Abstract
This study was aimed at investigating the mutations in colorectal cancer (CRC) for recurrent neoantigen identification. A total of 1779 samples with whole exome sequencing (WES) data were obtained from 7 published CRC cohorts. Common HLA genotypes were used to predict the probability of neoantigens at high-frequency mutants in the dataset. Based on the WES data, we not only obtained the most comprehensive CRC mutation landscape so far but also found 1550 mutations which could be identified in at least 5 patients, including KRAS G12D (8%), KRAS G12V (5.8%), PIK3CA E545K (3.5%), PIK3CA H1047R (2.5%), and BMPR2 N583Tfs∗44 (2.8%). These mutations can also be recognized by multiple common HLA molecules in Chinese and TCGA cohort as potential “public” neoantigens. Many of these mutations also have high mutation rates in metastatic pan-cancers, suggesting their value as therapeutic targets in different cancer types. Overall, our analysis provides recurrent neoantigens as potential cancer immunotherapy targets.
1. Introduction
Colorectal cancer (CRC) is the third most common malignancy in the world and the second leading cause of cancer-related mortality [1, 2]. Traditional treatments, such as surgery, chemotherapy, and radiation, have been important in prolonging patients' survival, but for patients with advanced CRC, especially those with metastatic disease, these treatments are limited and often intolerant [3].
In recent years, immunotherapy, including immune checkpoint inhibitors (ICIs), cancer vaccines, and neoantigen-based tumor-infiltrating lymphocytes (TILs), has played an increasingly important role in cancer therapy [4]. Certain CRC patients with high microsatellite instability (MSI-H) could potentially benefit from ICIs treatment [5]. However, not all CRC patients with MSI-H show clinical efficacy in ICI treatment. Neoantigen-based immunotherapy is complementary to ICIs since it has no specific requirement for patient's MSI status nor tumor mutation burden (TMB) [6, 7]. Previous studies on CRC genomics mainly focused on the mechanism of tumor development and metastasis and less involves neoantigen and neoantigen-based immunotherapy [8–13]. By integrating the mutation data of already existing CRC cohorts and combining the common HLA genotypes in these populations [14, 15], our study is expected to find the common neoantigens in CRC patients and facilitate further development of off-the-shelf neoantigen-based immunotherapy.
2. Materials and Methods
2.1. Genomic Data of CRC
This study was approved by the Institutional Review Board on Bioethics and Biosafety of BGI group. All somatic mutations, including single-nucleotide variants (SNVs) and short insertion/deletion (indels), were downloaded from the latest publications (Table 1 and Supplementary Table S1), which represent seven geographically diverse study groups involving 1779 CRC patients. Since all data used in this study were from public databases with informed consent from participants in the original genome study, no additional informed consent was required.
Table 1.
Summary of clinical information of CRC cohort, including patients from seven studies.
| Characteristic | Baylor | Beijing | COCA-CN | Genetech | Harvard | TCGA | Texas | Total |
|---|---|---|---|---|---|---|---|---|
| (n = 110) | (n = 98) | (n = 321) | (n = 74) | (n = 619) | (n = 528) | (n = 29) | (n = 1779) | |
| Age (years) | ||||||||
| <60 | 38 (34.5%) | 48 (49.0%) | 155 (48.3%) | 0 (0%) | 67 (10.8%) | 158 (29.9%) | 0 (0%) | 466 (26.2%) |
| ≥60 | 70 (63.6%) | 50 (51.0%) | 166 (51.7%) | 0 (0%) | 550 (88.9%) | 366 (69.3%) | 0 (0%) | 1202 (67.6%) |
| Unknown | 2 (1.8%) | 0 (0%) | 0 (0%) | 74 (100%) | 2 (0.3%) | 4 (0.8%) | 29 (100%) | 111 (6.2%) |
| Sex | ||||||||
| Female | 65 (59.1%) | 50 (51.0%) | 127 (39.6%) | 0 (0%) | 380 (61.4%) | 253 (47.9%) | 15 (51.7%) | 890 (50.0%) |
| Male | 45 (40.9%) | 48 (49.0%) | 194 (60.4%) | 0 (0%) | 239 (38.6%) | 273 (51.7%) | 14 (48.3%) | 813 (45.7%) |
| Unknown | 0 (0%) | 0 (0%) | 0 (0%) | 74 (100%) | 0 (0%) | 2 (0.4%) | 0 (0%) | 76 (4.3%) |
| Stage | ||||||||
| I | 12 (10.9%) | 10 (10.2%) | 40 (12.5%) | 0 (0%) | 152 (24.6%) | 94 (17.8%) | 0 (0%) | 308 (17.3%) |
| II | 42 (38.2%) | 44 (44.9%) | 94 (29.3%) | 0 (0%) | 187 (30.2%) | 196 (37.1%) | 0 (0%) | 563 (31.6%) |
| III | 48 (43.6%) | 39 (39.8%) | 130 (40.5%) | 0 (0%) | 159 (25.7%) | 150 (28.4%) | 1 (3.4%) | 527 (29.6%) |
| IV | 8 (7.3%) | 4 (4.1%) | 56 (17.4%) | 0 (0%) | 65 (10.5%) | 69 (13.1%) | 28 (96.6%) | 230 (12.9%) |
| Unknown | 0 (0%) | 1 (1.0%) | 1 (0.3%) | 74 (100%) | 56 (9.0%) | 19 (3.6%) | 0 (0%) | 151 (8.5%) |
| MSI status | ||||||||
| MSI-H | 24 (21.8%) | 8 (8.2%) | 0 (0%) | 15 (20.3%) | 91 (14.7%) | 65 (12.3%) | 0 (0%) | 203 (11.4%) |
| MSI-L | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 77 (14.6%) | 0 (0%) | 77 (4.3%) |
| MSS | 81 (73.6%) | 32 (32.7%) | 0 (0%) | 59 (79.7%) | 438 (70.8%) | 346 (65.5%) | 29 (100%) | 985 (55.4%) |
| Unknown | 5 (4.5%) | 58 (59.2%) | 321 (100%) | 0 (0%) | 90 (14.5%) | 40 (7.6%) | 0 (0%) | 514 (28.9%) |
2.2. Pipeline for Neoantigen Prediction
For neoantigen prediction, a total of 43 HLA genotypes were selected with frequencies greater than 5% in the Chinese or TCGA cohort. Mutations present in at least 5 patients, including 476 SNVs and 974 indels, were selected for subsequent neoantigen analysis. A total of five software are used for affinity prediction between neoantigen peptide and HLA alleles, which are NetMHC [16], NetMHCpan [17], PickPocket [18], PSSMHCpan [19], and SMM [20]. Candidate high-affinity peptides were further predicted by EPIC [21]. EPIC is a neoantigen prediction software based on mass spectrometry-derived motifs and tissue-specific expression profiles. It considers various complex factors in antigen presentation process, such as affinity and tumour-specific gene expression, and can accurately predict epitope presentation. If the sample is not quantified for gene expression, the software's default expression value (TPM = 4) is used. According to our previous research [22], neoantigen peptides need to meet the following four criteria: (1) between 8 and 11 meters in length; (2) affinity IC50 < 500 nM in at least two tools; (3) mutant (MT) peptides affinity score lower than the wild type (WT); and (4) the presentation score of EPIC > 0.5.
2.3. Statistical Analysis
The statistical analysis was done in R-studio and the mutation analysis and drawing were done with the maftools package [23]. If no special instructions were given, P < 0.05 was considered significant.
3. Results
3.1. The Integrated Mutation Landscape of CRC Patients
The mutation profiles of all CRC samples are shown in Figure 1 and Figure S1. In general, missense mutation is the main type of mutations. At the base substitution level, C>T is the dominant mutant type, followed by C>A (Figure S2), which is consistent with TCGA and previous reports [24, 25]. The median number of mutations in each sample was 110, and APC, MUC16, TP53, SYNE1, KRAS, and PIK3CA were the most frequently mutated genes. Apart from the samples without MSI status, there are 203 MSI-H samples in the combined cohort (Table 1), accounting for 11.4%. The mutation load of MSI-H samples was higher than that of MSS samples (Figure S3-S4). Interestingly, through the integration analysis of the MSS samples, we found the mutation of four genes, including TENM1, SOX9, PIK3CA, and KRAS, and the mutation of TP53 gene were mutually exclusive (Fisher's exact test, P < 0.05, Figure S5). This different mutation pattern may suggest that the carcinogenic mechanisms are different in CRC patients carrying mutations in these four genes and in those carrying TP53 mutation. Correspondingly, there is no such mutual exclusion effect in the MSI-H samples (Figure S6).
Figure 1.
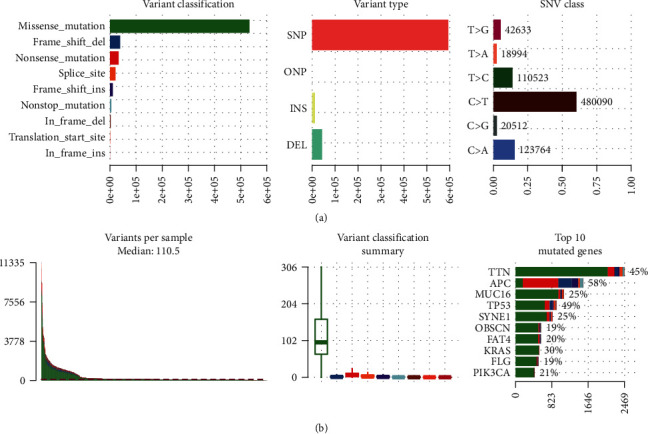
The mutation landscape in the CRC cohort. (a) From left to right, counts of each variant classification, counts of each variant type, and counts of each SNV class. (b) From left to right, variant number per sample, variant classification, and top 10 significantly mutated genes.
There are many hotspot mutations in CRC samples. Of these, 1550 recurrent mutations could be identified in at least 5 patients, with 476 SNVs and 974 indels. Previous studies have shown that common neoantigens in cancers could be used as potential immunotherapy targets [22, 26]. Therefore, in order to find out whether there are common neoantigens in CRC populations, we used these mutations in downstream analyses to predict tumour-specific neoantigens.
3.2. Neoantigens Shared among CRC Patients
Due to the difference in the frequency of HLA in different populations, in order to search for “public” neoantigens in CRC populations, we selected high-frequency HLA in Chinese (HLA frequency > 5% in Han Chinese [15]) and high-frequency HLA in TCGA (HLA frequency > 5% in TCGA [27]) for neoantigen analysis. Finally, a total of 43 HLA alleles were used for neoantigen prediction (Table S2).
We detected 274 SNV-derived neoantigens and 1269 indel-derived neoantigens (Table S3-4). Each SNV usually produces 1-2 high-affinity peptides, while each indel can produce multiple high-affinity peptides. The top ten high-frequency SNV and indel-related neoantigens are shown in Tables 2 and 3, respectively. In terms of SNV, mutations of KRAS, PIK3CA, PCBP1, and CHEK2 can produce 10 neoantigens with the highest frequency (Table 2 and Table S3). In terms of indel, although the mutation frequency is not as high as SNV's, generally one indel can produce about 5-10 neoantigen peptides (Table 3 and Table S4).
Table 2.
Top ten SNVs and the corresponding neoantigens in the CRC cohort.
| Chr | Location | Gene | AA change | Peptide | Frequency | HLA types |
|---|---|---|---|---|---|---|
| chr12 | 25398284 | KRAS | G12D | VVVGADGVGK | 143 | A11:01 |
| chr12 | 25398284 | KRAS | G12V | VVGAVGVGK | 104 | A11:01 |
| chr3 | 1.79E+08 | PIK3CA | E545K | STRDPLSEITK | 63 | A03:01; A11:01 |
| chr3 | 1.79E+08 | PIK3CA | E545K | ITKQEKDFLW | 63 | B57:01 |
| chr3 | 1.79E+08 | PIK3CA | H1047R | ARHGGWTTK | 45 | B27:05 |
| chr3 | 1.79E+08 | PIK3CA | R88Q | REEFFDETRQL | 30 | B40:01 |
| chr2 | 70315174 | PCBP1 | L100Q | RPPVTQRLVV | 28 | B07:02 |
| chr2 | 70315174 | PCBP1 | L100Q | SRPPVTQRL | 28 | C06:02; C07:01; C07:02 |
| chr22 | 29091840 | CHEK2 | K373E | SEILGETSL | 21 | B18:01; B40:01 |
| chr12 | 25398284 | KRAS | G12A | VVVGAAGVGK | 19 | A11:01 |
Table 3.
Top ten indels and the corresponding frequency in the CRC cohort.
| Chr | Location | Gene | AA change | Frequency |
|---|---|---|---|---|
| chr2 | 203420130 | BMPR2 | N583Tfs∗44 | 50 |
| chr10 | 890939 | LARP4B | T163Hfs∗47 | 37 |
| chr1 | 1290110 | MXRA8 | R301Gfs∗107 | 31 |
| chr18 | 34205516 | FHOD3 | S336Vfs∗138 | 29 |
| chr15 | 45003781 | B2M | L15Ffs∗41 | 28 |
| chr12 | 110019240 | MVK | A141Rfs∗18 | 27 |
| chr3 | 168833257 | MECOM | G614Efs∗30 | 26 |
| chr22 | 20130522 | ZDHHC8 | T459Rfs∗177 | 20 |
| chr8 | 103289349 | UBR5 | E2121Kfs∗28 | 20 |
| chr6 | 158508009 | SYNJ2 | P1113Lfs∗5 | 19 |
3.3. Comparison of Neoantigens in Different Subtypes of CRC
By comparing the neoantigen profiles of different subtypes of CRC, we found that there were more SNV- and indel-derived neoantigens in MSI-H patients than in MSS patients (Fisher's exact test, P < 0.01, Figure 2(a)). The 1269 indel-related neoantigens can cover 86.7% of patients with MSI-H CRC, but only 3.9% of MSS patients were covered, indicating that indel is the main source neoantigens of MSI-H CRC. SNV-derived neoantigens can cover 41% of MSS patients and 66% of MSI-H patients.
Figure 2.
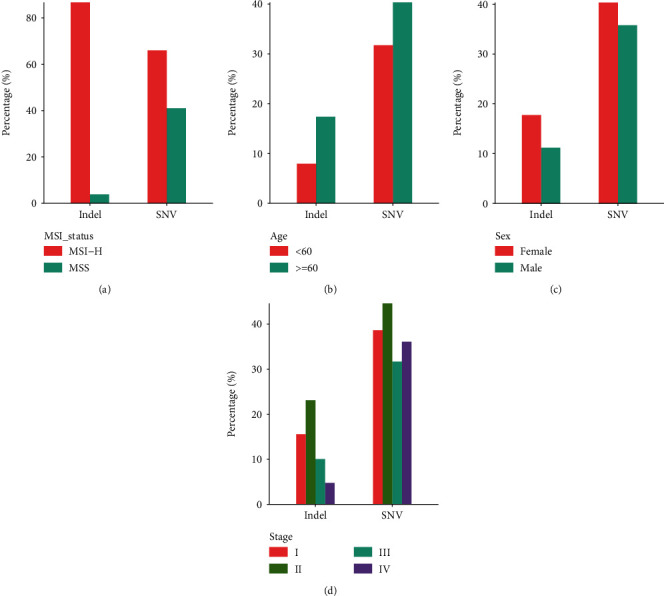
The comparison of neoantigens between different subgroups: (a) between different MSI statuses; (b) between age ≥ 60 and age < 60 groups; (c) between female and male groups; (d) between different stages. These analyses excluded patients with unknown subtypes.
Patients older than 60 carry more neoantigens than those under 60 years of age (Fisher's exact test, P < 0.01, Figure 2(b)). Women tend to carry more neoantigens than men, especially those derived from indels. Female patients accounted for 62.2% in the MSI-H cohort and 51.2% in the MSS cohort. The proportion of MSI-H subtypes was higher in female CRC patients (Fisher's exact test, P value = 0.007); this may partly explain why the results show that women have a higher neoantigen load than men. In terms of cancer stage, Stage II CRC patients carry the most abundant neoantigens, which may be related to the mutation load of the corresponding subgroup (Figures 2(c) and 2(d)). We compared the percentage of patients with MSI-H in Stage II and other stages, finding 28% of patients are MSI-H in Stage II while 10.1% in other stages (Fisher's exact test, P value < 0.001). We also analyzed the mutated rates of KRAS and PIK3CA in Stage II CRC patients. For KRAS, the mutation rate was 28.8% in Stage II while 28.4% in other stages, without significant difference. For PIK3CA, the mutation rate of PIK3CA in Stage II patients was higher than that in other stages, with the mutation rate of 25.6% and 17.1%, respectively (Fisher's exact test, P value < 0.001). According to the above analysis results, the higher neoantigen load of patients at Stage II may be due to the higher proportion of MSI-H or the higher mutation rate of PIK3CA in these patients.
3.4. Hotspot Mutation-Related Neoantigens That May Be a Potential Source of Immunotherapy Target in CRC and Pan-Cancer
To further investigate the potential significance of these high-frequency neoantigens, we focused on mutations with the highest frequency, including KRAS G12D, KRAS G12V, PIK3CA E545K, and PIK3CA H1047R, because these mutations not only produce recurrent neoantigens but also have a higher frequency in the CRC cohort (Figures 3(a) and 3(b)).
Figure 3.
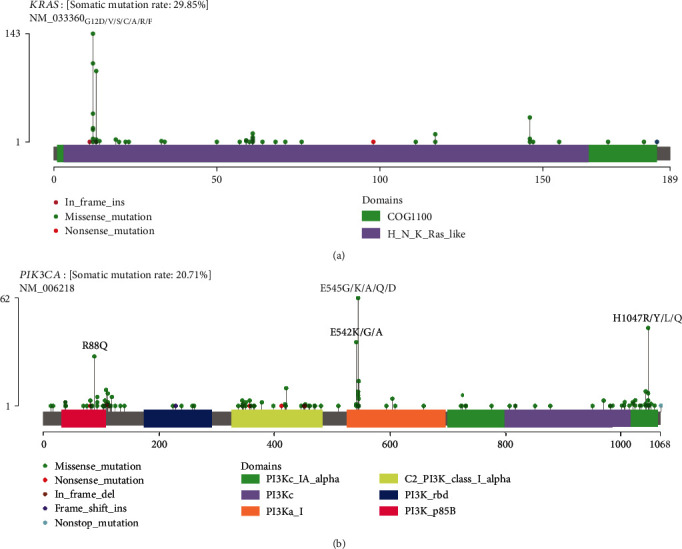
Mutational spectrum of KRAS (a) and PIK3CA (b) in 1179 CRC patients.
KRAS Gly12 (including G12V, G12C, and G12D) is a classic driver mutation that occurs more than 20% in the metastatic pancreatic and appendiceal cancers [24]. Both G12D and G12V are highly mutated in multiple metastatic cancers, including endometrial, CRC, and non-small cell lung cancer (Figures 4(a) and 4(b)). Mayakonda et al. have reported the high frequency of KRAS G12D mutations in pancreatic cancer [23, 25]. Liang et al. also demonstrated by mass spectrometry in 2019 that KRAS G12V-mutated neoantigen can be presented by HLA-A11:01 cell lines [28]. And as far as we know, clinical trials for KRAS G12V mutations in patients with HLA-A11:01 are already under way (NCT03190941).
Figure 4.
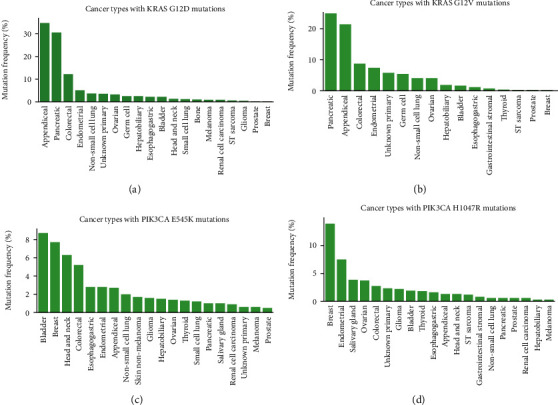
Mutation frequency in MSK-IMPACT cohorts. KRAS G12D (a), KRAS G12V (b), PIK3CA E545K (c), and PIK3CA H1047R (d) in MSK-IMPACT pan-cancer cohorts.
PIK3CA is one of the driver genes in gastrointestinal malignancies [29, 30]. PIK3CA E545K is a hotspot mutation in breast cancer and has corresponding first-line drugs (Alpelisib and Fulvestrant) [31]. In addition to breast cancer, this mutation could be found in more than 5% bladder, head and neck, and colorectal cancer patients (Figure 4(c)). PIK3CA H1047R is most commonly found in breast cancer [32] and is also frequently mutated among multiple tumor types in the MSK-IMPACT metastatic cancers (Figure 4(d)). Our previous work has shown that the epitopes of this mutation can be presented by multiple HLA molecules (e.g., HLA-C07:02, HLA-C 07:01, HLA-A30:01, and HLA-B58:01) and can be a potential neoantigen in patients with gastric cancer [22]. Combined with the results of this study, it is suggested that this mutation can be used as an important therapeutic target for patients with gastrointestinal tumors.
4. Discussion
There are many different types of antigens that can be used as targets for immunotherapy, such as tumor-associated antigens, cancer-testis antigens, and neoantigens [33]. Tumor-associated antigens (e.g., ERBB2/HER2) are highly expressed in tumor and poorly expressed in normal tissues, so they may serve as therapeutic targets for some tumor types, but the disadvantage is that they may cause nonspecific immune responses. Cancer-testis antigens (e.g., NY-ESO-1 antigen [34] and MAGE [35]) are not expressed in normal adult cells except in reproductive tissues (e.g., testis, fetal ovaries, and trophoblast cells). However, both tumor-associated antigens and cancer-testis antigens are prone to severe immune responses. For example, attempts to target melanoma-associated antigen 3 (MAGE-A3) with adoptive cell therapy have resulted in severe neurotoxicity and death, which may be related to the expression in the brain of MAGE-A family members that has not been previously recognized [36]. Compared with the above two antigen types, neoantigen has stronger immunogenicity and tumor specificity, so it is an ideal target for immunotherapy [37, 38]. However, due to the difference of neoantigen among patients, the current neoantigen-based immunotherapy is completely individualized. Since the human exome region is about 30 M, the probability of the same mutation between different individuals is relatively low, and the epitope of the mutation is presented by specific HLA allele, so the possibility of the same neoantigen epitope between different individuals is very low.
However, not all mutations in tumors are random. Current genomic studies have shown that there are many hotspot mutations in driver genes, and the neoantigen epitopes formed by these mutations are potential “public” immunotherapy targets, such as KRAS G12D/V and CDK4 R24C/L [39]. In CRC, the frequency of frameshift mutations is higher in patients with MSI-H subtype, and it has been reported that frameshift peptides have been used in the clinical treatment of CRC (NCT01461148). In our study, we also found that indel-related neoantigens can cover a large proportion of MSI-H CRC patients. At present, 10-20 neoantigen peptides are usually synthesized at the same time to prepare vaccines or other immunotherapy products [40, 41]. If combined with the neoantigens corresponding to hotspot point mutations and frameshift mutations, it is believed that these neoantigen can cover more CRC patients, so that more patients can benefit from neoantigen-based immunotherapy.
5. Conclusion
Based on the analysis of the published WES data of CRC, the most complete mutation landscape of CRC was obtained. We selected HLA subtypes with high frequency in Chinese and TCGA cohort to predict the common neoantigens in the population. The high-frequency mutations, including KRAS G12D (8%), KRAS G12V (5.8%), PIK3CA E545K (3.5%), PIK3CA H1047R (2.5%), and BMPR2 N583Tfs∗44 (2.8%), can be recognized and presented by many HLA genotypes, such as HLA-A1101, HLA-A03:01, and HLA-B57:01. These HLA genotypes are the main HLA subtypes in Chinese and Americans, indicating the broad spectrum of the neoantigens we identified. In conclusion, we have found a series of “public” neoantigens for CRC, which provide important resources for immunotherapy of CRC in the future.
Acknowledgments
We thank professor Xiuqing Zhang for her guidance and help in this project. This work is supported by the National Natural Science Foundation of China (No. 81702826), the Shenzhen Municipal Government of China (No. 20170731162715261), and the Science, Technology and Innovation Commission of Shenzhen Municipality (grant No. JCYJ20170817145845968).
Contributor Information
Chao Chen, Email: 409985846@qq.com.
Bo Li, Email: libo@genomics.cn.
Data Availability
Data supporting the results of this study are available from corresponding authors on request.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
Figure S1: the landscape of 25 significantly mutated genes in all the CRC patients. Figure S2: ratio of base conversion and transversion in mutations. Figure S3: the landscape of 25 significantly mutated genes in MSI-H CRC patients. Figure S4: the landscape of 25 significantly mutated genes in MSS CRC patients. Figure S5: somatic interaction analysis among the significantly mutated genes found four genes mutually exclusive with TP53 in MSS samples. Figure S6: somatic interaction analysis in MSI-H samples. Table S1: the list of clinical information of samples in previous seven studies of CRC. Table S2: the list of high-frequency HLA genotypes in Chinese and TCGA. Table S3: the list of SNV derived neoantigens. Table S4: the list of indel-derived neoantigens.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M., Sierra M. S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Hu Z., Ding J., Ma Z., et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nature Genetics. 2019;51(7):1113–1122. doi: 10.1038/s41588-019-0423-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klebanoff C. A., Rosenberg S. A., Restifo N. P. Prospects for gene-engineered T cell immunotherapy for solid cancers. Nature Medicine. 2016;22(1):26–36. doi: 10.1038/nm.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley J. C., Lin M. T., Le D. T., Eshleman J. R. Microsatellite instability as a biomarker for PD-1 blockade. Clinical Cancer Research : an official journal of the American Association for Cancer Research. 2016;22(4):813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 6.Malekzadeh P., Pasetto A., Robbins P. F., et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. Journal of Clinical Investigation. 2019;129(3):1109–1114. doi: 10.1172/JCI123791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren L., Leisegang M., Deng B., et al. Identification of neoantigen-specific T cells and their targets: implications for immunotherapy of head and neck squamous cell carcinoma. Oncoimmunology. 2019;8(4, article e1568813) doi: 10.1080/2162402X.2019.1568813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giannakis M., Mu X. J., Shukla S. A., et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Reports. 2016;15(4):857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guda K., Veigl M. L., Varadan V., et al. Novel recurrently mutated genes in African American colon cancers. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(4):1149–1154. doi: 10.1073/pnas.1417064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seshagiri S., Stawiski E. W., Durinck S., et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasaikar S., Huang C., Wang X., et al. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell. 2019;177(4):1035–1049.e19. doi: 10.1016/j.cell.2019.03.030. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z., Yang C., Li X., et al. The landscape of somatic mutation in sporadic Chinese colorectal cancer. Oncotarget. 2018;9(44):27412–27422. doi: 10.18632/oncotarget.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.dos Santos E. J. M., McCabe A., Gonzalez-Galarza F. F., Jones A. R., Middleton D. Allele frequencies net database: improvements for storage of individual genotypes and analysis of existing data. Human Immunology. 2016;77(3):238–248. doi: 10.1016/j.humimm.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Zhou F., Cao H., Zuo X., et al. Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nature Genetics. 2016;48(7):740–746. doi: 10.1038/ng.3576. [DOI] [PubMed] [Google Scholar]
- 16.Lundegaard C., Lamberth K., Harndahl M., Buus S., Lund O., Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Research. 2008;36(suppl_2):W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoof I., Peters B., Sidney J., et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61(1):1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H., Lund O., Nielsen M. The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics. 2009;25(10):1293–1299. doi: 10.1093/bioinformatics/btp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu G., Li D., Li Z., et al. PSSMHCpan: a novel PSSM-based software for predicting class I peptide-HLA binding affinity. GigaScience. 2017;6(5):1–11. doi: 10.1093/gigascience/gix017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters B., Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6(1):p. 132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu W., Qiu S., Li Y., et al. EPIC: MHC-I epitope prediction integrating mass spectrometry derived motifs and tissue-specific expression profiles. Biorxiv; 2019. [Google Scholar]
- 22.Chen C., Zhou Q., Wu R., et al. A comprehensive survey of genomic alterations in gastric cancer reveals recurrent neoantigens as potential therapeutic targets. BioMed Research International. 2019;2019:10. doi: 10.1155/2019/2183510.2183510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayakonda A., Lin D. C., Assenov Y., Plass C., Koeffler H. P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Research. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., Yuen S. T., Xu J., et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nature Genetics. 2014;46(6):573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 25.Zang Z. J., Cutcutache I., Poon S. L., et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nature Genetics. 2012;44(5):570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 26.Zhou J., Zhao W., Wu J., et al. Neoantigens derived from recurrently mutated genes as potential immunotherapy targets for gastric cancer. BioMed Research International. 2019;2019:11. doi: 10.1155/2019/8103142.8103142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charoentong P., Finotello F., Angelova M., et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Reports. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Liang Z., Qin L., Chen L., et al. The common neoantigens in colorectal cancer are predicted and validated to be presented or immunogenic. bioRxiv; 2019. [Google Scholar]
- 29.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C., Shi C., Huang X., et al. Molecular profiles and metastasis markers in Chinese patients with gastric carcinoma. Scientific Reports. 2019;9(1):p. 13995. doi: 10.1038/s41598-019-50171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juric D., Janku F., Rodón J., et al. Alpelisib plus Fulvestrant inPIK3CA-Altered andPIK3CA-Wild-Type estrogen receptor-positive advanced breast Cancer. JAMA Oncology. 2019;5(2, article e184475) doi: 10.1001/jamaoncol.2018.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engelman J. A., Chen L., Tan X., et al. Effective use of PI3K and MEK inhibitors to treat mutant _Kras_ G12D and _PIK3CA_ H1047R murine lung cancers. Nature Medicine. 2008;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarchoan M., Johnson B. A., III, Lutz E. R., Laheru D. A., Jaffee E. M. Targeting neoantigens to augment antitumour immunity. Nature Reviews Cancer. 2017;17(4):209–222. doi: 10.1038/nrc.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gnjatic S., Nishikawa H., Jungbluth A. A., et al. NY‐ESO‐1: review of an immunogenic tumor antigen. Advances in Cancer Research. 2006;95:1–30. doi: 10.1016/S0065-230X(06)95001-5. [DOI] [PubMed] [Google Scholar]
- 35.Chomez P., De Backer O., Bertrand M., De Plaen E., Boon T., Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Research. 2001;61(14):5544–5551. [PubMed] [Google Scholar]
- 36.Morgan R. A., Chinnasamy N., Abate-Daga D., et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. Journal of Immunotherapy. 2013;36(2):133–151. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richters M. M., Xia H., Campbell K. M., Gillanders W. E., Griffith O. L., Griffith M. Best practices for bioinformatic characterization of neoantigens for clinical utility. Genome Medicine. 2019;11(1):p. 56. doi: 10.1186/s13073-019-0666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maby P., Galon J., Latouche J. B. Frameshift mutations, neoantigens and tumor-specific CD8(+) T cells in microsatellite unstable colorectal cancers. Oncoimmunology. 2016;5(5, article e1115943) doi: 10.1080/2162402X.2015.1115943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klebanoff C. A., Wolchok J. D. Shared cancer neoantigens: making private matters public. Journal of Experimental Medicine. 2018;215(1):5–7. doi: 10.1084/jem.20172188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keskin D. B., Anandappa A. J., Sun J., et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565(7738):234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumacher T. N., Schreiber R. D. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: the landscape of 25 significantly mutated genes in all the CRC patients. Figure S2: ratio of base conversion and transversion in mutations. Figure S3: the landscape of 25 significantly mutated genes in MSI-H CRC patients. Figure S4: the landscape of 25 significantly mutated genes in MSS CRC patients. Figure S5: somatic interaction analysis among the significantly mutated genes found four genes mutually exclusive with TP53 in MSS samples. Figure S6: somatic interaction analysis in MSI-H samples. Table S1: the list of clinical information of samples in previous seven studies of CRC. Table S2: the list of high-frequency HLA genotypes in Chinese and TCGA. Table S3: the list of SNV derived neoantigens. Table S4: the list of indel-derived neoantigens.
Data Availability Statement
Data supporting the results of this study are available from corresponding authors on request.


