We compared the ability of 2 commercial molecular amplification assays (RealTime SARS-CoV-2 on the m2000 [abbreviated ACOV; Abbott] and ID Now COVID-19 [abbreviated IDNOW; Abbott]) and a laboratory-developed test (modified CDC 2019-nCoV reverse transcriptase PCR [RT-PCR] assay with RNA extraction by eMag [bioMérieux] and amplification on QuantStudio 6 or ABI 7500 real-time PCR system [abbreviated CDC COV]) to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in upper respiratory tract specimens.
KEYWORDS: COVID-19, RT-PCR, SARS-CoV-2, coronavirus, novel coronavirus
ABSTRACT
We compared the ability of 2 commercial molecular amplification assays (RealTime SARS-CoV-2 on the m2000 [abbreviated ACOV; Abbott] and ID Now COVID-19 [abbreviated IDNOW; Abbott]) and a laboratory-developed test (modified CDC 2019-nCoV reverse transcriptase PCR [RT-PCR] assay with RNA extraction by eMag [bioMérieux] and amplification on QuantStudio 6 or ABI 7500 real-time PCR system [abbreviated CDC COV]) to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in upper respiratory tract specimens. Discrepant results were adjudicated by medical record review. A total of 200 nasopharyngeal swab specimens in viral transport medium (VTM) were collected from symptomatic patients between 27 March and 9 April 2020. Results were concordant for 167 specimens (83.5% overall agreement), including 94 positive and 73 negative specimens. The ACOV assay yielded 33 additional positive results, 25 of which were also positive by the CDC COV assay but not by the IDNOW assay. In a follow-up evaluation, 97 patients for whom a dry nasal swab specimen yielded negative results by IDNOW had a paired nasopharyngeal swab specimen collected in VTM and tested by the ACOV assay; SARS-CoV-2 RNA was detected in 13 (13.4%) of these specimens. Medical record review deemed all discrepant results to be true positives. The IDNOW test was the easiest to perform and provided a result in the shortest time but detected fewer cases of COVID-19. The ACOV assay detected more cases of COVID-19 than the CDC COV or IDNOW assays.
INTRODUCTION
In December 2019, a cluster of patients with pneumonia of unknown origin was linked to exposure to a wet market in Wuhan, Hubei Province, China (1). Very quickly, a novel betacoronavirus was isolated from a lower respiratory tract sample from one patient, and the full genome of the virus was sequenced (2). This novel coronavirus, which was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) for its genetic homology to SARS-CoV, spread rapidly across the globe (3–10). As of 29 April 2020, more than 3 million cases of SARS-CoV-2 infection had been identified worldwide, with over 200,000 deaths; approximately one-third of cases have been identified in the United States.
Laboratory testing plays a critical role in defining the disease characteristics and epidemiology of an emerging infectious pathogen such as SARS-CoV-2 and in controlling its spread. Early on, laboratory testing for SARS-CoV-2 in the United States was performed only at the Centers for Disease Control and Prevention (CDC) laboratories in Atlanta, GA using a reverse transcriptase PCR (RT-PCR) assay that was developed there. Subsequently, the CDC test was to be implemented in all state public health laboratories, but rollout was slow due to technical problems. Following the declaration of a public health emergency, the U.S. Food and Drug Administration (FDA) moved to allow in vitro diagnostic assays under an emergency use authorization (EUA) in an attempt to expedite test development by commercial and clinical laboratories. The majority of assays approved through EUA are nucleic acid amplification tests that target conserved regions of the SARS-CoV-2 genome. Abbott Laboratories received authorization for the RealTime SARS-CoV-2 assay performed on the m2000 (ACOV) on 18 March 2020 (11). The Abbott ID Now COVID-19 (IDNOW) was granted EUA on 27 March 2020. In vitro diagnostic device (IVD) assays with EUA status from commercial manufacturers do not undergo usual FDA review under the de novo request or the 510(k) premarket notification; as such, limited data comparing these assays are available.
In this study, we compared the performance of the ACOV and IDNOW assays and a laboratory developed test that is a modification of the CDC 2019-nCoV assay (CDC COV) for the detection of SARS-CoV-2 RNA from upper respiratory tract specimens.
MATERIALS AND METHODS
Clinical samples.
For the initial evaluation of the three test systems, we collected nasopharyngeal swab specimens in 3-ml M4RT viral transport medium (VTM) (Remel, Lenexa, KS) from symptomatic (fever or cough or shortness of breath) adult and pediatric outpatients, emergency department (ED) patients, and inpatients at Rush University Medical Center or Rush Oak Park Hospital; both hospitals are in metropolitan Chicago, IL. Specimens were collected between 27 March and 9 April 2020 and tested within 72 h of collection. Specimens were held refrigerated at 4°C if all testing could not be completed on the same day. All samples in VTM were heat inactivated at 56°C for 35 ± 5 min prior to testing in order to reduce the risk of accidental transmission of SARS-CoV-2 to laboratory personnel.
In a separate follow-up evaluation, symptomatic patients who had a negative result on a dry nasal swab that was tested at the point of care by the IDNOW assay also had a paired nasopharyngeal swab sample collected and transported to the on-site clinical microbiology laboratory for testing by the ACOV assay.
Age, sex, and location of swab collection were extracted from the electronic medical record (EMR) for all patients. The study was reviewed and given expedited approval by the Rush University Medical Center (RUMC) institutional review board with a waiver of written informed consent.
Modified CDC 2019-nCoV RT-PCR assay.
We validated and implemented a modification of the CDC 2019-nCoV RT-PCR assay (12) for clinical use in our laboratory; this was the first SARS-CoV-2 RT-PCR assay we adopted during the COVID-19 pandemic. The original CDC assay received EUA approval on 4 February 2020. This assay targets two regions of the nucleocapsid (N) gene of the SARS-CoV-2 genome. The human RNase P (RP) gene target is also included and is used as an extraction and amplification control.
Nucleic acids were purified and extracted using the eMag automated nucleic acid sample extraction system (bioMérieux, Marcy l’Etoile, France). Briefly, total nucleic acids were extracted from VTM using an input sample volume of 200 μl into 2,000 μl of easyMag lysis buffer with the specific B protocol to which a final eluted volume of purified nucleic acids was 50 μl. The starting volume of sample used and the final elution volume were modified from the original CDC assay. We utilized the TaqPath 1-step reverse transcriptase quantitative PCR (RT-qPCR) master mix (Life Technologies, Frederick, MD) and the 2019-nCoV CDC EUA kit (Integrated DNA Technologies, Coralville, IA) for target detection. Amplification and real-time detection were performed on the ABI 7500 real-time PCR system (Life Technologies) with software version 2.3 or on the QuantStudio 6 Flex real-time PCR system (Life Technologies) using software version 1.4 (modification from original CDC assay). The total sample volume per reaction was 15 μl of master mix, combined primer/probe mix, and nuclease-free water and 5 μl of eluted sample. Assay run parameters were as described in the original CDC protocol (12). Samples that gave a cycle threshold (CT) value of <40 for both N1 and N2 targets were considered positive. Samples negative for both N1 and N2 targets had to have a positive amplification curve for the RP gene to be considered a valid negative result. Samples that gave a CT value of <40 for either N1 or N2 targets but not both were considered inconclusive and repeat testing was performed per original CDC protocol. If results were still inconclusive after repeat testing, a result of inconclusive was reported.
Abbott Molecular RealTime SARS-CoV-2 assay.
Next, we verified the RealTime SARS-CoV-2 assay (ACOV) (Abbott Molecular, Des Plaines, IL), which is a qualitative real-time assay performed on the Abbott m2000 platform (11). The system includes the m2000sp instrument with automated extraction of nucleic acids using the DNA (total nucleic acid) sample preparation kit in batches of up to 96 samples. The ACOV assay employs dual targets for sequences of the N and RNA-dependent RNA polymerase (RdRP) genes. Both probes utilize the same fluorophore; a second probe for the internal control—added to each specimen at the beginning of sample preparation—is included to assess overall performance, including nucleic acid extraction and possible PCR inhibition. Automated extraction was performed using a sample input volume of 500 μl VTM followed by automated addition of amplification pack reagents and extracts (40-μl volume used for RT-PCR amplification and detection). Two controls (one positive and one negative) provided by the manufacturer were included with each run. Amplification curves were interpreted by the m2000rt system and reported as detected or not detected.
ID Now COVID-19 assay.
The third assay introduced was the IDNOW (Abbott Diagnostics Scarborough, Inc., Scarborough, ME), an isothermal assay based on nicking enzyme amplification reaction technology that targets the RdRP gene (13). Samples can only be tested one at a time. Following an initial 3-min warm-up of the test system, 200 μl of VTM is added to elution buffer in the sample base using the provided transfer pipette and then mixed for 10 s. Using the sample transfer device, 200 μl of sample is transferred into the test cartridge, the lid is closed, and the instrument automatically initializes the assay, which runs for 10 min. The IDNOW does not report CT values to the user. The instrument software interprets amplification data, and final results are reported as positive, negative, or invalid. Samples that yield an initial invalid result are repeated. If an invalid result is generated twice, the final result is reported as invalid.
Estimation of SARS-CoV-2 RNA concentration in nasopharyngeal swab samples.
We tested purified genomic RNA from a reference strain of SARS-CoV-2, isolate USA-WA1/2020 (BEI Resources, Manassas, VA) to generate standard curves for the CDC COV and ACOV assays in order to estimate the concentration of SARS-CoV-2 genome equivalents in nasopharyngeal swab samples. We serially diluted the standard and tested extraction and amplification on both assays independently in triplicate. The IDNOW assay does not provide CT values to the user, so we could not generate a standard curve for this system.
Data analysis.
Because there is no reference standard for SARS-CoV-2 detection by RT-PCR, sensitivity and specificity of the assays could not be determined. Instead, we calculated overall, positive, and negative agreement. Analysis comparing CT values between groups and estimating RNA concentration was performed using Prism 8 (GraphPad, San Diego, CA). Discordant results were adjudicated by medical record review (completed by M. K. Hayden) to assess whether the patients’ clinical courses were consistent with COVID-19 infection. Statistical significance of agreement across tests was determined by contingency table analysis using SPSS v 22 (IBM, Armonk, NY).
RESULTS
Clinical overview.
Specimens from 200 unique patients were included. The first 94 samples tested were collected consecutively. Subsequently, we enriched for positive samples (n = 58) by including samples in which SARS-CoV-2 RNA was detected by ACOV—our standard assay during the study period—and included 48 additional negative samples. Mean patient age was 50 ± 17 years, and 54% of patients were women. Seventy-nine (40%) patients were hospitalized, 29 (36%) of whom were in an intensive care unit; 76 (38%) were evaluated in an ambulatory location, including 55 (72%) who were seen in a COVID-19 screening clinic and 45 (23%) who were seen in an ED.
Assay performance using nasopharyngeal swab samples in VTM.
There were 94 (47%) samples in which SARS-CoV-2 gene sequences were detected by all three assays and 73 (36.5%) samples in which SARS-CoV-2 RNA was not detected by any assay (Table 1). The median cycle threshold (CT) for positive samples by the ACOV assay was 15.34 (interquartile range [IQR], 11.27 to 18.13) or approximately 447 genome equivalents/μl (Fig. 1). Overall agreement among the three assays was 83.5% (95% confidence interval [CI], 77.7% to 88.0%). Two-way positive and negative agreements between results are shown in Table 2. Positive agreement ranged from 75.2% to 100%, with the lowest agreement observed between the ACOV and IDNOW assays. Negative agreement ranged from 92.4% (CDC COV versus ACOV) and 100% (CDC COV versus IDNOW and ACOV versus IDNOW).
TABLE 1.
Detection of SARS-CoV-2 RNA by laboratory-modified CDC COV assay, ACOV assay, and IDNOWa
| No. samples tested (n = 200) | CDC COV | ACOV | IDNOW |
|---|---|---|---|
| 94 | Detected | Detected | Detected |
| 73 | Not detected | Not detected | Not detected |
| 23 | Detected | Detected | Not detected |
| 2 | Detected | Detected | Invalidb |
| 6 | Not detected | Detected | Not detected |
| 2 | Inconclusivec | Detected | Not detected |
Categories with zero samples are not shown.
Invalid defined as a sample that gave neither a positive nor a negative result.
Inconclusive defined as a sample that gave a CT value of <40 for either N1 or N2 targets.
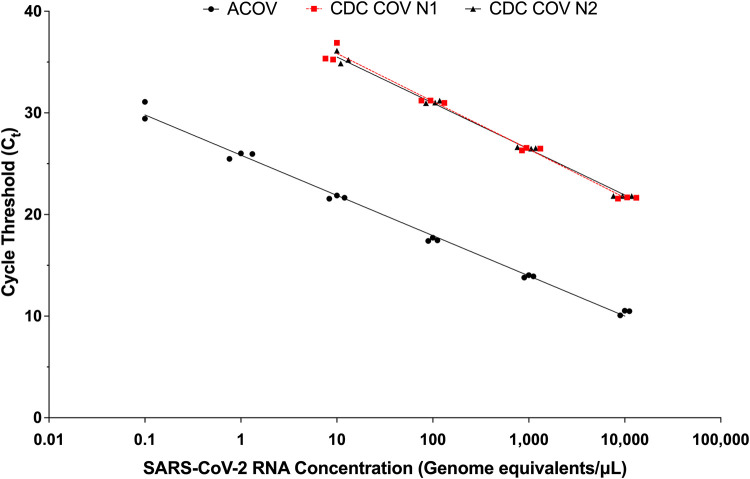
FIG 1.
SARS-CoV-2 standard curves for CDC COV and ACOV assays. Values shown represent results of testing independent replicates in triplicate. Trend line equations: CDC COV assay (N1), y = −2.054ln(x) + 40.585, R2 = 1.0; CDC assay (N2), y = −1.966ln(x) + 40.022, R2 = 0.99; ACOV assay, y = −1.729ln(x) + 25.899, R2 = 0.99.
TABLE 2.
Performance agreement for detection of SARS-CoV-2 RNA laboratory-modified CDC COV assay, ACOV assay, and IDNOW assaya
| Assay comparison (A vs B) | Positive percent agreement (95% CI) | Negative percent agreement (95% CI) |
|---|---|---|
| CDC COV+ vs ACOV+ | 100 (96.9–100) | |
| CDC COV+ vs IDNOW+ | 80.3 (71.9–87.1) | |
| ACOV+ vs IDNOW+ | 75.2 (66.7–82.5) | |
| CDC COV− vs ACOV− | 92.4 (84.2–97.2) | |
| CDC COV− vs IDNOW− | 100 (95.4–100) | |
| ACOV− vs IDNOW− | 100 (95.4–100) |
n = 200 samples. +, positive; −, negative.
For the CDC COV assay, SARS-CoV-2 target RNA sequences were detected in 119 (60%) samples. Six (3%) samples gave an initial inconclusive result. Upon repeat testing, 4 yielded valid results: SARS-CoV-2 RNA was detected in 3 samples and was not detected in 1 sample. The remaining 2 (1%) samples repeated as inconclusive (only one of the two targets amplified in the specimen) (Table 1). The median CT value for positive samples was 30.29 (IQR, 25.40 to 34.03) for N1 and 30.20 (IQR, 25.12 to 34.55) for N2, which corresponds to an RNA concentration of approximately 150 genome equivalents/μl of sample (calculated using N1 standard curve equation) (Fig. 1).
The ACOV assay yielded 127 (63.5%) positive results and no invalid results (Table 1). The median CT value for positive samples was 17.27 (IQR, 13.27 to 21.40), which corresponds to an RNA concentration of approximately 147 genome equivalents/μl (Fig. 1). The IDNOW assay yielded 94 (47%) positive results (Table 1). Five (2.5%) samples first gave invalid results; 3 resolved after repeat testing and the remaining 2 repeated as invalid.
Analysis of discordant results.
There were 33 (17%) samples that yielded discordant results across the three assays (Table 1). Eight discordant samples were not detected or gave inconclusive results by the CDC COV assay but were detected by the ACOV assay. The median CT value for these samples by the ACOV assay was 27.73 (IQR, 27.37 to 28.40) or approximately 0.34 genome equivalents/μl (Fig. 2). Thirty-three samples (including 2 invalid samples) were not detected by IDNOW but were detected by the ACOV assay; 25 of these were also detected by the CDC COV assay. The median CT value for these samples by the ACOV assay was 21.42 (IQR, 20.80 to 23.88) or approximately 13.3 genome equivalents/μl (Fig. 2).
FIG 2.
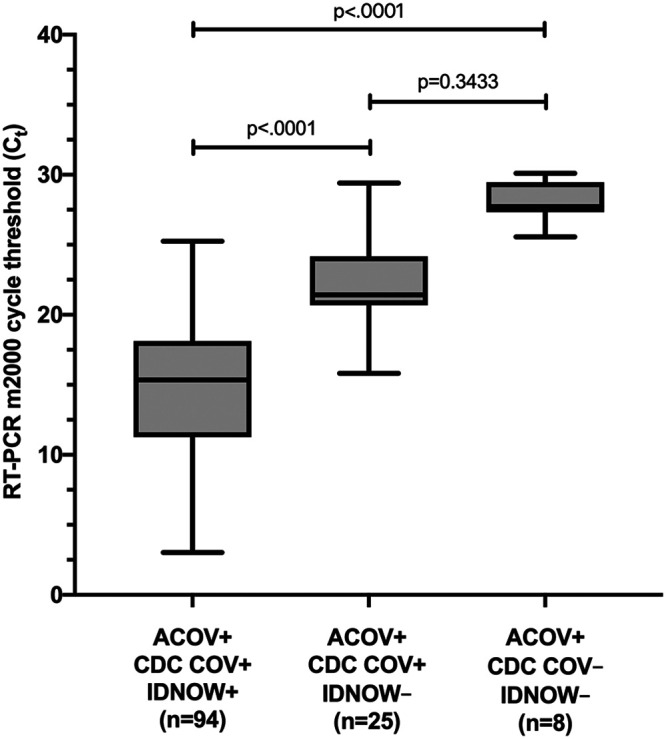
Comparison of CT values among samples detected by each of the three assays as measured by the ACOV assay. Median CT value differences were compared using the Kruskal-Wallis test and Dunn’s correction for multiple comparisons. CDC COV, modified CDC 2019-nCoV RT-PCR assay; ACOV, RealTime SARS-CoV-2 assay; IDNOW, ID Now COVID-19 assay; +, positive; −, negative.
Medical record review resolved all discrepant results in favor of the positive result (127 true positives). The ACOV assay detected significantly more cases of COVID-19 than either the CDC COV assay (8 [4%] undetected cases; standard deviation [SD], 0.014; 95% CI, 0.013 to 0.067) or the IDNOW (33 [16.5%] undetected cases; SD, 0.026; 95% CI, 0.11 to 0.22); the difference in detection between the CDC COV assay and IDNOW assay was also significant.
Assessment of dry nasal swabs that yielded negative results by the IDNOW assay.
We conducted a follow-up study to analyze the performance of the IDNOW assay using dry nasal swabs. Ninety-seven symptomatic ED patients who were not included in our original study and who had a negative result on a dry nasal swab that was tested at the point of care by the IDNOW assay also had a paired nasopharyngeal swab sample collected in VTM and tested by the ACOV assay. Mean age of patients was 59 ± 17 years, and 48 (48%) were women. SARS-CoV-2 RNA was detected in 13 (13.4%) paired nasopharyngeal swab samples by the ACOV assay; the median CT value was 19.82 (IQR, 16.08 to 26.19). SARS-CoV-2 RNA was not detected in the remaining 84 (86.6%) samples.
Workflow analysis.
Differences among key aspects of workflow for each of the three assays and platforms are summarized in Table 3. Batch testing and reporting of results can be completed for 58 samples using the CDC COV assay (27 patients + 4 controls per batch) and for 96 samples (94 patients + 2 controls) by the ACOV assay in an 8-h shift. The IDNOW assay, which was developed for point-of-care testing, requires the least hands-on time and provides the fastest results. However, throughput is limited (1 sample/instrument/5 to 15 min).
TABLE 3.
Workflow analysis comparing laboratory-modified CDC COV assay, ACOV assay, and IDNOW assay
| Parametera | CDC COV | ACOV | IDNOW |
|---|---|---|---|
| Off-board lysis | Yes | No | No |
| Specimen processing and set up | 1.75 | 1.00 | 0.03 |
| Instrument extraction time | 1.30 | 4.0 | 0.05 |
| Amplification and real-time detection | 1.25 | 2.25 | 0.22 |
| Manual interpretation and result entry | 0.5 | 0.75 | 0.02 |
| Total time (h) to resultb | 4.8 | 8.0 | 0.27 |
| No. samples processed in 8-h shift per instrument | 58c | 94d | 32e |
Times (hours) per batch for the in-house laboratory-developed test (LDT) and Abbott m2000 assays, and per sample for the ID Now assay.
Times (hours) from sample processing through result reporting.
A maximum of 58 patient samples, not including external positive control, negative control, negative template control, RNase P control.
A maximum of 94 patient samples, not including external positive and negative controls.
Assumes continuous processing of 32 patient samples, all with negative results. Results for positive samples may be generated within 5 min, and results for negative samples are generated within 13 min.
DISCUSSION
Rapid, accurate detection of COVID-19 is essential to ensure speedy and appropriate patient management, outbreak containment, and to better understand the global epidemiology of the virus. Laboratory testing to date has relied primarily on the amplification and detection of viral gene sequences in upper respiratory tract specimens. As new test kits are made available through the EUA pathway, laboratories are confronted with the dilemma of deciding which test or platform to adopt for SARS-CoV-2 detection. Additionally, laboratory directors are faced with numerous questions from clinicians regarding performance characteristics of the tests. Responding to these questions is difficult, since EUA requires only limited test validation (14); assays approved under EUA have not been evaluated in clinical trials, and robust performance data from real world assessments are lacking.
Results of the current study help to fill this knowledge gap. We found significant differences in detection of SARS-CoV-2 viral sequences among the ACOV, CDC COV, and IDNOW assays. The ACOV assay detected the most cases, followed by the CDC COV assay, and then by IDNOW. Discrepant results were observed almost exclusively in samples with higher CT values, i.e., lower viral titer. These findings suggest differences in lower limit of detection of the assays. For the CDC COV assay, this might be explained in part by smaller input sample volumes for extraction (200 μl) and amplification (5 μl) compared to the 500-μl extraction and 40-μl amplification volumes in the ACOV assay, i.e., there are more available targets for amplification and detection in the ACOV assay. Our results comparing the ACOV and IDNOW assays are concordant with those of Harrington et al., who reported increased detection of SARS-CoV-2 RNA gene sequences by ACOV compared to that by IDNOW (15).
In order to eliminate confounding that could have been introduced by testing different sample types on different systems, we evaluated aliquots of the same nasopharyngeal swab in VTM in all three assays. At the time of this study, nasopharyngeal swab specimens in VTM were deemed acceptable sample types for all 3 assays. Following the completion of our study, Abbott amended the package insert of IDNOW to state that testing VTM could lead to false-negative results. However, in our subsequent comparison of dry nasal swab samples tested by IDNOW and paired nasopharyngeal swabs tested by ACOV, the ACOV assay yielded significantly more positive results, suggesting that false-negative IDNOW results were not due entirely to dilution.
We observed differences in turnaround time, workflow, and throughput among the three tests. The ACOV had the longest runtime of the three assays, approximately 8 h for one full run of 94 patient samples. Total testing time for the CDC COV assay was shorter, but the throughput was less (58 samples in an 8-h shift, i.e., 2 batch runs). The IDNOW was the easiest to perform and yielded the fastest results; positive results were generated in as few as 5 min, which is faster than any other test system available currently in the United States, but the ability to test only a single sample significantly reduced throughput. Ease of use and speed are advantages in settings without laboratory expertise or when rapid results are needed. The assay platform is small and can be utilized at near-patient settings, thereby increasing the overall testing capacity for SARS-CoV-2 within health care facilities. Availability of different platforms provides flexibility to meet testing needs of different populations and different health care settings.
Our study has limitations. Because there is not a reference standard for SARS-CoV-2 infection, we were unable to calculate sensitivity or specificity of the assays. Instead, we calculated percent agreement, which is appropriate when a nonstandard reference method is utilized to compare assay performance (16). We resolved discrepant results through review of patient medical records, which may have introduced bias since concordant test results were not confirmed in the same way (17). We enriched for samples that were positive by the ACOV assay, which may have biased in favor of this test. Not all testing was performed on the same day due to workflow and personnel limitations, although all testing was completed within 72 h of sample collection. Storage of specimens at ambient room (22°C) or refrigerated (4°C) temperature has been shown to have little impact on detection of other RNA viruses by RT-PCR (18).
In conclusion, we found that the ACOV assay detected more cases of COVID-19 infection than the CDC COV assay or the IDNOW assay, and the CDC COV assay detected more cases than the IDNOW assay. The lesser sensitivity of the IDNOW should be weighed against its ease of use, speed, and small footprint when deciding on a test platform for a health care or community setting.
ACKNOWLEDGMENT
The following reagent was obtained through BEI Resources, NIAID, NIH: Genomic RNA from SARS-related coronavirus 2, isolate USA-WA1/2020, NR-52285.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, Ng OT, Marimuthu K, Ang LW, Mak TM, Lau SK, Anderson DE, Chan KS, Tan TY, Ng TY, Cui L, Said Z, Kurupatham L, Chen MI, Chan M, Vasoo S, Wang LF, Tan BH, Lin RTP, Lee VJM, Leo YS, Lye DC, Singapore Novel Coronavirus Outbreak Research Team. 2020. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard Stoecklin S, Rolland P, Silue Y, Mailles A, Campese C, Simondon A, Mechain M, Meurice L, Nguyen M, Bassi C, Yamani E, Behillil S, Ismael S, Nguyen D, Malvy D, Lescure FX, Georges S, Lazarus C, Tabai A, Stempfelet M, Enouf V, Coignard B, Levy-Bruhl D, Investigation T. 2020. First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Euro Surveill 25:pii=2000094. doi: 10.2807/1560-7917.ES.2020.25.6.2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spiteri G, Fielding J, Diercke M, Campese C, Enouf V, Gaymard A, Bella A, Sognamiglio P, Sierra Moros MJ, Riutort AN, Demina YV, Mahieu R, Broas M, Bengner M, Buda S, Schilling J, Filleul L, Lepoutre A, Saura C, Mailles A, Levy-Bruhl D, Coignard B, Bernard-Stoecklin S, Behillil S, van der Werf S, Valette M, Lina B, Riccardo F, Nicastri E, Casas I, Larrauri A, Salom Castell M, Pozo F, Maksyutov RA, Martin C, Van Ranst M, Bossuyt N, Siira L, Sane J, Tegmark-Wisell K, Palmerus M, Broberg EK, Beaute J, Jorgensen P, Bundle N, Pereyaslov D, Adlhoch C, Pukkila J, Pebody R, Olsen S, et al. 2020. First cases of coronavirus disease 2019 (COVID-19) in the WHO European Region, 24 January to 21 February 2020. Euro Surveill 25:2000178. doi: 10.2807/1560-7917.ES.2020.25.9.2000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mousavi SH, Shah J, Giang HTN, Al-Ahdal TMA, Zahid SU, Temory F, Paikan FM, Karimzadeh S, Huy NT. 2020. The first COVID-19 case in Afghanistan acquired from Iran. Lancet Infect Dis 20:657–658. doi: 10.1016/S1473-3099(20)30231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silverstein WK, Stroud L, Cleghorn GE, Leis JA. 2020. First imported case of 2019 novel coronavirus in Canada, presenting as mild pneumonia. Lancet 395:734. doi: 10.1016/S0140-6736(20)30370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK, Washington State 2019-nCoV Case Investigation Team. 2020. First case of 2019 novel coronavirus in the United States. N Engl J Med 382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JY, Choe PG, Oh Y, Oh KJ, Kim J, Park SJ, Park JH, Na HK, Oh MD. 2020. The first case of 2019 novel coronavirus pneumonia imported into Korea from Wuhan, China: implication for infection prevention and control measures. J Korean Med Sci 35:e61. doi: 10.3346/jkms.2020.35.e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghinai I, McPherson TD, Hunter JC, Kirking HL, Christiansen D, Joshi K, Rubin R, Morales-Estrada S, Black SR, Pacilli M, Fricchione MJ, Chugh RK, Walblay KA, Ahmed NS, Stoecker WC, Hasan NF, Burdsall DP, Reese HE, Wallace M, Wang C, Moeller D, Korpics J, Novosad SA, Benowitz I, Jacobs MW, Dasari VS, Patel MT, Kauerauf J, Charles EM, Ezike NO, Chu V, Midgley CM, Rolfes MA, Gerber SI, Lu X, Lindstrom S, Verani JR, Layden JE, Illinois COVID-19 Investigation Team. 2020. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet 395:1137–1144. doi: 10.1016/S0140-6736(20)30607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbott Laboratories. Abbott RealTime SARS-CoV-2 assay package insert. Abbott Laboratories, Chicago, IL: https://www.molecular.abbott/sal/9N77-095_SARS-CoV-2_US_EUA_Amp_PI.pdf. [Google Scholar]
- 12.Centers for Disease Control and Prevention. 2020. CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel services. Centers for Disease Control and Prevention, Atlanta, GA: https://www.fda.gov/media/134922/download. [Google Scholar]
- 13.Abbott Laboratories. ID Now COVID-19 package insert. Abbott Laboratories, Chicago, IL: https://www.fda.gov/media/136525/download. [Google Scholar]
- 14.Food and Drug Administration. Policy for diagnostics tests for coronavirus disease-2019 during the public health emergency. Food and Drug Administration, Washington, DC: https://www.fda.gov/media/135659/download. Accessed 17 March 2020. [Google Scholar]
- 15.Harrington A, Cox B, Snowdon J, Bakst J, Ley E, Grajales P, Maggiore J, Kahn S. 23 April 2020. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol doi: 10.1128/JCM.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. 2008. User protocol for evaluation of qualitative test performance; approved guideline—2nd ed CLSI document EP12-A2 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.McAdam AJ. 2000. Discrepant analysis: how can we test a test? J Clin Microbiol 38:2027–2029. doi: 10.1128/.38.6.2027-2029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Druce J, Garcia K, Tran T, Papadakis G, Birch C. 2012. Evaluation of swabs, transport media, and specimen transport conditions for optimal detection of viruses by PCR. J Clin Microbiol 50:1064–1065. doi: 10.1128/JCM.06551-11. [DOI] [PMC free article] [PubMed] [Google Scholar]