Abstract
Pachytene piRNAs, which comprise >80% of small RNAs in the adult mouse testis, have been proposed to bind and regulate target RNAs like miRNAs, cleave targets like siRNAs, or lack biological function altogether. Although piRNA pathway protein mutants are male sterile, no biological function has been identified for any mammalian piRNA-producing locus. Here, we report that males lacking piRNAs from a conserved mouse pachytene piRNA locus on chromosome 6 (pi6) produce sperm with defects in capacitation and egg fertilization. Moreover, heterozygous embryos sired by pi6−/− fathers show reduced viability in utero. Molecular analyses suggest that pi6 piRNAs repress gene expression by cleaving mRNAs encoding proteins required for sperm function, pi6 also participates in a network of piRNA-piRNA precursor interactions that initiate piRNA production from a second piRNA locus on chromosome 10 as well as pi6 itself. Our data establish a direct role for pachytene piRNAs in spermiogenesis and embryo viability.
Introduction
Only animals produce PIWI-interacting RNAs (piRNAs), 21–35-nt RNAs that form the most abundant small RNA class in the germline. piRNAs protect the germline genome from transposons and repetitive sequences, and, in some arthropods, fight viruses and transposons in somatic tissues1–5. The mammalian male germline makes three classes of piRNAs: (1) 26–28 nt transposon-silencing piRNAs predominate in the fetal testis1; (2) shortly after birth, 26–27 nt piRNAs derived from mRNA 3’ untranslated regions (UTRs) emerge6; and (3) at the pachytene stage of meiosis, ~30 nt, non-repetitive pachytene piRNAs appear. Pachytene piRNAs accumulate to comprise >80% of all small RNAs in the adult mouse testis, and they continue to be made throughout the male mouse reproductive lifespan. These piRNAs contain fewer transposon sequences than the genome as a whole7, and most pachytene piRNAs map only to the loci from which they are produced. The diversity of pachytene piRNAs is unparalleled in development, with >1 million distinct species routinely detected in spermatocytes or spermatids. Intriguingly, the sequences of pachytene piRNAs are not themselves conserved, but piRNA-producing loci have been maintained at the syntenic regions across eutherian mammals8,9, suggesting that the vast sequence diversity of pachytene piRNAs is itself biologically meaningful.
One-hundred mouse pachytene piRNA-producing loci have been annotated9–13. All are coordinately regulated by the transcription factor A-MYB (MYBL1), which also promotes expression of proteins that convert piRNA precursor transcripts into mature piRNAs, as well as proteins required for cell cycle progression and meiosis14. Of the 100 piRNA-producing loci, 15 pairs of pachytene piRNA-producing genes are divergently transcribed from bidirectional, A-MYB-binding promoters10. The contribution of pachytene piRNAs from each piRNA-producing locus is unequal, with just five loci located on five different chromosomes—pi2, pi6, pi7, pi9, and pi17—contributing >50% of all pachytene piRNAs at 17 days postpartum (dpp).
Loss of proteins required to make pachytene piRNAs, including the pachytene piRNA-binding protein, MIWI (PIWIL1), invariably arrests spermatogenesis without producing sperm, rendering males sterile10,15–19. Yet loss of the chromosome 17 pachytene piRNA-producing locus, 17-qA3.3-27363(−),26735(+) (henceforth, pi17), has no detectable phenotype or impact on male fertility20, even though pi17 produces ~16% of all pachytene piRNAs in pachytene spermatocytes. Similarly, mice with disrupted expression of a chromosome 2 piRNA locus are viable and fertile (P.-H.W., K.C., and P.D.Z, unpublished; and 21). Consequently, the function of pachytene piRNAs in mice is actively debated. One model proposes that pachytene piRNAs regulate meiotic progression of spermatocytes by cleaving mRNAs during meiosis22,23. Another posits that pachytene piRNAs direct degradation of specific mRNAs via a miRNA-like mechanism involving mRNA deadenylation24. A third model proposes that MIWI functions without piRNAs, and that piRNAs are byproducts without a critical function25. Compelling evidence supports each model.
In fact, direct demonstration of piRNA function in any animal has proven elusive. Only two piRNA-producing loci have been directly shown to have a biological function; both were identified genetically before the discovery of piRNAs and are found only in members of the melanogaster subgroup of flies8,26–31,31–33. In male D. melanogaster, piRNAs from Suppressor of Stellate, a multi-copy gene on the Y chromosome, silence the X-linked selfish gene Stellate; deletion of Suppressor of Stellate leads to Stellate protein crystals in spermatocytes34,35. In female flies, deletion of the piRNA-producing flamenco gene, which is expressed in somatic follicle cells that support oogenesis, leads to gypsy family transposon activation and female infertility36,37.
Here, we report that a promoter deletion in the chromosome 6 pachytene piRNA locus 6-qF3-28913(−),8009(+) (chr6:127,776,075–127,841,890, mm10; henceforth, pi6) disrupts male fertility. The pi6 locus generates 5.8% of pachytene piRNAs in the adult testis and is conserved among eutherian mammals. Mice lacking pi6-derived piRNAs produce normal numbers of sperm and continue to repress transposons. However, pi6 mutant sperm show defective capacitation and fail to penetrate the zona pellucida, a glycoprotein layer surrounding the egg. Consistent with these phenotypes, spermatids from pi6 mutant males show increased steady-state abundance of mRNAs encoding proteins involved in sperm acrosome function and penetration of the oocyte zona pellucida. In addition to decreasing specific mRNA abundance, pi6 piRNAs concurrently facilitate biogenesis of piRNAs from other loci. Our findings provide direct evidence for a biological function for pachytene piRNAs in male mouse fertility, and pi6 promoter deletions provide a new model for future studies of piRNA biogenesis and function.
Results
pi6 Promoter Deletion Eliminates pi6 Pachytene piRNAs
To eliminate production of pi6 pachytene piRNAs while minimizing the impact on adjacent genes, we used Cas9 and a pair of single-guide RNAs (sgRNAs) to delete a 227 bp sequence that encompasses the A-MYB-binding site and promoter (Ref. 10; Fig. 1, Extended Data Fig. 1a and 1b, and Supplementary Table 1). To test that the phenotype of pi6em1/em1 male mice reflects loss of the pi6 promoter—and not an off-target mutation elsewhere in the genome—we used a second pair of sgRNAs to generate a 117 bp pi6 promoter deletion, pi6em2 (Fig. 1, Extended Data Fig. 1a and 1c, and Supplementary Table 1). For comparison, we created an analogous 583 bp promoter deletion in pi17. We established stable pi6em1 mutant lines (pi6em1-1, -2, and -3 in Extended Data Fig. 1a and 1b) from three founders whose pi6 promoter deletion sizes range from 219 to 230 bp and differ at their deletion boundaries, reflecting imprecise DNA repair after Cas9 cleavage. All three deletions eliminated pi6 primary transcripts and mature pachytene piRNAs from both arms of the locus (Fig. 1). Because these lines were created using the same pair of sgRNA guides, we refer to all as the pi6em1 allele. Similarly, we refer to the stable mutant lines generated from mutant founders carrying pi6em2 (pi6em2-1 and -2 in Extended Data Fig. 1a and 1c) or pi17−/− (pi17−/−-1 and -2 in Extended Data Fig. 1a and 1d) deletions as pi6em2 and pi17−/− alleles, respectively.
Figure 1. pi6em1/em1, pi6em2/em2, and pi17−/− promoter deletion in mice.
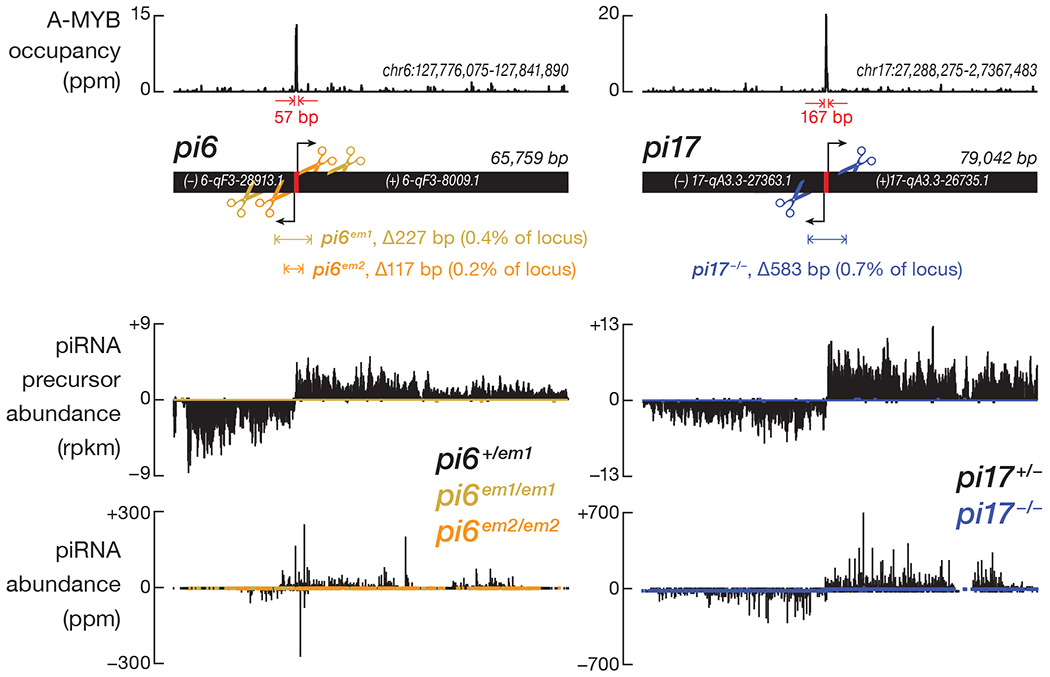
Scissors indicate sites targeted by sgRNAs used to guide the Cas9-catalyzed promoter deletions. RNA-seq was used to measure the steady-state abundance of piRNA primary transcripts, and sequencing of NaIO4 oxidation-resistant small RNA was used to measure the abundance of mature piRNAs in 17.5 dpp testes.
See also Extended Data Figure 1 and Supplementary Table 1.
pi6 is Required for Male Mouse Fertility
When paired with C57BL/6 females, 2–8 month-old pi6em1/em1 males produced fewer pups compared to their littermates, even at peak reproductive age (Fig. 2a and Extended Data Fig. 2a). In six months, C57BL/6 males produced 7 ± 1 (n = 5) litters, while pi6em1/em1 males produced 2 ± 2 (n = 6) litters. The significantly smaller number of progeny produced by pi6em1/em1 males over their reproductive lifetime reflects two abnormal aspects of their fertility (Fig. 2a and 2b; Supplementary Note). First, 29% of pi6em1/em1 males never produced pups. Second, the mutants that did sire pups did so less frequently. In contrast, males and females carrying a ~583-bp promoter deletion in pi17 were fully fertile, as observed previously for an independent, partial-loss-of-function pi17 promoter deletion20, despite loss of primary transcripts and mature piRNAs from both arms of the pi17 locus (Fig. 1).
Figure 2. Reduced fertility in pi6em1/em1 males by natural mating.
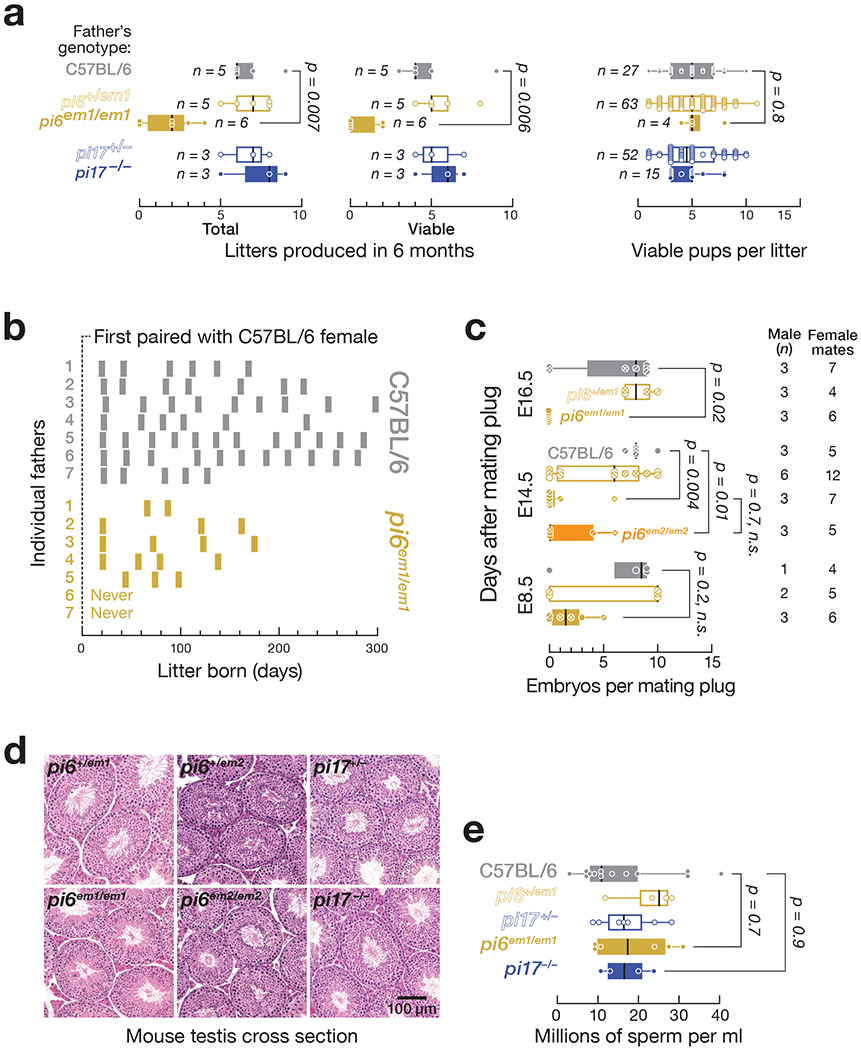
(a) Number of litters and pups per litter produced by male mice between 2–8 months of age. (b) Frequency and periodicity of litter production. Each bar represents a litter. (c) Number of embryos produced by males mated with C57BL/6 females. (d) Testis morphology analyzed by hematoxylin and eosin staining. (e) Concentration of sperm from the caudal epididymis of C57BL/6 (n = 13), pi6+/em1 (n = 4), pi6em1/em1 (n = 6), pi17+/− (n = 7), and pi17−/− (n = 4) males. In (a), (c), and (e), vertical black lines denote median; boxes indicate 75th and 25th percentiles; whiskers report the maximal and minimal values. Each dot represents an individual male.
See also Extended Data Figure 2.
Like pi6em1/em1 male mice, pi6em2/em2 males produced neither primary pi6 transcripts nor mature pi6 piRNAs and showed reduced fertility (Fig. 1 and Extended Data Fig. 2a). We conclude that pi6 piRNAs are required for male fertility in C57BL/6 mice.
pi6 Mutant Males Produce Fewer Embryos
pi6 mutant male matings produced fewer fully developed embryos. We examined the embryos produced by natural mating of C57BL/6 females housed with C57BL/6, pi6+/em1, or pi6em1/em1 males at 8.5, 14.5, or 16.5 days after occurrence of a mating plug. At 8.5 days after mating, C57BL/6 females housed with pi6em1/em1 males carried fewer embryos (2 ± 2, n = 3) compared to females paired with pi6+/em1 (6 ± 5, n = 2) or C57BL/6 control (7 ± 4, n = 1) males (Fig. 2c). At 14.5 and 16.5 days post-mating, female mice paired with pi6em1/em1 males had even fewer embryos. Female mice paired with pi6em2/em2 males similarly had fewer embryos at 14.5 days after mating (2 ± 3, n = 3). Naturally-born pups sired by pi6em1/em1 and pi6em2/em2 males were rare but healthy with no obvious abnormalities (Extended Data Fig. 2b and Supplementary Note).
pi6 Mutant Sperm Fail to Fertilize Wild-type Eggs
pi6emi/emi and pi6em2/em2 adult testes had normal gross histology (Fig. 2d, Extended Data Fig. 2b and Supplementary Note). The quantity of caudal epididymal sperm produced by pi6em1/em1 mice (19 ± 10 million sperm per ml; n = 6) was also comparable to that of their pi6+/em1 (23 ± 7 million sperm/ml; n = 4) or C57BL/6 (20 ± 10 million sperm per ml; n = 13) littermates (Fig. 2e and Extended Data Fig. 2c–e; Supplementary Note).
Because pi6−/− males are ineffectual at siring offspring, we used in vitro fertilization (IVF) to distinguish between defects in mating behavior and sperm function, incubating sperm from C57BL/6, pi6+/em1, or pi6em1/em1 males with wild-type oocytes and scoring for the presence of both male and female pronuclei and the subsequent development of the resulting bi-pronuclear zygotes into two-cell embryos 24 h later (Fig. 3a). The majority of oocytes incubated with sperm from C57BL/6 (86 ± 17%; 774 total oocytes; n = 6) or pi6+/em1 (60 ± 35%; 412 total oocytes; n = 3) males developed into two-cell embryos. By contrast, only 7 ± 5% (12-fold decrease compared to C57BL/6; Cohen’s d = 6.3; 1,026 total oocytes; n = 7) of oocytes incubated with pi6em1/em1 sperm reached the two-cell stage. Similarly, no oocytes incubated with pi6em2/em2 sperm developed into two-cell embryos by 24 h. The majority of these oocytes remained undivided, and few contained a male pronucleus, suggesting that pi6em1/em1 and pi6em2/em2 Sperm are defective in fertilization.
Figure 3. Fertilization defects of pi6em1/em1 and pi6em2/em2 sperm revealed by IVF and ICSI.
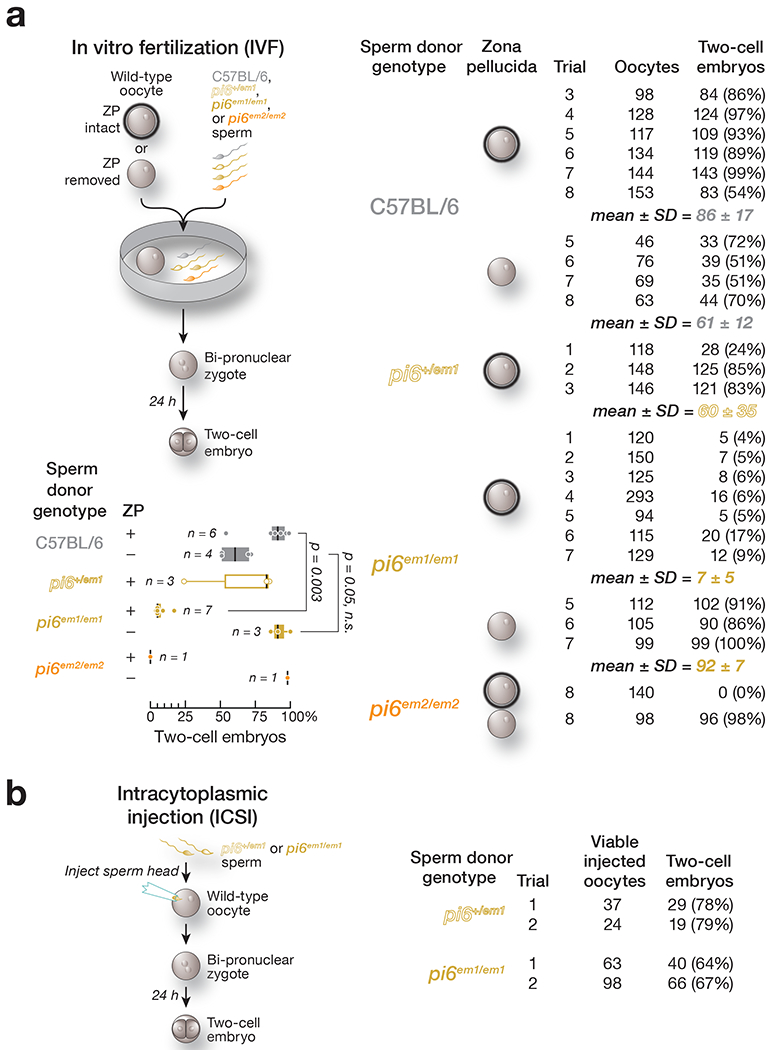
(a) Sperm function analyzed by in vitro fertilization (IVF) using oocytes with or without zona pellucida. Vertical black lines denote median; boxes indicate 75th and 25th percentiles; whiskers report the maximal and minimal values. Each dot represents the IVF result using sperm from an individual male. (b) Sperm function analyzed by intracytoplasmic sperm injection (ICSI).
See also Extended Data Figure 3
pi6em1/em1 Sperm Nuclei Support Fertilization
The best studied piRNA function is transposon silencing, and mouse pi2 has been proposed to be involved in LINE1 element silencing, although pi2 mutant males are fertile21. Moreover, LINE1 transcript abundance increases in mice bearing inactivating mutations in the catalytic site of MIWI16. Transposon activation can produce DNA damage, and genomic integrity is critical for fertilization38–41. However, pachytene piRNAs are depleted of repetitive sequences in contrast to other types of piRNA-producing genomic loci (Extended Data Fig. 3a)7,9,42.
We asked whether the defect in fertilization by pi6 mutant sperm might reflect DNA damage or epigenetic dysregulation of the sperm genome. pi6+/em1 or pi6em1/em1 sperm heads were individually injected into the cytoplasm of wild-type oocytes (intracytoplasmic sperm injection, ICSI; Fig. 3b), bypassing the requirement for sperm motility, acrosome reaction, egg binding, or sperm-egg membrane fusion43. pi6em1/em1 sperm heads delivered by ICSI fertilized the oocyte at a rate similar to that of pi6+/em1 sperm: 66% of oocytes (161 total viable oocytes from two separate trials) injected with homozygous mutant pi6em1/em1 sperm heads reached the two-cell stage, compared to 79% for pi6+/em1 (61 total viable oocytes from two separate trials). Thus, most pi6em1/em1 nuclei are capable of fertilization. We found no DNA damage or increased transposon expression in pi6em1/em1 and pi6em2/em2 spermatogenic cells, further evidence that indicates pi6 is not important for transposon silencing (Extended Data Fig. 3b and 3c; Supplementary Note).
pi6 Mutant Sperm Struggle to Penetrate the Zona Pellucida
Mammalian spermatozoa stored in the epididymis are immotile and dormant. Sperm capacitate, i.e., resume maturation, only upon entering the female reproductive tract44. Upon capacitation, sperm become capable of undergoing the acrosome reaction, which is required to bind and penetrate the outer oocyte glycoprotein layer, the zona pellucida44–46. To test whether the defect in fertilization by pi6 mutant sperm reflects impaired binding to or penetration of the zona pellucida, we compared IVF using unmanipulated oocytes to oocytes with the zona pellucida removed (Fig. 3a). Strikingly, removing the zona pellucida fully rescued the fertilization rate of pi6em1/em1 sperm: 92 ± 7% (316 total oocytes; n = 3) of zona pellucida-free oocytes incubated with pi6em1/em1 sperm reached the two-cell stage after 24 h, compared to 7 ± 5% for intact zona pellucida (1,026 total oocytes; n = 3). Similarly, 98% of zona pellucida-free oocytes incubated with pi6em2/em2 sperm developed into two-cell embryos after 24 h (98 total oocytes; n =1), in contrast to 0% of those with intact zona pellucida (140 total oocytes; n = 1).
Impaired Capacitation in pi6 Mutant Sperm
One hallmark of sperm capacitation is a switch to “hyperactivated motility,” a swimming pattern characterized by a high amplitude and non-symmetric beating of the flagellum that facilitates penetration of the zona pellucida47–50. To assess pi6 mutant sperm capacitation, we measured the motility of freshly extracted caudal epididymal sperm from pi6em1/em1, pi6em2/em2, or C57BL/6 mice using computer-assisted sperm analysis (CASA51; Fig. 4a). After 90 min incubation under capacitation-promoting conditions, pi6sm1/em1 and pi6em/em2 sperm populations had reduced path and progressive velocity, measures of sperm motility, compared to control sperm (Fig. 4b and 4c; Supplementary Movies 1–10; Supplementary Note).
Figure 4. Impaired sperm capacitation in pi6 mutant sperm.
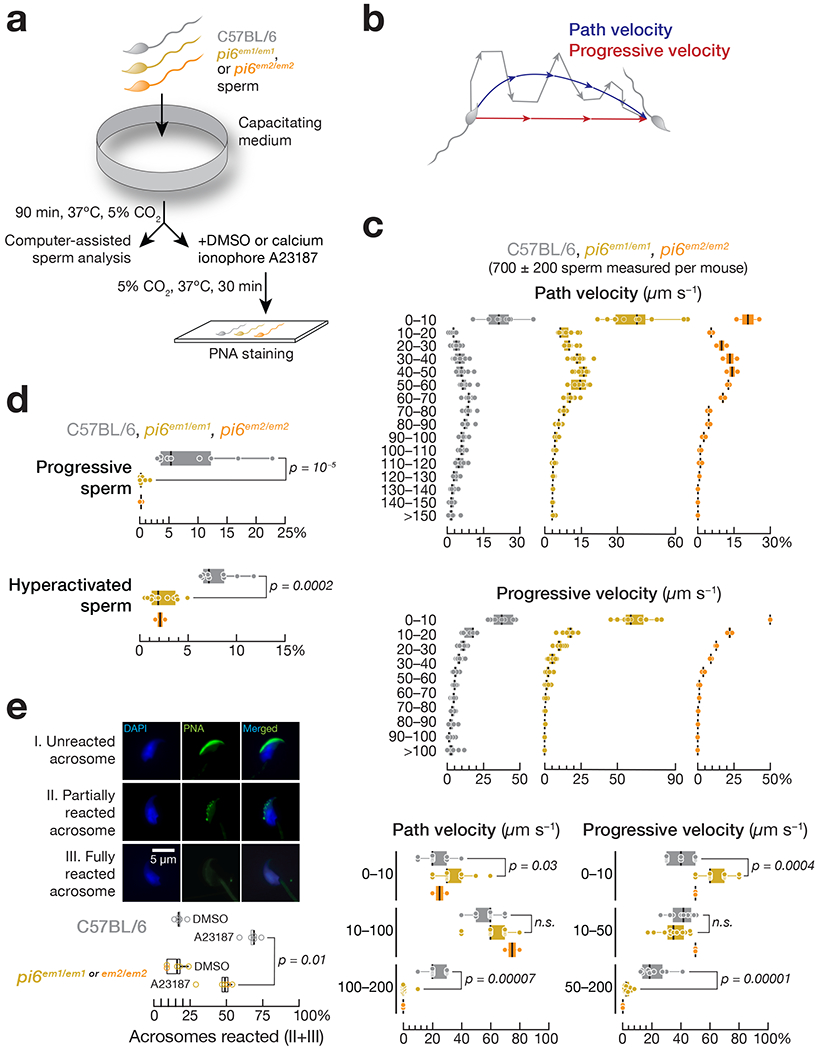
(a) Strategy to measure sperm motility and acrosome reaction triggered with Ca2+ ionophore A23187. (b) Definition of path and progressive velocities. (c) Distribution of path and progressive velocities for sperm from C57BL/6 (n = 9), pi6em1/em1 (n = 11), and pi6em2/em2 (n = 2). Top: 10 μm/sec bins; bottom: bins correspond to immotile or slow, intermediate, and vigorous motility. (d) Distribution of progressive and hyperactivated sperm from C57BL/6 (n = 9), pi6em1/em1 (n = 11), and pi6em2/em2 (n = 2) mice determined by CASAnova. (e) (Top panel) Acrosome status of representative wild-type caudal epididymal spermatozoa. Green, peanut agglutinin to detect the acrosome; blue, DAPI to detect DNA. (Bottom panel) Acrosome reaction rates for C57BL/6 (n = 5) and pi6 mutant (n = 5) sperm. The results using pi6em1/em1 and pi6em2/em2 sperm for acrosome reaction were combined as indicated. In (c), (d), and (e), vertical lines denote median; boxes indicate 75th and 25th percentiles; whiskers report the maximal and minimal values. Each dot represents an individual male.
See also Supplementary Movies 1–10.
To more rigorously evaluate progressive motility and hyperactivation, we used CASAnova, an unsupervised machine learning tool52, to analyze caudal epididymal sperm from pi6em1/em1, pi6em2/em2, and C57BL/6 control mice. After 90 min in capacitating conditions, CASAnova identified just 0.3 ± 0.5% of pi6em1/em1 (n = 11) and 0.2 ± 0.3% of pi6sm2/em2 (n = 2) sperm as progressive, compared to 9 ± 7% for C57BL/6 (n = 9; Fig. 4d). Similarly, only 2 ± 1% of pi6em1/em1 or pi6sm2/em2 sperm displayed hyperactivated motility, compared to 8 ± 2% for the control (Fig. 4d), a percentage typical for the C57BL/6 mouse strain52.
Acrosome reaction in sperm can be visualized and measured ex vivo (Fig. 4a and 4e). While the spontaneous acrosome reaction rates for C57BL/6 (18 ± 3%; n = 5) and pi6 mutant sperm were similar (15 ± 6%; n = 5), acrosome reaction triggered by ionophore-induced Ca2+ influx (i.e., ionophore-induced minus spontaneous) differed between the two genotypes: 46 ± 10% (31 ± 12%) of pi6 mutant sperm (n = 5) underwent partial or complete reaction, compared to 68 ± 6% (50 ± 7; n = 5) for C57BL/6 (p = 0.01; Cohen’s d = 2.78; Fig. 4e). Our data suggest that pi6 mutant sperm less effectively undergo an acrosome reaction triggered by ionophore-induced Ca2+ influx, a defect expected to impair binding and penetrating the zona pellucida. Together, our data indicate that insufficient capacitation is responsible for the poor fertilization capability of pi6 mutant sperm.
Potential Role of Paternal pi6 piRNAs in Embryo Development
Even when pi6 mutant sperm successfully fertilize an oocyte, the resulting heterozygous embryos are less likely to complete gestation. We monitored pre-implantation development in vitro for up to 96 h, a period during which the one-cell embryo develops into a blastocyst. Of the oocytes incubated with pi6em1/em1 sperm, 40% remained undivided without evidence of a male pronucleus, presumably because they were not fertilized. Among the remaining 60% oocytes that progressed to at least the two-cell stage, indicating successful fertilization by pi6em1/em1 sperm, 82% showed delayed development, requiring 48 h to reach the two-cell stage. None of these developed further. Just 3% of oocytes fertilized by pi6em1/em1 sperm progressed to the blastocyst stage by 96 h, compared to 98% for C57BL/6 sperm (Fig. 5a).
Figure 5. Embryos derived from pi6em1/em1 sperm fail to develop.
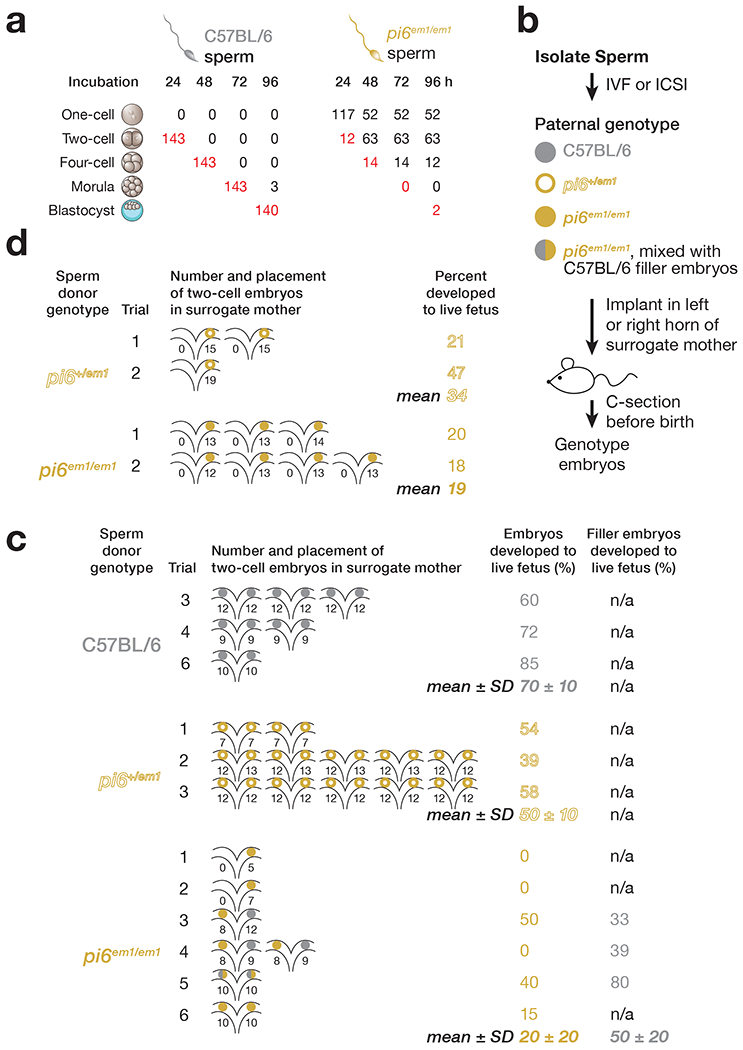
(a) Development of IVF-derived embryos. Red, number of embryos that developed to the stage appropriate for the elapsed time after fertilization. (b) Strategy for surgical transfer of fertilized two-cell embryos to surrogate mothers. (c) Percentages of IVF-derived two-cell embryos using sperm from C57BL/6 (n = 3), pi6+/em1 (n = 3), or pi6em1/em1 (n = 6) mice that developed to term. Each uterine cartoon represents one surrogate mother; colored circles depict embryos. The number of embryos transferred to each side of the oviduct is indicated. (d) Percentages of ICSI-derived two-cell embryos using sperm from pi6+/em1 (n = 2) or pi6em1/em1 (n = 2) mice that developed to term.
See also Extended Data Figure 4
Two-cell embryos generated by IVF using sperm from pi6+/em1, pi6em1/em1, or C57BL/6 control mice were transferred to wild-type surrogate mothers (Fig. 5b). Most embryos from pi6+/em1 (50 ± 10%; 23 ± 4 embryos per female; n = 3) or C57BL/6 control sperm (70 ± 10%; 21 ± 3 embryos per female; n = 3) developed to term (Fig. 5c and Extended Data Fig. 4a; Supplementary Note), a rate typical for this genetic background53. By contrast, only 20 ± 20% of two-cell embryos from pi6em1/em1 sperm developed to term (n = 6). Likewise, fewer ICSI-derived pi6+/em1 embryos developed to term in the surrogate females (Fig. 5d and Extended Data Fig. 4b; Supplementary Note). We conclude that paternal pi6 piRNAs play a direct or indirect role in early embryogenesis.
Changes in Spermatocyte and Spermatid mRNA Abundance Accompany Loss of pi6 piRNAs
Pachytene piRNAs repress their RNA targets at least in part by an siRNA-like cleavage mechanism. Mice bearing mutations that selectively inactivate the endonuclease activity of MIWI are phenotypically indistinguishable from those lacking MIWI altogether15,16. Moreover, ectopic expression in mice of the largest human piRNA-producing locus triggers cleavage and degradation of mouse Dpy19l2 mRNA, causing male sterility22. To begin to identify direct targets of pi6 piRNAs, we used RNA-seq to measure steady-state RNA abundance in pachytene spermatocytes, diplotene spermatocytes, secondary spermatocytes, and spermatids purified from pi6em1/em1, pi6em2/em2, and C57BL/6 adult testis (Fig. 6a).
Figure 6. mRNAs encoding proteins required for sperm capacitation and zona pellucida-binding are direct targets of pi6 piRNAs.
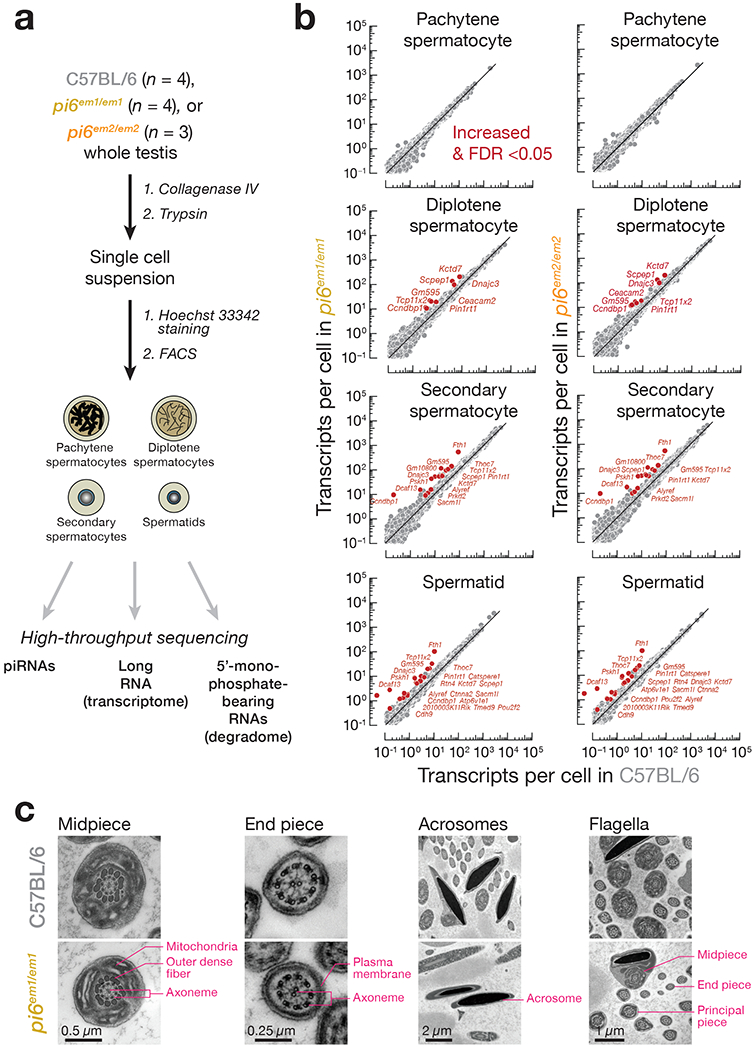
(a) Strategy for purifying specific male germ cell types. (b) Scatter plots of steady-state transcript abundance in sorted testicular germ cells. Each dot represents the mean abundance of an mRNA measured using four (wild-type and pi6em1/em1 cells) or three (pi6em2/em2 cells) individual males. Differentially expressed transcripts (>2 fold-change and FDR <0.05) were identified using DESeq2 (see also Methods) and are indicated, (c) Ultrastructure of caudal epididymal sperm flagella and acrosomes from mice of indicated genotypes by transmission electron microscopy.
See also Extended Data Figure 5 and 6, and Supplementary Tables 2, 3, and 4.
The steady-state abundance of the RNA targets of pi6 piRNA-guided cleavage are predicted to increase in pi6 mutants. We searched for transcripts whose steady-state abundance increased in both pi6em1/em1 (n = 4) and pi6em2/em2 (n = 3) cells compared to C57BL/6 controls (n = 4; Fig. 6b and Supplementary Table 2). pi6em1 and pi6em2 deletions increased the abundance of 8 diplotene spermatocyte mRNAs, 15 secondary spermatocyte mRNAs, and 21 spermatid mRNAs but did not affect genes neighboring pi6 (Supplementary Note). Although pi6 piRNAs first begin to accumulate in pachytene spermatocytes (Extended Data Fig. 3b), the abundance of no pachytene spermatocyte mRNA changed significantly (FDR <0.05) in both pi6em1/em1 and pi6em2/em2 mice, suggesting that pi6 piRNAs do not accumulate to functional levels until the diplotene phase of meiosis. In total, loss of pi6 piRNAs more than doubled the mRNA level of 24 genes in at least one spermatogenic cell type, 13 (54%) of which remained significantly altered in subsequent stages.
Genes Essential for Sperm Functions Are Regulated by pi6 piRNAs
Among the 24 genes with increased mRNA abundance in pi6 mutant cells, Atp6v1e1 and Catsperel encode proteins required for sperm function (Supplementary Table 3). ATP6V1E1, the testis-specific, catalytic subunit of the vacuolar-type FT ATPase, resides in the inner and outer-membranes of the acrosome and acts to acidify the acrosome, stabilizing enzymes required for sperm to penetrate the oocyte zona pellucida54,55. Although Atp6v1e1 overexpression has not been examined in mouse spermatogenesis, overexpression of Atp6v1e1 in somatic tissues is associated with cancer56. In pi6em1/em1 and pi6em2/em2 spermatids, Atp6v1e1 mRNA expression increased by 2.1- (FDR = 2 × 10−10) and 2.3-fold (FDR = 5 × 10−7), respectively (Fig. 6b and Supplementary Table 2). CATSPERE1 is one of the multi-subunits of the sperm-specific CatSper calcium channel, which resides in the flagellar membrane and is required for the transition to hyperactivated motility during capacitation57. In humans, a homozygous in-frame deletion of Catsperel prevents sperm from fertilizing oocytes, resulting in male infertility58. In addition to the well-defined sperm functions of Atp6v1e1 and Catsperel, Ceacam2, Pou2f2/Oct2, and Tcp11x2, genes whose mRNA abundance increases in pi6 mutants, have been proposed to function in spermatogenesis (Supplementary Note). Seventeen additional genes whose mRNA abundance increased in pi6 mutants have reported functions only in somatic cells but appear to regulate related cellular processes (Supplementary Note). The known and inferred functions of pi6-regulated genes suggest that the mechanism underlying pi6 mutant sperm defects reflects dysregulated ion homeostasis rather than aberrant sperm flagellar structure. Consistent with this, transmission electron microscopy detected no architectural abnormalities in the pi6em1/em1 Sperm flagellum or acrosome (Fig. 6c).
pi6 piRNAs Direct Cleavage of Their mRNA Targets
All known catalytically active Argonaute proteins cleave their targets at the phosphodiester bond linking nucleotides t10 and t11, the bases paired to guide nucleotides g10 and g11. Target cleavage generates a 5’ product bearing a 3’ hydroxyl terminus and a 3’ product beginning with a 5’ monophosphate. Thus, cleaved targets can be identified by high-throughput sequencing methods designed to capture long RNAs bearing a 5’ monophosphate (degradome-seq), coupled with computational identification of piRNAs capable of directing production of the putative 3’ cleavage products.
We performed small RNA-seq to define the piRNA repertoire and degradome- seq of C57BL/6 and pi6em1/em1 germ cells to identify candidate pi6 piRNA-directed target cleavage products. Because the specific rules for piRNA-guided target cleavage are poorly defined, we identified target candidates by first requiring g2-g7 seed complementarity between a pi6 piRNA and a cleaved RNA fragment. Then, we searched for seed-matched transcripts with a cleavage product whose 5’ end overlapped 10 nt with the piRNA (Extended Data Fig. 5a). Finally, we compared both the steady-state (RNA-seq) and cleaved fragment (degradome-seq) abundance of target candidate RNAs in wild-type and pi6 mutant germ cells. These criteria identified pi6 piRNA-dependent cleavage sites in six mRNAs whose abundance increased in pi6 mutants: Alyref, Catsperel, Dnajc3, Fth1, Kctd7, and Scpepl (Extended Data Fig. 5b and Supplementary Table 4; Supplementary Note).
pi6 piRNAs Reciprocally Facilitate Biogenesis of piRNAs from Other Loci
Because piRNA-directed cleavage of piRNA precursor transcripts generates 5’ monophosphorylated pre-pre-piRNAs, piRNAs play a central role in the initiation of piRNA production37,59–62. Consistent with this, pi6 piRNAs initiate piRNA production by cleaving pachytene piRNA precursor transcripts. In pi6em1/em1 diplotene spermatocytes, we detected a large reduction in the abundance of 3’ cleavage fragments from pachytene piRNA precursor transcripts targeted by pi6 piRNAs, but not for piRNA precursor transcripts targeted by pi17 piRNAs (Fig. 7a, left panel). Strings of head-to-tail piRNAs (“phased” or “trailing” piRNAs) beginning at the 5’ end of a piRNA-directed 3’ cleavage fragment are the hallmark of piRNA-initiated piRNA production. We detected such phased piRNAs downstream of pi6 piRNA-directed cleavage sites within pachytene piRNA precursors in wild-type diplotene spermatocytes (Fig. 7a, top right panel). In pi6 mutants, the abundance of these trailing piRNAs decreased, whereas the abundance of trailing piRNAs initiated by pi17 piRNAs was unchanged. Conversely, in pi17−/− mutants, the abundance of trailing piRNAs initiated by pi17 piRNAs, but not pi6 piRNAs, diminished (Fig. 7a, bottom right panel), indicating that initiation of piRNA production by piRNAs is not unique to pi6 piRNAs.
Figure 7. pi6 piRNAs and piRNAs from other loci form a network to repress mRNA expression and facilitate piRNA biogenesis.
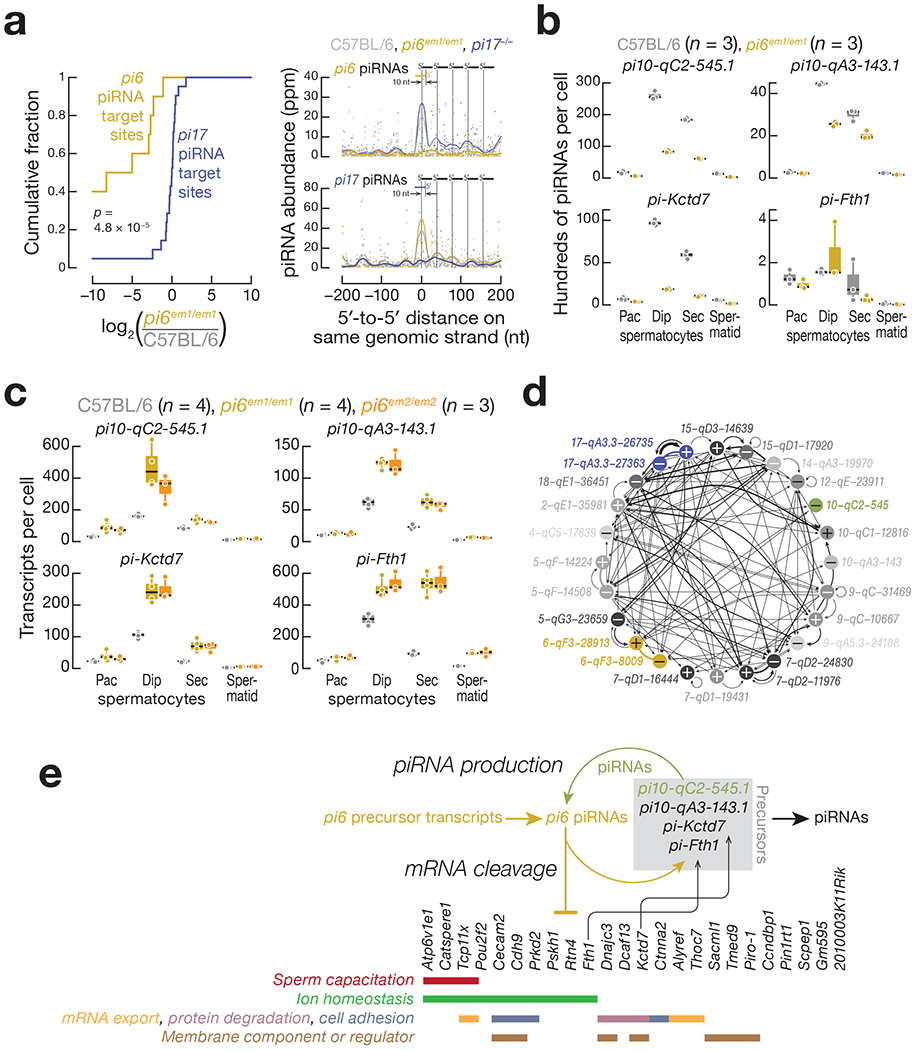
(a) (Left panel) Cumulative abundance of pi6 and pi17 piRNA-directed, 3′ cleavage products in pi6em1/em1 diplotene spermatocytes. (Right panel) Analysis of 5′ to 5′ distances for mature piRNAs derived from pachytene piRNA precursors cleaved by pi6 or pi17 piRNAs in diplotene spermatocytes. piRNA-directed cleavage sites were identified requiring uninterrupted base-pairing from position g2 to g16 between a pi6 (10 sites; top right panel) or pi17 (21 sites; bottom right panel) piRNA and the transcript, such that target cleavage between t10 and t11 would generate the 5′ monophosphorylated, 3′ cleavage fragment detected by degradome-seq. p value was computed using the Kolmogorov-Smirnov test, (b) Abundance of mature piRNAs measured by small RNA-seq. Each dot represents the abundance of uniquely mapping reads in one mouse. (c) Expression of piRNA precursors measured by RNA-seq. Each dot represents the abundance of transcripts in one mouse. In (b) and (c), horizontal black lines denote median; boxes report 75th and 25th percentiles; whiskers indicate the maximal and minimal values. (d) Pachytene piRNA-directed cleavage in pachytene piRNA precursors. The 22 loci producing the most abundant pachytene piRNAs plus pi10-qC2-545.1 and pi10-qA3-143.1 are shown. Each arrow points from the source of a piRNA towards the locus whose transcript it cleaves and the line thickness is proportional to the number of piRNA-directed cleavage sites shown in Supplementary Table 6. Thick lines link loci sharing a single divergently transcribed promoter. Plus and minus signs indicate the Watson and Crick strands, respectively. (e) A model for pi6 piRNA biogenesis and function.
See also Extended Data Figures 5 and 6, and Supplementary Tables 4, 5, and 6.
Intriguingly, loss of pi6 piRNAs specifically decreased the abundance of piRNAs and accumulated steady-state precursors from two pachytene piRNA-producing loci on chromosome 10, but not from any other loci, including the major piRNA loci pi2, pi7, pi9, or pi17 (Fig. 7b and 7c, Extended Data Fig. 3b and 6b, and Supplementary Table 5; Supplementary Note). Further supporting the idea that pi6 piRNA-directed cleavage initiates pachytene piRNA production from pi10-qC2-545.1, degradome sequencing detected two different pi6 piRNA-dependent cleavage sites in pi10-qC2-545.1 transcripts (Extended Data Fig. 5b and Supplementary Table 4). Each cleavage site can be explained by an extensively complementary pi6 piRNA predicted to direct MILI or MIWI to cut the pi10-qC2-545.1 precursor transcript immediately before the 5’ end of the 5’ monophosphorylated RNA identified by degradome sequencing. Remarkably, pi10-qC2-545.1 piRNAs, whose production required pi6 piRNAs, reciprocally promote pi6 piRNA biogenesis: three pi10-qC2-545.1 piRNAs map to pi6 precursor cleavage sites that initiate pi6 piRNA production in wild-type pachytene and diplotene spermatocytes (Extended Data Fig. 5c; Supplementary Table 4). pi10-qC2-545.1 is found in both mice and rats, but the gene produces a IncRNA in rats and a piRNA precursor in mice. The finding that pi10-qC2-545.1 generates piRNAs only in Mus musculus suggests it emerged recently as a pachytene piRNA-producing locus. Perhaps the fortuitous production of pi6 piRNAs with sufficient complementarity to direct cleavage of pi10-qC2-545.1 transcripts has converted the locus to a source of piRNAs that enhance piRNA production from the more ancient pi6 locus.
Reciprocal initiation of piRNA biogenesis between pi6 and pi10-qC2-545.1 is far from exceptional. In fact, precursors from all major pachytene piRNA-producing loci are cleaved by pachytene piRNAs produced by another locus (Fig. 7d and Supplementary Table 6; Supplementary Note). In addition to pachytene piRNA precursor targets, pi6 piRNAs also initiate piRNA biogenesis from two piRNA-regulated protein-coding genes, Kctd7 and Fth1 (Fig. 7b and 7c, Supplementary Table 2 and 4; Supplementary Note). Together, our data demonstrate that pi6 piRNAs not only repress mRNA expression but also initiate piRNA biogenesis in trans from other piRNA-producing loci (Fig. 7e).
Discussion
Deletion of the promoter of the mouse pi6 pachytene piRNA locus causes specific, quantifiable defects in male fertility. These include impaired sperm capacitation and a failure of sperm to bind and penetrate the zona pellucida. The male fertility defects accompanying loss of pi6 piRNAs are specific to this locus, as deletion of the promoter of pi17, which eliminates pi17 piRNAs, had no detectable effect on male or female fertility or viability, as reported previously20. The phenotypic defects of pi6 mutants reflect the molecular changes—increased steady-state abundance of mRNAs that encode proteins functioning in sperm capacitation, acrosome function, and other pathways with links to sperm biology.
Our finding that deletion of pi6, but not of pi17, the most prolific piRNA-producing locus, leads to male fertility defects, suggests that individual pachytene piRNA loci can regulate distinct sets of genes (Supplementary Note). Pachytene piRNAs have been proposed to act collectively in meiotic spermatocytes or post-meiotic spermatids to target mRNAs for destruction22,24. Our data argue against pachytene piRNAs acting en mass24,63, since not only does pi6 produce far fewer piRNAs than pi17, but just a tiny fraction of pi6 piRNAs can explain the effect of loss of pi6 piRNAs on the transcriptome. pi6 produces 80,354 distinct piRNA sequences, representing 10,943 unique g 1 —g21 piRNA sequences reproducibly (n = 3) present at >1 molecule per cell. Yet loss of pi6 piRNAs dysregulates just 24 mRNAs, consistent with its remarkably specific mutant phenotype. This finding calls to mind the mechanism of sex determination in the silkworm Bombyx mori: a single piRNA species derived from a piRNA precursor locus, Feminizer {Fern), on the W chromosome, targets the transcript of the Masculinizer gene on the Z chromosome. Among thousands of W chromosome-derived B. mori piRNAs, just a single piRNA species regulates the mRNA that plays a dominant role in female sex determination64.
Moreover, our data argue strongly against miRNA-like regulation by piRNAs. miRNA binding through little more than the seed match accelerates mRNA degradation (Ref. 65; Supplementary Note). siRNA directed target cleavage requires more extensive sequence complementarity but can tolerate a variety of mismatches—even a 7 nt insertion—around the cleavage site and at the 3’ end of the siRNA66,67. If pachytene piRNAs bound their target RNAs by a miRNA-like, seed-based mechanism, the predicted target repertoire of piRNAs produced by individual loci would be enormous: pi6 piRNAs encompass 9,880 distinct 7mer-m8 seed sequences (g2-g8)68, while pi17 generates 134,358 distinct piRNA sequences, encompassing 11,324 distinct g2-g8 seeds. Yet loss of pi17 piRNAs has no detectable phenotype, while loss of pi6 piRNAs causes specific defects in sperm motility, the acrosome reaction, and egg fertilization. Although 104 pi6 piRNAs are more abundant than miR-20a, the tenth most abundant miRNA in diplotene spermatocytes, loss of pi6 piRNAs reproducibly increases the abundance of just 24 mRNAs. Of these, just six mRNAs appear to be direct cleavage targets of pi6 piRNAs, consistent with pachytene piRNAs acting like long siRNAs: they find and cleave targets with extensive—but often incomplete—complementarity.
The current model for piRNA biogenesis posits that piRNA-directed cleavage of precursor transcripts facilitates the biogenesis of other piRNAs60–62. Consistent with this view, pi6 piRNAs are required for biogenesis of piRNAs from four other piRNA- producing loci: pi10-qC2-545.1 and pi10-qC2-143.1, both sources of pachytene piRNAs; and two protein-coding genes, Kctd7, a hybrid piRNA gene, and Fth1, a pre-pachytene piRNA gene. Despite producing just 3% as many piRNAs as pi6, pi10-qC2-545.1 makes piRNAs that can cleave pi6 transcripts, initiating biogenesis of pi6 piRNAs. Such positive feedback loops appear to operate among many piRNA-producing loci, suggesting that this is an important mechanism for pachytene piRNA biogenesis in mice and perhaps other Eutheria. Given the strikingly small number of mRNAs regulated by pi6 piRNAs, we speculate that the extreme sequence diversity of pachytene piRNAs may be a natural byproduct of the positive feedback loops among pachytene piRNA loci. That is, many piRNAs may serve to reinforce piRNA production rather than reflect evolutionarily pressure to regulate large numbers of mRNAs.
Beyond the requirement for pi6 piRNAs to produce fully functional sperm, pi6 piRNAs appear to play an additional role in embryo development. Our data suggest that the arrested development and reduced viability of embryos derived from pi6 mutant sperm reflects a paternal defect and not the embryonic genotype. Damaged sperm DNA, abnormal sperm chromatin structure, and failure to form a male pronucleus in fertilized embryos have been reported to be linked to retarded embryo development69,70. Our analysis of transposon RNA abundance in pi6 mutant germ cells argues against a role for pi6 piRNAs in transposon silencing during spermatogenesis, but we cannot currently exclude a direct or indirect role for pi6 piRNAs in silencing transposons in the early embryo71. Of course, DNA damage might reflect incomplete repair of the double- stranded DNA breaks required for recombination, rather than transposition or transposon-induced illegitimate recombination.
How pachytene piRNAs identify their targets remains poorly understood in part because of a lack of suitable biochemical or genetic model systems. The availability of a mouse mutant missing a specific set of piRNAs whose absence causes a readily detectable phenotype should provide an additional tool for understanding the base-pairing rules that govern the binding of piRNAs to their RNA targets and for unraveling the regulatory network created by pachytene piRNA. Finally, we note that in many placental mammals, the syntenic location corresponding to pi6 also produces piRNAs. Yet, despite the importance of pi6 piRNAs for mouse fertility, the actual sequence of the pi6 precursor transcript is conserved only among rodents7. Explaining how the essential function of fertility can rely on regulatory molecules whose sequence is so poorly conserved remains the central challenge of pachytene piRNA biology.
Methods
Mouse mutants
Mice were maintained and sacrificed according to guidelines approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School (A-2222-17).
Small guide RNAs (sgRNAs) flanking piRNA promoters were designed using CRISPR design tools (crispr.mit.edu/). DNA oligos containing guide sequences were cloned into pX330 vectors72, and their cleavage activity tested in NIH3T3 cells by cotransfecting pX330 constructs containing sgRNA sequences and puromycin-resistant plasmid (pPUR) using TransIT-X2 (Mirus Bio, Madison, WI). Puromycin (3 pg/pl) was added 24 h after transfection and DNA extracted 48 h afterwards. Promoter deletions were detected by PCR using primers flanking the predicted Cas9 cleavage sites.
For mice, sgRNAs were generated by in vitro transcription and purified by electrophoresis on 8% (w/v) polyacrylamide gels. To generate the pi6em1/em1 and pi7−/− lines used in this study, in vitro transcribed sgRNAs (10 ng/μl each) targeting pi6 and pi17 were mixed with Cas9 mRNA (40 ng/μl) and injected together into the cytoplasm of one-cell C57BL/6 zygotes (RNA only). For some founders, the sgRNA and Cas9 mRNA mixture was combined with pX330 plasmids expressing the same four sgRNAs and Cas9 and injected into both the cytoplasm and pronuclei of one-cell C57BL/6 zygotes (RNA + DNA). For pi6em2/em2, in vitro transcribed sgRNAs and Cas9 mRNA were injected into the cytoplasm of one-cell C57BL/6 embryos. Embryos were transferred to pseudopregnant females using standard methods. To screen for mutant founders, DNA was extracted from small pieces of tail clipped from three-week-old pups73. Deletions were detected by PCR, and PCR products purified and cloned into TOPO blunt vectors. Mutant sequences were determined by Sanger sequencing. Mouse mutant lines were established and maintained by mating mutant founders with C57BL/6 males or females. All mutant mice in this study were backcrossed for at least two generations before use.
Mouse fertility test
Each 2–8 month-old male mouse was housed with one 2–4 month-old C57BL/6 female, who was examined for the presence of a vaginal plug the following morning. When a plug was observed, the female was housed separately. For male mice who did not produce pups after 3 months (~3 cycles), the original female was replaced with a new female and the fertility test continued.
Testis histology, sperm count, and sperm morphology
Mouse testes were fixed in Bouin’s solution overnight, washed with 70% ethanol, embedded in paraffin, and sectioned at 5 μm thickness. Sections were stained with hematoxylin solution, countered stained with eosin solution, and imaged using Leica DMi8 brightfield microscope equipped with an 20× 0.4 N.A. objective (HC PL FL L 20×/0.40 CORR PH1, Leica Microbiosystems, Buffalo Grove, IL). To quantify sperm abundance, the cauda epididymides were collected from mice and placed in phosphate- buffered saline (PBS) containing 4% (w/v) bovine serum albumin. A few incisions were made in the epididymides with scissors to release the sperm, followed by incubation at 37°C and 5% CO2 for 20 min. A 20 μl aliquot of sperm suspension was diluted in 480 pl of 1% (w/v) paraformaldehyde (PFA), and sperm cells counted at 10× by brightfield microscopy. To assess sperm morphology, caudal epididymal sperm were fixed in 1% (w/v) PFA, stained with trypan blue, and a Leica DMi8 brightfield microscope equipped with an 63× 1.4 N.A. oil immersion objective (HC PL APO; Leica Microbiosystems, Buffalo Grove, IL). Sperm stained with Alexa 488-conjugated PNA (see below) were also used to assess sperm morphology.
Meiotic chromosome spreads
Meiotic chromosome spreads were prepared as described74. Mouse testes were incubated in hypotonic buffer (30 mM Tris-CI, pH 8.2, 50 mM sucrose, 17 mM sodium citrate, 5 mM EDTA, 0.5 mM DTT) for 30 min on ice, then small fragments of seminiferous tubules were moved to 100 mM sucrose solution and pulled apart with forceps to release germ cells. A drop of sucrose solution containing germ cells was pipetted onto a glass slide with a thin layer of 1× PBS containing 1% PFA and 0.15% (v/v) Triton-X100 (pH 9.2) and spread by swirling. Slides were placed in a humidifying chamber for 2.5 h, air-dried, and washed twice with 1× PBS with 0.4% Photo-Flo 200 (Kodak, Rochester, NY) and once with water with 0.4% Photo-Flo 200, and air-dried. For immunostaining of meiotic chromosomes, slides were sequentially washed with (1) 1× PBS with 0.4% Photo-Flo 200, (2) 1× PBS containing 0.1% (v/v) Triton-X, and (3) blocked with PBS containing 3% (w/v) BSA, 0.05% (v/v) Triton X-100, and 10% (v/v) goat serum in 1× PBS at room temperature. The slides were then incubated with primary antibodies, anti-SCP1 (1:1000 dilution) and anti-SCP3 (1:1000 dilution), in a humidifying chamber overnight at room temperature. Washing and blocking steps were repeated the next day, and the slides were incubated with Alexa 488- or Alexa 594-conjugated secondary antibodies (1:10,000 dilution) for 1 h at room temperature. Slides were washed thrice with 1× PBS containing 0.4% (v/v) Photo-Flo 200, once with water containing 0.4% Photo-Flo 200 mixture, air-dried in the dark, mounted by incubation in ProLong Gold Antifade Mountant with DAPI (4′,6′-diamidino-2-phenylindole; Thermo Fisher Scientific, Waltham, MA) overnight in the dark, and imaged using a Leica DMi8 fluorescence microscope equipped with an 63× 1.4 N.A. oil immersion objective (HC PL APO; Leica Microbiosystems, Buffalo Grove, IL).
Cell sorting by FACS
Testicular cell sorting was performed as described75. Testes were collected, decapsulated, and incubated in 0.4 mg/ml collagenase type IV (Worthington LS004188) in 1× Grey’s Balanced Salt Solution (GBSS, Sigma, G9779) at 33°C rotating at 150 rpm for 15 min. Separated seminiferous tubules were washed with 1× GBSS and incubated in 0.5 mg/ml Trypsin and 1 μg/ml DNase I in 1× GBSS at 33°C rotated at 150 rpm for 15 min. Tubules were dissociated on ice by gentle pipetting, and then 7.5% (v/v) fetal bovine serum (f.c.) was added to inactivate trypsin. The cell suspension was filtered through a pre-wetted 70 pm cell strainer, and cells pelleted at 300× g for 10 min at 4°C. Cells were resuspended in 1× GBSS containing 5% (v/v) FBS, 1 μg/ml DNase I, and 5 μg/ml Hoechst 33342 (Thermo Fisher Scientific, Waltham, MA) and rotated at 150 rpm at 33°C for 45 min. Propidium iodide (0.2 μg/ml, f.c.; Thermo Fisher Scientific, Waltham, MA) was added, and cells strained through a pre-wetted 40 μm cell strainer. Cell sorting was performed on a FACSAria II (BD Biosciences, Franklin Lakes, NJ). The purity of sorted fractions was assessed by immunostaining. Secondary spermatocyte and spermatid populations were >90% pure, and the pachytene spermatocytes and diplotene spermatocytes were >80% pure.
In vitro fertilization (IVF) and embryo transfer
In vitro fertilization was performed as previously described76 using spermatozoa from caudal epididymis of C57BL/6, pi6+/em1, or pi6em1/em1 mice. Spermatozoa were incubated in complete human tubal fluid media (HTF; 101.6 mM NaCl, 4.69 mM KCI, 0.37mM KH2PO4, 0.2 mM MgSO4·7H2O, 21.4 mM Na-lactate, 0.33 mM Na-pyruvate, 2.78 mM glucose, 25 mM NaHCO3, 2.04 mM CaCl2-2H2O, 0.075 mg/ml Penicillin-G, 0.05 mg/ml streptomycin sulfate, 0.02% (v/v) phenol red, 4 mg/ml BSA) with oocytes (98–146 for control sperm and 120–293 for pi6em1/em1 sperm) from B6SJLF1/J mice for 3–4 h at 37°C with constant 5% O2, 90% N2, and 5% CO2 concentration. Oocyte viability and the presence of pronuclei were assessed under a Nikon SMZ-2B (Nikon, Tokyo, Japan) dissecting microscope. To observe embryo development, embryos were moved into potassium-supplemented simplex optimized media (KSOM; 95 mM NaCl, 2.5 mM KCl, 0.35 mM KH2PO4, 0.2 mM MgSO4·7H2O, 10 mM Na-lactate, 0.2 mM Na-pyruvate, 0.2 mM glucose, 25 mM NaHCO3, 1.71 mM CaCl2-2H2O, 1 mM L-glutamine, 0.01 mM EDTA, 0.075 mg/ml Penicillin-G, 0.05 mg/ml streptomycin sulfate, 0.02% (v/v) phenol red, 1 mg/ml BSA; Millipore Sigma, Burlington, MA) after IVF and assessed every 24 h. To measure birth rates, two-cell embryos were transferred to Swiss Webster pseudopregnant females, and fetuses isolated by cesarean section 18.5 d after embryo transfer.
For zona-free IVF, the zona pellucida of oocytes was removed with acid Tyrode’s solution as described77,78.
Intracytoplasmic sperm injection (ICSI)
Frozen caudal epididymal spermatozoa were thawed, the sperm tails detached76, and individual pi6+/em1 or pi6em1/em1 sperm heads injected into B6D2F1/J oocytes in Chatot- Ziomek-Bavister media (CZB; 81.62 mM NaCl, 4.83 mM KCl, 1.18 mM KH2PO4, 1.18 mM MgSO4-7H2O, 25 mM Na2HCO3, 1.70 mM CaCl2-2H2O, 0.11 mM Na2-ETDA-2H2O, 1 mM L-glutamine, 28 mM Na-lactate, 0.27 mM Na-pyruvate, 5.55 mM glucose, Penicillin-G 0.05 mg/ml, 0.07 mg/ml streptomycin sulfate, 4 mg/ml BSA; Millipore Sigma, Burlington, MA) using the PiezoXpert (Eppendorf, Hamburg, Germany; Cat#5194000024). Surviving oocytes were counted, collected, and cultured in KSOM (Millipore Sigma, Burlington, MA) at 37°C and 5% CO2 for 24 h. Two-cell embryos were surgically transferred unilaterally into the oviducts of pseudopregnant Swiss Webster females. At 16.5 days after the surgery, live fetus isolated by cesarean section.
Sperm motility
Cauda epidydimal sperm were collected from mice and placed in 37°C HTF media containing 4 mg/ml BSA in an incubator with 5% CO2. A drop of sperm was removed from the suspension and pipetted into a sperm counting glass chamber, then assayed by CASA or video acquisition. CASA was conducted using an IVOS II instrument (Hamilton Thorne, Beverly, MA) with the following settings: 100 frames acquired at 60 Hz; minimal contrast = 50; 4 pixel minimal cell size; minimal static contrast = 5; 0%straightness (STR) threshold; 10 μm/s VAP Cutoff; prog, min VAP, 20 μm/s; 10 μm/s VSL Cutoff; 5 pixel cell size; cell intensity = 90; static head size = 0.30–2.69; static head intensity = 0.10–1.75; static elongation = 10–94; slow cells motile = yes; 0.68 magnification; LED illumination intensity = 3000; IDENT illumination intensity = 3603; 37°C. The raw data files (i.e., .dbt files for motile sperm and .dbx files for static sperm) were used for sperm motility analysis. For the motile sperm, only those whose movement was captured with ≥45 consecutive frames were analyzed. For the boxplots, the number of static sperm was re-calculated for each mouse according to the percentage of motile sperm with ≥ 45 frames. For hyperactivated motility analysis, .dbt files of motile sperm were used as input for CASAnova, as previously described52. For movie acquisition, a Nikon Diaphot 200 microscope (Nikon, Tokyo, Japan) with darkfield optics equipped with Nikon E Plan 10×/0.25 160/- Ph1 DL objective (Nikon, Tokyo, Japan), ZWO ASI 174mm Monochrome CMOS Imaging camera (ZWO, SuZhou, China), and the SharpCap software (https://docs.sharpcap.co.Uk/2.9/) using darkfield at 10× magnification were used to record sperm movement at 37°C.
Ex vivo acrosome reaction and capacitation assay
Ex vivo, the acrosome reaction occurs spontaneously in some sperm and can also be triggered by the Ca2+ ionophore A2318779, which results in an acrosome reaction visually indistinguishable from that triggered by natural ligands such as progesterone80 or ZP381, while bypassing signaling pathways essential for acrosome reaction in vivo82. Sperm capacitation was induced and acrosome reaction was assessed as described79. Cauda epididymides were collected from mice, placed in HTF media containing 4 mg/ml BSA pre-warmed for at least 2 h in a 37°C incubator at 5% CO2. A few incisions were made in the epididymides with scissors to release the sperm, followed by incubation at 37°C in 5% CO2 for 90 min. Calcium ionophore A23187 (10 μm f.c. in DMSO) was added, and incubation continued for 30 min. Sperm were fixed at room temperature for 10 min by adding two volumes of 4% (w/v) PFA, pelleting at 1,000 × g for 5 min, washed with 1× PBS, resuspended in fresh 1× PBS, spotted on a glass slide, and air-dried. Methanol was pipetted onto the sperm to permeabilize the cells, followed by washing with 1× PBS. Slides were incubated overnight in 10 pg/ml Alexa Fluor 488-conjugated peanut agglutinin (PNA) in 1× PBS83, washed with 1× PBS, air-dried, and mounted with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, Waltham, MA). Sperm were imaged using a Leica DMi8 fluorescence microscope equipped with a 63× 1.4 N.A. oil immersion objective (HC PL APO; Leica Microbiosystems, Buffalo Grove, IL) and analyzed using ImageJ (version 2.0.0-rc-68/1.52e; https://fiji.sc/).
Transmission electron microscopy
Mouse caudal epididymides were dissected and immediately fixed by immersion in Karnovsky’s fixative (2% formaldehyde (v/v) and 3% glutaraldehyde (v/v) in 0.1M sodium phosphate buffer, pH 7.4; Electron Microscopy Sciences, Hatfield, PA) overnight at 4°C, and washed three times in 0.1M phosphate buffer. Following the third wash, the tissues were post-fixed in 1% osmium tetroxide (w/v; Electron Microscopy Sciences, Hatfield, PA) for 1 h at room temperature, washed three more times with water for 10 min each, and dehydrated using a graded series of 30%, 50%, 70%, 85%, 95%, 100% (3 changes) ethanol and 100% propylene oxide (two changes) and a mixture of 50% propylene oxide (v/v) and 50% SPI-Pon 812 resin mixture (v/v; SPI Supplies, West Chester, PA). The sample was incubated in seven successive changes of SPI-Pon 812 resin over three days, polymerized at 68°C in flat molds, and reoriented to allow cross- sectioning of spermatozoa in the lumen of epididymis. 70nm sections were cut on a Leica EM UC7 ultramicrotome (Leica Microsystems, Wetzlar, Germany) using a diamond knife, collected on copper mesh grids, and stained with 3% lead citrate (w/v) and 0.1% uranyl acetate (w/v) to increase contrast. Finally, sections were examined using Philips CM10 transmission electron microscope (Philips Electron Optics, Eindhoven, The Netherlands) at 100 KV. Images were recorded using the Erlangshen digital camera system (Gatan Inc., Pleasanton, CA).
RNA-seq and small RNA-seq analysis
Small RNA-seq and RNA-seq libraries were constructed incorporating unique molecular identifiers for removal of PCR duplicates and sequenced using NextSeq 500 (lllumina, San Diego, CA) as described62,84. To sequence mature piRNAs, small RNA was oxidized with 25 mM NaIO4 in 30 mM sodium borate, 30 mM boric acid (pH 8.6; Sigma Aldrich, St. Louis, MO) at 25°C for 30 min. RNA was precipitated with ethanol before adapter ligation. A set of 9 synthetic 2′-O-methylated RNA oligonucleotides was added to each RNA sample to allow measurement of molecules per cell. Small RNA-seq and RNA-seq reads were mapped to mouse genome assembly mm 10 using piPipes85. For small RNA quantification, sequences of synthetic spike-in oligonucleotides were identified allowing no mismatches and the number of molecules of small RNAs per library was calculated based on the read abundance of the spike-in oligonucleotides. For long transcript quantification, 1 μL of 1:100 dilution of ERCC spike-in mix 1 (Thermo Fisher, 4456740, LOT00418382) was added to 1 μg of total RNA in the first step. For Figure 6, Extended Data Figure 3, and Supplementary Tables 2 and 5, differentially expressed transcripts were determined using DESeq286. Statistical testing was performed first using the Wald test and the derived p values were adjusted for multiple testing using the Benjamini Hochberg procedure, as previously described. Transcript abundance between pi6+/em1 and C57BL/6 testes were indistinguishable (<2-fold change and FDR >0.05). See also Life Sciences Reporting Summary.
Analysis of piRNA cleavage sites
Cleaved RNA fragments bearing 5′ monophosphates (“degradome” sequences) were cloned as previously described87,88 and sequenced using a paired-end sequencing kit on NextSeq 500 (Iliumina, San Diego, CA). Briefly, 5′ monophosphate-bearing RNAs were enriched using an adapter with a 3′ hydroxyl and T4 RNA ligase. cDNA was generated using random primers conjugated with the sequence of the 3′ adapter and PCR-amplified using primers containing multiplex barcodes. piRNAs (>1 ppm) and degradome sequences from the same cell types were used for piRNA target analysis. Degradome sequences were extended 3′ to 5′ based on the mouse reference genome mm10, and putative targeting piRNAs were identified first by their pairing with specific degradome sequences at g2-g7 and a minimum of 8 additional base-pairs 5′ of g7. Only degradome sequences that begin at g11 of the matching piRNAs were used. Lastly, cleavage sites for which the read abundance significantly decreased (>2-fold and FDR <0.05) in pi6 mutants were extracted. Analysis of 5′ to 5′ distance for mature pi6 piRNAs was performed as described62. Briefly, 5′ monophosphorylated pachytene piRNA precursor fragments from wild-type diplotene spermatocyte were detected by degradome-seq. Those fragments whose 5′ ends could be by explained by cleavage directed by complementary (g2—g15) pi6 piRNAs were identified. Only fragments whose abundance decreased in pi6 mutant diplotene spermatocytes were retained. See also Code Availability.
qRT-PCR RNA quantification
Isolated total RNA from sorted germ cells was treated with Turbo DNase (Thermo Fisher Scientific, Waltham, MA) at 37°C for 30 min and purified using RNA Clean & Concentrator (Zymo Research, Irvine, CA). First strand cDNA was synthesized using oligo dT20 (for full-length transcripts) or random hexamers (for all transcript fragments) and Superscript III (Invitrogen, Carlsbad, CA). Quantitative PCR was performed for each sample using SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA) with technical triplicates and sorted spermatids from three individual males for each genotype. Relative transc ript abundance was calculated using the ΔΔCt method. Gapdhs, a testis-specific mRNA that remains unchanged in pi6 mutants based on RNA-seq analysis, was used for normalization. Statistical significance was calculated using the unpaired two-sided t-test.
Transposon mapping
RNA-seq reads were intersected using BEDtools89 with Repeat Masker annotation from UCSC (downloaded from https://genome.ucsc.edu/cgi-bin/hgTables). Reads mapping to multiple genomic locations were apportioned. Reads for individual repeats were aggregated to obtain reads counts for repeat families. See also Life Sciences Reporting Summary.
Statistics and Reproducibility
All statistics were performed using R v1.2.5042 (https://www.rstudio.com/) and graphs were generated using Igor Pro v7.08 (WaveMetrics) or ggplot2 v3.1.0 (https://ggplot2.tidyverse.org/). Unless otherwise stated in the figure legends or Methods, Mann-Whitney-Wilcoxon two-sided test was used to calculate p values. For Figures 2d, 6c, and 7a, Extended Data Figure 2c, and Supplementary Movies 1–10, representative data (reproducible in ≥3 individual mice) from a single mouse are shown. See also Life Sciences Reporting Summary.
Data Availability
All sequencing data are available through the National Center for Biotechnology Information Sequence Read Archive using accession number PRJNA634688.
Code Availability
The code used for identifying piRNA-directed cleavage sites is available at https://qithub.com/wenq-lab/GTBuster. All other codes used in this study are described in Methods and Life Sciences Reporting Summary.
Extended Data
Extended Data Fig. 1. Confirmation of mutant founder genotypes.
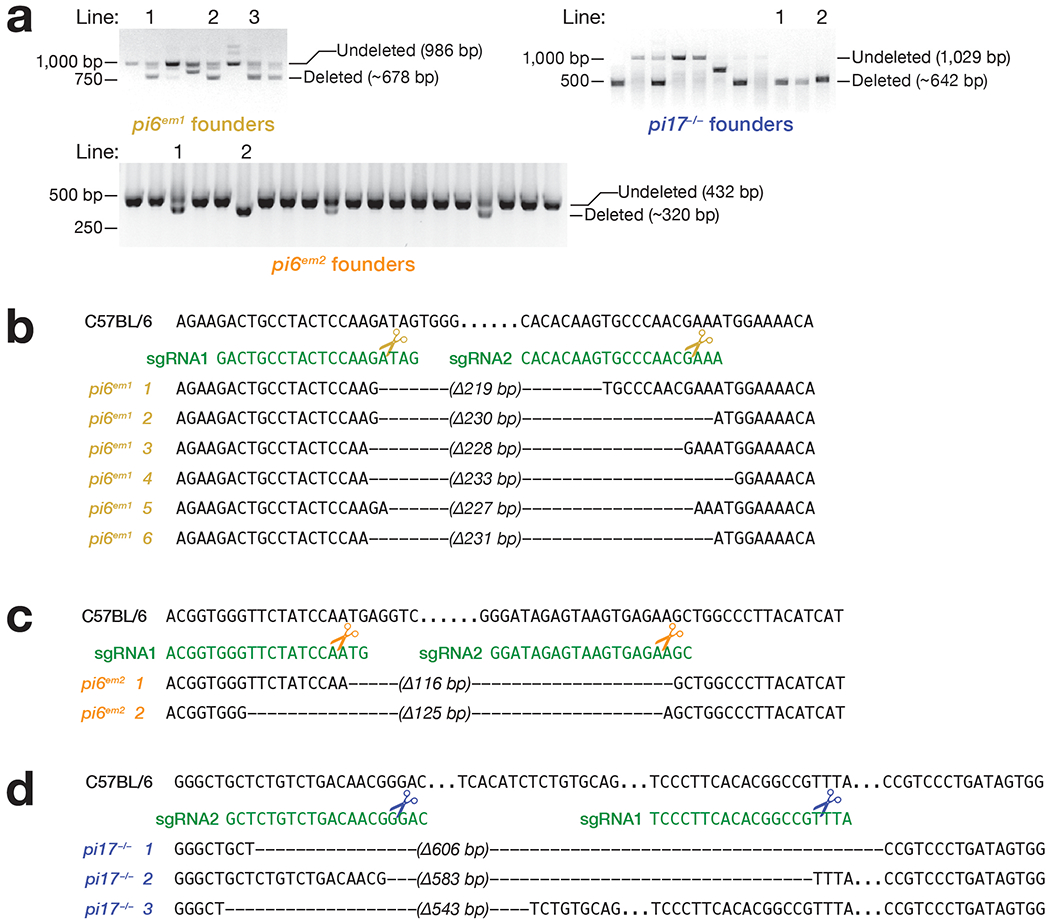
(a) Genotyping of mutant founders by PCR. Mutant founders were generated by injecting sgRNAs and Cas9 mRNAs into C57BL/6 one-cell zygotes, which were transferred to surrogate mothers and screened after birth. Gel images were cropped for clarity (see also Source Data). Genomic sequences of pi6 promoter region in pi6em1 (b) and pi6em2 (c) mouse lines. The presence of both deleted and undeleted PCR products indicate a heterozygous mutant founder that carries just one CRISPR-edited chromosome. (d) Genomic sequences of pi17 promoter region in pi17−/− mouse lines. Dashes, genomic sequences deleted by CRISPR; dots, unaltered sequence omitted for clarity.
Extended Data Fig. 2. pi6em1/em1 adult male phenotype.
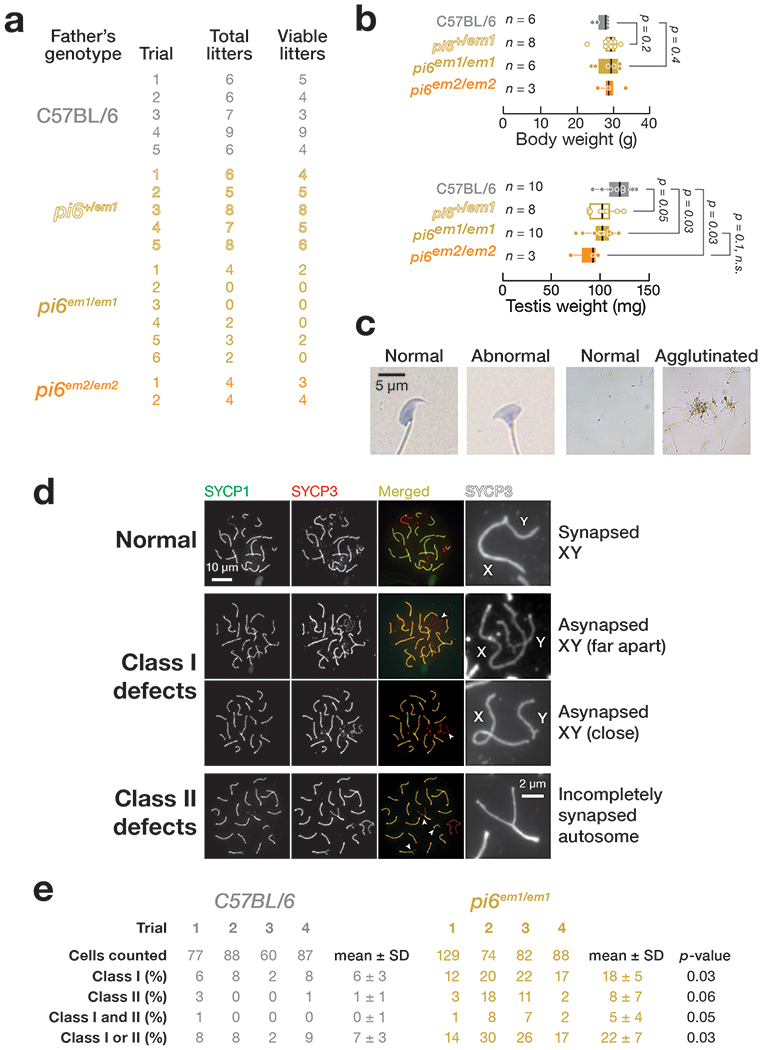
(a) Number of litters produced in 6 months by 2–8 month-old males. (b) Body and testis weight of 2–4 month-old pi6em1/em1 and pi6em2/em2 males. Each dot represents an individual mouse. Vertical lines denote median; boxes report 75th and 25th percentiles; whiskers indicate the maximal and minimal values. (c) Representative spermatozoa from C57BL/6 and pi6em1/em1 males. (d) Representative patterns of meiotic chromosome synapsis in pi6em1/em1 pachytene spermatocytes. SYCP1, Synaptonemal complex protein 1; SYCP3, Synaptonemal complex protein 3. (e) Quantification of patterns of meiotic chromosome synapsis depicted in (d) from C57BL/6 (n = 4) and pi6em1/em1 (n = 4) males.
Extended Data Fig. 3. Abundance of transposons in pi6em1/em1 and pi6em2/em2 germ cells.
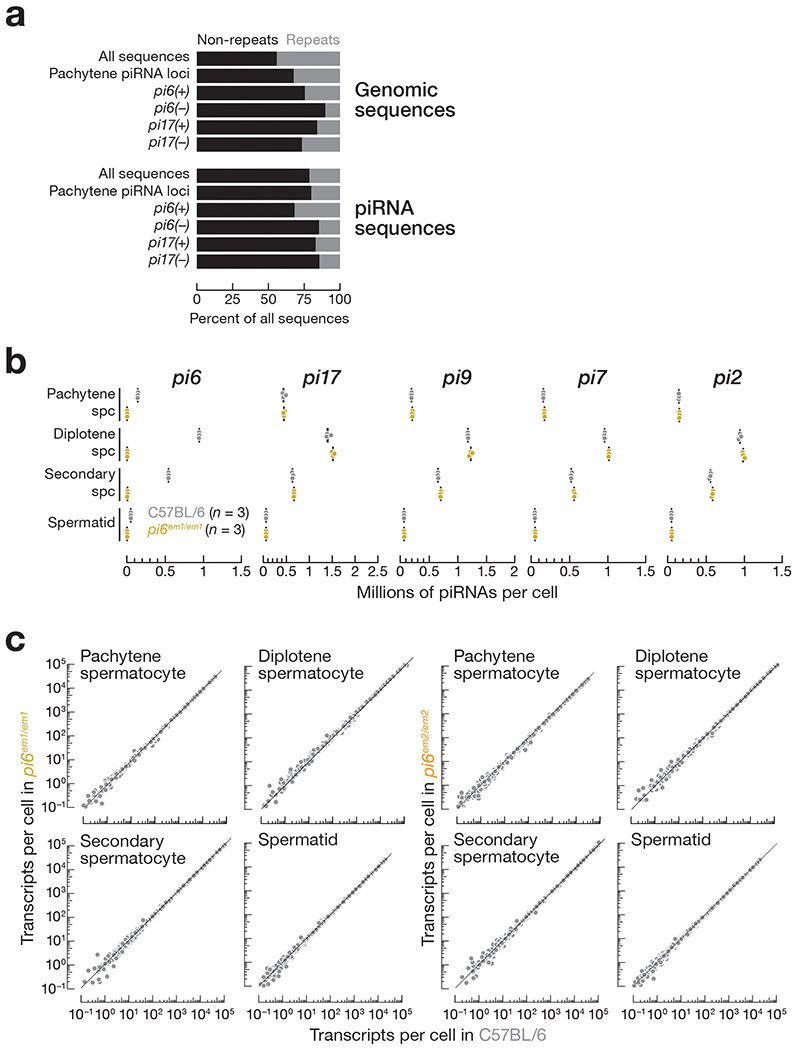
(a) Proportions of the whole genome or piRNA sequences composed of repetitive sequences. (b) Abundance of mature piRNAs from the top five major pachytene piRNA-producing loci in indicated cell types measured by small RNA-seq. Each dot represents the abundance of uniquemapping reads in one C57BL/6 (n = 3) or pi6em1/em1 (n = 3) male. Vertical black lines denote median; boxes report 75th and 25th percentiles; whiskers indicate the maximal and minimal values. (c) Abundance of transposon-derived RNAs in mouse germ cells. Each dot represents the mean of four (wild-type and pi6em1/em1) or three (pi6em2/em2) biologically independent RNA-seq experiments. Gray dots indicate change in abundance <2-fold and/or FDR >0.05 determined by DESeq2 (see also Methods).
Extended Data Fig. 4. Pregnancy rate of surrogate mothers in IVF and ICSI experiments.
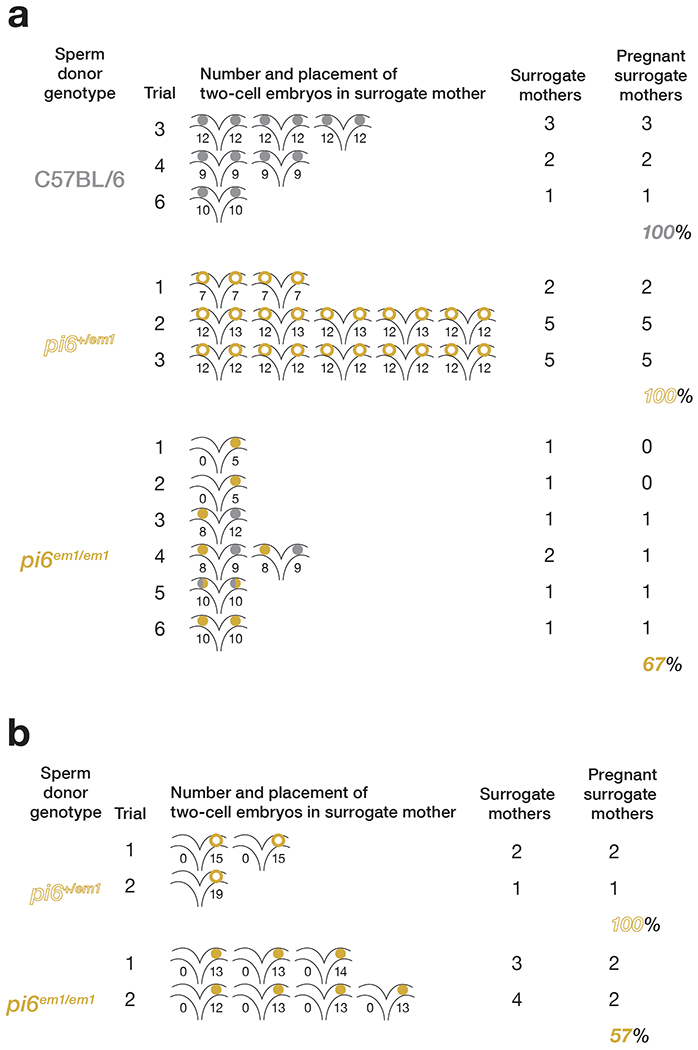
Percent of pregnant surrogate mothers in IVF (a) and ICSI (b).
Extended Data Fig. 5. Transcripts directly cleaved by pi6 and pi10-qC2-545.1 piRNAs.
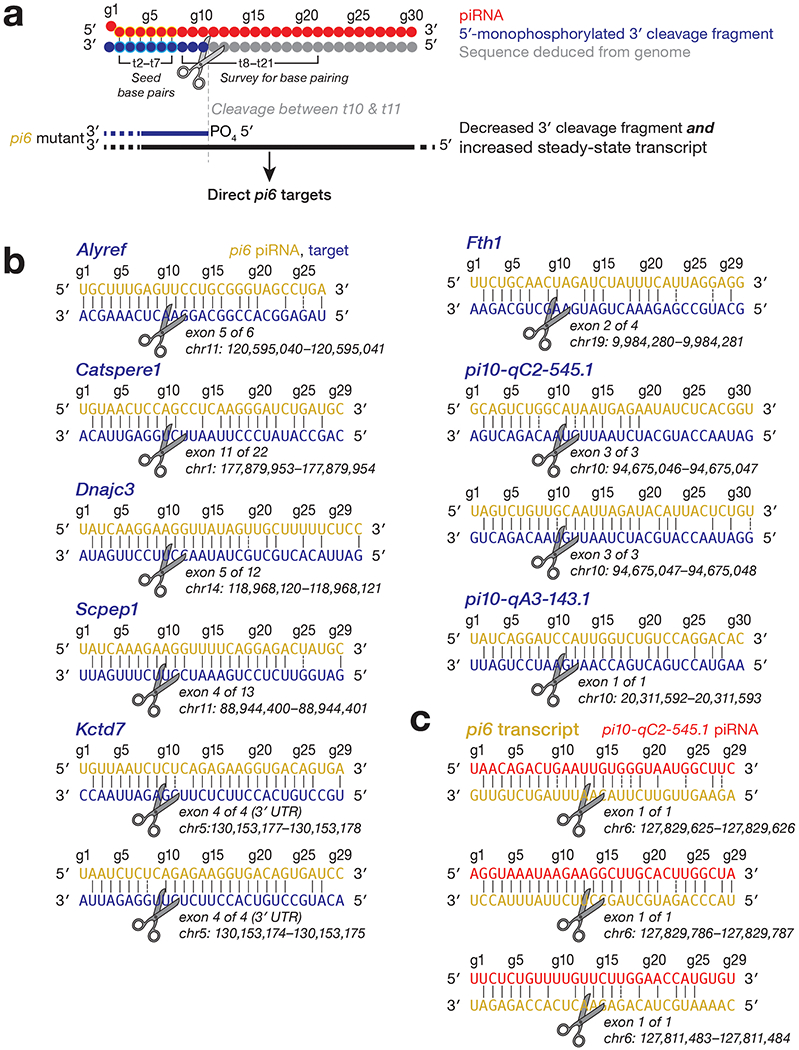
(a) Strategy to identify piRNA-directed cleavage sites. (b) pi6-dependent cleavage sites in mRNAs or pachytene piRNA precursors from pi10-qC2-545.1 and pi10-qA3-143.1 showing inferred base pairing with the corresponding pi6 piRNA guides. An exemplary piRNA guide is shown where more than one piRNA can direct the same cleavage. (c) Cleavage sites in pi6 precursors explained by pi10-qC2-545.1 piRNAs. An exemplary piRNA guide is shown.
Extended Data Fig. 6. Transcriptome changes in pi6em1/em1 and pi6em2/em2 cells.
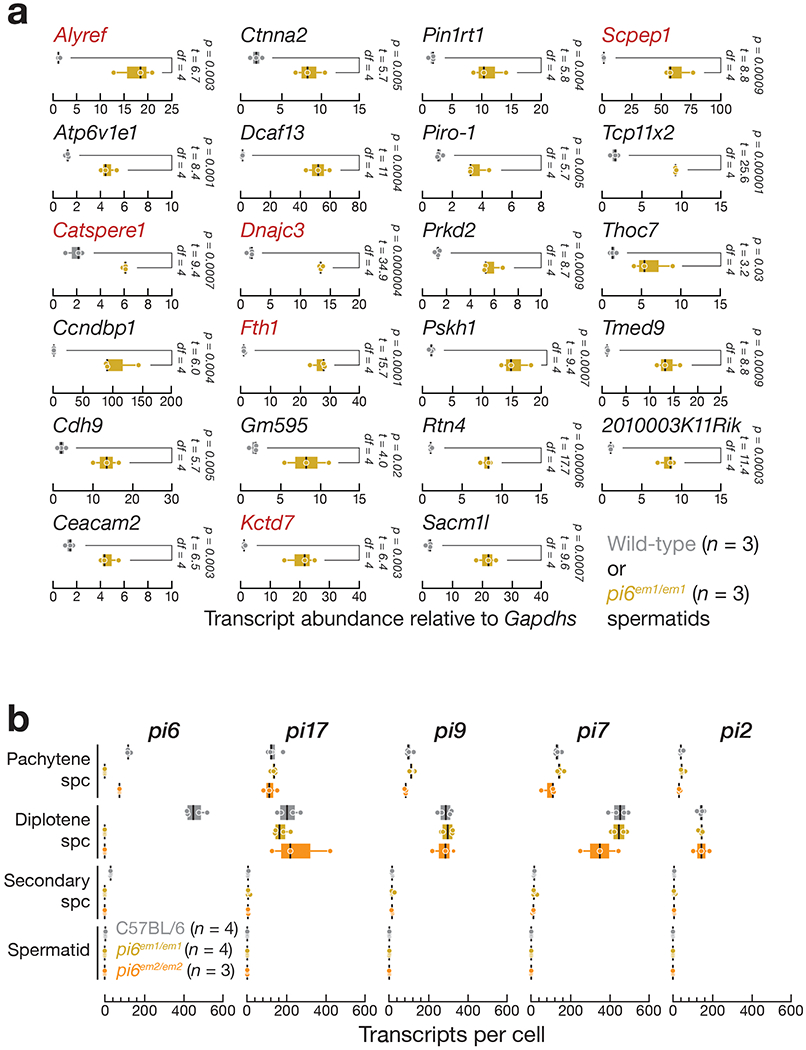
(a) Expression of mRNAs measured by qRT-PCR using oligo dT(20) to prime cDNA synthesis and PCR primers spanning pi6 piRNA-directed cleavage sites (gene names in red) or designed to detect full-length RNA (gene names in black). Pou2f2 mRNA abundance in spermatids was below the limit of detection by qRTPCR. (b) Abundance of piRNA precursors from the top five major pachytene piRNA-producing loci in indicated cell types measured by RNA-seq. For (a) and (b), thick vertical lines denote median, boxes report 75th and 25th percentiles, and whiskers indicate the maximal and minimal values. Each dot represents an individual mouse.
Supplementary Material
Unprocessed gel image
C57BL/6 sperm motility at 10 min
pi6em1/em1 sperm motility at 10 min
C57BL/6 sperm motility at 90 min
pi6em1/em1 sperm motility at 90 min
C57BL/6 sperm motility at 3 h
pi6em1/em1 sperm motility at 3 h
C57BL/6 sperm motility at 4 h
pi6em1/em1 sperm motility at 4 h
C57BL/6 sperm motility at 5 h
pi6em1/em1 sperm motility at 5 h
Supplementary Tables 1-6
Supplementary Note
Acknowledgements
We thank P. Cohen, K. Grive, and E. Crate at Cornell University for generously sharing protocols and advice on germ cell sorting and meiotic chromosome studies; H. Florman, P. Visconti, and M. Gervasi for sharing protocols and advice on sperm studies; the UMMS Transgenic Animal Modeling Core for advice on fertility test and embryo phenotype; the UMMS FACS core for advice on and help with germ cell sorting; the UMMS EM Core (supported by National Center for Research Resources Award SI0OD021580) for advice on and help with sperm transmission electron microscopy; and members of our laboratories for critical comments on the manuscript. This work was supported in part by National Institutes of Health grants GM65236 to P.D.Z. and P01HD078253 to P.D.Z. and Z.W.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Aravin AA et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31, 785–799 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis SH et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol 2, 174–181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista PJ et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31, 67–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das PP et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell 31, 79–90 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houwing S et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69–82 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Robine N et al. A broadly conserved pathway generates 3′ UTR-directed primary piRNAs. Curr Biol 19, 2066–2076 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozata DM et al. Evolutionarily Conserved Pachytene piRNA Loci are Highly Divergent among Modern Humans. Nature Ecology & Evolution in press (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chirn GW et al. Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals. PLoS Genet 11, e1005652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girard A, Sachidanandam R, Hannon GJ & Carmell MA A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Li XZ et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50, 67–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau NC et al. Characterization of the piRNA complex from rat testes. Science 313, 363–367 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Grivna ST, Beyret E, Wang Z & Lin H A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20, 1709–1714(2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ro S et al. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA 13, 1693–1702 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolcun-Filas E et al. A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 138, 3319–3330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W & Lin H Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2, 819–830 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Reuter M et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264–267 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Zheng K & Wang PJ Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet 8, e1003038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasik KA et al. RNF17 blocks promiscuous activity of PIWI proteins in mouse testes. Genes Dev 29, 1403–1415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castañeda J et al. Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J 33, 1999–2019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homolka D et al. PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep 12, 418–428 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Xu M et al. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol Reprod 79, 51–57 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Goh WS et al. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev 29, 1032–1044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res 25, 193–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gou LT et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res 24, 680–700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vourekas A et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 19, 773–781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palumbo G, Bonaccorsi S, Robbins LG & Pimpinelli S Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics 138, 1181–1197 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mével-Ninio M, Pelisson A, Kinder J, Campos AR & Bucheton A The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175, 1615–1624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bozzetti MP et al. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta subunit of casein kinase 2. Proc Natl Acad Sci U S A 92, 6067–6071 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics 124, 303–316 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics 107, 611–634 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robert V, Prud’homme N, Kim A, Bucheton A & Pélisson A Characterization of the flamenco region of the Drosophila melanogaster genome. Genetics 158, 701–713 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prud’homme N, Gans M, Masson M, Terzian C & Bucheton A flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139, 697–711 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pélisson A et al. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J 13, 4401–4411 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aravin AA et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11, 1017–1027 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Aravin AA et al. The small RNA profile during Drosophila melanogaster development. Dev Cell 5, 337–350 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Saito K et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461, 1296–1299 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Brennecke J et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Bourc’his D & Bestor TH Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Ahmadi A & Ng SC Fertilizing ability of DNA-damaged spermatozoa. J Exp Zool 284, 696–704 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Morris ID, Ilott S, Dixon L & Brison DR The spectrum of DNA damage in human sperm assessed by single cell gel electrophoresis (Comet assay) and its relationship to fertilization and embryo development. Hum Reprod 17, 990–998 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Lewis SE & Aitken RJ DNA damage to spermatozoa has impacts on fertilization and pregnancy. Cell Tissue Res 322, 33–41 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Aravin A et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Kuretake S, Kimura Y, Hoshi K & Yanagimachi R Fertilization and development of mouse oocytes injected with isolated sperm heads. Biol Reprod 55, 789–795 (1996). [DOI] [PubMed] [Google Scholar]
- 44.de Lamirande E, Leclerc P & Gagnon C Capacitation as a regulatory event that primes spermatozoa for the acrosome reaction and fertilization. Mol Hum Reprod 3, 175–194 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Florman HM & Storey BT Mouse gamete interactions: the zona pellucida is the site of the acrosome reaction leading to fertilization in vitro. Dev Biol 91, 121–130 (1982). [DOI] [PubMed] [Google Scholar]
- 46.Jin M et al. Most fertilizing mouse spermatozoa begin their acrosome reaction before contact with the zona pellucida during in vitro fertilization. Proc Natl Acad Sci U S A 108, 4892–4896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stauss CR, Votta TJ & Suarez SS Sperm motility hyperactivation facilitates penetration of the hamster zona pellucida. Biol Reprod 53, 1280–1285 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Suarez SS, Katz DF, Owen DH, Andrew JB & Powell RL Evidence for the function of hyperactivated motility in sperm. Biol Reprod 44, 375–381 (1991). [DOI] [PubMed] [Google Scholar]
- 49.Qi H et al. All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc Natl Acad Sci U S A 104, 1219–1223 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quill TA et al. Hyperactivated sperm motility driven by CatSper2 is required for fertilization. Proc Natl Acad Sci U S A 100, 14869–14874 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mortimer ST CASA—Practical Aspects. J. Androl 21, 515–524 (2000). [PubMed] [Google Scholar]
- 52.Goodson SG, Zhang Z, Tsuruta JK, Wang W & O’Brien DA Classification of mouse sperm motility patterns using an automated multiclass support vector machines model. Biol Reprod 84, 1207–1215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.González-Jara P et al. Optimization of the balance between effort and yield in unilateral surgical transfer of mouse embryos. Lab Anim 51, 622–628 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Sun-Wada GH et al. A proton pump ATPase with testis-specific E1-subunit isoform required for acrosome acidification. J Biol Chem 277, 18098–18105 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Huang TT et al. pH and protease control of acrosomal content stasis and release during the guinea pig sperm acrosome reaction. Biol Reprod 32, 451–462 (1985). [DOI] [PubMed] [Google Scholar]
- 56.Son SW et al. Prognostic significance and function of the vacuolar H+-ATPase subunit V1E1 in esophageal squamous cell carcinoma. Oncotarget 7, 49334–49348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung JJ et al. CatSpê regulates the structural continuity of sperm Ca2+ signaling domains and is required for normal fertility. Elife 6, e23082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown SG et al. Homozygous in-frame deletion in CATSPERE in a man producing spermatozoa with loss of CatSper function and compromised fertilizing capacity. Hum Reprod 33, 1812–1816(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gunawardane LS et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Mohn F, Handler D & Brennecke J piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348, 812–817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han BW, Wang W, Li C, Weng Z & Zamore PD Noncoding RNA. piRNA- guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gainetdinov I, Colpan C, Arif A, Cecchini K & Zamore PD A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol Cell 71, 775–790.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Post C, Clark JP, Sytnikova YA, Chirn GW & Lau NC The capacity of target silencing by Drosophila PIWI and piRNAs. RNA 20, 1977–1986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiuchi T et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509, 633–636 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Bartel DP Metazoan MicroRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Becker WR et al. High-Throughput Analysis Reveals Rules for Target RNA Binding and Cleavage byAG02. Mol Cell 75, 741–755.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wee LM, Flores-Jasso CF, Salomon WE & Zamore PD Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 151, 1055–1067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bartel DP MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borini A et al. Sperm DNA fragmentation: paternal effect on early post-implantation embryo development in ART. Hum Reprod 21, 2876–2881 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Sakkas D et al. Sperm nuclear DNA damage and altered chromatin structure: effect on fertilization and embryo development. Hum Reprod 13 Suppl 4, 11–19 (1998). [DOI] [PubMed] [Google Scholar]
- 71.Peaston AE et al. Retrotransposons regulate host genes in mouse oocytes and pre-implantation embryos. Dev Cell 7, 597–606 (2004). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 72.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Truett GE et al. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29, 52, 54 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Holloway JK, Sun X, Yokoo R, Villeneuve AM & Cohen PE Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J Cell Biol 205, 633–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cole F et al. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat Genet 46, 1072–1080 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nagy A, Gertsenstein MV, K & Behringer R Manipulating the mouse embryo, a laboratory manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2003). [Google Scholar]
- 77.Yanagimachi R, Yanagimachi H & Rogers BJ The use of zona-free animal ova as a test-system for the assessment of the fertilizing capacity of human spermatozoa. Biol Reprod 15, 471–476 (1976). [DOI] [PubMed] [Google Scholar]
- 78.Johnson A, Smith RG, Bassham B, Lipshultz LI & Lamb DJ The microsperm penetration assay: development of a sperm penetration assay suitable for oligospermic males. Fertil Steril 56, 528–534 (1991). [PubMed] [Google Scholar]
- 79.Talbot P, Summers RG, Hylander BL, Keough EM & Franklin LE The role of calcium in the acrosome reaction: an analysis using ionophore A23187. J Exp Zool 198, 383–392 (1976). [DOI] [PubMed] [Google Scholar]
- 80.Osman RA, Andria ML, Jones AD & Meizel S Steroid induced exocytosis: the human sperm acrosome reaction. Biochem Biophys Res Commun 160, 828–833 (1989). [DOI] [PubMed] [Google Scholar]
- 81.Arnoult C, Zeng Y & Florman HM ZP3-dependent activation of sperm cation channels regulates acrosomal secretion during mammalian fertilization. J Cell Biol 134, 637–645 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tateno H et al. Ca2+ ionophore A23187 can make mouse spermatozoa capable of fertilizing in vitro without activation of cAMP-dependent phosphorylation pathways. Proc Natl Acad Sci U S A 110, 18543–18548 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mortimer D, C. E. F. M. G. Specific labelling by peanut agglutinin of the outer acrosomal membrane of the human spermatozoon. J Reprod Fertil 81, 127–135 (1987). [DOI] [PubMed] [Google Scholar]
- 84.Fu Y, Wu PH, Beane T, Zamore PD & Weng Z Elimination of PCR duplicates in RNA-seq and small RNA-seq using unique molecular identifiers. BMC Genomics 19, 531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han BW, Wang W, Zamore PD & Weng Z piPipes: a set of pipelines for piRNA and transposon analysis via small RNA-seq, RNA-seq, degradome- and CAGE-seq, ChIP-seq and genomic DNA sequencing. Bioinformatics 31, 593–595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Addo-Quaye C, Eshoo TW, Bartel DP & Axtell MJ Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18, 758–762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang W et al. Slicing and binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol Cell 59, 819–830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Unprocessed gel image
C57BL/6 sperm motility at 10 min
pi6em1/em1 sperm motility at 10 min
C57BL/6 sperm motility at 90 min
pi6em1/em1 sperm motility at 90 min
C57BL/6 sperm motility at 3 h
pi6em1/em1 sperm motility at 3 h
C57BL/6 sperm motility at 4 h
pi6em1/em1 sperm motility at 4 h
C57BL/6 sperm motility at 5 h
pi6em1/em1 sperm motility at 5 h
Supplementary Tables 1-6
Supplementary Note
Data Availability Statement
All sequencing data are available through the National Center for Biotechnology Information Sequence Read Archive using accession number PRJNA634688.


