Abstract
Bacteria must rapidly respond to both intracellular and environmental changes to survive. One critical mechanism to rapidly detect and adapt to changes in environmental conditions is control of gene expression at the level of protein synthesis. At each of the three major steps of translation—initiation, elongation, and termination—cells use stimuli to tune translation rate and cellular protein concentrations. For example, changes in nutrient concentrations in the cell can lead to translational responses involving mechanisms such as dynamic folding of riboswitches during translation initiation or the synthesis of alarmones, which drastically alter cell physiology. Moreover, the cell can fine-tune the levels of specific protein products using programmed ribosome pausing or inducing frameshifting. Recent studies have improved understanding and revealed greater complexity regarding long-standing paradigms describing key regulatory steps of translation such as start-site selection and the coupling of transcription and translation. In this review, we describe how bacteria regulate their gene expression at the three translational steps and discuss how translation is used to detect and respond to changes in the cellular environment. Finally, we appraise the costs and benefits of regulation at the translational level in bacteria.
Keywords: translation control, ribosome, post-translational modification (PTM), gene regulation, environmental regulation, mRNA folding, stringent response, translational frameshifting, cellular adaptation, ribosome function
Bacterial cells face a wide variety of challenges from their outside environment, including rapid changes in temperatures and nutrient concentrations. To survive and remain competitive, bacteria use strategies to rapidly adapt to diverse stimuli, many of which require transcriptional changes that impact cellular concentration of adaptive protein factors (1, 2). For example, under heat stress, transcription of critical thermotolerance genes are controlled through the transcription factor σ32 (3). Another means to control transcription dependent on environmental stimuli is the use of two component systems. Two component systems transduce information from the environment, such as changes in nutrient levels, into rapid transcriptional responses that lead to adaptive processes like chemotaxis (4, 5). Regulation of gene expression through σ factors and two component systems are just two of the several mechanisms of adaptive transcriptional control.
Adaptation to external stimuli is not reliant on transcriptional regulation alone; regulation at the level of protein synthesis can also result in rapid and fine-tuned responses to challenges. Translation is a complex and dynamic process that includes a sequence of steps that must occur rapidly to optimally decode genetic information (6–8). Translation in bacteria can be roughly partitioned into three phases: initiation, elongation, and termination. Translation initiation comprises the small subunit of the ribosome binding to the correct site on the mRNA and assembling with support of initiation factors IF1, IF2, and IF3. During elongation tRNAs charged with amino acids are brought to the ribosome by the elongation factor EF-Tu and added to the growing peptide chain. Ribosomal translocation is then facilitated by EF-G. When the ribosome reaches a stop codon, translation termination is promoted by release factor 1 or 2, depending on the sequence identity of the stop codon.
Given the growing body of data elaborately describing the basic mechanistic aspects of each step of translation, recent studies have been able to focus on the intricacies of translational regulation. These studies have been aided by the advent of technologies such as ribosome profiling, allowing researchers to rapidly collect ribosomes from cells under many different conditions and calculate ribosome “footprints,” which indicate the location and density of ribosomes upon a given mRNA. Insights into translational events using ribosome profiling technology have significantly influenced the field's understanding of the three major steps of translation (initiation, elongation, and termination) at a single-ribosome scale. Recent studies have also begun to question long-standing paradigms in key regulatory steps of translation such as start-site selection and the coupling of transcription and translation. In this review, we will discuss specific points of regulation during the three steps of translation and how control at each of these steps can lead to rapid and effective adaptation to environmental stimuli.
Initiation
Protein synthesis places a significant demand on cellular energy pools, specifically through the use of ATP and GTP to activate amino acids for transfer onto tRNAs and hydrolysis by translational GTPases, respectively (9–11). To prevent the superfluous use of energy, translation must be tightly regulated at the point of initiation. The rate of translation initiation has also been observed to be rate-limiting in protein production (12). Another key function of translational regulation at the step of initiation is to maintain protein stoichiometry, allowing for the synthesis of functional complexes under variable conditions (13). Control of initiation can be achieved through mechanisms such as regulation of key initiation determinants, including sequestration of the ribosome-binding site and control of the number of available ribosomes. This section gives an overview of some of the most common ways bacteria regulate their physiology at the step of translation initiation.
Ribosome-binding site
In many bacteria, translation initiation sites in leading genes are recognized by the ribosome through interactions with specific nucleotide sequences near the 5′-end of the mRNA called Shine–Dalgarno motifs (SDs). This sequence pairs to an anti-SD (aSD) sequence found in the 16S rRNA of the ribosomal small subunit, beginning assembly of the translation initiation complex (14–16). Protein output is directly correlated with ribosome loading on a given transcript, and by changing the potential for the SD and aSD to pair through changing the accessibility or sequence identity of the SD, ribosome occupancy on a given transcript can be controlled (Fig. 1A) (17–19). Generally, highly translated mRNAs have SD sequences closer to the consensus sequence (AGGAGG) than those that are less well-translated because of more energetically favorable hybridization between the mRNA at the SD sequence and the aSD in the 16S rRNA, although this is not always the case. Further, not all transcripts contain identified SDs, and the proportion of mRNAs that encode SDs differs between organisms (14, 19–21). For example, in Escherichia coli ∼57% of all genes are preceded by a consensus SD, whereas Bacillus subtilis and Bacteroidetes encode identified SDs in ∼90% and less than 10% of genes, respectively (14, 23, 24).
Figure 1.
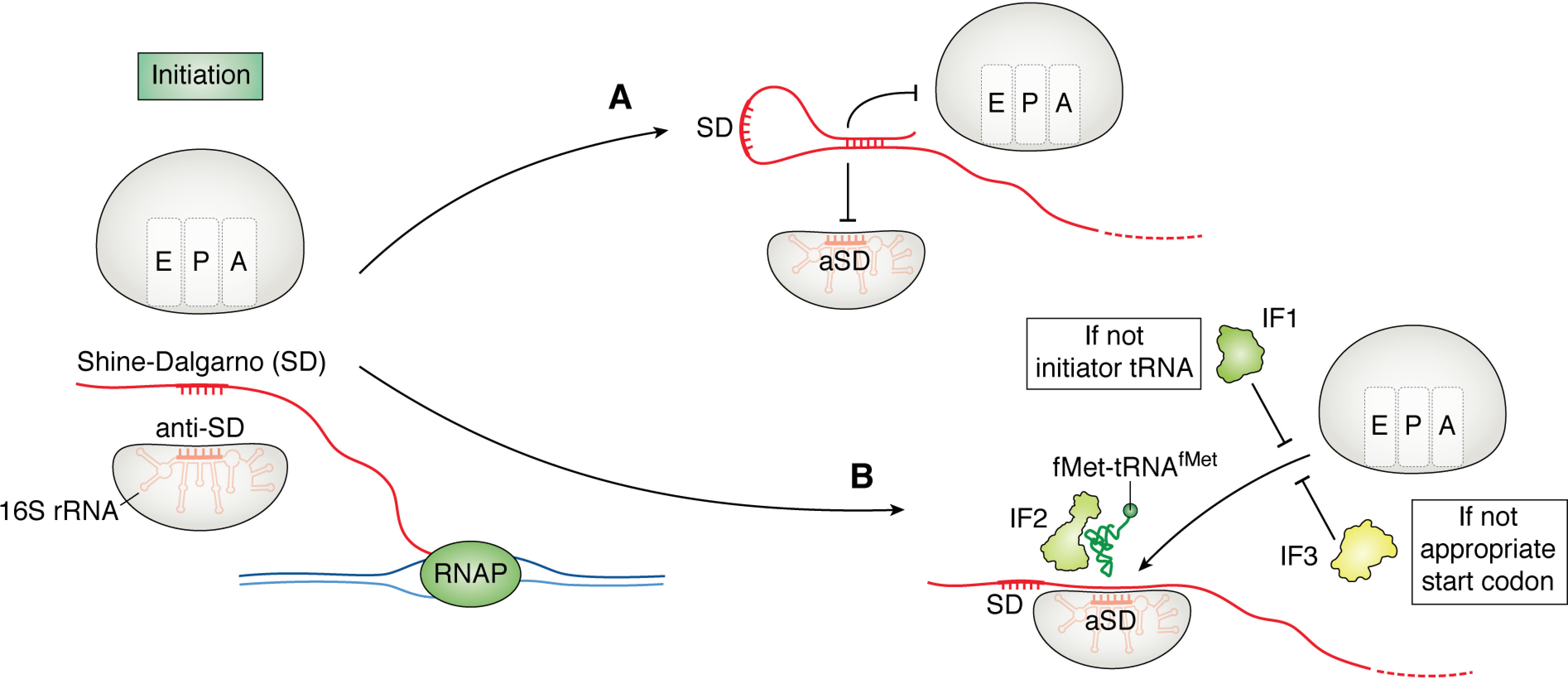
Regulation of translation initiation in bacteria. In many bacteria, translation initiation occurs downstream of a SD with the aid of initiation factors 1–3. A, one mechanism to regulate initiation is to inhibit ribosomal subunit joining to the mRNA by sequestering the SD through RNA folding events. B, when the process of translational initiation begins, IF1 inspects the tRNA that is brought to the ribosome by IF2. IF3 aids in initiation fidelity by ensuring initiation at the appropriate start codon.
Translation of genes without SDs depends on multiple factors. In E. coli, the ribosomal protein S1 has a role in recognition of mRNAs without an SD through its recognition of A/U-rich sequences (16). RNA folding also plays a critical role in translation of genes in which the SD is absent because the propensity for mRNA folding is inversely correlated with the initiation efficiency, where mRNA folding can occlude critical determinants for mRNA recognition by the initiating ribosome (25). Recently, ribosome profiling was performed in Flavobacterium johnsoniae, a species of Bacteroidetes in which SD sequences are absent (23). From the profiling data, it was found that F. johnsoniae utilizes mRNAs that contain an overrepresentation of adenine near the translation start site, specifically in the region 11–14 nucleotides upstream of the start codon. One consequence stemming from initiating translation without an SD is that F. johnsoniae also has underrepresentation of AUG codons within genes, a potential mechanism to prevent erroneous internal translation initiation. This overrepresentation of adenine is not unique to F. johnsoniae; E. coli also showed an enrichment of adenine in similar regions of the 5′-UTR of the mRNA. The presence of adenine was also seen to enhance translation of reporter sequences in both E. coli and F. johnsoniae, suggesting that bacteria may use “Kozak-like” consensus sequences. In eukaryotes, Kozak sequences are adenines 3–6 nucleotides upstream of the start codon and are strong positive determinant for translation initiation (15, 23).
When SDs are present, they may have a greater role in tuning initiation rate than in determining start codon selection. It was recently found that in E. coli, the aSD sequence in the 16S rRNA is not the main determinant for start codon selection; rather, the SD-aSD pairing functions as a mechanism to tune initiation efficiency in concert with mRNA folding and transacting factors (26). In this study, E. coli was also observed to use the A-rich sequences upstream of the translation start site, suggesting a universal role for Kozak-like sequences. These findings also support the model of ribosome “standby” sites. These sites have been proposed to function as locations upstream of the start codon where a ribosome can load on the mRNA upstream of an annotated SD. Standby sites could be used to allow for rapid responses to stimuli by allowing ribosome occupancy on an mRNA without dedicating resources to initiation. Once there is a stimulus, the ribosomes can immediately begin protein synthesis. Determinants such as the RNA structure and the presence of adenines with the proper spacing may facilitate the binding and initiation of a ribosome without the requirement of an available SD sequence.
Ribosome assembly and start codon selection
Ribosome levels in the cell can be regulated post-transcriptionally, either through changes during their assembly or by modulating the active pool of ribosomes. Ribosome assembly is a tightly controlled process in which ribosomal proteins and subunits must interact and coordinate in a manner that promotes the assembly of the mature 70S particle (27). Ribosomal protein binding is hierarchical and requires a host of quality control steps to properly assemble. One example of a ribosome assembly quality control factor is the translational GTPase LepA, which facilitates binding of ribosomal proteins to the small subunit of the ribosome (28, 29). Beyond ribosomal protein binding, LepA also has a role in the maturation of the 16S rRNA, in which an accumulation of 17S precursor rRNA is found in the absence of LepA. Another assembly factor, BipA, has been shown to bind to the stress response molecule ppGpp and potentially regulate ribosome assembly under stress conditions (30). These functions demonstrate the potential roles for LepA, BipA, and similar translational GTPases in controlling ribosome maturation and, in effect, the pool of active ribosomes in the cell.
Ribosomal subunit joining is also a critical step in translation initiation. Initiation factors coordinate the timing and location of initiation (Fig. 1B) (31). Initiation factor 1 (IF1) aids in initiator tRNAfMet selection within the 70S ribosome and shows both association and disassociation activity during initiation, along with other functions (32, 33). Initiation factor 2 (IF2) ensures selection of fMet-tRNAfMet to begin translation. Initiation factor 3 (IF3) serves in start codon selection by promoting dissociation of the ribosomal subunits if assembly is occurring at a noncanonical start codon and facilitating tRNAfMet interactions with the proper start codon.
Recently, evidence has been presented that after termination, the 70S ribosome is stimulated by IF1, IF3, and tRNAfMet to scan for the next start codon without the requirement of disassembly and reassembly (33). This mechanism of scanning may be used both to mitigate the energy requirements of de novo initiation and to sense the available pools of charged tRNAfMet.
Ribosome scanning allows for an intricate mechanism for translation reinitiation in which the stop codon of the upstream gene and the start codon of the downstream gene overlap and a potential SD can be found near the 3′-end of the upstream gene. Recently, translational efficiency of the upstream gene was observed to control the rate of reinitiation in polycistronic transcripts (24). Bioinformatics analyses have shown that ∼30% of genes in prokaryotes are structured in overlapping codirectional pairs in which the stop codon of the upstream gene and the start codon of the downstream gene overlap.
Overlapping gene structure is a potential regulatory point for bacterial cells; if the upstream gene does not get fully translated, translation of the downstream gene is inhibited. This would be an intriguing mechanism for maintaining stoichiometry in proteins found in operons. The efficiency of reinitiation is also dependent on the strength of the upstream SD, but only if the organism uses an SD in its leading genes. Bacteriodetes, for example, have a low propensity to use SD in their leading or monocistronic genes; therefore they have fewer reinitiating genes with an SD at the 3′-end of the leading gene. This mechanism of regulation of gene expression through translational coupling is conserved in bacteria and archaea, although there is evidence that the determinants for coupling have coevolved with SD usage between individual phyla (34).
mRNA folding
One of the most important properties of RNA is its ability to anneal to itself and other RNAs through base-pairing, forming secondary structures such as stem-loops, hairpins, and pseudoknots. RNA folding generally impacts translation at the initiation step by modulating the accessibility of the ribosome-binding site or start codon (35–38). Control of gene expression via mRNA folding allows the cell to rapidly adapt to chemical and physical stimuli without synthesizing or degrading mRNA. RNA folding can occur in response to various stimuli, including temperature, pH, small metabolites, and macromolecules such as tRNA (36, 39). For example, thermosensing mRNA can regulate cell adaptation upon entry into a host, controlling the expression of virulence determinants through changes in RNA folding caused by temperature shifts (1). A wide range of genes are controlled by differential mRNA folding and sequestration of the ribosome-binding site, including those involved in temperature adaptation, metabolite biosynthesis, and regulation of charged tRNA pools (1, 17, 40).
The synthesis of some proteins is controlled by the binding of ligands such as nucleotides, amino acids, or enzymatic cofactors to cis-acting RNAs called riboswitches (39, 41). Riboswitches bind to ligands through an aptamer domain, a strongly conserved nucleotide sequence found in the RNA. The aptamer domain allows flexibility of the surrounding nucleotide se-quence, permitting a variety of RNA folding responses to take place in response to aptamer-ligand binding. Riboswitch-controlling ligands are not limited to small molecules; for example, the 5′-UTR for many ribosomal proteins mimic rRNA structure and bind their own products (35). ThrRS, the threonine tRNA synthetase, is controlled in a similar fashion, in which the 5′-end of its mRNA takes the shape of tRNAThr (35). By mimicking tRNAThr, bacteria can use thrS to sense cellular concentrations of ThrRS and tune translation initiation accordingly. When ThrRS levels are high, the probability for ThrRS to bind thrS mRNA increases, which then prevents ribosome binding to thrS and decreases further ThrRS production. Inversely, if ThrRS concentrations in the cell are low, the ability for ribosomes to initiate translation of thrS increases, restoring ThrRS levels. In some instances, the ligand for riboswitches is another RNA such as tRNA. “T-box” riboswitches are able to bind the anticodon sequence of a tRNA and detect whether the tRNA is charged with an amino acid through recognition of its acceptor stem (39). Generally, T-box riboswitches control transcription of genes through transcription terminator/antiterminator regulation, but more recent work has shown that in Actinobacteria, this class of riboswitch can function through the sequestration of a ribosome-binding site (36).
An example of translational regulation of gene expression through thermosensing RNAs has been recently described in the cold shock response in E. coli (1). It was initially observed that when E. coli experiences a temperature shift from 37 °C (normal growth temperature) to 10 °C (cold shock), there is an acclimation phase in which cell growth and protein synthesis are stopped. After this acclimation phase, the cells begin to grow at a much slower rate. By using ribosome profiling to determine ribosome occupancy on mRNAs and dimethyl sulfate sequencing to probe mRNA structure, global increases in mRNA folding were found to correlate with a decrease in translation efficiency. Intriguingly, mRNAs encoding cold shock proteins make specific structures that permit maintenance of their translation at low temperatures. These proteins function to decrease total mRNA structure in the cell during the acclimation phase of the cold shock response. Once total cellular mRNA structure is decreased, the acclimation phase ends, and cells begin to grow.
Ribosome multiplicity
The majority of the cell's resources are allocated toward the synthesis of rRNA during exponential growth (42). In early studies, it was found that the relative number of ribosomes in a cell is directly proportional to the growth rate and that rRNA synthesis is controlled through a feedback-inhibition mechanism (42). Ribosomes can be synthesized at such high quantities because of the multiplicity of rRNA operons (rrns). These operons can vary in number, from 14 in the Clostridioides to only 1 in Mycobacterium tuberculosis. This multiplicity was found to be essential for E. coli (7 rrns) to adapt to changes in environmental conditions (43, 44). Under nonstressed conditions, up to 3 rrns can be deleted without a substantial growth defect because transcription initiation and elongation of the other operons increases to counteract the loss of rrns (42). When there are fewer than 4 rrns, the transcription machinery is not able to compensate for the depleted ribosome pools, which decreases the probability of translation initiation at genes that are required for adaptation to changing conditions. The correlation between rrn number and adaptive potential has also been observed using computational approaches, in which bacteria that live in more dynamic environments tend to have larger genomes and a greater rrn copy number, whereas organisms that live in more stable environments (such as intracellular parasites) have reduced genomes and subsequently reduced rrns copies (44). The number of rrns is also controlled so that the copy number is not cumbersome to the cell, because increasing the copy number of rrns can lead to growth defects through the allotment of limited cellular resources for rRNA synthesis (44).
Elongation
During translation elongation, the ribosome catalyzes peptide bond formation between amino acids. Amino acids are ligated onto tRNAs by aminoacyl tRNA synthetases and delivered to the ribosome by the translation elongation factor EF-Tu. Once peptide bond formation is complete, the ribosome is translocated by EF-G. In E. coli, translation elongation occurs at ∼20 amino acids/s under optimal conditions (45). The intrinsic rate of translation elongation does not vary drastically under many different conditions. Instead, other processes are impacted that can control gene expression during elongation, such as tRNA availability or the pool of active ribosomes in the cell (7, 8, 42, 43). In this section, we describe some of the methods bacterial cells utilize to modulate translation elongation to regulate synthesis of their proteome.
Programmed translational arrest
During translation elongation, specific strings of amino acids can lead to ribosome pausing through various mechanisms (46–51). These strings of amino acids can be used to regulate translation rate over different protein regions such as interdomain linker regions (46, 52).
Programmed translational arrest is probably most well-known as the regulatory mechanism in the Sec pathway. In the Sec pathway, the secretion monitor peptide secM encodes for a peptide (FXXXXWIXXXXXGIRAGP, where X is any amino acid), which induces ribosome stalling. This stalling is alleviated by pulling through the secretory system, allowing elongation to resume (49, 53, 54). If concentrations of secretory factors are low or if secretion of SecM is defective for other reasons, ribosome pausing at the leader region is pervasive. Ribosome stalling in the secM gene is caused by interactions between the nascent SecM peptide and the ribosome exit tunnel, specifically with the 23S rRNA and ribosomal protein L22, preventing proper ribosome translocation (54). Ribosome pausing at this sequence makes the SD for the gene encoding the downstream translocase, secA, more accessible for translation initiation. By monitoring translocation of SecM peptide, cells can achieve proper stoichiometry between their secretory proteins. If the SecM stall sequence is mutated to prevent ribosome stalling, the resulting depletion of SecA is lethal in E. coli (48, 53). Ribosomal mutants that lose the ability to interact with SecM within the exit tunnel also show defects associated with loss of SecA homeostasis.
Beyond the SecM stall peptide sequence, other specific motifs in nascent peptides can cause ribosome pausing (48, 49). For example, when a ribosome reaches a stretch of two or more prolines in a row (a polyproline (PPX) motif), slowed peptide bond formation can cause the ribosome to pause (55–57). Ribosome pausing at PPX motifs occurs because proline is the slowest of all canonical amino acids in peptide bond formation, being both a poor peptide bond donor and acceptor because of its unique pyrrolidyl ring structure (58). When a ribosome is paused at a PPX motif, the A- and P-site tRNAs are unstable, further decreasing the rate of translation elongation. Pausing at PPX motifs leads to decreased protein synthesis of PPX-containing proteins and causes ribosome queuing on PPX-encoding transcripts, which can lead to defects in cell physiology (56, 59, 60).
The translation elongation factor EF-P alleviates ribosome pausing at PPX motifs by facilitating peptide bond formation between proline residues (Fig. 2A) (61). EF-P enters the ribosome from the E-site and interacts with the P-site tRNA between the P- and E-sites, stabilizing the A- and P-site tRNAs and increasing the protein synthesis rate (61). EF-P also displays high specificity for Pro-tRNAPro. The major determinant for tRNAPro recognition is the unique D-arm that is shared between tRNAPro and tRNAfMet, and structural experiments demonstrate that EF-P is in direct contact with the D-arm of the P-site tRNAPro within the ribosome, suggesting that EF-P function is directly linked to polyproline translation and is a sensitive target for regulation via these stalling motifs (61, 62).
Figure 2.
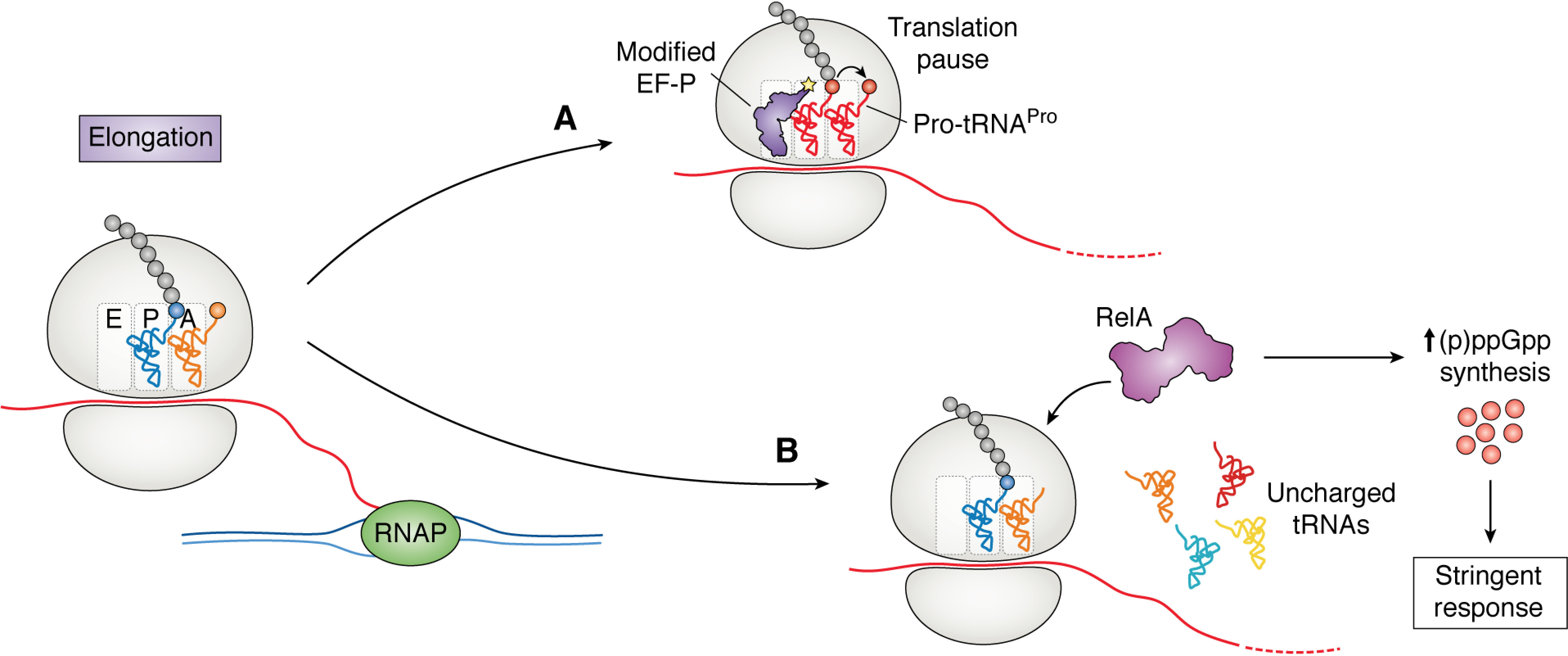
Regulation of gene expression at the level of translation elongation. A, translation pausing can be induced by the slow peptide bond formation between proline residues (red). These pauses are relieved by the translational elongation factor EF-P, which facilitates proline–proline bond formation. B, when uncharged tRNA enters the A-site of the ribosome under starvation conditions, it is recognized by RelA. RelA synthesizes (p)ppGpp, inducing the stringent response. The stringent response stimulates widespread regulatory functions in the cell, including regulation translation through the control of ribosome biosynthesis.
One potential mechanism for regulating translation elongation at PPX motifs is through EF-P post-translational modification (PTM). In most cases, EF-P must be post-translationally modified for full function as modification aids in both stabilizing the P-site tRNA and increasing the dwell time of EF-P within the ribosome (63). Until recently, EF-P function was believed to be dependent on its PTM in all bacteria (57, 64, 65). It is now known that in some Actinobacteria, EF-P does not require PTM for function. This may be a result of the high number of PPX motifs in Actinobacteria, where there is >1 PPX motif per protein on average (compared with 0.5 in E. coli and 0.4 in B. subtilis) (66). The high polyproline burden present in these organisms may prevent the fine-tuning of translation elongation based upon PPX motifs. Although the PTM is essential for EF-P function in many organisms, the identity of the PTM is varied, ranging from (R)-β-lysine in E. coli to rhamnose in Pseudomonas aeruginosa (64, 67).
Not only are the structures varied, but the pathways for modification also are divergent. β-Lysine is synthesized by the lysine aminomutase EpmB (YjeK), ligated to EF-P using a lysyl-tRNA synthetase paralog, EpmA (PoxA), and hydroxylated by EpmC (YfcM) (67). Rhamnose is synthesized by the Rml pathway in Pseudomonas, Shewanella, and Neisseria and is attached to EF-P using the glycosylase EarP (64, 68). The synthesis and ligation of the post-translational modification in B. subtilis, 5-aminopentanol, is substantially different from that of the other known modifications (65). The attachment of 5-aminopentanol to EF-P occurs modularly; the modification is built upon EF-P in multiple steps (69). It was recently found that structurally distinct modifications had different effects on enhancing peptide bond formation depending on the X residue of a PPX motif (69). For example, although in a Δefp strain a Pro-Pro-Trp motif causes the strongest pause, in some modification mutants Pro-Pro-Pro had the greatest defect in peptide bond formation. These results suggest that there may be instances in which B. subtilis requires EF-P with only a partial modification to translationally control the output of a protein product. Interestingly, in all known cases of EF-P modification, the moiety is derived from a core metabolic pathway (β-lysine from amino acid biosynthesis, rhamnose from sugar metabolism, and 5-aminopentanol from fatty acid biosynthesis), which may be indicative of mechanisms to sense the levels of these key metabolites and regulate translation elongation rates accordingly.
Because translation elongation generally occurs at a constant rate in a WT cell, one way to translationally control gene expression is to modulate the number of ribosomes that can load on a certain transcript, as mentioned in previous sections of this review (18, 21). Ribosome profiling data reveal that two transcripts can have the same PPX motif with similar context within the gene; however, one would require EF-P for optimal protein output, whereas the other would not (70, 71). This was discovered to be an effect of translation initiation, in which a transcript with a strong SD would be more EF-P–dependent than a transcript with a weak SD. One example of this phenomenon is the regulation of the ATP synthase proteins AtpA and AtpD. In Gammaproteobacteria, both proteins have the same motif encoded in a region of the protein, which would imply similar dependences on EF-P for translation (72). Surprisingly, AtpA does not require EF-P for translation, whereas AtpD translation depends on EF-P (18). This is a result of AtpA having a weaker SD than AtpD; therefore translation of AtpA does not induce ribosome queuing in the absence of EF-P. If the dwell time of a ribosome on a PPX motif is shorter than it takes for the next ribosome to initiate and reach the motif, there will be a minimal effect on total protein output. In this scenario both translation initiation and elongation have a role in modulating the synthesis of components of a core biosynthetic protein, the ATP synthase.
Interplay between translation elongation and cell homeostasis
Misregulation of translation has pleotropic consequences for the cell. For example, slowed translation rate leads to higher frequencies of RNA polymerase backtracking, and this results in a decreased frequency of head-on collisions between RNA polymerase and the replication machinery (73). Conflicts between transcription and replication can lead to the formation of DNA:RNA hybrids and cause genomic instability, requiring the function of collision-resolving factors such as Rep, UvrD, and Rho, which aid in preventing the deleterious effects that are associated with collisions between RNA polymerase and the replisome (74–77). It was seen that ribosome pausing caused by decreased charged tRNA pools or the loss of EF-P permitted cell survival in the absence of collision-resolving proteins (73). This study highlighted the intricacies in bacterial physiology, in which many factors must coordinate to allow for rapid translation and growth while resolving potential conflicts between the macromolecular complexes that perform replication and transcription.
Bacterial cells have a generally consistent translation elongation rate under a variety of conditions. Recently, a series of studies have shown that even under conditions such as low nutrients, elongation continued at an appreciable rate (∼8 amino acids/s) (7, 8). Surprisingly, in conditions that were previously considered to decrease translation such as in the presence of translational inhibitors, the elongation rate has been observed to stay constant, whereas the fraction of active ribosomes decrease (7). Control of elongation in starvation conditions is also dependent on the limiting substrate. Under carbon limitation, the number of active ribosomes is decreased because a fraction of the ribosomes in the cell do not initiate translation (78). When cells are starved for phosphorous, ribosome biogenesis is impaired, decreasing the ribosome concentration within the cell. The effect of carbon and phosphate limitation is similar to the effect of rrn multiplicity previously discussed, in which gene expression is not controlled at the level of translation elongation rate; rather, the number of actively translating ribosomes changes. When the cells are limited for nitrogen, it was found that the elongation rate is decreased through depletion of cellular pools of aminoacylated tRNAGln, inducing ribosome pausing at glutamine codons. Glutamine is a key molecule for nitrogen storage in bacteria and is used for many other processes that require nitrogen outside of translation. By sensing the levels of charged tRNAGln, the cells can both decrease the energetic burden of translation through reduced elongation and induce stress response pathways such as the stringent response, which will be discussed in the next section. It was also noted that observed elongation rate fluctuations were mainly caused by changes in availability of translational cofactors such as tRNAs or elongation factors.
The stringent response
A critical mechanism that bacterial cells utilize to adapt to changing environmental conditions is through the synthesis of the alarmone (p)ppGpp and the subsequent stringent response (79). Synthesis of ppGpp can occur as a response to a variety of stimuli, including ribosome pausing during translation elongation (79–81). When cells are in conditions with low amino acid availability, the fraction of uncharged tRNA increases, in turn raising the probability of uncharged tRNA entering the ribosomal A-site. When this occurs, the ppGpp synthase RelA binds the ribosome and begins to synthesize ppGpp, which binds and regulates the action of many divergent cellular processes, the most well-studied of which is directing transcription through binding to RNA polymerase (Fig. 2B). Because this regulation is specific to uncharged tRNA binding to the ribosome, mischarging of tRNA with noncognate amino acid prevents the stringent response in conditions of amino acid starvation (40).
Recent studies have shown that ppGpp has many binding targets. By capturing proteins using a cross-linkable ppGpp analog, it was observed that ppGpp has the potential to bind factors that have major roles in all aspects of cellular physiology (73, 82–84). Binding of ppGpp can regulate translation initiation, elongation, and termination, specifically at the levels of ribosome biogenesis and translational GTPase activity (30, 82, 85–87). Studies have shown that of the translation initiation factors, IF2 is specifically targeted by ppGpp-dependent regulation (86, 88). Control of cellular ribosome concentrations by ppGpp has also been observed directly by modulating the synthesis and degradation of ppGpp in E. coli (42, 79). When ppGpp levels are high, growth is suppressed by decreasing ribosome biosynthesis, potentially through binding and inhibiting ribosome assembly GTPases (84). When ppGpp concentrations are low, cell growth is also suppressed, but it is postulated that this is because of inappropriate resource allocation toward ribosome synthesis and assembly. By sensing nutrient concentrations through translation elongation, bacteria can regulate the synthesis of ppGpp and properly allocate resources to processes that will increase fitness under stress.
The effects of translation elongation on transcription
One of the major paradigms of bacterial physiology is the spatial and temporal coordination of transcription and translation. When mRNA is transcribed, it has been shown that ribosomes can immediately initiate once an appropriate ribosome-binding site becomes accessible (12, 15). Without a ribosome rapidly initiating translation, RNA polymerase shows a greater propensity to backtrack, which can lead to transcriptional stalling (73, 83). The coupling of transcription and translation was thought to be absolute in bacteria and required for transcription processivity. However, recent work has argued against the requirement of a dedicated coupling mechanism between the ribosome and RNA polymerase during protein synthesis (34, 89). This correlates with data showing that transcription elongation is not inhibited by the absence of a leading ribosome, despite an increased probability to backtrack (89).
Although transcription and translation do not require a dedicated coupling mechanism, the presence of a ribosome does have effects on transcription termination. When E. coli cells are treated with a translational inhibitor or translation prematurely terminates, Rho-dependent transcription termination events increase (73). Rho-dependent termination, known as transcriptional polarity, occurs when a Rho-utilization (rut) site in the mRNA is not protected by a ribosome (90–92). Polarity has classically been associated with the presence of a premature stop codon and the subsequent decrease in translation of downstream genes. Rho-dependent termination has been ob-served in ∼200 loci in the E. coli genome (93). Beyond occurrences of premature translation termination, ribosome pausing at PPX motifs has been seen to decouple transcription and translation, leading to the exposure of rut sites where Rho can bind and terminate transcription (94).
Translational pausing during elongation can also control downstream mRNA folding (Fig. 3). One of the most well-characterized mechanisms of translational control by RNA folding is through an mRNA leader region. Leader regions are sensitive mechanisms to sample the cellular concentrations and rapidly respond to changes in a specific metabolite (17). These regions are found upstream of the gene in which they control and can modulate transcription and translation of the downstream gene. The trp leader is a well-characterized example of translational control of gene expression, in which there are two consecutive tryptophan codons in a leader region (trpL) before the start codon of genes encoding Trp synthesis proteins in a polycistronic mRNA (17). When tryptophan levels in the cell are high, the charged pools of Trp-tRNATrp are sufficient for rapid translation of trpL. Rapid synthesis of the TrpL peptide leads to the formation of an attenuator stem-loop, which is recognized as a termination sequence by the RNA polymerase, leading to transcription termination before tryptophan biosynthesis genes are expressed (95). If the level of charged tRNATrp is low, the ribosome will pause at the Trp codons in trpL, allowing for the formation of a mutually exclusive antiterminator stem-loop, facilitating transcription and translation of the trp operon. This mechanism of control is not exclusive to the trp operon; another example of leader-mediated regulation in Escherichia coli includes translational control of PheS and PheT, the subunits of the phenylalanine tRNA synthetase, which are controlled by a leader region that senses charged tRNAPhe pools (96). By sensing the levels of charged tRNA through ribosome pausing, cells regulate both transcription and translation of core biosynthetic pathways. Further, the Mgt system in Salmonella is regulated by ribosome translocation speed through a proline-encoding stretch and is required for survival in macrophages and the uptake of magnesium (97). Ribosome pausing at mgtP greatly increases transcription of mgtC, a virulence factor, whereas pausing at mgtL up-regulates the magnesium transporter mgtA. Salmonella EF-P is down-regulated when there are depleted levels of Pro-tRNAPro, which is found in the intramacrophage environment. Depletion of EF-P concurrent with depleted Pro-tRNAPro levels causes ribosome pausing at the PPP motif found in mgtP, enhancing mgtC transcription in a similar fashion as the previously discussed trp operon (17). Unlike the sec operon, which utilizes the function of the Sec protein products to test for their concentrations within the cell, the trp operon and other leader region-containing operons use translation to directly assess the availability of a metabolite and regulate biosynthesis genes accordingly.
Figure 3.
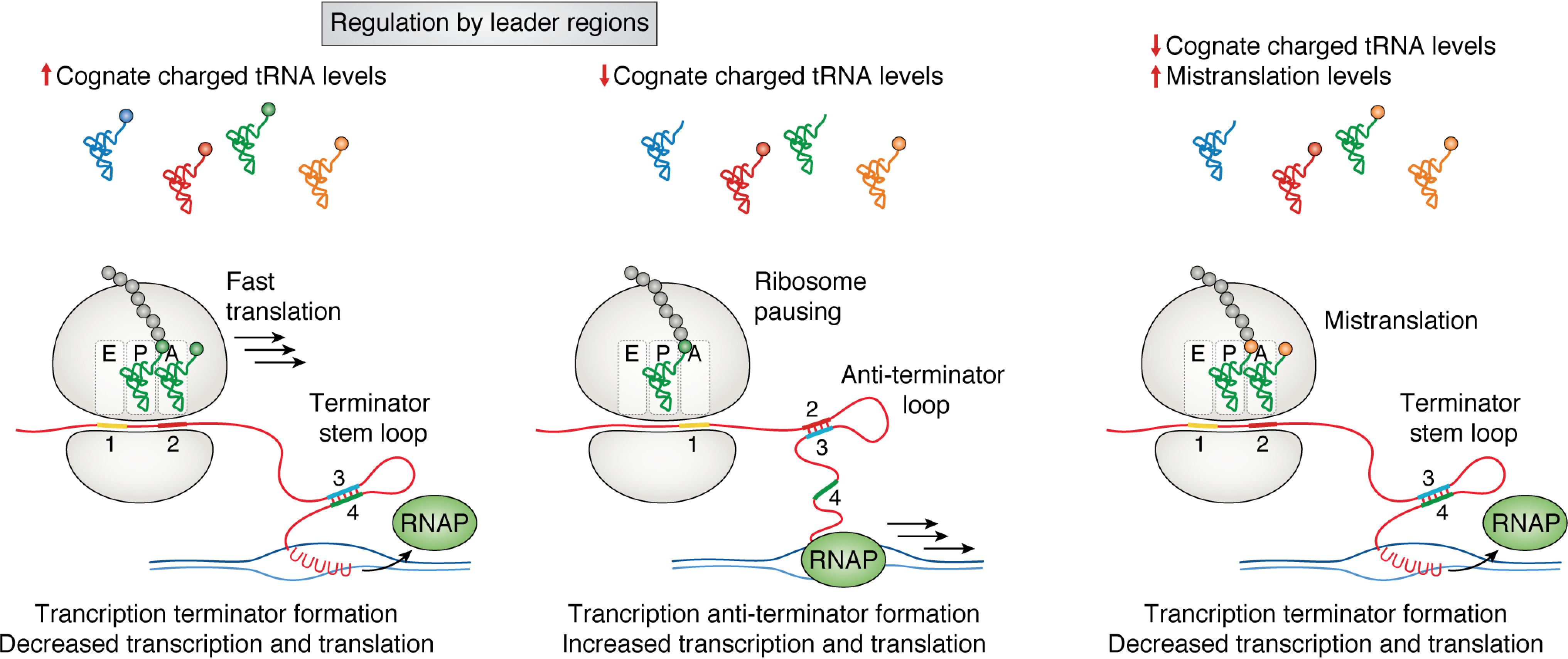
Translational regulation by leader regions. Core biosynthetic pathways can contain mRNA leader regions that sample charged tRNA pools to control gene expression. Left panel, when charged cognate tRNA levels are high, rapid translation of the leader peptide leads to the formation of a terminator stem-loop. This stem-loop leads to transcription termination. Middle panel, when the levels of charged tRNA are low, ribosome pausing leads to the formation of a mutually exclusive antiterminator loop that facilitates transcription of the downstream gene. Right panel, in the event of mistranslation, mischarged tRNA can lead to premature termination of transcription when cognate amino acid levels are low.
Termination and frameshifting
Translation termination in bacteria is performed by three translation release factors: RF1, RF2, and RF3 (31, 98). RF1 recognizes UGA/UAA codons through its PXT motif, whereas RF2 recognizes UAG/UAA stop codons through its SPF motif (99). Both RF1 and RF2 have a conserved GGQ motif that is essential for peptide release through hydrolysis of the P-site tRNA ester linkage (99). RF3 aids in dissociation of the release factors from the ribosome and begins the process of ribosome recycling through conformational changes induced by GTP hydrolysis (100). Without functional release factors, ribosomes stall at stop codons, and the active concentration of ribosomes in the cell decreases (101). Translational regulation of gene expression at the point of termination generally occurs in the form of frameshifting and stop codon readthrough (102). By regulating the translational reading frame, bacterial cells can maintain proper protein stoichiometries through the use of premature stop codons and control the amino acid sequence of protein products to increase proteome diversity (102–104).
Quality control during termination
When ribosomes are unable to terminate, such as during translation of a nonstop mRNA, they are rescued by transfer-mRNA (tmRNA). Ribosome release from stalling by tmRNA is mediated by trans-translation, in which tmRNA functions as a tRNA mimic and is brought to the ribosome by SmpB. The ribosome then uses tmRNA both as a tRNA and as a new reading frame that encodes a degradation tag for the nascent peptide (105). This functions as a method to both replenish ribosome pools and remove potentially aggregate-prone mistranslation products (106–108). Although this is an efficient mechanism to rescue ribosomes that are unable to terminate, further studies based upon release factor mutants have given insight into the importance of rapid peptide release in bacterial cells. In a recent ribosome profiling study, it was observed that if termination was impaired, some operons that contained leader regions became misregulated. This is likely due to ribosome loading on the mRNA without termination leading to formation of attenuator mRNA secondary structures and premature transcription termination (Fig. 4B). Because many important biosynthesis genes contain leader regions for their regulation, this is one mechanism in which translation termination is critical for the maintenance of cellular homeostasis.
Figure 4.
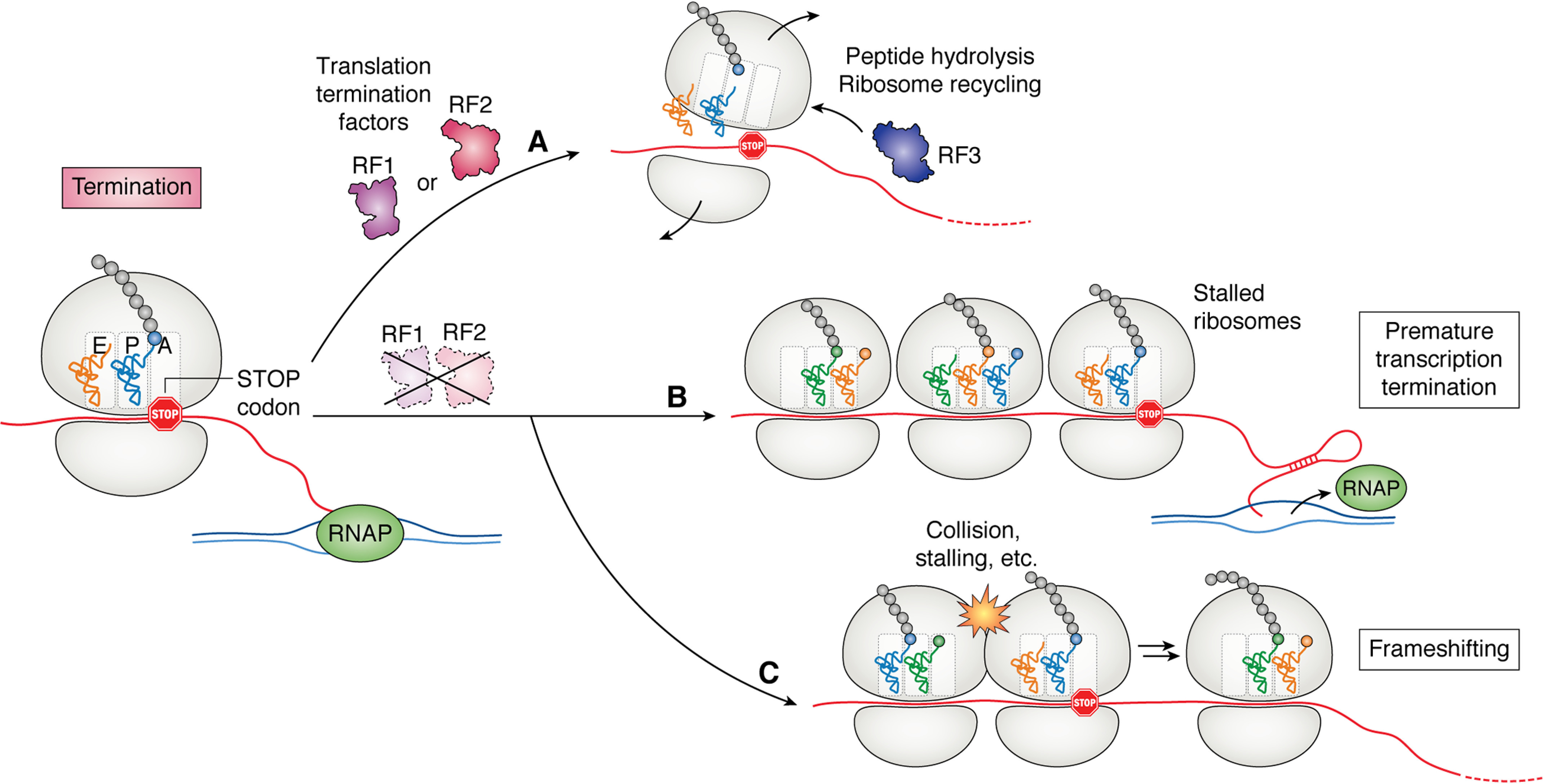
Translation termination factors RF1 and RF2 are required for termination of translation in bacteria. A, when a ribosome reaches a stop codon in the presence of RF1 and RF2, these factors aid in peptide hydrolysis and ribosome recycling. RF1 releases ribosomes at UAG and UAA codons, whereas RF2 releases ribosomes at UGA and UAA codons. RF3 facilitates ribosome recycling and the release of RF1 or RF2 from the ribosome. B and C, in the absence of these factors, stalled ribosomes at stop codons can lead to premature transcription termination (B) or frameshifting events (C).
Translation termination is also a critical step for protein quality control. For example, if a mismatched tRNA is incorporated to the P-site of the ribosome, RF2 hydrolyzes the peptide with the aid of RF3 (100, 109). This mechanism of quality control has been observed both in vitro and in vivo and prevents the accumulation of potentially deleterious peptides (106, 107). When RF3 is deleted, cells display decreased fitness that is associated with greater translational error rates. Translational fidelity mediated by RF3 was also observed to control protein levels and mRNA stability. In the absence of translational proofreading, the cells may be insensitive to mistranslation-induced changes in amino acid pools, which would allow for readthrough of sensor sequences. This also impacts mRNA stability because of a greater ribosome occupancy on mistranslated mRNAs without RF3.
Proteome regulation through frameshifting
In the absence of timely ribosome release or rescue, ribosomes can spontaneously change reading frame and alter the resulting protein product (Fig. 4C). A well-studied programmed frameshifting event regulates levels of RF2 in bacteria, where there is a programmed frameshift in the RF2 gene (prfB) (103). The in-frame coded sequence of RF2 contains a premature stop codon that produces a truncated protein product that is not functional in translation termination. When RF2 levels are low, however, ribosomes queue at the stop codon without release. This can then lead to a frameshift in which the complete RF2 protein is made. This mechanism effectively senses the levels of RF2 in the cell through its mechanism of action and utilizes a programmed frameshift to regulate expression of the prfB gene.
Frameshifting has also been observed in the regulation of iron uptake and other virulence determinants. In meningococci, frameshifting occurs in iron uptake genes and depends on a stretch of guanines that suppress downstream nonsense codons (104). It has been postulated that both translational frameshifting and the propensity of the DNA replication errors at these sequences aid the cells in antigenic variation and evasion of the host immune system. In M. tuberculosis, some rifampicin-resistant strains contain an insertion mutation in rpoB, the β subunit of RNA polymerase (110). This insertion is suppressed by a subsequent translational +1 frameshift that leads to a 3-amino acid change in RpoB, resulting in high levels of rifampicin resistance. By programming frameshifts to make variable protein products from the same mRNA, bacterial cells can add an element of heterogeneity to the proteome and potentially increase fitness under stress conditions (104, 111).
There are also mechanisms to prevent unwanted frameshifting in bacteria. For example, the CCC codon encoding for proline is considered highly prone to frameshifting because of the 0 and +1 frameshift reading identically when taking account of the wobble base pair. To prevent pervasive frameshifting at these codons, the UGG isoacceptor for proline, which can read all Pro codons, is methylated at position G37 by TrmD (97, 112). Using reporter assays, it was observed that without fully functional TrmD, frameshifting at CCC-C is increased by 8-fold. Not only is the proline codon itself prone to frameshifting, the slow formation rate of Pro-Pro peptide bonds also contributes to frameshifting. The ribosomal protein bL9 is critical for reducing compaction between ribosomes during translational stalling events; decreased ribosome spacing during elongation can block the E-site and prevent release of the E-site tRNA (22, 113). As a result, bL9 is essential when bacterial cells lose EF-P function because of the increased rate of ribosome collisions at PPX motifs (22, 47, 113). The effect of frameshifting also appears to be dependent on the rate of ribosome loading on the transcript (22). If the strength of the ribosome-binding site is changed, the increase in ribosome loading impacts the rate of frameshifting, although the effect depends on the genetic context.
Outlook
Translational control of gene expression is pervasive in bacteria and can occur using a variety of mechanisms including differential mRNA folding or post-translational modification of translation factors. At each step of translation, there are mechanisms for the cellular machinery to sample and respond to internal and environmental stimuli. Translation regulation has also been observed to work in concert with transcriptional control of gene expression to precisely tune protein output through events such as attenuation and acclimatization. In the future, it will be critical to further account for the breadth of stimuli to which a bacterial cell must respond, and the mechanisms that drive the responses. As mentioned in previous sections, translation is an energetically expensive task. It will be important to elucidate how the cell senses its energy levels and regulates translation accordingly. Work is currently ongoing in the field of bacterial persistence linking cellular energy concentration and dormancy; for example, do bacteria use a functional translational machinery as a proxy for energy pool sufficiency with respect to ATP and GTP availability? Another way in which there can be better insight into translational control of gene regulation is to perform studies under conditions in which stress may cause mistranslation. For example, T-box riboswitches have the ability to discriminate between tRNAs based upon their anticodon, but are they sensitive to tRNA charged with an incorrect amino acid? Understanding translational regulation in bacteria is also critical for clinical research. Is there a way to target regulatory functions that help bacteria evade the immune system such as translational frameshifting to prevent the rise of antimicrobial resistance? It is also known that without EF-P, many bacteria become avirulent. Are there ways to target EF-P as a combinatorial therapeutic? By continuing studies into the intricacies of translational regulation, it is possible to gain insight into the regulatory pathways and responses that lead to cellular adaptation and competitiveness.
Acknowledgments
We thank members of the Ibba lab for helpful discussions about the article.
Author contributions—R. T. and M. I. conceptualization; R. T. writing-original draft; R. T. and M. I. writing-review and editing; M. I. funding acquisition.
Funding and additional information—This work was supported by National Institutes of Health Grant GM065183 (to M. I.) and the Ohio State Presidential Fellowship (to R. T.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflict of interest with the contents of this article.
- SD
- Shine–Dalgarno motif
- aSD
- anti-SD
- EF
- elongation factor
- IF
- initiation factor
- PPX
- polyproline
- PTM
- post-translational modification
- tmRNA
- transfer-mRNA.
References
- 1. Zhang Y., Burkhardt D. H., Rouskin S., Li G.-W., Weissman J. S., and Gross C. A. (2018) A stress response that monitors and regulates mRNA structure is central to cold shock adaptation. Mol. Cell 70, 274–286.e7 10.1016/j.molcel.2018.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Riehle M. M., Bennett A. F., Lenski R. E., and Long A. D. (2003) Evolutionary changes in heat-inducible gene expression in lines of Escherichia coli adapted to high temperature. Physiol. Genomics 14, 47–58 10.1152/physiolgenomics.00034.2002 [DOI] [PubMed] [Google Scholar]
- 3. Grossman A. D., Straus D. B., Walter W. A., and Gross C. A. (1987) σ32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes Dev. 1, 179–184 10.1101/gad.1.2.179 [DOI] [PubMed] [Google Scholar]
- 4. Groisman E. A. (2016) Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 70, 103–124 10.1146/annurev-micro-102215-095331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capra E. J., and Laub M. T. (2012) Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347 10.1146/annurev-micro-092611-150039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morgan G. J., Burkhardt D. H., Kelly J. W., and Powers E. T. (2018) Translation efficiency is maintained at elevated temperature in Escherichia coli. J. Biol. Chem. 293, 777–793 10.1074/jbc.RA117.000284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dai X., Zhu M., Warren M., Balakrishnan R., Patsalo V., Okano H., Williamson J. R., Fredrick K., Wang Y.-P., and Hwa T. (2016) Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2, 16231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dai X., Zhu M., Warren M., Balakrishnan R., Okano H., Williamson J. R., Fredrick K., and Hwa T. (2018) Slowdown of translational elongation in Escherichia coli under hyperosmotic stress. MBio 9, e02375–17 10.1128/mBio.02375-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maracci C., and Rodnina M. V. (2016) Review: translational GTPases. Biopolymers 105, 463–475 10.1002/bip.22832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gibbs M. R., and Fredrick K. (2018) Roles of elusive translational GTPases come to light and inform on the process of ribosome biogenesis in bacteria. Mol. Microbiol. 107, 445–454 10.1111/mmi.13895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ling J., Reynolds N., and Ibba M. (2009) Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63, 61–78 10.1146/annurev.micro.091208.073210 [DOI] [PubMed] [Google Scholar]
- 12. Laursen B. S., Sørensen H. P., Mortensen K. K., and Sperling-Petersen H. U. (2005) Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69, 101–123 10.1128/MMBR.69.1.101-123.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oppenheim D. S., and Yanofsky C. (1980) Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics 95, 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma J., Campbell A., and Karlin S. (2002) Correlations between Shine–Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184, 5733–5745 10.1128/jb.184.20.5733-5745.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234, 187–208 10.1016/S0378-1119(99)00210-3 [DOI] [PubMed] [Google Scholar]
- 16. Nakagawa S., Niimura Y., Miura K., and Gojobori T. (2010) Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 107, 6382–6387 10.1073/pnas.1002036107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee F., and Yanofsky C. (1977) Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc. Natl. Acad. Sci. U. S. A 74, 4365–4369 10.1073/pnas.74.10.4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hersch S. J., Elgamal S., Katz A., Ibba M., and Navarre W. W. (2014) Translation initiation rate determines the impact of ribosome stalling on bacterial protein synthesis. J. Biol. Chem. 289, 28160–28171 10.1074/jbc.M114.593277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vind J., Sørensen M. A., Rasmussen M. D., and Pedersen S. (1993) Synthesis of proteins in Escherichia coli is limited by the concentration of free ribosomes: expression from reporter genes does not always reflect functional mRNA levels. J. Mol. Biol. 231, 678–688 10.1006/jmbi.1993.1319 [DOI] [PubMed] [Google Scholar]
- 20. Chevance F. F. V., Le Guyon S., and Hughes K. T. (2014) The effects of codon context on in vivo translation speed. PLoS Genet. 10, e1004392 10.1371/journal.pgen.1004392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cortes T., Schubert O. T., Rose G., Arnvig K. B., Comas I., Aebersold R., and Young D. B. (2013) Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 5, 1121–1131 10.1016/j.celrep.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith A. M., Costello M. S., Kettring A. H., Wingo R. J., and Moore S. D. (2019) Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs. Proc. Natl. Acad. Sci. U.S.A. 116, 21769–21779 10.1073/pnas.1910613116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baez W. D., Roy B., McNutt Z. A., Shatoff E. A., Chen S., Bundschuh R., and Fredrick K. (2019) Global analysis of protein synthesis in Flavobacterium johnsoniae reveals the use of Kozak-like sequences in diverse bacteria. Nucleic Acids Res. 47, 10477–10488 10.1093/nar/gkz855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huber M., Faure G., Laass S., Kolbe E., Seitz K., Wehrheim C., Wolf Y. I., Koonin E. V., and Soppa J. (2019) Translational coupling via termination–reinitiation in archaea and bacteria. Nat. Commun. 10, 4066 10.1038/s41467-019-12040-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakagawa S., Niimura Y., and Gojobori T. (2017) Comparative genomic analysis of translation initiation mechanisms for genes lacking the Shine–Dalgarno sequence in prokaryotes. Nucleic Acids Res. 45, 3922–3931 10.1093/nar/gkx124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saito K., Green R., and Buskirk A. R. (2020) Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing. Elife 9, e55002 10.7554/eLife.55002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pestova T. V., Kolupaeva V. G., Lomakin I. B., Pilipenko E. V., Shatsky I. N., Agol V. I., and Hellen C. U. (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U. S.A. 98, 7029–7036 10.1073/pnas.111145798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balakrishnan R., Oman K., Shoji S., Bundschuh R., and Fredrick K. (2014) The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 42, 13370–13383 10.1093/nar/gku1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gibbs M. R., Moon K.-M., Chen M., Balakrishnan R., Foster L. J., and Fredrick K. (2017) Conserved GTPase LepA (elongation factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome. Proc. Natl. Acad. Sci. U. S. A 114, 980–985 10.1073/pnas.1613665114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fan H., Hahm J., Diggs S., Perry J. J. P., and Blaha G. (2015) Structural and functional analysis of BipA, a regulator of virulence in enteropathogenic Escherichia coli. J. Biol. Chem. 290, 20856–20864 10.1074/jbc.M115.659136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodnina M. V. (2018) Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 10, a032664 10.1101/cshperspect.a032664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cummings H. S., and Hershey J. W. B. (1994) Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J. Bacteriol. 176, 198–205 10.1128/jb.176.1.198-205.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamamoto H., Wittek D., Gupta R., Qin B., Ueda T., Krause R., Yamamoto K., Albrecht R., Pech M., and Nierhaus K. H. (2016) 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc. Natl. Acad. Sci. U. S. A 113, E1180–E1189 10.1073/pnas.1524554113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen M., and Fredrick K. (2018) Measures of single-versus multiple-round translation argue against a mechanism to ensure coupling of transcription and translation. Proc. Natl. Acad. Sci. U.S.A. 115, 10774–10779 10.1073/pnas.1812940115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meyer M. M. (2017) The role of mRNA structure in bacterial translational regulation. Wiley Interdiscip. Rev. RNA 8, e1370 10.1002/wrna.1370 [DOI] [PubMed] [Google Scholar]
- 36. Sherwood A. V., and Henkin T. M. (2016) Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374 10.1146/annurev-micro-091014-104306 [DOI] [PubMed] [Google Scholar]
- 37. Breaker R. R. (2018) Riboswitches and translation control. Cold Spring Harb. Perspect. Biol 10, a032797 10.1101/cshperspect.a032797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winkler W. C., and Breaker R. R. (2005) Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59, 487–517 10.1146/annurev.micro.59.030804.121336 [DOI] [PubMed] [Google Scholar]
- 39. Williams-Wagner R. N., Grundy F. J., Raina M., Ibba M., and Henkin T. M. (2015) The Bacillus subtilis tyrZ gene encodes a highly selective tyrosyl tRNA synthetase and is regulated by a MarR regulator and T box riboswitch. J. Bacteriol. 197, 1624–1631 10.1128/JB.00008-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bullwinkle T. J., and Ibba M. (2016) Translation quality control is critical for bacterial responses to amino acid stress. Proc. Natl. Acad. Sci. U.S.A. 113, 2252–2257 10.1073/pnas.1525206113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang L., Serganov A., and Patel D. J. (2010) Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol. Cell 40, 774–786 10.1016/j.molcel.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Condon C., French S., Squires C., and Squires C. L. (1993) Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 12, 4305–4315 10.1002/j.1460-2075.1993.tb06115.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Condon C., Liveris D., Squires C., Schwartz I., and Squires C. L. (1995) rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177, 4152–4156 10.1128/jb.177.14.4152-4156.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gyorfy Z., Draskovits G., Vernyik V., Blattner F. F., Gaal T., and Posfai G. (2015) Engineered ribosomal RNA operon copy-number variants of E. coli reveal the evolutionary trade-offs shaping rRNA operon number. Nucleic Acids Res. 43, 1783–1794 10.1093/nar/gkv040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu M., Dai X., and Wang Y.-P. (2016) Real time determination of bacterial in vivo ribosome translation elongation speed based on LacZα complementation system. Nucleic Acids Res 44, e155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vasquez K. A., Hatridge T. A., Curtis N. C., and Contreras L. (2016) Slowing translation between protein domains by increasing affinity between mRNAs and the ribosomal anti–Shine–Dalgarno sequence improves solubility. ACS Synth. Biol. 5, 133–145 10.1021/acssynbio.5b00193 [DOI] [PubMed] [Google Scholar]
- 47. Woolstenhulme C. J., Guydosh N. R., Green R., and Buskirk A. R. (2015) High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 11, 13–21 10.1016/j.celrep.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Woolstenhulme C. J., Parajuli S., Healey D. W., Valverde D. P., Petersen E. N., Starosta A. L., Guydosh N. R., Johnson W. E., Wilson D. N., and Buskirk A. R. (2013) Nascent peptides that block protein synthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, E878–E887 10.1073/pnas.1219536110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tanner D. R., Cariello D. A., Woolstenhulme C. J., Broadbent M. A., and Buskirk A. R. (2009) Genetic identification of nascent peptides that induce ribosome stalling. J. Biol. Chem. 284, 34809–34818 10.1074/jbc.M109.039040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsai C.-J., Sauna Z. E., Kimchi-Sarfaty C., Ambudkar S. V., Gottesman M. M., and Nussinov R. (2008) Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 383, 281–291 10.1016/j.jmb.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alejo J. L., and Blanchard S. C. (2017) Miscoding-induced stalling of substrate translocation on the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 114, E8603–E8610 10.1073/pnas.1707539114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Qi F., Motz M., Jung K., Lassak J., and Frishman D. (2018) Evolutionary analysis of polyproline motifs in Escherichia coli reveals their regulatory role in translation. PLoS Comput. Biol. 14, e1005987 10.1371/journal.pcbi.1005987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murakami A., Nakatogawa H., and Ito K. (2004) Translation arrest of SecM is essential for the basal and regulated expression of SecA. Proc. Natl. Acad. Sci. U.S.A. 101, 12330–12335 10.1073/pnas.0404907101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Butkus M. E., Prundeanu L. B., and Oliver D. B. (2003) Translocon “pulling” of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. J. Bacteriol. 185, 6719–6722 10.1128/jb.185.22.6719-6722.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bullwinkle T. J., Zou S. B., Rajkovic A., Hersch S. J., Elgamal S., Robinson N., Smil D., Bolshan Y., Navarre W. W., and Ibba M. (2013) (R)-β-Lysine–modified elongation factor P functions in translation elongation. J. Biol. Chem. 288, 4416–4423 10.1074/jbc.M112.438879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ude S., Lassak J., Starosta A. L., Kraxenberger T., Wilson D. N., and Jung K. (2013) Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339, 82–85 10.1126/science.1228985 [DOI] [PubMed] [Google Scholar]
- 57. Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., and Rodnina M. V. (2013) EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 10.1126/science.1229017 [DOI] [PubMed] [Google Scholar]
- 58. Pavlov M. Y., Watts R. E., Tan Z., Cornish V. W., Ehrenberg M., and Forster A. C. (2009) Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. U.S.A. 106, 50–54 10.1073/pnas.0809211106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hersch S. J., Wang M., Zou S. B., Moon K.-M., Foster L. J., Ibba M., and Navarre W. W. (2013) Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. MBio 4, e00180–13 10.1128/mBio.00180-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tollerson R. 2nd, Witzky A., and Ibba M. (2018) Elongation factor P is required to maintain proteome homeostasis at high growth rate. Proc. Natl. Acad. Sci. U.S.A. 115, 11072–11077 10.1073/pnas.1812025115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huter P., Arenz S., Bock L. V., Graf M., Frister J. O., Heuer A., Peil L., Starosta A. L., Wohlgemuth I., Peske F., Nováček J., Berninghausen O., Grubmüller H., Tenson T., Beckmann R., et al. (2017) Structural basis for polyproline-mediated ribosome stalling and rescue by the translation elongation factor EF-P. Mol. Cell 68, 515–527.e6 10.1016/j.molcel.2017.10.014 [DOI] [PubMed] [Google Scholar]
- 62. Katoh T., Wohlgemuth I., Nagano M., Rodnina M. V., and Suga H. (2016) Essential structural elements in tRNA Pro for EF-P–mediated alleviation of translation stalling. Nat. Commun. 7, 11657 10.1038/ncomms11657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mohapatra S., Choi H., Ge X., Sanyal S., and Weisshaar J. C. (2017) Spatial distribution and ribosome-binding dynamics of EF-P in live Escherichia coli. MBio 8, e00300–17 10.1128/mBio.00300-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lassak J., Keilhauer E. C., Fürst M., Wuichet K., Gödeke J., Starosta A. L., Chen J.-M., Søgaard-Andersen L., Rohr J., Wilson D. N., Häussler S., Mann M., and Jung K. (2015) Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat. Chem. Biol. 11, 266–270 10.1038/nchembio.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rajkovic A., Hummels K. R., Witzky A., Erickson S., Gafken P. R., Whitelegge J. P., Faull K. F., Kearns D. B., and Ibba M. (2016) Translation control of swarming proficiency in Bacillus subtilis by 5-amino-pentanolylated elongation factor P. J. Biol. Chem. 291, 10976–10985 10.1074/jbc.M115.712091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pinheiro B., Scheidler C. M., Kielkowski P., Schmid M., Forné I., Ye S., Reiling N., Takano E., Imhof A., Sieber S. A., Schneider S., and Jung K. (2020) Structure and function of an elongation factor P subfamily in actinobacteria. Cell Rep. 30, 4332–4342.e5 10.1016/j.celrep.2020.03.009 [DOI] [PubMed] [Google Scholar]
- 67. Roy H., Zou S. B., Bullwinkle T. J., Wolfe B. S., Gilreath M. S., Forsyth C. J., Navarre W. W., and Ibba M. (2011) The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine. Nat. Chem. Biol. 7, 667–669 10.1038/nchembio.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rajkovic A., Erickson S., Witzky A., Branson O. E., Seo J., Gafken P. R., Frietas M. A., Whitelegge J. P., Faull K. F., Navarre W., Darwin A. J., and Ibba M. (2015) Cyclic rhamnosylated elongation factor P establishes antibiotic resistance in Pseudomonas aeruginosa. MBio 6, e00823 10.1128/mBio.00823-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Witzky A., Hummels K. R., Tollerson R. 2nd, Rajkovic A., Jones L. A., Kearns D. B., and Ibba M. (2018) EF-P posttranslational modification has variable impact on polyproline translation in Bacillus subtilis. MBio 9, e00306–18 10.1128/mBio.00306-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Elgamal S., Katz A., Hersch S. J., Newsom D., White P., Navarre W. W., and Ibba M. (2014) EF-P dependent pauses integrate proximal and distal signals during translation. PLoS Genet. 10, e1004553 10.1371/journal.pgen.1004553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mohammad F., Woolstenhulme C. J., Green R., and Buskirk A. R. (2016) Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 14, 686–694 10.1016/j.celrep.2015.12.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peil L., Starosta A. L., Lassak J., Atkinson G. C., Virumäe K., Spitzer M., Tenson T., Jung K., Remme J., and Wilson D. N. (2013) Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc. Natl. Acad. Sci. U.S.A. 110, 15265–15270 10.1073/pnas.1310642110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Myka K. K., Hawkins M., Syeda A. H., Gupta M. K., Meharg C., Dillingham M. S., Savery N. J., Lloyd R. G., and McGlynn P. (2017) Inhibiting translation elongation can aid genome duplication in Escherichia coli. Nucleic Acids Res. 45, 2571–2584 10.1093/nar/gkw1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lang K. S., Hall A. N., Merrikh C. N., Ragheb M., Tabakh H., Pollock A. J., Woodward J. J., Dreifus J. E., and Merrikh H. (2017) Replication–transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell 170, 787–799.e18 10.1016/j.cell.2017.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Aguilera A., and García-Muse T. (2012) R Loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 10.1016/j.molcel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 76. Chen X., Yang J. R., and Zhang J. (2016) Nascent RNA folding mitigates transcription-associated mutagenesis. Genome Res. 26, 50–59 10.1101/gr.195164.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lang K. S., and Merrikh H. (2018) The clash of macromolecular titans: replication–transcription conflicts in bacteria. Annu. Rev. Microbiol. 72, 71–88 10.1146/annurev-micro-090817-062514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li S. H.-J., Li Z., Park J. O., King C. G., Rabinowitz J. D., Wingreen N. S., and Gitai Z. (2018) Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat. Microbiol. 3, 939–947 10.1038/s41564-018-0199-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Traxler M. F., Summers S. M., Nguyen H.-T., Zacharia V. M., Hightower G. A., Smith J. T., and Conway T. (2008) The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148 10.1111/j.1365-2958.2008.06229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Brown A., Fernández I. S., Gordiyenko Y., and Ramakrishnan V. (2016) Ribosome-dependent activation of stringent control. Nature 534, 277–280 10.1038/nature17675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Starosta A. L., Lassak J., Jung K., and Wilson D. N. (2014) The bacterial translation stress response. FEMS Microbiol. Rev. 38, 1172–1201 10.1111/1574-6976.12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang B., Dai P., Ding D., Del Rosario A., Grant R. A., Pentelute B. L., and Laub M. T. (2019) Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150 10.1038/s41589-018-0183-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kamarthapu V., Epshtein V., Benjamin B., Proshkin S., Mironov A., Cashel M., and Nudler E. (2016) ppGpp couples transcription to DNA repair in E. coli. Science 352, 993–996 10.1126/science.aad6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhu M., and Dai X. (2019) Growth suppression by altered (p)ppGpp levels results from non-optimal resource allocation in Escherichia coli. Nucleic Acids Res. 47, 4684–4693 10.1093/nar/gkz211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sanchez-Vazquez P., Dewey C. N., Kitten N., Ross W., and Gourse R. L. (2019) Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 116, 8310–8319 10.1073/pnas.1819682116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vinogradova D. S., Zegarra V., Maksimova E., Nakamoto J. A., Kasatsky P., Paleskava A., Konevega A. L., and Milón P. (2020) How the initiating ribosome copes with ppGpp to translate mRNAs. PLoS Biol. 18, e3000593 10.1371/journal.pbio.3000593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mitkevich V. A., Ermakov A., Kulikova A. A., Tankov S., Shyp V., Soosaar A., Tenson T., Makarov A. A., Ehrenberg M., and Hauryliuk V. (2010) Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J. Mol. Biol. 402, 838–846 10.1016/j.jmb.2010.08.016 [DOI] [PubMed] [Google Scholar]
- 88. Milon P., Tischenko E., Tomsic J., Caserta E., Folkers G., La Teana A., Rodnina M. V., Pon C. L., Boelens R., and Gualerzi C. O. (2006) The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl. Acad. Sci. U.S.A. 103, 13962–13967 10.1073/pnas.0606384103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhu M., Mori M., Hwa T., and Dai X. (2019) Disruption of transcription–translation coordination in Escherichia coli leads to premature transcriptional termination. Nat. Microbiol. 4, 2347–2356 10.1038/s41564-019-0543-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Richardson J. P., Grimley C., and Lowery C. (1975) Transcription termination factor rho activity is altered in Escherichia coli with suA gene mutations. Proc. Natl. Acad. Sci. U.S.A. 72, 1725–1728 10.1073/pnas.72.5.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yarchuk O., Jacques N., Guillerez J., and Dreyfus M. (1992) Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J. Mol. Biol. 226, 581–596 10.1016/0022-2836(92)90617-S [DOI] [PubMed] [Google Scholar]
- 92. Hu K., and Artsimovitch I. (2017) A screen for rfaH suppressors reveals a key role for a connector region of termination factor rho. MBio 8, e00753–17 10.1128/mBio.00753-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Peters J. M., Mooney R. A., Kuan P. F., Rowland J. L., Keles S., and Landick R. (2009) Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 106, 15406–15411 10.1073/pnas.0903846106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Elgamal S., Artsimovitch I., and Ibba M. (2016) Maintenance of transcription–translation coupling by elongation factor P. MBio 7, e01373–16 10.1128/mBio.01373-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kolter R., and Yanofsky C. (1982) Attenuation in amino acid biosynthetic operons. Annu. Rev. Genet. 16, 113–134 10.1146/annurev.ge.16.120182.000553 [DOI] [PubMed] [Google Scholar]
- 96. Ryckelynck M., Giegé R., and Frugier M. (2005) tRNAs and tRNA mimics as cornerstones of aminoacyl-tRNA synthetase regulations. Biochimie 87, 835–845 10.1016/j.biochi.2005.02.014 [DOI] [PubMed] [Google Scholar]
- 97. Gall A. R., Datsenko K. A., Figueroa-Bossi N., Bossi L., Masuda I., Hou Y.-M., and Csonka L. N. (2016) Mg2+ regulates transcription of mgtA in Salmonella typhimurium via translation of proline codons during synthesis of the MgtL peptide. Proc. Natl. Acad. Sci. U.S.A. 113, 15096–15101 10.1073/pnas.1612268113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Burroughs A. M., and Aravind L. (2019) The origin and evolution of release factors: Implications for translation termination, ribosome rescue, and quality control pathways. Int. J. Mol. Sci. 20, 1981 10.3390/ijms20081981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Korostelev A. A. (2011) Structural aspects of translation termination on the ribosome. RNA 17, 1409–1421 10.1261/rna.2733411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zaher H. S., and Green R. (2011) A primary role for release factor 3 in quality control during translation elongation in Escherichia coli. Cell 147, 396–408 10.1016/j.cell.2011.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Florin T., Maracci C., Graf M., Karki P., Klepacki D., Berninghausen O., Beckmann R., Vázquez-Laslop N., Wilson D. N., Rodnina M. V., and Mankin A. S. (2017) An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat. Struct. Mol. Biol. 24, 752–757 10.1038/nsmb.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Baranov P. V., Gesteland R. F., and Atkins J. F. (2002) Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 3, 373–377 10.1093/embo-reports/kvf065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Márquez V., Wilson D. N., Tate W. P., Triana-Alonso F., and Nierhaus K. H. (2004) Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell 118, 45–55 10.1016/j.cell.2004.06.012 [DOI] [PubMed] [Google Scholar]
- 104. Richardson A. R., and Stojiljkovic I. (1999) HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J. Bacteriol. 181, 2067–2074 10.1128/JB.181.7.2067-2074.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Moore S. D., and Sauer R. T. (2007) The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76, 101–124 10.1146/annurev.biochem.75.103004.142733 [DOI] [PubMed] [Google Scholar]
- 106. Choe Y.-J., Park S.-H., Hassemer T., Körner R., Vincenz-Donnelly L., Hayer-Hartl M., and Hartl F. U. (2016) Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531, 191–195 10.1038/nature16973 [DOI] [PubMed] [Google Scholar]
- 107. Ling J., Cho C., Guo L.-T., Aerni H. R., Rinehart J., and Söll D. (2012) Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol. Cell 48, 713–722 10.1016/j.molcel.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Liu Y., Satz J. S., Vo M.-N., Nangle L. A., Schimmel P., and Ackerman S. L. (2014) Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc. Natl. Acad. Sci. U.S.A. 111, 17570–17575 10.1073/pnas.1420196111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Petropoulos A. D., McDonald M. E., Green R., and Zaher H. S. (2014) Distinct roles for release factor 1 and release factor 2 in translational quality control. J. Biol. Chem. 289, 17589–17596 10.1074/jbc.M114.564989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Huseby D. L., Brandis G., Alzrigat L. P., and Hughes D. (2020) Antibiotic resistance by high-level intrinsic suppression of a frameshift mutation in an essential gene. Proc. Natl. Acad. Sci. U.S.A. 117, 3185–3191 10.1073/pnas.1919390117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Fan Y., Evans C. R., Barber K. W., Banerjee K., Weiss K. J., Margolin W., Igoshin O. A., Rinehart J., and Ling J. (2017) Heterogeneity of stop codon readthrough in single bacterial cells and implications for population fitness. Mol. Cell 67, 826–836.e5 10.1016/j.molcel.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gamper H. B., Masuda I., Frenkel-Morgenstern M., and Hou Y. (2015) Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat. Commun. 6, 7226 10.1038/ncomms8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Naganathan A., Wood M. P., and Moore S. D. (2015) The large ribosomal subunit protein L9 enables the growth of EF-P deficient cells and enhances small subunit maturation. PLoS One 10, e0120060 10.1371/journal.pone.0120060 [DOI] [PMC free article] [PubMed] [Google Scholar]


