Abstract
Pseudomonas aeruginosa exhibits a high requirement for iron, which it can acquire via several mechanisms, including the acquisition and utilization of heme. The P. aeruginosa genome encodes two heme uptake systems, the heme assimilation system (Has) and the Pseudomonas heme utilization (Phu) system. Extracellular heme is sensed via the Has system, which encodes an extracytoplasmic function (ECF) σ factor system. Previous studies have shown that the transfer of heme from the extracellular hemophore HasAp to the outer membrane receptor HasR is required for activation of the σ factor HasI and upregulation of has operon expression. Here, employing site-directed mutagenesis, allelic exchange, quantitative PCR analyses, immunoblotting, and 13C-heme uptake experiments, we delineated the differential contributions of the extracellular FRAP/PNPNL loop residue His-624 in HasR and of His-221 in its N-terminal plug domain required for heme capture to heme transport and signaling, respectively. Specifically, we show that substitution of the N-terminal plug His-221 disrupts both signaling and transport, leading to dysregulation of both the Has and Phu uptake systems. Our results are consistent with a model wherein heme release from HasAp to the N-terminal plug of HasR is required to initiate signaling, whereas His-624 is required for simultaneously closing off the heme transport channel from the extracellular medium and triggering heme transport. Our results provide critical insight into heme release, signaling, and transport in P. aeruginosa and suggest a functional link between the ECF σ factor and Phu heme uptake system.
Keywords: heme coordination, heme assimilation system (Has), cell surface signaling, transcriptional activation, heme uptake, HasR, iron uptake, Pseudomonas heme utilization (Phu), extracytoplasmic function (ECF), outer membrane, heme, bacterial signal transduction, bacterial transcription, cell surface receptor, heme assimilation system
Iron is an essential micronutrient that is required for the survival and virulence of almost all bacterial pathogens. Iron is extremely limited in mammalian hosts because of sequestration by the innate immune system through upregulation of iron storage proteins, downregulation of ferritin, and the secretion of iron-chelating proteins such as lipocalin-2 (1). During infection, bacterial pathogens, including Pseudomonas aeruginosa, circumvent iron deficiency by utilizing a variety of iron acquisition systems (2–4). In response to iron starvation, P. aeruginosa deploys two siderophore systems, pyoverdine (Pvd) and pyochelin (Pch) (5, 6), as well as the ferrous uptake system (Feo) (7). P. aeruginosa also encodes two nonredundant heme acquisition systems, the Pseudomonas heme uptake (Phu) and heme assimilation (Has) systems, that utilize heme as an iron source (8, 9). Therefore, bacteria must be able to adapt to utilize the most readily available iron source. One mechanism by which bacteria respond and adapt to extracellular nutrients is through cell surface signaling (CSS) systems, which are coupled to extracytoplasmic function (ECF) σ factors (10, 11). These alternative σ factors complex with the core RNA polymerase, direct binding to the promoter of a target gene, and activate transcription. P. aeruginosa encodes several iron-responsive ECF σ factors, including PvdS and FpvI, which are coupled to the pyoverdine uptake system (12–14). Heme uptake systems associated with ECF σ factor systems are conserved across a number of bacterial pathogens, including Serratia marcescens (15, 16), Bordetella pertussis (17), and P. aeruginosa (8).
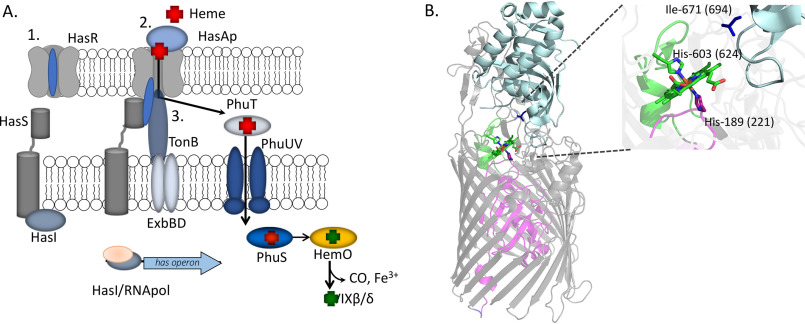
The P. aeruginosa Has system senses heme through the interaction of a secreted extracellular hemophore, HasAp, which scavenges and releases heme to the outer membrane receptor HasR (Fig. 1A). Capture of the heme by the N-terminal plug domain of HasR results in inactivation of the anti-σ factor HasS and release of the σ factor HasI. HasI then binds to the hasR promoter, recruits the core RNA polymerase, and upregulates transcription of the has operon (Fig. 1A). Simultaneously, heme released to HasR is transported through the receptor by the TonB-dependent coupling of the proton-motive force of the cytoplasmic membrane. The Has system does not encode a periplasmic transport system, so heme is sequestered and translocated to the cytoplasm by the PhuT-PhuUV periplasmic ATP-dependent binding cassette (ABC) transport system (Fig. 1A). Within the cytoplasm, heme is trafficked to the iron-dependent heme oxygenase (HemO) by the cytoplasmic heme-binding protein PhuS. HemO catalyzes the oxidative cleavage of heme to release CO, iron, and the heme metabolites biliverdin IXβ (BVIXβ) and IXδ (BVIXδ) (18). In addition to heme-dependent transcriptional activation of the Has system, the hasAp transcript is subject to posttranscriptional regulation by the heme metabolites BVIXβ and/or BVIXδ (19). The several layers of transcriptional and posttranscriptional regulation over the Has system allow the bacteria to rapidly fine-tune its response to changes in the host environment (19, 20). We have further shown the P. aeruginosa Has and Phu systems have nonredundant roles, where the Has system is primarily required for sensing, whereas the Phu system is the major transporter (9). The significance of heme sensing and uptake to P. aeruginosa pathogenesis is evident in the fact that clinical isolates from patients with chronic lung infection adapt to utilize heme while decreasing their ability to biosynthesize pyoverdine (21). In an attempt to dissect the mechanism of heme signaling and transport by the P. aeruginosa Has system, we have employed site-directed mutagenesis, allelic exchange, quantitative PCR (qPCR), immunoblotting, and 13C-heme uptake studies to determine the contributions of the heme coordinating ligands of HasR (Fig. 1). Previous studies employing similar approaches with variants of the extracellular hemophore HasAp showed heme release to HasR is required to activate the cell surface signaling cascade (20).
Figure 1.
Heme-dependent cell surface signaling and transport. A, schematic of ECF factor activation and heme transport. 1, in the absence of heme, the HasR receptor N-terminal plug occludes the periplasm from the extracellular environment. 2, holo-HasAp, on interaction with HasR, releases heme, triggering conformational rearrangement of the N-terminal plug driving interaction with the anti-σ factor HasS. This event triggers the release of the σ factor HasI, recruitment of the core RNA polymerase, and transcriptional activation of the has operon. 3, TonB on interaction with the heme-loaded HasR partially displaces the N-terminal plug, facilitating heme transport to the periplasm. Subsequently, heme is sequestered by PhuT and translocated to the cytoplasm by PhuUV, where it is trafficked by PhuS to HemO for degradation and iron release. B, structure of the S. marcescens holo-HasAp-HasR complex showing HasAp (light blue) and HasR with the N-terminal plug (magenta) and β-barrel (gray). The heme (green) and heme-coordinating residues His-189 (magenta), His-603 on FRAP/PNPNL loop (green), and Ile-671 (dark blue) are shown as sticks. The numbers in parentheses represent the corresponding residues in P. aeruginosa HasR. The image was generated from PDB file 3CSL in PyMOL (30).
The P. aeruginosa HasR protein has significant sequence identity (50%) to that of S. marcescens. The S. marcescens HasAs-HasR structure revealed His-603 of the FRAP/PNPNL loop (so called for the conserved amino acid motifs in the heme receptors) and His-189 of the N-terminal plug as the heme coordinating residues of HasR (Fig. 1B). Following heme release and dissociation of HasAp, heme coordination through His-603 was proposed to close off the extracellular milieu from the HasR heme transport channel. Despite structural determination of the HasAs-HasR complex, the molecular mechanism of heme signaling and transport has remained elusive. Here, we show the corresponding FRAP/PNPNL loop residue in P. aeruginosa HasAp His-624 is required for heme transport (Fig. 1B). In contrast, the N-terminal plug His-221 is critical for both heme signaling and transport. Based on the HasAs-HasR structure, Ile-671, located on extracellular loop L8, was proposed to prevent heme backsliding to HasAp. We further show that the corresponding Ile-694 in P. aeruginosa HasR is also essential for heme transport (Fig. 1B). Interestingly, heme bound to HasAp is not accessible to the Phu system, as shown by the significant growth defect in the transport-deficient HasR mutants when supplemented with holo-HasAp rather than heme. Interestingly, the N-terminal plug hasRH221R strain, which is unable to either activate the CSS cascade or transport heme, revealed a global dysregulation in both the Has and Phu system, indicating a direct or indirect link between the ECF σ factor HasI and the Phu heme uptake system.
Results
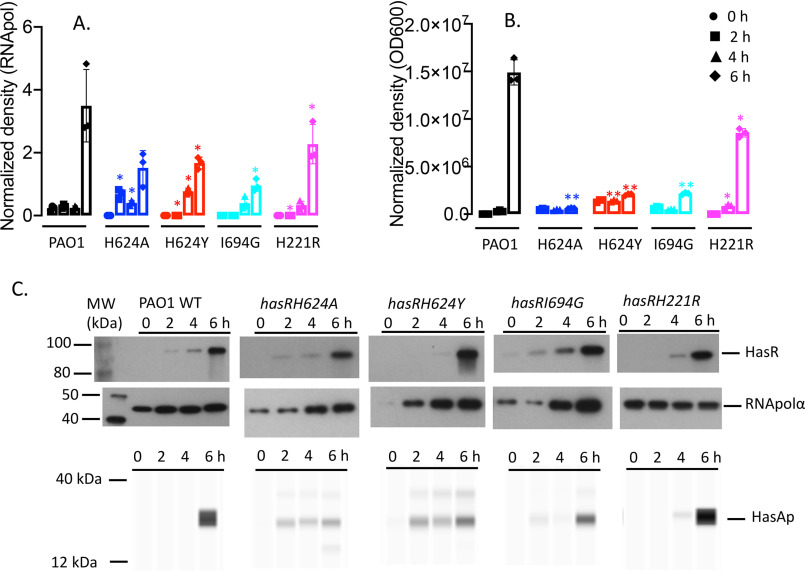
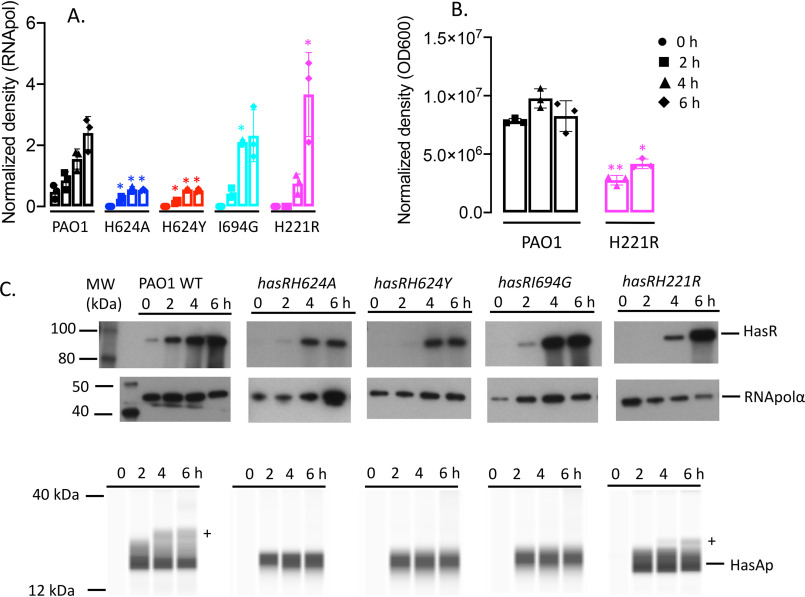
The hasR allelic strains show similar HasR and HasAp expression profiles in low iron
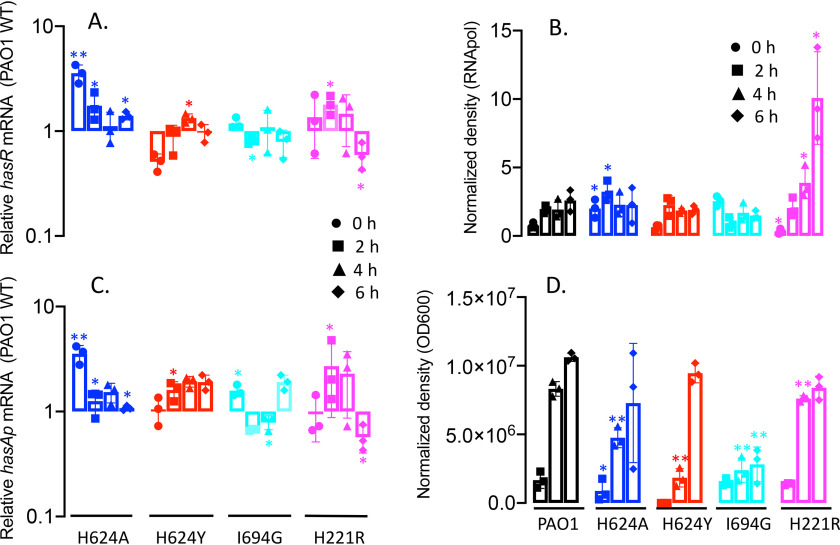
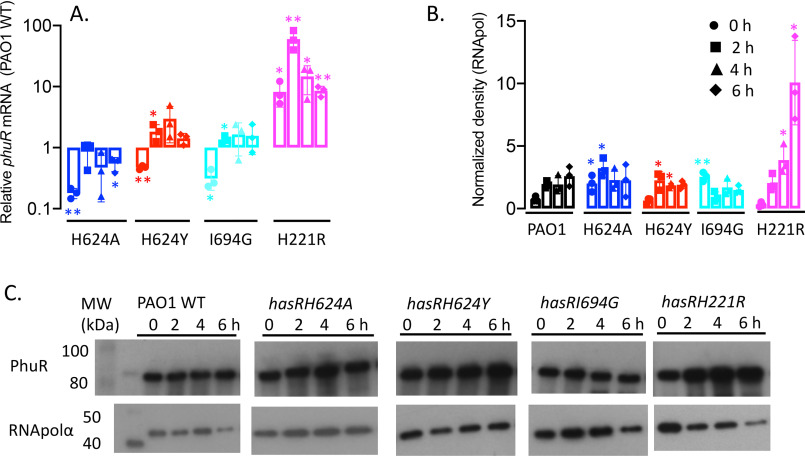
The expression profiles of the hasR allelic strains were first analyzed under iron-depleted conditions. The hasR variant strains all show a growth profile similar to that of PAO1 WT (Fig. 2A). For the hasR loop mutant strains, the hasR mRNA levels at each time point were all within 2-fold of the parent PAO1 strain, with negligible differences in protein expression (Fig. 3, A and B, and Fig. S1A). The data confirm the HasR loop variants are folded and stable. Similarly, the hasAp mRNA and protein levels for the hasR loop variants were very similar to those in the PAO1 WT strain (Fig. 3, C and D, and Fig. S1B). In contrast to the hasR loop variants, the N-terminal plug hasRH221R strain showed a significant increase in HasR protein at the later time points (Fig. 3B and Fig. S1A). To confirm that the expression of the Phu system was not compromised in the hasR variant strains, we analyzed the mRNA and protein levels by qPCR and Western blotting, respectively. The loop hasRH624A, H624Y, and I694G variants showed mRNA and protein profiles similar to that of the PAO1 WT strain (Fig. 4). Interestingly, the relative mRNA levels of phuR in the hasRH221R strain revealed a significant increase over the PAO1 WT (Fig. 4A). Furthermore, this increase in relative mRNA levels translated to protein expression (Fig. 4B). Overall, the data suggest that the hasRH221R variant, even under low-iron conditions, has a phenotype distinct from that of the hasR loop variants.
Figure 2.
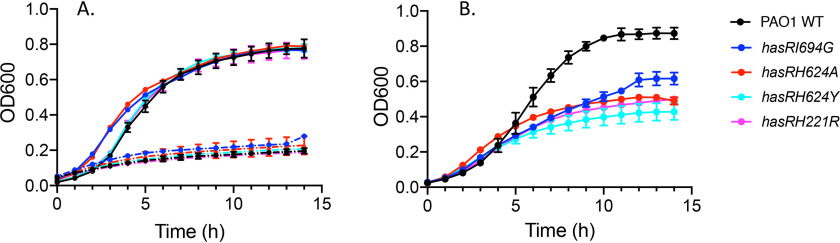
Growth curves of PAO1 WT and the hasR allelic strains. A, strains were grown in M9 minimal media (dashed line) or M9 supplemented with 1 μm heme (solid line). B, cells grown in M9 minimal media supplemented with 1 μm holo-HasAp. Strains are color coded as shown.
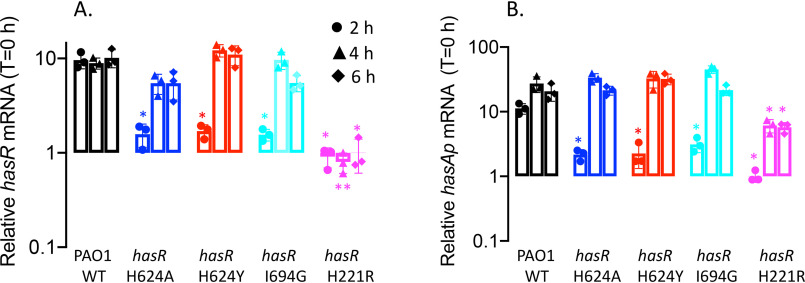
Figure 3.
Relative HasR and HasAp mRNA and protein levels for the hasR allelic strains under iron-depleted conditions. A, hasR mRNA isolated at 0, 2, 4, and 6 h following growth in M9 minimal media. mRNA values represent the mean from three biological experiments, each performed in triplicate and normalized to PAO1 WT at the same time point. Error bars represent the standard deviations from three independent experiments performed in triplicate. The indicated p values were normalized to mRNA levels of PAO1 WT at the same time point: *, p < 0.05. B, Western blot analysis of PAO1 WT and the hasR allelic strains. For HasR, total protein (5 μg) was loaded in each well. RNApolα was used as a loading control. Normalized density (n = 3) was performed for three separate biological replicates. The indicated p values were normalized to PAO1 at the same time point: *, p < 0.05; **, p < 0.005. C, hasAp mRNA analyzed as in panel A. D, Western blot analysis of HasAp as in panel B. Extracellular supernatant (4 μl) was loaded and run on the automated Wes capillary Western system as described in Experimental procedures. Representative Western blot images are shown in the supporting information (Fig. S1).
Figure 4.
Relative phuR mRNA and protein levels for the hasR allelic strains under iron-depleted conditions. A, mRNA isolated at 0, 2, 4, and 6 h following growth in M9 minimal media. mRNA values represent the means from three biological experiments, each performed in triplicate and normalized to PAO1 WT at the same time point. Error bars represent the standard deviations from three independent experiments performed in triplicate. The indicated p values were normalized to mRNA levels of PAO1 WT at the same time point: *, p < 0.05; **, p < 0.005. B, normalized density (n = 3) was determined on Western blots for three separate biological replicates. The indicated p values were normalized to PAO1 WT at the same time point: *, p < 0.05; **, p < 0.005. C, representative Western blotting of PAO1 WT and the hasR allelic strains. Total protein (5 μg) was loaded in each well. RNApolα was used as a loading control.
The hasR allelic strains are deficient in heme uptake
The growth of the hasRH624A, hasH624Y, hasI694G, and hasRH221R strains was monitored following supplementation with either 1 μm heme or 1 μm holo-HasAp. All of the mutant strains were competent to utilize free heme as an iron source compared with the growth rates in iron-depleted media (Fig. 2A). However, when heme was supplied in the form of holo-HasAp, a significant inhibition in growth was observed for all of the variant strains compared with that of PAO1 WT (Fig. 2B). These results suggest that heme, but not heme complexed to HasAp, is accessible to the hasR variant strains via the Phu heme uptake system. qPCR analysis of phuR on heme supplementation showed that the relative mRNA levels of the hasR loop variants are similar to that of PAO1 WT at all time points (Fig. S2). However, the relative mRNA levels in the hasRH221R variant are 10-fold higher than those of PAO1 at all time points, consistent with the profile under iron-depleted conditions. The PhuR protein levels for PAO1 WT and all of the hasR variant strains follow a similar profile, where the protein is induced at 2 h, followed by a decrease at 4 h and a subsequent increase again at 6 h. This pattern of expression we interpret as a shock response on the addition of free heme, as we do not observe this effect when heme is complexed to HasAp. However, despite statistically significant differences in protein expression levels, as determined by Western blotting, it is clear that all of the hasR variant strains express PhuR protein within 2-fold of the level observed for PAO1 WT (Fig. S2, B and C).
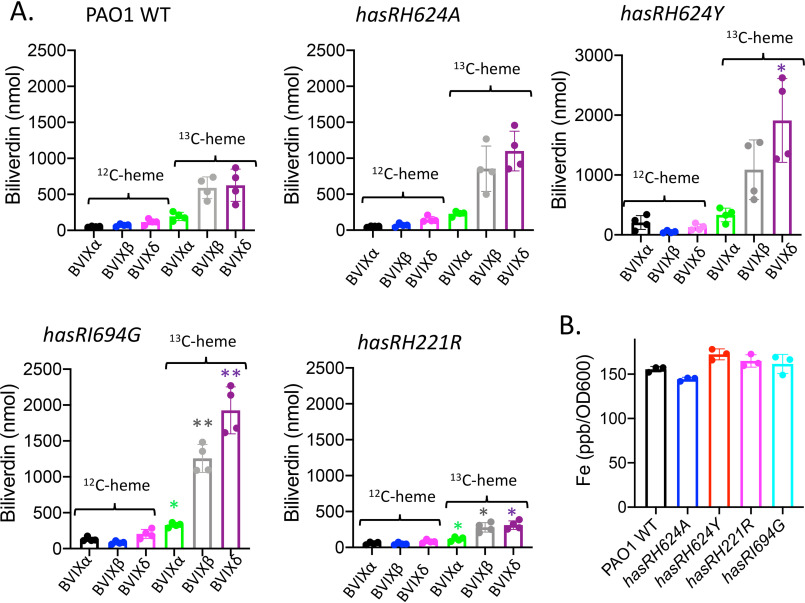
The ability of the hasR variant strains to utilize heme but not holo-HasAp was confirmed by isotopically labeled 13C-heme uptake and inductively coupled plasma mass spectrometry (ICP-MS) experiments. Supernatants were collected 4 h following supplementation of PAO1 WT or the hasR allelic strains with either 13C-heme or 13C-heme-HasAp. The resulting BVIX metabolites were extracted and quantified by LC-MS/MS. When supplemented with 13C-heme, the hasR variant strains were all capable of taking up and degrading heme, as judged by the similar levels of 13C-BVIXβ and 13C-BVIXδ compared that of PAO1 WT (Fig. 5A). Consistent with the ability to utilize exogenously added 13C-heme, ICP-MS analysis confirmed the intracellular iron content of the hasR variant strains is similar to that of PAO1 WT (Fig. 5B). Taken together, the data suggest that in the absence of a functional Has system, free heme is readily acquired by the major transporter PhuR.
Figure 5.
Heme utilization by the hasR allelic strains when supplemented with 13C-heme. A, LC-MS/MS analysis of the BVIX isomers supplemented with 1 μm 13C-heme at 4 h. Biliverdin values represent the standard deviations from four biological replicates. The indicated p values are for each strain compared to the PAO1 WT for the respective BVIX isomers: *, p < 0.05; **, p < 0.005. B, ICP-MS analysis of intracellular iron levels. The indicated p values are for each strain compared with PAO1 WT: *, p < 0.05; **, p < 0.005.
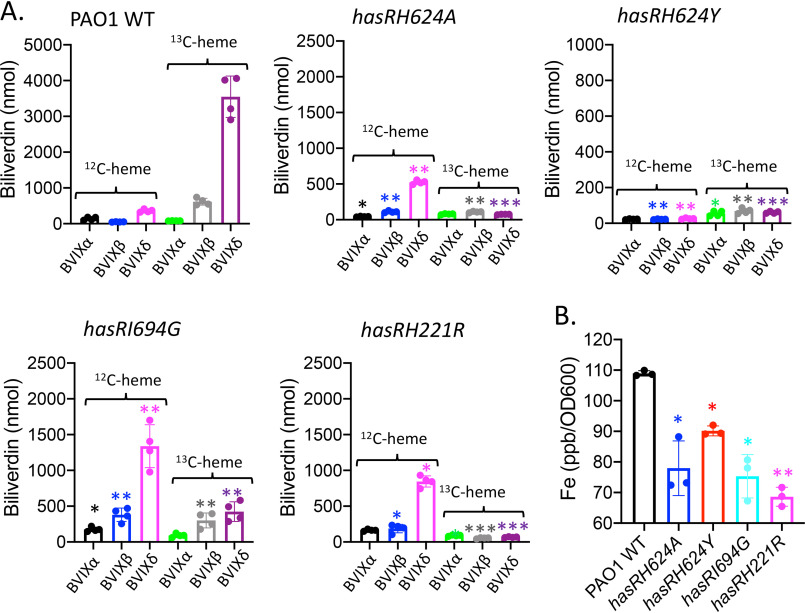
In contrast, the hasR variant strains were unable to utilize the 13C-heme–HasAp complex as an iron source, as indicated by the lack of 13C-BVIXβ and BVIXδ metabolites (Fig. 6A). Concomitant with the decreased ability to utilize extracellular heme, we see a slight but significant increase in endogenously derived BVIX (12C-heme) metabolites. Such redistribution of endogenous heme iron is consistent with the lower growth rate and dysregulation of iron homeostasis (Fig. 2B). The inability to utilize heme bound to HasAp was further confirmed on measurement of the intracellular iron levels by ICP-MS (Fig. 6B). The hasR variant strains all showed decreased intracellular iron content, which is particularly noticeable for the H221R N-terminal plug mutant. Analysis of phuR mRNA levels in the hasR loop variants are within 2-fold of those for PAO1 WT (Fig. S3). Western blotting further confirmed protein levels mirror those of PAO1 (Fig. S3, B and C). However, as can be seen from the qPCR analysis, the relative mRNA levels for all time points are significantly higher for the hasRH221R plug variant (Fig. S3). This difference is not solely because of the decreased iron levels, as iron depletion is also observed for the hasR loop variants (Fig. 6B). Furthermore, we observed a similar mRNA profile for the hasR variant strains in cultures supplemented with heme, where the intracellular iron levels were identical to that of PAO1 WT (Fig. 4B). However, the increase in mRNA levels when supplemented with holo-HasAp is not reflected in significant changes in protein levels (Fig. S3). Taken together, the data clearly show heme bound to HasAp, in the absence of a functional HasR transporter, is not accessible to PhuR. This is consistent with previous studies in which we have shown the Has and Phu systems have distinct nonredundant roles in heme signaling and transport, respectively (9).
Figure 6.
Heme utilization by the hasR allelic strains when supplemented with 13C-heme-HasAp. A, LC-MS/MS analysis of the BVIX isomers supplemented with 1 μm 13C-heme-HasAp at 4 h. Biliverdin values represent the standard deviations from four biological replicates. The indicated p values are for each strain compared with PAO1 WT for the respective BVIX isomers: *, p < 0.05; **, p < 0.005. B, ICP-MS analysis of intracellular iron levels. The indicated p values are for each strain compared with PAO1 WT: *, p < 0.05; **, p < 0.005.
Activation of ECF σ factor system by the hasR H624A, H624Y, I694G, and H221R allelic strains
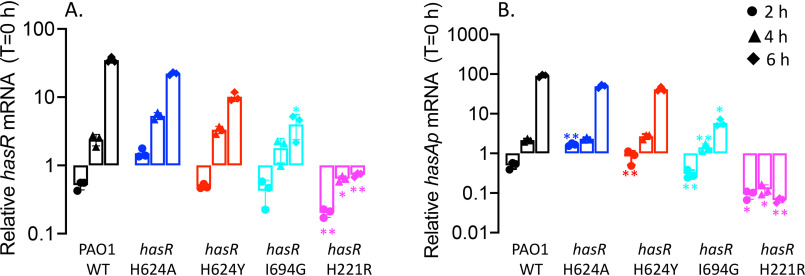
We next sought to determine the contributions of the extracellular loop and N-terminal plug residues on activation of the CSS cascade. HasI-dependent transcriptional activation of hasR and hasAp following supplementation with heme or holo-HasAp was monitored over time by qPCR analysis and Western blotting. As previously reported for supplementation of PAO1 WT with 1 μm heme, following an initial decrease in mRNA, we observed an increase in hasR mRNA levels over time as extracellular HasAp levels accumulate (Fig. 7A). The hasAp mRNA profiles mirror those of hasR; however, as previously reported, the relative mRNA levels are greater than those of hasR because of increased mRNA stability following processing of the hasRAp transcript (Fig. 7B) (20). Western blot analysis of HasR and HasAp protein levels are consistent with the respective mRNA profiles (Fig. 8).
Figure 7.
Relative hasR and hasAp mRNA levels for the hasR allelic strains under heme-supplemented conditions. A, hasR relative mRNA levels. B, hasAp relative mRNA levels. mRNA isolated at 0, 2, 4, and 6 h following growth in M9 supplemented with 1 μm heme. mRNA values represent the mean from three biological experiments, each performed in triplicate and normalized to PAO1 WT at the same time point. Error bars represent the standard deviations from three independent experiments performed in triplicate. The indicated p values were normalized to mRNA levels of PAO1 WT at the same time point: *, p < 0.05; **, p < 0.005.
Figure 8.
Relative HasR and HasAp protein levels for the hasR allelic strains under heme-supplemented conditions. A, HasR protein at 0, 2, 4, and 6 h following growth in M9 minimal media supplemented with 1 μm heme. Total protein (5 μg) was loaded in each well. RNApolα was used as a loading control. Normalized density (n = 3) was performed on Western blots for three separate biological replicates. B, as in panel A but for HasAp. Extracellular supernatant (4 μl) was loaded for analysis on the automated Wes capillary system as described in Experimental procedures. Normalized density (n = 3) was corrected for differences in OD600 for three biological replicates. Error bars represent the standard deviations from three independent experiments performed in triplicate. Indicated p values for the hasR allelic strains normalized to PAO1 WT at the same time point: *, p < 0.05; **, p < 0.005. C, representative Western blotting of PAO1 WT and the hasR allelic strains for HasR and digital HasAp images generated by capillary Western blot analysis on an automated Wes system.
The hasR loop variants show a similar profile but slightly lower hasR and hasAp mRNA and protein levels compared with those of the WT (Fig. 7 and 8). The increase in relative mRNA levels on addition of heme suggests that despite an inability to transport heme, they retain the ability to activate the CSS cascade (Fig. 7). In contrast, the hasRH221R strain showed no heme-dependent transcriptional activation of hasR and hasAp (Fig. 7). This lack of transcriptional activation is not solely the result of decreased HasAp levels in the extracellular medium, as we observed significant protein levels by Western blotting (Fig. 8). Previous studies from our laboratory have shown hasAp is subject to positive posttranscriptional regulation by the heme metabolites BVIXβ and/or BVIXδ (19, 20). Therefore, the discrepancy between mRNA and protein levels can be rationalized by the fact that heme actively taken up by the Phu system is further metabolized to BVIXβ and BVIXδ.
We further analyzed the transcriptional activation of the has operon on supplementation with a fixed concentration of holo-HasAp. In these studies, we utilized a previously characterized functional but truncated HasAp to distinguish, by Wes analysis, the supplemented holo-HasAp from the endogenously produced protein (19). In contrast to the lag on addition of heme (∼4 h; Fig. 7), addition of holo-HasAp to PAO1 immediately activates transcription from the hasR promoter (Fig. 9A). For PAO1 WT, transcriptional activation peaks at the earlier 2-h time point and is maintained over 6 h. In contrast, the hasR loop variants show decreased transcriptional activation at the early time points. The lag in transcriptional activation for the loop variants is further manifested at the protein level, where significantly higher HasR expression is observed for PAO1 WT at the earlier time points (Fig. 10A). Whereas the hasAp mRNA levels for the hasR loop variants follow a trend similar to those of hasR (Fig. 9B), we do not detect any endogenous HasAp protein (Fig. 10B). This lack of protein expression can be rationalized by the inability of the hasR loop variants to take up heme, leading to lower intracellular BVIX levels (Fig. 6A) and decreased HasAp translational efficiency. For the N-terminal plug hasRH221R strain, we observed no transcriptional activation of the hasR operon, as shown by the lack of increase in hasR mRNA over time (Fig. 9A). As expected, we also observed no transcriptional activation of hasAp at the 2-h time point (Fig. 9B). However, the accumulation of hasAp mRNA at the later time points is a consequence of both the inherent stability of the hasAp mRNA transcript versus hasR and derepression of Fur as a consequence of iron deficiency (Fig. 6B). Indeed, the relative increase in hasR and hasAp mRNA levels over time is also observed for the hasR loop variants, suggesting iron deficiency also contributes to some degree to the increase in transcription. Furthermore, this translates to HasR expression, where we observe an increase in protein levels at the 4- and 6-h time points (Fig. 10A). Particularly noticeable in the hasRH221R strain are the detectable levels of HasAp (Fig. 10B) despite the fact that we see no accumulation of BVIX metabolites (Fig. 6A). Although this could be attributed to the lower intracellular iron levels (Fig. 6B), the lack of detectable HasAp protein in the hasR loop variants (that are also iron deficient) does not support this. Interestingly, we initially attempted to construct the hasRH221A variant strain; however, on sequencing we noted an inversion of the DNA sequence within this region that led to a significant change in the amino acid sequence (data not shown). This suggested the hasRH221A mutation introduced structural changes within the plug that most likely resulted in the strain not being viable. These observations may account for the detectable levels of HasAp in the hasRH221R variant, where subtle conformational changes within the N-terminal plug may lead to destabilization of the anti-σ factor HasS. Furthermore, the increase in PhuR mRNA and protein levels in the hasRH221R strain over both the WT and hasR loop variants under all conditions is consistent with a global dysregulation in iron homeostasis on disruption of the CSS cascade.
Figure 9.
Relative hasR and hasAp mRNA levels for the hasR allelic strains under holo-HasAp-supplemented conditions. A, hasR relative mRNA levels. B, hasAp relative mRNA levels. mRNA was isolated at 0, 2, 4, and 6 h following growth in M9 supplemented with 1 μm holo-HasAp. mRNA values represent the means from three biological experiments, each performed in triplicate and normalized to 0 h for each strain. Error bars represent the standard deviations from three independent experiments performed in triplicate. The indicated p values were normalized to mRNA levels of PAO1 WT at the same time point: *, p < 0.05; **, p < 0.005.
Figure 10.
Relative HasR and HasAp protein levels for the hasR allelic strains under holo-HasAp-supplemented conditions. A, HasR protein at 0, 2, 4, and 6 h following growth in M9 minimal media supplemented with 1 μm holo-HasAp. B, as in panel A but for HasAp. Normalized density (n = 3) was performed on Western blots for three separate biological replicates. Error bars represent the standard deviations from three independent experiments performed in triplicate. The indicated p values for HasR were normalized to the RNApolα subunit as a loading control and to the OD600 for HasAp. Indicated p values are for the hasR allelic strains normalized to PAO1 WT at the same time point: *, p < 0.05; **, p < 0.005. C, representative Western blotting of PAO1 WT and the hasR allelic strains for HasR and HasAp. HasAp digital images were generated by capillary Western blot analysis on an automated Wes system. Endogenously produced HasAp is denoted by a plus sign. The lower band represents exogenously added truncated holo-HasAp.
Discussion
The crystal structure of the S. marcescens HasAs-HasR complex revealed His-603 of the extracellular FRAP/PNPNL loop (L7) and His-189 on the N-terminal plug axially coordinated the heme (Fig. 1) (22). Based on the crystal structure and previous NMR studies of HasAs, heme transfer was proposed to occur through a transient intermediate where His-32 of HasAs is displaced on protein–protein interaction with HasR, whereas the stronger Tyr-75 ligand remains coordinated because of deprotonation of Tyr-75 by the nearby His-83 (22, 23). In subsequent steps, steric hindrance by Ile-671 of HasR and transient protonation of Tyr-75 by His-83 promotes release of heme to HasR. It was also suggested that Ile-671 sterically hinders heme from backsliding into the HasAs hydrophobic pocket (Fig. 1). However, our recent studies with the P. aeruginosa ΔhasAp strain supplemented with exogenous holo-HasAp H32A, Y75A, or H83A mutant proteins showed an increase in the transcriptional activation of the CSS cascade compared with the WT strain (20). The heme content in the holo-HasAp WT and mutant proteins was identical, suggesting that the increase in transcriptional activation by the HasAp mutants resulted from a kinetically trapped intermediate. Therefore, the data do not support the sequential release of His-32 followed by protonation and release of Tyr-75 but rather a concerted mechanism where both His-32 and Tyr-75 are simultaneously released through a conformational rearrangement and free energy gain on interaction with HasR (20). To further study the mechanism of heme signaling and uptake, we turned our attention to the role of the outer membrane receptor HasR. Specifically, we have interrogated the role of the heme ligands His-221 of the N-terminal plug, His-624 of the FRAP/PNPNL loop, and the extracellular loop (L8) Ile-694, proposed to promote heme release from HasAp.
The hasRH624A strain, on supplementation with holo-HasAp, was unable to utilize heme, as judged by a lack of 13C-heme metabolites and decreased intracellular iron levels (Fig. 6), but still was able to trigger the activation of the CSS cascade (Fig. 9). The data indicate that heme coordination by the N-terminal plug His-221 is sufficient to trigger the CSS cascade, although significantly less efficiently than the WT strain. The data further suggest heme coordination with HasR via the FRAP/PNPNL loop His-624 and N-terminal plug His-221 is required to trigger conformational rearrangement of the plug to facilitate heme transport while simultaneously closing off the receptor channel from the extracellular environment.
To further test the role of heme coordination on heme signaling and transport, we mutated the FRAP/PNPNL loop His-624 to a Tyr mimicking the heme coordination motif of PhuR (24). Similarly, we observed the hasRH624Y strain on supplementation with holo-HasAp was capable of triggering the CSS cascade but deficient in heme uptake. Given that heme complexed to HasAp is not accessible to the PhuR receptor (Fig. 2 and 6), one might have expected making the receptor more “PhuR-like” would disrupt heme signaling and transport. However, the fact that we observed the activation of the CSS cascade (Fig. 9) shows heme can be displaced from holo-HasAp. Therefore, the inability of PhuR to extract heme from HasAp is more likely because of the lack of a direct protein–protein interaction. However, somewhat surprisingly, we observed no active heme uptake, indicating either (i) Tyr-624 is sterically hindered from coordinating with the heme and facilitating loop closure or (ii) the Tyr-His coordination is not able to trigger the conformational rearrangement of the N-terminal plug required for transport. Biochemical and spectroscopic characterization of the purified HasR variants reconstituted in lipid nanodiscs is currently underway to determine their respective heme coordination properties.
Interestingly, despite the fact that the FRAP/PNPNL heme coordinating residue His-624 and the N-terminal plug His-221 remain intact, the hasRI694G strain, when supplemented with holo-HasAp, is unable to transport heme (Fig. 6). Similar to the His-624 mutants, qPCR analysis revealed the hasRI694G strain still was capable of activating the CSS cascade, albeit with an apparent decrease in efficiency, as judged by the lower relative mRNA levels than that of the WT (Fig. 7 and 9). The data are consistent with the loss of Ile-694 allowing for transient release of heme from HasAp to the receptor, such that heme signaling is triggered, but heme is not sterically constrained to allow loop closure and transport. We conclude that both His-624 and Ile-694 are essential for facilitating heme release, capture, and transport. Ongoing biochemical and spectroscopic characterization of the purified HasR variants reconstituted in lipid nanodiscs will shed more light on the effect of the FRAP/PNPNL loop mutations on heme binding and coordination.
In contrast to the hasR loop variant strains, the hasRH221R strain was deficient in both heme signaling (Fig. 7 and 9) and transport (Fig. 6). It is unlikely that this is because holo-HasAp does not interact with the HasR H221R receptor, given that the hasR loop variants are functional in activating the signaling cascade. Therefore, heme coordination to His-221 is absolutely required for transduction of the signal from the periplasmic face of the N-terminal plug to the cytoplasmic signaling domain. Taken together, the data are consistent with a model where conformational rearrangement on interaction of holo-HasAp with HasR, facilitated by the steric clash of Ile-694, triggers heme release to HasR. Following release, heme binding to the HasR N-terminal plug via His-221 triggers the signal through the N-terminal domain, inactivating the anti-σ factor HasS, releasing the σ factor HasI, and transcriptionally activating the has operon (Fig. 1A). Simultaneous release of HasAp and loop closure activates heme transport through the receptor channel to the periplasmic face.
Interestingly, even in the hasRH221R strain, we observe a phenotype distinct from that of the hasR extracellular loop variant strains, including significant differences in the expression profile of PhuR. As previously mentioned, under iron-depleted conditions the relative mRNA and protein levels of PhuR are significantly higher than those of either the WT or hasR extracellular loop variants (Fig. 3). Furthermore, on closer analysis of the PhuR Western blots supplemented with either heme or holo-HasAp, a doublet can be observed in what appears to be posttranscriptional or proteolytic processing (Fig. S2 and S3). Taken together, the data suggest that the inactivation of the CSS cascade, and the cell's ability to sense extracellular heme, has a direct or indirect effect on the Phu system. Although the Has and Phu systems have distinct nonredundant roles in heme sensing and transport, it is reasonable to assume that there is a regulatory link given their distinct roles in adapting to changes in physiological environments. The nature of the regulatory link between the Has and Phu systems is not clear, but it is possible that the ECF σ factor HasI has undetermined targets beyond autoregulation of the has operon. The insight into heme release, signaling, and transport gained from these studies provides a platform for future structural and mechanistic studies of the heme transport channels of both HasR and PhuR.
Experimental procedures
Bacterial strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are listed in Table S1, and oligonucleotide primers and probes are listed in Table S2. Escherichia coli strains were routinely grown in Luria-Bertani (LB) broth (American Bioanalytical) or on LB agar plates, and P. aeruginosa strains were freshly streaked and maintained on Pseudomonas isolation agar (PIA) (BD Biosciences). Brain heart infusion agar and LB were used in generating the hasR mutant strains as described previously (25). All strains were stored frozen at −80 °C in LB broth with 20% glycerol. For qPCR and Western blotting, singly isolated colonies from each Pseudomonas strain were picked, inoculated into 10 ml of LB broth, and grown overnight at 37 °C with shaking (210 rpm). The bacteria then were harvested and washed in 10 ml of M9 minimal medium (Nalgene). The iron levels in M9 medium were determined by inductively coupled plasma-MS to be less than 1 nM. Following centrifugation, the bacterial pellet was resuspended in 10 ml of M9 medium and used to inoculate 50 ml of fresh M9 low-iron medium to a starting A600 of 0.04. Cultures were grown at 37 °C with shaking for 3 h before the addition of supplements (0 h) and incubated for a further 6 h. When required, antibiotics were used at the following final concentrations (μg ml−1): ampicillin, 100; tetracycline (Tc), 10 (for E. coli) and 150 (for P. aeruginosa); gentamicin, 250; and carbenicillin, 500.
Gene expression analysis using quantitative real-time PCR
To analyze gene expression, total RNA was purified from 1-ml aliquots collected at several time points from cultures grown under various conditions. RNA was stabilized by the addition of 250 μl of RNALater solution (Ambion), and the samples were stored at −80 °C until further use. Total RNA was isolated from each cell pellet using the RNeasy minispin columns according to the manufacturer's directions (Qiagen). 6 µg of total RNA was treated with RNase-free DNase I (New England Biolabs) for 3 h at 37 °C to remove contaminating chromosomal DNA and precipitated with 0.1 volume of 3 m sodium acetate (pH 5.2) and 2 volumes of 100% (v/v) ethanol. RNA quantity and quality were assessed by UV absorption at 260 nm in a NanoDrop 2000c spectrophotometer (ThermoFisher Scientific). cDNA was generated using the GoScriptTM reverse transcriptase kit (Promega) from RNA (250 ng) and random primers (0.5 µg). cDNA (10 ng) was analyzed with gene-specific primers (Table S2) using the StepOnePlus real-time PCR system (Applied Biosystems) and FastStart universal probe master mix (Roche Applied Sciences). The relative gene expression was calculated using the ΔΔCT method, and the cycle threshold values at each time point were normalized to the constitutively expressed rRNA 16s gene. mRNA values represent the standard deviation from three independent experiments performed in triplicate.
SDS-PAGE and Western blot analysis
Aliquots (1 ml) from PAO1 and hasR mutant cultures were collected at the 0-, 2-, 4-, and 6-h time points. Samples were harvested at 14,800 rpm for 5 min at room temperature. Cell pellets were resuspended in 200 μl per 1.0 A600 unit of BugBuster (Novagen). Cells were incubated at room temperature for 30 min with occasional agitation to ensure complete cell lysis, and total protein concentrations were determined using the Bio-Rad RC DC assay. Samples of the cell lysate (5 µg of total protein) in SDS-PAGE loading buffer were run on a 7.5% SDS-PAGE for HasR and PhuR detection. Following normalization between strains for differences in A600 at each time point, the supernatants (final volume, 1–8 μl) were loaded in SDS-PAGE loading buffer. Proteins were transferred by electrophoresis to polyvinylidene difluoride membranes (Bio-Rad) for Western blot analysis. Membranes were blocked with blocking buffer (5%, w/v, skim milk in TBS with 0.2%, v/v, Tween 20), washed, and probed with a 1:500 dilution of anti-HasR or anti-PhuR primary antibody in hybridization buffer (1%, w/v, skim milk in TBS with 0.2%, v/v, Tween 20). Antibodies were obtained from Covance custom antibodies and generated from purified proteins supplied by our laboratory. Antibody specificity and sensitivity was previously determined with the respective purified proteins (19), and all experimental Western blots were run with molecular weight markers as standards. Membranes were rinsed three times in TBS with 0.2% (v/v) Tween 20 and probed with goat anti-rabbit or goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (KPL) at a dilution of 1:10,000 in hybridization buffer. Proteins were visualized and enhanced by chemiluminescent detection using the SuperSignal chemiluminescence kit (Pierce) and Hyperfilm ECL (Amersham Biosciences). The normalized density represents the relative abundance of each protein compared with RNA polymerase α subunit (RNApolα) as the loading control for n = 3 independent biological replicates. The RNApolα subunit was detected with the anti-E. coli antibody (BioLegend). Densitometry analysis was performed on an AlphaImager HP system using the manufacturer's AlphaView software. The HasAp supernatant samples were analyzed on the Protein Simple Wes capillary Western system. No more than 4 μl of each sample was loaded with an antibody dilution of 1:25 in antibody diluent. The samples were separated by size on a 12- to 230-kDa capillary tray. The separation time, separation voltage, antibody diluent time, primary antibody time, and secondary antibody time were 25 min, 375 V, 30 min, 30 min, and 30 min, respectively. Samples were processed and quantification performed using the Compass for SW (ProteinSimple) software, where the area of each peak based on molecular weight was calculated and normalized to the optical density at 600 nm (OD600) for (n = 3) independent biological replicates.
Construction of the PAO1 hasR allelic strains
The in-frame P. aeruginosa hasR mutant allelic strains were constructed using two primers, hasR upstream and hasR downstream (Table S2), designed to amplify the 5′ upstream and 3′ downstream regions of the gene from the genomic DNA. The resulting Taq PCR product was ligated into pCR2.1-TOPO-TA vector (Invitrogen) for subcloning. Mutagenesis was performed by PCR using the QuikChange mutagenesis kit (Agilent Technologies) and primers specified in Table S2. The sequence of all PCR products and amino acid changes was confirmed by DNA sequencing (Eurofins MWG Operon). Following KpnI–XbaI digestion, the hasR mutant genes were cloned into the counterselective suicide plasmid pEX18Tc (25). The resulting pEX18TchasR was transformed into E. coli S17-1-λpir cells and conjugated into P. aeruginosa PAO1. The resulting colonies that underwent the first recombination event were screened on PIA plus Tc plates at 37 °C overnight. Isolated colonies were patched several times to ensure they were Tcr. To remove the pEX18Tc backbone, isolated colonies were grown in 1 ml LB shaken overnight at 210 rpm at 37 °C. A 10-μl aliquot of cells was used to inoculate fresh LB cultures and was repeated twice more to induce the second recombination event. Lastly, a small dilution of cells was streaked onto a PIA plus 5% sucrose plate and incubated at 37 °C overnight. Several isolated colonies were patched onto PIA plus 5% sucrose and PIA plus Tc plates. Only the Tcs colonies were selected for sequencing to confirm complementation with the mutant allele at the original locus.
Preparation and isolation of 13C-heme
Labeled [4-13C]δ-aminolaevulinic acid (ALA) was purchased from Frontier Scientific. [4-13C]δ-ALA was used as a biosynthetic precursor to produce 13C-labeled heme. Expression of cytochrome b5 in the presence of [4-13C]δ-ALA induces heme biosynthesis in E. coli, where the produced cytochrome b5 captures and acts as a reservoir for the synthesized heme (26). 13C-heme was prepared as previously reported (9, 27). Briefly, rat outer mitochondrial cytochrome b5 was expressed in E. coli BL21(DE3) cells in M9 minimal medium with the following supplements: 2 mm MgSO4, 100 μm CaCl2, 150 nM (NH4)6Mo7O24, 40 μm FeSO4 (acidified with 1 N HCl), 17 μm EDTA, 3 μm CuSO4, 2 μm Co(NO3)2, 7.6 μm ZnSO4, 9.4 μm Na2B4O7·10H2O, and 1 mm thiamine. Following lysis and centrifugation, the cellular lysate was loaded onto a Q-Sepharose anion exchange column (2.5 by 10 cm) equilibrated in 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 50 mm NaCl. The column was washed with the same buffer, followed by an additional wash with 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 125 mm NaCl (5 column volumes). Cytochrome b5 was eluted in 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 250 mm NaCl and dialyzed overnight in 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 50 mm NaCl. Heme extraction from cytochrome b5 was performed by the acid/butanone method as previously described (28). Heme concentrations were determined by the pyridine hemochrome assay (29). Heme stocks were prepared immediately prior to use by dissolving in 0.1 N NaOH and buffered to pH 7.4 with 1 m Tris-HCl, and the final concentration was determined by pyridine hemochrome (pH 7.4). The labeling pattern of biosynthesized heme using [4-13C]δ-ALA was as previously reported (9).
Extraction of BVIX isomers from P. aeruginosa supernatants
PAO1 WT and allelic strains were grown as described above and supplemented with either 1 μm 13C-labeled heme or HasAp bound to 13C-labeled heme. 50-ml cultures were shaken at 210 rpm for 3 h at 37 °C in 250-ml baffled flasks. After 3 h they were supplemented and allowed to shake for an additional 4 h. Cells were harvested by centrifugation in 50-ml conical tubes at 6000 rpm for 20 min at 4 °C. The BVIX isomers were extracted as previously described (19). Briefly, the supernatant was collected and filtered by a Nalgene vacuum filtration unit with a 0.22-μm PVDF membrane. Supernatants were acidified to pH ∼2.5 with 10% TFA, supplemented with 10 nM (final concentration in 45 ml) dimethyl ester BVIXα, and loaded over a C18 Sep-Pak column purchased from Waters. The column was prepared by flushing with 2 ml of acetonitrile (ACN), H2O, 0.1% TFA in H2O and methanol–0.1% TFA (10:90). After sample application, the column was washed with 4 ml 0.1% TFA, 4 ml ACN–0.1% TFA (20:80), 2 ml methanol–0.1% TFA (50:50) and eluted with 1 ml methanol. Purified BVIX isomers were speed vacuumed dry and stored for up to 1 week at −80 °C prior to LC-MS/MS analysis.
LC-MS/MS analysis of BVIX isomers
Samples were resuspended in 10 μl DMSO, diluted to 100 μl with mobile-phase ACN (50:50, v/v), and centrifuged at 14,000 rpm for 5 min at room temperature to remove particulates. The BVIX isomers (2 μl) were separated and analyzed by LC-MS/MS (Waters TQ-XS triple quadrupole mass spectrometer with AQUITY H-Class UPLC) as previously described, with slight modifications (16). BVIX isomers were separated on an Ascentis RP-amide 2.7-μm C18 column (10 cm by 2.1 mm) at a flow rate of 0.4 ml/min. The mobile phase consisted of A, H2O:0.1% formic acid, and B, ACN–0.1% formic acid. The initial gradient is 64% A and 36% B and then 5 min 55% A and 45% B, 8 min 40% A and 60% B, 8.5 min 5% A and 95% B, and 10 min 64% A and 36% B. Fragmentation patterns of the precursor ions were detected at 583.21 (12C-BVIX) and 591.21 (13C-BVIX) using multiple-reaction monitoring (MRM). The source temperature was set to 150 °C, the capillary voltage to 3.60 kV, and the cone voltage to 43V. The column was kept at 30 °C during separation. The precursor ion used in MRM for 12C-BVIXα, -β, and -δ isomers was 297.1, 343.1, and 402.2 with a collision energy of 38, 36, and 30 V, respectively. The product ion used in MRM for 13C-BVIXα, -β, and -δ isomers was 301.1, 347.1, and 408.2, respectively, with a collision energy of 34 V for each.
ICP-MS analysis of intracellular iron content
Aliquots (2 ml) were removed from cultures grown as described for the heme uptake studies at 2, 5, and 7 h, pelleted, and washed in fresh M9 media. The pellets were dissolved in trace metal-free ultrapure 20% HNO3 and boiled overnight at 100 °C. Samples were further diluted with ultrapure water to a final concentration of 2% HNO3 and subjected to ICP-MS on an Agilent 7700 ICP-MS (Agilent Technologies). ICP-MS runs were calibrated with high-purity iron standard solution (Sigma-Aldrich), and rawICP-MS data (ppb) were corrected for drift using values for scandium and germanium (CPI International) as internal standards and added to samples during processing. Corrected values were then normalized to culture density as determined by the absorbance at 600 nm. Experimental values and reported standard deviations were averages from three biological replicates.
Data availability
All data are contained within the manuscript.
Supplementary Material
Acknowledgments
We thank the University of Maryland, School of Pharmacy, Mass Spectrometry Center, including the Center Director Maureen Kane and Stephanie Shiffka, for their technical expertise. A. T. D. also thanks Susana Mouriño for helpful discussion and reading of the manuscript.
This article contains supporting information.
Author contributions—A. T. D. and A. W. conceptualization; A. T. D. and A. W. supervision; A. T. D. and A. W. funding acquisition; A. T. D. and A. W. investigation; A. T. D. and A. W. methodology; A. T. D. and A. W. writing-original draft; A. T. D. and A. W. project administration; A. T. D. and A. W. writing-review and editing.
Funding and additional information—This work was supported by National Institutes of Health grant R01 AI134886 (to A. W.). A. T. D. was funded by National Institutes of Health Ruth L. Kirschstein (NRSA) Individual Predoctoral Fellowship GM126860. A. T. D. also acknowledges previous support from a Chemistry/Biology Interface Training Program (NIGMS/NIH T32GM066706). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare they have no conflict of interest with the contents of this article.
- Phu
- Pseudomonas heme uptake
- PAO1
- Pseudomonas aeruginosa O1
- Has
- heme assimilation system
- HasAs
- Serratia marcescens HasA
- HasAp
- Pseudomonas aeruginosa HasA
- BVIX
- biliverdin
- ECF
- extracytoplasmic function
- CSS
- cell surface signaling
- ABC
- ATP-dependent binding cassette
- qPCR
- quantitative PCR
- PIA
- Pseudomonas isolation agar
- Tc
- tetracycline
- RNApolα
- RNA polymerase α
- OD600
- optical density at 600 nm
- ACN
- acetonitrile
- ICP-MS
- inductively coupled plasma mass spectrometry
- MRM
- multiple-reaction monitoring.
References
- 1. Ganz T. (2009) Iron in innate immunity: starve the invaders. Curr. Opin. Immunol. 21, 63–67 10.1016/j.coi.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sheldon J. R., and Heinrichs D. E. (2015) Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiol. Rev. 39, 592–630 10.1093/femsre/fuv009 [DOI] [PubMed] [Google Scholar]
- 3. Huang W., and Wilks A. (2017) Extracellular heme uptake and the challenge of bacterial cell membranes. Annu. Rev. Biochem. 86, 799–823 10.1146/annurev-biochem-060815-014214 [DOI] [PubMed] [Google Scholar]
- 4. Contreras H., Chim N., Credali A., and Goulding C. W. (2014) Heme uptake in bacterial pathogens. Curr. Opin. Chem. Biol 19, 34–41 10.1016/j.cbpa.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heinrichs D. E., Young L., and Poole K. (1991) Pyochelin-mediated iron transport in Pseudomonas aeruginosa: involvement of a high-molecular-mass outer membrane protein. Infect. Immun. 59, 3680–3684 10.1128/IAI.59.10.3680-3684.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Royt P. W. (1990) Pyoverdine-mediated iron transport. Fate of iron and ligand in Pseudomonas aeruginosa. Biol. Met. 3, 28–33 10.1007/BF01141174 [DOI] [PubMed] [Google Scholar]
- 7. Hunter R. C., Asfour F., Dingemans J., Osuna B. L., Samad T., Malfroot A., Cornelis P., and Newman D. K. (2013) Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. mBio 4, e00557–13 10.1128/mBio.00557-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ochsner U. A., Johnson Z., and Vasil M. L. (2000) Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146, 185–198 10.1099/00221287-146-1-185 [DOI] [PubMed] [Google Scholar]
- 9. Smith A. D., and Wilks A. (2015) Differential contributions of the outer membrane receptors PhuR and HasR to heme acquisition in Pseudomonas aeruginosa. J. Biol. Chem. 290, 7756–7766 10.1074/jbc.M114.633495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Helmann J. D. (2002) The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46, 47–110 10.1016/S0065-2911(02)46002-X [DOI] [PubMed] [Google Scholar]
- 11. Mascher T. (2013) Signaling diversity and evolution of extracytoplasmic function (ECF) sigma factors. Curr. Opin. Microbiol. 16, 148–155 10.1016/j.mib.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 12. Potvin E., Sanschagrin F., and Levesque R. C. (2008) Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32, 38–55 10.1111/j.1574-6976.2007.00092.x [DOI] [PubMed] [Google Scholar]
- 13. Llamas M. A., Mooij M. J., Sparrius M., Vandenbroucke-Grauls C. M., Ratledge C., and Bitter W. (2008) Characterization of five novel Pseudomonas aeruginosa cell-surface signalling systems. Mol. Microbiol. 67, 458–472 10.1111/j.1365-2958.2007.06061.x [DOI] [PubMed] [Google Scholar]
- 14. Llamas M. A., Imperi F., Visca P., and Lamont I. L. (2014) Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol. Rev. 38, 569–597 10.1111/1574-6976.12078 [DOI] [PubMed] [Google Scholar]
- 15. Ghigo J. M., Letoffe S., and Wandersman C. (1997) A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J. Bacteriol. 179, 3572–3579 10.1128/jb.179.11.3572-3579.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Biville F., Cwerman H., Letoffe S., Rossi M. S., Drouet V., Ghigo J. M., and Wandersman C. (2004) Haemophore-mediated signalling in Serratia marcescens: a new mode of regulation for an extra cytoplasmic function (ECF) sigma factor involved in haem acquisition. Mol. Microbiol. 53, 1267–1277 10.1111/j.1365-2958.2004.04207.x [DOI] [PubMed] [Google Scholar]
- 17. Vanderpool C. K., and Armstrong S. K. (2003) Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185, 909–917 10.1128/jb.185.3.909-917.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ratliff M., Zhu W., Deshmukh R., Wilks A., and Stojiljkovic I. (2001) Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403 10.1128/JB.183.21.6394-6403.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mourino S., Giardina B. J., Reyes-Caballero H., and Wilks A. (2016) Metabolite-driven regulation of heme uptake by the biliverdin IXbeta/delta-selective heme oxygenase (HemO) of Pseudomonas aeruginosa. J. Biol. Chem. 291, 20503–20515 10.1074/jbc.M116.728527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dent A. T., Mourino S., Huang W., and Wilks A. (2019) Post-transcriptional regulation of the Pseudomonas aeruginosa heme assimilation system (Has) fine-tunes extracellular heme sensing. J. Biol. Chem. 294, 2771–2785 10.1074/jbc.RA118.006185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen A. T., O'Neill M. J., Watts A. M., Robson C. L., Lamont I. L., Wilks A., and Oglesby-Sherrouse A. G. (2014) Adaptation of iron homeostasis pathways by a Pseudomonas aeruginosa pyoverdine mutant in the cystic fibrosis lung. J. Bacteriol. 196, 2265–2276 10.1128/JB.01491-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krieg S., Huche F., Diederichs K., Izadi-Pruneyre N., Lecroisey A., Wandersman C., Delepelaire P., and Welte W. (2009) Heme uptake across the outer membrane as revealed by crystal structures of the receptor-hemophore complex. Proc. Natl. Acad. Sci. U S A 106, 1045–1050 10.1073/pnas.0809406106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caillet-Saguy C., Piccioli M., Turano P., Lukat-Rodgers G., Wolff N., Rodgers K. R., Izadi-Pruneyre N., Delepierre M., and Lecroisey A. (2012) Role of the iron axial ligands of heme carrrier HasA in heme uptake and release. J. Biol. Chem. 287, 26932–26943 10.1074/jbc.M112.366385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith A. D., Modi A. R., Sun S., Dawson J. H., and Wilks A. (2015) Spectroscopic determination of distinct heme ligands in outer-membrane receptors PhuR and HasR of Pseudomonas aeruginosa. Biochemistry 54, 2601–2612 10.1021/acs.biochem.5b00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., and Schweizer H. P. (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- 26. Rivera M., and Walker F. A. (1995) Biosynthetic preparation of isotopically labeled heme. Anal. Biochem. 230, 295–302 10.1006/abio.1995.1477 [DOI] [PubMed] [Google Scholar]
- 27. O'Neill M. J., and Wilks A. (2013) The P. aeruginosa heme binding protein PhuS is a heme oxygenase titratable regulator of heme uptake. ACS Chem. Biol. 8, 1794–1802 10.1021/cb400165b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teale F. W. (1959) Cleavage of the haem-protein link by acid methylethylketone. Biochim. Biophys. Acta 35, 543 10.1016/0006-3002(59)90407-X [DOI] [PubMed] [Google Scholar]
- 29. Fuhrhop J. H., and Smith K. M. (1975) Laboratory methods in porphyrin and metalloporphyrin research. Elsevier Scientific Publishing Company, New York [Google Scholar]
- 30. Anonymous. 2020. The PyMOL Molecular Graphics System, version 1.2r3pre. Schrödinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.