Abstract
As the most prevalent and abundant transcriptional modification in the eukaryotic genome, the continuous and dynamic regulation of N6-methyladenosine (m6A) has been shown to play a vital role in physiological and pathological processes of cardiovascular diseases (CVDs), such as ischemic heart failure (HF), myocardial hypertrophy, myocardial infarction (MI), and cardiomyogenesis. Regulation is achieved by modulating the expression of m6A enzymes and their downstream cardiac genes. In addition, this process has a major impact on different aspects of internal biological metabolism and several other external environmental effects associated with the development of CVDs. However, the exact molecular mechanism of m6A epigenetic regulation has not been fully elucidated. In this review, we outline recent advances and discuss potential therapeutic strategies for managing m6A in relation to several common CVD-related metabolic disorders and external environmental factors. Note that an appropriate understanding of the biological function of m6A in the cardiovascular system will pave the way towards exploring the mechanisms responsible for the development of other CVDs and their associated symptoms. Finally, it can provide new insights for the development of novel therapeutic agents for use in clinical practice.
Keywords: N6-methyladenosine (m6A), RNA methylation, Cardiovascular system, Metabolic disorder
1. Introduction
Epigenetics has attracted considerable attention in the biomedical field. Accumulating evidence has shown that epigenetic alterations can affect a variety of common pathological reactions, including ischemia, inflammation, aging, and tumorigenesis (He et al., 2013). Epigenetics can also have a vital impact on gene expression and function without altering the base sequence of DNA. Moreover, these effects are reversible, heritable and are influenced by the external environment (Gibney and Nolan, 2010; Mazzio and Soliman, 2012).
Current epigenetic mechanisms include mainly DNA methylation, post-translational histone modification, chromatin remodeling, and deployment of non-coding RNA. Recent advances in the field of epigenetics can aid our understanding of a series of complicated biological processes associated with aging (Rodriguez-Rodero et al., 2010), development (Smith et al., 2012), inflammation (Bayarsaihan, 2011), immunity (Fernández-Morera et al., 2010), stem cell biology (Calvanese and Fraga, 2012), and angiogenesis (Buysschaert et al., 2008).
The role of reversible RNA methylation has just begun to attract worldwide scientific attention compared with the processes of DNA methylation and histone modification which are now well-established in genetics and cell biology (He, 2010; Panneerdoss et al., 2018). As an important part of developing RNA epigenetics, the modification of N 6-methyladenosine (m6A) has recently been explored in the cardiovascular field (Liu and Pan, 2015). It has been shown that the m6A modification is the most universal and abundant internal modification of eukaryotic messenger RNA (mRNA) and long non-coding RNA (lncRNA) and occurs mainly in the 3'-untranslated regions (3'-UTRs) and nearby the stop codons of the mRNA molecules (Parashar et al., 2018; Gan et al., 2019). Furthermore, several independent lines of evidence have revealed that the continuous and dynamic regulation of m6A may have a far-reaching impact on modulating the expression of specific genes (Niu et al., 2013). In this review, we outline recent advances and discuss potential therapeutic strategies for managing m6A in relation to several common cardiovascular diseases (CVDs), related metabolic disorders, and external environmental factors.
2. Biological function of m6A modification
2.1. Methyltransferase and demethylases of m6A
It has been shown that m6A methylation is catalyzed by the multicomponent RNA methyltransferase complex, RNA demethylases, and m6A readers (Liu et al., 2014). The core components of the RNA methyltransferase complex include methyltransferase-like 3/14 (METTL3/14) and Wilms tumor 1 associated protein (WTAP) (Liu et al., 2014). Among them, the spliceosome-associated protein WTAP has recently been found to play its role by recruiting RNA METTL3/14 to sites of methylation in the nucleus (Liu et al., 2014). Although the methyltransferase complex of m6A methylation has not been fully elucidated, remarkable progress including the discovery of ALKB homolog 5 (ALKBH5) and fat mass-and obesity-related proteins (FTOs) has recently been made. The so-called erasers of specific markers operate on target transcripts via an α-ketoglutarate-and Fe2+-dependent way in the m6A and N 6,2'-O-dimethyladenosine (m6Am) of mammalian poly-adenylated RNA (Gerken et al., 2007; Niu et al., 2013; Mauer et al., 2017). In addition to the methyltransferase complex and demethylase enzyme, m6A is also regulated by the activity of m6A readers, such as YT521-B homology (YTH) m6A RNA-binding proteins (YTHDF), which can regulate the stability and degradation of RNAs and affect the efficiency of target mRNA translation (Wang and He, 2014; Kennedy et al., 2016). Therefore, m6A regulates its effects mainly by recruiting a variety of proteins containing the YTH domains and via its assembly by methyltransferases that are located in relative consensus motifs following transcription (Wei and Moss, 1977; Patil et al., 2018). The m6A modification may not only affect the secondary structure of mRNA and the interactions between mRNA-protein and mRNA-small nuclear RNA (snRNA), but may also be involved in modulating translatability, RNA transport and splicing, and susceptibility to post-transcriptional silencing (Zhong et al., 2008).
2.2. m6A regulation in the eukaryotic genome
It is believed that m6A methylation plays an important role in the regulation of gene expression by affecting the stability, degradation, and translation of RNA (Fu et al., 2014; Meyer et al., 2015). Several studies have confirmed that alteration of m6A levels may have a significant impact on various biological processes in mammals, such as the maintenance and differentiation of embryonic stem cells (ESCs), transcriptional splicing, nuclear RNA export, protein translation control, cell fate determination, circadian rhythm modification, heat shock response, meiotic progression, and neuronal function (Guo et al., 2017; Roignant and Soller, 2017; Wei et al., 2017). Moreover, a lack of m6A modification can lead to the development of several diseases, such as obesity, cancer, type 2 diabetes mellitus (T2DM), infertility, and developmental arrest (Wei et al., 2017).
Therefore, attention has been focused on the role and importance of m6A regulation in the eukaryotic genome in relation to physiological and pathological processes. The m6A profiles of full transcriptomes have shown that m6A modifications occur in countless RNA transcripts with unique patterns of distribution. Although the role of m6A editors in the processing, synthesis, and degradation of mRNAs and proteins has been discovered (Patil et al., 2018), the functions and mechanisms of most RNA modifications found in mRNA molecules remain poorly understood, especially with regard to the development of CVDs.
3. m6A RNA modification in CVDs
Due to improvements in the standard of living and medical care, CVD mortality has declined markedly in certain high-income countries of Western Europe and North America (Libby et al., 2013). However, CVD remains the leading cause of death worldwide in both developed and developing countries (Iyen et al., 2019). Accumulating evidence has shown the contribution of several important but underappreciated issues, such as air pollution, to the genesis and development of CVDs (Cosselman et al., 2015; Peña and Rollins, 2017). Epigenetics plays an effective and leading regulatory role in modulating cardiovascular repair functions (Jin et al., 2019). Therefore, new concepts and therapeutic measures must be developed for myocardial repair and regeneration to improve cardiac function. However, although certain transcription factors and co-activators have been studied in heart failure (HF), post-transcriptional modifications that mediate specific mRNAs affecting the main protein expression and cardiac function have not been explored in detail.
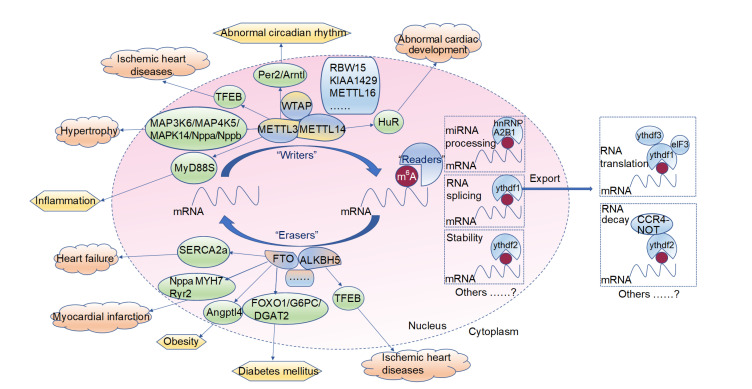
As a key enzyme of m6A, FTO is widely expressed in cardiac ventricular tissues, with a high level of expression in humans (Boissel et al., 2009). Therefore, particular attention must be paid to the possible associations between m6A and the physiological and pathological processes of different CVDs. Emerging evidence has identified epigenetic regulation of m6A and suggested possible therapeutic strategies for treating the development of cardiac defects, including ventricular septal and atrio-ventricular defects (Boissel et al., 2009), arrhythmias (Carnevali et al., 2014), coronary heart disease (Gustavsson et al., 2014), and cardiomyocyte hypertrophy (Dorn et al., 2019) (Fig. 1).
Fig. 1.
Molecular roles of N 6-methyladenosine (m6A) enzymes in different cardiovascular diseases, related metabolic disorders, and external environmental factors
m6A methylation is catalyzed by the multicomponent RNA methyltransferase complex (methyltransferase-like 3/14 (METTL3/14), Wilms tumor 1 associated protein (WTAP), and KIAA1429), RNA demethylases (ALKB homolog 5 (ALKBH5) and fat mass-and obesity-related protein (FTO)), and m6A readers (YT521-B homology (YTH) domain family 1/2/3 (YTHDF1/2/3), YTH domain containing 1 (YTHDC1), heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1), and E74-like factor 3 (elF3)). Modification of m6A may not only affect the secondary structure of messenger RNA (mRNA) and the interactions between mRNA-protein and mRNA-small nuclear RNA (snRNA), but may also be involved in modulating translatability, RNA transport and splicing, and susceptibility to post-transcriptional silencing. Furthermore, emerging evidence has identified the epigenetic regulation of m6A and suggested possible therapeutic strategies for treating the development of cardiac defects and internal metabolic disorders, including heart failure, myocardial infarction, cardiomyocyte hypertrophy, obesity, and diabetics mellitus. TFEB: transcription factor EB; MAP3K6: mitogen-activated protein kinase (MAPK) kinase kinase 6; MAP4K5: MAPK kinase kinase kinase 5; Nppa: natriuretic peptide precursor (Npp)-A; MyD88S: splice variant of myeloid differentiation factor 88; SERCA2a: sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 2a; MYH7: β-myosin heavy chain 7; Ryr2: ryanodine receptor 2; Angptl4: angiopoietin-like 4; Per2: period 2; Arntl: aryl hydrocarbon receptor nuclear translocator like; FOXO1: forkhead box O1; G6PC: glucose-6-phosphatase (G6Pase) catalytic subunit; DGAT2: diacylglycerol acyltransferase 2; RBW15: RNA-binding motif protein 15; HuR: human antigen R; hnRN: heterogeneous nuclear ribonucleoprotein; CCR4-NOT: carbon catabolite repressor protein 4-negative on TATA
3.1. Ischemic heart diseases
By altering the oxygenation capacity of cardiomyocytes directly, ischemic heart disease caused by stenosis or occlusion of the myocardial coronary artery is considered one of the main causes of morbidity and mortality worldwide, including myocardial infarction (MI) and HF (Ge et al., 2019).
Several studies have shown that cardiomyocytes under chronic anoxic conditions, such as anemia, chronic intermittent hypoxia, and sleep apnea, can retain the homeostasis of heart contractile function and energy metabolism by triggering the corresponding regulation pathways and mediating cardiac metabolic remodeling via induction of transcription (Essop, 2007; Cole et al., 2016).
As one of the main drivers of metabolic disturbance in ischemic disease and MI, hypoxia is the result of multiple physiological and pathological stimuli (da Luz Sousa Fialho et al., 2019). Previous studies of cardiac ventricular biopsies from patients who had undergone coronary artery bypass surgery have shown that hypoxia inducible factor (HIF) is a major transcription factor with significantly increased activity in the early period of post-MI (Lee et al., 2000; Smith et al., 2008). It is well-established that HIF-1 activates most of the regulatory factors involved in oxygen homeostasis in mammals by binding to hypoxia response elements (HREs) and activating the transcription of multiple target genes which may play a critical role in the hypoxic response at both the cellular and the systemic levels. Subsequently, this process regulates the relevant signal transduction pathways and the corresponding microenvironments of the pathological blood vessels induced by hypoxia and ischemia (Ortiz-Barahona et al., 2010).
Due to the deficiency in oxygen availability, cellular metabolism of cardiomyocytes is one of the major changes that occur during chronic adaptation to hypoxia (Dang and Semenza, 1999). In addition to metabolism, the binding between HIF and HREs can be affected by epigenetic mechanisms, such as CpG methylation and oxidative DNA damage caused by hypoxia and reactive oxygen species (ROS) (Rodriguez et al., 1997). It has also been reported that HIF-1 is associated with hypoxia and may be regulated by histone methylation directly. With regard to certain common methylases, several studies have shown that hypoxia induces the expression of ALKBH5 in various types of breast cancer cells (Essop, 2007). In addition, el Azzouzi et al. (2013) confirmed the binding sites of FTO mRNA for several hypoxia-induced cardiac microRNAs (miRs) well-known to be up-regulated in HF, such as miR-21, miR-24, miR-488, miR-224, and miR-489. These results suggest that epigenetic regulation of hypoxia may play a significant role in the pathogenesis of CVDs.
In hypoxia/reoxygenation (H/R)-treated cardiomyocytes and ischemia/reperfusion (I/R)-treated mouse heart tissues, the modification of m6A was increased by METTL3, which is the main factor of abnormal m6A regulation. This was caused by promoting the translation of specific mRNAs (Lin et al., 2016; Song et al., 2019).
The silencing of METTL3 can increase autophagic flux and inhibit apoptosis in H/R-treated cardiomyocytes, suggesting that METTL3 is a negative regulator of autophagy, which determines the cellular and systemic function of H/R-treated cardiomyocytes (Song et al., 2019). METTL3 methylates transcription factor EB (TFEB), a key gene involved in lysosomal biogenesis during the autophagic process, at two m6A residues in the 3'-UTR. METTL3 enhances the binding of the protein heterogeneous nuclear ribonucleoprotein D (hnRNPD) to TFEB pre-mRNA, thereby decreasing the expression level of TFEB (Pastore et al., 2016). Further experiments have shown that the autophagic flux enhanced by METTL3 deficiency is TFEB-dependent. Therefore, it is reasoned that METTL3 knockdown can increase the levels of the green fluorescent protein (GFP)-TFEB protein by up-regulating the endogenous levels of TFEB via the adenosine monophosphate (AMP)-activated protein kinase (AMPK)-mammalian target of rapamycin (mTOR) pathway (Song et al., 2019).
3.1.1 Heart failure
Current evidence shows that dysregulation of m6A levels in RNA can lead to the development of several pathological mechanisms, such as HF, at the molecular, cellular, and organic levels (Jia et al., 2011; Meyer et al., 2012). Mathiyalagan et al. (2019) confirmed that the increased expression of m6A induced by ischemia and hypoxia may be reduced by altering the expression of FTOs. This evidence was reported in a study of different models of HF and hypoxic primary cardiomyocytes (Mathiyalagan et al., 2019). In addition to improving cardiomyocyte function and attenuating cardiac remodeling in HF by restoring the expression of relative proteins, such as sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 2a (SERCA2a), experimental evidence has shown that FTO overexpression decreases cardiac fibrosis and promotes angiogenesis in models of MI (Mathiyalagan et al., 2019). Furthermore, it has been shown that cardiac contractile transcripts associated with ischemia and hypoxia are demethylated selectively by FTO, which can increase the stability of mRNA and protein expression (Hess et al., 2013).
3.1.2 Myocardial infarction
MI is a heart attack caused by the formation of plaque in the interior walls of the arteries that reduces blood flow to the heart and damages the heart muscles due to tissue inflammation and scarring (Lu et al., 2015). Epicardial specificity in response to injury occurs by down-regulating a group of genes expressed differentially in the epicardium (Bochmann et al., 2010). This is considered one of the main mechanisms of MI development.
During embryogenesis, the proepicardium differentiates into cardiac vasculature and interstitial cells and eventually becomes a single epithelial cell layer covering the heart, which is essential for normal development (Reese et al., 2002; Cai et al., 2008; Zhou et al., 2008; van Wijk et al., 2009). However, it remains controversial whether the epicardium can differentiate into endothelial cells of the vasculature (Smart et al., 2007; Winter and Groot, 2007) and whether it can contribute directly to the formation of the myocardium (Zhou et al., 2008). Although previous study has shown that the epicardium can regulate cardiac regeneration following injury (noted in lower vertebrates), it has also been reported that epicardial cell genes may be a promising therapeutic target following the injury of left coronary artery (LCA) ligation by the application of transcriptomics (Bochmann et al., 2010). Therefore, these studies suggest that the involvement of epigenetics should be explored with regard to MI.
Mathiyalagan et al. (2019) demonstrated that the levels of FTO in mice were down-regulated within 4 h following MI induction by LCA. Furthermore, they examined cardiac remodeling soon after MI and demonstrated that maintenance of FTO expression for nearly one week could be helpful in protecting and repairing the function of cardiomyocytes by improving cardiac homeostasis. Overexpression of FTO was caused by adenoviral transfection. These data suggested the use of FTO or FTO mimics for therapeutic targeting.
3.2. Hypertrophy
Hypertrophic cardiomyopathy is one of the common causes of myocardial injury accompanied by pathological phenomena, such as myocardial hypertrophy (Hou et al., 2017; Liu et al., 2019).
In addition to other common stimuli, Dorn et al. (2019) confirmed that methylation regulated by METTL3 on N 6-adenosines is essential for the pathological process of hypertrophy in vivo and in vitro. Although the increase in the expression level of METTL3, which acts with METTL14, WTAP, or other regulatory subunits, may cause cardiomyocyte remodeling, it is notable that the inhibition of its activity can abrogate the ability of cardiomyocytes to cause cardiac compensatory hypertrophy in response to hypertrophic stimuli, such as aging and stress. Furthermore, cardiac-specific METTL3 knockout mice were shown to promote eccentric cardiomyocyte remodeling and dysfunction in vivo (Liu et al., 2014; Dorn et al., 2019). These data highlight that the dynamic methylation and modification mediated by METTL3 are necessary for regulating the myocyte growth response.
Since both METTL3 and FTO are localized in the nucleus of isolated cardiac myocytes, FTO knockdown was found to decrease the augmented size of neonatal rat cardiac myocytes (NRCMs) induced by α-adrenergic stimulation in vitro (Kmietczyk et al., 2019). Taken collectively, these results suggest that m6A methylation may be a novel and vital epigenetic mechanism that should be studied in relation to the maintenance of cardiac homeostasis.
In addition, due to the tissue specificity of different ALKBH proteins, it is still debatable whether they can be used with similar therapeutic effects as those of the METTL3-containing protein, which is expressed ubiquitously (Fedeles et al., 2015).
3.3. Abnormal cardiac development
The consensus sequence contained in m6A is highly homologous between human and mouse species, and their mRNA levels are regulated in a tissue-specific manner from the embryonic period to adulthood (Dominissini et al., 2012; Meyer et al., 2012). Higher m6A expression in embryogenesis than in adult tissues has suggested the critical importance of m6A during growth and development at the cellular and organic levels.
Mouse ESCs (mESCs) are well-known for their roles in embryonic development. Wang et al. (2014) reported that METTL3 and METTL14 can modify m6A to maintain and even improve the self-renewal capability of mESCs. Their enrichment analysis indicated that m6A editing might be associated with the stability of RNA, which was in turn negatively associated with regulatory proteins by blocking the binding of human antigen R (HuR)’s RNA, thereby retaining the biological function of mESCs (Wang et al., 2014).
Moreover, in a study of growth retardation and developmental delay, FTO and its homozygous mutant were shown to play a vital role in the early development of the human central nervous and cardiovascular systems by maintaining the stability of 2-oxoglutarate turnover and N-methyl-nucleoside demethylase activity (Daoud et al., 2016).
3.4. Downstream gene expression
Alteration in the expression levels of myocardial genes in response to different pathological stimuli plays a major role in the occurrence and progression of HF in several animal models (Tan et al., 2002). A recent study has highlighted the importance of specific modifications that occur in reversible mRNA sequences that control gene expression at the epigenetic level (Tan et al., 2002). As an increase in the levels of m6A methylation is associated with cardiomyopathy, it is meaningful to investigate the role of internal m6A modification on the expression of the associated genes. Dynamic m6A methylation can influence the stability of transcription by regulating the efficiency of mRNA translation in stressed cardiomyocytes. For example, in addition to the size and cardiac function of cardiomyocytes, the expression of the downstream markers natriuretic peptide precursor (Npp)-A (Nppa) and -B (Nppb) that are associated with hypertrophy can be influenced by the enzymatic activity of METTL3. This suggests that m6A modification plays an integrative role in the regulation of cardiac gene expression and cellular growth response in vivo and in vitro (Kmietczyk et al., 2019).
Overall, the engineering and manipulation of m6A enzymes or any other type of small molecule at specific sites may be a novel therapeutic avenue for several cardiac diseases caused by dysregulation of m6A, such as HF and MI (James et al., 2003).
4. m6A RNA modification is an internal metabolic disorder caused by external environmental factors
Different studies have attempted to understand the important and necessary role of m6A in relation to metabolism and environmental effects that can, in turn, lead to the development of pathophysiology and abnormal heart function noted in CVDs (Table 1).
Table 1.
Multiple functions exerted by m6A RNA methylation in various cardiovascular diseases
| Disease | Molecule | Target gene | Function | Regulation | Mechanism | Reference |
| Ischemic heart diseases | METTL3 | TFEB | Writer | Up-regulation | Silencing METTL3 enhances autophagic flux and inhibits apoptosis in H/R-treated cardiomyocytes by TFEB | Pastore et al., 2016 |
| ALKBH5 | TFEB | Eraser | Down-regulation | ALKBH5 overexpression enhances autophagic flux and inhibits apoptosis in H/R-treated cardiomyocytes by TFEB | Pastore et al., 2016 | |
| Heart failure (HF) | FTO | SERCA2a | Eraser | Down-regulation | FTO overexpression improves cardiomyocyte function and attenuatis cardiac remodeling in HF by regulating SERCA2a | Mathiyalagan et al., 2019 |
| Myocardial infarction (MI) | FTO | Nppa/MYH7/Ryr2/SERCA2a | Eraser | Down-regulation | FTO overexpression protects and repairs cardiomyocyte function through improving cardiac homeostasis after MI | Mathiyalagan et al., 2019 |
| Hypertrophy | METTL3 | MAP3K6/MAP4K5/MAPK14/Nppa/Nppb | Writer | Up-regulation | Silencing METTL3 maintains normal cardiac function in response to hypertrophic stimuli through stress-response mechanism | Dorn et al., 2019 |
| FTO | Eraser | Up-regulation | Silencing FTO blunts hypertrophy of NRCM in response to α-adrenergic stimulation | Kmietczyk et al., 2019 | ||
| Abnormal cardiac development | METTL3/14 | HuR | Writer | Down-regulation | The level of METTL3 expression maintains and even improves the self-renewal capability of mESCs | Wang et al., 2014 |
| FTO | Eraser | Mutation | FTO plays an important role in early development of human central nervous and cardiovascular systems | Daoud et al., 2016 | ||
| Obesity | FTO | Angptl4 | Eraser | Up-regulation | FTO influences triglyceride metabolism in adipocytes and by post-transcriptional regulation of Angptl4 | Wang et al., 2015 |
| YTHDF2 | PPARγ/C/EBPα | Reader | Down-regulation | Adipogenesis of porcine adipocytes can be led by a loss of m6A on FAM134B through m6A-YTHDF2-dependent way | Cai et al.,2019 | |
| Diabetes mellitus | FTO | FOXO1/G6PC/DGAT2 | Eraser | Up-regulation | FTO expression is highly related tothe impairment of plasma glucose and even multiple complications by altering target genes | Yang et al., 2019 |
| Abnormal circadian rhythm | METTL3 | Per2/Arntl | Writer | Down-regulation | Silencing METTL3 can elicit circadian period elongation and RNA processing delay by distributing clock genes Per2 and Arntl | Fustin et al., 2013 |
| Inflammation | METTL3 | MyD88S | Writer | Up-regulation | Silencing METTL3 attenuates theinflammatory response via regulating MyD88S in LPS-induced hDPCs | Liu et al., 2019 |
m6A: N 6-methyladenosine; METTL3: methyltransferase-like 3; ALKBH5: ALKB homolog 5; FTO: fat mass-and obesity-related protein; YTHDF2: YT521-B homology (YTH) domain family 2; TFEB: transcription factor EB; SERCA2a: sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 2a; Nppa: natriuretic peptide precursor (Npp)-A; MYH7: β-myosin heavy chain 7; Ryr2: ryanodine receptor 2; MAP3K6: mitogen-activated protein kinase (MAPK) kinase kinase 6; MAP4K5: MAPK kinase kinase kinase 5; HuR: human antigen R; Angptl4: angiopoietin-like 4; PPARγ: peroxisome proliferator-activated receptor γ; C/EBPα: CCAAT enhancer-binding protein α; FOXO1: forkhead box O1; G6PC: glucose-6-phosphatase (G6Pase) catalytic subunit; DGAT2: diacylglycerol acyltransferase 2; Per2: period 2; Arntl: aryl hydrocarbon receptor nuclear translocator like; MyD88S: splice variant of myeloid differentiation factor 88; H/R: hypoxia/reoxygenation; NRCM: neonatal rat cardiac myocyte; mESCs: mouse embryonic stem cells; FAM134B: family with sequence similarity 134 member B; LPS: lipopolysaccharide; hDPCs: human dental pulp cells
4.1. Obesity
The improvement in the general standard of living has rendered obesity a sub-health symptom. Several clinical and basic research studies have shown that the increasing incidence and mortality of various CVDs and even cancers are closely associated with obesity, suggesting it contributes to the severe financial burden associated with the treatment of these conditions (Scuteri et al., 2007; Koliaki et al., 2019).
In addition to dietary habits, the FTO gene, which is closely linked to obesity, was identified by the present study as a positive regulator of energy homeostasis, for example via lipid metabolism, by modulating m6A levels in RNA and DNA sequences in humans (Bartosovic et al., 2017; Chen and Du, 2019). The physiological and pathological development of obesity may be associated with FTO variants, which carry the at-risk alleles of single-nucleotide polymorphisms (SNPs), including rs9939609, rs8050136, rs3751812, rs7202116, and rs9930506, in the first intron of the FTO gene (Church et al., 2010; Wang et al., 2015). Variants of FTO were reported to have enhanced FTO expression and/or activity in previous study performed on homozygous individuals carrying the at-risk alleles (Church et al., 2010). Moreover, most studies demonstrated that these at-risk SNPs appeared to promote energy intake and a preference for energy-dense foods (Cecil et al., 2008; Speakman et al., 2008; Timpson et al., 2008; Haupt et al., 2009), while in two independent genome-wide association studies, the FTO gene was shown to be a high risk factor contributing to early-onset and severe obesity of European and Indonesian subjects (Dina et al., 2007; Daya et al., 2019).
An analysis of different phenotypes of mice has shown that adipocyte-specific FTO can play an important role in influencing triglyceride metabolism in adipocytes and consequently body weight. This is caused partly by post-transcriptional regulation of downstream genes, such as Angptl4. This may decrease energy expenditure and promote fatty acid storage in adipose tissues, regardless of the intake from a high-fat or standard diet (Wang et al., 2015).
Furthermore, a loss of m6A on family with sequence similarity 134 member B (FAM134B) can lead to the adipogenesis of porcine adipocytes through the m6A-YTHDF2-dependent pathway (Cai et al., 2019).
The epigenetic modification of m6A can be a risk factor of CVDs related to obesity, although the exact molecular mechanism remains unclear.
4.2. Diabetes mellitus
Pathoglycemia induced by diabetes mellitus, including hypoglycemia and hyperglycemia, has been identified to be the main risk factor contributing to CVDs and all-cause mortality in diabetic patients. In addition to controlling blood glucose, an increase in insulin sensitivity has become a new auxiliary or main treatment strategy of diabetes, notably in cases with insulin resistance (Khunti et al., 2015; Abdul-Ghani et al., 2019).
Furthermore, hyperglycemia may lead to the down-regulation of m6A levels by enhancing the expression of FTO or other methyltransferases in patients with T2DM, suggesting an important epigenetic role of m6A in regulating glucose and lipid metabolism disorders (Gilbert and Liu, 2012; Yang et al., 2019). In addition to dysregulated oxidation of glucose, increased FTO expression induced by diabetes is highly associated with the impairment of plasma glucose and even multiple complications caused by altering the expression of target functional genes, such as forkhead box O1 (FOXO1), glucose-6-phosphatase (G6Pase) catalytic subunit (G6PC), and diacylglycerol acyltransferase 2 (DGAT2) (Souness et al., 1982; Kursawe et al., 2016; Li et al., 2017; Yang et al., 2019).
Recently, it has been shown that an altered content of m6A can be used as a novel biomarker of T2DM since significantly lower levels of m6A are noted in both T2DM patients and diabetic rats. At present, FTO expression is the only indicator found to be positively correlated with the risk of diabetes by correlation analysis (Shen et al., 2015).
Nevertheless, whether m6A modification in diabetes mellitus is locus-specific remains to be elucidated.
4.3. Abnormal circadian rhythm
Previous study has suggested that the circadian rhythm (day/night cycle) is one of the basic and vital features of the natural environment due to its contribution to the regulation of normal development and function of the cardiovascular system (Melkani and Panda, 2017). It affects the expression of related genes and the levels of hormones associated with cardiac function, such as angiotensin II, renin, aldosterone, growth hormone, and atrial natriuretic peptide (McNamara et al., 2001; Martino et al., 2004; Martino and Sole, 2009).
Moreover, it has been suggested that the occurrence and the severity of certain CVDs are highly associated with the circadian rhythm via specific intrinsic mechanisms that are currently unknown (Mukamal et al., 2000; Henriques et al., 2003).
The circadian clock may regulate metabolism in an epigenetic feedback loop by modifying the expression levels of certain genes coding for metabolic switches or other important components. This modification is caused at the level of transcription. The acetylation/deacetylation of histones occurs on the promoters related to protein complexes or on clock proteins directly. In addition to this process, methylation of histone proteins has been shown to play a vital role in maintaining the normal function of the circadian rhythm in vivo (Asher et al., 2008; Nakahata et al., 2008; Katada and Sassone-Corsi, 2010).
A recent study has shown that the transcriptional functions of several clock genes, such as period 2 (Per2) and aryl hydrocarbon receptor nuclear translocator like (Arntl), play an important role in extending the circadian rhythm period by inhibiting the m6A modification of methylase METTL3, thereby promoting the uncoupling between steady-state pre-mRNA and cytoplasmic mRNA rhythm (Fustin et al., 2013). It has also been predicted that the period of the circadian rhythm is positively associated with the efficiency of degradation and the nuclear import and export of Per2 mRNA, as determined by network model analysis (Wilkins et al., 2007; Miki et al., 2012). Moreover, Arntl is much more sensitive than Per2 with regard to the inhibition of METTL3 due to its higher mRNA nuclear accumulation and longer period of transcription (Fustin et al., 2013).
The decrease in RNA methylation induced by the pharmacological action of 3-deazaadenosine (3-DZA) might elongate the period of circadian rhythm in vivo and in vitro via the decreased hydrolysis of S-adenosylhomocysteine (SAH) (Fustin et al., 2013; Singhal et al., 2013; Lokody, 2014).
It has been suggested that m6A modification affects the circadian rhythm and may regulate cardiac function to a certain degree.
4.4. Inflammation
CVDs and diabetes mellitus are defined as metabolic syndromes and have recently been investigated as chronic inflammatory diseases triggered by long-term stimulation of abnormal levels of plasma glucose, free fatty acids, and blood pressure. These abnormal biochemical profiles are promoted via the oxidative stress pathway and the interaction between the cytokines and the renin-angiotensin system (Wong et al., 2008; Grandl and Wolfrum, 2018; Fiechter et al., 2019).
Inflammation, including innate and adaptive immunity, is one of the major and fundamental risk factors of all stages of atherosclerosis (Weber and Noels, 2011; Legein et al., 2015; Ketelhuth et al., 2019). Recent studies have shown that the pathological development of atherosclerosis is driven by immune reactions and is related mainly to immune effector mechanisms, which are involved in the infiltration and accumulation of lipoproteins in the arterial intima (Libby et al., 2011, 2013; Ketelhuth and Hansson, 2016).
In addition to the epigenetic regulation of SNPs that is related to different degrees of CVD risks, hyperacetylation at the 27th lysine residue of the histone H3 protein (H3K27ac) is linked to lower expression of proinflammatory genes in the reversible process of inflammation and aging. This may also explain certain unclear mechanisms in the link between inflammation and the progression of CVDs (Smith and Humphries, 2009; IL6R Genetics Consortium Emerging Risk Factors Collaboration, 2012; Xuan et al., 2016; Mitrokhin et al., 2017; Cheng et al., 2018).
Furthermore, telomere shortening and modulation of certain microRNAs associated with DNA damage are considered another epigenetic mechanism accounting for inflammation, since they can promote the secretion of proinflammatory cytokines through several proinflammatory pathways (Olivieri et al., 2015).
Since the specific mechanism of the association between inflammation and atherosclerosis has not been fully clarified, m6A has become a new focus for the development of novel clinical therapeutic strategies against CVD.
Recent studies have demonstrated that the loss of N 6-adenosine methyltransferase METTL3 can attenuate the increased expression levels of several inflammatory cytokines and various genes associated with the inflammatory response, mainly by changing the phosphorylation levels of related signaling pathways and by regulating the mRNA splicing of splice variant of myeloid differentiation factor 88 (MyD88S) in lipopolysaccharide (LPS)-induced human dental pulp cells (HDPCs). This suggests the possible role of epitranscriptomic modification of m6A during the process of inflammation in pathophysiological mechanisms leading to CVDs (Feng et al., 2018).
5. Conclusions
Several independent studies have confirmed that the continuous and dynamic regulation of m6A in different internal biological metabolic processes has a profound impact on modulating the expression of cardiac genes in the pathological development of CVDs. Moreover, these risk factors can be combined with several other environmental factors that affect the development of these diseases. This occurs by regulation of RNA transcription, splicing, processing, translation, and decay (Niu et al., 2013; Guo et al., 2017; Roignant and Soller, 2017; Wei et al., 2017). Therefore, certain m6A enzymes, including FTO and METTL3, have been shown to control epitranscriptomic regulation of specific cardiac defects caused by distinct mechanisms. However, the possible association between m6A and other CVDs and even between cardiovascular-associated diseases, such as high blood pressure and congenital heart disease, remains unexplored. Although the use of specific inhibitors in different models of animals or cardiocytes has established the cardioprotective role of RNA-modifying enzymes in m6A modification, additional therapeutic agents for CVDs should be assessed in future studies.
Footnotes
Project supported by the Grants-in-Aid from the Graduate Research and Innovation Projects of Jiangsu Province (No. KYCX18_1461), China
Contributors: Kun ZHAO wrote and edited the manuscript. Chuan-xi YANG and Peng LI collated the literature. Wei SUN edited and revised the manuscript. Kun ZHAO and Xiang-qing KONG designed the study. All authors have read and approved the final manuscript and, therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines: Kun ZHAO, Chuan-xi YANG, Peng LI, Wei SUN, and Xiang-qing KONG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Abdul-Ghani MA, Jayyousi A, Defronzo RA, et al. Insulin resistance the link between T2DM and CVD: basic mechanisms and clinical implications. Curr Vasc Pharmacol. 2019;17(2):153–163. doi: 10.2174/1570161115666171010115119. [DOI] [PubMed] [Google Scholar]
- 2.Asher G, Gatfield D, Stratmann M, et al. Sirt1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 3.Bartosovic M, Molares HC, Gregorova P, et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 2017;45(19):11356–11370. doi: 10.1093/nar/gkx778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayarsaihan D. Epigenetic mechanisms in inflammation. J Dent Res. 2011;90(1):9–17. doi: 10.1177/0022034510378683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochmann L, Sarathchandra P, Mori F, et al. Revealing new mouse epicardial cell markers through transcriptomics. PLoS ONE. 2010;5(6):e11429. doi: 10.1371/journal.pone.0011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boissel S, Reish O, Proulx K, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009;85(1):106–111. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buysschaert I, Schmidt T, Roncal C, et al. Genetics, epigenetics and pharmaco-(epi)genomics in angiogenesis. J Cell Mol Med. 2008;12(6b):2533–2551. doi: 10.1111/j.1582-4934.2008.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai CL, Martin JC, Sun YF, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454(7200):104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai M, Liu Q, Jiang Q, et al. Loss of m6A on FAM134B promotes adipogenesis in porcine adipocytes through m6A-YTHDF2-dependent way. IUBMB Life. 2019;71(5):580–586. doi: 10.1002/iub.1974. [DOI] [PubMed] [Google Scholar]
- 10.Calvanese V, Fraga MF. Epigenetics of embryonic stem cells. Adv Exp Med Biol. 2012;741:231–253. doi: 10.1007/978-1-4614-2098-9_16. [DOI] [PubMed] [Google Scholar]
- 11.Carnevali L, Graiani G, Rossi S, et al. Signs of cardiac autonomic imbalance and proarrhythmic remodeling in FTO deficient mice. PLoS ONE. 2014;9(4):e95499. doi: 10.1371/journal.pone.0095499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cecil JE, Tavendale R, Watt P, et al. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359(24):2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 13.Chen JL, Du B. Novel positioning from obesity to cancer: FTO, an m6A RNA demethylase, regulates tumour progression. J Cancer Res Clin Oncol. 2019;145(1):19–29. doi: 10.1007/s00432-018-2796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng H, Xuan HW, Green CD, et al. Repression of human and mouse brain inflammaging transcriptome by broad gene-body histone hyperacetylation. Proc Natl Acad Sci USA. 2018;115(29):7611–7616. doi: 10.1073/pnas.1800656115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Church C, Moir L, McMurray F, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42(12):1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole MA, Jamil AHA, Heather LC, et al. On the pivotal role of PPARα in adaptation of the heart to hypoxia and why fat in the diet increases hypoxic injury. FASEB J. 2016;30(8):2684–2697. doi: 10.1096/fj.201500094R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosselman KE, Navas-Acien A, Kaufman JD. Environmental factors in cardiovascular disease. Nat Rev Cardiol. 2015;12(11):627–642. doi: 10.1038/nrcardio.2015.152. [DOI] [PubMed] [Google Scholar]
- 18.da Luz Sousa Fialho M, Jamil AHA, Stannard GA, et al. Hypoxia-inducible factor 1 signalling, metabolism and its therapeutic potential in cardiovascular disease. Biochim Biophys Acta Mol Basis Dis. 2019;1865(4):831–843. doi: 10.1016/j.bbadis.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 19.Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24(2):68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- 20.Daoud H, Zhang D, McMurray F, et al. Identification of a pathogenic FTO mutation by next-generation sequencing in a newborn with growth retardation and developmental delay. J Med Genet. 2016;53(3):200–207. doi: 10.1136/jmedgenet-2015-103399. [DOI] [PubMed] [Google Scholar]
- 21.Daya M, Pujianto DA, Witjaksono F, et al. Obesity risk and preference for high dietary fat intake are determined by FTO rs9939609 gene polymorphism in selected Indonesian adults. Asia Pac J Clin Nutr. 2019;28(1):183–191. doi: 10.6133/apjcn.201903_28(1).0024. [DOI] [PubMed] [Google Scholar]
- 22.Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 23.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 24.Dorn LE, Lasman L, Chen J, et al. The N 6-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation. 2019;139(4):533–545. doi: 10.1161/circulationaha.118.036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.el Azzouzi H, Leptidis S, Dirkx E, et al. The hypoxia-inducible microRNA cluster miR-199a~214 targets myocardial PPARδ and impairs mitochondrial fatty acid oxidation. Cell Metab. 2013;18(3):341–354. doi: 10.1016/j.cmet.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Essop MF. Cardiac metabolic adaptations in response to chronic hypoxia. J Physiol. 2007;584(3):715–726. doi: 10.1113/jphysiol.2007.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fedeles BI, Singh V, Delaney JC, et al. The AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem. 2015;290(34):20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng ZH, Li QM, Meng RS, et al. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J Cell Mol Med. 2018;22(5):2558–2568. doi: 10.1111/jcmm.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández-Morera JL, Calvanese V, Rodríguez-Rodero S, et al. Epigenetic regulation of the immune system in health and disease. Tissue Antigens. 2010;76(6):431–439. doi: 10.1111/j.1399-0039.2010.01587.x. [DOI] [PubMed] [Google Scholar]
- 30.Fiechter M, Haider A, Bengs S, et al. Sex differences in the association between inflammation and ischemic heart disease. Thromb Haemost. 2019;119(9):1471–1480. doi: 10.1055/s-0039-1692442. [DOI] [PubMed] [Google Scholar]
- 31.Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 32.Fustin JM, Doi M, Yamaguchi Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 33.Gan HL, Hong L, Yang FL, et al. Progress in epigenetic modification of mRNA and the function of m6A modification. Chin J Biotechnol. 2019;35(5):775–783. doi: 10.13345/j.cjb.180416. (in Chinese) [DOI] [PubMed] [Google Scholar]
- 34.Ge L, Cai Y, Ying F, et al. miR-181c-5p exacerbates hypoxia/reoxygenation-induced cardiomyocyte apoptosis via targeting PTPN4. Oxid Med Cell Longev, 2019: 1957920. 2019 doi: 10.1155/2019/1957920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerken T, Girard CA, Tung YCL, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity (Edinb) 2010;105(1):4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 37.Gilbert ER, Liu DM. Epigenetics: the missing link to understanding β-cell dysfunction in the pathogenesis of type 2 diabetes. Epigenetics. 2012;7(8):841–852. doi: 10.4161/epi.21238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grandl G, Wolfrum C. Hemostasis, endothelial stress, inflammation, and the metabolic syndrome. Semin Immunopathol. 2018;40(2):215–224. doi: 10.1007/s00281-017-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo MJ, Liu XH, Zheng XT, et al. m6A RNA modification determines cell fate by regulating mRNA degradation. Cell Reprogram. 2017;19(4):225–231. doi: 10.1089/cell.2016.0041. [DOI] [PubMed] [Google Scholar]
- 40.Gustavsson J, Mehlig K, Leander K, et al. FTO genotype, physical activity, and coronary heart disease risk in Swedish men and women. Circ Cardiovasc Genet. 2014;7(2):171–177. doi: 10.1161/circgenetics.111.000007. [DOI] [PubMed] [Google Scholar]
- 41.Haupt A, Thamer C, Staiger H, et al. Variation in the FTO gene influences food intake but not energy expenditure. Exp Clin Endocrinol Diabetes. 2009;117(4):194–197. doi: 10.1055/s-0028-1087176. [DOI] [PubMed] [Google Scholar]
- 42.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6(12):863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 43.He SK, Li XH, Chan N, et al. Review: epigenetic mechanisms in ocular disease. Mol Vis. 2013;19:665–674. [PMC free article] [PubMed] [Google Scholar]
- 44.Henriques JPS, Haasdijk AP, Zijlstra F, et al. Outcome of primary angioplasty for acute myocardial infarction during routine duty hours versus during off-hours. J Am Coll Cardiol. 2003;41(12):2138–2142. doi: 10.1016/S0735-1097(03)00461-3. [DOI] [PubMed] [Google Scholar]
- 45.Hess ME, Hess S, Meyer KD, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16(8):1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 46.Hou N, Wen Y, Yuan X, et al. Activation of Yap1/Taz signaling in ischemic heart disease and dilated cardiomyopathy. Exp Mol Pathol. 2017;103(3):267–275. doi: 10.1016/j.yexmp.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.IL6R Genetics Consortium, Emerging Risk Factors Collaboration, et al. Interleukin-6 receptor pathways in coronary heart disease: a collaborative meta-analysis of 82 studies. Lancet. 2012;379(9822):1205–1213. doi: 10.1016/s0140-6736(11)61931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iyen B, Qureshi N, Kai J, et al. Risk of cardiovascular disease outcomes in primary care subjects with familial hypercholesterolaemia: a cohort study. Atherosclerosis. 2019;287:8–15. doi: 10.1016/j.atherosclerosis.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 49.James K, Weitzel LRB, Engelman CD, et al. Genome scan linkage results for longitudinal systolic blood pressure phenotypes in subjects from the Framingham Heart Study. BMC Genet, 4:S83. 2003 doi: 10.1186/1471-2156-4-s1-s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia GF, Fu Y, Zhao X, et al. N 6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin J, Liu YF, Huang LH, et al. Advances in epigenetic regulation of vascular aging. Rev Cardiovasc Med. 2019;20(1):19–25. doi: 10.31083/j.rcm.2019.01.3189. [DOI] [PubMed] [Google Scholar]
- 52.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17(12):1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kennedy EM, Bogerd HP, Kornepati AVR, et al. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19(5):675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ketelhuth DFJ, Hansson GK. Adaptive response of T and B cells in atherosclerosis. Circ Res. 2016;118(4):668–678. doi: 10.1161/circresaha.115.306427. [DOI] [PubMed] [Google Scholar]
- 55.Ketelhuth DFJ, Lutgens E, Bäck M, et al. Immunometabolism and atherosclerosis: perspectives and clinical significance: a position paper from the Working Group on Atherosclerosis and Vascular Biology of the European Society of Cardiology. Cardiovasc Res. 2019;115(9):1385–1392. doi: 10.1093/cvr/cvz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khunti K, Davies M, Majeed A, et al. Hypoglycemia and risk of cardiovascular disease and all-cause mortality in insulin-treated people with type 1 and type 2 diabetes: a cohort study. Diabetes Care. 2015;38(2):316–322. doi: 10.2337/dc14-0920. [DOI] [PubMed] [Google Scholar]
- 57.Kmietczyk V, Riechert E, Kalinski L, et al. m6A-mRNA methylation regulates cardiac gene expression and cellular growth. Life Sci Alliance. 2019;2(2):e201800233. doi: 10.26508/lsa.201800233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koliaki C, Liatis S, Kokkinos A. Obesity and cardiovascular disease: revisiting an old relationship. Metabolism. 2019;92:98–107. doi: 10.1016/j.metabol.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 59.Kursawe R, Dixit VD, Scherer PE, et al. A role of the inflammasome in the low storage capacity of the abdominal subcutaneous adipose tissue in obese adolescents. Diabetes. 2016;65(3):610–618. doi: 10.2337/db15-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee SH, Wolf PL, Escudero R, et al. Early expression of angiogenesis factors in acute myocardial ischemia and infarction. N Engl J Med. 2000;342(9):626–633. doi: 10.1056/nejm200003023420904. [DOI] [PubMed] [Google Scholar]
- 61.Legein B, Janssen EM, Theelen TL, et al. Ablation of CD8α+ dendritic cell mediated cross-presentation does not impact atherosclerosis in hyperlipidemic mice. Sci Rep, 5:15414. 2015 doi: 10.1038/srep15414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Ma ZQ, Jiang S, et al. A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog Lipid Res. 2017;66:42–49. doi: 10.1016/j.plipres.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 63.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473(7347):317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 64.Libby P, Lichtman AH, Hansson GK. Immune effector mechanisms implicated in atherosclerosis: from mice to humans. Immunity. 2013;38(6):1092–1104. doi: 10.1016/j.immuni.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin SB, Choe J, Du P, et al. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu JZ, Yue YN, Han DL, et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N 6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu N, Pan T. RNA epigenetics. Transl Res. 2015;165(1):28–35. doi: 10.1016/j.trsl.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Lin L, Li Q, et al. ERK1/2 communicates GPCR and EGFR signaling pathways to promote CTGF-mediated hypertrophic cardiomyopathy upon Ang-II stimulation. BMC Mol Cell Biol, 20:14. 2019 doi: 10.1186/s12860-019-0202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lokody I. Gene regulation: RNA methylation regulates the circadian clock. Nat Rev Genet. 2014;15(1):3. doi: 10.1038/nrg3638. [DOI] [PubMed] [Google Scholar]
- 70.Lu L, Liu M, Sun RR, et al. Myocardial infarction: symptoms and treatments. Cell Biochem Biophys. 2015;72(3):865–867. doi: 10.1007/s12013-015-0553-4. [DOI] [PubMed] [Google Scholar]
- 71.Martino T, Arab S, Straume M, et al. Day/night rhythms in gene expression of the normal murine heart. J Mol Med (Berl) 2004;82(4):256–264. doi: 10.1007/s00109-003-0520-1. [DOI] [PubMed] [Google Scholar]
- 72.Martino TA, Sole MJ. Molecular time: an often overlooked dimension to cardiovascular disease. Circ Res. 2009;105(11):1047–1061. doi: 10.1161/circresaha.109.206201. [DOI] [PubMed] [Google Scholar]
- 73.Mathiyalagan P, Adamiak M, Mayourian J, et al. FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair. Circulation. 2019;139(4):518–532. doi: 10.1161/circulationaha.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mauer J, Luo XB, Blanjoie A, et al. Reversible methylation of m6Am in the 5' cap controls mRNA stability. Nature. 2017;541(7637):371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mazzio EA, Soliman KFA. Basic concepts of epigenetics: impact of environmental signals on gene expression. Epigenetics. 2012;7(2):119–130. doi: 10.4161/epi.7.2.18764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McNamara P, Seo SB, Rudic RD, et al. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105(7):877–889. doi: 10.1016/S0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- 77.Melkani GC, Panda S. Time-restricted feeding for prevention and treatment of cardiometabolic disorders. J Physiol. 2017;595(12):3691–3700. doi: 10.1113/jp273094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meyer KD, Patil DP, Zhou J, et al. 5' UTR m6A promotes cap-independent translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miki T, Xu ZX, Chen-Goodspeed M, et al. PML regulates PER2 nuclear localization and circadian function. EMBO J. 2012;31(6):1427–1439. doi: 10.1038/emboj.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mitrokhin V, Nikitin A, Brovkina O, et al. Association between interleukin-6/6R gene polymorphisms and coronary artery disease in Russian population: influence of interleukin-6/6R gene polymorphisms on inflammatory markers. J Inflamm Res. 2017;10:151–160. doi: 10.2147/jir.s141682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mukamal KJ, Muller JE, Maclure M, et al. Increased risk of congestive heart failure among infarctions with nighttime onset. Am Heart J. 2000;140(3):438–442. doi: 10.1067/mhj.2000.108830. [DOI] [PubMed] [Google Scholar]
- 83.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niu YM, Zhao X, Wu YS, et al. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11(1):8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olivieri F, Albertini MC, Orciani M, et al. DNA damage response (DDR) and senescence: shuttled inflamma-miRNAs on the stage of inflammaging. Oncotarget. 2015;6(34):35509–35521. doi: 10.18632/oncotarget.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ortiz-Barahona A, Villar D, Pescador N, et al. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010;38(7):2332–2345. doi: 10.1093/nar/gkp1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Panneerdoss S, Eedunuri VK, Yadav P, et al. Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Sci Adv. 2018;4(10):eaar8263. doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parashar NC, Parashar G, Nayyar H, et al. N 6-adenine DNA methylation demystified in eukaryotic genome: from biology to pathology. Biochimie. 2018;144:56–62. doi: 10.1016/j.biochi.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 89.Pastore N, Brady OA, Diab HI, et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy. 2016;12(8):1240–1258. doi: 10.1080/15548627.2016.1179405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patil DP, Pickering BF, Jaffrey SR. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 2018;28(2):113–127. doi: 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peña MSB, Rollins A. Environmental exposures and cardiovascular disease: a challenge for health and development in low-and middle-income countries. Cardiol Clin. 2017;35(1):71–86. doi: 10.1016/j.ccl.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reese DE, Mikawa T, Bader DM. Development of the coronary vessel system. Circ Res. 2002;91(9):761–768. doi: 10.1161/01.RES.0000038961.53759.3C. [DOI] [PubMed] [Google Scholar]
- 93.Rodriguez H, Drouin R, Holmquist GP, et al. A hot spot for hydrogen peroxide-induced damage in the human hypoxia-inducible factor 1 binding site of the PGK 1 gene. Arch Biochem Biophys. 1997;338(2):207–212. doi: 10.1006/abbi.1996.9820. [DOI] [PubMed] [Google Scholar]
- 94.Rodriguez-Rodero S, Fernández-Morera JL, Fernandez AF, et al. Epigenetic regulation of aging. Discov Med. 2010;10(52):225–233. [PubMed] [Google Scholar]
- 95.Roignant JY, Soller M. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33(6):380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 96.Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shen F, Huang W, Huang JT, et al. Decreased N 6-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J Clin Endocrinol Metab. 2015;100(1):E148–E154. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Singhal A, Arora G, Sajid A, et al. Regulation of homocysteine metabolism by Mycobacterium tuberculosis S-adenosylhomocysteine hydrolase. Sci Rep, 3:2264. 2013 doi: 10.1038/srep02264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smart N, Risebro CA, Melville AAD, et al. Thymosin β-4 is essential for coronary vessel development and promotes neovascularization via adult epicardium. Ann N Y Acad Sci. 2007;1112(1):171–188. doi: 10.1196/annals.1415.000. [DOI] [PubMed] [Google Scholar]
- 100.Smith AJP, Humphries SE. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev. 2009;20(1):43–59. doi: 10.1016/j.cytogfr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 101.Smith TG, Robbins PA, Ratcliffe PJ. The human side of hypoxia-inducible factor. Br J Haematol. 2008;141(3):325–334. doi: 10.1111/j.1365-2141.2008.07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith ZD, Chan MM, Mikkelsen TS, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484(7394):339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Song HW, Feng X, Zhang H, et al. METTL3 and ALKBH5 oppositely regulate m6A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15(8):1419–1437. doi: 10.1080/15548627.2019.1586246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Souness JE, Stouffer JE, de Sanchez VC. Effect of N 6-methyladenosine on fat-cell glucose metabolism: evidence for two modes of action. Biochem Pharmacol. 1982;31(24):3961–3971. doi: 10.1016/0006-2952(82)90642-6. [DOI] [PubMed] [Google Scholar]
- 105.Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring) 2008;16(8):1961–1965. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- 106.Tan FL, Moravec CS, Li JB, et al. The gene expression fingerprint of human heart failure. Proc Natl Acad Sci USA. 2002;99(17):11387–11392. doi: 10.1073/pnas.162370099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Timpson NJ, Emmett PM, Frayling TM, et al. The fat mass- and obesity-associated locus and dietary intake in children. Am J Clin Nutr. 2008;88(4):971–978. doi: 10.1093/ajcn/88.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.van Wijk B, van den Berg G, Abu-Issa R, et al. Epicardium and myocardium separate from a common precursor pool by crosstalk between bone morphogenetic protein-and fibroblast growth factor-signaling pathways. Circ Res. 2009;105(5):431–441. doi: 10.1161/circresaha.109.203083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang CY, Shie SS, Wen MS, et al. Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism. Sci Signal. 2015;8(407):ra127. doi: 10.1126/scisignal.aab3357. [DOI] [PubMed] [Google Scholar]
- 110.Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014;11(6):669–672. doi: 10.4161/rna.28829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang Y, Li Y, Toth JI, et al. N 6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med. 2011;17(11):1410–1422. doi: 10.1038/nm.2538. [DOI] [PubMed] [Google Scholar]
- 113.Wei CM, Moss B. Nucleotide sequences at the N 6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16(8):1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 114.Wei WQ, Ji XY, Guo XQ, et al. Regulatory role of N6-methyladenosine (m6A) methylation in RNA processing and human diseases. J Cell Biochem. 2017;118(9):2534–2543. doi: 10.1002/jcb.25967. [DOI] [PubMed] [Google Scholar]
- 115.Wilkins AK, Barton PI, Tidor B. The Per2 negative feedback loop sets the period in the mammalian circadian clock mechanism. PLoS Comput Biol. 2007;3(12):e242. doi: 10.1371/journal.pcbi.0030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Winter EM, Groot ACGD. Epicardium-derived cells in cardiogenesis and cardiac regeneration. Cell Mol Life Sci. 2007;64(6):692–703. doi: 10.1007/s00018-007-6522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wong C, Kanetsky P, Raj D. Genetic polymorphisms of the RAS-cytokine pathway and chronic kidney disease. Pediatr Nephrol. 2008;23(7):1037–1051. doi: 10.1007/s00467-008-0816-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xuan Y, Wang LN, Zhi H, et al. Association between 3 IL-10 gene polymorphisms and cardiovascular disease risk: systematic review with meta-analysis and trial sequential analysis. Medicine (Baltimore) 2016;95(6):e2846. doi: 10.1097/md.0000000000002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang Y, Shen F, Huang W, et al. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J Clin Endocrinol Metab. 2019;104(3):665–673. doi: 10.1210/jc.2018-00619. [DOI] [PubMed] [Google Scholar]
- 120.Zhong SL, Li HY, Bodi Z, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20(5):1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou B, Ma Q, Rajagopal S, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454(7200):109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]