ specifies the total number of patients involved in the study, in brackets split by training and validation cohorts if applicable and specifying other cancer types of cohorts if applicable. This table has been simplified to clarify presentation – more details for each study are available in Supplementary Table 3.
| Reference | Stage | Endpoint | Data (training + validation) | Radiomic features in final model | Result |
|---|---|---|---|---|---|
| Genomics | |||||
| Aerts et al. 2016 [56] | Early stage | EGFR | 47 diagnostic CT and follow-up | Shape and texture | AUC = 0.74−0.91 |
| Rios Velazquez et al. 2017 [57] | 1−4 | EGFR, KRAS | 705 diagnostic CT (353 + 352) |
Shape, first order and texture | AUC = 0.69−0.80 |
| Mei et al. 2018 [58] | Not specified | EGFR | 296 CT | Texture | AUC = 0.664 |
| Digumarthy et al. 2019 [59] | Not specified | EGFR | 93 CT | First order | AUC = 0.713 |
| Jia et al. 2019 [60] | 1−4 | EGFR | 504 CT (345 + 158) |
Shape, first order and texture | AUC = 0.802 |
| Li et al. 2019 [61] | 1−4 | EGFR subtypes (19Del and L858R) | 312 CT (236 + 76) |
Shape, first order and texture | AUC = 0.775−0.793 |
| Tu et al. 2019 [62] | 1−4 | EGFR | 404 CT (243 + 161) |
First order and texture | AUC = 0.775 |
| Yang et al. 2019 [63] | Not specified | EGFR | 467 CT (306 + 161) |
Shape, first order and texture | AUC = 0.789 |
| Wang et al. 2019 [64] | 1−2 | EGFR, TP53 | 61 CT (41 + 20) |
First order and texture | AUC = 0.604, 0.586 |
| Wang et al. 2019 [64] | 1−2 | Tumor mutation burden | 61 CT (41 + 20) |
Texture | AUC = 0.606 |
| Signaling pathways | |||||
| Grossman et al. 2017 [15] | 1−3 | Various | 351 CT (262 + 89) |
Shape, first order and texture | AUC = 0.62−0.72 |
| Bak et al. 2018 [65] | 1−4 | Various | 57 CT | First order and texture | OR = 0.08−23.94 |
| Histopathology | |||||
| Patil et al. 2016 [66] | Not specified | ADC, LCC, SCC, NOS | 317 pCT | Shape, first order and texture | 88 % accuracy |
| Wu et al. 2016 [67] | 1−4 | ADC, SCC | 350 pCT (198 + 152) |
First order and texture | AUC = 0.72 |
| Ferreira Junior et al. 2018 [38] | 1−4 | ADC, SCC | 68 CT (52 + 16) |
Not specified | AUC = 0.81 |
| Zhu et al. 2018 [68] | Not specified | ADC, SCC | 129 CT (81 + 48) |
First order and texture | AUC = 0.893 |
| Digumarthy et al. 2019 [59] | Not specified | ADC, SCC | 93 CT | First order | AUC = 0.744 |
| E et al. 2019 [69] | Not specified | ADC, SCC, SCLC | 229 CT | Shape, first order and texture | AUC = 0.657−0.875 |
| Ferreira-Junior et al. 2019 [45] | 1−4 | ADC, SCC | 85 CT | Shape, first order, texture | AUC = 0.88 |
| Liu et al. 2019 [70] | Not specified | ADC, LCC, SCC, NOS | 349 CT (278 + 71) |
Not specified | AUC = 0.86 |
| Zhou et al. 2018 [71] | 1−4 | Ki-67 | 110 CT | Shape and texture | AUC = 0.61−0.77 |
| Gu et al. 2019 [72] | Not specified | Ki-67 | 245 CT | First order and texture | AUC = 0.776 |
| Song et al. 2017 [73] | 1−3 | Micropapillary pattern | 339 CT | First order | AUC = 0.751 |
| Chen et al. 2018 [74] | Not specified | Degree of differentiation | 487 CT (303 + 184) |
First order and texture | AUC = 0.782 |
| She et al. 2018 [75] | Not specified | Invasive vs non-invasive adenocarcinoma | 402 CT (207 + 195) |
Shape, first order and texture | AUC = 0.89 |
| Yang et al. 2019 [76] | Not specified | Invasive vs non-invasive adenocarcinoma | 192 CT (116 + 76) |
First order and texture | AUC = 0.77 |
Abbreviations: ADC, adenocarcinoma; AUC, area under the curve; CI, concordance index; EGFR, epidermal growth factor receptor; KRAS, Kirsten rat sarcoma viral oncogene homolog; LCC, large cell carcinoma; NOS, not otherwise specified; OR, odds ratio; SCC, squamous cell carcinoma.
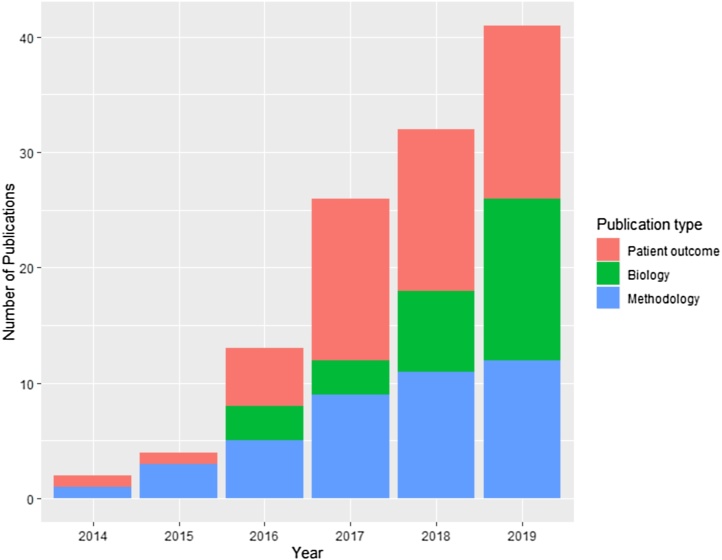
In addition, 42 studies reported on radiomics methodological limitations, potential problems, and possible solutions in CT based studies using data from NSCLC patients or imaging phantoms. The frequency of publications, for all types of NSCLC radiomics study, has markedly increased over the last six years (Fig. 2). Our search strategies are described in detail in Supplementary Materials.
Fig. 2.
Frequency of CT NSCLC radiomics studies published from 2014 to 2019. Publications are categorized as those investigating radiomics methodological concerns, those evaluating radiomic signatures as prognostic or predictive biomarkers of patient outcome, and those evaluating radiomic signatures as biomarkers of tumor biology.
The initial studies labelled as ‘radiomics’ were published in 2014 and 2015. Aerts and colleagues showed that a radiomic signature based on shape and texture metrics was associated with overall survival, validating the signature in patients with NSCLC and patients with head and neck cancers [13]. The study also found positive associations between the radiomic signature and gene expression. Coroller and colleagues showed that a different set of texture metrics were associated with the subsequent development of distant metastases [33]. The hypothesized mechanism was that tumor heterogeneity, identified by the radiomics analyses, drives worse outcomes. Both studies were performed using radiotherapy planning CT data.
Over the next four years (2015–2019), 41 CT studies were published that linked radiomics to lung cancer patient outcome. In general, studies sought to evaluate whether or not radiomic signatures could outperform existing methods for patient risk stratification. 20 studies related radiomics to overall survival [[13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32]], 18 to the likelihood of local or metastatic recurrence [[18], [19], [20],[32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46]], 6 to response, disease-free or progression-free survival [32,[47], [48], [49], [50], [51]], and 2 to staging [54,55]. Two further studies focused on the association of radiomics signatures to lung toxicity [52,53], Four studies investigated multiple endpoints.
The majority of studies derived radiomics signatures in radiotherapy planning or diagnostic images acquired prior to therapy. Nearly all studies evaluated patients undergoing treatment with cytotoxic chemo-radiotherapy. More recently, a number of studies have evaluated the potential of radiomics to improve patient stratification for targeted therapies and immunotherapy agents [21,51,56]. For example, Tang and colleagues linked radiomic features to a tumor immune phenotype in patients with stage I-III NSCLC, finding patients with heterogeneous tumors, which correlated with low PD-L1 and high CD3 cell count, had better prognosis [21].
There are 24 CT studies evaluating how radiomic signatures of NSCLC relate to genomics [[56], [57], [58], [59], [60], [61], [62], [63], [64]], signalling pathways [15,65] and histopathology [38,45,59,[66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76]]. For example, Rios Velazquez and colleagues found distinct imaging phenotypes for EGFR and KRAS mutations from CT images of patients with NSCLC [57]. Some of the studies that relate radiomics to patient outcome also relate their radiomic signature to genomics [13] or biological markers [28].
Collectively, these 64 studies present a positive view of the potential for radiomics signatures to deliver personalized medicine. However, two important limitations are readily apparent. Firstly, while nearly all studies report at least one positive association between CT radiomic signature and either outcome (OS, PFS, recurrence or toxicity) or tumor biology (genomic or pathology biomarkers and signalling pathways), the particular radiomic signature derived varies substantially between studies. Consequently, few study signatures are directly comparable with one another, and so the literature does not identify specific candidate radiomic signatures for further large multicenter evaluation.
Secondly, it has become clear that studies can suffer from significant technical limitations. Studies of these limitations have also increased over the last five years, although at a slower pace than the patient outcome studies (Fig. 2).
3. Reported methodological limitations of CT based radiomics studies
All biomarkers, including radiomic signatures, must undergo technical and biological validation to become robust tools used to guide clinical decision-making. These validation steps take a biomarker from discovery to research assay where the biomarker can be used with confidence to determine an outcome in a research setting (termed ‘crossing translational gap 1′). The regulatory approval process (through e.g. the FDA or EMA) then takes the biomarker from research assay to clinically approved assay for use in decision-making in patients (termed ‘crossing translational gap 2′) [12].
To date, very few radiomics signatures have crossed either of these translational gaps. The first radiology product with radiomics capabilities to receive such approvals was QuantX for detection of breast abnormalities based on MRI, receiving FDA approval in 2017 [77]. Soon afterwards, Feedback Medical received CE approval for TexRAD Lung, a quantitative image texture analysis technology [78].
In this section, we evaluate the methodological limitations preventing CT based radiomics signatures from crossing these translational gaps. We review the potential problems and proffered solutions identified in 42 studies of imaging phantoms or patients with NSCLC (summarized in Table 3 and expanded in Supplementary Table 1).
Table 3.
Potential problems at each step of the radiomics workflow along with possible solutions offered by the literature. Each workflow step with potential problems and solutions identified by the literature is labelled with a letter A-H to reference in-text. Note: Modelling does not have a letter associated with since there is no consensus on the best statistical modelling strategies.
| Problem area | Potential problems | Potential solutions | |
|---|---|---|---|
| Image acquisition | A | Different scanners and acquisition protocols affect feature reproducibility [[79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91]] | Image phantoms on different scanners to provide baseline [79], establish credibility of scanners and protocols [84], catalogue reproducible features [86,90], model a correction algorithm [89], harmonize data [91]. |
| B | Patient motion affects feature reproducibility [80,92,93] | Set motion tolerances, reduce ROI boundaries [80], use single phase from 4D images [92], find robust features using 4DCT data [93]. | |
| Image acquisition and reconstruction | C | Image resolution parameters (voxel size, slice thickness) affect feature values [79,88,[94], [95], [96], [97], [98]] model performance [99]. | Control resolution [79] parameters in prospective studies, resample to common resolution and voxel depth [[94], [95], [96],98], apply smoothing image filters [95], apply deep learning methods [100]. |
| Image reconstruction | D | Image reconstruction algorithm and reconstruction parameters (kernel) affects features [97,101,102] | Pre-processing image correction [101] and harmonization of acquisition techniques [97,102]. |
| Segmentation | E | Delineation variability [90,[103], [104], [105], [106], [107]] affects features and is time consuming [106,107]. Results from one disease site are not necessarily transferrable to another [108]. | Expert ROI definition [103], multiple observers [103,104,108], identification of stable features with respect to delineation [90,104,105], automated segmentation [106,107], image filtering [108] |
| Pre-processing | F | Number of grey levels used to discretize histogram and texture features affects feature values [96,98,109], as does bin width [94]. | Texture features can be normalized to reduce dependency on the number of grey levels [98], number of grey levels used for discretization should be recorded with feature formula. 128 grey levels may be optimal for texture features, along with thresholding [109] |
| Feature extraction | No studies found in the literature search. | ||
| Feature correlation | G | Strong correlations between tumor volume and radiomic features exist [98,[110], [111], [112]] | Normalization of features to volume [98], bit depth resampling [110], feature redesign [110], more robust statistics to check added value of radiomics signatures [111]. |
| Test re-test | H | Radiomic features may not be repeatable over multiple measurements [[113], [114], [115]], repeatable features are not generalizable to other disease sites [116]. | Test-retest data acquisition [113,116], use of multiple 4D phases [113,115], use of simulated retest by image perturbation [114]. |
| Modelling clinical outcome | Different modelling strategies affect model performance [[117], [118], [119], [120]] | Sample sizes above 50 give better predictive performance [118], as does normalizing features [117]. No consensus on best modelling strategies to use. |
3.1. Image acquisition
Many radiomics studies are retrospective evaluations of CT images, often with data acquired at multiple different institutions and on different CT scanner vendor platforms. Consequently, nearly all studies contend with variations in image acquisition and reconstruction protocols.
Studies assessing the impact of different CT scanners and protocols on radiomic features have shown some features have poor reproducibility [79,80,[84], [85], [86], [87]]. Performing phantom studies on different scanners as a quality assurance step may ensure a level of feature consistency [84]. Indeed, one study showed that using a controlled protocol across different CT scanners reduced feature variability by over 50 % compared to using local protocols [79]. Other studies used post-extraction deep learning [100] or correction factors [89] to reduce feature variability.
Restricting study data to one scanner make and model along with one set of acquisition parameters, to reduce variability in image capture acquisition, is seldom feasible for a multicentre research study. Therefore, many of these issues still remain when setting up a well powered prospective clinical trial with radiomic signatures as exploratory endpoints.
3.2. Image reconstruction
Retrospective data analyses are constrained by image reconstruction parameters determined by clinical department protocols, chosen to optimize image anatomical quality. While variations in image reconstruction, slice thickness and in plane pixel dimensions may have negligible effect for clinical interpretation, they can induce variability in radiomic feature values, since many features correlate to these parameters [79,[94], [95], [96], [97], [98]].
Resampling the image to an equal voxel size has reduced feature dependency on acquisition in some studies [94,96] but not others [79,95]. Smoothing filters have also been suggested as a method for reducing voxel size dependency [95], as has limiting inclusion criteria to particular resolution ranges. For example, Lu et al. found that features calculated from images with 1.25 mm and 2.5 mm thick slices were comparable to each other but that both differed from those calculated on 5 mm slice thickness images [97].
Reconstruction techniques also influence feature values with studies demonstrating differences between features calculated on images reconstructed with soft or sharp kernels [97,102]. Potential solutions include the application of correction factors based on the image noise power spectrum [101]. Solutions that balance feature robustness with the need to make image inclusion criteria as permissive as possible are vital given the small cohorts size issues that blight many studies.
3.3. Segmentation
The ROI definition for feature extraction is known to be a particularly sensitive step in the radiomics pipeline [[103], [104], [105], [106], [107]]. Radiomics studies are popular in radiotherapy given the ready availability of pre-defined ROIs on treatment planning scans, typically using the clinically defined Gross Tumor Volume (GTV). The subjectivity of GTV definition can depend on the operator, as expert delineations may generate features with better predictive power than those from a non-specialist [103].
Frequently suggested solutions include the inclusion of multiple observers or the use of semi-automated delineation tools [106,107]. However, few studies have adopted these solutions, most likely due to the difficulty of getting clinically qualified staff to delineate ROIs. In studies not using radiotherapy planning CT scans, the ROIs must be drawn specifically for the purpose of the radiomics analysis and will suffer from all of the same issues discussed above.
3.4. Pre-processing
The preparation of images for feature extraction has a marked effect on feature value. Reducing the number of image grey-levels (voxel depth re-binning) is a commonly used method to supress image noise. However, studies have shown that radiomic features are not comparable when computed with a differing intensity bin sizes [94,96,98]. This has led to the proposed use of standardized bin resolution [98].
3.5. Feature extraction
Radiomics features span a range of calculation classes. Shape features contain information about the ROI morphology (such as volume and measures of sphericity). First-order image intensity features assess properties of the intensity histogram of voxels within the ROI (e.g. the mean intensity and other statistical moments of the histogram). Texture features summarize different measures of the way in which voxel intensities change across the ROI (e.g. voxel variation coarseness and homogeneity). These features may be calculated on the original image or derived after various filters have been applied that modify particular aspects of it, for example to enhance the edges where image intensity changes [11].
Many different software platforms exist for performing the feature extraction step, including free open-source software, commercial software, and software developed in-house by individual institutions. The Image Biomarker Standardization Initiative (IBSI) is an international collaboration between research groups with the aim of standardizing image biomarker extraction [121]. To date only one study has investigated whether feature extraction software influences radiomic features from CT scans of patients with NSCLC [122], which shows, consistent with data from other cancer types [123,124], that this can have substantial impact on feature values.
3.6. Feature correlation
Since many tens to thousands of features are calculated from images in radiomics, it is unsurprising that many features often correlate with one another. However, the fact that features often correlate strongly with tumor volume and clinical factors [98,110,111] is not well appreciated. While it has been suggested that radiomic feature calculations formulae should be modified to be account for tumor volume [98], it is crucial that studies also include transparent and robust feature reduction steps to account for other clinical prognostic and predictive factors. Robust feature reduction is also crucial in limiting the risk of model overfitting.
3.7. Test-retest
As highlighted by several studies, [113,116] and by consensus statements on imaging biomarkers [12], radiomics studies usually lack an assessment of the signatures’ single centre repeatability or multicentre reproducibility. The use of test-retest datasets in which multiple images of the same subjects or phantom have been acquired in quick succession have been proposed as a means to assess repeatability [113,116]. Alternative options include the use of multiple 4D image phases [113] and the simulation of retest data by image perturbation [114] where test-retest data are not available. Few radiomic studies incorporate any of these approaches.
3.8. Modelling clinical outcome
Typically, studies derive between tens to a few thousand image features in development datasets [125]. Dimensionality reduction to remove highly correlated and unstable radiomic features is often employed before finding the most informative features for a specific outcome, such as overall survival, treatment-related toxicities or cancer recurrence in a test dataset. Many different statistical options exist for deriving a model based on radiomic features. The choice of model and statistical methods can influence results [[118], [119], [120]].
Random forests have been found by some authors to give higher performance compared to other methods for classification tasks using radiomics features [118,120], with Naïve Bayes and Support Vector Machines also reported to perform well [118]. For radiomic feature based time-to-event analyses, one study found cox regression with gradient boost performed better than traditional cox regression (0.614 versus 0.660 concordance index) [119]. In terms of feature selection, there is no consensus on the best method to use. Optimal performance of feature selection techniques depend on the outcome of interest [118]. A contemporary non-radiomics study of classifier performance in radiotherapy datasets found that random forest and elastic net logistic regression performed best, but that classification accuracy depended on the specific dataset [126]. To summarize, there is limited consensus as to the best machine learning methods to employ for radiomics studies, and that the optimum choice may depend on the specific dataset used in the study.
Regardless of feature selection and modelling methodology, the resulting model (often termed a ‘radiomic signature’) should be robustly validated in line with the TRIPOD guidelines to ascertain if it is reproducible across different clinical datasets. This tests if the observed signature relates to the desired outcome in a different patient group, and aims to reduce the risk of overfitting in the training cohort [125].
Lastly, whatever approach is taken it is vital that investigators test whether incorporating radiomic features into a clinical model adds any benefit to well-known clinical prognostic factors such as tumor stage and performance status. Radiomic features will only have clinical utility if they provide more predictive information than is currently available in the clinic.
4. Assessing the quality of radiomics studies in NSCLC
We evaluated the quality of the 43 radiomics studies we identified that report a relationship between a CT defined radiomic signature and clinical outcome in patients with NSCLC (Supplementary Table 2) using both established assessment tools and the results of our review of methodological limitations reported above. We then applied the same tools to the 24 studies that evaluated the relationship between CT radiomic signatures and genomic, protein expression, and pathology biomarkers in patients with NSCLC (Supplementary Table 3). Some studies investigated multiple endpoints, so in total we evaluated 75 outcomes. The four tools we use to interpret the technical validation of these studies are:
-
1
The strength of the validation in each study, assessed by the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) guidelines [127]. TRIPOD provides an ordinal score (1−4, with 4 being the most robust). These guidelines are not specific to radiomics studies, but provide insight into the level of validation in a study (details in Supplementary Table 4).
-
2
The Radiomics Quality Score (RQS) developed by Lambin and colleagues [128]. RQS provides a checklist to evaluate aspects of study design, by assessing various technical and statistical aspects of the radiomics pipeline. It consists of 16 components, each of which award or penalize points, to provide the RQS. The total number of points available range from -8 to 36 (the more points the better) and are often presented as a percentage (Supplementary Table 5).
-
3
Qualitative assessment of radiomics methodological limitations resulting from our literature review and labelled as A–H and listed in Table 3.
-
4
The reported evidence for added value of the radiomics signature to a clinical model of outcome tested in the study (for the patient outcome studies only). This provides an assessment of clinical utility.
5. Interpreting the quality of radiomics studies in NSCLC
Studies linking CT radiomics signatures to clinical outcome and tumor biology were found to have a high incidence of methodological limitations (summarized in Table 4). Overall, half of studies had a TRIPOD type of either 1a or 1b (meaning the results were not validated or validated within the same dataset). Only 13/75 studies had TRIPOD type of 3 or 4 (meaning the results were validated in an external dataset). The median RQS was 6 (range of -8 to 36). Details on RQS and TRIPOD are found in Supplementary Material. We found that 70 % of studies (52 of 75) had six or more methodological limitations, and no study had less than three methodological limitations. Finally, over half of studies relating radiomics to patient outcome did test the added benefit of the radiomic signature to a clinical model.
Table 4.
Summary of the 4 assessment criteria - TRIPOD score, RQS, number of methodological limitations and testing the added value of radiomics to a clinical model. The added value of radiomics to a clinical model was only tested for the patient outcome studies (N = 50).
| N = 75 | |
|---|---|
| TRIPOD type (n (%)) | |
| 1a – no validation | 10 (13) |
| 1b – internal validation | 27 (36) |
| 2a – dataset randomly split for validation | 18 (24) |
| 2b – dataset non-randomly split for validation | 7 (9) |
| 3 – external validation | 10 (13) |
| 4 – validation only | 3 (4) |
| RQS (median, [IQR]) | 6 [2−12.25] |
| Number of methodological limitations (n (%)) | |
| 0−2 | 0 (0) |
| 3 | 4 (5) |
| 4 | 4 (5) |
| 5 | 15 (20) |
| 6 | 21 (28) |
| 7 | 23 (31) |
| 8 | 8 (11) |
| N = 50 | |
| Added value of radiomics to clinical model tested? (n (%)) | |
| Yes | 32 (64) |
| No | 18 (36) |
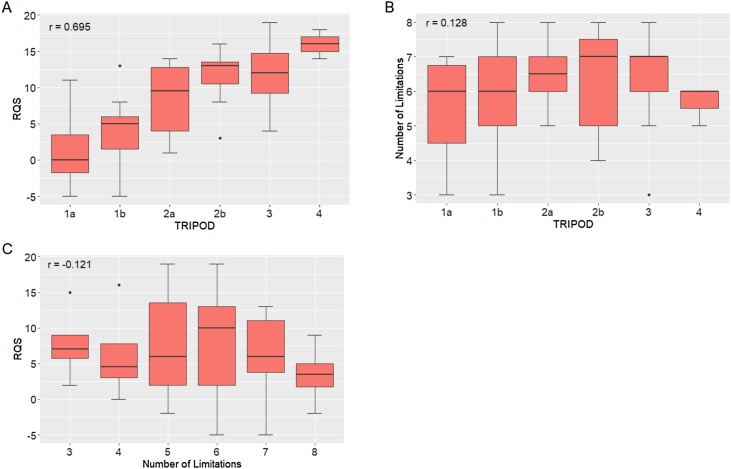
Our analysis suggests that the four assessment tools provide useful and complimentary critiques. Fig. 3A shows that the TRIPOD ordinal score focusing on validation and the RQS score focusing on study reporting are correlated (Pearson correlation coefficient 0.70). This reflects the importance the RQS places on study validation. However, both the TRIPOD score and RQS score were relatively independent of our assessment of study methodological limitations (Fig. 3B-C, Pearson correlation coefficients -0.12 and 0.13). Indeed, some studies with high TRIPOD and RQS scores had several technical limitations listed. For example, two studies with a TRIPOD score of 4 and the highest reported RQS scores (16 and 18 respectively) [14,15], had five and six identified methodological limitations respectively. In contrast, one study with a low TRIPOD score of 1b and a moderate RQS score (of 7) had just three pipeline technical limitations [18].
Fig. 3.
The assessment of the literature plotted against each other as boxplots. (A) RQS versus TRIPOD, (B) RQS versus the number of methodological limitations found in this review and (C) TRIPOD versus the number of methodological limitations found in this review.
An illustrative example is given by three studies [14,15,23] that externally validated the landmark radiomic signature developed by Aerts and colleagues in 2014 [13]. However, subsequent work [111,129] has suggested that the prognostic value of the signature reflected the correlation of the signature with tumor volume, rather than reflecting underlying tumor heterogeneity. An important workflow step our review identified is the assessment of feature correlations and potential confounders (G). While the RQS recommends performing multivariable analysis and testing the benefit of the radiomics signature to a gold standard, it does not explicitly recommend testing for feature correlations or confounders.
Study quality depends not only on quality of reporting, but also on ensuring that features used are robust against potential problems. There is a raised recognition of the methodological issues that limit the potential utility of the radiomics concept, as shown by the increase in studies in this area (Fig. 2). However, we find that only 39 % of the patient outcome studies and 50 % of the biology studies we identified cite methodology papers. This suggests that there is still limited appreciation of the need to employ more rigorous radiomics workflows. The IBSI guidelines and RQS are aimed at addressing these issues. For example the IBSI reference manual gives recommendations for image processing techniques as well as suggesting standardized feature definitions, nomenclature, and guidelines for reporting [121]. The RQS rewards the use of test-retest approaches, multiple segmentation analyses, and the use of phantoms to resolve inter-scanner differences.
However, our review of limitations highlights further concerns, such as differing slice thickness or voxel size (C) and the specification of grey-level binning size (F). These are not included in RQS (only 58 % of studies in Table 1, Table 2 specified the grey-level binning method or size). The IBSI guidelines, the RQS and TRIPOD assessment schemes are important steps that should improve the technical quality of radiomics studies. However, they are not sufficient alone and review of the literature suggests a need to either update them to include more granular limitations or to use them alongside other assessment tools.
One result of the increase prevalence of studies investigating methodological limitations that would accelerate clinical translation would be the identification of a subset of robust features that should be used in outcome studies. Unfortunately, comparing results across studies is difficult. In addition to the risks to reliability listed in Table 3, the software used for feature extraction often uses different nomenclature (one of issues the IBSI addresses) and can calculate ostensibly similar features in different ways and with different parameter settings so that they are not comparable [122]. Software use varied greatly across all studies included in this review. Of the patient outcome and biology studies, 15 % did not specify the software used, 48 % used in-house developed software and just 37 % used free or commercial options. These numbers are similar for the methodology studies; 14 % did not specify the software used, 40 % used in-house developed software and 47 % used free or commercial options. Four of the patient outcome and biology studies did not specify the features in the final radiomic signature at all. The result is that there is no consensus on which particular features or feature signatures should be used for clinical studies. However, there are now increasing numbers of studies that employ the techniques used to determine which features are reliable. Table 4 and Supplementary Tables 2 and 3 list the remaining limitations for each clinical and biological study - 42 % of the assessed studies applied at least one of the suggested solutions to methodological limitations to increase feature robustness. Of these studies, 46 % used a test re-test dataset, 58 % used multiple segmentations and 4% tested CT model dependence.
A further important step in the radiomics workflow where community consensus would increase the comparability of studies is that of the optimal machine learning techniques that should be used to develop the resulting statistical models. We found that the top feature reduction technique used in all studies was univariable analysis (53 %) followed by LASSO (27 %). The most common modelling technique was logistic regression (39 %) followed by cox regression (34 %). 16 % of studies used random forest and 11 % SVM, both of which were highlighted as high performing by the methodology studies [118,120]. The techniques used in each study are listed in Supplementary Tables 2 and 3. Four outcome studies used multiple modelling techniques to determine which one performed best on their data; a recommended method as model performance is dataset-dependent [126]. Out of these four studies, the best performing classifiers were random forest [72] and Naïve Bayes [38,67]. One study did not reveal the best performing model [69].
The lack of consensus in how to address limitations to the reliability of radiomics features, or of a preferred way to conduct the subsequent statistical modelling, means there is still significant variability in approach, with each finely tuned to its own particular dataset. Progress along the imaging biomarker translation roadmap [12] is dependent on the development of reliable measures that can be used to test clinical hypotheses. These findings agree with those of previous authors [121,128] and show there is still an unmet need to move away from the current heterogeneous landscape to one that is more standardized. The validation of existing signatures in different datasets [14,15,23] discussed above is a vital part of this effort.
Lastly, in addition to the assessment of technical quality, radiomic signatures need to be evaluated for clinical relevance. It is important to test whether incorporating radiomic features into a clinical model improves performance over known prognostic or predictive factors. This need is well-recognized with 64 % of the studies in in Table 1 making its assessment. Future studies will be most impactful if they explicitly evaluate the clinical utility of a radiomic signature as part of data reporting.
In summary, use of the four different assessment tools allows us to draw three conclusions. Firstly, there is a high prevalence of methodological limitations among CT radiomics studies exploring the potential of the approach to guide personalized medicine. Secondly, there remains considerable variability in the approach to addressing these limitations, and that modelling approaches are likely tuned to specific datasets. Thirdly, different assessment tools provided complementary information, which taken together provided the greatest insight into how study data could be improved.
6. Future directions
Personalized medicine is of great potential benefit to patients, but this vision is dependent on the identification of stratification and predictive biomarkers [5]. Imaging biomarkers, derived from routinely acquired patient images, have enormous translational potential given the ubiquity of imaging in clinical workflows. Evaluation of the radiomics literature in NSCLC reveals the exponential rate of publication of new radiomics studies, which, in their conclusions, present a very positive view of the potential for radiomics to deliver this goal.
This review puts these findings in context for NSCLC, but the messages are likely to be generic to all cancer types. All published studies are at risk of translational hurdles due to technical and methodological issues. Importantly, some of these limitations are well recognized, well investigated and have solutions proposed that are beginning to be applied to clinical studies. In distinction, other limitations are poorly understood or researched, and so substantial barriers to translation remain. In addition, wider concerns surrounding over-fitting data and biological validation persist. Lastly, no single radiomic signature or methodological approach is used widely, so further work is required to identify candidates to take forward in larger multicenter studies.
The fact that all the radiomics studies identified in the NSCLC literature have some limitations should not infer that the published data and conclusions are incorrect; rather that risk exists in interpreting their findings at face value. Standardization issues, variability in methodology and a general lack of reporting hinders comparison of results across studies. Identifying limitations, by employing recognized assessment methodology tools, can help inform and educate design of future radiomics studies in NSCLC and beyond. This will improve study quality and expedite the translation of radiomic biomarkers as tools in personalized medicine.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by Cancer Research UK through the Cancer Research UK Manchester Centre: [C147/A18083] and [C147/A25254]; and an Advanced Clinician Scientist Fellowship to Professor James P B O’Connor [C19221/A22746]. Professor Corinne Faivre-Finn and Professor James P B O’Connor are supported by the NIHR Manchester Biomedical Research Centre.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.lungcan.2020.05.028.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Institute SEER Cancer Statistics Review, 1975 - 2011. https://seer.cancer.gov/archive/csr/1975_2011/ Accessed September 9, 2019.
- 3.Cancer Research UK . 2010. Lung Cancer Survival Statistics.https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/lung-cancer/survival Accessed February 19, 2020. [Google Scholar]
- 4.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 5.Schilsky R.L. Personalized medicine in oncology: the future is now. Nat. Rev. Drug Discov. 2010;9(5):363–366. doi: 10.1038/nrd3181. [DOI] [PubMed] [Google Scholar]
- 6.Salem A., Asselin M.C., Reymen B. Targeting hypoxia to improve non–small cell lung cancer outcome. J. Natl. Cancer Inst. 2018;110(1):14–29. doi: 10.1093/jnci/djx160. [DOI] [PubMed] [Google Scholar]
- 7.de Langen A.J., van den Boogaart V., Lubberink M. Monitoring response to antiangiogenic therapy in non-small cell lung cancer using imaging markers derived from PET and dynamic contrast-enhanced MRI. J. Nucl. Med. 2011;52(1):48–55. doi: 10.2967/jnumed.110.078261. [DOI] [PubMed] [Google Scholar]
- 8.Nishino M., Hatabu H., Johnson B.E., McLoud T.C. State of the art: response assessment in lung cancer in the era of genomic medicine. Radiology. 2014;271(1):6–27. doi: 10.1148/radiol.14122524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillies R.J., Anderson A.R., Gatenby R.A., Morse D.L. The biology underlying molecular imaging in oncology: from genome to anatome and back again. Clin. Radiol. 2010;65(7):517–521. doi: 10.1016/j.crad.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aerts H.J.W.L. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2(12):1636–1642. doi: 10.1001/jamaoncol.2016.2631. [DOI] [PubMed] [Google Scholar]
- 12.O’Connor J.P.B., Aboagye E.O., Adams J.E. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 2017;14(3):169–186. doi: 10.1038/nrclinonc.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aerts H.J.W.L., Velazquez E.R., Leijenaar R.T.H. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5(4006):4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Timmeren J.E., Leijenaar R.T.H., van Elmpt W. Survival prediction of non-small cell lung cancer patients using radiomics analyses of cone-beam CT images. Radiother. Oncol. 2017;123(3):363–369. doi: 10.1016/j.radonc.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Grossmann P., Stringfield O., El-Hachem N. Defining the biological basis of radiomic phenotypes in lung cancer. Elife. 2017;6:1–22. doi: 10.7554/eLife.23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu W., Tang C., Hobbs B.P. Development and validation of a predictive radiomics model for clinical outcomes in stage I non-small cell lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 2017;102(4):1090–1097. doi: 10.1016/j.ijrobp.2017.10.046. [DOI] [PubMed] [Google Scholar]
- 17.Chaddad A., Desrosiers C., Toews M., Abdulkarim B. Predicting survival time of lung cancer patients using radiomic analysis. Oncotarget. 2017;8(61):104393–104407. doi: 10.18632/oncotarget.22251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fave X., Zhang L., Yang J. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci. Rep. 2017;7(1):588. doi: 10.1038/s41598-017-00665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q., Kim J., Balagurunathan Y. CT imaging features associated with recurrence in non-small cell lung cancer patients after stereotactic body radiotherapy. Radiat. Oncol. 2017;12(1):158. doi: 10.1186/s13014-017-0892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q., Kim J., Balagurunathan Y. Imaging features from pretreatment CT scans are associated with clinical outcomes in nonsmall-cell lung cancer patients treated with stereotactic body radiotherapy. Med. Phys. 2017;44(8):4341–4349. doi: 10.1002/mp.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang C., Hobbs B., Amer A. Development of an immune-pathology informed radiomics model for non-small cell lung cancer. Sci. Rep. 2018;8(1):1922. doi: 10.1038/s41598-018-20471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianconi F., Fravolini M.L., Bello-Cerezo R., Minestrini M., Scialpi M., Palumbo B. Evaluation of shape and textural features from CT as prognostic biomarkers in non-small cell lung cancer. Anticancer Res. 2018;38(4):2155–2160. doi: 10.21873/anticanres.12456. [DOI] [PubMed] [Google Scholar]
- 23.de Jong E.E.C., van Elmpt W., Rizzo S. Applicability of a prognostic CT-based radiomic signature model trained on stage I-III non-small cell lung cancer in stage IV non-small cell lung cancer. Lung Cancer. 2018;124:6–11. doi: 10.1016/j.lungcan.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 24.Lee G., Park H., Sohn I. Comprehensive computed tomography radiomics analysis of lung adenocarcinoma for prognostication. Oncologist. 2018;23(7):806–813. doi: 10.1634/theoncologist.2017-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He B., Zhao W., Pi J.Y. A biomarker basing on radiomics for the prediction of overall survival in non-small cell lung cancer patients. Respir. Res. 2018;19(1):199. doi: 10.1186/s12931-018-0887-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starkov P., Aguilera T.A., Golden D.I. The use of texture-based radiomics CT analysis to predict outcomes in early-stage non-small cell lung cancer treated with stereotactic ablative radiotherapy. Br. J. Radiol. 2019;92(1094):20180228. doi: 10.1259/bjr.20180228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Yang J., Zhou X. Development of a radiomics nomogram based on the 2D and 3D CT features to predict the survival of non-small cell lung cancer patients. Eur. Radiol. 2019;29(5):2196–2206. doi: 10.1007/s00330-018-5770-y. [DOI] [PubMed] [Google Scholar]
- 28.Wang L., Dong T., Xin B. Integrative nomogram of CT imaging, clinical, and hematological features for survival prediction of patients with locally advanced non-small cell lung cancer. Eur. Radiol. 2019;29(6):2958–2967. doi: 10.1007/s00330-018-5949-2. [DOI] [PubMed] [Google Scholar]
- 29.Shi L., Rong Y., Daly M. Cone-beam computed tomography-based delta-radiomics for early response assessment in radiotherapy for locally advanced lung cancer. Phys. Med. Biol. 2019;65(1):15009. doi: 10.1088/1361-6560/ab3247. [DOI] [PubMed] [Google Scholar]
- 30.van Timmeren J.E., van Elmpt W., Leijenaar R.T.H. Longitudinal radiomics of cone-beam CT images from non-small cell lung cancer patients: evaluation of the added prognostic value for overall survival and locoregional recurrence. Radiother. Oncol. 2019;136:78–85. doi: 10.1016/j.radonc.2019.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang L., Chen J., Hu W. Assessment of a radiomic signature developed in a general NSCLC cohort for predicting overall survival of ALK-positive patients with different treatment types. Clin. Lung Cancer. 2019;20(6):e638–e651. doi: 10.1016/j.cllc.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Franceschini D., Cozzi L., De Rose F. A radiomic approach to predicting nodal relapse and disease-specific survival in patients treated with stereotactic body radiation therapy for early-stage non-small cell lung cancer. Strahlenther. Onkol. 2019;(November) doi: 10.1007/s00066-019-01542-6. [DOI] [PubMed] [Google Scholar]
- 33.Coroller T.P., Grossmann P., Hou Y. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 2015;114(3):345–350. doi: 10.1016/j.radonc.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattonen S.A., Palma D.A., Johnson C. Detection of local cancer recurrence after stereotactic ablative radiation therapy for lung cancer: physician performance versus radiomic assessment. Int. J. Radiat. Oncol. Biol. Phys. 2016;94(5):1121–1128. doi: 10.1016/j.ijrobp.2015.12.369. [DOI] [PubMed] [Google Scholar]
- 35.Huynh E., Coroller T.P., Narayan V. CT-based radiomic analysis of stereotactic body radiation therapy patients with lung cancer. Radiother. Oncol. 2016;120(2):258–266. doi: 10.1016/j.radonc.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 36.Huynh E., Coroller T.P., Narayan V. Associations of radiomic data extracted from static and respiratory-gated CT scans with disease recurrence in lung cancer patients treated with SBRT. PLoS One. 2017;12(1):e0169172. doi: 10.1371/journal.pone.0169172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou T.H., Coroller T.P., van Griethuysen J.J.M., Mak R.H., Aerts H.J.W.L. Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. PLoS One. 2018;13(11):e0206108. doi: 10.1371/journal.pone.0206108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira Junior J.R., Koenigkam-Santos M., Cipriano F.E.G., Fabro A.T., de Azevedo-Marques P.M. Radiomics-based features for pattern recognition of lung cancer histopathology and metastases. Comput. Methods Programs Biomed. 2018;159:23–30. doi: 10.1016/j.cmpb.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Yang X., Pan X., Liu H. A new approach to predict lymph node metastasis in solid lung adenocarcinoma: a radiomics nomogram. J. Thorac. Dis. 2018;10(Suppl 7):S807–S819. doi: 10.21037/jtd.2018.03.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong Y., Yuan M., Zhang T., Zhang Y.D., Li H., Yu T.F. Radiomics approach to prediction of occult mediastinal lymph node metastasis of lung adenocarcinoma. Am. J. Roentgenol. 2018;211(1):109–113. doi: 10.2214/AJR.17.19074. [DOI] [PubMed] [Google Scholar]
- 41.Lafata K.J., Hong J.C., Geng R. Association of pre-treatment radiomic features with lung cancer recurrence following stereotactic body radiation therapy. Phys. Med. Biol. 2019;64(2) doi: 10.1088/1361-6560/aaf5a5. [DOI] [PubMed] [Google Scholar]
- 42.Akinci D’Antonoli T., Farchione A., Lenkowicz J. CT radiomics signature of tumor and peritumoral lung parenchyma to predict nonsmall cell lung cancer postsurgical recurrence risk. Acad. Radiol. 2020;27(4):497–507. doi: 10.1016/j.acra.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 43.He L., Huang Y., Yan L., Zheng J., Liang C., Liu Z. Radiomics-based predictive risk score: a scoring system for preoperatively predicting risk of lymph node metastasis in patients with resectable non-small cell lung cancer. Chin. J. Cancer Res. 2019;31(4):641–652. doi: 10.21147/j.issn.1000-9604.2019.04.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X., Huang L., Chen J. Application of radiomics signature captured from pretreatment thoracic CT to predict brain metastases in stage III/IV ALK-positive non-small cell lung cancer patients. J. Thorac. Dis. 2019;11(11):4516–4528. doi: 10.21037/jtd.2019.11.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreira-Junior J.R., Koenigkam-Santos M., Magalhães Tenório A.P. CT-based radiomics for prediction of histologic subtype and metastatic disease in primary malignant lung neoplasms. Int. J. Comput. Assist. Radiol. Surg. 2020;15(1):163–172. doi: 10.1007/s11548-019-02093-y. [DOI] [PubMed] [Google Scholar]
- 46.Cong M., Feng H., Ren J.L. Development of a predictive radiomics model for lymph node metastases in pre-surgical CT-based stage IA non-small cell lung cancer. Lung Cancer. 2020;139:73–79. doi: 10.1016/j.lungcan.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 47.Coroller T.P., Agrawal V., Narayan V. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother. Oncol. 2016;119(3):480–486. doi: 10.1016/j.radonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Y., Liu Z., He L. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non—small cell lung cancer. Radiology. 2016;281(3):947–957. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 49.Song J., Dong D., Huang Y., Zang Y., Liu Z., Tian J. Association between tumor heterogeneity and progression-free survival in non-small cell lung cancer patients with EGFR mutations undergoing tyrosine kinase inhibitors therapy. Conf Proc IEEE Eng Med Biol Soc. 2016:1268–1271. doi: 10.1109/EMBC.2016.7590937. [DOI] [PubMed] [Google Scholar]
- 50.Coroller T.P., Agrawal V., Huynh E. Radiomic-based pathological response prediction from primary tumors and lymph nodes in NSCLC. J. Thorac. Oncol. 2017;12(3):467–476. doi: 10.1016/j.jtho.2016.11.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tunali I., Gray J.E., Qi J. Novel clinical and radiomic predictors of rapid disease progression phenotypes among lung cancer patients treated with immunotherapy: an early report. Lung Cancer. 2019;129:75–79. doi: 10.1016/j.lungcan.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moran A., Daly M.E., Yip S.S.F., Yamamoto T. Radiomics-based assessment of radiation-induced lung injury after stereotactic body radiotherapy. Clin. Lung Cancer. 2017;18(6):e425–e431. doi: 10.1016/j.cllc.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 53.Krafft S.P., Rao A., Stingo F. The utility of quantitative CT radiomics features for improved prediction of radiation pneumonitis. Med. Phys. 2018;45(11):5317–5324. doi: 10.1002/mp.13150. [DOI] [PubMed] [Google Scholar]
- 54.Yuan M., Liu J.Y., Zhang T., Zhang Y.D., Li H., Yu T.F. Prognostic impact of the findings on thin-section computed tomography in stage 1 lung adenocarcinoma with visceral pleural invasion. Sci. Rep. 2018;8(1):4743. doi: 10.1038/s41598-018-22853-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang M., Ren Y., She Y. Imaging phenotype using radiomics to predict dry pleural dissemination in non-small cell lung cancer. Ann. Transl. Med. 2019;7(12):259. doi: 10.21037/atm.2019.05.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aerts H.J.W.L., Grossmann P., Tan Y. Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC. Sci. Rep. 2016;6:33860. doi: 10.1038/srep33860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rios Velazquez E., Parmar C., Liu Y. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017;77(14):3922–3930. doi: 10.1158/0008-5472.CAN-17-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mei D., Luo Y., Wang Y., Gong J. CT texture analysis of lung adenocarcinoma: can radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging. 2018;18(1):52. doi: 10.1186/s40644-018-0184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Digumarthy S.R., Padole A.M., Gullo R.L., Sequist L.V., Kalra M.K. Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Medicine (Baltimore) 2019;98(1):e13963. doi: 10.1097/MD.0000000000013963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia T.Y., Xiong J.F., Li X.Y. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modelling. Eur. Radiol. 2019;29(9):4742–4750. doi: 10.1007/s00330-019-06024-y. [DOI] [PubMed] [Google Scholar]
- 61.Li S., Ding C., Zhang H., Song J., Wu L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med. Phys. 2019;46(10):4545–4552. doi: 10.1002/mp.13747. [DOI] [PubMed] [Google Scholar]
- 62.Tu W., Sun G., Fan L. Radiomics signature: a potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer. 2019;132:28–35. doi: 10.1016/j.lungcan.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 63.Yang X., Dong X., Wang J. Computed tomography-based radiomics signature: a potential Indicator of epidermal growth factor receptor mutation in pulmonary adenocarcinoma appearing as a subsolid nodule. Oncologist. 2019;24(11):e1156–e1164. doi: 10.1634/theoncologist.2018-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X., Kong C., Xu W. Decoding tumor mutation burden and driver mutations in early stage lung adenocarcinoma using CT-based radiomics signature. Thorac. Cancer. 2019;10(10):1904–1912. doi: 10.1111/1759-7714.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bak S.H., Park H., Lee H.Y. Imaging genotyping of functional signaling pathways in lung squamous cell carcinoma using a radiomics approach. Sci. Rep. 2018;8(1):1–9. doi: 10.1038/s41598-018-21706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patil R., Mahadevaiah G., Dekker A. An approach toward automatic classification of tumor histopathology of non–small cell lung cancer based on radiomic features. Tomography. 2016;2(4):374–377. doi: 10.18383/j.tom.2016.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu W., Parmar C., Grossmann P. Exploratory study to identify radiomics classifiers for lung cancer histology. Front. Oncol. 2016;6(71):1–11. doi: 10.3389/fonc.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu X., Dong D., Chen Z. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur. Radiol. 2018;28(7):2772–2778. doi: 10.1007/s00330-017-5221-1. [DOI] [PubMed] [Google Scholar]
- 69.E L., Lu L., Li L., Yang H., Schwartz L.H., Zhao B. Radiomics for classifying histological subtypes of lung cancer based on multiphasic contrast-enhanced computed tomography. J. Comput. Assist. Tomogr. 2019;43(2):300–306. doi: 10.1097/RCT.0000000000000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu J., Cui J., Liu F., Yuan Y., Guo F., Zhang G. Multi-subtype classification model for non-small cell lung cancer based on radiomics: SLS model. Med. Phys. 2019;46(7):3091–3100. doi: 10.1002/mp.13551. [DOI] [PubMed] [Google Scholar]
- 71.Zhou B., Xu J., Tian Y., Yuan S., Li X. Correlation between radiomic features based on contrast-enhanced computed tomography images and Ki-67 proliferation index in lung cancer: a preliminary study. Thorac. Cancer. 2018;9(10):1235–1240. doi: 10.1111/1759-7714.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu Q., Feng Z., Liang Q. Machine learning-based radiomics strategy for prediction of cell proliferation in non-small cell lung cancer. Eur. J. Radiol. 2019;118:32–37. doi: 10.1016/j.ejrad.2019.06.025. [DOI] [PubMed] [Google Scholar]
- 73.Song S.H., Park H., Lee G. Imaging phenotyping using radiomics to predict micropapillary pattern within lung adenocarcinoma. J. Thorac. Oncol. 2017;12(4):624–632. doi: 10.1016/j.jtho.2016.11.2230. [DOI] [PubMed] [Google Scholar]
- 74.Chen X., Fang M., Dong D. A radiomics signature in preoperative predicting degree of tumor differentiation in patients with non–small cell lung cancer. Acad. Radiol. 2018;25(12):1548–1555. doi: 10.1016/j.acra.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 75.She Y., Zhang L., Zhu H. The predictive value of CT-based radiomics in differentiating indolent from invasive lung adenocarcinoma in patients with pulmonary nodules. Eur. Radiol. 2018;28(12):5121–5128. doi: 10.1007/s00330-018-5509-9. [DOI] [PubMed] [Google Scholar]
- 76.Yang B., Guo L., Lu G., Shan W., Duan L., Duan S. Radiomic signature: a non-invasive biomarker for discriminating invasive and non-invasive cases of lung adenocarcinoma. Cancer Manag. Res. 2019;11:7825–7834. doi: 10.2147/CMAR.S217887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.2017. FDA Device Classification. QuantX.https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/denovo.cfm?ID=DEN170022 Accessed February 19, 2020. [Google Scholar]
- 78.2020. Medical F. TexRAD.https://fbkmed.com/texrad-landing-2/ Accessed February 22, 2020. [Google Scholar]
- 79.Ger R.B., Zhou S., Chi P.C.M. Comprehensive investigation on controlling for CT imaging variabilities in radiomics studies. Sci. Rep. 2018;8(1):13047. doi: 10.1038/s41598-018-31509-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fave X., Mackin D., Yang J. Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer? Med. Phys. 2015;42(12):6784–6797. doi: 10.1118/1.4934826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hepp T., Othman A., Liebgott A., Kim J.H., Pfannenberg C., Gatidis S. Effects of simulated dose variation on contrast-enhanced CT-based radiomic analysis for non-small cell lung cancer. Eur. J. Radiol. 2020;124:108804. doi: 10.1016/j.ejrad.2019.108804. [DOI] [PubMed] [Google Scholar]
- 82.Kakino R., Nakamura M., Mitsuyoshi T. Comparison of radiomic features in diagnostic CT images with and without contrast enhancement in the delayed phase for NSCLC patients. Phys. Med. 2020;69:176–182. doi: 10.1016/j.ejmp.2019.12.019. [DOI] [PubMed] [Google Scholar]
- 83.Mackin D., Ger R., Dodge C. Effect of tube current on computed tomography radiomic features. Sci. Rep. 2018;8(1):2354. doi: 10.1038/s41598-018-20713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mackin D., Fave X., Zhang L. Measuring computed tomography scanner variability of radiomics features. Invest. Radiol. 2015;50(11):757–765. doi: 10.1097/RLI.0000000000000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mahmood U., Apte A.P., Deasy J.O., Schmidtlein C.R., Shukla-Dave A. Investigating the robustness neighborhood gray tone difference matrix and gray level co-occurrence matrix radiomic features on clinical computed tomography systems using anthropomorphic phantoms: evidence from a multivendor study. J. Comput. Assist. Tomogr. 2017;41(6):995–1001. doi: 10.1097/RCT.0000000000000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Midya A., Chakraborty J., Gönen M., Do R.K.G., Simpson A.L. Influence of CT acquisition and reconstruction parameters on radiomic feature reproducibility. J. Med. Imaging. 2018;5(01):011020. doi: 10.1117/1.jmi.5.1.011020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yasaka K., Akai H., Mackin D. Precision of quantitative computed tomography texture analysis using image filtering. Med. (United States) 2017;96(21):e6993. doi: 10.1097/MD.0000000000006993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim Y.J., Lee H.J., Kim K.G., Lee S.H. The effect of CT scan parameters on the measurement of CT radiomic features: a lung nodule phantom study. Comput. Math. Methods Med. 2019;2019:8790694. doi: 10.1155/2019/8790694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhovannik I., Bussink J., Traverso A. Learning from scanners: Bias reduction and feature correction in radiomics. Clin. Transl. Radiat. Oncol. 2019;19:33–38. doi: 10.1016/j.ctro.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tunali I., Hall L.O., Napel S. Stability and reproducibility of computed tomography radiomic features extracted from peritumoral regions of lung cancer lesions. Med. Phys. 2019;46(11):5075–5085. doi: 10.1002/mp.13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mahon R.N., Ghita M., Hugo G.D., Weiss E. ComBat harmonization for radiomic features in independent phantom and lung cancer patient computed tomography datasets. Phys. Med. Biol. 2020;65(1):15010. doi: 10.1088/1361-6560/ab6177. [DOI] [PubMed] [Google Scholar]
- 92.Lafata K., Cai J., Wang C., Hong J., Kelsey C.R., Yin F.F. Spatialoral variability of radiomic features and its effect on the classification of lung cancer histology. Phys. Med. Biol. 2018;63(22):225003. doi: 10.1088/1361-6560/aae56a. [DOI] [PubMed] [Google Scholar]
- 93.Du Q., Baine M., Bavitz K. Radiomic feature stability across 4D respiratory phases and its impact on lung tumor prognosis prediction. PLoS One. 2019;14(5):e0216480. doi: 10.1371/journal.pone.0216480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Larue R.T.H.M., van Timmeren J.E., de Jong E.E.C. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. (Madr) 2017;56(11):1544–1553. doi: 10.1080/0284186X.2017.1351624. [DOI] [PubMed] [Google Scholar]
- 95.Mackin D., Fave X., Zhang L. Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLoS One. 2017;12(9):e0178524. doi: 10.1371/journal.pone.0178524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shafiq-ul-hassan M., Zhang G.G., Latifi K. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017;44(3):1050–1062. doi: 10.1002/mp.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu L., Ehmke R.C., Schwartz L.H., Zhao B. Assessing agreement between radiomic features computed for multiple CT imaging settings. PLoS One. 2016;11(12):e0166550. doi: 10.1371/journal.pone.0166550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shafiq-Ul-Hassan M., Latifi K., Zhang G., Ullah G., Gillies R., Moros E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci. Rep. 2018;8(1):10545. doi: 10.1038/s41598-018-28895-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Y., Lu L., Xiao M. CT slice thickness and convolution kernel affect performance of a radiomic model for predicting EGFR status in non-small cell lung cancer: a preliminary study. Sci. Rep. 2018;8(1):17913. doi: 10.1038/s41598-018-36421-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park S., Lee S.M., Do K.H. Deep learning algorithm for reducing CT slice thickness: effect on reproducibility of radiomic features in lung cancer. Korean J. Radiol. 2019;20(10):1431. doi: 10.3348/kjr.2019.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shafiq-ul-Hassan M., Zhang G.G., Hunt D.C. Accounting for reconstruction kernel-induced variability in CT radiomic features using noise power spectra. J. Med. Imaging. 2017;5(1):11013. doi: 10.1117/1.JMI.5.1.011013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao B., Tan Y., Tsai W.Y. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci. Rep. 2016;6(1):23428. doi: 10.1038/srep23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haga A., Takahashi W., Aoki S. Classification of early stage non-small cell lung cancers on computed tomographic images into histological types using radiomic features: interobserver delineation variability analysis. Radiol. Phys. Technol. 2018;11(1):27–35. doi: 10.1007/s12194-017-0433-2. [DOI] [PubMed] [Google Scholar]
- 104.Huang Q., Lu L., Dercle L. Interobserver variability in tumor contouring affects the use of radiomics to predict mutational status. J. Med. Imaging. 2017;5(1):011005. doi: 10.1117/1.jmi.5.1.011005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kalpathy-Cramer J., Mamomov A., Zhao B. Radiomics of lung nodules: a multi-institutional study of robustness and agreement of quantitative imaging features. Tomography. 2016;2(4):430–437. doi: 10.18383/j.tom.2016.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Owens C.A., Peterson C.B., Tang C. Lung tumor segmentation methods: impact on the uncertainty of radiomics features for non-small cell lung cancer. PLoS One. 2018;13(10):1–22. doi: 10.1371/journal.pone.0205003. Fan Y, ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parmar C., Rios Velazquez E., Leijenaar R. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One. 2014;9(7):e102107. doi: 10.1371/journal.pone.0102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pavic M., Bogowicz M., Wurms X. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol. (Madr) 2018;57(8):1070–1074. doi: 10.1080/0284186X.2018.1445283. [DOI] [PubMed] [Google Scholar]
- 109.Wang H.Y.C., Donovan E.M., Nisbet A. The stability of imaging biomarkers in radiomics: a framework for evaluation. Phys. Med. Biol. 2019;64(16):165012. doi: 10.1088/1361-6560/ab23a7. [DOI] [PubMed] [Google Scholar]
- 110.Fave X., Zhang L., Yang J. Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non-small cell lung cancer. Transl. Cancer Res. 2016;5(4):349–363. doi: 10.21037/tcr.2016.07.11. [DOI] [Google Scholar]
- 111.Welch M.L., McIntosh C., Haibe-Kains B. Vulnerabilities of radiomic signature development: the need for safeguards. Radiother. Oncol. 2019;130:2–9. doi: 10.1016/j.radonc.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 112.Choi W., Riyahi S., Kligerman S.J., Liu C.-J., Mechalakos J.G., Lu W. Technical note: identification of CT texture features robust to tumor size variations for normal lung texture analysis. Int. J. Med. Phys. Clin. Eng. Radiat. Oncol. 2018;7(3):330–338. doi: 10.4236/ijmpcero.2018.73027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Larue R.T.H.M., Van De Voorde L., Van Timmeren J.E. 4DCT imaging to assess radiomics feature stability: an investigation for thoracic cancers. Radiother. Oncol. 2017;125(1):147–153. doi: 10.1016/j.radonc.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 114.Zwanenburg A., Leger S., Agolli L. Assessing robustness of radiomic features by image perturbation. Sci. Rep. 2019;9(1):614. doi: 10.1038/s41598-018-36938-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tanaka S., Kadoya N., Kajikawa T. Investigation of thoracic four-dimensional CT-based dimension reduction technique for extracting the robust radiomic features. Phys. Med. 2019;58:141–148. doi: 10.1016/j.ejmp.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 116.Van Timmeren J.E., Leijenaar R.T.H., van Elmpt W. Test–retest data for radiomics feature stability analysis: generalizable or study-specific? Tomography. 2016;2(4):361–365. doi: 10.18383/j.tom.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haga A., Takahashi W., Aoki S. Standardization of imaging features for radiomics analysis. J. Med. Invest. 2019;66(12):35–37. doi: 10.2152/jmi.66.35. [DOI] [PubMed] [Google Scholar]
- 118.Zhang Y., Oikonomou A., Wong A., Haider M.A., Khalvati F. Radiomics-based prognosis analysis for non-small cell lung cancer. Sci. Rep. 2017;7(1):46349. doi: 10.1038/srep46349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun W., Jiang M., Dang J., Chang P., Yin F.F. Effect of machine learning methods on predicting NSCLC overall survival time based on Radiomics analysis. Radiat. Oncol. 2018;13(1):197. doi: 10.1186/s13014-018-1140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Parmar C., Grossmann P., Bussink J., Lambin P., Aerts H.J.W.L. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 2015;5(1):13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zwanenburg A., Leger S., Vallières M., Löck S. Image biomarker standardisation initiative. arXiv Prepr. 2020 arXiv161207003. [Google Scholar]
- 122.Fornacon-Wood I., Mistry H., Ackermann C.J. Reliability and prognostic value of radiomic features are highly dependent on choice of feature extraction platform. Eur. Radiol. 2020 doi: 10.1007/s00330-020-06957-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bogowicz M., Leijenaar R.T.H., Tanadini-Lang S. Post-radiochemotherapy PET radiomics in head and neck cancer – the influence of radiomics implementation on the reproducibility of local control tumor models. Radiother. Oncol. 2017;125(3):385–391. doi: 10.1016/j.radonc.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 124.Foy J.J., Armato S.G., Al-Hallaq H.A. Effects of variability in radiomics software packages on classifying patients with radiation pneumonitis. J. Med. Imaging. 2020;7(01):1. doi: 10.1117/1.jmi.7.1.014504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.O’Connor J.P.B. Rethinking the role of clinical imaging. Elife. 2017;6:e30563. doi: 10.7554/eLife.30563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Deist T.M., Dankers F.J.W.M., Valdes G. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: an empirical comparison of classifiers. Med. Phys. 2018;45(7):3449–3459. doi: 10.1002/mp.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Collins G.S., Reitsma J.B., Altman D.G., Moons K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. Eur. Urol. 2015;67(6):1142–1151. doi: 10.1016/j.eururo.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 128.Lambin P., Leijenaar R.T.H., Deist T.M. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 129.Vallières M., Kay-Rivest E., Perrin L.J. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci. Rep. 2017;7(1):10117. doi: 10.1038/s41598-017-10371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.