Abstract
Cold atmospheric plasma (CAP) has been demonstrated to be a successful antiseptic for chronic and infected wounds. Although experimental work has focused on elucidation of the curative power of CAP for wound healing, the molecular mechanisms behind this ability are less understood. To date, the direct effect of CAP on the activity of microbial virulence factors has not been investigated. In the present study, we therefore examined whether CAP can modulate the detrimental activity of M1 protein, one of the most studied Streptococcus pyogenes virulence determinant. Our results show that CAP abolishes the ability of M1 protein to trigger inflammatory host responses. Subsequent mass spectrometric analysis revealed that this effect was caused by oxidation of Met81 and Trp128 located at the sub-N-terminal region of M1 protein provoking a conformational change. Notably, our results also show that CAP has an insignificant effect on the host immune system, supporting the benefits of using CAP to combat infections. Considering the growing number of antibiotic-resistant bacteria, novel antimicrobial therapeutic approaches are urgently needed that do not bear the risk of inducing additional resistance. Our study therefore may open new research avenues for the development of novel approaches for the treatment of skin and wound infections caused by S. pyogenes
Keywords: Cold atmospheric plasma, Oxidation, Virulence factor, Streptococcus pyogenes, M1 protein
Introduction
Cold atmospheric plasma (CAP) is emerging as a promising tool for medical applications as an antiseptic for infections and wound management. Though its therapeutic application has been investigated for many years, progress on deciphering the molecular mechanisms of CAP was only recently reported [1]. CAP is a partially ionised gas producing reactive oxygen or nitrogen species, ions, and charged particles as well as thermal and ultraviolet radiation [2, 3]. CAP is also an interesting clinical treatment option because of its antimicrobial activity towards many bacterial species. In addition, CAP has been found to inactivate viruses and to promote wound healing in vivo [4, 5, 6, 7, 8, 9, 10, 11, 12]. Subsequent risk assessment studies have demonstrated that CAP is non-mutagenic and well tolerated by healthy tissues because of its temperature of <40°C [13, 14]. Thus, CAP is an interesting tool to search for novel antimicrobial therapeutic concepts.
The process of wound healing from skin infections is complex and highly organised to control homeostasis, inflammation, proliferation, and remodelling [15]. Immediate immune responses are important as soon as the skin barrier is disrupted and microorganisms can invade the host in order to establish infection. A substantial portion of such infections are attributed to the gram-positive bacterium Streptococcus pyogenes (group A streptococci) [16, 17]. Group A streptococci can evoke a variety of skin diseases ranging from minor infections, such as impetigo or pyoderma, to serious deep infections, such as cellulitis and necrotising fasciitis [18]. In severe circumstances, streptococcal skin infections can further develop into life-threatening systemic infections, such as bacteraemia and streptococcal toxic shock syndrome, often associated with high rates of morbidity and mortality [16, 19]. To establish skin infection, group A streptococci have evolved several mechanisms involving expression of a number of virulence factors that can modulate the host environment [20]. Among the most important and extensively studied are the M and M-like proteins [21]. M proteins are composed of two polypeptide chains complexed in an alpha-helical coiled-coil structure attached to the bacterial cell wall. Notably, M and M-like proteins are used for the classification of streptococcal serotypes. The M1 serotype is still the most prevalent serotype in streptococcal infections and overrepresented in infections evoking invasive disease [22]. M1 protein has previously been reported to be a potent activator of neutrophils, monocytes, and T cells [23, 24, 25, 26]. Activation of these cells will result in massive release of inflammatory cytokines, a clinical feature found in streptococcal toxic shock syndrome and septic shock [26]. Under inflammatory conditions, M1 protein can be cleaved off from the bacterial surface by endogenous or host-derived proteinases [23, 27]. In solution, the streptococcal protein can form complexes with fibrinogen which can for instance deposit in tissue biopsies of a patient suffering from necrotising fasciitis and streptococcal toxic shock syndrome [23, 27].
The growing number of antibiotic-resistant bacteria constitutes a significant clinical problem that is expected to increase further. Thus, there is an urgent need for new treatment approaches with novel non-antibiotic modes of action. This said, a great deal of work has been undertaken to study the antimicrobial activity of CAP and to provide molecular explanations of the improved wound healing observed upon CAP treatment in vivo. Though CAP treatment is antiseptic towards all types of bacterial species tested, little is known on whether the effect of CAP treatment can abolish the detrimental activity of bacterial virulence factors. To our knowledge, only one article on this topic has been published so far [28].
We therefore decided to investigate whether CAP can modulate the pro-inflammatory activities of M1 protein. We used the kINPen plasma jet, operated with pure argon as gas supply, to generate CAP [29]. Our results show that CAP-treated M1 protein lost its capacity to induce an inflammatory response. Consequently, wound healing improved and inflammation from keratinocytes and immune cells was diminished. We found that oxidation of Met81 and Trp128 was responsible for the loss of the detrimental effects of M1 protein. Based on our results, we speculate that CAP can also destroy the activity of other virulence factors and that our findings can thus be used for the development of novel antimicrobial treatment options.
Materials and Methods
Cell Culture and Reagents
The spontaneously immortalised human keratinocyte cell line HaCaT, purified leukocytes in RPMI (Gibco, Life Technologies, Carlsbad, CA, USA), and whole blood diluted 1:1 in RPMI were used in this study. HaCaT cells were cultured in serum-free keratinocyte medium without calcium chloride (Invitrogen, Carlsbad, CA, USA) and supplemented with 25 μg/mL bovine pituitary extract, 0.2 ng/mL epidermal growth factor, and 1× Gibco's 100× Antibiotic-Antimycotic (Life Technologies) [30]. The streptococcal M1 protein used in this study was purified from an isogenic S. pyogenes AP1-mutant strain expressing an M1 protein lacking the cell wall-anchoring region [31]. This M1 protein construct therefore accumulates in the growth medium, from which it was purified by affinity chromatography using immobilised human fibrinogen coupled to Sepharose. A detailed description of the purification procedure has previously been published [32]. When working with wild-type bacteria, overnight cultures of the S. pyogenes M1 serotype AP1 were cultivated to mid-exponential growth phase in Todd-Hewitt broth (Difco, Becton Dickinson, Franklin Lakes, NJ, USA) supplemented with 0.2% yeast extract at 37°C in a 5% CO2 atmosphere. The bacteria were then used for determination of bacterial killing by CAP treatment. Cultures were diluted 1:20 before seeding onto agar plates.
Non-Thermal/CAP Source and Plasma Treatments
In order to create CAP the kINPen MED (neoplas tools GmbH, Greifswald, Germany) was employed. Plasma is generated as a jet with 1 mm diameter by a voltage being applied to air between the walls of the device and a central electrode. Argon gas is used to expulse the plasma [29]. The argon gas flow was constant at 3 L/min, 2.5 bar gas pressure, and at a fixed distance in all experiments. The streptococcal M1 protein was subjected to CAP for 2, 5, and 15 s or indicated time points in cell culture medium containing 0.1% FBS or PBS before addition to cells. The same procedure was repeated for cell culture medium containing 0.1% FBS, without M1 protein, which served as control together with untreated cells. Cells in suspension or human whole blood were pretreated with CAP for indicated times and incubated 30 min before the addition of M1 protein or CAP-treated M1 protein to the cell suspensions, cell culture media, and whole blood (diluted 1:1 in RPMI) and incubated for 30 min at 37°C on rotation. Samples were centrifuged and supernatants or plasma were collected. Untreated cells served as controls. For the in vivo experiment, 0.5 mL of M1 protein (250 μg/mL) were treated with CAP for 15 s. The streptococcal M1 serotype AP1 was cultured on agar plates and treated with CAP for 5, 15, and 30 s from a fixed distance (8–10 mm, recommended in vivo treatment distance from the manufacturer) and incubated overnight at 37°C. Colony-free zones were measured (mm) and colony-forming units within the colony-free zones were counted.
In vitro Wound Healing Assay
Wound scratch assays were performed with HaCaT cells grown to 85–90% confluence. Scratches were made using a 200-μL pipette tip across the diameter of each well. Cells were washed with PBS twice to remove cellular debris. Cell culture medium containing 0.1% FBS with or without CAP pretreatment, or supplemented with M1 protein (5 μg/mL) with or without CAP pretreatment, were added to cells and incubated at 37°C in a 5% CO2 incubator for 96 h after scratching. After 48 h of incubation, cells were washed with PBS and the cell culture medium was exchanged without CAP treatment, supplementation of M1 protein, or CAP-treated M1 protein in order to reproduce a recovery phase. Untreated scratched cells served as control. Cell images were monitored every 24 h using an Olympus CKX41 microscope with an Olympus SC30 camera and the cellSens Entry software (Olympus, Tokyo, Japan). Cell images were presented in grey scale with minor contrast adjustments applied to the entire image area of all images. Quantitative analysis of the wound area from three independent experiments was performed using the ImageJ software with the macro MRI Wound Healing Tool and presented as percentage of initial wound area: wound area (%) = wound area T / wound area T0 × 100, where T is the treatment time and T0 is the time when the scratch was initially created.
Enzyme-Linked Immunosorbent Assay
IL-8 levels in the keratinocyte culture supernatants or IL-6 levels in human and mouse plasma were measured by a sandwich enzyme-linked immunosorbent assay (ELISA; DuoSet ELISA development kit; R&D Systems) according to the manufacturer's protocol. Briefly, the plates were incubated overnight at 4°C with 100 μL capture antibody diluted in coating buffer. The contents of the wells were blocked with coating buffer containing 0.5% bovine serum albumin. Standards and samples diluted in assay buffer were added to the wells and incubated. Wells were then washed, followed by the addition of detection antibody diluted in assay buffer. The contents of the wells were incubated with streptavidin conjugated to horseradish peroxidase before the visualisation of antibody binding by adding 100 μL 3,3′,5,5′-tetramethylbenzidine solution, and the reaction was stopped by addition of 1.8 M H2SO4. Absorbance was measured at 450 nm using a microplate reader (iMarkTM microplate absorbance reader, Bio-Rad Laboratories, Hercules, CA, USA). To address neutrophil activation and degranulation, heparin-binding protein (HBP) levels in plasma were measured using ELISA as described above. Microtiter plates were coated with a mouse monoclonal antibody directed against human HBP (2F23A; 0.3 μg/mL in coating buffer) [33]. A polyclonal rabbit antiserum towards human HBP, diluted 1:7,000, was used as detection antibody. The ability of M1 protein to bind fibrinogen following CAP treatment was analysed with ELISA by coating microtiter plates with fibrinogen (5 μg/mL) in coating buffer. Dilution series of M1 protein and CAP-treated M1 protein were added to wells. A polyclonal rabbit anti-M1 antibody, diluted 1:10,000, was used as primary antibody. Antibody bindings were later completed by addition of a goat anti-rabbit IgG HRP conjugated antibody (1:3,000, Bio-Rad Laboratories) and visualised by addition of 0.05% ABTS (Sigma Aldrich) and 0.01% H2O2 in substrate buffer. Absorbance was measured at 405 nm. All measurements were performed in duplicate. Data are presented as mean cytokine expression (pg/mL), normalised to that of control or original mean absorbance values ± SD from at least three independent experiments.
Apoptosis Assay
HaCaT cells were cultured to confluence in 24-well plates (Invitrogen) before treatment with CAP, M1 protein, or CAP-treated M1 protein and incubated for 24 h at 37°C in a 5% CO2 incubator. Apoptotic cells detached from the wells were collected by adsorbing the supernatants, concentrated by centrifugation, and manually counted by the use of a Burker chamber and an Olympus CKX41 microscope. The data represent the mean percentage of dead cells compared to the total number of cells (confluent 24-well plates from Invitrogen are considered to be 0.2 × 106 cells/well) ± SD from three independent experiments.
Electron Microscopy
For scanning electron microscopy, HaCaT cells were incubated with M1 protein (5 μg/mL) or CAP-treated M1 protein for 15 s (5 μg/mL) and fixed overnight at room temperature using 2.5% glutaraldehyde in cacodylate buffer. Cells were washed with cacodylate buffer and dehydrated with an ascending ethanol series from 50% (v/v) to absolute ethanol. The cells were then subjected to critical point drying with carbon dioxide, and absolute ethanol was used as an intermediate solvent. Samples were mounted on aluminum holders, sputtered with 20 nm palladium/gold, and examined in a JEOL JSM-350 scanning electron microscope using a secondary electron detector (Everhart-Tornley detector, JEOL style).
SDS-PAGE/Gel Electrophoresis
Streptococcal M1 protein and CAP-treated M1 protein were added to SDS sample buffer and heated for 5 min at 95°C. Samples were cooled on ice before equal amounts of protein were loaded to a TGX mini protean gel for any kDa and run in TBS buffer at 200 V as constant until the sample buffer almost passed through the gel. The gel was stained with GelCode Blue Safe protein stain (Thermo Scientific, Waltham, MA, USA) for 1 h at room temperature and later destained with ultra-pure water on shake at room temperature overnight. Images of the gels were captured by a GelDocTM EZ Imager system (Bio-Rad Laboratories). One representative gel is presented out of three independent experiments.
Sample Preparation and HDX-MS Analysis
The hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments were carried out using automated sample preparation on a LEAP H/D-X PALTM platform interfaced to an liquid chromatography-MS analysis system, comprising an Ultimate 3000 micro-LC coupled to an Orbitrap Q Exactive+ MS.
Pepsin Digestion and HDX-MS
Samples of 5 μL untreated M1 protein (control) and CAP-treated (15 s) M1 protein (CAP) at a concentration of 1 mg/mL were diluted 1:5 ratio with pure PBS or an HDX labelling buffer prepared in D2O, dPBS, pH 7.0. HDX labelling reactions were carried out at 0, 30, 300, and 3,000 s at 20°C. The labelling was quenched by addition of 1% FA and 3 M urea, pH 2.5 at 1°C. Quenched samples of 50 μL were injected and subjected to online pepsin digestion at 4°C (Life Technologies, pepsin column, 2.1 × 30 mm). The loading pump used for online digestion and trapping was run at 50 µL/min using a mobile phase of 0.1% formic acid, pH 2.5, corresponding to approximately 3 min of digestion. The digested peptides were collected on a PepMap300 trapping column (1 mm × 15 mm) and subjected to a 60-s trap wash. Thereafter, the trap column was switched in over a reversed-phase analytical column, Hypersil GOLD, particle size 1.9 µm, 1 × 50 mm, and separation was performed at 1°C using a gradient of 5–40% B over 13 min; the mobile phases were 0.1% formic acid (A) and 90% acetonitrile/10% water/0.1% formic acid (B). Following the shallow gradient, the trap and column were washed using a saw tooth gradient of 5–90% B, 1 min followed by 1 min of equilibration at the highest and lowest organic content. The needle port and sample loop were cleaned between injections with mobile phase B followed by mobile phase A. Between each sample injection the pepsin column was washed by injecting 90 μL of wash solution, 1% TFA/3 M urea/4% ACN. In order to control carryover, a full blank was run between all sample injections. Separated peptides were analysed on a Q Exactive Plus MS, equipped with a HESI source, operated at a capillary temperature of 250°C. For non-deuterated samples (t = 0 s) data-dependent MS/MS HCD spectra of resulting peptides were also acquired. For HDX analysis (labelled samples) MS full-scan spectra were collected at a resolution setting of 70K.
Data Analysis
PEAKS Studio 8.5 Bioinformatics Solutions Inc. (BSI, Waterloo, ON, Canada) was used for peptide identification after pepsin digestion of non-deuterated samples (i.e., time point 0 s for both the control and CAP-treated samples). The search was done on a database containing only M1 protein, with a mass error tolerance of 15 ppm and a fragment mass error tolerance of 0.05 Da, allowing for fully unspecific cleavage. Variable modifications allowed: dioxidation (M): lysine oxidation to aminoadipic semialdehyde; oxidation (HWM): oxidation or hydroxylation (C), oxidation to pyroglutamic acid, and tryptophan oxidation to hydroxykynurenine.
In order to obtain a quantitative measurement of the oxidations observed in the non-deuterated pepsin-digested samples, the MS data were also searched against the M1 FASTA file using Thermo Biopharma Finder v3.0. The Biopharma Finder software was used to facilitate in-depth characterisation of M1 by peptide mapping (using both ms and ms/ms spectra). The search parameters included S/N ratio set at default value 32, low-specificity pepsin, and a mass accuracy set to 5 ppm with a minimum confidence of 0.8 to ensure a low level of false-positive identifications. The quantitative values are presented as percent abundance based on the sum of all modified components divided by the sum of all components [34].
HDX-MS Analysis
Peptides identified by PEAKS in both non-deuterated control and CAP samples, having a peptide score value of log p > 30, were used to generate a peptide list for the HDX analysis. For the CAP-treated sample, Met81 oxidation was added as a point modification in the sequence. HDX data analysis and visualisation was performed using HDExaminer version 2.5 (Sierra Analytics, Modesto, CA, USA). Due to the comparative nature of the study the deuterium incorporation levels for the peptic peptides were directly derived from the observed mass difference between the deuterated and non-deuterated peptides without back-exchange correction using a fully deuterated sample.
Chemotactic Activity Assay
The chemotactic ability of supernatants from keratinocytes, stimulated with CAP treatment and M1 protein, was assessed by applying a Boyden chamber assay. Cell culture inserts with 0.3-μm polycarbonate membrane (Corning Inc., Corning, NY, USA) were used. Whole blood was collected in citrate tubes and leukocytes were purified using a density gradient (PolymorphprepTM, Axis-Shield PoC AS, Oslo, Norway) according to the manufacturer's protocol. A mixed population of leukocytes was added to the inner chambers of cell culture inserts in a 24-well plate containing HaCaT supernatant incubated for 24 h without treatment (control), treated with 15 s of CAP, pretreated with 15 s CAP 1 h before the addition of M1 protein (5 μg/mL), M1 protein (5 μg/mL), or just M1 protein in cell culture medium (serum-free medium + M1 protein). Cells were incubated for 4 h at 37°C and 5% CO2. Cells that migrated through the membrane were quantified by cell counting without gating using flow cytometry during 1 min/sample. The chemotactic index was calculated by dividing counted cell numbers observed with counted cell numbers to that of control (untreated HaCaT supernatant). p values were determined by comparing the means of the chemotactic indices for three independent experiments using different donors.
In vitro Permeability Assay
Keratinocyte monolayer permeability, induced by M1 protein or CAP treatment, was studied using the In Vitro Vascular Permeability Assay kit (Millipore) according to the manufacturer's protocol. Briefly, HaCaT cells were seeded onto 24-well plate inserts containing membranes with 1-μm pores pre-coated with type 1 rat tail collagen. The HaCaT cells were cultured for 72 h to ensure 100% confluence and formation of tight junctions. Inserts without a cell monolayer were included as positive control. Cell culture media were changed to fresh media (control), media containing M1 protein (5 µg/mL), or media containing 15 s of CAP-treated M1 protein (5 µg/mL) and incubated for 48 h at 37°C and 5% CO2. For CAP treatment of cells, media were exchanged for fresh media pretreated with CAP for 15 s 30 min before the end of incubation in order to study the immediate effects of CAP application. Culture inserts were then washed once with PBS and transferred to a new 24-well plate containing 0.5 mL PBS and added 150 µL FITC-dextran into the inserts and incubated covered from light at room temperature for 20 min. Inserts were removed and 100 µL from the receiver wells were transferred to a black 96-well plate in four replicates/sample. The amount of FITC-dextran that passed through the cell monolayer was measured by a fluorescence plate reader (Viktor3 model 1420; PerkinElmer, Waltham, MA, USA) at 485/535 nm. Data are presented as relative permeability to that of control (%) ± SD from three independent experiments.
In vitro Phagocytosis Assay
Polymorphonuclear leukocytes were purified from whole blood and collected in citrate tubes by using a density gradient (PolymorphprepTM) according to the manufacturer's protocol. Leukocytes (100,000 cells/well) were plated into a black 96-well plate containing RPMI without phenol red (Gibco, Life Technologies). Cells were treated with CAP for 2, 5, and 15 s, added M1 protein (1 µg/mL) or CAP-pretreated M1 protein (15 s, 1 µg/mL), and incubated for 30 min at 37°C. pHodoTM Green Staphylococcus aureus BioParticles® Conjugate (Life Technologies) were prepared in RPMI without phenol red according to the manufacturer's protocol. Cell culture medium was removed by centrifugation and aspiration using a multi-pipette and leukocytes were resuspended in 100 µL pHodoTM BioParticles® suspension. Cells were incubated and covered from light for 2 h at 37°C in order for phagocytosis of the fluorescent bio-particles to take place. Results were obtained by reading the fluorescence intensity using a fluorescence plate reader (Viktor3 model 1420) at 485/535 nm. Relative phagocytosis activity was calculated from the mean fluorescent intensity of three replicates per sample, subtracted background from bio-particle suspension and cell suspension, and calculated as a fraction (percent effect) of the control phagocytosis (untreated leukocytes). Data are represented as the average phagocytosis effect (percent of control) ± SD from three independent experiments using different donors.
Statistical Analysis
One-way ANOVA was used for comparisons of IL-8, IL-6, and HBP levels as well as for the apoptosis assay, followed by Tukey's multiple comparisons test. The chemotactic activity and phagocytosis assays was analysed by one-way ANOVA and Dunnett's multiple comparisons test. Two-way ANOVA and Tukey's multiple comparisons test were used to analyse scratch areas. All statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA). p values were considered statistically significant at p < 0.05.
Results
CAP Treatment of the Streptococcal M1 Protein Improves Wound Healing Recovery
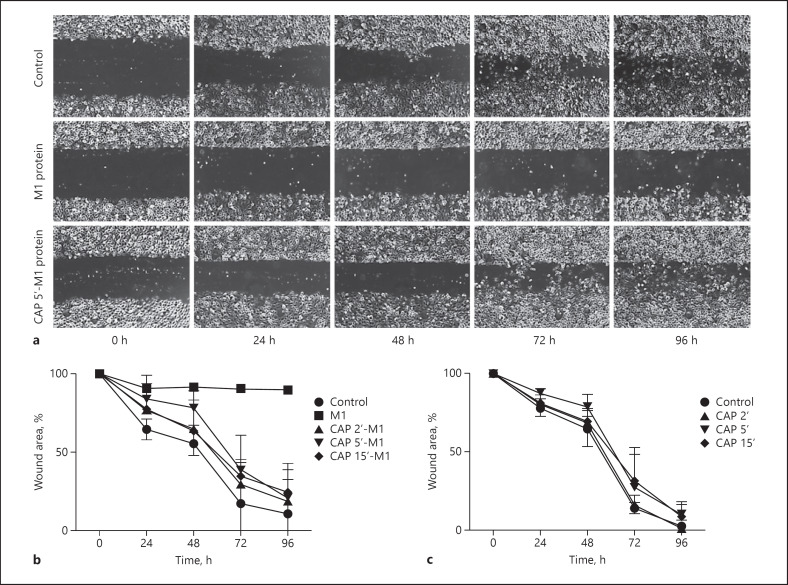
CAP has been demonstrated to have antimicrobial effects. In order to study whether CAP can also modulate the activity of important virulence factors, several analyses were performed using the streptococcal M1 protein. We have previously reported that M1 protein can induce inflammatory signalling in keratinocytes as well as abolish wound healing activities in vitro [35, 36]. We started by investigating whether CAP, in low tissue-tolerable doses, had any effect on M1 protein and wound healing activities by employing a wound scratch assay. Figure 1a shows that M1 protein-treated HaCaT cells lose their ability to proliferate upon scratching compared to control, reaching full confluence 96 h after scratching. This impairment persists even when the bacterial protein is removed from the cells during cell culture media exchange after 48 h of incubation. When M1 protein was pretreated with short CAP treatment during 2, 5, and 15 s, the wound healing activity was improved, and after removal of the bacterial protein, full recovery and confluence of the HaCaT cells were observed. In order to measure the effect on wound healing, the non-recovered wound areas were quantified using the different experimental conditions. As depicted in Figure 1b, no recovery was monitored when keratinocytes were incubated with untreated M1 protein and also after the removal of the streptococcal protein cell proliferation was completely inhibited. On the contrary, keratinocytes incubated with CAP-treated M1 protein adopted a similar wound healing pattern as seen in controls. Though some minor detention was observed, subsequent analysis revealed that the delays were not statistically significant (Fig. 1b). To exclude that the CAP-treated culture medium was responsible for any impaired wound healing, the experiments were also performed in the absence of M1 protein. As depicted in Figure 1c, no changes in the recovery phase were noted under these experimental conditions. Together our data suggest that CAP modulates the bacterial virulence factor in favour of the host.
Fig. 1.
M1 protein-triggered inhibition of cell recovery processes is abolished upon CAP treatment. a Representative cell images of the ability of keratinocytes to heal a scratch after treatment with M1 protein (5 μg/mL), CAP-treated M1 protein (5 μg/mL), or left untreated, followed over 96 h. After 48 h, cell culture media were exchanged without addition/supplementation of bacterial protein. Cell images were monitored every 24 h and wound areas were quantified by the ImageJ software utilising the macro MRI Wound Healing Tool. b Summary of scratch assays supplemented with M1 protein and CAP-treated M1 protein at indicated time points and HaCaT cells added (c) with CAP-treated cell culture media without bacterial protein at indicated time points. Untreated cells served as control. All results were analysed by two-way ANOVA and Tukey's multiple comparisons test, resulting in a significant difference (p < 0.01) of M1 protein-treated cells compared to control after 48 h as well as compared to all groups (p < 0.0001) after 72 and 96 h. The remaining comparisons were statistically insignificant. The data represent the average percentage of initial wound area ± SD from three independent experiments for every group. CAP, cold atmospheric plasma.
Reduced Toxicity of CAP-Treated M1 Protein
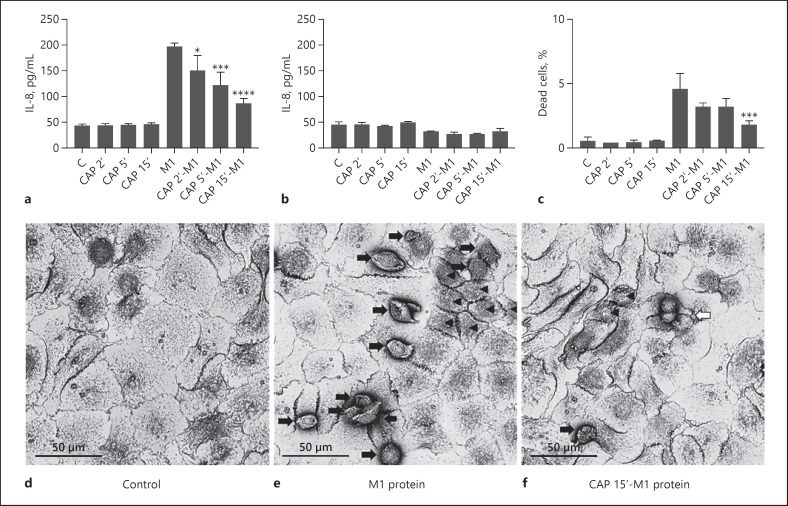
Our previous work has shown that HaCaT cells respond to M1 protein by release of the inflammatory cytokine IL-8 [35]. Thus, we next studied whether CAP can alter the inflammatory response of these cells. Figure 2a illustrates that IL-8 release was down-regulated when cells were incubated with CAP-treated M1. Notably, when M1 protein was omitted in these experiments, CAP-treated cell culture medium was not able to trigger the mobilisation of the cytokine (Fig. 2a). In the next series of experiments, HaCaT cells were stimulated under the same experimental conditions, followed by a washing step to remove the bacterial protein from samples. Cells were then incubated in culture medium for another 24 h before the release of IL-8 was measured. Figure 2b shows that no increase in the levels of the cytokine was measured using these experimental settings. To further investigate the cellular viability when challenged with M1 protein and CAP-treated M1 protein, apoptosis assays were carried out and scanning electron microscopy was employed. Figure 2c shows that treatment with M1 protein significantly increased the number of dead cells (p < 0.0001), while CAP-treated M1 protein mediated less apoptotic turnover. With increasing time, CAP treatment sequentially prevented M1 protein-induced cell death compared to untreated M1 protein, reaching statistical significance at 15 s (p = 0.0002). Using CAP-treated growth medium in the absence of M1 protein as a negative control, no cell death was recorded under the same experimental settings. Scanning electron microscopy assessed the cellular morphology of HaCaT cells challenged with M1 protein, CAP-treated M1 protein, or left untreated (Fig. 2d–f). The micrographs revealed that M1 protein-treated cells were shrunken in size with dissolved tight junctions and more cell death. These features were not seen in the untreated control group having an intact monolayer. Cells challenged with CAP-treated M1 protein were less shrunken, maintaining the cell monolayer, and had replicating cells, indicating a much better cellular status compared to cells treated with native M1 protein. The cellular improvement on HaCaT cells caused by CAP treatment of M1 protein prompted us to evaluate the antimicrobial activity of CAP on living streptococci (online suppl. Fig. S1; for all online suppl. material, see www.karger.com/doi/10.1159/000502959). A dose-dependent, almost complete killing of streptococci was demonstrated with tissue-tolerable CAP treatment. The antibacterial effect also conveyed from the plasma jet of 1 mm diameter up to 12 mm, although with declining bacterial killing efficiency the further away from the plasma jet. Taken together, short CAP treatment, capable of killing S. pyogenes, can also abolish detrimental effects of M1 protein on keratinocytes.
Fig. 2.
CAP treatment reduces M1 protein-induced toxicity of keratinocytes. a IL-8 release into HaCaT supernatants stimulated with M1 protein (5 μg/mL), CAP-treated M1 protein (5 μg/mL) at indicated time points, or CAP-pretreated cell culture media, incubated for 24 h. b IL-8 release into HaCaT supernatants 24 h after cell culture media exchange without the addition of treatments. The data represent mean expression values ± SD from three independent experiments. c The number of detached apoptotic cells in cell culture medium from HaCaT cells stimulated with M1 protein, CAP-treated M1 protein, and CAP-treated cell culture media without bacterial protein at indicated time points during 24 h. Untreated cells served as control. The data represent the percentage of dead cells ± SD from three independent experiments. All results were analysed by one-way ANOVA and Tukey's multiple comparisons test; * p < 0.05, *** p < 0.001, ****p < 0.0001. d–f Representative images from scanning electron microscopy of untreated HaCaT cells (d) and HaCaT cells incubated with M1 protein (e) or CAP-treated (15 s) M1 protein (f) for 48 h. Black arrows indicate apoptotic cells, arrowheads indicate detached, shrunken cells, and the white arrow indicates replicating cells. CAP, cold atmospheric plasma.
Protein Chemical and Structural Analysis of CAP-Treated M1 Protein
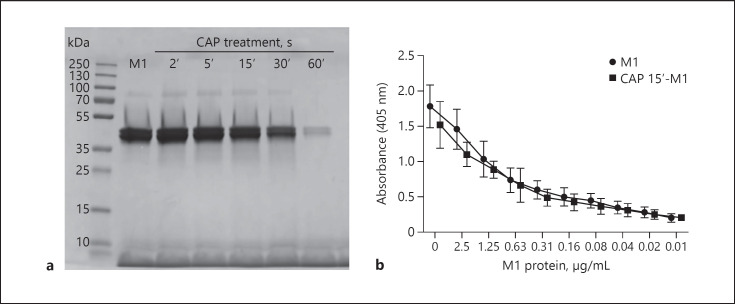
Having shown that CAP reduces M1 protein-induced toxicity on keratinocytes, the next set of experiments was carried out to investigate what alterations CAP introduces to the streptococcal M1 protein. First, we performed SDS-PAGE analysis of M1 protein and CAP-treated M1 protein to study whether longer CAP treatment, such as 30 or 60 s, can evoke degradation or vaporisation of the bacterial protein (Fig. 3a). For the shorter CAP treatments (2–15 s), cleavage or fragmentation were ruled out as explanation for the disarmed M1 protein (Fig. 3a). We next tested the ability of M1 protein and CAP-treated M1 protein to bind to fibrinogen. To this end, we performed ELISA analysis, revealing no biological difference in fibrinogen binding (Fig. 3b). Both results indicate that M1 protein, when treated for <30 s, undergoes no major structural conformational changes. Based on these results, we concluded that a variable region of the M1 protein was preserved without protein degradation, although short CAP treatment has been applied to the virulence factor.
Fig. 3.
M1 protein is not degraded upon short-time CAP treatment. a Representative image of SDS-PAGE with M1 protein treated with CAP at indicated time points, showing intact M1 protein but reduced quantity with longer CAP treatment. M1 protein has an apparent molecular weight in the range of 38–40 kDa. b Binding of M1 protein and CAP-treated M1 protein (15 s) to fibrinogen coated to ELISA plates. The data represent mean absorbance values ± SD from three independent experiments. CAP, cold atmospheric plasma; ELISA, enzyme-linked immunosorbent assay.
CAP Induces Oxidation of M1 Protein
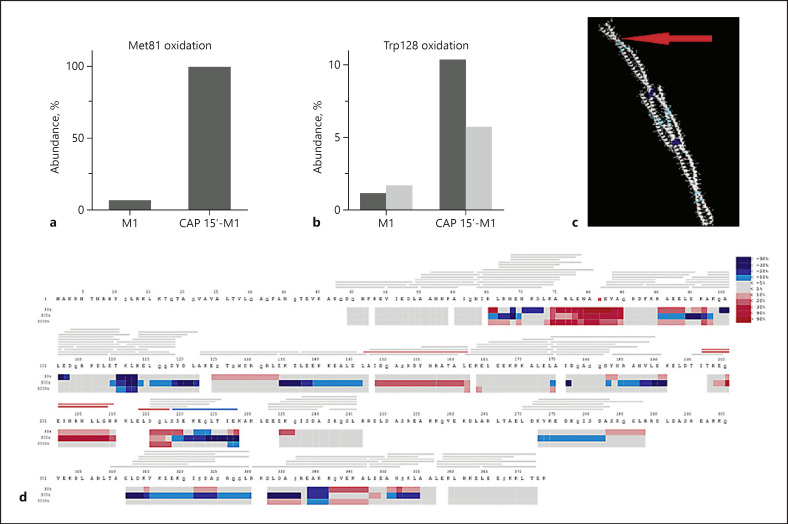
Cold plasma treatment of proteins can be expected to induce a multitude of different chemical modifications. The most likely modification is different forms of oxidations. It has been shown that the relative reactivity of amino acids exposed to plasma treatment follows the order Met > Cys > Trp > Phe > Tyr > His [37]. The high-sequence coverage and redundancy obtained by pepsin digestion as applied in HDX-MS provides a suitable technique for investigation of oxidative modifications introduced by CAP treatment of the streptococcal virulence factor M1. The sequence coverage and total ion current (online suppl. Fig. S2) for the control (native M1 protein) and the CAP-treated sample (M1 protein treated with CAP for 15 s) was almost identical. However, in the CAP-treated sample all ten peptides spanning Met81 were identified with oxidation, while for the control sample only two peptides were identified with Met81 oxidised (Fig. 4). An additional oxidation site was identified at Trp128, where both single and double oxidation of one peptide could be followed in both samples. Data analysis with Biopharma Finder revealed that for the CAP-treated sample, oxidation at Met81 was almost complete (>99.9%), compared to 6.7% for the untreated control sample (Fig. 5a). The single oxidation state observed at Trp128 was 10.4% for the CAP-treated sample and 1.2% for the control sample, and double oxidation of Trp128 reached levels of 5.7% (CAP) and 1.7% (control) (Fig. 5b). Considering the high level of oxidation observed after CAP treatment, especially for Met81, it is very likely that both conformation and function of the M1 protein are affected (Fig. 5c). The obtained HDX data indicate that the CAP-treated M1 protein undergoes complex conformational changes. The main observations were an apparent increased flexibility in the region 74–84 for the CAP-treated sample (Fig. 5d). Here, all observed oxidised Met81-spanning peptides have a high deuterium uptake already at the 30-s time point (Fig. 5c and supplementary uptake plots). Note that there is a degree of uncertainty due to the low HDX coverage for the control sample in this region. In most native proteins, the deuterium uptake will follow EX2 kinetics, a gradual increase with time due to fast conformational fluctuations. The HDX data for the CAP-treated sample showed regions with an increased/changed bimodal isotope distribution (online suppl. Fig. S3); this kinetic behaviour can be “true EX1” or “false EX1” due to aggregation or multimerization, abnormal complete back exchange, or carryover [38]. While carryover could be excluded as a source for the observed EX1 kinetics, it is well known that protein oxidation can result in denaturation and aggregation, which could have been a source for the observed EX1 kinetics [39, 40, 41]. Further, more extensive HDX studies are required in order to more accurately pinpoint involved regions and understand the nature of the structural changes introduced to the M1 protein by CAP treatment.
Fig. 4.
Oxidation of M1 protein following CAP treatment measured by HDX-MS. M1 sequence coverage after pepsin digestion of M1 protein (a) and CAP-treated M1 protein (b). For the CAP-treated sample, all peptides spanning Met81 showed considerable oxidation (yellow boxes) compared to the untreated control sample. CAP, cold atmospheric plasma; HDX-MS, hydrogen deuterium exchange mass spectrometry.
Fig. 5.
CAP treatment of M1 protein leads to oxidation of Met81 and Trp128. a Relative abundance of peptides with Met81 oxidation as reported by Biopharma Finder. In the CAP-treated sample all observed peptides with Met81 were oxidised, while in the untreated control sample the peak area corresponding to peptides with oxidised Met81 was <7%. b Relative abundance of oxidised Trp128 showing that 1.2% of Trp128 was oxidised and 1.7% of Trp128 was double oxidised for untreated M1 protein. When treating M1 protein with CAP, these oxidations increased to 10.4 and 5.7%, respectively. c HDX differential deuterium uptake at 30 s, overlaid on PDB 2OTO. The red arrow indicates the position of Met81 and the blue areas the structures that have lower deuterium uptake in CAP-treated samples. d The HDX heat-map shows the difference in deuterium uptake between the untreated M1 protein and the CAP-treated samples. Bars show peptide coverage, coloured as the average of all time points, and heat-map regions coloured. It is evident that CAP treatment leads to significantly increased uptake (red colours) in the region 74–85 flexible/disordered as in free Spx. Individual uptake curves for observed peptides can be found in online supplementary Figure S5. CAP, cold atmospheric plasma; HDX, hydrogen deuterium exchange.
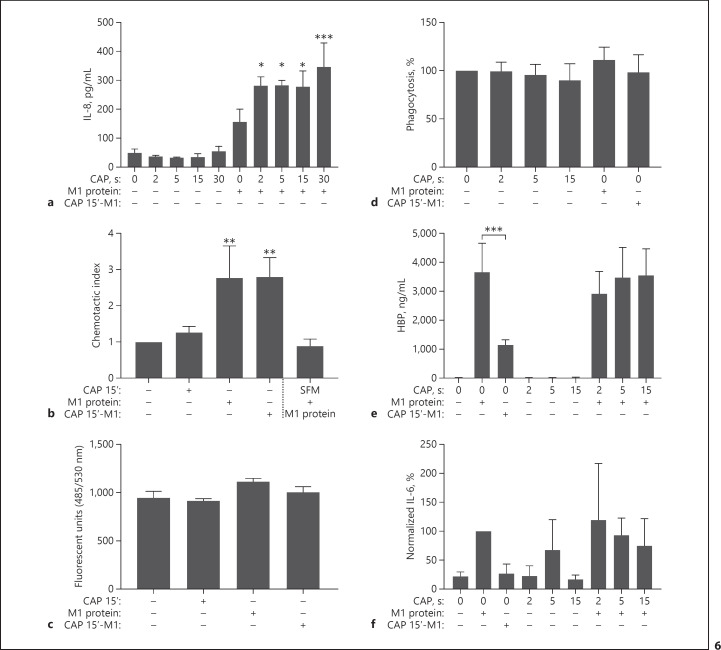
CAP Does Not Abrogate Important Host Immune Responses
The next set of experiments aimed to elucidate how CAP interferes with other important functions present in infected wounds. Figure 6a shows that keratinocytes pretreated with CAP are potentiated in their IL-8 response towards M1 protein compared to unstimulated cells. However, the CAP-induced rise in IL-8 levels did not result in an increased chemotactic activity, as shown in Figure 6b. For these experiments, HaCaT cells were treated with CAP followed by stimulation with M1 protein. After 24 h, IL-8-containing supernatants were collected and their chemotactic activity was tested as described in Materials and Methods. M1 protein in serum-free medium served as a negative control. The results show that supernatants from CAP-treated HaCaT cells only exerted chemoattractive activity when they were collected from M1-stimulated cells. The signal obtained was not distinguishable from supernatants of non-CAP-treated cells stimulated with M1 protein (Fig. 6b), suggesting that the CAP had no influence on chemotaxis under these experimental conditions. Figure 6c demonstrates that the permeability of keratinocytes remains stable, as no significant differences were seen when HaCaT cells were challenged with M1 protein, CAP-treated M1 protein, or 15 s of CAP treatment. Together, these data show that CAP treatment did not impair essential and initial functions of the innate immune response.
Fig. 6.
Innate immune responses triggered by M1 protein are reduced upon CAP-treated M1 protein but not altered by CAP in the absence of M1 protein. a Increased IL-8 expression in HaCaT supernatants pretreated with CAP at indicated time points before the addition of M1 protein (5 μg/mL), incubated for 24 h. Untreated cells served as control. The data represent mean expression values ± SD from three independent experiments. Results were analysed by one-way ANOVA and Tukey's multiple comparisons test; * p < 0.05, *** p < 0.001. b Chemotactic activity of culture supernatants from HaCaT cells, untreated or pretreated with CAP for 15 s, incubated with or without M1 protein for 24 h and soluble M1 protein (5 μg/mL) in serum-free media were assessed using Boyden chamber assay. Migrated leukocytes were quantified using flow cytometry. The data represent the average chemotactic indices ± SD from three independent experiments, using different donors. Results were analysed by one-way ANOVA and Dunnett's multiple comparisons test; ** p < 0.01. c The ability of HaCaT cells to sustain a barrier when challenged with CAP (15 s), M1 protein, or CAP-treated M1 protein was addressed in a permeability assay. The results are expressed as the number of fluorescent units that passed through the HaCaT cell monolayer. The data represent the mean fluorescent units ± SD from three independent experiments. d No altered ability in phagocytosis was seen when purified leukocytes were treated with CAP at indicated time points, M1 protein (1 μg/mL), or CAP-treated (15 s) M1 protein (1 μg/mL). The data represent the average percentage of phagocytosis to that of control ± SD from three independent experiments using different donors. e, f HBP (e) and IL-6 (f) secretion levels in whole blood, diluted 1:1 in RPMI, stimulated with M1 protein (1 μg/mL), CAP-treated (15 s) M1 protein (1 μg/mL), or CAP treatment for indicated time points, incubated for 24 h. Three independent experiments were carried out using different donors. HBP levels are expressed as mean expression values ± SD and were analysed by one-way ANOVA and Tukey's multiple comparisons test; *** p < 0.001. IL-6 levels were normalised to M1-induced IL-6 levels and expressed as average percentage ± SD, due to donor-dependent variation in expression levels. The original data can be found in online supplementary Figure S4. CAP, cold atmospheric plasma; HBP, heparin-binding protein.
In the infected wound, leukocytes are recruited to combat intruding bacteria. A phagocytosis assay was carried out in order to investigate whether CAP treatment or M1 protein would favour or deteriorate phagocytosis function (Fig. 6d). Briefly, bio-particles from S. aureus, conjugated to a fluorescent dye, were added to purified leukocytes followed by incubation with M1 protein, CAP-treated M1 protein, and CAP-treated medium. Upon digestion by leukocytes the particles translocated to phagosomes. With the acidification in the phagosome it is possible to measure emission of the pH-sensitive dye. As depicted in Figure 6d, no significant changes in phagocytosis activity were observed with the different treatments compared to control. Furthermore, the release of inflammatory products such as HBP and the cytokine IL-6 in response to CAP treatment, M1 protein, and CAP-treated M1 protein in whole blood were analysed using ELISA (Fig. 6e–f). Untreated M1 protein significantly increased the release of HBP and IL-6 compared to control, while CAP-treated M1 protein was less potent, resulting in a three-fold reduction in mobilised HBP and IL-6. Pretreatment with CAP before addition of M1 protein did not affect HBP levels, and CAP itself did not induce any HBP release in whole blood (Fig. 6e). As the levels of secreted IL-6 in response to M1 protein and CAP treatment differed significantly among the donors tested, the values were normalised for comparison (Fig. 6f; original data are depicted in online suppl. Fig. S4). M1 protein increased IL-6 responses in the blood samples from all donors, and induction of IL-6 using CAP-treated M1 protein was absent. Two donors showed increased IL-6 levels in response to CAP treatment, especially during 5 s of treatment, although none responded to the longer treatment time (15 s). Together, our findings support the safety of using CAP as an antiseptic treatment without aggravating inflammation and early molecular immune responses.
To conclude, short CAP treatments (2–15 s), sufficient for bacterial killing and tissue tolerable, can also modify and disarm the detrimental activities of M1 protein, reduce M1 protein-triggered inflammation, and improve host recovery during wound healing. To our knowledge, this is the first time CAP has been shown to modulate a specific bacterial virulence factor in favour of the host, and this novelty might be transferable to other virulence factors across different bacterial species.
Discussion
The use of CAP as an enhancer of wound healing is a novel treatment concept that has not yet received much attention. The first article on CAP devices for medical issues was released as early as 2013 [42]. Since then, evidence that confirms the beneficial effect of CAP on healing of normal and chronic wounds and skin infections has accumulated [6, 9, 10, 11, 12]. The latter findings are supported by reports that CAP exerts antimicrobial activity towards many different bacterial species, while it is rather harmless to human cells upon short exposure times [6, 7, 43, 44]. Importantly, no microbial resistance towards CAP treatment has been reported so far, which is an important feature at a time when antibiotic resistance is an emerging serious clinical problem. While the functional antiseptic effect of CAP treatment has been demonstrated, the molecular mechanisms behind the benefits of CAP treatment remain to be clarified. Also, no studies have been conducted analysing the molecular modulation of microbial virulence factors caused by CAP.
Our study was carried out to address this issue, and we therefore investigated whether CAP can modulate the activity of a specific virulence factor. To unravel this question, we chose to focus on M1 protein, an important streptococcal virulence factor. Previous work has demonstrated that the protein can be shed from the bacterial surface, impair wound healing activities, and aggravate inflammation [23, 24, 25, 36, 45]. Our results show that CAP treatment not only abolished the detrimental activity of the streptococcal virulence factor towards keratinocytes, but also led to better wound healing recovery, less IL-8 release, and improved cellular viability compared to treatment with the native M1 protein.
Trying to decipher the molecular mechanisms of CAP treatment that caused the inactivation of M1 protein, HDX-MS was employed, a highly versatile technique to study the conformation, dynamics, and interaction of proteins. Since CAP is known to create reactive oxygen and nitrogen species, we hypothesised that CAP treatment can chemically introduce amino acid modifications of the M1 protein sequence, which in turn down-regulate its virulent activity. MS analysis did indeed reveal oxidation of Met81 and Trp128 of the M1 protein. These oxidations seem to introduce structural changes affecting the function of the M1 protein, seen as increased uptake (flexibility), lower uptake (oligomerization, aggregation), and changes in the HDX isotope uptake pattern indicative of alterations in the conformational population. The conformational state of a protein is well known to be crucial for protein function and has previously been described for the streptococcal M1 protein by McNamara et al. [46].
Many novel antimicrobial substances cannot be used in clinical settings because of their toxic side effects. Thus, experiments were performed to study whether CAP treatment can trigger deleterious host responses. However, throughout our study, we did not find any overwhelming or disturbed immune reactions due to CAP treatment. Instead, we found that short tissue-tolerable CAP treatments disarm the important virulence factor, supporting the usefulness of CAP application to combat skin- or surgery-related bacterial infections. To our knowledge, this is the first time such CAP-related utility towards a specific bacterial virulence factor is shown. We believe that CAP modulation can be used to destroy the pernicious activity of other virulence factors across different bacterial species. Thus, the data presented in this study may lead to novel concepts for the development of new antimicrobial therapies.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
All authors declare that they have no conflict of interests.
Author Contributions
S.T. Persson performed the research, analysed the data, and wrote the paper. S. Ekström performed the HDX-MS and data analysis. P. Papareddy contributed with experiments and oversaw the paper. H. Herwald supervised the study.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
Support from the Swedish National Infrastructure for Biological Mass Spectrometry (BioMS) is gratefully acknowledged. We wish to thank Pia Andersson (Department of Clinical Sciences, Lund University) for excellent technical assistance. We also thank Maria Baumgarten and the IQ Biotechnology platform for their excellence in electron microscopy. This work was supported in part by the foundations of Alfred Österlund, Crafoord, the Knut and Alice Wallenberg Foundation, the Ragnar Söderberg Foundation, the Medical Faculty at Lund University, the Swedish Foundation for Strategic Research, and the Swedish Research Council.
References
- 1.Graves DB. The emerging role of reactive oxygen and nitrogen species in redox biology and some implications for plasma applications to medicine and biology. J Phys D Appl Phys. 2012;45((26)):45. [Google Scholar]
- 2.Weltmann KD, Kindel E, Brandenburg R, Meyer C, Bussiahn R, Wilke C, et al. Atmospheric Pressure Plasma Jet for Medical Therapy: Plasma Parameters and Risk Estimation. Contrib Plasma Phys. 2009;49((9)):631–40. [Google Scholar]
- 3.Lackmann JW, Schneider S, Edengeiser E, Jarzina F, Brinckmann S, Steinborn E, et al. Photons and particles emitted from cold atmospheric-pressure plasma inactivate bacteria and biomolecules independently and synergistically. J R Soc Interface. 2013 Sep;10((89)):20130591. doi: 10.1098/rsif.2013.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daeschlein G, Scholz S, Ahmed R, von Woedtke T, Haase H, Niggemeier M, et al. Skin decontamination by low-temperature atmospheric pressure plasma jet and dielectric barrier discharge plasma. J Hosp Infect. 2012 Jul;81((3)):177–83. doi: 10.1016/j.jhin.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Daeschlein G, Scholz S, Arnold A, von Podewils S, Haase H, Emmert S, et al. In Vitro Susceptibility of Important Skin and Wound Pathogens Against Low Temperature Atmospheric Pressure Plasma Jet (APPJ) and Dielectric Barrier Discharge Plasma (DBD) Plasma Process Polym. 2012;9((4)):380–9. [Google Scholar]
- 6.Daeschlein G, von Woedtke T, Kindel E, Brandenburg R, Weltmann KD, Junger M. Antibacterial Activity of an Atmospheric Pressure Plasma Jet Against Relevant Wound Pathogens in vitro on a Simulated Wound Environment. Plasma Process Polym. 2010;7((3-4)):224–30. [Google Scholar]
- 7.Alkawareek MY, Gorman SP, Graham WG, Gilmore BF. Potential cellular targets and antibacterial efficacy of atmospheric pressure non-thermal plasma. Int J Antimicrob Agents. 2014 Feb;43((2)):154–60. doi: 10.1016/j.ijantimicag.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 8.Weiss M, Daeschlein G, Kramer A, Burchardt M, Brucker S, Wallwiener D, et al. Virucide properties of cold atmospheric plasma for future clinical applications. J Med Virol. 2017 Jun;89((6)):952–9. doi: 10.1002/jmv.24701. [DOI] [PubMed] [Google Scholar]
- 9.Ulrich C, Kluschke F, Patzelt A, Vandersee S, Czaika VA, Richter H, et al. Clinical use of cold atmospheric pressure argon plasma in chronic leg ulcers: A pilot study. J Wound Care. 2015 May;24((5)):196–203. doi: 10.12968/jowc.2015.24.5.196. [DOI] [PubMed] [Google Scholar]
- 10.Isbary G, Heinlin J, Shimizu T, Zimmermann JL, Morfill G, Schmidt HU, et al. Successful and safe use of 2 min cold atmospheric argon plasma in chronic wounds: results of a randomized controlled trial. Br J Dermatol. 2012 Aug;167((2)):404–10. doi: 10.1111/j.1365-2133.2012.10923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinlin J, Zimmermann JL, Zeman F, Bunk W, Isbary G, Landthaler M, et al. Randomized placebo-controlled human pilot study of cold atmospheric argon plasma on skin graft donor sites. Wound Repair Regen. 2013 Nov-Dec;21((6)):800–7. doi: 10.1111/wrr.12078. [DOI] [PubMed] [Google Scholar]
- 12.Hartwig S, Preissner S, Voss JO, Hertel M, Doll C, Waluga R, et al. The feasibility of cold atmospheric plasma in the treatment of complicated wounds in cranio-maxillo-facial surgery. J Craniomaxillofac Surg. 2017 Oct;45((10)):1724–30. doi: 10.1016/j.jcms.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Wende K, Bekeschus S, Schmidt A, Jatsch L, Hasse S, Weltmann KD, et al. Risk assessment of a cold argon plasma jet in respect to its mutagenicity. Mutat Res Genet Toxicol Environ Mutagen. 2016 Mar;798-799:48–54. doi: 10.1016/j.mrgentox.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Kluge S, Bekeschus S, Bender C, Benkhai H, Sckell A, Below H, et al. Investigating the Mutagenicity of a Cold Argon-Plasma Jet in an HET-MN Model. PLoS One. 2016 Sep;11((9)):e0160667. doi: 10.1371/journal.pone.0160667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med. 1999 Sep;341((10)):738–46. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 16.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005 Nov;5((11)):685–94. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 17.DiNubile MJ, Lipsky BA. Complicated infections of skin and skin structures: when the infection is more than skin deep. J Antimicrob Chemother. 2004 Jun;53(Suppl 2):ii37–50. doi: 10.1093/jac/dkh202. [DOI] [PubMed] [Google Scholar]
- 18.Johansson L, Thulin P, Low DE, Norrby-Teglund A. Getting under the skin: the immunopathogenesis of Streptococcus pyogenes deep tissue infections. Clin Infect Dis. 2010 Jul;51((1)):58–65. doi: 10.1086/653116. [DOI] [PubMed] [Google Scholar]
- 19.Stevens DL. Streptococcal toxic shock syndrome associated with necrotizing fasciitis. Annu Rev Med. 2000;51((1)):271–88. doi: 10.1146/annurev.med.51.1.271. [DOI] [PubMed] [Google Scholar]
- 20.Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000 Jul;13((3)):470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oehmcke S, Shannon O, Mörgelin M, Herwald H. Streptococcal M proteins and their role as virulence determinants. Clin Chim Acta. 2010 Sep;411((17-18)):1172–80. doi: 10.1016/j.cca.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 22.Gherardi G, Vitali LA, Creti R. Prevalent emm Types among Invasive GAS in Europe and North America since Year 2000. Front Public Health. 2018 Mar;6:59. doi: 10.3389/fpubh.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herwald H, Cramer H, Mörgelin M, Russell W, Sollenberg U, Norrby-Teglund A, et al. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell. 2004 Feb;116((3)):367–79. doi: 10.1016/s0092-8674(04)00057-1. [DOI] [PubMed] [Google Scholar]
- 24.Påhlman LI, Mörgelin M, Eckert J, Johansson L, Russell W, Riesbeck K, et al. Streptococcal M protein: a multipotent and powerful inducer of inflammation. J Immunol. 2006 Jul;177((2)):1221–8. doi: 10.4049/jimmunol.177.2.1221. [DOI] [PubMed] [Google Scholar]
- 25.Påhlman LI, Olin AI, Darenberg J, Mörgelin M, Kotb M, Herwald H, et al. Soluble M1 protein of Streptococcus pyogenes triggers potent T cell activation. Cell Microbiol. 2008 Feb;10((2)):404–14. doi: 10.1111/j.1462-5822.2007.01053.x. [DOI] [PubMed] [Google Scholar]
- 26.Bjorck V, Pahlman LI, Tornebrant J, Bodelsson M. Streptococcal M1 protein induces hyporesponsiveness and cytokine release from human arteries in a fibrinogendependent manner: a translational study. Scand J Trauma Resus; 2018. p. p. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995 Apr;270((17)):9862–7. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 28.Ziuzina D, Boehm D, Patil S, Cullen PJ, Bourke P. Cold Plasma Inactivation of Bacterial Biofilms and Reduction of Quorum Sensing Regulated Virulence Factors. PLoS One. 2015 Sep;10((9)):e0138209. doi: 10.1371/journal.pone.0138209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bekeschus S, Schmidt A, Weltmann KD, von Woedtke T. The plasma jet kINPen – A powerful tool for wound healing. Clin Plasma Med. 2016;4((1)):19–28. [Google Scholar]
- 30.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988 Mar;106((3)):761–71. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collin M, Olsén A. Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol Microbiol. 2000 Jun;36((6)):1306–18. doi: 10.1046/j.1365-2958.2000.01942.x. [DOI] [PubMed] [Google Scholar]
- 32.Akesson P, Schmidt KH, Cooney J, Björck L. M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem J. 1994 Jun;300((Pt 3)):877–86. doi: 10.1042/bj3000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tapper H, Karlsson A, Mörgelin M, Flodgaard H, Herwald H. Secretion of heparin-binding protein from human neutrophils is determined by its localization in azurophilic granules and secretory vesicles. Blood. 2002 Mar;99((5)):1785–93. doi: 10.1182/blood.v99.5.1785. [DOI] [PubMed] [Google Scholar]
- 34.BioPharma Finder, User Guide, Software Version 3.0, ThermoFisher Scientific, XCALI- 97898, Revision A, November 2017. pp. 211–2015.
- 35.Persson ST, Wilk L, Mörgelin M, Herwald H. Vigilant keratinocytes trigger pathogen-associated molecular pattern signaling in response to streptococcal M1 protein. Infect Immun. 2015 Dec;83((12)):4673–81. doi: 10.1128/IAI.00887-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Persson ST, Hauri S, Malmstrom J, Herwald H. Leukocyte recruitment and molecular fortification of keratinocytes triggered by streptococcal M1 protein. Cell Microbiol. 2017;20((1)):e12792. doi: 10.1111/cmi.12792. [DOI] [PubMed] [Google Scholar]
- 37.Zhou R, Zhou R, Zhuang J, Zong Z, Zhang X, Liu D, et al. Interaction of Atmospheric-Pressure Air Microplasmas with Amino Acids as Fundamental Processes in Aqueous Solution. PLoS One. 2016 May;11((5)):e0155584. doi: 10.1371/journal.pone.0155584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang J, Rand KD, Beuning PJ, Engen JR. False EX1 signatures caused by sample carryover during HX MS analyses. Int J Mass Spectrom. 2011 Apr;302((1-3)):19–25. doi: 10.1016/j.ijms.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirzaei H, Regnier F. Protein:protein aggregation induced by protein oxidation. J Chromatogr B Analyt Technol Biomed Life Sci. 2008 Sep;873((1)):8–14. doi: 10.1016/j.jchromb.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 40.Weids AJ, Ibstedt S, Tamas MJ, Grant CM. Distinct stress conditions result in aggregation of proteins with similar properties. Sci Rep-Uk; 2016. p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo Q, Joubert MK, Stevenson R, Ketchem RR, Narhi LO, Wypych J. Chemical modifications in therapeutic protein aggregates generated under different stress conditions. J Biol Chem. 2011 Jul;286((28)):25134–44. doi: 10.1074/jbc.M110.160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isbary G, Shimizu T, Li YF, Stolz W, Thomas HM, Morfill GE, et al. Cold atmospheric plasma devices for medical issues. Expert Rev Med Devices. 2013 May;10((3)):367–77. doi: 10.1586/erd.13.4. [DOI] [PubMed] [Google Scholar]
- 43.Ermolaeva SA, Varfolomeev AF, Chernukha MY, Yurov DS, Vasiliev MM, Kaminskaya AA, et al. Bactericidal effects of non-thermal argon plasma in vitro, in biofilms and in the animal model of infected wounds. J Med Microbiol. 2011 Jan;60((Pt 1)):75–83. doi: 10.1099/jmm.0.020263-0. [DOI] [PubMed] [Google Scholar]
- 44.Heinlin J, Isbary G, Stolz W, Morfill G, Landthaler M, Shimizu T, et al. Plasma applications in medicine with a special focus on dermatology. J Eur Acad Dermatol Venereol. 2011 Jan;25((1)):1–11. doi: 10.1111/j.1468-3083.2010.03702.x. [DOI] [PubMed] [Google Scholar]
- 45.Kramer A, Dissemond J, Kim S, Willy C, Mayer D, Papke R, et al. Consensus on Wound Antisepsis: Update 2018. Skin Pharmacol Physiol. 2018;31((1)):28–58. doi: 10.1159/000481545. [DOI] [PubMed] [Google Scholar]
- 46.McNamara C, Zinkernagel AS, Macheboeuf P, Cunningham MW, Nizet V, Ghosh P. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science. 2008 Mar;319((5868)):1405–8. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data