Abstract
The widely successful use of synthetic herbicides over the past 70 years has imposed strong and widespread selection pressure, leading to the evolution of herbicide resistance in hundreds of weed species. Both target-site resistance (TSR) and nontarget-site resistance (NTSR) mechanisms have evolved to most herbicide classes. TSR often involves mutations in genes encoding the protein targets of herbicides, affecting the binding of the herbicide either at or near catalytic domains or in regions affecting access to them. Most of these mutations are nonsynonymous SNPs, but polymorphisms in more than one codon or entire codon deletions have also evolved. Some herbicides bind multiple proteins, making the evolution of TSR mechanisms more difficult. Increased amounts of protein target, by increased gene expression or by gene duplication, are an important, albeit less common, TSR mechanism. NTSR mechanisms include reduced absorption or translocation and increased sequestration or metabolic degradation. The mechanisms that can contribute to NTSR are complex and often involve genes that are members of large gene families. For example, enzymes involved in herbicide metabolism–based resistances include cytochromes P450, GSH S-transferases, glucosyl and other transferases, aryl acylamidase, and others. Both TSR and NTSR mechanisms can combine at the individual level to produce higher resistance levels. The vast array of herbicide-resistance mechanisms for generalist (NTSR) and specialist (TSR and some NTSR) adaptations that have evolved over a few decades illustrate the evolutionary resilience of weed populations to extreme selection pressures. These evolutionary processes drive herbicide and herbicide-resistant crop development and resistance management strategies.
Keywords: Cytochrome P450, glutathione S-transferase, herbicide metabolism, reduced translocation, target-site resistance, nontarget-site resistance, cross-resistance, multiple resistance, plant evolution, selection pressure, plant defense, plant physiology, plant molecular biology, plant biochemistry, mutant, evolution, xenobiotic, herbicide, resistance mechanism
Plants that are not wanted at a particular time and/or place (weeds) have been managed mostly with synthetic herbicides for more than 70 years. Before that time, weeds were largely controlled with laborious manual weeding and often environmentally damaging tillage. Adoption of synthetic herbicides reduced the cost and increased the efficacy of weeding, thereby contributing to the yield increases and efficiency of agriculture seen since the middle of the last century. However, as with antibiotics, the utility of synthetic weed killers is being threatened by widespread evolution of resistance to most chemical classes of herbicides that act on most of the more than 25 molecular targets of current commercial herbicides (1).
The main goal of this review is to provide an update on the rapidly evolving topic of mechanisms of evolved herbicide resistance in weeds, as there has been no recent comprehensive review on this topic. We hope that this review will inspire plant molecular biologists and biochemists to determine more clearly how these resistance mechanisms evolve and the biochemical and physiological changes in weeds imparted by resistance mutations. Such information will be useful in resistance management and in the design of herbicide molecules for which evolution of resistance is more problematic for weeds. We provide discussions of the implications of herbicide-resistance mechanisms for the development of new herbicides and herbicide-resistant crops.
We summarize the wide array of resistance mechanisms that weeds have evolved to survive the intense selection pressure imparted by commercial herbicides. Herbicide-resistance mechanisms can be broadly divided into two categories, referred to as target-site resistance (TSR) mechanisms and nontarget-site resistance (NTSR) mechanisms. Herbicide efficacy is generally dependent on how much of the herbicide enters a plant cell and how long its active form remains available to interact with its site of action (also called the target site). A full understanding of the mechanism of resistance to a herbicide requires understanding that herbicide's mechanism of action (MOA). The MOA of herbicides is not discussed in detail in this review, but we have provided a summary of the molecular targets and MOAs of the herbicides mentioned in this review (Table 1). There are 26 molecular target sites of the more than 260 commercial herbicide active ingredients that are recognized by the Herbicide Resistance Action Committee, an industry organization that monitors herbicide resistance (Herbicide Resistance Action Committee (2020) HRAC Mode of Action Classification 2020 Map, https://hracglobal.com/tools/hrac-mode-of-action-classification-2020-map; accessed April 22, 2020). Of these target sites, resistance has evolved globally (in 92 crops in 70 countries) to 167 herbicides representing about 23 of these targets, with 512 weed species evolving resistance to one of more herbicides (Heap, I. (2020) The international survey of herbicide resistant weeds; available at www.weedscience.com; accessed February 25, 2020). This review will not provide an encyclopedic elaboration for each of these cases but will give examples of different mechanisms of resistance that often cross herbicide classes.
Table 1.
Herbicides mentioned in the text with their molecular targets and mechanisms of resistance
Only the mechanisms provided in the text are listed. Additional herbicides to which resistance has evolved are found in Heap, I. (2020) The international survey of herbicide resistant weeds; available at www.weedscience.com; accessed February 25, 2020.
| Inhibited process/molecular target | Herbicide chemical class/herbicidea | Mechanism(s) of resistance |
|---|---|---|
| Amino acid synthesis | ||
| ALS; branched-chain amino acids | Imidazolinones | |
| Imazethapyr | TSR and NTSR | |
| Imazaquin | TSR | |
| Sulfonylureas | ||
| Benzulfuron | TRS and NTSR | |
| Chlorimuron | TSR and NTSR | |
| Chlorsulfuron | TSR and NTSR | |
| Pyrimidinyl benzolates | ||
| Bispyribac | TSR and NTSR | |
| Triazolopyrimidine | ||
| Penoxsulam | NTSR | |
| EPSPS; shikimate pathway function | Glyphosate | TSR and NTSR |
| Glutamine synthase | Glufosinate | NTSR |
| Auxin mimics | ||
| F-box proteins | Phenoxycarboxylates | |
| 2,4-D | TSR and NTSR | |
| Dicamba | TSR and NTSR | |
| Quinclorac | NTSR | |
| Picolinates | ||
| Picloram | TSR | |
| Carotenoid synthesis | ||
| HPPD | Isoxaflutole | NTSR |
| Triketones | ||
| Mesotrione | NTSR | |
| Tembotrione | NTSR | |
| Lycopene cyclase | Amitrole | NTSR |
| Phytoene desaturase | Diphenyl heterocycles | |
| Fluridone | TSR | |
| Phenyl ethers | ||
| Beflubutamid | No resistance | |
| Diflufenican | No resistance | |
| Picolinafen | No resistance | |
| N-Phenyl heterocycles | ||
| Norflurazon | TSR | |
| Deoxy-d-xylulose phosphate synthase | Clomazone | NTSR |
| Cell division | ||
| α-Tubulin | Dinitroanilines | |
| Oryzalin | TSR | |
| Trifluralin | TRS | |
| Chlorophyll synthesis | ||
| PPO | Diphenyl ethers | |
| Lactofen | TSR | |
| Fomesafen | NTSR | |
| Fatty acid synthesis | ||
| ACCase | Aryloxyphenoxypropionates | |
| Diclofop-methyl | TSR and NTSR | |
| Fenoxaprop-P-ethyl | TSR and NTSR | |
| Clodinofop | NTSR | |
| Quizalofop | TSR | |
| Cyclohexanediones | ||
| Sethoxydim | TSR | |
| Tralkoxydim | TSR and NTSR | |
| Pinoxaden | NTSR | |
| Fatty acid thioesterase | Cinmethylin | NTSR |
| Very long-chain fatty acid synthases | Thiobencarb | NTSR |
| Flufenacet | NTSR | |
| Chloroacetamides | ||
| Alachlor | NTSR | |
| Metolachlor | NTSR | |
| Photosynthesis | ||
| Photosystem II D1 protein | Amides | |
| Propanil | TSR and NTSR | |
| Nitriles | ||
| Bromoxynil | NTSR | |
| Triazines | ||
| Atrazine | TSR and NTSR | |
| Ureas | ||
| Chlorotoluron | NTSR | |
| Bentazon | NTSR | |
| Photosystem I energy diversion | Paraquat | NTSR |
aThe chemical classes of herbicides underlined are not provided, as they are the only representatives of their chemical class.
NTSR mechanisms include all mechanisms that reduce the concentration of active herbicide remaining available to interact with the target site protein, as well as mechanisms that allow the plant to cope with inhibition of the target site. NTSR mechanisms include reduced herbicide uptake and translocation, increased herbicide sequestration, and enhanced degradation or metabolism of the herbicide to less toxic compounds. On the other hand, TSR mechanisms alter the amino acid sequence and/or expression level of the target enzyme, reducing the herbicide's ability to inhibit the enzyme or requiring a greater herbicide concentration to achieve adequate inhibition.
Under intense selection pressure from highly effective herbicides, all possible mechanisms conferring a greater chance of survival and reproduction to the individual may be selected. More than one mechanism may be operating to confer resistance, including combinations of TSR and NTSR mechanisms. Several resistance mechanisms can co-exist within a species, within a population, and even within a single individual. Different resistance mechanisms combine through cross-pollination between individuals. Species with high levels of cross-pollination are more likely to accumulate diverse resistance mechanisms (to a single herbicide or to multiple herbicides), and this process can occur more rapidly than in self-pollinated species.
Target-site mechanisms
A single nucleotide mutation in the gene encoding a protein bound by a herbicide can result in a single amino acid change, disrupting the ability of the herbicide to bind to the protein without disabling the enzyme function. Generally, there are few amino acids in or near the herbicide-binding site of most target-site proteins where an amino acid substitution will result in TSR. Most target-site mutations occur in or near the herbicide-binding site, but some mutations occur elsewhere in the protein structure. Target-site mutations are identified by the amino acid and its position in the protein, numbered from the protein's start codon. In some cases, the mutation can confer very high-level resistance, and in other cases, the mutation confers lower level (but significant) resistance. Some target-site mutations reduce normal enzymatic function, and other mutations retain nearly full enzymatic function. In addition to single nucleotide substitutions, whole-codon deletions can also reduce herbicide binding to the target-site enzyme. It is important to note that the same molecular mechanism (e.g. an SNP leading to an amino acid change) forms the basis of resistance for many different herbicide sites of action.
TSR mechanisms are specialist mechanisms, specific to a single site of action. Whether a specific target-site mutation that confers resistance to a given herbicide also confers resistance to different chemical families within the same site-of-action group varies, depending on how the specific herbicides interact with the target protein. TSR can also be due to increased expression of the target-site gene, producing more enzyme than can be substantially inhibited by typical herbicide application rates. Increased gene expression can be due to regulatory changes increasing transcription and/or increased genomic copy number of the target-site gene, also resulting in increased transcription.
SNP
Nonsynonymous SNPs imparting resistance to a herbicide target site is the most common mechanism of TSR. These mutations may be within or in the proximity of the catalytic domain of an enzyme and affect a herbicide's ability to compete for the binding of a substrate, or these mutations may affect other domains of enzymes and proteins.
Mutations affecting herbicide binding
The first discovered target-site mutation was for the photosystem II (PSII)-inhibiting herbicides, which compete with plastoquinone for binding on the D1 protein encoded by the psbA gene and thereby inhibit PSII electron transport (2). Amino acid substitutions in the psbA gene typically confer high-level resistance to a single chemical family, but not to herbicides from other families or groups. For example, the single amino acid change S264G confers high-level resistance to the triazine herbicides in numerous species around the world (e.g. Ref. 3) but moderate to no resistance to the other families of PSII inhibitors, including the triazinones, which are in the same group as the triazines. The substitution of Gly for Ser at position 264 prevents triazine binding but also compromises plastoquinone binding and impairs photosynthesis, resulting in a strong fitness penalty (2). A S264T substitution confers resistance to triazines and ureas, but not to the nitrile or triazinone families. Additional psbA resistance-imparting mutations include V219I, N266T, F255I, and A251V.
The dinitroaniline herbicides (such as trifluralin and oryzalin) bind to plant tubulin protein and disrupt meristem development by depolymerizing microtubules (1). The first reported mutation affecting binding of dinitroaniline herbicides to plant microtubules was found in an α-tubulin gene transcript from goosegrass (Eleusine indica) that encoded a T239I substitution (4). This substitution conferred resistance to trifluralin and oryzalin when transformed in maize, demonstrating that this mutation was the molecular basis of dinitroaniline herbicide resistance (4). Substitutions of V202F (5) and Arg-243 to Met or Lys (6) have been reported for dinitroaniline resistance in annual ryegrass (Lolium rigidum). A substitution of M268T has also been identified in E. indica, and both T239I and L136F have been identified in green foxtail (Setaria viridis) (7), demonstrating that substitutions at multiple amino acid positions in a target site gene can confer resistance, depending on the molecular structure of the target site protein and where the herbicide binds to the protein.
Acetyl-CoA carboxylase (ACCase; EC 6.4.1.2) is a key enzyme for fatty acid biosynthesis pathways. The plastidic form of ACCase in grasses is inhibited by the aryloxyphenoxypropionate (APP), cyclohexanedione (CHD), and phenylpyrazoline (PPZ; pinoxaden is the only member) herbicide chemical families (1). Eight mutations have been reported, all contained within the carboxyl transferase domain of the ACCase enzyme. Known mutations occur at seven positions: I1781L or I1781V, W1999C or W1999L, W2027C, I2041N or I2041V, D2078G, C2088R, and G2096A or G2096S (8). Substitutions at positions Ile-1781 and Asp-2078 have been reported most often, and these substitutions confer resistance to APP, CHD, and PPZ herbicides. Ile-1781 is within the binding site for the three ACCase herbicide chemical families, explaining the resistance pattern to all three classes. Asp-2078 is not within the binding site but occurs next to Ile-1781, so the substitution of Gly for Asp at 2078 can cause a large effect on resistance level due to the substantial change in the structure of the binding site. Mutations at positions Ile-2041 and Gly-2096 confer resistance only to APP herbicides, whereas mutations at Trp-2027 can confer resistance to APP and PPZ herbicides (9).
Somatic mutations
Target site mutations have also been identified in the invasive aquatic weed Hydrilla verticillata conferring resistance to fluridone, an inhibitor of phytoene desaturase (PDS; EC 1.3.99.31), an enzyme essential for carotenoid synthesis (10). H. verticillata is a dioecious plant (male and female flowers are on separate plants), and only the female form was introduced to the United States. Consequently, resistance has evolved through the selection of a mutation in meristematic tissue that was able to regenerate into whole plants and spread to other lakes (11). The specific mutations in PDS are Arg-304 to Ser, Cys, or His. These mutations have arisen through somatic variation, with the consequence that H. verticillata populations within a confined body of water normally contain a single resistance mutation. The three mutations confer cross-resistance to the PDS-inhibiting herbicide norflurazon, but also confer negative cross-resistance (i.e. increased sensitivity) to three different PDS inhibitors (beflubutamid, picolinafen, and diflufenican) (12). This finding further emphasizes that the effects of amino acid substitutions are dependent on the target site protein structure and vary across different herbicide site-of-action groups.
An interesting aspect of somatic mutations imparting resistance to herbicides is that the lack of sexual reproduction may make the resistance trait more stable in a population than in species dependent on sexual reproduction due to a lack of genetic recombination. Consequently, the three distinct populations with variable resistance to fluridone have remained in the same abundance in some of these lakes despite an 8-year period with no fluridone selection pressure (13). Whereas the aquatic environment in which this case evolved may be partly responsible for this occurring, some terrestrial weeds also can reproduce vegetatively, suggesting the possibility of somatic mutations creating herbicide resistance in these species.
Mutations affecting access to target site
Acetolactate synthase (ALS; EC 2.2.1.6) (also called acetohydroxyacid synthase or AHAS) is a key enzyme in the synthesis of branched-chain amino acids. Several chemical families have ALS as their site of action, including the sulfonylureas (SUs) and imidazolinones (IMIs). Twenty-one combinations of weed species by ALS inhibitor resistance–endowing amino acid substitutions have been reported to date, with 127 total unique (by species) occurrences of the different substitutions (Heap, I. (2020) The international survey of herbicide resistant weeds; available at www.weedscience.com; accessed February 25, 2020). Resistance-imparting substitutions at Pro-197 have been reported most frequently, followed by mutations at Trp-574. As with ACCase target site mutations, some of the ALS mutations confer very high-level resistance, and the resistance spectrum across chemical families varies by mutation. General patterns are that the Trp-574 mutation confers resistance to SUs and IMIs, the Ser-653 mutation confers resistance to IMIs but not SUs, and the Pro-197 mutation confers resistance to SUs but not IMIs (depending on the specific amino acid substitution; some Pro-197 mutations do confer resistance to IMIs). The known ALS resistance mutations do not occur at the substrate-binding site but instead occur at amino acid positions where the ALS herbicides can bind to and block an access channel within the enzyme through which the ALS substrates must move (14). This is the biochemical reason why ALS mutations generally have little effect on the normal catalytic activity, in contrast to mutations in the psbA gene that significantly affect normal biochemical function. The SUs and IMIs bind to partially overlapping sites in the ALS enzyme but have different modes of binding (Fig. 1). This is important from an evolutionary perspective, because rotating different ALS chemical families in the field will likely select for the same mutation, whereas rotating among different PSII-inhibiting chemical families may select for different mutations. In the case of ALS-inhibiting herbicides, rotations to other ALS-inhibiting chemical families provide the same selection pressure from an evolutionary perspective and are not functional in slowing the evolution of resistance. In support of this concept, resistance to ALS-inhibiting herbicides has occurred rapidly following introduction of these herbicides. Rotations among different PSII-inhibiting chemical families may in some cases slow the evolution of TSR, because the amino acid substitution conferring resistance to one family does not confer resistance to another family.
Figure 1.
Crystal structure of Arabidopsis ALS. A, view of the homodimer with chain A (gold) and chain B (slate colors) (adapted from McCourt et al. (14)). The herbicide imazaquin is located at the entrance of the channel leading to the catalytic domain of the enzyme. B, closer view of the interface between the two ALS monomers with imazaquin positioned at the entrance. The area in red highlights the position of Ser-653 imparting resistance to imidazolines but not to sulfonylureas. This research was originally published in the Proceedings of the National Academy of Sciences of the United States of America. McCourt, J. A., Pang, S. S., King-Scott, J., Guddat, L. W., and Duggleby, R. G. Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:569–573. © United States National Academy of Sciences.
The rarer a mutation is within a population prior to herbicide selection, the longer it will take for the mutation to be selected and reach a high frequency within the population. A study of a herbicide-susceptible L. rigidum population found that SU target site resistance allele frequency within previously untreated populations was as high as 1.2 × 10−4, and IMI target site resistance allele frequency was as high as 5.8 × 10−5 (15). Similarly, a mutagenesis experiment in Arabidopsis found ALS resistance (SU and IMI) at a frequency of 3.2 × 10−5 in progeny of M1 lines (first generation after chemical mutagenesis) with no detectable glyphosate resistance in 250,000 M1 progeny screened (16). These experimental results are corroborated by the relatively fast initial evolution of resistance to ALS inhibitors following their introduction and the relatively slower initial evolution of resistance to glyphosate following its introduction (Heap, I. (2020) The international survey of herbicide resistant weeds; available at www.weedscience.com; accessed February 25, 2020).
The relatively higher number of mutations imparting resistance to various classes of ALS-inhibiting herbicides compared with other herbicide groups is because these molecules do not compete with the substrate for the catalytic domain of the enzyme. Instead, they block the opening of the channel leading to the catalytic domain (Fig. 1). Consequently, mutations imparting resistance often have no impact on the kinetic properties of ALS.
Glyphosate inhibits 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS; EC 2.5.1.19), a key enzyme in the shikimate pathway, and mutations in the EPSPS gene have been reported in weeds. Most described EPSPS target site mutations in weeds are located at the Pro-106 residue, using the Arabidopsis numbering system, from the start of the mature enzyme (reviewed by in Ref. 17). Known resistance mutations in weeds include Pro-106 to Ser, Thr, Ala, or Leu (17). Glyphosate inhibits EPSPS by competing with the normal substrate phosphoenolpyruvate (PEP) for binding to the enzyme (Fig. 2). Glyphosate binding is almost irreversible, so once bound, that unit of EPSPS is blocked. The Pro-106 residue is not directly involved in a molecular interaction with glyphosate or PEP, but it provides part of the molecular structure at the active site (18), and changing the Pro-106 to a different residue changes the spacing in the active site. This increases the inhibitory constant (Ki, the concentration of inhibitor required to decrease reaction rate to half of the uninhibited value) for glyphosate. This increase in Ki for EPSPS with the mutation is the reason for resistance, as more glyphosate is required to inhibit an equivalent amount of enzyme; however, the structural change also increases the Michaelis constant for PEP (Km, the substrate concentration required for effective catalysis to occur).
Figure 2.
Interaction of glyphosate with EPSPS. A, interaction between S3P and glyphosate (GLY) (yellow dotted line) within the catalytic domain of EPSPS. B, location of the TIPS double mutation in glyphosate-resistant EPSPS relative to the S3P-glyphosate complex. Leucine is shown in pink, and serine is shown in slate. C, mutation-induced structural changes in EPSPS. In the ternary complex, the mutations cause a shift of the Cα atom of Gly-96 toward the phosphonate moiety of glyphosate, seen most drastically in the TIPS enzyme (pink), thereby narrowing the inhibitor-binding site (residue numbers are for E. coli EPSPS and equivalent to Gly-101, Thr-102, and Pro-106 in plants). Adapted from Funke et al. (21). This research was originally published in the Journal of Biological Chemistry. Funke, T., Yang, Y., Han, H., Healy-Fried, M., Olesen, S., Becker, A., and Schönbrunn, E. Structural basis of glyphosate resistance resulting from the double mutation Thr97 → Ile and Pro101 → Ser in 5-enolpyruvylshikimate-3-phosphate synthase from Escherichia coli. J. Biol. Chem. 2009; 284:9854–9860. © the American Society for Biochemistry and Molecular Biology.
An EPSPS mutant with a higher Km requires higher PEP concentration to achieve the same reaction velocity as the WT EPSPS with normal Km. This means that the selectivity factor for PEP binding over glyphosate binding is affected, due to changed affinity for PEP, and a higher PEP concentration is necessary under normal conditions to maintain the same reaction rate. This effectively reduces the catalytic activity of EPSPS, a possible reason why this target site mutation may be rarer within populations compared with ALS target site mutations (which do not generally affect the Km or catalytic activity of ALS). A mutation at T102S conferred glyphosate resistance in tridax daisy (Tridax procumbens) (19). This mutation imparted lower affinity to glyphosate but also higher affinity for PEP.
Multiple nucleotide polymorphisms
A concomitant mutation, generated by point mutation of the maize EPSPS and commercialized as GA21 glyphosate-resistant maize (20), of both T102I and P106S (TIPS) resulted in structural changes that retained high affinity for PEP and made the enzyme insensitive to glyphosate inhibition (21) (Fig. 2). The first known naturally occurring case of this TIPS double mutation evolved in E. indica (22), in which the P106S had previously evolved. This double mutation confers a much higher level of glyphosate resistance and maintains the affinity for PEP at a similar level as the WT enzyme (23), although the TIPS mutation was found to have a high fitness cost in E. indica (24). The double TIPS mutation has also been reported in hairy beggarticks (Bidens pilosa) from Mexico (25). A double T102I and P106T (TIPT) mutation was found in greater beggarticks (Bidens subalternans) from Brazil, a tetraploid species in which the TIPT mutation was found in only one of the two genomes (26). Recently, a triple amino acid substitution in EPSPS was found in smooth pigweed (Amaranthus hybridus) in Argentina, with a T102I, A103V, and P106S (TAP-IVS) allele conferring high resistance to glyphosate (27, 28). This stepwise evolution of mutations is an excellent example of how herbicide resistance is rapid evolution, as many gene families over evolutionary time have evolved by this same process involving the incremental accumulation of mutations that alter and improve enzymatic activity and efficiency. When a herbicide with a single site of action is used repeatedly, evolutionary processes leading to higher resistance levels and more efficient resistance mechanisms are expected to occur.
Receptor/co-receptor interactions
Synthetic auxin herbicides mimic the endogenous auxin hormone indole-3-acetic acid (IAA) and deregulate growth and development processes. Synthetic auxins bind to a receptor protein (auxin F-box, or AFB) and to a co-receptor protein (Aux/IAA) to deregulate gene expression controlling plant growth, as well as binding to other auxin-binding proteins. In the case of synthetic auxin herbicides, mutations that reduce herbicide binding to auxin-binding proteins, AFB proteins, or Aux/IAA proteins (the sites of action for synthetic auxins) could have roles in conferring resistance in weeds. For example, assays of auxin-binding protein preparations isolated from auxinic herbicide-susceptible and resistant wild mustard (Sinapsis arvensis) found similar binding for the normal substrate IAA, but differences in binding were found for several auxinic herbicides, and these differences correlated with the whole-plant resistance phenotypes (29).
Multiple auxin-binding proteins interact with native auxins and synthetic auxin herbicides, and evidence indicates that some mutations confer resistance specifically to certain chemical families of synthetic auxins. For example, mutations in the Arabidopsis TIR1 homolog, an AFB protein, confer resistance to the picolinate class of synthetic auxins, such as picloram, but not to 2,4-D (30).
Aux/IAA transcriptional repressors are co-receptors for synthetic auxin herbicides and part of the target site complex. A double-nucleotide TSR substitution was discovered in the IAA16 gene of kochia (Bassia scoparia), changing a GGT (Gly) at amino acid position 127 to an AAT (Asn) (31). This double mutation is located in the conserved degron region II of the Aux/IAA protein and confers resistance to dicamba.
From an evolutionary perspective, rotating different synthetic auxin herbicide chemical families may be an effective resistance management practice, as TSR mechanisms may be highly specific to certain chemical families and may not necessarily confer resistance across different families.
Codon deletion affecting topology of target site
A codon deletion is the removal of three nucleotides from the coding sequence of a gene. The coding frame is unaffected by a codon deletion, and a single amino acid is removed from the protein encoded by the allele carrying the codon deletion. Codon deletions conferring herbicide resistance are rare relative to single nucleotide substitutions, with the only known example to date applying to herbicides that inhibit protoporphyrinogen oxidase (PPO; EC 1.3.3.4), a key enzyme in chlorophyll and heme biosynthesis.
Common waterhemp (Amaranthus tuberculatus) resistant to PPO inhibitors (including lactofen, fomesafen, and others) had a three-nucleotide deletion in its PPX2 gene, which encodes a PPO enzyme (32). It is thought that the three-nucleotide deletion was fostered by its occurrence within a region containing bi-repeats of three nucleotides. These short simple repeat (SSR) regions are typically associated with insertion and deletion events. Most plant species do not have an SSR region in the homologous location of PPX2, suggesting that they are not predisposed for the same mutation (33). Palmer amaranth (Amaranthus palmeri), however, is one of the few weeds with a homologous SSR region (34), and, in fact, resistance to PPO inhibitors due to the same codon deletion was subsequently documented in this species (35). The codon deletion, which specifically results in removal of a glycine at position 210 was functionally validated to confer resistance using a transgenic E. coli system (32) and also has been subjected to biochemical and structural analysis (36). Deletion of glycine 210 is proposed to partially unravel an α-helix adjacent to the PPO active site, raising the Ki for the herbicide and enlarging the active-site cavity (Fig. 3). This structural change confers broad cross-resistance to PPO-inhibiting herbicides at both the enzyme and whole-plant level, although resistance magnitudes vary among herbicides by up to 10-fold (37). Efforts are under way to develop new PPO-inhibiting herbicides that overcome the Gly-210 PPO deletion (38). To date, a codon deletion has not been reported to confer herbicide resistance in any weed species other than A. tuberculatus and A. palmeri.
Figure 3.
View of the catalytic domain of PPO. The porphyrin substrate is centered on top of α-helix 8 (green) and stabilized by several interactions with residues lining the pocket. The yellow spheres represent the position of Gly-210, the deletion of which confers TSR. The two groups of pink spheres represent Arg-128 and Gly-399, which can be substituted to impart TSR.
Increased expression of target site genes
Up-regulation
Baerson et al. (39) found elevated EPSPS expression (2.5–3 times higher) in glyphosate-resistant L. rigidum. The EPSPS protein extracted from resistant and susceptible individuals was equally sensitive to glyphosate inhibition, but basal enzyme activity was higher in the resistant population. The authors concluded that higher EPSPS mRNA expression was resulting in higher EPSPS production, but they were uncertain whether the magnitude of overexpression accounted for the observed resistance level. No evidence was found to indicate EPSPS gene duplication in the glyphosate-resistant population using DNA blot hybridization, despite the observed 2.5–3 times higher EPSPS expression (39). In glyphosate-resistant populations of horseweed (Conyza canadensis) and hairy fleabane (C. bonariensis), basal EPSPS mRNA expression was 2-fold higher than in glyphosate-susceptible populations when measured using Northern blots.
Other examples of target site gene up-regulation have been reported. In three ALS-resistant barleygrass (Hordeum leporinum) populations from Western Australia, ALS enzyme activity was 3-fold higher than in an ALS-susceptible population (40). In addition, the three populations also had a mutation at Pro-197 resulting in a serine substitution. Whereas this mutation confers ALS resistance on its own, the increased ALS expression may also contribute to the observed resistance level. It is not known whether the up-regulation of ALS protein activity is due to altered gene expression regulation, gene duplication, and/or reduced enzyme turnover rates. A johnsongrass (Sorghum halepense) population resistant to the ACCase-inhibiting herbicides sethoxydim and quizalofop had 2–3-fold higher ACCase enzyme activity relative to an ACCase-susceptible population (41). The I50 (herbicide concentration required to inhibit 50% of enzyme activity in vitro) was similar between resistant and susceptible populations, but the higher activity was maintained across a range of herbicide concentrations in vitro. However, the study did not determine whether the increased ACCase enzymatic activity was due to ACCase gene duplication or up-regulation of ACCase transcription.
Target site gene duplication
Gene duplication, the heritable replication of a coding segment of DNA resulting in one or more additional gene copies within the genome of an organism (42), is a common process in the evolutionary history of plants and is vital for generating genomic diversity (43). Increased gene expression at the mRNA level is the immediate result of gene duplication, and as mutations accumulate in duplicated gene copies over time, duplicated gene copies can begin to have variations in function or acquire new functions (42). The term gene amplification is also frequently used synonymously with gene duplication. Gene amplification has been defined in some literature as the nonheritable replication of a segment of DNA, such as in cases of cancer tumors where amplified gene copies are not inherited in the progeny of the individual, and gene duplication indicates the heritable replication of a segment of DNA (42).
The initial examples of EPSPS gene duplication related to glyphosate resistance were obtained in cell culture studies. The involvement of gene duplication in glyphosate-resistant A. palmeri (44) was the first case identified in a weed population (Fig. 4). The EPSPS gene in a glyphosate-resistant A. palmeri population was duplicated from 4- to over 100-fold relative to a susceptible population. Expression of EPSPS mRNA and EPSPS protein corresponded with the increased genomic EPSPS copy number. The increased EPSPS expression means that more EPSPS is present at the target site than the typical applied concentration of glyphosate can inhibit (17). Enough uninhibited EPSPS remains following typical glyphosate applications such that the plant can survive. In the studied A. palmeri population, duplicate copies of the EPSPS gene appeared to be present on all chromosomes (2n = 34).
Figure 4.
Summary of the mechanisms of resistance to glyphosate. Observed (normal type) and putative (italic type) glyphosate resistance mechanisms are shown. Glyphosate (red circles) crosses the plasma membrane (blue) to enter the cytoplasm and is transported into the chloroplast (green) to the target-site enzyme, EPSPS, in herbicide-sensitive plants. Expression of EPSPS variants with 1–3 amino acid differences can confer resistance to the herbicide (17, 24, 27). Target gene duplication of EPSPS produces more EPSPS protein that remains sensitive to glyphosate, requiring proportionally more glyphosate to cause complete inhibition of the extra enzyme (50). Other routes to resistance include sequestration in the vacuole and enhanced metabolism by aldo-keto reductases (77, 78, 145). Altered import/export from the chloroplast and/or cytoplasm may also alter glyphosate effectiveness, but these mechanisms remain hypothetical and have not been documented in weeds. The phoenix phenomenon shown in Fig. 6 results in reduced translocation and involves a currently unknown mechanism triggering cell death upon glyphosate application.
In a fascinating example of molecular genetic variation leading to adaptation, the duplicated EPSPS gene was contained within a >300-kb replicon containing multiple additional open reading frames for other genes and various types of repetitive DNA elements (45). The replicon was found to have a circular structure existing outside the chromosome, termed an extrachromosomal circular DNA (eccDNA) (46). The eccDNA can attach to the chromosomes to be transmitted both at mitosis and at meiosis, explaining the observed variation in heritability of EPSPS gene copy number in A. palmeri (47, 48). The eccDNA sequence across glyphosate-resistant A. palmeri populations from across the United States was nearly identical, suggesting the possibility of a single origin of the eccDNA for glyphosate resistance followed by seed- and pollen-mediated gene flow (45, 49). EPSPS gene duplication has been reported in many glyphosate-resistant weed species. Glyphosate resistance via EPSPS gene duplication can be viewed as an example of convergent evolution across these diverse plant species (50). Populations of B. scoparia and A. tuberculatus have fewer duplicated EPSPS copies than A. palmeri, in the range of 4–10-fold (Table 2).
Table 2.
EPSPS gene duplication and glyphosate resistance level (LD50, dose required to cause 50% mortality) reported in glyphosate-resistant weed species
Note that some values for LD50 were measured in different populations and reported in different studies than EPSPS copy number (adapted from Ref. 17). This research was originally published in Pest Management Science. Sammons, D. R., and Gaines, T. A. Glyphosate resistance: state of knowledge. Pest Manag. Sci. 2014; 70:1367–1377. © John Wiley & Sons, Inc.
| Species | EPSPS relative genomic copy number range | LD50 (resistant/susceptible) |
|---|---|---|
| Bassia scoparia | 3–9 | 2–8 |
| Chloris truncata | 32–48 (51) | 2.4–8.7 |
| Hordeum glaucum | 9–11 (52) | 2.8–6.6 |
| Bromus diandrus | 10–36 (53) | 4.7 |
| Amaranthus spinosus | 26–37 | 5 |
| Amaranthus tuberculatus | 2–8 | 5–19 |
| Lolium multiflorum | 15–25 | 12–13 |
| Amaranthus palmeri | 2–160 | 15–40 |
| Eleusine indica | 28 (54) |
Despite having a lower quantity of duplicated EPSPS copies, A. tuberculatus populations have a similarly high level of resistance as the A. palmeri populations (Table 2). Both EPSPS mRNA expression and EPSPS protein levels have been reported to have a linear correlation with EPSPS genomic copy number in B. scoparia and A. tuberculatus populations (55, 56), as well as in Italian ryegrass (Lolium multiflorum populations) (57). In the case of L. multiflorum, EPSPS protein expression level correlated very well with population level resistance; as EPSPS protein expression increased, so did the dose required to achieve 50% reduction in plant growth (57). Comparisons of resistance level across species are problematic, as the various reports were conducted under different experimental conditions, and estimated LD50 and GR50 (concentrations causing 50% mortality and growth reduction, respectively) parameters are not directly comparable. However, available evidence suggests that higher EPSPS copy number confers higher glyphosate resistance in A. palmeri (e.g. Ref. 48), L. multiflorum (57), and B. scoparia (58). Continued observation and monitoring of EPSPS copy number in populations over time will be needed to determine whether EPSPS copy number may increase following continued glyphosate selection pressure.
Recently, a population of glyphosate-resistant spiny amaranth (Amaranthus spinosus) was reported in which EPSPS gene duplication and sequence data revealed that the EPSPS gene in glyphosate-resistant A. spinosus individuals is identical to glyphosate-resistant A. palmeri EPSPS (59) and not glyphosate-susceptible A. spinosus. This result indicates that the EPSPS gene has transferred through interspecific cross-pollination. The two species are most closely related to each other among Amaranthus species (60), and the two species can hybridize and produce fertile hybrids (61). This is important from an evolutionary perspective, because a genetic trait that may be extremely rare within populations (such as the eccDNA-containing EPSPS) has a selective advantage in multiple species that are under similar selection environments (repeated exposure to glyphosate). Presumably, genetic transfer between A. palmeri and A. spinosus occurs at a low level in wild populations, but the selective advantage of the glyphosate resistance trait that initially evolved in A. palmeri enabled transfer of this trait, plus additional, unknown linked genetic traits, to A. spinosus. This genetic transfer may have evolutionary implications beyond loss of glyphosate effectiveness in A. spinosus.
Whereas EPSPS gene duplication in A. palmeri has occurred via eccDNA, in B. scoparia a tandem gene duplication has occurred (58). Several repeat units were identified in a bacterial artificial chromosome assembly of the duplicated locus, including a 56.1-kb repeat containing seven predicted genes and a 32.7-kb repeat containing four predicted genes (62). Some of these co-duplicated genes showed similar increased expression as EPSPS. The border of the duplicated region contained a mobile genetic element that may have provided the initial DNA break to initiate the tandem duplication process. The potential evolutionary consequences of duplicating and overexpressing other genes in addition to EPSPS need further exploration, as does the relative stability of duplicated EPSPS inheritance for both tandem duplication and eccDNA mechanisms (63).
In addition to EPSPS gene duplication, the target-site gene ACCase was found to have a 5–7-fold higher gene copy number in a large crabgrass (Digitaria sanguinalis) population resistant to five ACCase inhibitor herbicides, resulting in 3–9-fold higher ACCase transcript abundance (64).
Nontarget-site mechanisms
Reduced absorption
To be effective, herbicides must be absorbed into cells of plants through the roots, in the case of soil-applied herbicides, or from the leaves in the case of foliar-applied herbicides (Fig. 5). Menendez et al. (65) provide an excellent description of the factors involved in foliar absorption and root absorption of herbicides. Differences in root absorption of herbicides between species have been attributed to root morphology differences (66). There are no cases of evolved resistance to soil-applied herbicides due to reduced root absorption. Early work on differential foliar absorption of herbicides between species was attributed mainly to differences in cuticle thickness and/or composition (66), but the number and/or structures of leaf trichomes and hairs have also been implicated (67). Hirsute leaves are covered with hairy trichomes that can retain spray droplets better than smooth, hairless, or glandless cuticles, thereby facilitating absorption. Other leaves have lysigenous glands involved in the production and storage of oily secondary metabolites that can compartmentalize lipophilic herbicides, preventing them from reaching their site of action (68). Differences in foliar absorption of herbicides between plants have been attributed to leaf anatomical features rather than any biochemical differences.
Figure 5.
Summary of NTSR. Plants can evolve resistance to a herbicide by reducing its absorption, altering its translocation and/or sequestration, or developing a rapid necrosis of the foliage (phoenix phenomenon) or via degradation of the active ingredient through phases I, II, and III of metabolism.
Decreased absorption is not a common NTSR mechanism, but it has been reported with resistance of common sunflower (Helianthus annuus) to imazethapyr and chlorimuron (69), prickly lettuce (Lactuca serriola) to 2,4-D, annual bluegrass (Poa annua) to atrazine (70), and L. multiflorum and S. halepense to glyphosate (71, 72). No differences were found in cuticular wax amount per unit area of leaf surface between two biotypes of L. multiflorum with a 3-fold difference in glyphosate susceptibility and reduced absorption in the less sensitive biotype (73). When reduced absorption is implicated, it is most often only one contributing factor to the overall resistance mechanism. For example, resistance to glyphosate in A. tuberculatus biotypes was due to both reduced absorption and a herbicide-resistance allele of the glyphosate enzyme target EPSPS (74).
Reduced translocation and vacuolar sequestration
Many foliar-applied systemic herbicides rely on translocation through the phloem for optimal activity. These herbicides must cross the cuticle barrier and enter the cells of mature source leaves (symplast). This transport can involve active (i.e. protein-mediated) and/or passive diffusion processes. Once inside the symplast, systemic herbicides translocate from source leaves to younger sink leaves via the phloem, often along with the movement of photosynthetic sugars. Herbicide resistance due to reduced translocation occurs when the herbicide is retained in source leaves and prevented from translocating to the growing points (Fig. 5). Mechanisms that trap the herbicide in source leaves (e.g. through sequestration within vacuoles or leaf trichomes) or prevent its normal movement to the growing points across membrane barriers (through altered activity of active membrane transporters) will reduce the total amount of herbicide translocated, thus conferring resistance. Reduced absorption across the cuticle and reduced translocation out of source leaves sometimes work in concert.
Reduced translocation of glyphosate is the most prominent example of this NTSR mechanism (63). In these plants, the amount of glyphosate delivered to the meristems is lower than what is necessary to be phytotoxic. Reduced glyphosate translocation was first demonstrated in glyphosate-resistant L. rigidum from Australia, where glyphosate moved to the edges of treated leaves, and less glyphosate translocated to the meristems, relative to glyphosate-susceptible L. rigidum (75). Glyphosate-resistant C. canadensis had reduced translocation as well (76). This is due to differences in cellular distribution of glyphosate and subsequent phloem loading and translocation.
In these biotypes, glyphosate enters the symplasm of source leaves normally but cannot translocate to the meristems because it is rapidly sequestered within the vacuole (77) (Fig. 4). The vacuole sequestration process is temperature-dependent, with less sequestration occurring in C. canadensis under colder temperatures (78). The dependence on temperature suggests the involvement of active membrane transporters. Considerable research effort with both microarray and transcriptomic sequencing (79, 80) has been devoted to the search for a specific ABC transporter gene suspected to be responsible for the vacuolar sequestration, but no causative specific gene has yet been confirmed.
The effect of lower temperatures on the reduced translocation mechanism has also been reported in S. halepense and L. multiflorum (81). The temperature effect supports the hypothesis that vacuole sequestration is an active process and restricts glyphosate translocation by preventing normal glyphosate movement into the phloem.
Reduced translocation may result from other NTSR mechanisms in A. palmeri, A. tuberculatus, and S. halepense (17). In these reports, reduced cellular glyphosate absorption across the plasma membrane may be associated with altered rate of an active glyphosate transport process (Fig. 4). Reduced cellular uptake is not predicted to alter translocation at the whole-plant level, and generally no changes in glyphosate translocation have been observed in A. palmeri. Finally, reduced chloroplast absorption of glyphosate has been suggested (17) (Fig. 4). This phenotype is difficult to measure, as isolating intact chloroplasts is technically difficult and may have unknown effects on the physiological ability of the isolated chloroplasts to exhibit any reduced absorption. To date, reduced chloroplast glyphosate absorption as a glyphosate resistance mechanism has not been experimentally demonstrated.
Reduced translocation of paraquat from treated source leaves to sink leaves has also been reported for paraquat-resistant L. rigidum (82). In this population, there was no difference in the interaction of paraquat with the target site photosystem I complex (indicating a lack of TSR), and no differences were found in the antioxidant systems of superoxide dismutase or ascorbate peroxidase (increased antioxidant activity may allow a plant to tolerate the free radicals generated by diversion of electrons from photosystem I). Because the population had reduced translocation and because of the finding of Ge et al. (77) demonstrating vacuole sequestration of glyphosate, intact protoplasts of paraquat-resistant and -susceptible L. rigidum were isolated. The paraquat concentration was measured, as a method to determine whether the paraquat concentration within single cells was higher in paraquat-resistant protoplasts than in paraquat-susceptible protoplasts (83). Paraquat-resistant L. rigidum protoplasts contained more paraquat than susceptible protoplasts. Thus, the paraquat is likely sequestered in the vacuole, in a process similar to that reported for vacuolar sequestration of glyphosate. Reduced paraquat translocation attributed to vacuole sequestration has also been reported in L. multiflorum from California (84) and in two Conyza spp. from California (85). Characterization of a paraquat-resistant Arabidopsis mutant revealed a mutation in a gene called Paraquat Resistant 1 (PAR1), a putative amino acid transporter (86). The mutant, par1, had similar cellular paraquat absorption as the normal PAR1 genotype but had reduced paraquat concentration in the chloroplast. Overexpressing the PAR1 homologue in rice resulted in paraquat hypersensitivity, and silencing the rice PAR1 resulted in paraquat resistance, revealing that reduction in paraquat transport into the chloroplast can confer resistance. Natural polymorphic variations in a gene (LHR1) for a plasma membrane–localized polyamine transporter in Arabidopsis caused variations in uptake of paraquat that correlated with sensitivity to paraquat (87). Endogenous polyamines have been shown to play a role in paraquat-resistant E. indica (88). Evolved paraquat resistance in weeds has not yet been attributed to selection for orthologs of PAR1 or LHR1.
Reduced translocation of 2,4-D was observed in a 2,4-D–resistant population of L. serriola, relative to a susceptible population (89), as well as in 2,4-D–resistant wild radish (Raphanus raphanistrum) (90). Reduced dicamba translocation was found in a dicamba-resistant B. scoparia population (91). Naturally occurring auxins such as IAA are polar and readily translocate in the phloem, so it is reasonable that a synthetic auxin such as 2,4-D or dicamba also requires adequate translocation from the application site to the target site in growing meristems. Changes in auxin transport from cell to cell via active transporters could play a role in reduced 2,4-D translocation. Reduced translocation of 2,4-D or dicamba would presumably reduce the total concentration achieved at the target site to low enough levels to enable survival.
An unusual way for a plant to achieve reduced translocation of a herbicide to the meristems has been described as the phoenix phenomenon, which is the result of reduced translocation caused by more rapid action of the herbicide (Fig. 6). One of the assets of glyphosate as a herbicide is that it acts slowly, allowing it to translocate to and kill meristematic tissues. With herbicides that act rapidly, translocation from treated plant organs is limited because of the rapid action of the herbicide. Giant ragweed (Ambrosia trifida) evolved a rapid response to glyphosate that prevents translocation of the herbicide to meristems (92, 93). Light- and/or sucrose-dependent rapid withering and desiccation of treated foliage occurs, followed by regrowth of the plant from meristems that were not contacted by the foliar spray, hence the “phoenix phenomenon.” None of the other known mechanisms of glyphosate resistance were found (93). Reactive oxygen species (ROS) accumulated within 30 min of treatment, only in the older leaves of the resistant biotype of the weed. Treatment of the leaf tissue with exogenous phenylalanine and tyrosine (two aromatic amino acids that are products of the shikimate pathway) prevents the rapid effect, indicating that the effect may be associated with EPSPS inhibition or with physiological blocking of the rapid response by the amino acids. The mechanism for accelerated action of glyphosate in this biotype is unknown, but it might be explained by rapid cessation of carbon fixation due a rapid deregulation of the shikimate pathway removing enough erythrose 4-phosphate and phosphoenolpyruvate from the C3 carbon fixation pathway to stop it. In another species with a rapid response to glyphosate, sugar beet (Beta vulgaris), cessation of carbon fixation is rapid, and the symptoms are similar to those of the resistant A. trifida. Alternatively, perception of glyphosate in mature, green plant cells of this resistant biotype may trigger a rapid cell death defense mechanism.
Figure 6.
The phoenix phenomenon in plants treated with glyphosate. Both giant ragweed (Ambrosia trifida) biotypes were sprayed with 0.7 kg/hectare glyphosate. Shown is glyphosate-susceptible A. trifida at 2 days (A) and 21 days (B) after glyphosate treatment, behaving like most plants treated with glyphosate. Growth stops, but no injury is observed for the first few days. Shown is glyphosate-resistant A. trifida at 2 days (C) and 21 days (D) after glyphosate treatment. In plants exhibiting the phoenix phenomenon, older leaves desiccate very rapidly, trapping most of the glyphosate in dead tissues, and the new shoots emerge undamaged from the glyphosate treatment. Cover image from Ref. 93 with permission from John Wiley & Sons, Inc.
Rapid necrosis followed by regrowth was also identified as a resistance mechanism to 2,4-D in a Sumatran fleabane (Conyza sumatrensis) biotype from southern Brazil (94). The symptoms after 2,4-D application are similar to those observed in glyphosate-resistant A. trifida (i.e. necrosis in older leaves and absent in younger leaves, followed by regrowth from meristems). The 2,4-D–resistant biotype was also resistant to glyphosate, but not by the phoenix phenomenon. Rapid necrosis symptoms did not occur with six other auxinic herbicides (e.g. picloram). ROS accumulation was much higher within 30 min after 2,4-D treatment than in a susceptible accession and remained much higher for more than 7 h. The resistant biotype did not show the normal epinasty symptom (leaves bending downward) caused by 2,4-D, probably due to rapid cell death caused by ROS inhibiting herbicide translocation.
Metabolic alterations
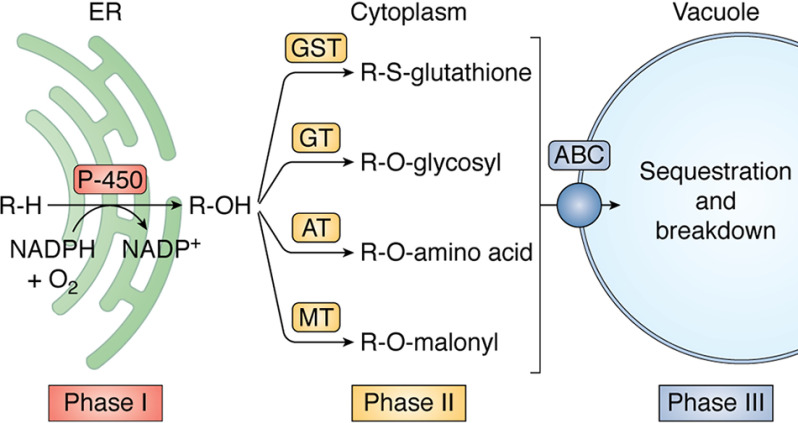
Plants contain large numbers of genes encoding enzymes that perform biochemical reactions for the synthesis of secondary metabolites and for detoxifying xenobiotic compounds (e.g. herbicides) (95). Serendipitously, some members of these gene families can also detoxify herbicides. The selective action of many herbicides (i.e. they control weeds without damaging crops) often depends on relatively rapid metabolism of the active ingredients into harmless breakdown products in crops compared with weeds. This biochemical feature (differential rates of detoxification) has been repeatedly exploited for selective chemical weed control. Herbicide detoxification is generally divided into three phases (Figs. 5 and 7). Phase I involves the addition of a functional group to the herbicide by oxidation, reduction, or hydrolysis, often mediated by cytochrome P450 monooxygenases (P450; EC 1.6.2.4). Phase II involves more complex changes to a herbicide, such as conjugation, either to GSH mediated by GSH S-transferases (GSTs; EC 2.5.1.18) or to glucose mediated by glucosyltransferases (GTs; EC 2.4). Note that phase II enzymes such as GSTs and GTs can directly detoxify some herbicides without depending on phase I activation. The final step in plants (phase III) involves compartmentalization of the herbicide metabolites in the vacuole or incorporation into cell walls (Fig. 7). In general, the same genes and biochemical mechanisms for herbicide detoxification often exist in weeds that are related to crops, with the critical difference being that expression of these genes is lower in the weeds. Thus, there is evolutionary potential to select for increased expression and/or mutations of these key genes in weeds, enabling enhanced herbicide metabolism to confer resistance.
Figure 7.
Summary of herbicide metabolism in a plant cell. Herbicides are normally taken through the three phases of metabolism as they are detoxified by plant cells. Typically, phase I introduces small functional groups on the structure of the active ingredient, phase II attaches a number of water-soluble metabolites via the action of several types of transferases, and phase III moves the conjugated metabolites to the vacuole (or the cell wall) for compartmentalization and further degradation. Active transport sometimes requires ABC transporters (or other transporter types) to move the herbicide metabolites across membranes.
Prominent examples of herbicides that are selective due to differential detoxification between crops and weeds include the selective ALS and ACCase inhibitors. Some of these, including chlorsulfuron, diclofop-methyl, and fenoxaprop-P-ethyl, are used to control grass weeds in wheat. For these herbicides, rapid metabolism and crop safety in wheat involves detoxification pathways including P450, GT, and GST. For example, metabolism of the ACCase inhibitor diclofop-methyl to nontoxic metabolites in wheat occurs via P450-mediated aryl hydroxylation, followed by glucose conjugation, likely mediated by GT (e.g. Ref. 96). These wheat-like metabolic pathways also exist in grass weeds that have considerable intraspecific variation for herbicide susceptibility. In susceptible weed populations, the detoxification rates are generally too slow to prevent herbicide phytotoxicity. However, some weed species, such as blackgrass (Alopecurus myosuroides) in Europe and L. rigidum in Australia, have evolved wheat-like rapid detoxification pathways (e.g. Refs. 97 and 98). These weed populations have long histories of repeated exposure to wheat-selective herbicides and have evolved enhanced metabolism-based resistance.
Enhanced herbicide metabolism is especially problematic from a weed management standpoint because the detoxification systems that confer resistance to one herbicide can sometimes have activity on other herbicides with the same or unrelated sites of action. The term cross-resistance is defined as when a single mechanism (such as enhanced metabolism) confers resistance to more than one herbicide with the same or of a different site-of-action group. The term multiple resistance is defined as when multiple, distinct mechanisms have combined within an individual (or population), and the individual (or population) is resistant to herbicides from more than one site-of-action group. When considering enhanced metabolism, it may often be difficult to distinguish phenotypically whether a single metabolic mechanism is conferring cross-resistance to other herbicide groups or if multiple distinct metabolic mechanisms are present. The key concept is that some enhanced metabolism mechanisms, including P450 and GST, can confer broad-spectrum resistance, with known examples of cross-resistance due to a single mechanism and multiple resistance due to accumulation of multiple, distinct mechanisms. From an evolutionary biology perspective, enhanced metabolism can be considered a broad-spectrum, generalist adaptive response. Critically, enhanced metabolism mechanisms can also combine with other mechanisms including TSR and reduced translocation to confer higher levels of resistance.
Cytochrome P450–mediated herbicide metabolism
Cytochrome P450 monooxygenases are membrane-bound proteins localized in the endoplasmic reticulum and are one of the largest gene families in all organisms (99). These enzymes have crucial roles in the synthesis of hormones, lipids, and metabolism of endogenous and exogenous substances (Fig. 8). The number and diversity of P450s in plants is higher than in other organisms. Whereas in humans there are ∼54 P450 genes (0.1% of genome), in plants the number is higher, with 246 in Arabidopsis (1% of genome) and 328 in rice (0.5-1% of genome). Weeds have been found to have high diversity in P450 genes, with 917, 323, and 277 in barnyard grass (Echinochloa crus-galli), C. canadensis, and L. rigidum, respectively. The greater diversity of the P450 genes in plants has likely evolved for chemical defense, mainly for degradation of many xenobiotics, and by chance these P450 genes also detoxify herbicides (100). These genes have gained importance in agriculture as the basis for herbicide selectivity in crops due to natural metabolic processes. For example, different P450 monooxygenases metabolize ALS inhibitors in rice (101), cinmethylin in wheat (102), 4-hydroxyphenylpyruvate dioxygenase (HPPD; EC 1.13.11.27) inhibitors in maize (103), and other mode-of-action herbicides in different crops (104) at a faster rate than in weeds. The implication is that genetic variation exists in plants for the expression level of P450 genes, and crops have higher expression of P450 genes that metabolize herbicides. It should be noted that the herbicide discovery process often identifies candidate herbicides that are metabolized faster in some weeds than in crops; these herbicides would not be effective and are not commercialized. However, the wide use of herbicides for weed control has selected biotypes with the same ability to inactivate herbicide molecules, threatening weed management worldwide. The most important reactions catalyzed by these enzymes are either aryl or alkyl hydroxylation, the first step in the metabolism of xenobiotics (105). In general, P450s insert molecular oxygen on a herbicide molecule to be more reactive or more soluble using an electron from NADPH P450 reductase. As a result, herbicide molecules are metabolized to products with reduced or modified phytotoxicity in weeds with metabolism-based resistance mechanisms.
Figure 8.

Examples of the reactions catalyzed by plant cytochrome P450 monooxygenases involved in herbicide metabolism. Functional groups either on the substrates of P450 monooxygenases or on the products of the reactions catalyzed by these enzymes are shown in red. ER, endoplasmic reticulum.
Extensive work with P450 inhibitors provides strong evidence for the role of various P450 isozymes in herbicide metabolism. Application of a P450 inhibitor prior to herbicide exposure will block P450 activity. When the resistance mechanism is due to enhanced metabolism by P450s, inhibition of the P450 activity enables the herbicide to reach the target site in a high enough concentration to cause normal phytotoxicity. P450 inhibitors have been used in various resistant weeds to test for P450 herbicide metabolism. In L. rigidum populations resistant to multiple herbicides through metabolism-based mechanisms, the P450 inhibitor malathion restores chlorsulfuron activity (106). The herbicide amitrole, structurally similar to the P450 inhibitor 1-aminobenzotriazole, restores diclofop-methyl activity. However, the P450 inhibitors have specificity for P450 isoforms. Pretreatment with piperonyl butoxide (PBO), but not malathion, increased control of resistant P. annua with fenoxaprop (107), indicating that PBO inhibited specific P450 enzymes critical for fenoxaprop metabolism. Although P450 genes found in the past decade were mostly related to metabolism of ACCase, ALS, and PSII inhibitors, more recently P450 inhibitors have inhibited the P450 metabolism of several classes of herbicides (i.e. HPPD, PPO, synthetic auxins, and carotenoid synthesis inhibitors) (108–111). Critically, P450s have the potential to metabolize herbicides from different mode-of-action groups, creating unpredictable patterns of cross-resistance.
Successful isolation of microsomes with herbicide metabolism activity from weeds was achieved in late watergrass (Echinochloa phyllopogon) by first inducing P450 activity with a sublethal herbicide dose, demonstrating P450 activity in metabolizing bispyribac-sodium, fenoxaprop-P-ethyl, and thiobencarb (112). E. phyllopogon populations are also resistant to clomazone and penoxsulam via enhanced oxidative metabolism from P450 activity (113, 114). Expression of several P450 genes was measured using quantitative PCR in a metabolism-based resistant E. phyllopogon population relative to a susceptible population, and substantial induction of the P450 genes occurred following sublethal herbicide treatment (115). Analysis of 12 candidate P450 genes from resistant E. phyllopogon revealed that CYP81A12 and CYP81A21 had the highest transcription levels in the resistant biotype (116). These P450 genes were transformed into Arabidopsis, and transgenic plants exhibited resistance to bensulfuron-methyl and penoxsulam (116). Recently, P450 genes were cloned from E. phyllopogon and transformed into both Arabidopsis and E. coli (117). The ability to assay the membrane-bound P450 proteins in bacteria is a major advance in available tools to functionally validate candidate resistance genes from weeds. The E. phyllopogon CYP81As metabolized 18 herbicides from 13 different chemical classes. The recombinant expression of CYP81As in E. coli metabolized different herbicides by demethylation or hydroxylation reactions in unrelated herbicide groups belonging to ALS, ACCase, PDS, PSII, PPO, HPPD, and 1-deoxy-d-xylulose-5-phosphate synthase inhibitors (117), demonstrating the incredible potential of P450 enzymes to metabolize herbicides and confer cross-resistance.
The generally broad substrate recognition of plant P450 enzymes poses a threat for weed management due to potentially unpredictable cross-resistance patterns. Whereas CYP81A12 and CYP81A21 can metabolize ALS inhibitors through demethylation, the same genes are also involved in cross-resistance to ACCase inhibitors such as diclofop-methyl, tralkoxydim, and pinoxaden through hydroxylation (118). The same CYP81A12 and CYP81A21 genes, as well as CYP81A15 and CYP81A24, confer resistance to clomazone (119). Other enzymes from this P450 family, such as CYP81A6, were found to metabolize bentazon, sulfonylureas, and also quinclorac in rice. The P450 enzymes of family CYP81A appear to be “super-P450s” able to metabolize different chemical classes within different herbicide mechanisms of action in the same weed species. However, other P450 enzymes of families CYP72, CYP71, CYP70, and CYP96 have also been found to metabolize different herbicides.
Fewer examples of enhanced P450 metabolism have been identified in eudicot species (typically broadleaf species). The reasons may be biological, such as lower P450 activity or fewer P450 genes in eudicots, or the reason may be due to less research into enhanced herbicide metabolism for eudicots. Recently, reports of metabolism-based herbicide resistance in eudicots are increasing. More rapid initial metabolism of chlorimuron was found in an ALS-resistant A. hybridus population (120). A survey of ALS-resistant A. palmeri in Georgia found that more than half of the populations had no ALS target-site mutations and exhibited enhanced ALS herbicide metabolism (121). Resistance to HPPD inhibitors has been reported in A. tuberculatus (122, 123), in A. palmeri (108, 124), and recently in R. raphanistrum (125). The resistant biotypes metabolized HPPD inhibitors at a faster rate than the susceptible populations through hydroxylation reactions, indicating a P450 role in the resistance mechanism in the broadleaf species (108, 125, 126). Recently, A. tuberculatus biotypes from Nebraska with resistance to tembotrione and 2,4-D, as well as A. palmeri from Arkansas with resistance to fomesafen, were controlled when a P450 inhibitor was applied (109, 111, 127). As research efforts into metabolism-based resistance expand, more examples of broadleaf species with enhanced herbicide metabolism will likely be reported, and the candidate genes in eudicot species can be identified.
More work is needed to improve understanding of P450 evolution and regulation for herbicide resistance. The role of cytochrome P450 gene copy number variation should be studied, especially in polyploid weeds, as a pathway to generate novel allelic variation and increased expression. Chromosome duplication in polyploid species may generate more potential variation for the evolution of metabolic resistance through P450s. Little is known about the structure-activity relationship between the many P450s enzymes and which substrates (herbicides) they may degrade. Biochemical modeling predicting the tertiary structure of known P450s from weeds would enable docking simulations with different herbicides and anticipate classes of herbicides more likely to succumb to metabolic degradation. Transcriptional regulation of P450 genes is another area where more research is needed. Although a trans-element was proposed to control the expression of both CYP81A12 and CYP81A21 genes in the resistant E. phyllopogon (116), we lack studies to functionally demonstrate transcription factors, activator elements, repressor elements, or epigenetic modifications that may be regulating P450 gene expression in metabolic resistant weeds.
GSTs
The enzyme superfamily of GSTs is involved in herbicide detoxification by conjugating GSH to the herbicide molecule, rendering the herbicide nontoxic. This conjugation reaction can occur directly to the active herbicide or following the activity of other enzymes, such as P450s. GST activity was first identified on triazine herbicides, including atrazine in corn (128). Additional herbicides known to be GSH-conjugated by GSTs include chloroacetamides (such as alachlor and metolachlor), sulfonylureas (such as chlorimuron ethyl), diphenylethers (such as fluorodifen), and aryloxyphenoxypropionates (such as fenoxaprop-ethyl) (129). E. phyllopogon populations from California are resistant to fenoxaprop-ethyl via enhanced GST-mediated GSH conjugation (130), as are A. myosuroides populations from the United Kingdom (131). Not all herbicides within a given chemical group are equally susceptible to GST-mediated GSH conjugation. For example, whereas GST acts directly on the ACCase inhibitor fenoxaprop-ethyl (131), structurally similar ACCase inhibitor herbicides, such as diclofop-methyl, have no chemical features that can be GSH-conjugated and are instead able to be ring-hydroxylated by P450s (96). The chemical structure of the herbicide determines whether GST is able to conjugate the herbicide with GSH. Recently, a GST (AmGSTF1), from multiple herbicide–resistant A. myosuroides with enhanced GST activity, was cloned and expressed in transgenic Arabidopsis (132). The transgenic lines had increased resistance to chlorotoluron, alachlor, and atrazine and also had increased antioxidant flavonoid and anthocyanin content. In additional experiments, a GST inhibitor (4-chloro-7-nitro-benzoxadiazole) synergized with the herbicides fenoxaprop and clodinafop in multiple-resistant A. myosuroides populations and restored herbicide activity (132), providing additional evidence for the role of increased GST expression as a resistance mechanism. A. myosuroides populations in Europe were found to be resistant to flufenacet via enhanced GST-mediated metabolism (133). Populations of Lolium spp. resistant to flufenacet were found to conjugate GSH to flufenacet for metabolic detoxification, and overall GST activity was increased in protein extracts (134).
GTs and other transferases
The enzyme family of GTs has roles in phase II of herbicide metabolism, via conjugation of glucose to herbicide metabolites after initial phase I modification (typically hydroxylation or demethylation) (135). For example, monocot species (most often grasses) are tolerant to synthetic auxins in part because glycosylation of hydroxylated rings of auxinic herbicides tends to be irreversible, highlighting the importance of GT in protecting a plant from herbicide activity (67). To date, no reports are known of herbicides for which the first step of metabolism involves GT; however, increased GT expression may be necessary as a secondary biochemical step for metabolic resistance in weeds (136, 137).
On the other hand, sensitive eudicots tend to utilize other transferases, such as amino acid transferases, to catalyze a reversible conjugation of amino acid residues to auxinic herbicides, but a herbicidally active metabolite may be recovered upon hydrolysis (138).
Similarly, several other classes of transferases may be involved in phase II metabolism. However, enzymes such as malonyl-transferase typically impart herbicide selectivity but have not been demonstrated to be directly involved in evolved herbicide resistance (139).
Aryl acylamidase
The mechanism of natural tolerance of rice (Oryza spp.) to the PSII inhibitor herbicide propanil is high levels of aryl acylamidase (AA; EC 3.5.1.13), an enzyme that hydrolyzes propanil to 3,4-dinitroaniline, a nonphytotoxic compound. Studies of a WT rice and a rice mutant with no AA activity indicated that the normal role of this enzyme in plants is in nitrogen metabolism relating to asparagine. Weedy rice (feral forms of cultivated rice, often called red rice) is also tolerant to propanil by the same mechanism. Among Oryza species, tolerance to propanil correlates with AA activity (140). Resistance to propanil by elevated AA activity has evolved in jungle rice (Echinochloa colona) (141) and E. crus-galli (142). The AA inhibitors anilofos, piperophos, and carbaryl synergize with propanil activity in propanil-resistant E. crus-galli (142). Not all evolved resistance to propanil is by enhanced AA activity, as resistance to propanil in rice sedge (Cyperus difformis) is a single amino acid change of the D1 protein of PSII (143).
Aldo-keto reductase
Some plant species, especially legumes, metabolize glyphosate to aminomethyl phosphonic acid (AMPA) and glyoxylate, whereas others (grasses in particular) apparently have little capacity for this degradation pathway or any other transformation of the herbicide (144). Neither the enzyme nor the gene for the glyphosate oxidoreductase (GOX) that is considered responsible for glyphosate degradation in plants has been identified. AMPA is very weakly phytotoxic, so sufficiently rapid degradation of glyphosate to AMPA should provide resistance. Because glyphosate is a very slow-acting herbicide, evolution of such a resistance mechanism would seem likely. However, numerous studies have found no differences in glyphosate degradation in glyphosate-resistant weeds. Pan et al. (145), however, recently found that the mechanism of glyphosate resistance of an E. colona biotype involves elevated levels of aldo-keto reductase (AKR; EC 1.1) due to up-regulation of two AKR genes, resulting in more rapid metabolism of glyphosate to AMPA. Earlier, overexpression of an AKR gene from rice provided glyphosate resistance to tobacco, and silencing this gene in rice caused hypersensitivity to glyphosate (146). Whether AKRs are the GOX enzymes that cause accumulation of AMPA in other weeds and crops remains to be determined (144).
β-Cyanoalanine synthase
An example of the broad-spectrum resistance that can be conferred by enhanced metabolic activity is quinclorac resistance in E. phyllopogon. Quinclorac is a unique auxin-type herbicide because it has activity on grasses. Normally, most synthetic auxin herbicides have excellent activity on eudicot species but no activity or very little activity on grasses. Quinclorac stimulates ethylene synthesis in sensitive grass species, which results in accumulation of cyanide and subsequent toxicity (147). The enzyme β-cyanoalanine synthase (β-CAS; EC 4.4.1.9) detoxifies the cyanide generated upon ethylene synthesis, but normally β-CAS activity is insufficient to protect against the toxic accumulation of cyanide. Surprisingly, E. phyllopogon populations in California exhibited quinclorac resistance before quinclorac had been used commercially (148), due to their selection history with multiple other herbicides and resulting enrichment for enhanced metabolism mechanisms. These quinclorac-resistant populations were found to produce less ethylene after quinclorac treatment than susceptible populations. The P450 inhibitor malathion synergized with quinclorac on the resistant populations, suggesting that enhanced P450 activity detoxified quinclorac and inhibition of this P450 metabolism enabled quinclorac to reach phytotoxic concentrations. The mechanism is even more complicated, however, as β-CAS activity was 2–3-fold higher in resistant populations. The malathion treatment did not change ethylene production in either the resistant or the susceptible populations, but malathion treatment did inhibit β-CAS activity in the resistant population. Therefore, malathion is also an inhibitor of β-CAS activity in addition to P450 activity. β-CAS functions to process accumulated cyanide and metabolize it to nontoxic forms. The enhanced activity of this protective mechanism in the resistant population was inhibited by malathion, demonstrating that enhanced β-CAS activity can confer metabolic resistance to quinclorac. An additional, unknown mechanism makes the resistant populations less sensitive to the stimulation of ethylene production by quinclorac.
NTSR summary
The diverse mechanisms that can contribute to NTSR are complex and involve several different gene types, many of which exist in plants as gene families. This means that identifying the specific genes involved in a particular case of resistance can be difficult. Gene family members can be difficult to distinguish using traditional sequencing techniques and can be subject to specific expression regulation at various developmental stages or following herbicide treatments. Sequence variation among gene family members occurs normally and plays an important evolutionary role in functional diversification, so mutations altering substrate specificity for herbicide detoxification may also occur. The most important gene families for NTSR characterized to date are P450s and GSTs. Unraveling the complex mechanisms involved in NTSR and understanding their relationship to overall plant stress response pathways is a clear priority for future research (149). To that end, ongoing research is utilizing next-generation sequencing technology and transcriptomics to help unravel the molecular and genetic basis of NTSR, particularly for metabolism-based resistance (136, 150). An improved understanding of the evolution and regulation of NTSR could guide an additional classification system for herbicides, old and new, according to their metabolism or sequestration by specific NTSR mechanisms.
Importance of genomics and transcriptomics in understanding NTSR
Understanding the molecular and genetic basis of resistance mechanisms can help explain patterns of cross-resistance across different herbicide modes of action due to a single mechanism. Target site mutations are specific for herbicides within the same mode of action. NTSR mechanisms, however, can confer resistance across dissimilar modes of action. For example, similarities in the resistance mechanism could provide an explanation for the concomitant evolution of paraquat resistance following recurrent selection with low glyphosate rates in L. rigidum (151). A more thorough understanding of cross-resistance patterns across different modes of action would be highly beneficial for developing improved herbicide rotation recommendations, avoiding repeated selection with herbicides that are vulnerable to shared resistance mechanisms despite having different modes of action.
“Omics” approaches (genomics, transcriptomics, proteomics, and metabolomics) hold great potential for making rapid advances in our understanding of NTSR mechanisms (152). Of these omics approaches, transcriptomics has provided the most insights thus far. It is generally thought that NTSR is often mediated by increased expression of one or more genes (e.g. a gene encoding a P450 or an ABC transporter), and consequently, a transcriptomics approach is ideally suited to identify such genes (153). One of the downfalls of a transcriptomic approach, however, is that it usually yields several “false positives,” requiring significant downstream research to validate candidate genes. Consequently, scientists typically opt to select the most obvious candidates (such as genes annotated as coding potential metabolism enzymes) for follow-up study, reducing the likelihood that novel NTSR genes will be discovered. Additionally, identification of a gene that is overexpressed and confers herbicide resistance provides insight into the physiology of the resistance mechanism but not directly into the evolution of the mechanism. Specifically, transcriptomics is not well-suited to determine the molecular change that caused overexpression of the gene.
Draft genomes have recently been obtained for some of our most important herbicide-resistant weeds, including A. tuberculatus (154), C. canadensis (155), B. scoparia (62), R. raphanistrum (156), E. crus-galli (157), and E. indica (158). Although some of these genomes are still highly fragmented, those of A. tuberculatus, C. canadensis, and E. crus-galli have N50s greater than 1.5 Mb (N50 is the length of a computationally assembled contiguous sequence for which 50% of the sequences in an assembly are longer and 50% are shorter; longer N50 indicates a better assembly). Such high-quality genomes enable identification of candidate resistance genes through approaches such as genetic mapping of quantitative trait loci in biparental mapping populations and genome-wide association studies in existing populations (159). Such genomic approaches could identify herbicide-resistance genes even if they are not overexpressed, as well as potentially identify trans-acting factors that cause overexpression of genes identified through transcriptomics approaches.
Genetics of herbicide resistance
The inheritance and genetic basis of different resistance mechanisms have evolutionary implications, such as how rapidly selection can occur and how frequent mechanisms may be in unselected populations. As described above, many herbicide-resistance traits, particularly TSR, require a change of a single enzyme and, consequently, are inherited as single-gene traits. Furthermore, herbicide-resistance alleles often act in an additive to dominant fashion. Because of this single-gene, additive-to-dominant nature of many herbicide resistances, their evolution can occur rapidly, and they can spread effectively by both seed and pollen (160).
The additive-to-dominant nature of many herbicide-resistance traits means that the resistance allele often will be selected in heterozygous plants, particularly in cases where the magnitude of resistance is high. For example, single ALS amino acid changes can confer resistance ratios of 100-fold or more, relative to sensitive biotypes (e.g. Ref. 161). In such cases, heterozygotes and homozygous-resistant plants will have similar responses to typical field doses of the herbicide, even if the resistance allele is not completely dominant. Resistance in this case can be considered “functionally” dominant. In contrast, single amino acid changes to EPSPS confer very modest levels of resistance (typically <10-fold (17)). In E. indica, an EPSPS target-site mutation was additive (162). Consequently, in this case, plants heterozygous for an EPSPS mutation potentially could be controlled by full recommended rates of glyphosate, and using reduced rates would foster resistance evolution.
Known examples of herbicide resistance inherited as a recessive trait are restricted to TSR to herbicides that bind tubulin (105) and to a few cases of resistance to auxinic herbicides (163). As expected, because of the recessive nature of these traits, they have been observed almost exclusively in self-pollinated weed species. However, outcrossed weeds that have evolved recessive resistance include yellow starthistle (Centaurea solstitialis) resistant to some auxinic herbicides (164) and L. rigidum resistant to dinitroaniline herbicides (165). Population-genetics theory predicts that in an outcrossed population, the probability of a resistant plant occurring for a recessive mutation in a reasonably sized field (e.g. 30 hectares) is essentially nil, even with a relatively high mutation rate (1 × 10−6 gametes/locus/generation) and high weed density (500 m−2) (160). So how were C. solstitialis and L. rigidum able to evolve recessive resistance? In the case of C. solstitialis, it was reported that the species is not completely outcrossed, and even the very low selfing rate (<0.1%) would be sufficient for generating a few plants with the homozygous-recessive mutation (164). Lolium spp., although predominantly self-incompatible (female flowers on the plant reject pollen shed from the same plant, requiring pollination from a different individual), also can exhibit some degree of self-pollination (166). In addition, dinitroaniline herbicide resistance in L. rigidum was not completely recessive; heterozygous plants exhibited some resistance to low herbicide doses (165).
The discussion of herbicide-resistance inheritance so far has included only nuclear genes. An important exception to nuclear inheritance of herbicide resistance is maternal inheritance of resistance to PSII inhibitors, reported over 40 years ago (167). The basis for this maternal inheritance is now known to be a herbicide-insensitive D1 protein, which is encoded by a chloroplastic gene and, therefore, transmitted only maternally in most species (3). For species that are predominantly self-pollinated, the lack of pollen-mediated dispersal of this resistance trait is probably of limited consequence. In outcrossing species, however, dissemination of nucleus-inherited traits would be favored over maternally inherited traits. This may explain why nucleus-encoded NTSR to atrazine is more common in A. tuberculatus (a dioecious—and, therefore, outcrossed—species) than is maternally inherited atrazine TSR (168). However, TSR to PSII inhibitors also is known to incur a significant fitness penalty (2), and this also would disfavor the evolution of this resistance.
Inheritance of glyphosate resistance conferred by EPSPS duplication in some cases is observed to be consistent with a single-gene model. This apparent single-gene inheritance occurs because the multiple EPSPS copies are found together in the genome as tandem repeats (58). In other cases, notably in A. palmeri, the EPSPS copies are present as eccDNA and, consequently, inheritance does not follow a single-gene model. In fact, the EPSPS copy number appears to be unstable in genetic transmission, as segregating F2 A. palmeri progeny were highly variable for EPSPS copy number, from less than the parental copy number to greater than the sum of both parents (48).
NTSR traits, such as enhanced herbicide metabolism and reduced translocation, sometimes show inheritance consistent with a single-gene model. Examples include atrazine resistance in velvetleaf (Abutilon theophrasti) and A. tuberculatus, paraquat resistance in L. rigidum, glyphosate resistance in C. canadensis, and 2,4-D resistance in oriental mustard (Sisymbrium orientale) (169–172). More often, however, NTSR appears to be mediated by multiple genes.
The multigenic nature of NTSR often has been described as “creeping” resistance, in which the magnitude of resistance slowly increases over repeated herbicide selection, presumably due to the stacking of small-effect resistance alleles in the population (173). Empirical support for creeping resistance is provided by low-dose selection of experimental populations. Such studies, conducted with different species and different herbicides, have consistently demonstrated the ability of plants to evolve increased herbicide resistance (in some cases by many-fold) after only two or three generations (174–176). That such a shift in herbicide response was due to the accumulation of multiple alleles was supported by a follow-up study on one of the selected populations that indicated at least three loci were involved. It is also noteworthy that when similar selection protocols were followed on an outcrossed weed (L. rigidum) and selfed weeds (wild oat, Avena fatua), the selfed weed's shift was only about 5% of that of the outcrossed weed (176). This is consistent with the idea that the resistance evolution occurred by stacking together diverse alleles initially distributed among different plants.
The evolution of NTSR also could involve epigenetic factors (177). For example, changes in the methylation status of a gene could affect its expression. Epigenetic involvement in evolved herbicide resistance is an understudied topic, although differential methylation of EPSPS recently was reported between glyphosate-resistant and -sensitive biotypes of C. canadensis (178). It is not yet clear, however, whether this differential methylation contributes to resistance.
Predicting and mitigating herbicide resistance requires understanding not only the genetics of the resistance traits, but also the origins of the genetic diversity that fuels resistance evolution (179). We are now moving into an era in which population genomics approaches should be able to shed increased light on, for example, the importance of new mutations versus standing genetic variation in the evolutionary history of herbicide resistance (180).
Influences of ploidy
Understanding resistance mechanisms in polyploid species is complicated by the presence of multiple genomes and associated regulatory processes, ranging from whole-genome to single allele, that are not a consideration in diploids. In particular, allele dosage, or the relative contribution of orthologous or homoeologous genes (homoeologs are genes with homologous sequence/function that are present in the different genomes of a polyploid) in a polyploid individual, can influence the effect of target site mutations in weed species.
The influence of ploidy has been documented for herbicide resistance in multiple species. An ACCase inhibitor–resistant population of hexaploid A. fatua had three different mutations in ACCase (at positions 1781, 2078, and 2088) that segregated independently, indicating one mutation present for each homoeologous loci (181). When occurring alone, each allele conferred a lower ACCase resistance level than the same mutation confers in diploid species. The mutations showed an additive effect when co-expressed in vitro, suggesting in single mutant lines the contributions of susceptible ACCase alleles were diluting the effect of the resistance allele. Similar allele dose effects were observed in IMI-resistant hexaploid wheat (182). Resistance mutations on ALS genes on two of the three subgenomes (referring to one of the multiple genomes within a polyploid) resulted in higher resistance than when the mutation was present on only one subgenome.
More recently, an allotetraploid P. annua population was identified with resistance to glyphosate via a P106L mutation in EPSPS and 7-fold increased EPSPS copy number (183). Interestingly, both the target site mutation and duplicated EPSPS genes were present in homoeologs only from the supine bluegrass (Poa supina) progenitor subgenome. The P. supina subgenome is known to contain higher nucleotide substitution rates than the early meadow grass (Poa infirma) progenitor subgenome (184), and this expression dominance toward a more dynamic subgenome may represent a resistance evolution process present in this species. Despite these examples just described, studies are limited on the influence of ploidy on resistance mechanisms, and further research is needed to understand the complexities of herbicide resistance and evolution in polyploid weeds.
Implications for herbicide-resistant crops
The wide range of mechanisms of evolved resistance to herbicides by weeds provides a wealth of potential genes for production of herbicide-resistant crops. However, not since the production of triazine-resistant canola by backcrossing canola with an evolved triazine-resistant weed (185) has a gene from a weed been used to produce a commercial herbicide-resistant crop. The rapid evolution of resistance to ALS and ACCase inhibitors by SNPs provided a clue that crops resistant to these herbicides could be generated by mutagenesis and breeding. This approach has been especially successful in producing imidazolinone-resistant rice, wheat, oilseed rape, sorghum, maize, sunflower, and sugar beet. Imidazolinone-resistant crops have been the only significant commercial success with this technology, even though crops resistant to ACCase inhibitors and sulfonylurea ALS inhibitors have been generated by such mutation breeding approaches (186).
Relatively few herbicide-resistance genes have been utilized to produce commercial, transgenic, herbicide-resistant crops, and most of them have been of bacterial origin. These include bacterial genes for a glyphosate-resistant form of EPSPS and those that degrade glyphosate, glufosinate, 2,4-D, bromoxynil, and dicamba. A nonbacterial exception is a TIPS form of EPSPS that was engineered from a plant EPSPS gene and is used in some glyphosate-resistant maize cultivars (20). It took 15 more years after use as a transgene for the TIPS form of EPSPS to evolve in a weed (22). A bacterial gene with an SNP encoding an isoxaflutole-resistant HPPD is being used as a transgene for isoxaflutole-resistant crops (187). However, another company is developing crops resistant to triketone HPPD inhibitor herbicides through use of a mutant triketone-resistant HPPD gene from oats (188).
For the most part, researchers developing transgenic herbicide-resistant crops became involved with crops resistant to a particular herbicide before a weed had evolved resistance to that herbicide or the genes for weed resistance had been discovered. A major consideration in development of herbicide-resistant crops is the strong benefit of the transgene providing resistance to only one herbicide or class of herbicides (e.g. glyphosate or imidazolinones) controlled by the same company. The very high cost of developing and obtaining regulatory approval of such a crop can preclude production of a crop that is resistant to products belonging to a competing company. If transgenic, herbicide-resistant crops continue to be developed, the burgeoning information on the molecular biology of resistance to herbicides evolved by weeds might influence the choice of future transgenes. For example, targeted mutagenesis on a codon involved in resistance to certain PDS inhibitors identified a double mutation not likely to occur in nature that imparts even greater resistance to these herbicides (12). Interestingly, this mutation provided negative cross-resistance to other classes of PDS inhibitors. This may be particularly useful in managing volunteer plants as well as weeds that may evolve resistance by selection for this mutation.
What we know about the evolutionary biology of herbicide-resistant weeds will influence what herbicides are chosen for future herbicide-resistant crops and how resulting crops are managed. An impetus to the development of glyphosate-resistant crops in the early 1990s was the false view that evolution of weeds resistant to glyphosate would be almost impossible because more than one codon in EPSPS would have to mutate simultaneously to provide robust TSR (189). The unparalleled success of glyphosate-resistant crops and the resulting massive selection pressure for glyphosate resistance helped to overcome the difficulty that weeds had in evolving resistance. Thus, even if a herbicide is selected because mutations imparting resistance are less likely to evolve, resistance management must be part of the stewardship of future herbicide-resistant crops because sustained, strong selection pressure over a wide area can overcome any perceived genetic barrier.
Implications for herbicide discovery
Decades of nondiversified chemical weed control and the resistance challenges that arose from it have taught us several hard lessons about the importance of herbicide stewardship. Research into the evolution of resistance provides valuable insights for future herbicide discovery, as certain resistance mechanisms are more prevalent within certain modes of action than others. These findings can be used to proactively influence the development of less resistance-prone herbicides.
For TSR, the rate and likelihood at which a point mutation conferring resistance occurs is determined by both the target protein and the inhibitor. Many herbicidal molecules compete with substrates that are crucial for plant survival by emulating their volume, surface geometry, and binding properties. Compounds such as glyphosate and transition state analogues such as glufosinate tightly fit into the active site of the target enzyme (competitive binding), where they form strong hydrogen bridges. Point mutations in this highly conserved area would render the enzyme dysfunctional. In contrast, herbicides inhibiting ACCase, ALS, PPO, or PDS adopt different binding modes than the substrate so that a mutation can have differential effects on inhibitor and substrate binding. The enzyme's reaction would still take place despite an amino acid change in the inhibitor-binding domain, where genetic diversity within and among species is more likely. However, changes can come at a cost of a reduced turnover rate (kcat) for the substrate, as seen with the Gly-210 PPX2 deletion. Therefore, the vast majority of resistance-conferring point mutations are found in hydrophobic binding sites away from the active reaction site. ALS point mutations, for example, are found in the nonconserved terminal ends of the ALS sequences (190). Not all point mutations confer resistance to all compounds from the same chemical class. For example, whereas the 1781 mutation in A. myosuroides confers resistance to all ACCase inhibitors, the 2027 mutation confers resistance to aryloxyphenoxypropionates, but not to cyclohexanediones (8). The closer the mutation is to the active site, the more likely it is for a point mutation to negatively affect the enzyme's catalytic rate but also to confer resistance to all molecules of a site of action rather than to just one chemical class.
Sometimes, several homologues for the same target exist in a plant. Resistance via point mutations would be less likely to evolve if these isoforms were expressed at the same time, were nonredundant in function, and could be inhibited by the same molecule. In the case of a point mutation in one of the target enzyme isoforms, the inhibition of the remaining enzymes would then still lead to the death of the plant. Alternatively, one herbicide may inhibit two different target proteins (e.g. auxinic herbicides). The probability of an individual to evolve two coincidental, independent, and catalytically competent point mutations in each target enzyme is equal to the product of the frequencies of the single mutation. Hence, molecules that fully inhibit several targets that individually would lead to plant death are preferred. Likewise, target site mutations are less likely to occur if they need to be homozygous to confer resistance (recessive inheritance). In that case, an identical mutation would need to be present in both the paternal and maternal alleles. They are also less likely to spread if they impart a fitness penalty for the plant and if compensatory mutations rarely occur, as in the case of TSR to PSII inhibitors (191, 192). Duplication of the gene for the herbicide target protein, likely guided by transposons or nonhomologous recombination, is a resistance mechanism that is particularly difficult to predict in herbicide discovery strategies.
The physicochemical properties of a molecule define how it binds the protein target and strongly influence its uptake, translocation, and degradation pathway before it reaches the target site. For example, a major pathway of herbicide metabolism is through oxidation via P450s, which can add hydroxy groups to a molecule. This reaction is more difficult for small and polar molecules, which allow for strong hydrogen bridges, inhibiting the active site of the target only. In contrast, hydrophobic parts of the molecule, like aryl or alkyl moieties, are more prone to oxidation, as there is a positive correlation between lipophilicity and Km for P450-catalyzed reactions (193). Prevention of this metabolic degradation can be achieved by introduction of halogens into strategic positions of the molecules. A second major pathway of metabolic degradation is the nucleophilic attack of electrophilic centers in the inhibitor molecule, such as through GSTs. Such soft regions can be circumvented by the introduction of heteroatoms such as oxygen at the respective place of the molecule. However, these changes can alter a molecule's binding properties. Once phase I metabolism has taken place, phase II metabolism, such as glycosylation, cannot be prevented. The biggest challenge in pesticide discovery lies in creating a biologically active molecule that is toxicologically benign to nontarget organisms and is degraded in the environment as well as by nontarget organisms.
Normalized by use frequency, strong differences in the rates of herbicide-resistance evolution between different modes of action have been observed (194). Glyphosate has been such an excellent herbicide because it tightly occupies the active site of the EPSPS-S3P complex only, outcompeting the substrate PEP. Its low dissociation rate allows for almost irreversible inhibition of the enzyme. Heteroatoms and phosphonates that cannot be oxidized leave it inert for metabolism by most metabolic enzymes. Hence, resistant plants have generally had to “resort” to the fairly complicated and comparably slow-to-evolve mechanism of EPSPS gene duplication and even slower-to-evolve double codon mutations in EPSPS to survive. In the end, any type of persistent selection pressure will be eventually met by adaptation, and the potential number of resistance mechanisms are many. Even if all currently known resistance mechanisms could be addressed, new, unpredictable mechanisms may evolve, such as the “phoenix phenomenon” mechanism of glyphosate and 2,4-D resistance. Therefore, it is futile to set a goal of finding a “resistance-proof” herbicide. Nevertheless, the features of an ideal herbicide from a resistance management perspective listed below should be considered in herbicide discovery strategies.
Inhibits several different targets
Inhibits several homologous and nonredundant target genes
Fits tightly and deep into an enzyme's active site
Only fits into an enzyme's active site
Does not bind to hydrophobic pockets around the active site in case the natural substrate does not contain any lipophilic parts
Builds strong hydrogen bridges
Small polar molecule with low structural complexity
Low lipophilicity
Nonreversible binding, strong enzyme affinity (low Km or P50 values)
High-volume overlap with substrate for good competition
Weed biology, herbicide application rates, and herbicide mixtures all influence the type and rate at which resistance mechanisms evolve (195). We can aim to delay the evolution of resistance to newly developed molecules by choosing sensible targets and optimized molecules. We must recognize, however, that doing so will not replace the necessity to practice integrated resistance management.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- TSR
- target-site resistance
- NTSR
- nontarget-site resistance
- S3P
- shikimate 3-phosphate
- MOA
- mechanism of action
- PSII
- photosystem II
- ACCase
- acetyl-CoA carboxylase
- APP
- aryloxyphenoxypropionate
- CHD
- cyclohexanedione
- PPZ
- phenylpyrazoline
- PDS
- phytoene desaturase
- ALS
- acetolactate synthase
- SU
- sulfonylurea
- IMI
- imidazolinone
- EPSPS
- 5-enolpyruvylshikimate-3-phosphate synthase
- PEP
- phosphoenolpyruvate
- TIPS
- mutation of both T102I and P106S
- IAA
- indole-3-acetic acid
- AFB
- auxin F-box
- PPO
- protoporphyrinogen oxidase
- SSR
- short simple repeat
- eccDNA
- extrachromosomal circular DNA
- 2,4-D
- 2,4-dichlorophenoxyacetic acid
- ROS
- reactive oxygen species
- GST
- GSH S-transferase
- GT
- glucosyltransferase
- HPPD
- hydroxyphenylpyruvate dioxygenase
- AA
- aryl acylamidase
- AMPA
- aminomethyl phosphonic acid
- GOX
- glyphosate oxidoreductase
- AhR
- aldo-keto reductase
- β-CAS
- β-cyanoalanine synthase.
References
- 1. Dayan F. E., Barker A., Bough R., Ortiz M., Takano H., and Duke S. O. (2019) Herbicide mechanisms of action and resistance. In Comprehensive Biotechnology, 3rd Ed. (Moo-Young M., ed) pp. 36–48, Pergamon Press, Oxford [Google Scholar]
- 2. Gronwald J. W. (1994) Resistance to photosystem II inhibiting herbicides. In Herbicide Resistance in Plants: Biology and Biochemistry (Powles S. B., and Holtum J. A. M., eds) pp. 27–60, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 3. Hirschberg J., and McIntosh L. (1983) Molecular basis of herbicide resistance in Amaranthus hybridus. Science 222, 1346–1349 10.1126/science.222.4630.1346 [DOI] [PubMed] [Google Scholar]
- 4. Anthony R. G., Waldin T. R., Ray J. A., Bright S. W. J., and Hussey P. J. (1998) Herbicide resistance caused by spontaneous mutation of the cytoskeletal protein tubulin. Nature 393, 260–263 10.1038/30484 [DOI] [PubMed] [Google Scholar]
- 5. Chen J., Chu Z., Han H., Goggin D. E., Yu Q., Sayer C., and Powles S. B. (2020) A Val-202-Phe α-tubulin mutation and enhanced metabolism confer dinitroaniline resistance in a single Lolium rigidum population. Pest Manag. Sci. 76, 645–652 10.1002/ps.5561 [DOI] [PubMed] [Google Scholar]
- 6. Chu Z., Chen J., Nyporko A., Han H., Yu Q., and Powles S. (2018) Novel α-tubulin mutations conferring resistance to dinitroaniline herbicides in Lolium rigidum. Front. Plant Sci. 9, 97 10.3389/fpls.2018.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Délye C., Menchari Y., Michel S., and Darmency H. (2004) Molecular bases for sensitivity to tubulin-binding herbicides in green foxtail. Plant Physiol. 136, 3920–3932 10.1104/pp.103.037432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaundun S. S. (2014) Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest Manag. Sci. 70, 1405–1417 10.1002/ps.3790 [DOI] [PubMed] [Google Scholar]
- 9. Beckie H. J., and Tardif F. J. (2012) Herbicide cross resistance in weeds. Crop Protect. 35, 15–28 10.1016/j.cropro.2011.12.018 [DOI] [Google Scholar]
- 10. Michel A., Arias R. S., Scheffler B. E., Duke S. O., Netherland M. D., and Dayan F. E. (2004) Somatic mutation-mediated evolution of herbicide resistance in the nonindigenous invasive plant hydrilla (Hydrilla verticillata). Mol. Ecol. 13, 3229–3237 10.1111/j.1365-294X.2004.02280.x [DOI] [PubMed] [Google Scholar]
- 11. Dayan F. E., and Netherland M. D. (2005) Hydrilla, the perfect aquatic weed, becomes more noxious than ever. Outlooks Pest Manag. 16, 277–282 10.1564/16dec11 [DOI] [Google Scholar]
- 12. Arias R. S., Dayan F. E., Michel A., Howell J. L., and Scheffler B. E. (2006) Characterization of a higher plant herbicide-resistant phytoene desaturase and its use as a selectable marker. Plant Biotechnol. J. 4, 263–273 10.1111/j.1467-7652.2006.00179.x [DOI] [PubMed] [Google Scholar]
- 13. Netherland M. D., and Jones D. (2015) Fluridone-resistant hydrilla (Hydrilla verticillata) is still dominant in the Kissimmee chain of lakes. Invasive Plant Sci. Manag. 8, 212–218 10.1614/IPSM-D-14-00071.1 [DOI] [Google Scholar]
- 14. McCourt J. A., Pang S. S., King-Scott J., Guddat L. W., and Duggleby R. G. (2006) Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc. Natl. Acad. Sci. U.S.A. 103, 569–573 10.1073/pnas.0508701103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Preston C., and Powles S. B. (2002) Evolution of herbicide resistance in weeds: Initial frequency of target site-based resistance to acetolactate synthase-inhibiting herbicides in Lolium rigidum. Heredity 88, 8–13 10.1038/sj.hdy.6800004 [DOI] [PubMed] [Google Scholar]
- 16. Jander G., Baerson S. R., Hudak J. A., Gonzalez K. A., Gruys K. J., and Last R. L. (2003) Ethylmethanesulfonate saturation mutagenesis in Arabidopsis to determine frequency of herbicide resistance. Plant Physiol. 131, 139–146 10.1104/pp.102.010397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sammons D. R., and Gaines T. A. (2014) Glyphosate resistance: state of knowledge. Pest Manag. Sci. 70, 1367–1377 10.1002/ps.3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Healy-Fried M. L., Funke T., Priestman M. A., Han H., and Schönbrunn E. (2007) Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase. J. Biol. Chem. 282, 32949–32955 10.1074/jbc.M705624200 [DOI] [PubMed] [Google Scholar]
- 19. Li J., Peng Q., Han H., Nyporko A., Kulynych T., Yu Q., and Powles S. (2018) Glyphosate resistance in Tridax procumbens via a novel EPSPS Thr-102-Ser substitution. J. Agric. Food Chem. 66, 7880–7888 10.1021/acs.jafc.8b01651 [DOI] [PubMed] [Google Scholar]
- 20. Sidhu R. S., Hammond B. G., Fuchs R. L., Mutz J.-N., Holden L. R., George B., and Olson T. (2000) Glyphosate-tolerant corn: the composition and feeding value of grain from glyphosate-tolerant corn is equivalent to that of conventional corn (Zea mays L.). J. Agric. Food Chem. 48, 2305–2312 10.1021/jf000172f [DOI] [PubMed] [Google Scholar]
- 21. Funke T., Yang Y., Han H., Healy-Fried M., Olesen S., Becker A., and Schönbrunn E. (2009) Structural basis of glyphosate resistance resulting from the double mutation Thr97 → Ile and Pro101 → Ser in 5-enolpyruvylshikimate-3-phosphate synthase from Escherichia coli. J. Biol. Chem. 284, 9854–9860 10.1074/jbc.M809771200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Q., Jalaludin A., Han H., Chen M., Sammons R. D., and Powles S. B. (2015) Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance. Plant Physiol. 167, 1440–1447 10.1104/pp.15.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dill G. M. (2005) Glyphosate-resistant crops: history, status and future. Pest Manag. Sci. 61, 219–224 10.1002/ps.1008 [DOI] [PubMed] [Google Scholar]
- 24. Han H., Vila-Aiub M. M., Jalaludin A., Yu Q., and Powles S. B. (2017) A double EPSPS gene mutation endowing glyphosate resistance shows a remarkably high resistance cost. Plant Cell Environ. 40, 3031–3042 10.1111/pce.13067 [DOI] [PubMed] [Google Scholar]
- 25. Alcántara-de la Cruz R., Fernández-Moreno P. T., Ozuna C. V., Rojano-Delgado A. M., Cruz-Hipolito H. E., Domínguez-Valenzuela J. A., Barro F., and De Prado R. (2016) Target and non-target site mechanisms developed by glyphosate-resistant hairy beggarticks (Bidens pilosa L.) populations from Mexico. Front. Plant Sci. 7, 1492 10.3389/fpls.2016.01492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takano H. K., Fernandes V. N. A., Adegas F. S., Oliveira R. S. Jr., Westra P., Gaines T. A., and Dayan F. E. (2020) A novel TIPT double mutation in EPSPS conferring glyphosate resistance in tetraploid Bidens subalternans. Pest Manag. Sci. 76, 95–102 10.1002/ps.5535 [DOI] [PubMed] [Google Scholar]
- 27. Perotti V. E., Larran A. S., Palmieri V. E., Martinatto A. K., Alvarez C. E., Tuesca D., and Permingeat H. R. (2019) A novel triple amino acid substitution in the EPSPS found in a high-level glyphosate resistant Amaranthus hybridus population from Argentina. Pest Manag. Sci. 75, 1242–1251 10.1002/ps.5303 [DOI] [PubMed] [Google Scholar]
- 28. García M. J., Palma-Bautista C., Rojano-Delgado A. M., Bracamonte E., Portugal J., Alcántara-de la Cruz R., and De Prado R. (2019) The triple amino acid substitution TAP-IVS in the EPSPS gene confers high glyphosate resistance to the superweed Amaranthus hybridus. Internat. J. Mol. Sci. 20, 2396 10.3390/ijms20102396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Webb S. R., and Hall J. C. (1995) Auxinic herbicide-resistant and herbicide-susceptible wild mustard (Sinapis arvensis L.) biotypes: effect of auxinic herbicides on seedling growth and auxin-binding activity. Pestic. Biochem. Physiol. 52, 137–148 10.1006/pest.1995.1038 [DOI] [Google Scholar]
- 30. Walsh T. A., Neal R., Merlo A. O., Honma M., Hicks G. R., Wolff K., Matsumura W., and Davies J. P. (2006) Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 142, 542–552 10.1104/pp.106.085969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. LeClere S., Wu C., Westra P., and Sammons R. D. (2018) Cross-resistance to dicamba, 2,4-D, and fluroxypyr in Kochia scoparia is endowed by a mutation in an AUX/IAA gene. Proc. Nat. Acad. Sci. U.S.A. 115, E2911–E2920 10.1073/pnas.1712372115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patzoldt W. L., Hager A. G., McCormick J. S., and Tranel P. J. (2006) A codon deletion confers resistance to herbicides inhibiting protoporphyrinogen oxidase. Proc. Nat. Acad. Sci. U.S.A. 103, 12329–12334 10.1073/pnas.0603137103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dayan F. E., Barker A., and Tranel P. J. (2018) Origins and structure of chloroplastic and mitochondrial plant protoporphyrinogen oxidases: implications for the evolution of herbicide resistance. Pest Manag. Sci. 74, 2226–2234 10.1002/ps.4744 [DOI] [PubMed] [Google Scholar]
- 34. Riggins C. W., and Tranel P. J. (2012) Will the Amaranthus tuberculatus resistance mechanism to PPO-inhibiting herbicides evolve in other Amaranthus species?. Int. J. Agron. 2012, 1–7 10.1155/2012/305764 [DOI] [Google Scholar]
- 35. Salas R. A., Burgos N. R., Tranel P. J., Singh S., Glasgow L., Scott R. C., and Nichols R. L. (2016) Resistance to PPO-inhibiting herbicide in Palmer amaranth from Arkansas. Pest Manag. Sci. 72, 864–869 10.1002/ps.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dayan F. E., Daga P. R., Duke S. O., Lee R. M., Tranel P. J., and Doerksen R. J. (2010) Biochemical and structural consequences of a glycine deletion in the α-8 helix of protoporphyrinogen oxidase. Biochim. Biophys. Acta 1804, 1548–1556 10.1016/j.bbapap.2010.04.004 [DOI] [PubMed] [Google Scholar]
- 37. Patzoldt W. L., Tranel P. J., and Hager A. G. (2005) A waterhemp (Amaranthus tuberculatus) biotype with multiple resistance across three herbicide sites of action. Weed Sci. 53, 30–36 10.1614/WS-04-087R [DOI] [Google Scholar]
- 38. Armel G. R., Nielson R. L., Liebl R. A., Bowe S., Hennigh D. S., Francis I. K., Oostlander M. D., and Ramos R. A. (2018) Trifludimoxazin: a global perspective on a versatile PPO herbicide. 2018 Weed Science Society of America Annual Meeting, Arlington, VA, January 29, 2018 to February 1, 2018, Abstract 196 [Google Scholar]
- 39. Baerson S. R., Rodriguez D. J., Biest N. A., Tran M., You J. S., Kreuger R. W., Dill G. M., Pratley J. E., and Gruys K. J. (2002) Investigating the mechanism of glyphosate resistance in rigid ryegrass (Lolium ridigum). Weed Sci. 50, 721–730 10.1614/0043-1745(2002)050[0721:ITMOGR]2.0.CO;2 [DOI] [Google Scholar]
- 40. Yu Q., Nelson J. K., Zheng M. Q., Jackson M., and Powles S. B. (2007) Molecular characterisation of resistance to ALS-inhibiting herbicides in Hordeum leporinum biotypes. Pest Manag. Sci. 63, 918–927 10.1002/ps.1429 [DOI] [PubMed] [Google Scholar]
- 41. Bradley K. W., Wu J. R., Hatzios K. K., and Hagood E. S. (2001) The mechanism of resistance to aryloxyphenoxypropionate and cyclohexanedione herbicides in a Johnsongrass biotype. Weed Sci. 49, 477–484 10.1614/0043-1745(2001)049[0477:TMORTA]2.0.CO;2 [DOI] [Google Scholar]
- 42. Innan H., and Kondrashov F. (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108 10.1038/nrg2689 [DOI] [PubMed] [Google Scholar]
- 43. Flagel L. E., and Wendel J. F. (2009) Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564 10.1111/j.1469-8137.2009.02923.x [DOI] [PubMed] [Google Scholar]
- 44. Gaines T. A., Zhang W., Wang D., Bukun B., Chisholm S. T., Shaner D. L., Nissen S. J., Patzoldt W. L., Tranel P. J., Culpepper A. S., Grey T. L., Webster T. M., Vencill W. K., Sammons R. D., Jiang J. M., et al. (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Nat. Acad. Sci. U.S.A. 107, 1029–1034 10.1073/pnas.0906649107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Molin W. T., Wright A. A., Lawton-Rauh A., and Saski C. A. (2017) The unique genomic landscape surrounding the EPSPS gene in glyphosate resistant Amaranthus palmeri: a repetitive path to resistance. BMC Genom. 18, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koo D.-H., Molin W. T., Saski C. A., Jiang J., Putta K., Jugulam M., Friebe B., and Gill B. S. (2018) Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. Proc. Nat. Acad. Sci. U.S.A. 115, 3332–3337 10.1073/pnas.1719354115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Giacomini D. A., Westra P., and Ward S. M. (2019) Variable inheritance of amplified EPSPS gene copies in glyphosate-resistant Palmer amaranth (Amaranthus palmeri). Weed Sci. 67, 176–182 10.1017/wsc.2018.65 [DOI] [Google Scholar]
- 48. Gaines T. A., Shaner D. L., Ward S. M., Leach J. E., Preston C., and Westra P. (2011) Mechanism of resistance of evolved glyphosate-resistant Palmer amaranth (Amaranthus palmeri). J. Agric. Food Chem. 59, 5886–5889 10.1021/jf104719k [DOI] [PubMed] [Google Scholar]
- 49. Molin W. T., Wright A. A., VanGessel M. J., McCloskey W. B., Jugulam M., and Hoagland R. E. (2018) Survey of the genomic landscape surrounding the 5‐enolpyruvylshikimate‐3‐phosphate synthase (EPSPS) gene in glyphosate-resistant Amaranthus palmeri from geographically distant populations in the United States. Pest Manag. Sci. 74, 1109–1117 10.1002/ps.4659 [DOI] [PubMed] [Google Scholar]
- 50. Patterson E. L., Pettinga D. J., Ravet K., Neve P., and Gaines T. A. (2018) Glyphosate resistance and EPSPS gene duplication: convergent evolution in multiple plant species. J. Hered. 109, 117–125 10.1093/jhered/esx087 [DOI] [PubMed] [Google Scholar]
- 51. Ngo T. D., Malone J. M., Boutsalis P., Gill G., and Preston C. (2018) EPSPS gene amplification conferring resistance to glyphosate in windmill grass (Chloris truncata) in Australia. Pest Manag. Sci. 74, 1101–1108 10.1002/ps.4573 [DOI] [PubMed] [Google Scholar]
- 52. Adu-Yeboah P., Malone J. M., Fleet B., Gill G., and Preston C. (2020) EPSPS gene amplification confers resistance to glyphosate resistant populations of Hordeum glaucum Stued (northern barley grass) in South Australia. Pest Manag. Sci. 76, 1214–1221 10.1002/ps.5671 [DOI] [PubMed] [Google Scholar]
- 53. Malone J. M., Morran S., Shirley N., Boutsalis P., and Preston C. (2016) EPSPS gene amplification in glyphosate‐resistant Bromus diandrus. Pest Manag. Sci. 72, 81–88 10.1002/ps.4019 [DOI] [PubMed] [Google Scholar]
- 54. Chen J., Huang H., Zhang C., Wei S., Huang Z., Chen J., and Wang X. (2015) Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 242, 859–868 10.1007/s00425-015-2324-2 [DOI] [PubMed] [Google Scholar]
- 55. Lorentz L., Gaines T. A., Nissen S. J., Westra P., Strek H., Dehne H. W., Ruiz-Santaella J. P., and Beffa R. (2014) Characterization of glyphosate resistance in Amaranthus tuberculatus populations. J. Agric. Food Chem. 62, 8134–8142 10.1021/jf501040x [DOI] [PubMed] [Google Scholar]
- 56. Wiersma A. T., Gaines T. A., Preston C., Hamilton J. P., Giacomini D., Buell C. R., Leach J. E., and Westra P. (2015) Gene amplification of 5-enol-pyruvylshikimate-3-phosphate synthase in glyphosate-resistant Kochia scoparia. Planta 241, 463–474 10.1007/s00425-014-2197-9 [DOI] [PubMed] [Google Scholar]
- 57. Salas R. A., Dayan F. E., Pan Z., Watson S. B., Dickson J. W., Scott R. C., and Burgos N. R. (2012) EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manag. Sci. 68, 1223–1230 10.1002/ps.3342 [DOI] [PubMed] [Google Scholar]
- 58. Jugulam M., Niehues K., Godar A. S., Koo D.-H., Danilova T., Friebe B., Sehgal S., Varanasi V. K., Wiersma A., Westra P., Stahlman P. W., and Gill B. S. (2014) Tandem amplification of a chromosomal segment harboring EPSPS locus confers glyphosate resistance in Kochia scoparia. Plant Physiol. 166, 1200–1207 10.1104/pp.114.242826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nandula V. K., Wright A. A., Bond J. A., Ray J. D., Eubank T. W., and Molin W. T. (2014) EPSPS amplification in glyphosate-resistant spiny amaranth (Amaranthus spinosus): a case of gene transfer via interspecific hybridization from glyphosate-resistant Palmer amaranth (Amaranthus palmeri). Pest Manag. Sci. 70, 1902–1909 10.1002/ps.3754 [DOI] [PubMed] [Google Scholar]
- 60. Wassom J. J., and Tranel P. J. (2005) Amplified fragment length polymorphism-based genetic relationships among weedy Amaranthus species. J. Hered. 96, 410–416 10.1093/jhered/esi065 [DOI] [PubMed] [Google Scholar]
- 61. Gaines T. A., Ward S. M., Bukun B., Preston C., Leach J. E., and Westra P. (2012) Interspecific hybridization transfers a previously unknown glyphosate resistance mechanism in Amaranthus species. Evolution. Applic. 5, 29–38 10.1111/j.1752-4571.2011.00204.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Patterson E. L., Saski C. A., Sloan D. B., Tranel P. J., Westra P., and Gaines T. A. (2019) The draft genome of Kochia scoparia and the mechanism of glyphosate resistance via transposon-mediated EPSPS tandem gene duplication. Genome Biol. E 11, 2927–2940 10.1093/gbe/evz198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gaines T. A., Patterson E. L., and Neve P. (2019) Molecular mechanisms of adaptive evolution revealed by global selection for glyphosate resistance. New Phytol. 223, 1770–1775 10.1111/nph.15858 [DOI] [PubMed] [Google Scholar]
- 64. Laforest M., Soufiane B., Simard M.-J., Obeid K., Page E., and Nurse R. E. (2017) Acetyl-CoA carboxylase overexpression in herbicide resistant large crabgrass (Digitaria sanguinalis). Pest Manag. Sci. 73, 2227–2235 10.1002/ps.4675 [DOI] [PubMed] [Google Scholar]
- 65. Menendez J., Rojano-Delgado M. A., and De Prado R. (2014) Differences in herbicide uptake, translocation, and distribution as sources of herbicide resistance in weeds. Am. Chem. Soc. Symp. Ser. 1171, 141–157 [Google Scholar]
- 66. Hess F. E. (1985) Herbicide absorption and translocation and their relationship to plant tolerance and susceptibility. In Weed Physiology (Duke S. O., ed) pp. 191–214, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 67. Devine M., Duke S. O., and Fedtke C. (1992) Physiology of Herbicide Action, Prentice Hall, Englewood Cliffs, NJ [Google Scholar]
- 68. Stegink S. J., and Vaughn K. C. (1988) Norflurazon (SAN-9789) reduces abscisic acid levels in cotton seedlings: a glandless isoline is more sensitive than its glanded counterpart. Pestic. Biochem. Physiol. 31, 269–275 10.1016/0048-3575(88)90132-0 [DOI] [Google Scholar]
- 69. White A. D., Owen M. D. K., Hartzler R. G., and Cardina J. (2002) Common sunflower resistance to acetolactate synthase–inhibiting herbicides. Weed Sci. 50, 432–437 10.1614/0043-1745(2002)050[0432:CSRTAS]2.0.CO;2 [DOI] [Google Scholar]
- 70. Svyantek A. W., Aldahir P., Chen S., Flessner M. L., McCullough P. E., Sidhu S. S., and McElroy J. S. (2016) Target and nontarget resistance mechanisms induce annual bluegrass (Poa annua) resistance to atrazine, amicarbazone, and diuron. Weed Technol. 30, 773–782 10.1614/WT-D-15-00173.1 [DOI] [Google Scholar]
- 71. Michitte P., De Prado R., Espinoza N., Ruiz-Santaella J. P., and Gauvrit C. (2007) Mechanisms of resistance to glyphosate in a ryegrass (Lolium multiflorum) biotype from Chile. Weed Sci. 55, 435–440 10.1614/WS-06-167.1 [DOI] [Google Scholar]
- 72. Vila-Aiub M. M., Balbi M. C., Distéfano A. J., Fernandez L., Hopp E., Yu Q., and Powles S. B. (2012) Glyphosate resistance in perennial Sorghum halepense (Johnsongrass), endowed by reduced glyphosate translocation and leaf uptake. Pest Manag. Sci. 68, 430–436 10.1002/ps.2286 [DOI] [PubMed] [Google Scholar]
- 73. Nandula V. K., Reddy K. N., Poston D. H., Rimando A. M., and Duke S. O. (2008) Glyphosate tolerance mechanism in Italian ryegrass (Lolium multiflorum) from Mississippi. Weed Sci. 56, 344–349 10.1614/WS-07-115.1 [DOI] [Google Scholar]
- 74. Nandula V. K., Ray J. D., Ribeiro D. N., Pan Z., and Reddy K. N. (2013) Glyphosate resistance in tall waterhemp (Amaranthus tuberculatus) from Mississippi is due to both altered target-site and nontarget-site mechanisms. Weed Sci. 61, 374–383 10.1614/WS-D-12-00155.1 [DOI] [Google Scholar]
- 75. Lorraine-Colwill D. F., Powles S. B., Hawkes T. R., Hollinshead P. H., Warner S. A. J., and Preston C. (2002) Investigations into the mechanism of glyphosate resistance in Lolium rigidum. Pestic. Biochem. Physiol. 74, 62–72 10.1016/S0048-3575(03)00007-5 [DOI] [Google Scholar]
- 76. Feng P. C. C., Tran M., Chiu T., Sammons R. D., Heck G. R., and CaJacob C. A. (2004) Investigations into glyphosate-resistant horseweed (Conyza canadensis): retention, uptake, translocation, and metabolism. Weed Sci. 52, 498–505 10.1614/WS-03-137R [DOI] [Google Scholar]
- 77. Ge X., d'Avignon D. A., Ackerman J. J. H., and Sammons R. D. (2010) Rapid vacuolar sequestration: the horseweed glyphosate resistance mechanism. Pest Manag. Sci. 66, 345–348 10.1002/ps.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ge X., d'Avignon D. A., Ackerman J. J. H., Duncan B., Spaur M. B., and Sammons R. D. (2011) Glyphosate-resistant horseweed made sensitive to glyphosate: low-temperature suppression of glyphosate vacuolar sequestration revealed by P-31 NMR. Pest Manag. Sci. 67, 1215–1221 10.1002/ps.2169 [DOI] [PubMed] [Google Scholar]
- 79. Peng Y., Abercrombie L. L. G., Yuan J. S., Riggins C. W., Sammons R. D., Tranel P. J., and Stewart C. N. (2010) Characterization of the horseweed (Conyza canadensis) transcriptome using GS-FLX 454 pyrosequencing and its application for expression analysis of candidate non-target herbicide resistance genes. Pest Manag. Sci. 66, 1053–1062 10.1002/ps.2004 [DOI] [PubMed] [Google Scholar]
- 80. Yuan J. S., Abercrombie L. L. G., Cao Y., Halfhill M. D., Zhou X., Peng Y., Hu J., Rao M. R., Heck G. R., Larosa T. J., Sammons R. D., Wang X., Ranjan P., Johnson D. H., Wadl P. A., et al. (2010) Functional genomics analysis of horseweed (Conyza canadensis) with special reference to the evolution of non-target-site glyphosate resistance. Weed Sci. 58, 109–117 10.1614/WS-D-09-00037.1 [DOI] [Google Scholar]
- 81. Vila-Aiub M. M., Gundel P. E., Yu Q., and Powles S. B. (2013) Glyphosate resistance in Sorghum halepense and Lolium rigidum is reduced at suboptimal growing temperatures. Pest Manag. Sci. 69, 228–232 10.1002/ps.3464 [DOI] [PubMed] [Google Scholar]
- 82. Yu Q., Cairns A., and Powles S. B. (2004) Paraquat resistance in a population of Lolium rigidum. Funct. Plant Biol. 31, 247–254 10.1071/FP03234 [DOI] [PubMed] [Google Scholar]
- 83. Yu Q., Huang S., and Powles S. (2010) Direct measurement of paraquat in leaf protoplasts indicates vacuolar paraquat sequestration as a resistance mechanism in Lolium rigidum. Pestic. Biochem. Physiol. 98, 104–109 10.1016/j.pestbp.2010.05.007 [DOI] [Google Scholar]
- 84. Brunharo C. A. C. G., and Hanson B. D. (2017) Vacuolar sequestration of paraquat is involved in the resistance mechanism in Lolium perenne L. spp. multiflorum. Front. Plant Sci. 8, 1485 10.3389/fpls.2017.01485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Moretti M., and Hanson B. (2017) Reduced translocation is involved in resistance to glyphosate and paraquat in Conyza bonariensis and Conyza canadensis from California. Weed Res. 57, 25–34 10.1111/wre.12230 [DOI] [Google Scholar]
- 86. Li J., Mu J., Bai J., Fu F., Zou T., An F., Zhang J., Jing H., Wang Q., Li Z., Yang S., and Zuo J. (2013) PARAQUAT RESISTANT1, a Golgi-localized putative transporter protein, is involved in intracellular transport of paraquat. Plant Physiol. 162, 470–483 10.1104/pp.113.213892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fujita M., Fujita Y., Iuchi S., Yamada K., Kobayashi Y., Urano K., Kobayashi M., Yamaguchi-Shinozaki K., and Shinozaki K. (2012) Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 6343–6347 10.1073/pnas.1121406109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Luo Q., Wei J., Dong Z., Shen X., and Chen Y. (2019) Differences of endogenous polyamines and putative genes associated with paraquat resistance in goosegrass (Eleusine indica L. ). PLoS ONE 14, e0216513 10.1371/journal.pone.0216513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Riar D. S., Burke I. C., Yenish J. P., Bell J., and Gill K. (2011) Inheritance and physiological basis for 2,4-D resistance in prickly lettuce (Lactuca serriola L.). J. Agric. Food Chem. 59, 9417–9423 10.1021/jf2019616 [DOI] [PubMed] [Google Scholar]
- 90. Goggin D. E., Cawthray G. R., and Powles S. B. (2016) 2,4-D resistance in wild radish: reduced herbicide translocation via inhibition of cellular transport. J. Exp. Bot. 67, 3223–3235 10.1093/jxb/erw120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pettinga D. J., Ou J., Patterson E. L., Jugulam M., Westra P., and Gaines T. A. (2018) Increased chalcone synthase (CHS) expression is associated with dicamba resistance in Kochia scoparia. Pest Manag. Sci. 74, 2306–2315 10.1002/ps.4778 [DOI] [PubMed] [Google Scholar]
- 92. Moretti M. L., Van Horn C. R., Robertson R., Segobye K., Weller S. C., Young B. G., Johnson W. G., Douglas Sammons R., Wang D., Ge X., d'Avignon A., Gaines T. A., Westra P., Green A. C., Jeffery T., et al. (2018) Glyphosate resistance in Ambrosia trifida: Part 2. Rapid response physiology and non‐target‐site resistance. Pest Manag. Sci. 74, 1079–1088 10.1002/ps.4569 [DOI] [PubMed] [Google Scholar]
- 93. Van Horn C. R., Moretti M. L., Robertson R. R., Segobye K., Weller S. C., Young B. G., Johnson W. G., Schulz B., Green A. C., Jeffery T., Lespérance M., Tardif F., Sikkema P., Hall J. C., McLean M., et al. (2018) Glyphosate resistance in Ambrosia trifida: Part 1. Novel rapid cell death response to glyphosate. Pest Manag. Sci. 74, 1071–1078 10.1002/ps.4567 [DOI] [PubMed] [Google Scholar]
- 94. de Queiroz A. R. S., Delatorre C. A., Lucio F. R., Rossi C. V. S., Zobiole L. H. S., and Merotto A. (2020) Rapid necrosis: a novel plant resistance mechanism to 2,4-D. Weed Sci. 68, 6–18 [Google Scholar]
- 95. Yuan J. S., Tranel P. J., and Stewart C. N. (2007) Non-target-site herbicide resistance: a family business. Trends Plant Sci. 12, 6–13 10.1016/j.tplants.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 96. Zimmerlin A., and Durst F. (1992) Aryl hydroxylation of the herbicide diclofop by a wheat cytochrome-P-450 monooxygenase: substrate-specificity and physiological activity. Plant Physiol. 100, 874–881 10.1104/pp.100.2.874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hall L. M., Moss S. R., and Powles S. B. (1995) Mechanism of resistance to chlorotoluron in two biotypes of the grass weed Alopecurus myosuroides. Pestic. Biochem. Physiol. 53, 180–192 10.1006/pest.1995.1066 [DOI] [Google Scholar]
- 98. Christopher J. T., Powles S. B., Liljegren D. R., and Holtum J. A. M. (1991) Cross-resistance to herbicides in annual ryegrass (Lolium rigidum). II. Chlorsulfuron resistance involves a wheat-like detoxification system. Plant Physiol. 95, 1036–1043 10.1104/pp.95.4.1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Werck-Reichhart D., and Feyereisen R. (2000) Cytochromes P450: a success story. Genom. Biol. 1, 1–9 10.1186/gb-2000-1-6-reviews3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Werck-Reichhart D., Hehn A., and Didierjean L. (2000) Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 5, 116–123 10.1016/S1360-1385(00)01567-3 [DOI] [PubMed] [Google Scholar]
- 101. Deng F., and Hatzios K. K. (2002) Characterization of cytochrome P450-mediated bensulfuron-methyl O-demethylation in rice. Pestic. Biochem. Physiol. 74, 102–115 10.1016/S0048-3575(02)00151-7 [DOI] [Google Scholar]
- 102. Busi R., Dayan F. E., Francis I., Goggin D., Lerchl J., Porri A., Powles S. B., Sun C., and Beckie H. J. (2020) Cinmethylin controls multiple herbicide-resistant Lolium rigidum and its wheat selectivity is P450-based. Pest Manag. Sci. 76, 2601–2608 10.1002/ps.5798 [DOI] [PubMed] [Google Scholar]
- 103. Grossmann K., and Ehrhardt T. (2007) On the mechanism of action and selectivity of the corn herbicide topramezone: a new inhibitor of 4-hydroxyphenylpyruvate dioxygenase. Pest Manag. Sci. 63, 429–439 10.1002/ps.1341 [DOI] [PubMed] [Google Scholar]
- 104. Siminszky B. (2006) Plant cytochrome P450-mediated herbicide metabolism. Phytochem. Rev. 5, 445–458 10.1007/s11101-006-9011-7 [DOI] [Google Scholar]
- 105. Powles S. B., and Yu Q. (2010) Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347 10.1146/annurev-arplant-042809-112119 [DOI] [PubMed] [Google Scholar]
- 106. Christopher J. T., Preston C., and Powles S. B. (1994) Malathion antagonizes metabolism-based chlorsulfuron resistance in Lolium rigidum. Pestic. Biochem. Physiol. 49, 172–182 10.1006/pest.1994.1045 [DOI] [Google Scholar]
- 107. Wang H. C., Li J., Lv B., Lou Y. L., and Dong L. Y. (2013) The role of cytochrome P450 monooxygenase in the different responses to fenoxaprop-P-ethyl in annual bluegrass (Poa annua L.) and short awned foxtail (Alopecurus aequalis Sobol.). Pest. Biochem. Physiol. 107, 334–342 10.1016/j.pestbp.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 108. Küpper A., Peter F., Zöllner P., Lorentz L., Tranel P. J., Beffa R., and Gaines T. A. (2018) Tembotrione detoxification in 4-hydroxyphenylpyruvate dioxygenase (HPPD) inhibitor-resistant Palmer amaranth (Amaranthus palmeri S. Wats.). Pest Manag. Sci. 74, 2325–2334 10.1002/ps.4786 [DOI] [PubMed] [Google Scholar]
- 109. Figueiredo M. R. A., Leibhart L. J., Reicher Z. J., Tranel P. J., Nissen S. J., Westra P., Bernards M. L., Kruger G. R., Gaines T. A., and Jugulam M. (2018) Metabolism of 2,4-dichlorophenoxyacetic acid contributes to resistance in a common waterhemp (Amaranthus tuberculatus) population. Pest Manag. Sci. 74, 2356–2362 10.1002/ps.4811 [DOI] [PubMed] [Google Scholar]
- 110. Lu H., Yu Q., Han H., Owen M. J., and Powles S. B. (2020) Non-target-site resistance to PDS-inhibiting herbicides in a wild radish (Raphanus raphanistrum) population. Pest Manag. Sci. 76, 2015–2020 10.1002/ps.5733 [DOI] [PubMed] [Google Scholar]
- 111. Varanasi V. K., Brabham C., and Norsworthy J. K. (2018) Confirmation and characterization of non–target site resistance to fomesafen in Palmer amaranth (Amaranthus palmeri). Weed Sci. 66, 702–709 10.1017/wsc.2018.60 [DOI] [Google Scholar]
- 112. Yun M. S., Yogo Y., Miura R., Yamasue Y., and Fischer A. J. (2005) Cytochrome P-450 monooxygenase activity in herbicide-resistant and -susceptible late watergrass (Echinochloa phyllopogon). Pest. Biochem. Physiol. 83, 107–114 10.1016/j.pestbp.2005.04.002 [DOI] [Google Scholar]
- 113. Yasuor H., Osuna M. D., Ortiz A., Saldaín N. E., Eckert J. W., and Fischer A. J. (2009) Mechanism of resistance to penoxsulam in late watergrass (Echinochloa phyllopogon (Stapf) Koss.). J. Agric. Food Chem. 57, 3653–3660 10.1021/jf8039999 [DOI] [PubMed] [Google Scholar]
- 114. Yasuor H., Zou W., Tolstikov V. V., Tjeerdema R. S., and Fischer A. J. (2010) Differential oxidative metabolism and 5-ketoclomazone accumulation are involved in Echinochloa phyllopogon resistance to clomazone. Plant Physiol. 153, 319–326 10.1104/pp.110.153296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Iwakami S., Uchino A., Kataoka Y., Shibaike H., Watanabe H., and Inamura T. (2014) Cytochrome P450 genes induced by bispyribac-sodium treatment in a multiple-herbicide resistant biotype of Echinochloa phyllopogon. Pest Manag. Sci. 70, 549–558 10.1002/ps.3572 [DOI] [PubMed] [Google Scholar]
- 116. Iwakami S., Endo M., Saika H., Okuno J., Nakamura N., Yokoyama M., Watanabe H., Toki S., Uchino A., and Inamura T. (2014) Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant Physiol. 165, 618–629 10.1104/pp.113.232843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Dimaano N. G., Yamaguchi T., Fukunishi K., Tominaga T., and Iwakami S. (2020) Functional characterization of cytochrome P450 CYP81A subfamily to disclose the pattern of cross-resistance in Echinochloa phyllopogon. Plant Mol. Biol. 102, 403–416 10.1007/s11103-019-00954-3 [DOI] [PubMed] [Google Scholar]
- 118. Iwakami S., Kamidate Y., Yamaguchi T., Ishizaka M., Endo M., Suda H., Nagai K., Sunohara Y., Toki S., Uchino A., Tominaga T., and Matsumoto H. (2019) CYP81A P450s are involved in concomitant cross-resistance to acetolactate synthase and acetyl-CoA carboxylase herbicides in Echinochloa phyllopogon. New Phytol. 221, 2112–2122 10.1111/nph.15552 [DOI] [PubMed] [Google Scholar]
- 119. Guo F., Iwakami S., Yamaguchi T., Uchino A., Sunohara Y., and Matsumoto H. (2019) Role of CYP81A cytochrome P450s in clomazone metabolism in Echinochloa phyllopogon. Plant Sci. 283, 321–328 10.1016/j.plantsci.2019.02.010 [DOI] [PubMed] [Google Scholar]
- 120. Manley B. S., Hatzios K. K., and Wilson H. P. (1999) Absorption, translocation, and metabolism of chlorimuron and nicosulfuron in imidazolinone-resistant and -susceptible smooth pigweed (Amaranthus hybridus). Weed Technol. 13, 759–764 10.1017/S0890037X00042196 [DOI] [Google Scholar]
- 121. Vencill W. K., Li X., and Grey T. L. (2013) Multiple mechanisms of Palmer amaranth (Amaranthus palmeri) resistance to ALS-inhibiting herbicides. Proc. Weed Sci. Soc. Am. 53, 363 [Google Scholar]
- 122. Hausman N. E., Singh S., Tranel P. J., Riechers D. E., Kaundun S. S., Polge N. D., Thomas D. A., and Hager A. G. (2011) Resistance to HPPD-inhibiting herbicides in a population of waterhemp (Amaranthus tuberculatus) from Illinois, United States. Pest Manag. Sci. 67, 258–261 10.1002/ps.2100 [DOI] [PubMed] [Google Scholar]
- 123. McMullan P. M., and Green J. M. (2011) Identification of a tall waterhemp (Amaranthus tuberculatus) biotype resistant to HPPD-inhibiting herbicides, atrazine, and thifensulfuron in Iowa. Weed Technol. 25, 514–518 10.1614/WT-D-10-00150.1 [DOI] [Google Scholar]
- 124. Wilde T., Beffa R. S., Kleven T., Philbrook B., and Strek H. (2013) HPPD resistance testing in the U.S.A. - Preliminary bioassay results. Proc. Weed Sci. Soc. Am. 53, 131 [Google Scholar]
- 125. Lu H., Yu Q., Han H., Owen M. J., and Powles S. B. (2020) Evolution of resistance to HPPD-inhibiting herbicides in a wild radish population via enhanced herbicide metabolism. Pest Manag. Sci. 76, 1929–1937 10.1002/ps.5725 [DOI] [PubMed] [Google Scholar]
- 126. Ma R., McGinness D., Hausman N. E., Tranel P. J., Hager A., Kaundun S. S., Hawkes T., Vail G. D., and Riechers D. E. (2015) Investigation of resistance mechanisms to mesotrione in a waterhemp (Amaranthus tuberculatus) population from Illinois. Proc. Weed Sci. Soc. Am. 63, 799–809 [Google Scholar]
- 127. Oliveira M. C., Gaines T. A., Dayan F. E., Patterson E. L., Jhala A. J., and Knezevic S. Z. (2018) Reversing resistance to tembotrione in an Amaranthus tuberculatus (var. rudis) population from Nebraska, U.S.A. with cytochrome P450 inhibitors. Pest Manag. Sci. 74, 2296–2305 10.1002/ps.4697 [DOI] [PubMed] [Google Scholar]
- 128. Shimabukuro R. H., Frear D. S., Swanson H. R., and Walsh W. C. (1971) Glutathione conjugation: enzymatic basis for atrazine resistance in corn. Plant Physiol. 47, 10–14 10.1104/pp.47.1.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cummins I., Dixon D. P., Freitag-Pohl S., Skipsey M., and Edwards R. (2011) Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metabol. Rev. 43, 266–280 10.3109/03602532.2011.552910 [DOI] [PubMed] [Google Scholar]
- 130. Bakkali Y., Ruiz-Santaella J. P., Osuna M. D., Wagner J., Fischer A. J., and De Prado R. (2007) Late watergrass (Echinochloa phyllopogon): Mechanisms involved in the resistance to fenoxaprop-P-ethyl. J. Agric. Food Chem. 55, 4052–4058 10.1021/jf0624749 [DOI] [PubMed] [Google Scholar]
- 131. Cummins I., Moss S., Cole D. J., and Edwards R. (1997) Glutathione transferases in herbicide-resistant and herbicide-susceptible black-grass (Alopecurus myosuroides). Pestic. Sci. 51, 244–250 [DOI] [Google Scholar]
- 132. Cummins I., Wortley D. J., Sabbadin F., He Z., Coxon C. R., Straker H. E., Sellars J. D., Knight K., Edwards L., Hughes D., Kaundun S. S., Hutchings S. J., Steel P. G., and Edwards R. (2013) Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc. Natl. Acad. Sci. U.S.A. 110, 5812–5817 10.1073/pnas.1221179110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Dücker R., Zöllner P., Parcharidou E., Ries S., Lorentz L., and Beffa R. (2019) Enhanced metabolism causes reduced flufenacet sensitivity in black-grass (Alopecurus myosuroides Huds.) field populations. Pest Manag. Sci. 75, 2996–3004 10.1002/ps.5414 [DOI] [PubMed] [Google Scholar]
- 134. Dücker R., Zöllner P., Lümmen P., Ries S., Collavo A., and Beffa R. (2019) Glutathione transferase plays a major role in flufenacet resistance of ryegrass (Lolium spp.) field populations. Pest Manag. Sci. 75, 3084–3092 10.1002/ps.5425 [DOI] [PubMed] [Google Scholar]
- 135. Nandula V. K., Riechers D. E., Ferhatoglu Y., Barrett M., Duke S. O., Dayan F. E., Goldberg-Cavalleri A., Tétard-Jones C., Wortley D. J., Onkokesung N., Brazier-Hicks M., Edwards R., Gaines T., Iwakami S., Jugulam M., et al. (2019) Herbicide metabolism: crop selectivity, bioactivation, weed resistance, and regulation. Weed Sci. 67, 149–175 10.1017/wsc.2018.88 [DOI] [Google Scholar]
- 136. Gaines T. A., Lorentz L., Figge A., Herrmann J., Maiwald F., Ott M. C., Han H., Busi R., Yu Q., Powles S. B., and Beffa R. (2014) RNA-Seq transcriptome analysis to identify genes involved in metabolism-based diclofop resistance in Lolium rigidum. Plant J. 78, 865–876 10.1111/tpj.12514 [DOI] [PubMed] [Google Scholar]
- 137. Duhoux A., Carrère S., Gouzy J., Bonin L., and Délye C. (2015) RNA-Seq analysis of rye-grass transcriptomic response to an herbicide inhibiting acetolactate-synthase identifies transcripts linked to non-target-site-based resistance. Plant Mol. Biol. 87, 473–487 10.1007/s11103-015-0292-3 [DOI] [PubMed] [Google Scholar]
- 138. Scheel D., and Sandermann H. (1981) Metabolism of 2,4-dichlorophenoxyacetic acid in cell suspension cultures of soybean (Glycine max L.) and wheat (Triticum aestivum L.). Planta 152, 253–258 10.1007/BF00385153 [DOI] [PubMed] [Google Scholar]
- 139. Sandermann H. J., Haas M., Messner B., Pflumacher S., Schroder P., and Wetzel A. (1997) The role of glucosyl and malonyl conjugation in herbicide selectivity. In Regulation of Enzymatic Systems Detoxifying Xenobiotics in Plants (Hatzios K. K., ed) pp. 211–231, Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]
- 140. Chen J. J., and Matsunaka S. (1990) The propanil hydrolyzing enzyme aryl acylamidase in the wild rices of genus Oryza. Pestic. Biochem. Physiol. 38, 26–33 10.1016/0048-3575(90)90144-Q [DOI] [Google Scholar]
- 141. Leah J. M., Caseley J. C., Riches C. R., and Valverde B. (1994) Association between elevated activity of aryl acylamidase and propanil resistance in Jungle-rice, Echinochloa colona. Pestic. Sci. 42, 281–289 10.1002/ps.2780420405 [DOI] [Google Scholar]
- 142. Hirase K., and Hoagland R. E. (2006) Characterization of aryl acylamidase activity from propanil-resistant barnyardgrass (Echinochloa crus-galli [L.] Beauv. ). Weed Biol. Manag. 6, 197–203 10.1111/j.1445-6664.2006.00218.x [DOI] [Google Scholar]
- 143. Pedroso R. M., Al-Khatib K., Alarcón-Reverte R., and Fischer A. J. (2016) A psbA mutation (Val219 to Ile) causes resistance to propanil and increased susceptibility to bentazon in Cyperus difformis. Pest Manag. Sci. 72, 1673–1680 10.1002/ps.4267 [DOI] [PubMed] [Google Scholar]
- 144. Duke S. O. (2011) Glyphosate degradation in glyphosate-resistant and -susceptible crops and weeds. J. Agric. Food Chem. 59, 5835–5841 10.1021/jf102704x [DOI] [PubMed] [Google Scholar]
- 145. Pan L., Yu Q., Han H., Mao L., Nyporko A., Fan L., Bai L., and Powles S. (2019) Aldo-keto reductase metabolizes glyphosate and confers glyphosate resistance in Echinochloa colona. Plant Physiol. 181, 1519–1534 10.1104/pp.19.00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Vemanna R. S., Vennapusa A. R., Easwaran M., Chandrashekar B. K., Rao H., Ghanti K., Sudhakar C., Mysore K. S., and Makarla U. (2017) Aldo-keto reductase enzymes detoxify glyphosate and improve herbicide resistance in plants. Plant Biotechnol. J. 15, 794–804 10.1111/pbi.12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Grossmann K. (2010) Auxin herbicides: current status of mechanism and mode of action. Pest Manag. Sci. 66, 113–120 10.1002/ps.1860 [DOI] [PubMed] [Google Scholar]
- 148. Yasuor H., Milan M., Eckert J. W., and Fischer A. J. (2012) Quinclorac resistance: a concerted hormonal and enzymatic effort in Echinochloa phyllopogon. Pest Manag. Sci. 68, 108–115 10.1002/ps.2230 [DOI] [PubMed] [Google Scholar]
- 149. Délye C., Gardin J. A. C., Boucansaud K., Chauvel B., and Petit C. (2011) Non-target-site-based resistance should be the centre of attention for herbicide resistance research: Alopecurus myosuroides as an illustration. Weed Res. 51, 433–437 10.1111/j.1365-3180.2011.00864.x [DOI] [Google Scholar]
- 150. Gardin J., Beffa R., Gouzy J., Carrere S., and Delye C. (2013) A transcriptomics-based approach enables the first identification of candidate genes involved in non-target-site-based resistance to herbicides in a grass weed (Alopecurus myosuroides). In Global Herbicide Resistance Challenge, p. 68, Australian Herbicide Resistance Initiative, Perth, Australia [Google Scholar]
- 151. Busi R., and Powles S. B. (2011) Reduced sensitivity to paraquat evolves under selection with low glyphosate doses in Lolium rigidum. Agron. Sustain. Develop. 31, 525–531 10.1007/s13593-011-0012-6 [DOI] [Google Scholar]
- 152. Maroli A. S., Gaines T. A., Foley M. E., Duke S. O., Doğramacı M., Anderson J. V., Horvath D. P., Chao W. S., and Tharayil N. (2018) Omics in weed science: a perspective from genomics, transcriptomics, and metabolomics approaches. Weed Sci. 66, 681–695 10.1017/wsc.2018.33 [DOI] [Google Scholar]
- 153. Giacomini D. A., Gaines T., Beffa R., and Tranel P. J. (2018) Optimizing RNA-seq studies to investigate herbicide resistance. Pest Manag. Sci. 74, 2260–2264 10.1002/ps.4822 [DOI] [PubMed] [Google Scholar]
- 154. Kreiner J. M., Giacomini D. A., Bemm F., Waithaka B., Regalado J., Lanz C., Hildebrandt J., Sikkema P. H., Tranel P. J., Weigel D., Stinchcombe J. R., and Wright S. I. (2019) Multiple modes of convergent adaptation in the spread of glyphosate-resistant Amaranthus tuberculatus. Proc. Natl. Acad. Sci. U.S.A. 116, 21076–21084 10.1073/pnas.1900870116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Laforest M., Martin S. L., Bisaillon K., Soufiane B., Meloche S., and Page E. (2020) A chromosome-scale draft sequence of the Canada fleabane genome. Pest Manag. Sci. 76, 2158–2169 10.1002/ps.5753 [DOI] [PubMed] [Google Scholar]
- 156. Moghe G. D., Hufnagel D. E., Tang H., Xiao Y., Dworkin I., Town C. D., Conner J. K., and Shiu S.-H. (2014) Consequences of whole genome triplication as revealed by comparative genomic analyses of the wild radish Raphanus raphanistrum and three other brassicaceae species. Plant Cell 26, 1925–1937 10.1105/tpc.114.124297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Guo L., Qiu J., Ye C., Jin G., Mao L., Zhang H., Yang X., Peng Q., Wang Y., Jia L., Lin Z., Li G., Fu F., Liu C., Chen L., et al. (2017) Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 8, 1031 10.1038/s41467-017-01067-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zhang H., Hall N., Goertzen L. R., Bi B., Chen C. Y., Peatman E., Lowe E. K., Patel J., and McElroy J. S. (2019) Development of a goosegrass (Eleusine indica) draft genome and application to weed science research. Pest Manag. Sci. 75, 2776–2784 10.1002/ps.5389 [DOI] [PubMed] [Google Scholar]
- 159. Jamann T. M., Balint-Kurti P. J., and Holland J. B. (2015) QTL mapping using high-throughput sequencing. In Plant Functional Genomics: Methods and Protocols (Alonso J. M., and Stepanova A. N., eds) pp. 257–285, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 160. Jasieniuk M., Brûlé-Babel A. L., and Morrison I. N. (1996) The evolution and genetics of herbicide resistance in weeds. Weed Sci. 44, 176–193 10.1017/S0043174500093747 [DOI] [Google Scholar]
- 161. Foes M. J., Liu L., Tranel P. J., Wax L. M., and Stoller E. W. (1998) A biotype of common waterhemp (Amaranthus rudis) resistant to triazine and ALS herbicides. Weed Sci. 46, 514–520 10.1017/S0043174500091013 [DOI] [Google Scholar]
- 162. Huffman J. L., Riggins C. W., Steckel L. E., and Tranel P. J. (2016) The EPSPS Pro106Ser substitution solely accounts for glyphosate resistance in a goosegrass (Eleusine indica) population from Tennessee, United States. J. Integr. Agric. 15, 1304–1312 10.1016/S2095-3119(15)61220-5 [DOI] [Google Scholar]
- 163. Jugulam M., and Godar A. S. (2013) Understanding genetics of herbicide resistance in weeds: implications for weed management. Adv. Crop Sci. Technol. 1, 115 10.4172/2329-8863.1000115 [DOI] [Google Scholar]
- 164. Sabba R. P., Ray I. M., Lownds N., and Sterling T. M. (2003) Inheritance of resistance to clopyralid and picloram in yellow starthistle (Centaurea solstitialis L.) is controlled by a single nuclear recessive gene. J. Hered. 94, 523–527 10.1093/jhered/esg101 [DOI] [PubMed] [Google Scholar]
- 165. Chen J., Lu H., Han H., Yu Q., Sayer C., and Powles S. (2019) Genetic inheritance of dinitroaniline resistance in an annual ryegrass population. Plant Sci. 283, 189–194 10.1016/j.plantsci.2019.02.019 [DOI] [PubMed] [Google Scholar]
- 166. Fearon C. H., Hayward M. D., and Lawrence M. J. (1983) Self-incompatibility in ryegrass. Heredity 50, 35–45 10.1038/hdy.1983.5 [DOI] [Google Scholar]
- 167. Machado V. S., Bandeen J. D., Stephenson G. R., and Lavigne P. (1978) Uniparental inheritance of chloroplast atrazine tolerance in Brassica campetris. Can. J. Plant Sci. 58, 977–981 10.4141/cjps78-150 [DOI] [Google Scholar]
- 168. Patzoldt W. L., Dixon B. S., and Tranel P. J. (2003) Triazine resistance in Amaranthus tuberculatus (Moq) Sauer that is not site-of-action mediated. Pest Manag. Sci. 59, 1134–1142 10.1002/ps.743 [DOI] [PubMed] [Google Scholar]
- 169. Huffman J., Hausman N. E., Hager A. G., Riechers D. E., and Tranel P. J. (2015) Genetics and inheritance of nontarget-site resistances to atrazine and mesotrione in a waterhemp (Amaranthus tuberculatus) population from Illinois. Weed Sci. 63, 799–809 10.1614/WS-D-15-00055.1 [DOI] [Google Scholar]
- 170. Andersen R. N., and Gronwald J. W. (1987) Noncytoplasmic inheritance of atrazine tolerance in velvetleaf (Abutilon theophrasti). Weed Sci. 35, 496–498 10.1017/S0043174500060446 [DOI] [Google Scholar]
- 171. Dang H. T., Malone J. M., Boutsalis P., Krishnan M., Gill G., and Preston C. (2018) Reduced translocation in 2,4-D-resistant oriental mustard populations (Sisymbrium orientale L.) from Australia. Pest Manag. Sci. 74, 1524–1532 10.1002/ps.4845 [DOI] [PubMed] [Google Scholar]
- 172. Zelaya I. A., Owen M. D. K., and VanGessel M. J. (2004) Inheritance of evolved glyphosate resistance in Conyza canadensis (L.) Cronq. Theor. Appl. Genet. 110, 58–70 10.1007/s00122-004-1804-8 [DOI] [PubMed] [Google Scholar]
- 173. Gressel J. (2009) Evolving understanding of the evolution of herbicide resistance. Pest Manag. Sci. 65, 1164–1173 10.1002/ps.1842 [DOI] [PubMed] [Google Scholar]
- 174. Vieira B. C., Luck J. D., Amundsen K. L., Werle R., Gaines T. A., and Kruger G. R. (2020) Herbicide drift exposure leads to reduced herbicide sensitivity in Amaranthus spp. Sci. Rep. 10, 2146 10.1038/s41598-020-59126-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Tehranchian P., Norsworthy J. K., Powles S., Bararpour M. T., Bagavathiannan M. V., Barber T., and Scott R. C. (2017) Recurrent sublethal-dose selection for reduced susceptibility of Palmer amaranth (Amaranthus palmeri) to dicamba. Weed Sci. 65, 206–212 10.1017/wsc.2016.27 [DOI] [Google Scholar]
- 176. Busi R., Girotto M., and Powles S. B. (2016) Response to low-dose herbicide selection in self-pollinated Avena fatua. Pest Manag. Sci. 72, 603–608 10.1002/ps.4032 [DOI] [PubMed] [Google Scholar]
- 177. Markus C., Pecinka A., Karan R., Barney J. N., and Merotto A. (2018) Epigenetic regulation—contribution to herbicide resistance in weeds? Pest Manag. Sci. 74, 275–281 10.1002/ps.4727 [DOI] [PubMed] [Google Scholar]
- 178. Margaritopoulou T., Tani E., Chachalis D., and Travlos I. (2018) Involvement of epigenetic mechanisms in herbicide resistance: the case of Conyza canadensis. Agriculture 8, 17 10.3390/agriculture8010017 [DOI] [Google Scholar]
- 179. Hawkins N. J., Bass C., Dixon A., and Neve P. (2019) The evolutionary origins of pesticide resistance. Biol. Rev. 94, 135–155 10.1111/brv.12440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Kreiner J. M., Stinchcombe J. R., and Wright S. I. (2018) Population genomics of herbicide resistance: Adaptation via evolutionary rescue. Annu. Rev. Plant Biol. 69, 611–635 10.1146/annurev-arplant-042817-040038 [DOI] [PubMed] [Google Scholar]
- 181. Yu Q., Ahmad-Hamdani M. S., Han H., Christoffers M. J., and Powles S. B. (2013) Herbicide resistance-endowing ACCase gene mutations in hexaploid wild oat (Avena fatua): insights into resistance evolution in a hexaploid species. Heredity 110, 220–231 10.1038/hdy.2012.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Hanson B. D., Shaner D. L., Westra P., and Nissen S. J. (2006) Response of selected hard red wheat lines to imazamox as affected by number and location of resistance genes, parental background, and growth habit. Crop Sci. 46, 1206–1211 10.2135/cropsci2005.10-0392 [DOI] [Google Scholar]
- 183. Brunharo C. A. D C. G., Morran S., Martin K., Moretti M. L., and Hanson B. D. (2019) EPSPS duplication and mutation involved in glyphosate resistance in the allotetraploid weed species Poa annua L. Pest Manag. Sci. 75, 1663–1670 10.1002/ps.5284 [DOI] [PubMed] [Google Scholar]
- 184. Chen S., McElroy J. S., Dane F., and Goertzen L. R. (2016) Transcriptome assembly and comparison of an allotetraploid weed species, annual bluegrass, with its two diploid progenitor species, Poa supina Schrad and Poa infirma Kunth. Plant Genet. 9, 10.3835/plantgenome2015.06.0050 [DOI] [PubMed] [Google Scholar]
- 185. Beversdorf W. D., and Kott L. S. (1987) Development of triazine resistance in crops by classical plant breeding. Weed Sci. 35, 9–11 10.1017/S0043174500060975 [DOI] [Google Scholar]
- 186. Duke S. O. (2014) Biotechnology: herbicide-resistant crops. In Encyclopedia of Agriculture and Food Systems (Van Alfen N., ed) pp. 94–116, Elsevier, San Diego [Google Scholar]
- 187. Dreesen R., Capt A., Oberdoerfer R., Coats I., and Pallett K. E. (2018) Characterization and safety evaluation of HPPD W336, a modified 4-hydroxyphenylpyruvate dioxygenase protein, and the impact of its expression on plant metabolism in herbicide-tolerant MST-FGØ72-2 soybean. Regul. Toxicol. Pharmacol. 97, 170–185 10.1016/j.yrtph.2018.06.002 [DOI] [PubMed] [Google Scholar]
- 188. Hawkes T. R., Langford M. P., Viner R., Blain R. E., Callaghan F. M., Mackay E. A., Hogg B. V., Singh S., and Dale R. P. (2019) Characterization of 4-hydroxyphenylpyruvate dioxygenases, inhibition by herbicides and engineering for herbicide tolerance in crops. Pestic. Biochem. Physiol. 156, 9–28 10.1016/j.pestbp.2019.01.006 [DOI] [PubMed] [Google Scholar]
- 189. Bradshaw L. D., Padgette S. R., Kimball S. L., and Wells B. H. (1997) Perspectives on glyphosate resistance. Weed Technol. 11, 189–198 10.1017/S0890037X00041567 [DOI] [Google Scholar]
- 190. Tranel P. J., and Wright T. R. (2002) Resistance of weeds to ALS-inhibiting herbicides: what have we learned? Weed Sci. 50, 700–712 10.1614/0043-1745(2002)050[0700:RROWTA]2.0.CO;2 [DOI] [Google Scholar]
- 191. Ahrens W. H., and Stoller E. W. (1983) Competition, growth rate, and CO2 fixation in triazine-susceptible and -resistant smooth pigweed (Amaranthus hybridus). Weed Sci. 31, 438–444 10.1017/S0043174500069356 [DOI] [Google Scholar]
- 192. Conard S. G., and Radosevich S. R. (1979) Ecological fitness of Senecio vulgaris and Amaranthus retroflexus biotypes susceptible or resistant to atrazine. J. Appl. Ecol. 16, 171–177 10.2307/2402736 [DOI] [Google Scholar]
- 193. de Montellano P. R. O. (2015) Substrate oxidation by cytochrome P450 enzymes. In Cytochrome P450 (de Montellano P. R. O., ed) pp. 111–176, Springer, New York [Google Scholar]
- 194. Kniss A. R. (2018) Genetically engineered herbicide-resistant crops and herbicide resistant weed evolution in the United States. Weed Sci 66, 260–273 10.1017/wsc.2017.70 [DOI] [Google Scholar]
- 195. Powles S. B., and Holtum J. (1994) Herbicide Resistance in Plants: Biology and Biochemistry, CRC Press, Inc., Boca Raton, FL [Google Scholar]