Abstract
Multisubunit-tethering complexes (MTCs) are large (250 to >750 kDa), conserved macromolecular machines that are essential for soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE)–mediated membrane fusion in all eukaryotes. MTCs are thought to organize membrane trafficking by mediating the initial long-range interaction between a vesicle and its target membrane and promoting the formation of membrane-bridging SNARE complexes. Previously, we reported the structure of the yeast Dsl1 complex, the simplest known MTC, which is essential for coat protein I (COPI) mediated transport from the Golgi to the endoplasmic reticulum (ER). This structure suggests how the Dsl1 complex might tether a vesicle to its target membrane by binding at one end to the COPI coat and at the other to ER-associated SNAREs. Here, we used X-ray crystallography to investigate these Dsl1–SNARE interactions in greater detail. The Dsl1 complex comprises three subunits that together form a two-legged structure with a central hinge. We found that distal regions of each leg bind N-terminal Habc domains of the ER SNAREs Sec20 (a Qb-SNARE) and Use1 (a Qc-SNARE). The observed binding modes appear to anchor the Dsl1 complex to the ER target membrane while simultaneously ensuring that both SNAREs are in open conformations, with their SNARE motifs available for assembly. The proximity of the two SNARE motifs, and therefore their ability to enter the same SNARE complex, will depend on the relative orientation of the two Dsl1 legs. These results underscore the critical roles of SNARE N-terminal domains in mediating interactions with other elements of the vesicle docking and fusion machinery.
Keywords: membrane trafficking, SNARE protein, X-ray crystallography, COPI, vesicle, yeast, multisubunit tethering complex, Dsl1 complex, CATCHR complex
Eukaryotic cells use vesicles to transport cargo between organelles and to the plasma membrane for exocytosis. These transport vesicles bear tail-anchored SNARE proteins that, in concert with complementary SNAREs in the target membrane, draw the two membranes into close apposition and facilitate membrane fusion. Each SNARE contains at least one SNARE motif, and sequence features within these motifs define four groups of SNAREs: Qa, Qb, Qc, and R (1, 2). Fusogenic SNARE complexes contain one SNARE motif of each type and form via the coupled folding and assembly of cognate SNARE motifs into parallel four-helical bundles. In addition to C-terminal transmembrane anchors and adjacent SNARE motifs, most SNAREs contain N-terminal regions that can regulate SNARE assembly and/or interact with other proteins. Many R-SNAREs contain N-terminal longin domains (3), whereas many Q-SNAREs contain N-terminal three-helix bundles known as Habc domains (4–7). Some Qa-SNAREs exhibit “closed” conformations in which the Habc domain folds back onto the SNARE motif, preventing it from entering into a SNARE complex (8–11). Habc domains are also common within the N-terminal regions of Qb- and Qc-SNAREs (6, 7, 12, 13), where the available evidence suggests that they function as protein–protein interaction modules (7, 14).
In vivo, SNAREs collaborate with other factors including Sec1/Munc18 (SM) proteins and multisubunit-tethering complexes (MTCs). SM proteins function as SNARE chaperones, regulating the assembly of SNARE complexes (11, 15–17), whereas MTCs act upstream of SNARE complex assembly, mediating the initial attachment of a vesicle to its target membrane (17–19). Vesicle tethering canonically involves interactions between MTCs and membrane-associated Rab proteins but can also involve MTC•SNARE, MTC•coat, MTC•golgin, and/or MTC•phospholipid interactions. MTCs also appear to participate directly in SNARE complex assembly and in formation of the fusion pore (20–24). As a step toward understanding these roles in greater detail, we focus here on structural studies of MTC•SNARE complexes. Only a few such structures have previously been reported, and in no case has it been possible to integrate them into a complete structure of the MTC (6, 12, 22, 25).
The largest family of MTCs is the CATCHR (complexes associated with tethering containing helical rods) family, whose members function largely in trafficking to and from the Golgi (19, 26, 27). The CATCHR-family complexes—GARP/EARP, exocyst, conserved oligomeric Golgi, and Dsl1—contain 3–8 subunits each. Many of these subunits share a similar tertiary structure, the so-called CATCHR fold, comprising a series of helical bundles (28). The CATCHR fold is also found in the synaptic vesicle priming protein Munc13 (29). With the exception of the exocyst complex, the CATCHR complexes are multilegged and flexible (20, 30, 31). These attributes seem well-suited for roles in vesicle docking and SNARE complex assembly.
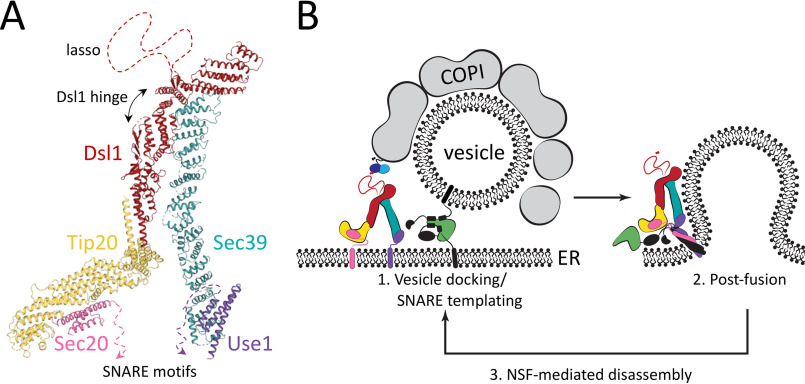
Of the five CATCHR-family MTCs, the Dsl1 complex is the smallest (∼250 kDa in Saccharomyces cerevisiae), the simplest (just three subunits), and the only one for which an essentially complete high-resolution structural model (based on overlapping crystal structures at resolutions ranging from 1.9 to 3.0 Å) is available (20, 32). The central subunit, Dsl1, bridges the other two subunits, Tip20 and Sec39, which each form an elongated leg. These legs, because of a flexible hinge within the Dsl1 subunit, are able to adopt a broad range of relative orientations (20). A distal portion of the Tip20 leg binds to the ER Qb-SNARE Sec20, whereas a distal portion of the Sec39 leg binds to the ER Qc-SNARE Use1 (20, 32). The three Dsl1 complex subunits and the two SNAREs form a stoichiometric complex in vitro and in vivo (20, 33, 34), consistent with the idea that the Dsl1 complex is stably anchored to the ER membrane via interactions of its two legs with two different ER-resident SNAREs. With its legs in a parallel orientation, the Dsl1 complex is ∼20 nm tall. At the apex of the complex, near the hinge, is an intrinsically disordered segment of the Dsl1 subunit called the lasso. The lasso contains singleton and ditryptophan motifs that bind to the COPI subunits α- and δ-COP, respectively (35–39). Thus, the Dsl1 complex, by interacting at its base with ER SNAREs and at its tip with COPI vesicles, may mediate vesicle capture upstream of SNARE complex formation.
The interactions between the Dsl1 lasso and COPI coat subunits have been relatively well-studied by methods including X-ray crystallography (38, 39). Here, we have used structural and biochemical experiments to reveal the molecular nature of the interactions between the two legs of the Dsl1 complex and the ER SNAREs Sec20 and Use1. We find that each interaction involves a trihelical region of the corresponding SNARE. The observed binding modes would prevent each SNARE from adopting the closed conformation that has been observed for Qa-SNAREs, thereby leaving its SNARE motif free to engage other SNAREs. Placed in the context of the intact Dsl1 complex, the structures we report are consistent with the previous observations that the Dsl1 complex accelerates SNARE complex assembly, albeit modestly, and can bind fully assembled SNARE complexes (20). More generally, our results highlight the critical roles of SNARE N-terminal domains in mediating interactions with other elements of the vesicle docking and fusion machinery.
Results
Tip20•Sec20NTD structure
The S. cerevisiae Dsl1 complex subunit Tip20 was discovered as a cytoplasmic protein that interacts with the cytoplasmic domain of the ER Qb-SNARE Sec20 (40). Biochemical experiments established that Tip20 binds an N-terminal region (residues 1–175), but not the SNARE motif, of Sec20 (20). Conversely, the N-terminal region of Tip20 (residues 1–81), which mediates binding to Dsl1, was not needed for binding to Sec20 (32). Therefore, we initially conducted crystallization screens using both full-length S. cerevisiae Tip20 and an N-terminally truncated version in combination with various constructs representing the N-terminal region of Sec20. Although a number of crystals were obtained, none of them diffracted well enough to allow structure determination. As an alternative approach, we screened orthologous Tip20•Sec20 complexes from other yeasts, co-expressed in bacteria. For this screen we used full-length Tip20 and an N-terminal fragment of Sec20 terminating just before the SNARE motif (Sec20NTD). Although most of the Tip20•Sec20NTD complexes were stable and soluble, only the Eremothecium gossypii complex yielded crystals. These initial crystals diffracted poorly, but replacing full-length Tip20 with an N-terminally truncated variant (Tip20A–E; Fig. 1A) led to the discovery of an additional crystal form that diffracted X-rays to 3.2 Å resolution.
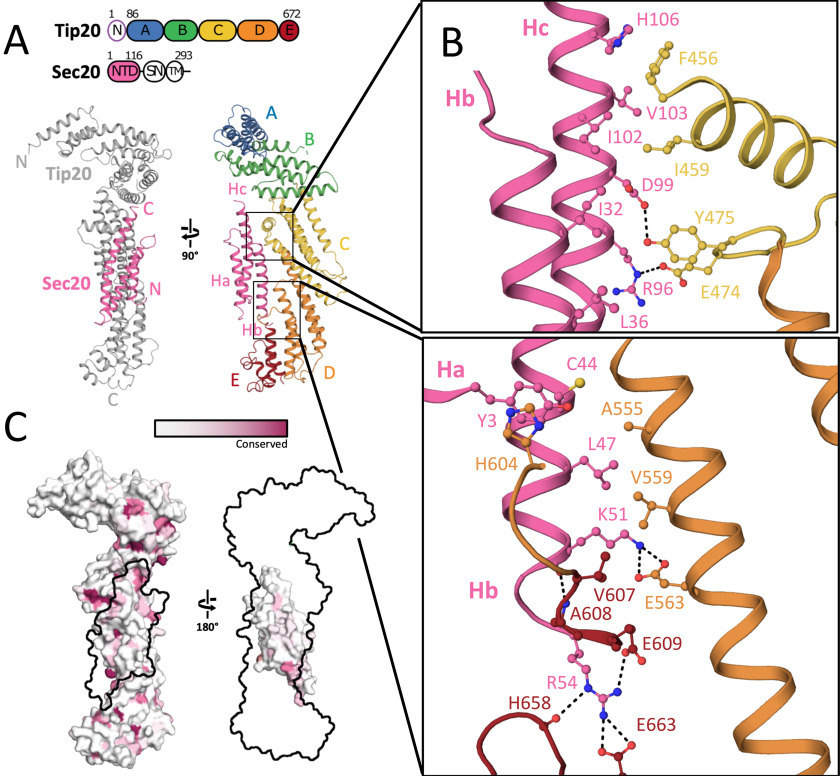
Figure 1.
Structure of E. gossypii Tip20A–E•Sec20NTD complex. A, Sec20NTD adopts an Habc fold, interacting with Tip20 domains C–E. The full domain architecture of the two proteins is cartooned above. The Dsl1 complex subunit Tip20 consists of an N-terminal Dsl1-interacting domain (N), followed by five CATCHR domains (A–E). The SNARE protein Sec20 consists of an N-terminal Habc domain (NTD), SNARE motif (SN), and transmembrane helix (T). B, details of the Tip20•Sec20 interface. C, the interacting surfaces of Tip20 and Sec20 are not highly conserved.
E. gossypii Tip20A–E•Sec20NTD was phased by molecular replacement using the previously reported S. cerevisiae Tip20 structure as a search model (32). In the resulting structure (Fig. 1A and Table S1), E. gossypii Tip20A–E displays the characteristic hooked structure that appears to set Tip20 apart from other CATCHR-fold proteins (32) (Fig. S1A). The greatest difference between E. gossypii Tip20 (in complex with Sec20NTD) and S. cerevisiae Tip20 (uncomplexed) lies in the angle between domains B and C (Fig. S1A). Although this might reflect an inherent difference between the two orthologues, it could also reflect intrinsic flexibility at the B/C domain junction. A comparable degree of flexibility near the B/C junction was also observed for the exocyst subunit Exo70 (41), whereas dramatic flexing near the B/C junction of the Dsl1 subunit gives rise to the hinge motion that allows the entire Dsl1 complex to open and close (20).
In addition to Tip20A–E, we observed electron density for three additional α-helices. Side-chain density allowed these helices to be unambiguously assigned to Sec20NTD (Fig. S1B). The helices adopt a canonical Habc fold, with a disordered loop (residues 59–77) connecting Hb and Hc. Also disordered is the C-terminal region of the crystallized Sec20 fragment (residues 113–136), corresponding to the linker between the Habc domain and the SNARE motif. Previously, structures of Habc domains from several Qa-SNAREs, one Qb-SNARE, and one Qc-SNARE have been reported (4–7, 12–14, 42). Among these, the Habc domain of the Qb-SNARE Vti1 (13) bears the closest resemblance to that of Sec20, with a root-mean-square deviation of 2.3 Å over 78 aligned Cα atoms (Fig. S1C). This structural similarity hints that Habc domains may be a broadly conserved feature of Qb-SNAREs, as they are of Qa-SNAREs.
Sec20NTD makes direct contact with Tip20 domains C–E, burying a surface of 1300 Å2 (Fig. 1A). The interface is split into two parts: one a contact between domain C of Tip20 and helices Ha and Hb of Sec20NTD and the other a contact between domains D and E of Tip20 and helices Hb and Hc of Sec20NTD (Fig. 1B). The centers of the two contact patches are formed by hydrophobic contacts between Ile-459 of Tip20 and Ile-102 of Sec20 and between Ala-555 of Tip20 and Leu-47 of Sec20, respectively. These residues are comparatively well-conserved, although Ala-555 is frequently replaced with a bulkier hydrophobic residue. The E. gossypii Tip20•Sec20 interface also features six salt bridges and ten hydrogen bonds (Fig. 1B).
Tip20•Sec20NTD binding
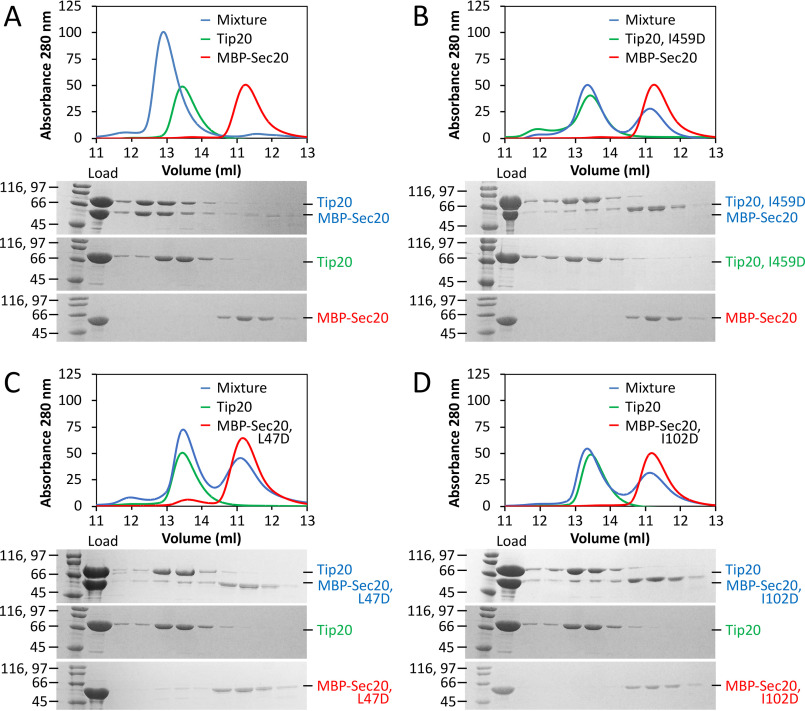
Unexpectedly, residues in the Tip20•Sec20NTD binding interface are not especially well-conserved, compared with the Tip20 and Sec20 surfaces overall (Fig. 1C), nor are they distinctive in terms of electrostatic surface potential or hydrophobicity (Fig. S2A). Therefore, we sought to verify that the interface observed in our crystal structure is indeed responsible for complex formation in solution. The PISA server (43) predicted that the observed interface has a high probability of being biologically relevant and further identified three of the four residues cited above (Ile-459 in Tip20 and Leu-47 and Ile-102 in Sec20; Fig. 1B) as among the residues contributing most favorably to the free energy of binding. We therefore substituted each of these residues, one at a time, with aspartate and used size-exclusion chromatography to evaluate Tip20•Sec20NTD complex formation. Binding of E. gossypii Tip20 to maltose-binding protein (MBP)–Sec20NTD was robust (Fig. 2A). Each of the three substitutions, however, abolished binding (Fig. 2, B–D). These results demonstrate that the interface we observe is not a crystallization artifact. We also characterized binding of the WT partners by isothermal titration calorimetry (ITC) and found that the dissociation constant is ∼100 nm (Table S2 and Fig. S2B).
Figure 2.
Structure-guided disruption of E. gossypii Tip20•Sec20 complex formation. A, size-exclusion chromatography demonstrates robust binding (blue) between WT E. gossypii Tip20 (green) and MBP-Sec20NTD (red). B–D, substitution of Tip20 Ile-459, Sec20 Leu-47, or Sec20 Ile-102 with aspartate abolishes binding of Tip20 to MBP-Sec20. The data presented for MBP-Sec20 alone in B and the data for Tip20 alone in C and D are identical to those in A.
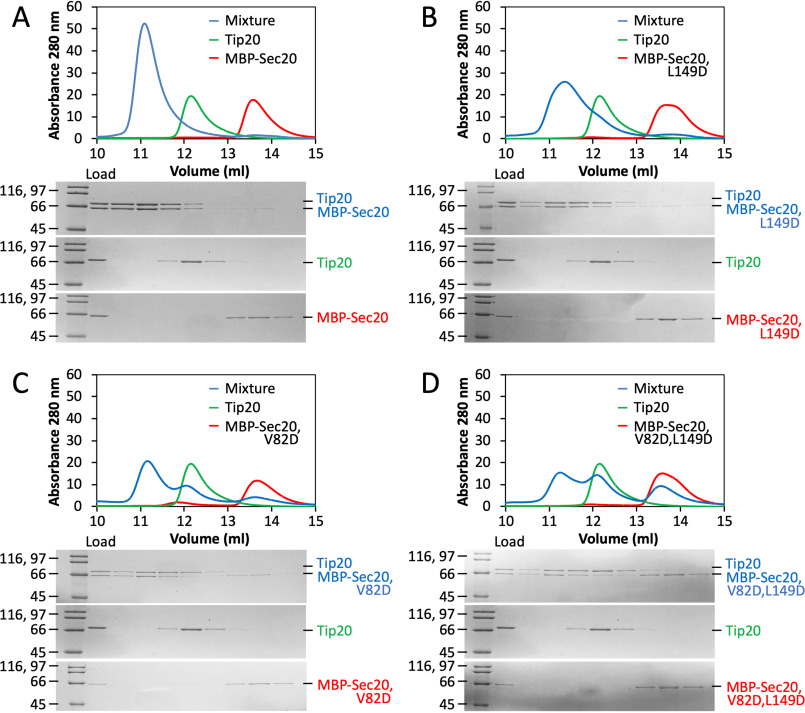
As noted above, Tip20 and Sec20NTD orthologues from a broad sampling of yeast species form stable complexes, suggesting that the interface may be conserved in spite of the modest sequence conservation of the interfacial residues. To test this more directly, we conducted binding experiments using S. cerevisiae proteins. Structurally equivalent residues in S. cerevisiae and E. gossypii Tip20 were identified on the basis of their X-ray structures. Unexpectedly, replacing S. cerevisiae Tip20 residues Ile-481 and Leu-585 (equivalent to E. gossypii Ile-459 and Ala-555) with aspartate, individually or in combination, had little effect on Tip20•Sec20NTD complex formation as judged by size-exclusion chromatography (Fig. S3). A more quantitative analysis using ITC revealed, however, that the double replacement led to a 15-fold increase in the equilibrium dissociation constant (Table S2 and Fig. S4). Predicting interface residues in S. cerevisiae Sec20NTD was more challenging; not only is the sequence homology low, but we lack a S. cerevisiae Sec20NTD structure to compare with E. gossypii Sec20NTD. Nonetheless, we found that changing S. cerevisiae Sec20 residue Val-82 (likely equivalent to E. gossypii Leu-47), alone or in combination with Leu-149 (likely equivalent to E. gossypii Ile-102), to aspartate modestly compromised Tip20•Sec20NTD complex formation as judged by size-exclusion chromatography (Fig. 3). Overall, our data suggest that interface mutations designed on the basis of the E. gossypii X-ray structure destabilized the S. cerevisiae complex, implying that in spite of the lack of strong sequence conservation, the interface is conserved. These results are suggestive of relatively rapid co-evolution of the interfacial residues.
Figure 3.
Structure-guided disruption of S. cerevisiae Tip20•Sec20 complex formation. A, size-exclusion chromatography demonstrates robust binding (blue) between WT S. cerevisiae Tip20 (green) and MBP-Sec20NTD (red). B, substitution of Sec20 Leu-149 with aspartate has little effect on complex formation. C, substitution of Sec20 Val-82 with aspartate partially compromises binding to Tip20. D, the two substitutions in combination have a stronger effect on binding. The data presented for Tip20 alone in B–D are identical to those in A.
To test the effect of weakening the Tip20•Sec20 interface in vivo, we used a plasmid shuffling strategy in S. cerevisiae to replace WT Tip20 or Sec20 with mutant versions. Deleting the N-terminal domain of Sec20 was lethal (Fig. 4). Although this result is consistent with the model that the Tip20•Sec20NTD interaction is essential in vivo, it is also possible that the Sec20 NTD plays an essential role that is independent of its binding to Tip20. Therefore, we attempted to confirm the model by replacing WT Tip20 or Sec20 with the double mutants characterized above. These substitutions did not, however, give rise to noticeable growth defects (Fig. 4 and Fig. S5). Thus, the affinity of Tip20 for Sec20 can be substantially reduced (∼15-fold for the structure-guided Tip20 mutations) without causing a major reduction in growth rate.
Figure 4.
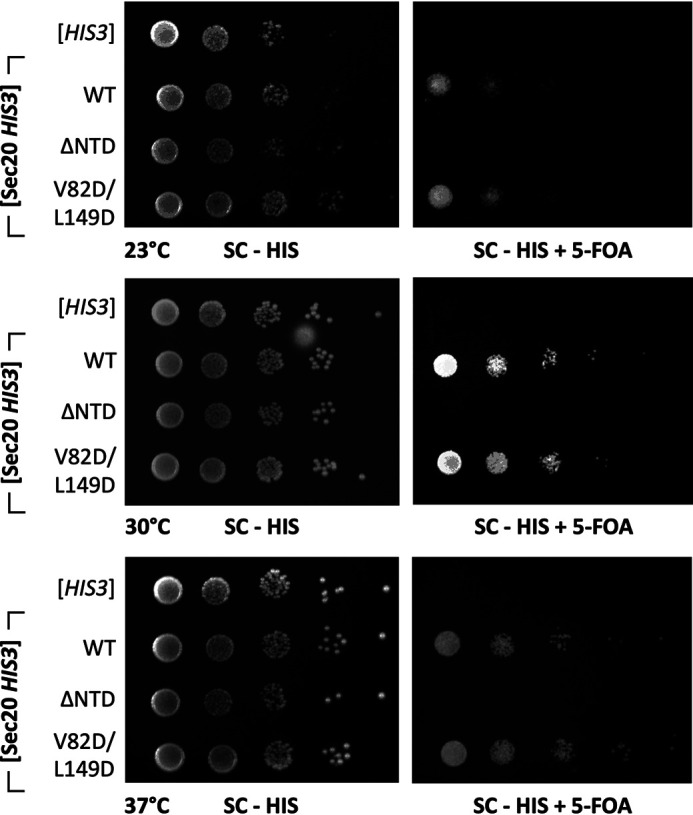
The N-terminal domain of Sec20 is essential for yeast viability. Yeast strains lacking endogenous Sec20 were maintained using a WT Sec20 covering plasmid marked with Ura3 and a second plasmid with the His3-linked Sec20 allele (SC - HIS) indicated at left. When the Ura3 plasmid is lost on 5-fluoroorotic acid (5-FOA) selective plates (right), yeast harboring Sec20 lacking its N-terminal domain (ΔNTD) were inviable at all temperatures tested. Structure-based mutations in the Tip20•Sec20 interface (Tip20 V82D/L149D), which increased the equilibrium dissociation constant by ∼15-fold (Table S2), did not have an evident impact on viability.
Low-resolution Sec39•Dsl1C–E•Use1NTD structure
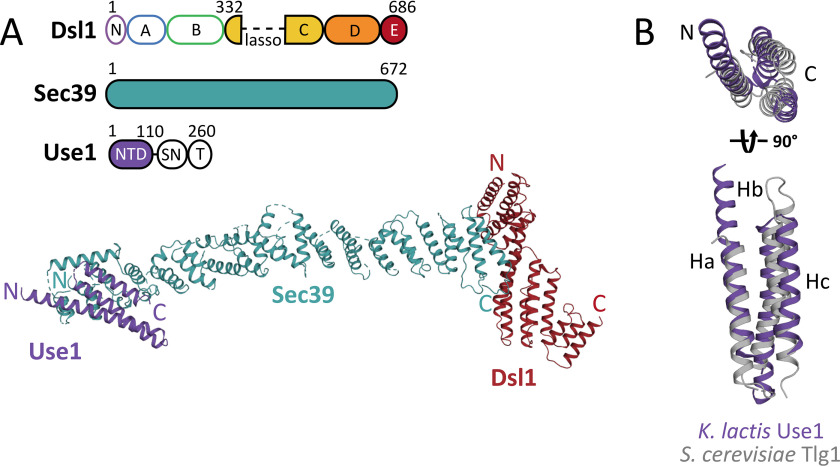
The Sec39 leg of the Dsl1 complex is anchored to the ER membrane through an interaction with an N-terminal region of the Qc-SNARE Use1. Previous work (20) demonstrated that the first 35 residues of S. cerevisiae Use1 are required for binding to Sec39 in vitro. After extensive orthologue screening and construct optimization, we were able to generate crystals of a complex containing Kluyveromyces lactis Sec39, Use1 (residues 1–110), and Dsl1 (domains C–E, with the lasso deleted) that diffracted to 6.5 Å resolution. The diffraction data were postprocessed using an anisotropic resolution cutoff (STARANISO, RRID:SCR_018362), which included data extending to 5.9 Å resolution (Table S1).The K. lactis Sec39•Dsl1C–E•Use1NTD data were phased using molecular replacement based on our previous Sec39•Dsl1C–E structure, which contained S. cerevisiae Sec39 (30% sequence identity with K. lactis Sec39) and K. lactis Dsl1C–E (20). In spite of the limited resolution of the data, a convincing molecular replacement solution was found in two steps: first placing the C-terminal portion of Sec39 and Dsl1C–E and then placing the N-terminal portion of Sec39. In the resulting model, the N- and C-terminal portions of Sec39 form a continuous α-solenoid (Fig. 5A).
Figure 5.
6.5 Å-resolution X-ray structure of K. lactis Sec39•Use1NTD•Dsl1C–E. A, the upper panel depicts the domain architecture of the three proteins crystallized. Dsl1 consists of an N-terminal Tip20-interacting domain (N) followed by five CATCHR domains (A–E). Sec39 consists of a long α-solenoid. For further details, see Fig. S6. The SNARE protein Use1 consists of an N-terminal domain (NTD), a SNARE motif (SN), and a transmembrane helix (T). In the lower panel, Use1 (purple) binds to the extreme N terminus of Sec39 (blue). B, K. lactis Use1 (purple) superimposes on its closest homologue, S. cerevisiae Tlg1 (gray, PDB accession code 2C5K, chain T) with an root-mean-square deviation of 4.3 Å over 75 residues of the structure.
Our earlier Sec39•Dsl1C–E structure lacked two regions near the N terminus of Sec39: residues 1–29 and 63–100. Re-examination of the earlier electron density map, however, revealed weak density consistent with two additional helical hairpins. These previously unmodeled hairpins were also visible in the Sec39•Use1NTD•Dsl1C–E map (hairpins 1 and 3 in (Fig. S6). Because side chains cannot be seen at this resolution, the corresponding α-helices (α1, α2, α5, and α6), as well as an extension of α7, were modeled as polyalanine. Also visible in the Sec39•Dsl1C–E•Use1NTD map was clear electron density for three additional α-helices located adjacent to the N-terminal portion of Sec39 (Fig. 5A and Fig. S7). This density has no counterpart in the earlier Sec39•Dsl1C–E map, where its position is blocked by a crystal contact, and was consistent with the secondary structure prediction that K. lactis Use1NTD is highly α-helical (Fig. 5B, Fig. S8). The closest homologue of Use1 with a known structure is the Habc domain of S. cerevisiae Tlg1, another Qc-SNARE (6). Therefore, we attempted molecular replacement with various Habc domains and obtained the highest-scoring solution by using S. cerevisiae Vti1 as a search model (13) (Fig. 5B, Fig. S7 and Table S3). Because it is based on low-resolution data, the resulting model of the complete K. lactis Sec39•Dsl1C–E•Use1NTD complex must be treated as preliminary. Nonetheless, it suggests that the interface between K. lactis Sec39 and Use1NTD primarily involves helices α1–2 and α5–7 of Sec39 and Hb of Use1. Overall, our results unambiguously demonstrate that Use1 binds the distal tip of Sec39 using a trihelical domain that is very likely to be an Habc domain.
Discussion
The Tip20•Sec20NTD and low-resolution Sec39•Dsl1C–E•Use1NTD structures are, to our knowledge, the first structures of full-length MTC subunits bound to SNAREs. These and our previous structures (20, 32) enabled us to construct a model for the complete Dsl1 complex anchored to the ER membrane by direct interactions between distal leg segments and ER SNAREs (Fig. 6A). The participating subunits—one (Tip20) a member of the CATCHR fold family, the other (Sec39) an α-solenoid—bind to N-terminal Habc domains of the Qb-SNARE (Sec20) and Qc-SNARE (Use1), respectively. In both the Tip20•Sec20NTD and low-resolution Sec39•Dsl1C–E•Use1NTD structures, the Hb/Hc groove of the SNARE NTD appears to be occluded by binding to the tethering complex subunit. It is not known whether uncomplexed Qb- and Qc-SNAREs adopt closed conformations analogous to those observed for Qa-SNAREs, in which the SNARE motif occupies the groove between the Hb and Hc helices of the Habc domain (9, 10, 22). However, in vivo the Dsl1 complex-anchoring SNAREs are almost certainly in open conformations, with their SNARE motifs accessible for SNARE assembly.
Figure 6.
Proposed model for Dsl1 complex–mediated COPI vesicle tethering and fusion. A, a composite model of the complete Dsl1 complex in complex with the ER SNAREs Sec20 and Use1 was generated by combining the current structures with previously reported structures of S. cerevisiae Dsl1 and Tip20 (PDB accession codes 3FHN and 3ETV). At the base of the complex, the two SNARE motifs can extend toward one another and toward the ER membrane. At the top of the complex are the COPI-interacting lasso and the Dsl1 flexible hinge. B, the Dsl1 complex, colored as in A, can adopt a range of conformations, positioning Sec20 and Use1 close enough to enter into the same SNARE bundle (right) or at a distance. Incoming vesicles are tethered by the Dsl1 lasso, and at later stages, a membrane-bridging complex is formed by the SM protein Sly1 (green) binding to SNAREs on the vesicle and ER membrane. The Dsl1 complex may remain associated with the same SNAREs through multiple cycles of SNARE assembly and Sec17/18–mediated disassembly.
The Dsl1 subunit hinge angle would appear to be the most important variable determining whether the Sec20 and Use1 SNARE motifs are sufficiently close to allow assembly. Conformations in which the legs are roughly parallel will hold the SNARE motifs in relatively close proximity and would be expected to facilitate their entry into a SNARE complex together. On the other hand, with its legs in a sufficiently splayed conformation, the Dsl1 complex could prevent the bound Sec20 and Use1 SNAREs from participating in the same SNARE complex. An intriguing alternative is that the bound SNAREs might instead enter into two different SNARE complexes; multiple Dsl1 complexes might even generate an array (for example, a ring) of SNARE complexes. In any case, it is clear that factors that influence the Dsl1 hinge angle—or otherwise affect the distance between Sec20NTD and Use1NTD—could have a major effect on SNARE assembly.
Testing the physiological significance of the two tether–SNARE interactions was complicated by our inability to abolish them via the structure-guided design of interface mutations. In addition, the bivalent attachment of the Dsl1 complex to the ER membrane via binding to two different ER SNAREs may allow for vesicle tethering even when one of the anchoring interactions is compromised. Nonetheless, potential support for the importance of each Dsl1–SNARE interaction in vivo comes from the lethal effect of deleting the Sec20 Habc domain (Fig. 4) and the strong temperature-sensitive growth defect caused by deleting the first 35 residues of Use1 (20). Both of these manipulations eliminate binding to the Dsl1 complex in vitro (20) but could in principle affect the localization and/or stability of the SNARE in vivo. In spite of this limitation, our results highlight the functional importance of N-terminal domains not only for Qa-SNAREs (8, 10, 45), but for Qb- and Qc-SNAREs as well.
In their unfolded states, the Sec20 and Use1 SNARE motifs represent flexible connectors between the Dsl1 complex and the ER membrane. Anchoring each leg of the Dsl1 complex via a flexible segment would appear to extend the reach of the Dsl1 complex as a vesicle tethering factor. With its legs in a parallel orientation, the Dsl1 complex itself has a height of ∼20 nm, and the disordered lasso at its tip extends this range further. The unfolded portion of the SNARE, at least 70 residues in length, would readily permit excursions of 10–15 nm from the ER membrane. Thus, the SNARE-anchored Dsl1 complex may be capable of capturing COPI vesicles that are 40 nm or more from the ER surface. Although this is a greater distance than SNAREs alone can reach (10–15 nm for unfolded SNARE motifs zippering from their N-terminal ends), it is far less than the tethering distance spanned by homodimeric coiled-coil tethering factors, which in fully extended conformations could reach 200 nm or more (46). Flexible regions within the Golgi coiled-coil tethering protein GCC185, however, allow its N and C termini to approach within ∼40 nm of one another (47). Another well-studied coiled-coil tethering protein, EEA1, undergoes entropic collapse upon vesicle binding (48), which could serve to pull the vesicle close enough to engage an MTC. In the case of the Dsl1 complex, retrograde trafficking from the Golgi to the ER has not been shown to require a coiled-coil tethering protein. It is possible that the need for long-range tethering is obviated by the close association of ER export and arrival sites. In the yeast Pichia pastoris, this creates bidirectional transport portals at which the Dsl1 complex may be able to capture COPI vesicles as they are budding from the Golgi (49).
What of SNARE assembly itself? The Dsl1 complex forms a stoichiometric complex in vivo with Sec20 and Use1 (33). Analogous complexes (containing the cytoplasmic portions of the SNAREs) are stable in solution and, in the additional presence of the Qa-SNARE Ufe1 and the R-SNARE Sec22, assemble to form a heptameric complex containing all four SNAREs and all three Dsl1 subunits (20). Based on these results and our current model for the anchored Dsl1 complex, we suggest that SNAREs undergo cycles of assembly and Sec17/18-catalyzed disassembly while remaining associated with the Dsl1 complex (Fig. 6B). In this way the Dsl1 complex, by directing vesicles to sites in which at least two of the three Q-SNAREs are already present, may be able sequentially to facilitate both tethering and SNARE assembly. It would, however, fall to other factors, such as the SM protein Sly1, to protect the assembling SNARE complexes from the disassembly activity of Sec17/18. Indeed, our results hint at a possible division of labor between two classes of protein—MTCs and SM proteins—known to chaperone SNARE complex assembly. As described here, the Dsl1 complex binds to Qb- and Qc-SNAREs in such a manner that their SNARE motifs are available for assembly. SM proteins, on the other hand, bind Qa-SNAREs, and recent studies of several SM proteins suggest that they bind R-SNAREs at an adjacent site and thereby function as templates to initiate SNARE assembly (16, 25). Thus, it is attractive to speculate that, upon close approach of the vesicle, its R-SNARE Sec22 engages the Sly1-bound Qa-SNARE Ufe1, to which the Dsl1 complex then presents the SNARE motifs of Sec20 and Use1 for the formation of a membrane-bridging trans-SNARE complex and subsequent membrane fusion.
Materials and methods
Protein expression and purification
A DNA fragment corresponding to E. gossypii Tip20 (residues 1–672) was amplified from genomic DNA by PCR and cloned directionally into the pQLinkH expression vector (50) using the BamHI/NotI restriction sites. Similarly, E. gossypii Sec20NTD (residues 1–136) was inserted into the pQLinkN vector. The two vectors were concatenated for co-expression using the LINK strategy (51). Subsequently, E. gossypii Tip20A–E (residues 86–676) was subcloned into pQLinkH and concatenated with Sec20NTD. The K. lactis Sec39•Use1NTD•Dsl1C–E expression vector was generated similarly, incorporating Sec39 into pQLinkH, Use1NTD (residues 1–110) into pQLinkN, and Dsl1C-E (residues 332–686, harboring an internal deletion of residues 367–423 replaced with the sequence Gly-Asp-Gly-Asp-Gly) into pQLinkN. The S. cerevisiae Tip20 and Sec201–174 expression constructs have been previously described (20, 32). For binding assays, E. gossypii Sec20NTD was subcloned into a modified pQLinkH expression vector containing an N-terminal MBP (25). All expression constructs were validated by DNA sequencing. Mutations were introduced using a modified QuikChange mutagenesis protocol (52).
All proteins were overproduced in BL21-CodonPlus(DE3) RIL E. coli (Agilent) grown in high salt Luria broth (Sigma) to an A600 of 0.5–0.7 and induced by the addition of 0.5–1 mm isopropyl β-d-1-thiogalactopyranoside. The cells were harvested after 16 h of induction at 15 °C; resuspended in 20 mm HEPES, pH 7.5, 150 mm NaCl (HBS) supplemented with 5 mm β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml DNase I (Roche); and lysed using a cell disrupter. After clarification, target proteins were purified from cell lysate by nickel–iminodiacetic acid affinity chromatography (Clontech). However, S. cerevisiae Tip20 was purified using TALON affinity chromatography (Clontech) in 20 mm Tris, pH 8.0, 200 mm NaCl (TBS). All proteins except S. cerevisiae Tip20 were further purified by anion-exchange chromatography (MonoQ, GE Healthcare). Finally, proteins were purified by size-exclusion chromatography (Superdex200 Increase, GE Healthcare). Purified proteins were concentrated, flash-frozen, and stored at −80 °C in TBS (S. cerevisiae Tip20) or HBS (all other proteins) supplemented with 1 mm DTT.
Crystallization and data collection
After screening orthologous complexes from Candida glabrata, Chaetomium thermophilum, E. gossypii, K. lactis, Kluyveromyces polyspora, S. cerevisiae, Schizosaccharomyces japonicus, Schizosaccharomyces pombe, Yarrowia lipolytica, and Zygosaccharomyces rouxii, crystals were obtained of the E. gossypii Tip20A-E•Sec20NTD heterodimeric complex by hanging-drop vapor diffusion at 20 °C, mixing 1 μl of protein (10 mg/ml) and 1 μl of well buffer (100 mm sodium citrate, pH 6.0, 725 mm ammonium sulfate, 1 mm DTT). After 3 days, rounded hexagonal crystals 100 × 100 × 100 μm were obtained and cryoprotected using a 1:1 mixture of formulation buffer to well buffer supplemented with 30% (v/v) glycerol before flash-freezing in liquid nitrogen. X-ray diffraction data were collected at the U.S. National Synchrotron Light Source II AMX Beamline.
After screening complexes from C. glabrata, K. lactis, K. polyspora, S. cerevisiae, S. japonicus, Schizosaccharomyces octosporus, S. pombe, Y. lipolytica, and Z. rouxii, crystals were obtained of the K. lactis Sec39•Use1NTD•Dsl1C–E heterotrimeric complex by hanging-drop vapor diffusion at 20 °C, mixing 1 μl of protein (5 mg/ml) and 1 μl of well buffer (100 mm Tris, pH 7.5, 100 mm NaCl, 8% (w/v) PEG 3,350, 10% (v/v) ethylene glycol, 1 mm DTT). After 1 or 2 days, thin plates 200 × 200 × 20 μm were obtained and cryoprotected in a solution containing 50 mm Tris, pH 7.5, 10 mm HEPES, pH 7.5, 125 mm NaCl, 11% (w/v) PEG 3,350, 30% (v/v) ethylene glycol and flash-frozen in liquid nitrogen. Diffraction data were collected at the National Synchrotron Light Source II FMX Beamline.
Structure determination and refinement
Tip20A–E•Sec20NTD data were processed using XDS (53). Initial search models for molecular replacement were derived from the S. cerevisiae Tip20 structure (PDB code 3FHN) (32) with nonconserved loops deleted and side chains pruned by the program CCP4 program CHAINSAW. Molecular replacement was carried out in PHASER (54), searching first for domains C–E and then separately for domain B. Rounds of refinement and manual building were performed using PHENIX.refine (55) and COOT, respectively (56).
Sec39•Dsl1C–E•Use1NTD data were processed in XDS, and an anisotropic resolution cutoff was applied (STARANISO, RRID:SCR_018362). Initial search models for molecular replacement were derived from the structure of S. cerevisiae Sec39 bound to K. lactis Dsl1332–686 (PDB code 3K8P) (20), modified by the CHAINSAW program. Molecular replacement in PHASER was carried in out in two steps: first searching for S. cerevisiae Sec39380–672 bound to K. lactis Dsl1332–686 and then searching for S. cerevisiae Sec391–379. Sec39 was maintained as two chains, one encompassing α1–α19 and the other encompassing α20–α34. One round of bulk solvent scaling, rigid body, and translation–libration–screw (TLS) refinement was performed in PHENIX.refine, assigning each chain as a single rigid body and a single TLS group. Four additional helices of Sec39 were built (α1, α2, α5, and α6) into unbuilt density in the K. lactis map, with reference to unbuilt regions of S. cerevisiae map. In addition, helices were extended or trimmed to fit the map and real-space rigid-body refinement was performed on poorly fit hairpins in COOT.
After a second round of refinement in PHENIX.refine, the updated model and map were used as a starting point for a new molecular replacement search in Phaser for each of the deposited Habc domain structures. The best solution, as judged by expected log likelihood gain, was found using S. cerevisiae Vti1 (13) (PDB code 3ONJ) (Table S3). The Vti1 model was adapted in COOT by extending the N-terminal helix and trimming the C-terminal helix to better reflect the density present in the K. lactis map. Each helix was rigid body–fit separately using COOT. Sec39 side chains were not modeled beyond Cα unless identical to S. cerevisiae Sec39, in which case the S. cerevisiae conformation was used. A final round of single-chain rigid body and TLS refinement was performed as above. To verify the position of Use1, an omit map was calculated omitting the entire Use1 chain.
Gel filtration binding assays
Proteins were diluted to 20 μm in HBS (E. gossypii proteins) or 8 μm in TBS (S. cerevisiae proteins) supplemented with 1 mm DTT in a volume of 200 μl, incubated at 4 °C for 1 h, and then loaded onto a Superdex 200 Increase 10/300 column. The fractions were collected and analyzed by 12% Tris/glycine SDS-PAGE.
Isothermal titration calorimetry
The proteins were exchanged into HBS (E. gossypii proteins) or TBS (S. cerevisiae proteins) supplemented with 1 mm DTT using pre-equilibrated BioSpin-6 spin columns (Bio-Rad) or, for Tip20L585D and Tip20I481D,L585D, 3,500-Da molecular mass cutoff Slide-A-Lyzer mini dialysis units (Thermo Scientific). The samples were then back-diluted to the indicated concentration. Tip20 was loaded into the sample cell, and Sec20 orthologues were loaded into the titration syringe. The experiments were performed using a MicroCal PEAQ–ITC (Malvern) and analyzed using the MicroCal PEAQ-ITC analysis software package (Malvern).
Yeast methods
A diploid strain heterozygous for the deletion of Sec20, marked with KanMX, was purchased from the essential gene knockout collection (Dharmacon). A Sec20-covering plasmid, containing the coding sequence of Sec20 and 500 bases upstream, was created based on the pRS416 Ura3-containing plasmid (57). After transformation of the covering plasmid (58), diploids were sporulated in sporulation medium containing 0.3% (w/v) potassium acetate and 0.02% (w/v) raffinose at 23 °C for 7 days. Haploid spores bearing a chromosomal deletion of Sec20 covered by the Ura3 plasmid were screened based on prototrophic and antibiotic resistance markers. Plasmids containing mutant alleles of Sec20, marked with His3, were then transformed into this haploid strain. Transformants were grown overnight at 30 °C in synthetic complete medium lacking histidine and then plated on synthetic complete agar lacking histidine, supplemented with 0.1% (w/v) 5-fluoroorotic acid (GoldBio) as indicated. The Tip20 deletion strain with Ura3-covering plasmid was created in an analogous manner. Mutants of Tip20 were introduced on a His3 plasmid and analyzed via 5-fluoroorotic acid counterselection.
Figures
Sequence alignments were generated using ClustalW (59) and rendered using JalView (60). Secondary structure prediction was performed in Jpred (61). Superimposition was performed using the CCP4 utility Superpose with secondary structure matching. Structures were rendered using PyMOL (Schrodinger), with conservation scores imported from ConSurf (62) where applicable. Hydrophobicity was assigned using the Eisenberg hydrophobicity scale (44).
Data availability
The structures presented within this work have been deposited in the Protein Data Bank (PDB) with the following accession codes: 6WC3 for Tip20A–E•Sec20NTD and 6WC4 for Sec39•Dsl1C–E•Use1NTD. All other data are available within the article.
Supplementary Material
Acknowledgments
We thank Angela Chan, Carly Geronimo, Venu Vandavasi, V. A. Zakian, and members of the Hughson laboratory past and present for valuable advice and discussions. The Princeton Biophysics and Macromolecular Crystallography core facilities provided essential assistance with isothermal titration calorimetry and X-ray crystallography, respectively. We gratefully acknowledge the assistance provided by the AMX and FMX Beamline staff at National Synchrotron Light Source II.
This article contains supporting information.
Author contributions—S. M. T., I.-M. Y., C. M., and F. M. H. conceptualization; S. M. T. data curation; S. M. T. formal analysis; S. M. T. and F. M. H. funding acquisition; S. M. T., K. D., I.-M. Y., C. M., S. H., G. R.-A., and P. D. J. investigation; S. M. T., P. D. J., and F. M. H. methodology; S. M. T. and F. M. H. writing-original draft; S. M. T., K. D., I.-M. Y., S. H., P. D. J., and F. M. H. writing-review and editing; F. M. H. project administration.
Funding and additional information—This work was supported by National Institutes of Health Grants T32GM007388 (to S. M. T. and K. D.), F31GM12676 (to S. M. T.), and R01GM071574 (to F. M. H.). This research used the AMX and FMX Beamlines of the National Synchrotron Light Source II, a U.S. Department of Energy Office of Science User Facility operated by Brookhaven National Laboratory under Contract DE-SC0012704. The Life Science Biomedical Technology Research resource, which supports AMX and FMX, is primarily supported by the NIGMS, National Institutes of Health through Biomedical Technology Research Resource P41 Grant P41GM111244, and by U.S. Department of Energy Office of Biological and Environmental Research Grant KP1605010. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- MTC
- multisubunit-tethering complex
- PDB
- Protein Data Bank
- SNARE
- soluble N-ethylmaleimide–sensitive factor attachment protein receptor
- COPI
- coat protein I
- ER
- endoplasmic reticulum
- SM
- Sec1/Munc18
- MBP
- maltose-binding protein
- ITC
- isothermal titration calorimetry
- TLS
- translation–libration–screw.
References
- 1. Fasshauer D., Sutton R. B., Brunger A. T., and Jahn R. (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. U.S.A. 95, 15781–15786 10.1073/pnas.95.26.15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kloepper T. H., Kienle C. N., and Fasshauer D. (2007) An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell 18, 3463–3471 10.1091/mbc.e07-03-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daste F., Galli T., and Tareste D. (2015) Structure and function of longin SNAREs. J. Cell Sci. 128, 4263–4272 10.1242/jcs.178574 [DOI] [PubMed] [Google Scholar]
- 4. Fernandez I., Ubach J., Dulubova I., Zhang X., Südhof T. C., and Rizo J. (1998) Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell 94, 841–849 10.1016/S0092-8674(00)81742-0 [DOI] [PubMed] [Google Scholar]
- 5. Dulubova I., Yamaguchi T., Wang Y., Südhof T. C., and Rizo J. (2001) Vam3p structure reveals conserved and divergent properties of syntaxins. Nat. Struct. Biol. 8, 258–264 10.1038/85012 [DOI] [PubMed] [Google Scholar]
- 6. Fridmann-Sirkis Y., Kent H. M., Lewis M. J., Evans P. R., and Pelham H. R. (2006) Structural analysis of the interaction between the SNARE Tlg1 and Vps51. Traffic 7, 182–190 10.1111/j.1600-0854.2005.00374.x [DOI] [PubMed] [Google Scholar]
- 7. Miller S. E., Collins B. M., McCoy A. J., Robinson M. S., and Owen D. J. (2007) A SNARE–adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature 450, 570–574 10.1038/nature06353 [DOI] [PubMed] [Google Scholar]
- 8. Nicholson K. L., Munson M., Miller R. B., Filip T. J., Fairman R., and Hughson F. M. (1998) Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat. Struct. Biol. 5, 793–802 10.1038/1834 [DOI] [PubMed] [Google Scholar]
- 9. Misura K. M., Scheller R. H., and Weis W. I. (2000) Three-dimensional structure of the neuronal–Sec1–syntaxin 1a complex. Nature 404, 355–362 10.1038/35006120 [DOI] [PubMed] [Google Scholar]
- 10. Munson M., Chen X., Cocina A. E., Schultz S. M., and Hughson F. M. (2000) Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat. Struct. Biol. 7, 894–902 10.1038/79659 [DOI] [PubMed] [Google Scholar]
- 11. Demircioglu F. E., Burkhardt P., and Fasshauer D. (2014) The SM protein Sly1 accelerates assembly of the ER–Golgi SNARE complex. Proc. Natl. Acad. Sci. U.S.A. 111, 13828–13833 10.1073/pnas.1408254111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abascal-Palacios G., Schindler C., Rojas A. L., Bonifacino J. S., and Hierro A. (2013) Structural basis for the interaction of the Golgi-associated retrograde protein complex with the t-SNARE syntaxin 6. Structure 21, 1698–1706 10.1016/j.str.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J., Gossing M., Fang P., Zimmermann J., Li X., von Mollard G. F., Niu L., and Teng M. (2011) Epsin N-terminal homology domains bind on opposite sides of two SNAREs. Proc. Natl. Acad. Sci. U.S.A. 108, 12277–12282 10.1073/pnas.1013101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Antonin W., Dulubova I., Arac D., Pabst S., Plitzner J., Rizo J., and Jahn R. (2002) The N-terminal domains of syntaxin 7 and vti1b form three-helix bundles that differ in their ability to regulate SNARE complex assembly. J. Biol. Chem. 277, 36449–36456 10.1074/jbc.M204369200 [DOI] [PubMed] [Google Scholar]
- 15. Ma C., Su L., Seven A. B., Xu Y., and Rizo J. (2013) Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 10.1126/science.1230473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiao J., He M., Port S. A., Baker R. W., Xu Y., Qu H., Xiong Y., Wang Y., Jin H., Eisemann T. J., Hughson F. M., and Zhang Y. (2018) Munc18-1 catalyzes neuronal SNARE assembly by templating SNARE association. eLife 7, e41771 10.7554/eLife.41771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baker R. W., and Hughson F. M. (2016) Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol. 17, 465–479 10.1038/nrm.2016.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gillingham A. K., and Munro S. (2019) Transport carrier tethering: how vesicles are captured by organelles. Curr. Opin. Cell Biol. 59, 140–146 10.1016/j.ceb.2019.04.010 [DOI] [PubMed] [Google Scholar]
- 19. Ungermann C., and Kümmel D. (2019) Structure of membrane tethers and their role in fusion. Traffic 20, 479–490 10.1111/tra.12655 [DOI] [PubMed] [Google Scholar]
- 20. Ren Y., Yip C. K., Tripathi A., Huie D., Jeffrey P. D., Walz T., and Hughson F. M. (2009) A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell 139, 1119–1129 10.1016/j.cell.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D'Agostino M., Risselada H. J., Lürick A., Ungermann C., and Mayer A. (2017) A tethering complex drives the terminal stage of SNARE-dependent membrane fusion. Nature 551, 634–638 10.1038/nature24469 [DOI] [PubMed] [Google Scholar]
- 22. Yue P., Zhang Y., Mei K., Wang S., Lesigang J., Zhu Y., Dong G., and Guo W. (2017) Sec3 promotes the initial binary t-SNARE complex assembly and membrane fusion. Nat. Commun. 8, 14236 10.1038/ncomms14236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dubuke M. L., Maniatis S., Shaffer S. A., and Munson M. (2015) The exocyst subunit Sec6 interacts with assembled exocytic SNARE complexes. J. Biol. Chem. 290, 28245–28256 10.1074/jbc.M115.673806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song H., Orr A. S., Lee M., Harner M. E., and Wickner W. T. (2020) HOPS recognizes each SNARE, assembling ternary trans-complexes for rapid fusion upon engagement with the 4th SNARE. eLife 9, e53559 10.7554/eLife.53559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baker R. W., Jeffrey P. D., Zick M., Phillips B. P., Wickner W. T., and Hughson F. M. (2015) A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 10.1126/science.aac7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whyte J. R., and Munro S. (2002) Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115, 2627–2637 [DOI] [PubMed] [Google Scholar]
- 27. Yu I. M., and Hughson F. M. (2010) Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 26, 137–156 10.1146/annurev.cellbio.042308.113327 [DOI] [PubMed] [Google Scholar]
- 28. Dong G., Hutagalung A. H., Fu C., Novick P., and Reinisch K. M. (2005) The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat. Struct. Mol. Biol. 12, 1094–1100 10.1038/nsmb1017 [DOI] [PubMed] [Google Scholar]
- 29. Pei J., Ma C., Rizo J., and Grishin N. V. (2009) Remote homology between Munc13 MUN domain and vesicle tethering complexes. J. Mol. Biol. 391, 509–517 10.1016/j.jmb.2009.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ha J. Y., Chou H. T., Ungar D., Yip C. K., Walz T., and Hughson F. M. (2016) Molecular architecture of the complete COG tethering complex. Nat. Struct. Mol. Biol. 23, 758–760 10.1038/nsmb.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chou H. T., Dukovski D., Chambers M. G., Reinisch K., and Walz T. (2016) CATCHR and HOPS/CORVET tethering complexes share a similar building plan. Nat. Struct. Mol. Biol. 23, 761–763 10.1038/nsmb.3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tripathi A., Ren Y., Jeffrey P. D., and Hughson F. M. (2009) Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat. Struct. Mol. Biol. 16, 114–123 10.1038/nsmb.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kraynack B. A., Chan A., Rosenthal E., Essid M., Umansky B., Waters M. G., and Schmitt H. D. (2005) Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol. Biol. Cell 16, 3963–3977 10.1091/mbc.e05-01-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meiringer C. T., Rethmeier R., Auffarth K., Wilson J., Perz A., Barlowe C., Schmitt H. D., and Ungermann C. (2011) The Dsl1 protein tethering complex is a resident endoplasmic reticulum complex, which interacts with five soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptors (SNAREs): implications for fusion and fusion regulation. J. Biol. Chem. 286, 25039–25046 10.1074/jbc.M110.215327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Andag U., and Schmitt H. D. (2003) Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J. Biol. Chem. 278, 51722–51734 10.1074/jbc.M308740200 [DOI] [PubMed] [Google Scholar]
- 36. Zink S., Wenzel D., Wurm C. A., and Schmitt H. D. (2009) A link between ER tethering and COP-I vesicle uncoating. Dev. Cell 17, 403–416 10.1016/j.devcel.2009.07.012 [DOI] [PubMed] [Google Scholar]
- 37. Hsia K. C., and Hoelz A. (2010) Crystal structure of α-COP in complex with ε-COP provides insight into the architecture of the COPI vesicular coat. Proc. Natl. Acad. Sci. U.S.A. 107, 11271–11276 10.1073/pnas.1006297107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suckling R. J., Poon P. P., Travis S. M., Majoul I. V., Hughson F. M., Evans P. R., Duden R., and Owen D. J. (2015) Structural basis for the binding of tryptophan-based motifs by δ-COP. Proc. Natl. Acad. Sci. U.S.A. 112, 14242–14247 10.1073/pnas.1506186112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Travis S. M., Kokona B., Fairman R., and Hughson F. M. (2019) Roles of singleton tryptophan motifs in COPI coat stability and vesicle tethering. Proc. Natl. Acad. Sci. U.S.A. 116, 24031–24040 10.1073/pnas.1909697116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sweet D. J., and Pelham H. R. (1993) The TIP1 gene of Saccharomyces cerevisiae encodes an 80 kDa cytoplasmic protein that interacts with the cytoplasmic domain of Sec20p. EMBO J. 12, 2831–2840 10.1002/j.1460-2075.1993.tb05944.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hamburger Z. A., Hamburger A. E., West A. P. Jr., and Weis W. I. (2006) Crystal structure of the S. cerevisiae exocyst component Exo70p. J. Mol. Biol. 356, 9–21 10.1016/j.jmb.2005.09.099 [DOI] [PubMed] [Google Scholar]
- 42. Lerman J. C., Robblee J., Fairman R., and Hughson F. M. (2000) Structural analysis of the neuronal SNARE protein syntaxin-1A. Biochemistry 39, 8470–8479 10.1021/bi0003994 [DOI] [PubMed] [Google Scholar]
- 43. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 44. Eisenberg D., Schwarz E., Komaromy M., and Wall R. (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179, 125–142 10.1016/0022-2836(84)90309-7 [DOI] [PubMed] [Google Scholar]
- 45. Gao G., and Banfield D. K. (2020) Multiple features within the syntaxin Sed5p mediate its Golgi localization. Traffic 21, 274–296 10.1111/tra.12720 [DOI] [PubMed] [Google Scholar]
- 46. Gillingham A. K., and Munro S. (2016) Finding the Golgi: golgin coiled-coil proteins show the way. Trends Cell Biol. 26, 399–408 10.1016/j.tcb.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 47. Cheung P. Y., Limouse C., Mabuchi H., and Pfeffer S. R. (2015) Protein flexibility is required for vesicle tethering at the Golgi. eLife 4, e12790 10.7554/eLife.12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murray D. H., Jahnel M., Lauer J., Avellaneda M. J., Brouilly N., Cezanne A., Morales-Navarrete H., Perini E. D., Ferguson C., Lupas A. N., Kalaidzidis Y., Parton R. G., Grill S. W., and Zerial M. (2016) An endosomal tether undergoes an entropic collapse to bring vesicles together. Nature 537, 107–111 10.1038/nature19326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roy Chowdhury S. R., Bhattacharjee C., Casler J. C., Jain B. K., Glick B. S., and Bhattacharyya D. (2020) ER arrival sites associate with ER exit sites to create bidirectional transport portals. J. Cell Biol. 219, e201902114 10.1083/jcb.201902114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Scheich C., Kümmel D., Soumailakakis D., Heinemann U., and Büssow K. (2007) Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 35, e43 10.1093/nar/gkm067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alexandrov A., Vignali M., LaCount D. J., Quartley E., de Vries C., De Rosa D., Babulski J., Mitchell S. F., Schoenfeld L. W., Fields S., Hol W. G., Dumont M. E., Phizicky E. M., and Grayhack E. J. (2004) A facile method for high-throughput co-expression of protein pairs. Mol. Cell. Proteomics 3, 934–938 10.1074/mcp.T400008-MCP200 [DOI] [PubMed] [Google Scholar]
- 52. Liu H., and Naismith J. H. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 10.1186/1472-6750-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 57. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., and Hieter P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 10.1016/0378-1119(92)90454-W [DOI] [PubMed] [Google Scholar]
- 58. Gietz R. D., and Schiestl R. H. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 10.1038/nprot.2007.13 [DOI] [PubMed] [Google Scholar]
- 59. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Drozdetskiy A., Cole C., Procter J., and Barton G. J. (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 10.1093/nar/gkv332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., and Ben-Tal N. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 10.1093/nar/gkw408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures presented within this work have been deposited in the Protein Data Bank (PDB) with the following accession codes: 6WC3 for Tip20A–E•Sec20NTD and 6WC4 for Sec39•Dsl1C–E•Use1NTD. All other data are available within the article.