Abstract
Porcine circovirus 3 (PCV3) has been detected in major pig-producing countries around the world since its first report in the US in 2016. Most current studies have focused on epidemiological investigations and detection methods of PCV3 because of lack of live virus strains for research on its pathogenesis in porcine cells or even in pigs. We constructed a recombinant plasmid pCMV-Cap carrying the PCV3 orf2 gene to investigate the effects of capsid (Cap) protein expression on autophagic response in human embryonic kidney cell line 293T (HEK293T). We demonstrate that PCV3 Cap protein induced complete autophagy shown as formation of autophagosomes and autophagosome-like vesicles as well as LC3-II conversion from LC3-I via inhibiting phosphorylation of the mammalian target of rapamycin (mTOR) in HEK293T cells. The ubiquitin–proteasome pathway is also involved in the autophagy process. These findings provide insight for further exploration of PCV3 pathogenetic mechanisms in porcine cells.
Keywords: Porcine circovirus 3 (PCV3), Capsid (Cap) protein, Autophagy
1. Introduction
Porcine circoviruses (PCVs) are single-stranded, non-enveloped circular DNA viruses, belonging to the genus Circovirus in the family Circoviridae (Ellis, 2014). Three types of PCVs (PCV1 (Tischer et al., 1974), PCV2 (Allan et al., 1998), and PCV3 (Phan et al., 2016; Palinski et al., 2017)) have been reported in pigs. PCV1 was a contaminant of the PK-15 cells and nonpathogenic to pigs (Tischer et al., 1974), while PCV2 is associated with porcine circovirus-associated diseases (PCVADs) and poses a serious threat to the pig industry (Ellis, 2014). Since the first report in the US in 2016 (Phan et al., 2016), PCV3 has been discovered in major pig-producing countries (Faccini et al., 2017; Ku et al., 2017; Kwon et al., 2017; Stadejek et al., 2017; Franzo et al., 2018; Hayashi et al., 2018; Kedkovid et al., 2018; Tochetto et al., 2018; Ye et al., 2018; Yuzhakov et al., 2018).
The PCV3 genome is 2000 nucleotides in length with three predicted open reading frames (ORFs). The gene orf1 encodes the replicase (Rep) protein of 296 amino acids (aa) and orf2, the capsid (Cap) protein of 241 aa, while the initiation codon of orf3 and the function of its encoded protein are not yet clear (Phan et al., 2016). Cap is the major structural protein of PCV3 and shares 24% and 26% identity to that of PCV1 and PCV2, respectively, at the amino acid level (Phan et al., 2016; Palinski et al., 2017). A recent report indicates that PCV3 Cap inhibited type I interferon signaling by interacting with signal transducer and activator of transcription 2 (STAT2) (Shen et al., 2020). Pigs infected with PCV3 are associated with clinical symptoms, such as porcine dermatitis and nephropathy syndrome (PDNS), reproductive failure, multi-systemic inflammation, and cardiac pathology (Phan et al., 2016; Palinski et al., 2017). However, PCV3 is also found in pigs with no clinical signs nor disease conditions (Stadejek et al., 2017; Zheng et al., 2017).
Autophagy is a highly conserved recycling process in eukaryotes, in which double-membrane autophagosomes sequester proteins, cytoplasmic organelles, and macromolecules, and fuse with lysosomes or vacuoles for degradation and turnover (He and Klionsky, 2009; Parzych and Klionsky, 2014). It is a dynamic multi-step cellular process from nucleation to formation of autophagosomes and to degradation upon fusion with lysosomes that is termed “autophagy flux” to reflect the progression of the complete autophagic process (Mizushima et al., 2010; Parzych and Klionsky, 2014). Autophagy is induced by intra-or extra-cellular stresses or signals such as endoplasmic reticulum (ER) stress, starvation, and bacterial or viral infections (He and Klionsky, 2009), and regulated by mammalian target of rapamycin (mTOR)-dependent pathway and mTOR-independent pathway (He and Klionsky, 2009; Ravikumar et al., 2010; Sarkar, 2013; Parzych and Klionsky, 2014). mTOR is a serine/threonine kinase belonging to the phosphoinositide-3-kinase (PI3K)-related kinase (PIKK) family and forms two distinct protein complexes, namely mTOR complex 1 (mTORC1) and 2 (mTORC2) (Saxton and Sabatini, 2017). mTORC1 is the master regulator of autophagy, and inhibition of mTORC1 is needed for initiation of the autophagy process, including nucleation, elongation, and maturation of autophagosomes via downstream targets, such as uncoordinated (UNC)‐ 51-like-kinase 1 and 2 (ULK1/2), autophagy-related gene 13 (ATG13), and others (Dossou and Basu, 2019).
Viruses have evolved diverse strategies to interact with the autophagic machinery or subvert autophagy to counteract the host antiviral effects (Yin et al., 2019). Some viruses, such as classical swine fever virus (Pei et al., 2014) and pseudorabies virus (Xu et al., 2018), can induce complete autophagy during infection. There are viruses that induce incomplete autophagy. Rabies virus infection, for instance, induced incomplete autophagy shown as increased accumulation of autophagosomes due to inhibition of the autophagy flux (Liu et al., 2017). Hepatitis B virus infection led to incomplete autophagy by impairing lysosomal acidification (Zhong et al., 2015). Coxsackievirus B3 infection inhibited autophagic flux by limiting the fusion of autophagosomes with lysosomes (Mohamud et al., 2018). In addition, some viruses can inhibit autophagy to facilitate viral replication. African swine fever virus infection inhibited autophagosome formation in Vero cells by interaction of its A179L protein, a viral homolog of B-cell lymphoma 2 (Bcl2), with Beclin 1 (Hernaez et al., 2013). Thus the autophagy process in the host varies among viruses. A better understanding of the interactions between host autophagic responses and viruses will provide insight into the pathogenesis of viruses and an avenue for the development of antiviral drugs.
Our laboratory and other researchers have recently proved that PCV2 induces complete autophagy, most likely by its Cap protein, and the virus employs autophagy machinery to enhance its replication in PK-15 cells (Zhu et al., 2012a, 2012b; Qian et al., 2017; Zhai et al., 2019). Although a recombinant PCV3 virus has been rescued (Jiang HJ et al., 2019), most of the recent studies on PCV3 focus on epidemiological investigations and detection methods, for the reason that there are no live PCV3 virus strains for research. There has been no report so far on the involvement of autophagy in PCV3 pathogenesis. In the absence of PCV3 virus for research, we constructed a recombinant plasmid pCMV-Cap carrying the orf2 gene of PCV3 to investigate whether the Cap protein induces a cellular autophagic response. Here, we provide the first evidence that the Cap protein of PCV3 induced autophagy by inhibiting phosphorylation of mTOR in human embryonic kidney cell line 293T (HEK293T).
2. Materials and methods
2.1. Cell culture
HEK293T cell line was used and the cells were cultured at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Thermo Scientific, MA, USA) containing 10% heat-inactivated fetal bovine serum (FBS; Gibco, Thermo Scientific) with 1% L-glutamine, 1% nonessential amino acids, and antibiotics (100 U/mL penicillin G and 100 μg/mL streptomycin).
2.2. Plasmid construction and transfection
The full-length of orf2 gene encoding Cap of the PCV3 strain (PCV3-US/MO2015, GenBank accession No. KX778720.1) was synthesized by BioSune Co., Ltd. (Shanghai, China), cloned into the pCMV plasmid (Clontech, CA, USA) using ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China), and confirmed by DNA sequencing (BioSune). Delivery of the recombinant and control plasmids into HEK293T cells was performed by using Lipofectamine 3000 (Invitrogen, CA, USA) according to the manufacturer’s instructions. Construction of the plasmid pcDNA3.1-EGFP-LC3B (pEGFP-LC3B) has been described elsewhere (Zhu et al., 2012a). All the primers used in this study are listed in Table 1.
Table 1.
Primers used for cloning
| Primer | Sequence (5'→3') |
| Cap-F | CCCAGGCCCGAATTCATGAGACACAGAGCTATATTCAGAA |
| Cap-R | CCCCGCGGCCGCTTACTTATCGTCGTCATCCTTGTAATCTCCACCGCTTCCACCGAGAACGGACTTGTAACGAATC |
Cap: capsid; F: forward; R: reverse
2.3. Antibodies and chemicals
Rabbit anti-PCV3 Cap polyclonal antibody was produced in our laboratory. Antibodies against β-actin, sequestosome 1 (SQSTM1/p62) and mTOR were purchased from DiagBio Technology Co., Ltd. (Hangzhou, China). Antibodies against lipid-conjugated 3 (LC3A/B) and phosphorylated mTOR (p-mTOR; phospho S2448) were purchased from Cell Signaling Technology (Danvers, MA, USA). Alexa fluor 555-conjugated goat anti-rabbit IgG, goat anti-rabbit IgG, and goat anti-mouse IgG secondary antibodies conjugated to horseradish peroxidase (HRP) were purchased from Sangon Biotech (Shanghai, China).
The chemicals rapamycin (Rapa; as autophagy inducer; Merck, Germany), chloroquine (CQ; as lysosomal inhibitor; MCE, USA), and MG132 (as proteasome inhibitor; MCE) were used in appropriate experiments. Nuclear stain 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) was purchased from Beyotime (Hangzhou, China).
2.4. Protein extraction and western blotting
Western blotting was performed to detect changes of the expression of target proteins in HEK293T cells transfected with plasmids or treated with chemicals. Briefly, cells in 24-well plates were harvested at indicated time points by using a pre-cooled lysis buffer (Beyotime) supplemented with protease inhibitor cocktail (Roche, Mannheim, Germany). The cell lysates were centrifuged and the supernatant samples were collected. Protein concentration in the samples was quantified using the Bradford assay kit (Beyotime). Equal amounts of the protein samples were loaded and separated on 8%–15% (8% (0.08 g/mL) for mTOR and p-mTOR; 12% (0.12 g/mL) for β-actin, p62, and Cap; 15% (0.15 g/mL) for LC3) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels after boiling for 10 min in the presence of the loading buffer. The proteins in the gels were transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Darmstadt, Germany), blocked at 37 °C for 1 h with Tris-buffered saline (25 mmol/L Tris at pH 7.5, 150 mmol/L NaCl) containing 0.05% Tween-20 (TBST) and 5% skim milk, and incubated with different primary antibodies at 4 °C overnight. The membranes were washed five times with TBST and incubated at 37 °C for 1 h with HRP-conjugated goat anti-rabbit or goat anti-mouse antibodies. The blots were visualized using the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific), and images were captured with the Gel 3100 Chemiluminescent Imaging System (Sagecreation, Beijing, China) as described (He et al., 2019). The bands were quantified by densitometric analysis using the ImageJ software (National Institutes of Health, USA). Ratios of the target proteins to the reference proteins were used for relative quantitative purposes.
2.5. Confocal microscopy
For detection of autophagosomes by confocal microscopy, HEK293T cells were cultured on coverslips in 29-mm dishes (Cellvis, USA). Cells were co-transfected with the plasmids pEGFP-LC3B and pCMV-Cap or the control plasmid pCMV, or transfected only with pEGFP-LC3B (mock) or treated with 2 μmol/L Rapa after 5 h of transfection with pEGFP-LC3B (positive control). After 36 h of transfection, cells were washed with phosphate-buffered saline (PBS; pH 7.4), fixed with 4% paraformaldehyde fix solution for 30 min at room temperature, permeabilized with 0.1% Triton X-100 for 15 min, and then blocked with 5% skim milk for 1 h at 37 °C. Following this, the cells were immuno-stained with rabbit anti-PCV3 Cap polyclonal antibody for 1 h at 37 °C, washed with PBS, and incubated with Alexa Fluor 555-conjugated goat anti-rabbit IgG for 1 h at 37 °C. Nuclei were counterstained with DAPI. Fluorescent EGFP-LC3B punctae were observed and captured by laser scanning confocal microscopy (IX81-FV1000; Olympus, Hamburg, Germany) as described (Shan et al., 2018).
2.6. Transmission electron microscopy
For ultrastructural visualization of autophagy, HEK293T cells transfected with pCMV-Cap, Rapa-or mock-treated were harvested by centrifugation and washed with PBS. Samples were fixed in 2.5% glutaraldehyde in phosphate buffer (PB, 0.1 mol/L, pH 7.0) at 4 °C overnight and post-fixed with 1% OsO4 in PB for 1 h. The samples were then dehydrated in graded ethanol (30%, 50%, 70%, 80%, 90%, 95%, and 100%) and transferred to pure acetone for 20 min. After placing in 1:1 mixture of Spurr resin (SPI-CHEM, PA, USA) and acetone for 1 h at room temperature, samples were transferred to 3:1 mixture of Spurr resin and acetone for 3 h and finally placed in pure Spurr resin overnight. Ultrathin sections (70–90 nm) were sliced by LEICA EM UC7 ultrathin slicer (Vienna, Austria) and stained with uranyl acetate and lead citrate. Autophagosomes were observed under a transmission electron microscope (TEM; H-7650, Hitachi, Tokyo, Japan).
2.7. Statistic analysis
All results are presented as mean±standard error of the mean (SEM) from three independent experiments. Statistical analyses were performed using GraphPad Prism 7 (GraphPad Inc., CA, USA). Differences were considered significant with P values of <0.05 and highly significant with P values of <0.01 using the Student’s t-test (Song et al., 2019).
3. Results
3.1. Effect of PCV3 Cap protein on induction of autophagy
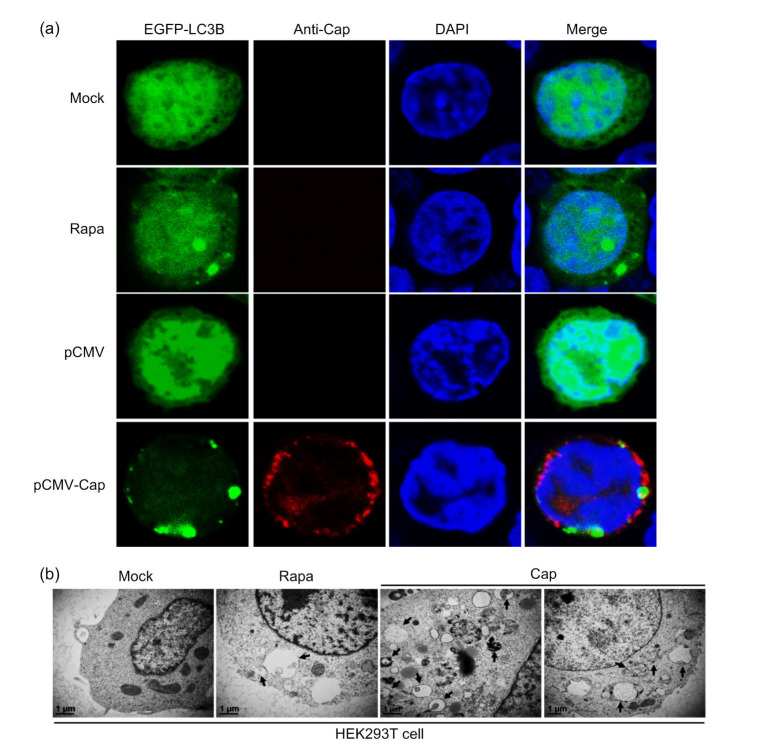
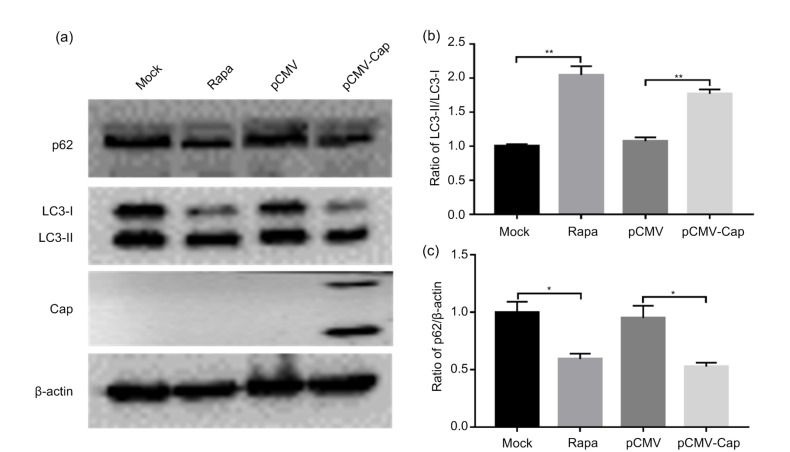
In order to investigate whether PCV3 Cap could trigger autophagic machinery, cells were co-transfected with recombinant plasmids expressing LC3B and PCV3-Cap for confocal imaging to visualize the LC3B punctae. Fig. 1a shows that overexpression of PCV3 Cap induced formation of autophagosomes shown as EGFP-LC3B punctae around the nuclei of HEK293T cells, similar to Rapa treatment, while the control cells did not show apparent LC3B punctae. To further confirm that PCV3 Cap could induce autophagy, we examined LC3 lipidation (LC3-II), a protein marker of autophagy (Mizushima et al., 2010), and SQSTM1/p62, known as autophagy substrate (Bjørkøy et al., 2005; Pankiv et al., 2007). Western blotting showed that PCV3 Cap expression and Rapa treatment significantly increased the conversion of LC3-I to LC3-II (P<0.01) and reduced the ratio of p62 to β-actin (P<0.05) relative to control cells, with statistical significance at 36 h post-transfection (hpt) (Fig. 2). TEM revealed typical autophagosome-like vesicles with double or single membrane in cells expressing PCV3 Cap or treated with Rapa. Autophagosome-like vesicles were rarely seen in the mock-treated cells (Fig. 1b). These results indicate that PCV3 Cap protein induced autophagy in HEK293T cells.
Fig. 1.
Formation of autophagosomes induced by PCV3 capsid protein shown as EGFP-LC3B punctae and autophagosome-like vesicles
(a) Cells were co-transfected with pEGFP-LC3B and pCMV-capsid (Cap) or the control plasmid pCMV, or transfected only with pEGFP-LC3B (mock), or treated with 2 μmol/L rapamycin (Rapa) after 5 h of transfection with pEGFP-LC3B (positive control). After 36 h of transfection, cells were immuno-stained with the rabbit anti-PCV3 Cap polyclonal antibody and counterstained with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) for nuclei, and then examined for formation of EGFP-LC3B punctae under the confocal microscope. (b) Cells were transfected with pCMV-Cap for 36 h or mock-treated (negative control) or treated with 2 μmol/L Rapa (positive control). Autophagosome-like vesicles were observed by transmission electron microscope. Black arrows indicate the characteristic structures of autophagosomes. Scale bar=1 μm
Fig. 2.
Increased conversion of LC3-I to LC3-II and downregulated ratio of p62 to β-actin caused by PCV3 capsid protein
Cells were first transfected with pCMV-capsid (Cap) or the control plasmid pCMV. Rapamycin (Rapa) was used as positive control. After 36 h of transfection, the whole cell lysates were subjected to western blotting for β-actin, LC3, p62, and PCV3 Cap. (a) A representative blot showing the effects of PCV3 Cap on target proteins. (b, c) Ratios of LC3-II to LC3-I and p62 to β-actin normalized to mock transfection set at 1.0. Data were expressed as mean±standard error of the mean (SEM), n=3. * P<0.05; ** P<0.01
3.2. Complete autophagy flux induced by PCV3 Cap protein
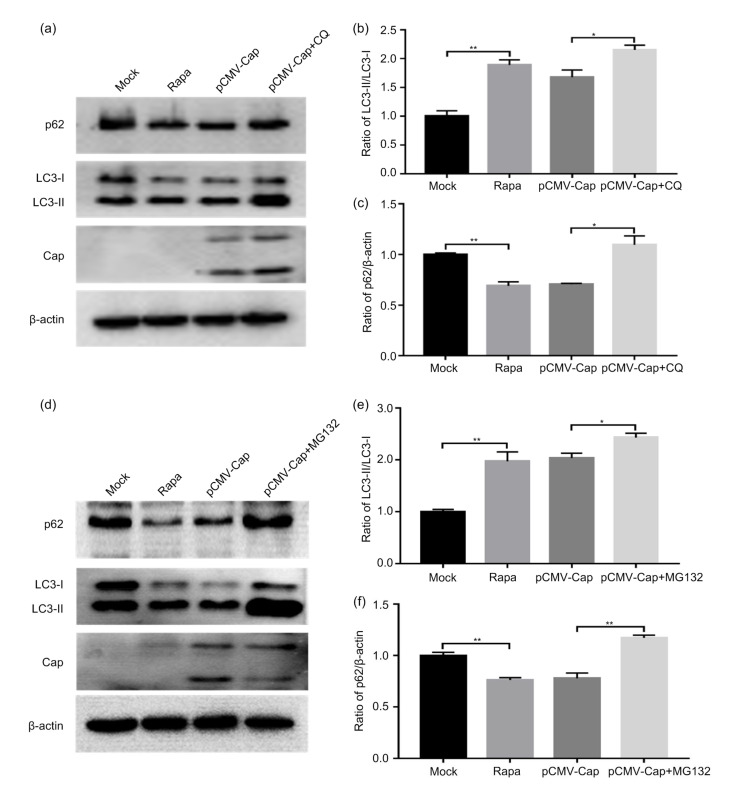
Autophagy flux reflects progression of the autophagic response (Mizushima et al., 2010). In order to investigate whether PCV3 Cap induces complete autophagy flux, we examined the protein level of p62 in the absence or presence of CQ, a downstream autophagy inhibitor, by inhibiting acidification of lysosomes (Poole and Ohkuma, 1981) and repressing lysosomal degradation of autophagic substrates such as LC3-II and p62 (Mizushima et al., 2010). In cells treated with CQ, the ratios of LC3-II to LC3-I and p62 to β-actin were significantly higher than those of the untreated cells (Figs. 3a–3c; P<0.05). These findings reveal that PCV3 Cap induced complete autophagy in HEK293T cells and p62 proteins were degraded via the autophagic lysosomal pathway.
Fig. 3.
Complete autophagy flux induced by PCV3 capsid protein and involvement of ubiquitin–proteasome pathway in the autophagy process
Cells were first transfected with pCMV-capsid (Cap) for 5 h and then treated either with 5 μmol/L chloroquine (CQ) or MG132, or left untreated as control. Rapamycin (Rapa) was used as a positive control. After 36 h of transfection, the whole cell lysates were subjected to western blotting for β-actin, LC3, p62, and PCV3 Cap. (a, d) Representative blots showing the effects of CQ or MG132 on target proteins. (b, e) Ratio of LC3-II to LC3-I normalized to mock transfection set at 1.0. (c, f) Ratio of p62 to β-actin normalized to mock transfection set at 1.0. Data were expressed as mean±standard error of the mean (SEM), n=3. * P<0.05; ** P<0.01
3.3. Involvement of ubiquitin–proteasome pathway in the autophagy process induced by PCV3 Cap protein
Considering that the autophagy–lysosomal pathway and the ubiquitin–proteasome pathway are two main machineries for protein degradation in the eukaryote (Hara et al., 2006; Komatsu et al., 2006), we examined whether ubiquitin–proteasome pathway participated in the autophagy process induced by PCV3 Cap. In the ubiquitin–proteasome pathway, ubiquitinated proteins are degraded by the 26S proteasome complex and the degradation process can be blocked by the peptide-aldehyde proteasome inhibitor MG132 (Han et al., 2009; Wiegering et al., 2019). When cells were treated with MG132 after 5 h of transfection with pCMV-Cap, the ratios of LC3-II to LC3-I (Fig. 3e; P<0.05) and p62 to β-actin (Fig. 3f; P<0.01) were significantly increased relative to the untreated cells, indicating that the ubiquitin–proteasome pathway might be involved in the autophagy process induced by PCV3 Cap.
3.4. Decreased phosphorylation of mTOR by PCV3 Cap protein
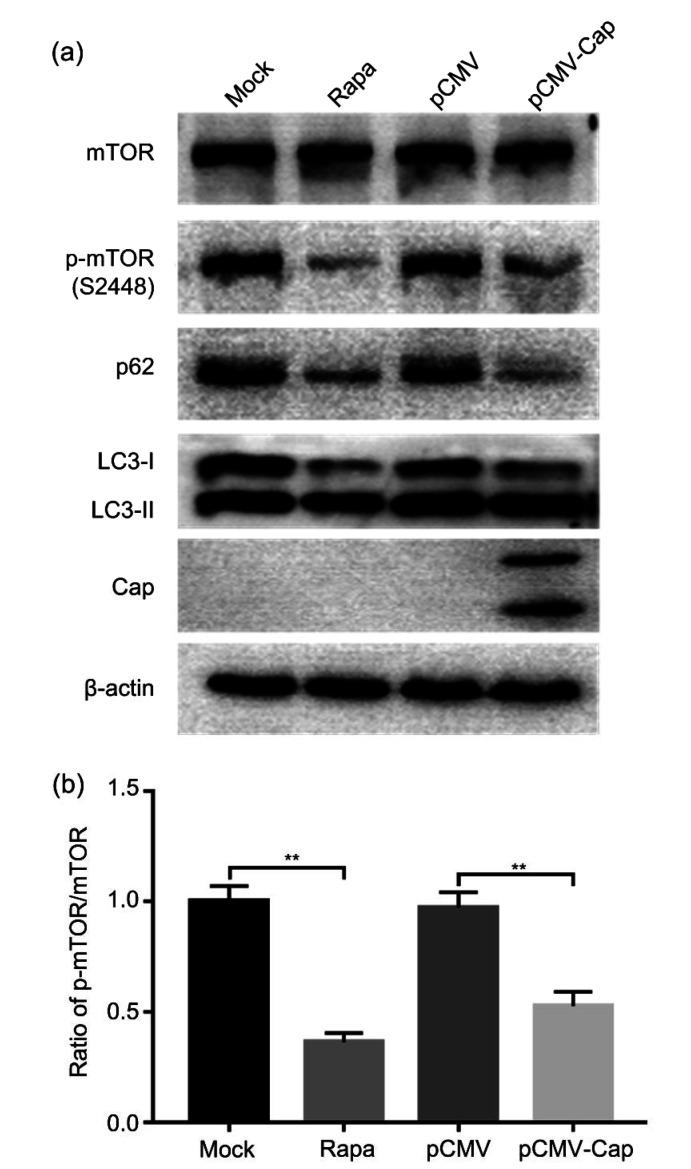
Since mTOR signaling is the main pathway that regulates autophagy (Díaz-Troya et al., 2008; Jung et al., 2010), we investigated whether mTOR was inhibited in HEK293T cells transfected with pCMV-Cap using Rapa as positive control. Fig. 4 shows that expression of PCV3 Cap or Rapa treatment downregulated the ratio of p-mTOR to the total mTOR (P<0.01), suggesting that mTOR signaling is involved in the autophagy process mediated by the PCV3 Cap protein.
Fig. 4.
Decreased phosphorylation of mTOR by PCV3 capsid protein
Cells were transfected with pCMV-capsid (Cap) or the control plasmid pCMV. Rapamycin (Rapa) was used as a positive control. After 36 h of transfection, the whole cell lysates were subjected to western blotting for mammalian target of rapamycin (mTOR), phosphorylated-mTOR (p-mTOR) (S2448), β-actin, LC3, p62, and PCV3 Cap. (a) A representative blot showing the effects of PCV3 Cap expression on target proteins. (b) Ratio of p-mTOR (S2448) to mTOR normalized to mock transfection set at 1.0. Data were expressed as mean±standard error of the mean (SEM), n=3. ** P<0.01
4. Discussion
Previous studies in our laboratory have shown that PCV2 induced autophagy, most probably by the Cap protein, and PCV2 exploited the autophagic machinery to enhance its replication in PK-15 cells (Zhu et al., 2012a, 2012b). Here, we provide evidence that the Cap protein from PCV3 induced autophagy in HEK293T cells.
Initially, the recombinant plasmid pCMV-Cap was transfected into PK-15 cells and porcine intestinal epithelial cells (IPEC-J2). However, the transfection efficiency was only 10%–20% in PK-15 and IPEC-J2 cells such that it prevented us from further study (data not shown). Low transfection efficiency may be related to the cell types. Considering that HEK293T cells have the advantage of high transfection efficiency, we attempted to explore whether PCV3 Cap expression could induce autophagy in this cell line.
By confocal imaging, TEM, and western blotting, we found that PCV3 Cap protein induced autophagy in HEK293T cells (Figs. 1 and 2). Autophagy flux is often used to monitor the process of autophagy (Mizushima et al., 2010; Yin et al., 2019). CQ is known to inhibit lysosome acidification (Poole and Ohkuma, 1981) and repress autophagy by inhibiting lysosome-mediated degradation of the autophagic debris. CQ used at 5 μmol/L for 36 h did not affect the viability of the HEK293T cells as determined by the Cell Counting Kit-8 (CCK-8; data not shown). The fact that the autophagy process induced by PCV3 Cap was blocked by CQ treatment (Fig. 3) indicates that complete autophagy flux had occurred, a finding similar to PCV2 (Zhu et al., 2012a).
One phenomenon that needs to be pointed out from the experiment is that, whenever the plasmid pCMV-Cap was transfected to PK-15, IPEC-J2, or HEK293T cells, two protein bands at 10–15 kDa and 15–25 kDa were always present by immunoblotting with the rabbit anti-PCV3 Cap polyclonal antibody or specific anti-Flag monoclonal antibody. In order to verify whether the Cap protein was partially degraded into two bands by the host proteasomes, MG132 was added to the cells transfected with pCMV-Cap to inhibit the activity of proteases. Two protein bands were still detected, indicating that these two bands were not degraded by the cellular proteasome pathway (Fig. 3). To further explore the two protein bands of the PCV3 Cap protein, we constructed two recombinant plasmids, one with full-length orf2 and the other with deletion of the region coding for the nuclear localization signal (NLS). Each peptide was fused on both ends: the N-terminus with 6×His tag and the C-terminus with Flag tag. HEK293T cells were transfected and collected for western blotting. Only one protein band (25–35 kDa) was seen in the cells expressing the His-tagged Cap as probed with the anti-His monoclonal antibody, while there were two bands in cells expressing the Flag-tagged Cap when probed with the anti-Flag monoclonal antibody (data not shown), similar to the results shown in Fig. 3. This indicates that the two in-frame initiation codons in the orf2 gene of PCV3 might functionally operate to generate two peptides, one as the full-length Cap protein and the other an N-terminally “truncated” peptide.
With 5 μmol/L of MG132 treatment, the cell viability was above 80% (data not shown), similar to published reports such as the one by Harhouri et al. (2017). Because MG132 treatment further increased the ratios of LC3-II to LC3-I and p62 to β-actin (Figs. 3d–3f), it is apparent that the ubiquitin–proteasome pathway is also involved in the autophagy process induced by PCV3 Cap protein. This is consistent with reports that exposure of MG132 impaired autophagic flux (Ji et al., 2016) and even led to activation of autophagy (Harhouri et al., 2017). LC3 could be degraded by the proteasomes in a stepwise manner (Gao et al., 2010). A recent report indicates that proteasomal degradation of LC3-I, but not directly of LC3-II, could be promoted by BRUCE, a protein containing ubiquitin-conjugating E2 domain with anti-apoptotic effect, leading to decreasing levels of LC3-II and autophagy (Jiang TX et al., 2019). This could be the reason why LC3-II was significantly increased in MG132-treated cells, because MG132 inhibited degradation of LC3-I by proteasomes without affecting its lipidation in the presence of an autophagy inducer, namely PCV3 Cap protein in this study. Because MG132 treatment could also induce autophagy (Harhouri et al., 2017), both PCV3 Cap protein and MG132 might have contributed to the increased level of LC3-II (Figs. 3c and 3d). Reduced expression of PCV3 Cap protein in MG132-treated cells (Fig. 3d) could be due to inhibition of protein synthesis because MG132 has been shown to inhibit general protein synthesis (Ding et al., 2006) possibly by increased phosphorylation of eukaryotic initiation factor 2α (eIF2α) (Jiang and Wek, 2005).
Since autophagy is regulated by a number of signaling pathways and the mTOR pathway is the major one (Díaz-Troya et al., 2008; Jung et al., 2010; Parzych and Klionsky, 2014), we examined whether PCV3 Cap expression affected mTOR phosphorylation, a key step in mTOR-mediated signaling of autophagy (Díaz-Troya et al., 2008; Jung et al., 2010). Fig. 4 shows that Cap expression reduced p-mTOR level, similar to treatment with Rapa known to induce autophagy while total mTOR remained essentially unchanged. For PCV2 and simian virus 40 small T antigen, mTOR was also inhibited in autophagy induction (Kumar and Rangarajan, 2009; Zhu et al., 2012b). We believe that the PCV3 Cap protein induces autophagy in HEK293T cells by inhibiting phosphorylation of mTOR.
5. Conclusions
In summary, PCV3 Cap protein induces complete autophagy in HEK293T cells by inhibiting mTOR phosphorylation. Further research is required to examine whether the PCV3 Cap protein is involved in autophagy in porcine cell lines including the primary cells from pigs once the recombinant PCV3 virus could be rescued and propagated in appropriate cell culture systems. Such studies could help better understanding of the PCV3 pathogenesis.
Acknowledgments
We thank Dr. Ying SHAN in the Core Facility Platform of the College of Animal Science, Zhejiang University (Hangzhou, China), for assistance with confocal microscopy, and the staff in the Bio-ultrastructure Analysis Center of the Agrobiology and Environmental Sciences, Zhejiang University, for assistance with transmission electron microscopy.
Footnotes
Project supported by the Zhejiang Provincial Department of Science and Technology (No. 2018C02028), China
Contributors: Shi-chao GENG contributed to the study design, performed the experiments and data analysis, wrote and edited the manuscript. Xiao-liang LI contributed to the study design, discussion, and revision of the manuscript. Wei-huan FANG conceived the study and contributed to study design, data verification, and revision and proofreading of the manuscript. All authors have read and approved the final manuscript and, therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines: Shi-chao GENG, Xiao-liang LI, and Wei-huan FANG have declared no conflict of interests.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Allan GM, McNeilly F, Kennedy S, et al. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10(1):3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 2.Bjørkøy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on Huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, et al. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4(7):851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 4.Ding QX, Dimayuga E, Markesbery WR, et al. Proteasome inhibition induces reversible impairments in protein synthesis. FASEB J. 2006;20(8):1055–1063. doi: 10.1096/fj.05-5495com. [DOI] [PubMed] [Google Scholar]
- 5.Dossou AS, Basu A. The emerging roles of mTORC1 in macromanaging autophagy. Cancers (Basel) 2019;11(10):1422. doi: 10.3390/cancers11101422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis J. Porcine circovirus: a historical perspective. Vet Pathol. 2014;51(2):315–327. doi: 10.1177/0300985814521245. [DOI] [PubMed] [Google Scholar]
- 7.Faccini S, Barbieri I, Gilioli A, et al. Detection and genetic characterization of porcine circovirus type 3 in Italy. Transbound Emerg Dis. 2017;64(6):1661–1664. doi: 10.1111/tbed.12714. [DOI] [PubMed] [Google Scholar]
- 8.Franzo G, Legnardi M, Hjulsager CK, et al. Full-genome sequencing of porcine circovirus 3 field strains from Denmark, Italy and Spain demonstrates a high within-Europe genetic heterogeneity. Transbound Emerg Dis. 2018;65(3):602–606. doi: 10.1111/tbed.12836. [DOI] [PubMed] [Google Scholar]
- 9.Gao ZH, Gammoh N, Wong PM, et al. Processing of autophagic protein LC3 by the 20S proteasome. Autophagy. 2010;6(1):126–137. doi: 10.4161/auto.6.1.10928. [DOI] [PubMed] [Google Scholar]
- 10.Han YH, Moon HJ, You BR, et al. The effect of MG132, a proteasome inhibitor on HeLa cells in relation to cell growth, reactive oxygen species and GSH. Oncol Rep. 2009;22(1):215–221. [PubMed] [Google Scholar]
- 11.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 12.Harhouri K, Navarro C, Depetris D, et al. MG132-induced progerin clearance is mediated by autophagy activation and splicing regulation. EMBO Mol Med. 2017;9(9):1294–1313. doi: 10.15252/emmm.201607315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi S, Ohshima Y, Furuya Y, et al. First detection of porcine circovirus type 3 in Japan. J Vet Med Sci. 2018;80(9):1468–1472. doi: 10.1292/jvms.18-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He CC, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43(1):67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He K, Xin YP, Shan Y, et al. Phosphorylation residue T175 in RsbR protein is required for efficient induction of sigma B factor and survival of Listeria monocytogenes under acidic stress. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2019;20(8):660–669. doi: 10.1631/jzus.B1800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernaez B, Cabezas M, Muñoz-Moreno R, et al. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr Mol Med. 2013;13(2):305–316. doi: 10.2174/156652413804810736. [DOI] [PubMed] [Google Scholar]
- 17.Ji MM, Lee JM, Mon H, et al. Proteasome inhibitor MG132 impairs autophagic flux through compromising formation of autophagosomes in Bombyx cells. Biochem Biophys Res Commun. 2016;479(4):690–696. doi: 10.1016/j.bbrc.2016.09.151. [DOI] [PubMed] [Google Scholar]
- 18.Jiang HJ, Wang D, Wang J, et al. Induction of porcine dermatitis and nephropathy syndrome in piglets by infection with porcine circovirus type 3. J Virol. 2019;93(4):e02045–18. doi: 10.1128/jvi.02045-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang HY, Wek RC. Phosphorylation of the α-subunit of the eukaryotic initiation factor-2 (eIF2α) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J Biol Chem. 2005;280(14):14189–14202. doi: 10.1074/jbc.M413660200. [DOI] [PubMed] [Google Scholar]
- 20.Jiang TX, Zou JB, Zhu QQ, et al. SIP/CacyBP promotes autophagy by regulating levels of BRUCE/Apollon, which stimulates LC3-I degradation. Proc Natl Acad Sci USA. 2019;116(27):13404–13413. doi: 10.1073/pnas.1901039116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung CH, Ro SH, Cao J, et al. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kedkovid R, Woonwong Y, Arunorat J, et al. Porcine circovirus type 3 (PCV3) infection in grower pigs from a Thai farm suffering from porcine respiratory disease complex (PRDC) Vet Microbiol. 2018;215:71–76. doi: 10.1016/j.vetmic.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 24.Ku X, Chen F, Li P, et al. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. 2017;64(3):703–708. doi: 10.1111/tbed.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar SH, Rangarajan A. Simian virus 40 small T antigen activates AMPK and triggers autophagy to protect cancer cells from nutrient deprivation. J Virol. 2009;83(17):8565–8574. doi: 10.1128/jvi.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon T, Yoo SJ, Park CK, et al. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet Microbiol. 2017;207:178–180. doi: 10.1016/j.vetmic.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Wang HL, Gu JY, et al. BECN1-dependent CASP2 incomplete autophagy induction by binding to rabies virus phosphoprotein. Autophagy. 2017;13(4):739–753. doi: 10.1080/15548627.2017.1280220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohamud Y, Shi JY, Qu JY, et al. Enteroviral infection inhibits autophagic flux via disruption of the snare complex to enhance viral replication. Cell Rep. 2018;22(12):3292–3303. doi: 10.1016/j.celrep.2018.02.090. [DOI] [PubMed] [Google Scholar]
- 30.Palinski R, Piñeyro P, Shang PC, et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. 2017;91(1):e01879–16. doi: 10.1128/jvi.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 32.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20(3):460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei JJ, Zhao MQ, Ye ZD, et al. Autophagy enhances the replication of classical swine fever virus in vitro. Autophagy. 2014;10(1):93–110. doi: 10.4161/auto.26843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phan TG, Giannitti F, Rossow S, et al. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. 2016;13(1):184. doi: 10.1186/s12985-016-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poole B, Ohkuma S. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J Cell Biol. 1981;90(3):665–669. doi: 10.1083/jcb.90.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian G, Liu DD, Hu JF, et al. Ochratoxin A-induced autophagy in vitro and in vivo promotes porcine circovirus type 2 replication. Cell Death Dis. 2017;8(6):e2909. doi: 10.1038/cddis.2017.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravikumar B, Sarkar S, Davies JE, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90(4):1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar S. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans. 2013;41(5):1103–1130. doi: 10.1042/bst20130134. [DOI] [PubMed] [Google Scholar]
- 39.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan Y, Liu ZQ, Li GW, et al. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-λ production by blocking the nuclear factor-κB nuclear translocation. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2018;19(7):570–580. doi: 10.1631/jzus.B1700283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen HQ, Liu XH, Zhang PF, et al. Porcine circovirus 3 Cap inhibits type I interferon signaling through interaction with STAT2. Virus Res, 275:197804. 2020 doi: 10.1016/j.virusres.2019.197804. [DOI] [PubMed] [Google Scholar]
- 42.Song ZQ, Liao ZJ, Hu YF, et al. Development and optimization of an intergeneric conjugation system and analysis of promoter activity in Streptomyces rimosus M527. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2019;20(11):891–900. doi: 10.1631/jzus.B1900270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadejek T, Woźniak A, Miłek D, et al. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg Dis. 2017;64(5):1350–1353. doi: 10.1111/tbed.12672. [DOI] [PubMed] [Google Scholar]
- 44.Tischer I, Rasch R, Tochtermann G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol Orig A. 1974;226(2):153–167. [PubMed] [Google Scholar]
- 45.Tochetto C, Lima DA, Varela APM, et al. Full-genome sequence of porcine circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transbound Emerg Dis. 2018;65(1):5–9. doi: 10.1111/tbed.12735. [DOI] [PubMed] [Google Scholar]
- 46.Wiegering A, Rüther U, Gerhardt C. The role of primary cilia in the crosstalk between the ubiquitin–proteasome system and autophagy. Cells. 2019;8(3):241. doi: 10.3390/cells8030241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu CM, Wang M, Song ZB, et al. Pseudorabies virus induces autophagy to enhance viral replication in mouse neuro-2a cells in vitro . Virus Res. 2018;248:44–52. doi: 10.1016/j.virusres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Ye XY, Berg M, Fossum C, et al. Detection and genetic characterisation of porcine circovirus 3 from pigs in Sweden. Virus Genes. 2018;54(3):466–469. doi: 10.1007/s11262-018-1553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin HC, Shao SL, Jiang XJ, et al. Interactions between autophagy and DNA viruses. Viruses. 2019;11(9):776. doi: 10.3390/v11090776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuzhakov AG, Raev SA, Alekseev KP, et al. First detection and full genome sequence of porcine circovirus type 3 in Russia. Virus Genes. 2018;54(4):608–611. doi: 10.1007/s11262-018-1582-z. [DOI] [PubMed] [Google Scholar]
- 51.Zhai NH, Liu K, Li H, et al. PCV2 replication promoted by oxidative stress is dependent on the regulation of autophagy on apoptosis. Vet Res. 2019;50(1):19. doi: 10.1186/s13567-019-0637-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng S, Wu X, Zhang L, et al. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound Emerg Dis. 2017;64(5):1337–1341. doi: 10.1111/tbed.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong L, Hu J, Shu W, et al. Epigallocatechin-3-gallate opposes HBV-induced incomplete autophagy by enhancing lysosomal acidification, which is unfavorable for HBV replication. Cell Death Dis, 6:e1770. 2015 doi: 10.1038/cddis.2015.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu BL, Xu F, Li J, et al. Porcine circovirus type 2 explores the autophagic machinery for replication in PK-15 cells. Virus Res. 2012;163(2):476–485. doi: 10.1016/j.virusres.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 55.Zhu BL, Zhou YS, Xu F, et al. Porcine circovirus type 2 induces autophagy via the AMPK/ERK/TSC2/mTOR signaling pathway in PK-15 cells. J Virol. 2012;86(22):12003–12012. doi: 10.1128/jvi.01434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]