The role of serologic testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in both the clinical and public health settings, will continue to evolve as we gain increasing insight into our immune response to the virus. Here, we evaluated four high-throughput serologic tests for detection of anti-SARS-CoV-2 IgG antibodies, from Abbott Laboratories (Abbott Park, IL), Epitope Diagnostics, Inc. (San Diego, CA), Euroimmun (Lubeck, Germany), and Ortho-Clinical Diagnostics (Rochester, NY), using a panel of serially collected serum samples (n = 224) from 56 patients with confirmed coronavirus disease 2019 (COVID-19), healthy donor sera from 2018, and a cross-reactivity serum panel collected in early 2020.
KEYWORDS: antibodies, COVID-19, IgG antibodies, SARS-CoV-2, serology
ABSTRACT
The role of serologic testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in both the clinical and public health settings, will continue to evolve as we gain increasing insight into our immune response to the virus. Here, we evaluated four high-throughput serologic tests for detection of anti-SARS-CoV-2 IgG antibodies, from Abbott Laboratories (Abbott Park, IL), Epitope Diagnostics, Inc. (San Diego, CA), Euroimmun (Lubeck, Germany), and Ortho-Clinical Diagnostics (Rochester, NY), using a panel of serially collected serum samples (n = 224) from 56 patients with confirmed coronavirus disease 2019 (COVID-19), healthy donor sera from 2018, and a cross-reactivity serum panel collected in early 2020. The sensitivities of the Abbott, Epitope, Euroimmun, and Ortho-Clinical IgG assays in convalescent-phase serum samples collected more than 14 days post-symptom onset or post-initial positive reverse transcriptase PCR (RT-PCR) result were 92.9% (78/84), 88.1% (74/84), 97.6% (82/84), and 98.8% (83/84), respectively. Among unique convalescent patients, sensitivities of the Abbott, Epitope, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG assays were 97.3% (36/37), 73% (27/37), 94.6% (35/37), and 97.3% (36/37), respectively. Overall assay specificity/positive predictive values based on a 5% prevalence rate were 99.6%/92.8%, 99.6%/90.6%, 98.0%/71.2%, and 99.6%/92.5%, respectively, for the Abbott, Epitope, Euroimmun, and Ortho-Clinical IgG assays. In conclusion, we show high sensitivity in convalescent-phase sera and high specificity for the Abbott, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG assays. With the unprecedented influx of commercially available serologic tests for detection of antibodies against SARS-CoV-2, it remains imperative that laboratories thoroughly evaluate such assays for accuracy prior to implementation.
INTRODUCTION
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was officially classified as a pandemic by the World Health Organization (WHO) on 11 March 2020. As of the writing of the manuscript, over 4 million cases have been confirmed worldwide, with over 1 million in the United States, nearly 100,000 of which have resulted in death (https://coronavirus.jhu.edu/map.html; accessed 19 May 2020). Diagnostic testing of patients who present with symptoms consistent with COVID-19 relies on detection of viral RNA using molecular methods, including reverse transcriptase PCR (RT-PCR), which can be performed on an array of different respiratory tract specimen types (1). Serologic assays to detect antibodies against SARS-CoV-2 are now also available as an additional tool in our global response to the COVID-19 pandemic. Currently, most infectious disease and microbiology organizations generally agree that the role for antibody testing is limited to use in seroprevalence and epidemiology studies, screening of potential convalescent plasma donors, assessment of candidate vaccine efficacy, and in select scenarios as an aid for the diagnosis of COVID-19 in SARS-CoV-2 RT-PCR negative patients who present later in disease course and for whom collection of a lower respiratory tract sample is not possible (2–4). The role of serologic testing for SARS-CoV-2 will likely evolve in the future as we gain a better understanding of our immune response to the virus, identify correlates of immunity, and determine the level and duration of such immunity following infection or vaccination. Until then, the impact of an antibody result at the individual patient level is limited and should not be used to guide decisions related to use of personal protective equipment or adherence to social distancing policies.
While the Food and Drug Administration (FDA) required Emergency Use Authorization (EUA) for SARS-CoV-2 molecular assays at the outset of the outbreak, until recently, serologic tests for SARS-CoV-2 fell under Section IV.D of the FDA’s “Policy for Diagnostic Tests for Coronavirus Disease-2019” (i.e., Pathway D), which required commercial manufacturers to only notify the FDA of their validated product, ultimately leading to the rapid influx of over 180 commercially available SARS-CoV-2 serologic test kits (https://www.fda.gov/medical-devices/emergency-situations-medical-devices/faqs-diagnostic-testing-sars-cov-2; accessed 1 May 2020). On 4 May 2020, the FDA amended these guidelines to require all commercial manufacturers to submit their validation data for EUA, and to date, 20 manufacturers or laboratories have received EUA for their serologic test. The commercially available serologic assays vary in the antibody detected (i.e., IgM, IgA, IgG, or total antibody), the targeted SARS-CoV-2 antigen (i.e., subunit 1 [S1] of the spike protein, the nucleocapsid protein [NP], or the receptor binding domain [RBD]), and the method used (e.g., lateral flow assay [LFA], enzyme-linked immunosorbent assay [ELISA], chemiluminescent immunoassay [CLIA], etc.), with few peer-reviewed studies currently available which independently evaluate their performance characteristics.
Here, we provide an assessment of four high-throughput serologic tests, anti-SARS-CoV-2 IgG assays from Abbott Laboratories (Abbott Park, IL), Epitope Diagnostics, Inc. (San Diego, CA), Euroimmun (Lubeck, Germany), and Ortho-Clinical Diagnostics (Rochester, NY), using a panel of serially collected serum samples from RT-PCR-confirmed patients with COVID-19, healthy donor sera from 2018, and a cross-reactivity antibody panel collected in early 2020.
MATERIALS AND METHODS
Specimens collected from COVID-19-confirmed patients.
A total of 224 serum samples were collected from March to April 2020 from 56 adult patients confirmed positive for COVID-19 by a laboratory-developed or commercially available FDA EUA SARS-CoV-2 RT-PCR assay, performed on a nasopharyngeal swab specimen (5). Among the 56 COVID-19 RT-PCR-confirmed patients, 33 were hospitalized (inpatient group) and 23 were treated as outpatients (outpatient group). Remnant, serial serum samples were collected as available throughout the hospital stay for the inpatient group until discharge, whereas prospective collection of acute (≤7 days post-first RT-PCR positive result) and convalescent (i.e., >20 days post-first RT-PCR positive result) serum samples were collected for outpatients. Medical records were reviewed to determine the date of symptom onset or, if unavailable, the date of first SARS-CoV-2 RT-PCR positive result and to identify whether the patient was hospitalized or treated as an outpatient. The focus of this study was to assess assay performance characteristics; therefore, although patient demographics, presenting symptoms, disease course, and outcomes were documented, they are not included in this article. This study was approved by the Mayo Clinic institutional review board.
Serum specimens collected for specificity studies.
A total of 149 healthy adult donor serum samples collected in 2018, prior to the SARS-CoV-2 outbreak (stored at –70°C) and 105 deidentified patient sera submitted for testing as part of routine clinical care in January and early February 2020 were evaluated using each assay to assess specificity. The 105 patient serum samples made up the cross-reactivity panel for this study, which included sera positive for IgM and/or IgG antibodies against cytomegalovirus (n = 15), influenza (n = 10), Mycoplasma pneumoniae (n = 15), and Chlamydophila pneumoniae (n = 21) and 12 serum samples from patients positive for Streptococcus pneumoniae urinary antigen. Additionally, sera were collected from patients found positive by a respiratory pathogen RT-PCR panel for commonly circulating coronaviruses (n = 6), influenza (n = 14), metapneumovirus (n = 4), respiratory syncytial virus (n = 3), adenovirus (n = 6), or rhinovirus/enterovirus (n = 1). Among these 254 serum samples, antibodies to hepatitis B virus (HBV) surface antigen, HIV, and hepatitis C virus (HCV) were detected in 90, 20, and 6 samples, respectively.
Serologic assays.
Euroimmun anti-SARS-CoV-2 IgG ELISA (Lubeck, Germany). The Euroimmun (EI) anti-SARS-CoV-2 IgG ELISA is based on a recombinant S1 protein from the SARS-CoV-2 spike protein. Testing was performed in accordance with the manufacturer’s instructions. Patient serum was diluted 1:101 in sample buffer, added to antigen-coated microtiter wells, and incubated at 37°C for 60 min alongside undiluted calibrator, negative and positive controls. The microtiter wells were washed, and 100 μl of peroxidase-labeled anti-human IgG antibody enzyme conjugate was added and allowed to incubate at 37°C for 30 min. After a second wash step, 100 μl of chromogen/substrate (3,3′,5,5′-tetramethylbenzidine [TMB]/H2O2) was added and incubated at room temperature for 30 min. Finally, 100 μl of 0.5 M sulfuric acid stop solution was added, and the optical density (OD) was spectrophotometrically measured at 450 nm with a 620-nm reference filter. Patient index values were calculated by dividing patient serum OD values by the mean of the duplicate calibrator OD values. Index values (signal to cutoff [S/Co] ratios) of <0.8, ≥0.8 to <1.1, and ≥1.1 were interpreted as negative, indeterminate, and positive, respectively, per the instructions for use. All testing was performed on Agility automated ELISA analyzers (Dynex Technologies, Inc., Chantilly, VA).
Epitope Diagnostics, Inc., novel coronavirus COVID-19 IgG ELISA (San Diego, CA). The Epitope Diagnostics, Inc. (EDI), COVID-19 IgG ELISA employs a full-length recombinant nucleocapsid protein from SARS-CoV-2. All samples were tested in singlet, and the assay was performed on the Agility automated ELISA analyzer per manufacturer instructions. Testing was performed at room temperature with 100 μl of undiluted negative- and positive-control and patient sera diluted 1:101 in sample diluent added to antigen-coated microwells and allowed to incubate for 30 min. Following a wash step, 100 μl of horseradish peroxidase-conjugated anti-human IgG was added and incubated for 30 min. After another wash step, 100 μl of chromogen/substrate (TMB/H2O2) was added and incubated for 20 min, followed by the addition of 100 μl of 0.5 M sulfuric acid stop solution and an OD measurement at 450 nm. Patient index values were determined by dividing the patient serum OD value by the cutoff OD, which is determined as the mean of the triplicate negative control OD value plus 0.18. Use of an index value approach is an alternative but equivalent calculation to that recommended by the manufacturer, which allows for ease of assay programing on the Agility instrument. Using this equivalent calculation approach, the manufacturer index value thresholds for positive, negative, and indeterminate results are ≥1.1, ≤0.9, and >0.9 and <1.1, respectively. To optimize assay specificity, these index thresholds were modified to ≥1.21, <1.01, and ≥1.01 and <1.21 for positive, negative, and indeterminate qualitative results, respectively.
Abbott Laboratories SARS-CoV-2 IgG chemiluminescent microparticle immunoassay (CMIA; Abbott Park, IL). The Abbott Laboratories SARS-CoV-2 IgG assay is a two-step qualitative CMIA which employs acridinium ester-mediated chemiluminescence and is performed on the Architect i2000SR automated immunoassay analyzer. Testing was performed in accordance with the manufacturer’s instructions. A combination of sample, SARS-CoV-2 nucleocapsid antigen-coated paramagnetic microparticles, and assay diluent were added to a reaction vessel which allowed specific antibodies present in the patient sample to bind to the antigen-coated microparticles. Following an incubation step, the mixture was washed, and anti-human IgG acridinium-labeled conjugate was added to create a reaction mixture. After incubation and a wash step, H2O2 and NaOH were added, and the resulting chemiluminescent reaction was measured using the Architect i2000SR system optics. The patient sample signal was divided by the calibrator signal, with calculated S/Co values of <1.4 and ≥1.4 reported as negative and positive, respectively.
Ortho-Clinical Diagnostics Vitros anti-SARS-CoV-2 IgG chemiluminescent immunoassay (CLIA; Rochester, NY). The Ortho-Clinical Diagnostics anti-SARS-CoV-2 IgG assay is a qualitative CLIA utilizing luminol-horseradish peroxidase (HRP)-mediated chemiluminescence that is performed on the Vitros 3600 automated immunoassay analyzer. Testing was performed in accordance with the manufacturer’s instructions. In the first stage, specific antibodies present in patient sample reacted with recombinant SARS-CoV-2 spike antigen coated to reaction wells, and after an incubation period, unbound materials were removed by a washing step. For the second stage, horseradish peroxidase (HRP)-labeled anti-human IgG antibody conjugate was added and allowed to incubate, and this was followed with a wash step to remove unbound conjugate. Finally, luminogenic substrate reagent and electron transfer agents were added to the reaction wells. The HRP in the bound conjugate catalyzed oxidation of the luminol, which produced a chemiluminescent reaction measured by a luminometer. The patient sample signal was divided by the calibrator signal, with the resulting S/Co values of <1.00 and ≥1.00 corresponding to nonreactive and reactive results, respectively.
A summary of the attributes of these four assays, including assay principle, EUA status, timing to first result, and throughput, are summarized in Table 1.
TABLE 1.
Attributes for the evaluated anti-SARS-CoV-2 IgG serologic assaysa
| Attribute | Data for: |
|||
|---|---|---|---|---|
| Abbott | Epitope | Euroimmun | Ortho-Clinical | |
| Assay principleb | CMIA | ELISA | ELISA | CLIA |
| Solid-phase antigen | Nucleocapsid | Nucleocapsid | Spike S1 | Spike |
| Specimen type | Serum, plasma | Serum | Serum | Serum |
| Sample volume | 25 μl | 10 μl | 10 μl | 20 μl |
| EUA status | Granted | Submitted | Granted | Granted |
| Result calculation | Index (S/Co) | Index (S/Co) | Index (S/Co) | Index (S/Co) |
| Positive cutoff threshold | ≥1.4 | ≥1.21c | ≥1.1 | ≥1.00 |
| Indeterminate cutoff range | NA | ≥1.01 to <1.21c | ≥0.8 to <1.1 | NA |
| Operational type | Continuous, random access | Batch | Batch | Continuous, random access |
| Time to first result | 29 min | 80 min | 120 min | 48 min |
| Test throughput per 8 h | 1,600 | 630c | 630d | 1,040 to 1,200 |
Abbreviations: CMIA, chemiluminescent microparticle immunoassay; ELISA, enzyme-linked immunosorbent assay; CLIA, chemiluminescence immunoassay; EUA, emergency use authorization; S/Co, signal-to-cutoff; NA, not applicable.
All assays are indirect in format.
Laboratory-determined cutoff threshold.
Testing performed on the Dynex Agility automated ELISA processor (Chantilly, VA).
Statistics.
Sensitivity, specificity, and 95% confidence intervals (CIs) were determined using GraphPad QuickCalcs software (La Jolla, CA, USA). All graphs and figures were created in Microsoft Excel. For statistical analysis, indeterminate results by the Euroimmun and Epitope anti-SARS-CoV-2 IgG ELISAs were considered negative.
RESULTS
COVID-19 patient demographics and serial serum collection.
The median age of the 33 inpatients was 61 years (range, 24 to 90 years), and 61% (20/33) were male. A total of 190 residual serum samples were collected from these inpatients, ranging in time frame from 0 to 26 days post-symptom onset, and 26 patients had two or more serial serum samples collected (range, 2 to 17 serial serum samples) (Table 2). Among the 23 outpatients, the median age was 37 years (range, 21 to 64 years), and 43% (10/23) were male. Review of medical records for this group revealed that the date of symptom onset was not routinely documented; therefore, the date of serum sample collection was compared to the date of first positive SARS-CoV-2 RT-PCR result. In total, 34 serum samples were prospectively collected from the outpatient group; 11 patients had both baseline and convalescent-phase serum samples collected at 3 to 7 days and 20 to 31 days post-initial positive SARS-CoV-2 RT-PCR result, respectively, and the remaining 12 outpatients only had a convalescent-phase sample collected (Table 2).
TABLE 2.
COVID-19-confirmed patient demographics (n = 56) and collected serum samples (n = 224)a
| Patient or sample characteristic | Data for: |
|
|---|---|---|
| Inpatients | Outpatients | |
| No. of patients | 33 | 23 |
| Median age in yrs (range) | 61 (24–90) | 37 (21–64) |
| No. male (%) | 20 (61%) | 10 (43%) |
| No. of serum samples collected at days post-symptom onset | ||
| 0–7 days | 38 | NA |
| 8–14 days | 91 | NA |
| 15–26 days | 61 | NA |
| No. of serum samples collected at days post-first positive SARS-CoV-2 RT-PCR result | ||
| 0–7 days | NA | 11 |
| 20–31 days | NA | 23 |
NA, not applicable.
Sensitivity of the anti-SARS-CoV-2 IgG serologic assays.
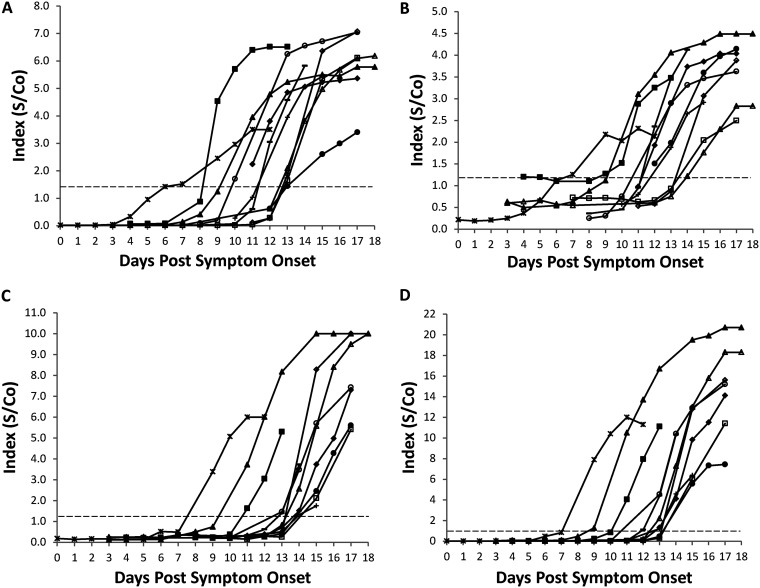
Among the samples collected from hospitalized patients, the Abbott, Epitope, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG assays were positive in 10.5% (4/38), 2.6% (1/38), 0% (0/38), and 2.6% (1/38), respectively, of samples collected 7 days or less post-reported symptom onset (Table 3). Sensitivity increased for all four assays in samples collected 8 to 14 days post-symptom onset (range, 27.5% to 49.5%), and all except the Abbott assay achieved 100% sensitivity in samples collected at least 15 days following initial disease manifestation. The sensitivity of the Abbott assay was 91.8% (56/61) for samples collected 15 days or more post-symptom onset; however, the 5 negative serum samples were all from a single patient who seroconverted at day 20 postonset. At the individual patient level, all inpatients who had a serum sample available after 14 days of symptoms (n = 14) seroconverted according to all four IgG assays. The temporal kinetics of the anti-SARS-CoV-2 IgG antibody response for each of the four evaluated assays is shown in Fig. 1 for inpatients who seroconverted during their hospital stay and who had at least five serial draws available (n = 11). The S/Co ratios increased over time for all patients and assays, and while seroconversion was observed as early as days 6 to 9 by all methods, the median time to seroconversion was day 12 for the Abbott and Epitope assays, day 13 for the Ortho-Clinical assay, and day 14 for the Euroimmun assay.
TABLE 3.
Performance characteristics of four commercially available anti-SARS-CoV-2 IgG assays
| Assay | % Sensitivity (no. of serum samples) |
% Specificity (no. of serum samples) |
||||||
|---|---|---|---|---|---|---|---|---|
| Inpatients, days post-symptom onseta
|
Outpatients, days post-first RT-PCR positive resultb
|
Healthy donors | Cross-reactivity panel | Overall (95% CI) | ||||
| ≤7 | 8–14 | ≥15 | ≤7 | ≥20 | ||||
| Abbott | 10.5% (4/38) | 49.5% (45/91) | 91.8% (56/61) | 18.2% (2/11) | 95.7% (22/23) | 100% (149/149) | 99% (104/105) | 99.6% (97.6%–100%) |
| Epitope | 2.6% (1/38) | 45.1% (41/91) | 100% (61/61) | 9.1% (1/11) | 56.5% (13/23) | 100% (149/149) | 99% (104/105) | 99.6% (97.6%–100%)c |
| Euroimmun | 0% (0/38) | 27.5% (25/91) | 100% (61/61) | 18.2% (2/11) | 91.3% (21/23) | 99.3% (148/149) | 96.2% (101/105) | 98% (95.3%–99.3%)d |
| Ortho-Clinical | 2.6% (1/38) | 38.5% (35/91) | 100% (61/61) | 9.1% (1/11) | 95.7% (22/23) | 99.3% (148/149) | 100% (105/105) | 99.6% (97.6%–100%) |
The number of unique patients providing samples collected ≤7, 8–14, and ≥15 days post-symptom onset was 11, 28, and 14, respectively.
The number of unique patients providing samples collected ≤7 and ≥15 days post-first positive RT-PCR result was 11 and 23, respectively.
Overall specificity is 98% (95% CI, 95.3%–99.3%) if indeterminate results are counted as positive.
Overall specificity is 97.2% (95% CI, 94.3%–98.8%) if indeterminate results are counted as positive.
FIG 1.
Anti-SARS-CoV-2 IgG kinetics in COVID-19 RT-PCR-confirmed inpatients. (A to D) Signal to cutoff (S/Co) values for the SARS-CoV-2 IgG assays from (A) Abbott Laboratories, (B) Epitope Diagnostics, Inc., (C) Euroimmun, and (D) Ortho-Clinical Diagnostics, were plotted against days post-symptom onset for all inpatients with at least five serial serum samples. Each symbol indicates an individual patient (n = 11).
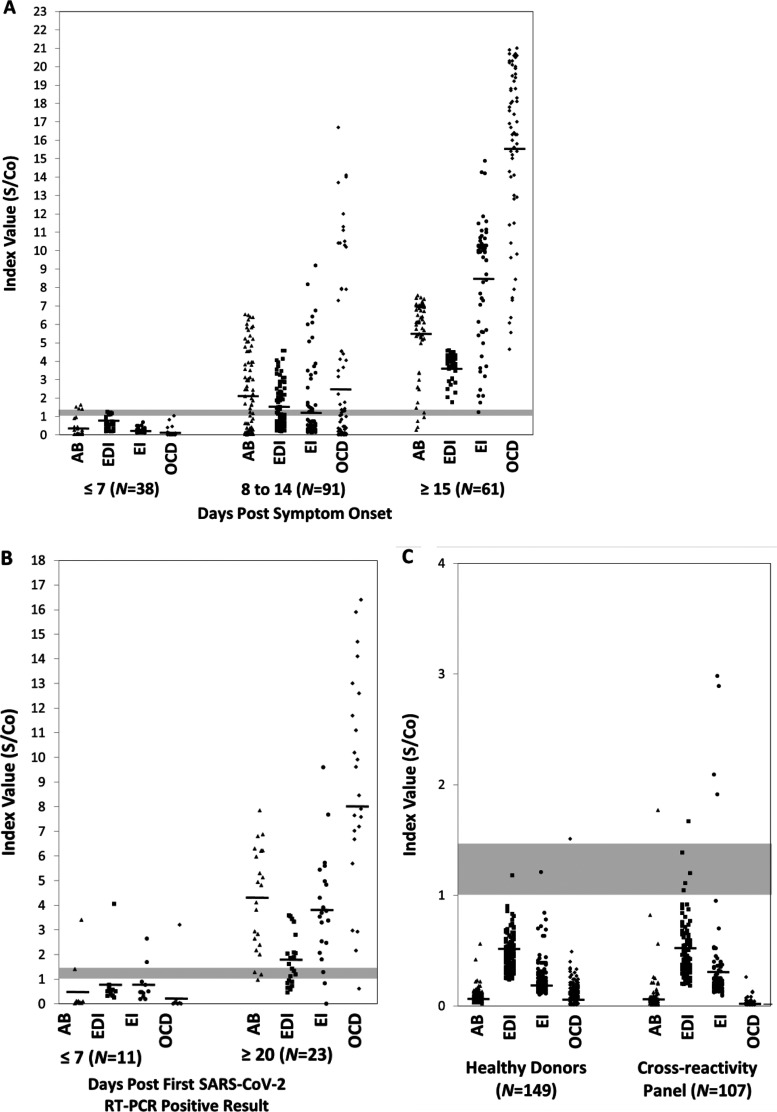
Among the 23 outpatients, the Abbott, Epitope, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG assays were positive in 18.2% (2/11), 9.1% (1/11), 18.2% (2/11), and 9.1% (1/11), respectively, of samples collected 7 days or fewer post-initial positive SARS-CoV-2 RT-PCR assay (Table 3). Similar to the inpatient samples, the sensitivity of the Abbott, Euroimmun, and Ortho-Clinical IgG assays increased significantly to 95.7% (22/23), 91.3% (21/23), and 95.7% (22/23), respectively, in samples collected 20 to 31 days following an initial positive molecular test (Table 3). While sensitivity of the Epitope anti-SARS-CoV-2 IgG ELISA also increased in this convalescent-phase sample set, to 56.5% (13/23), it was notably lower than that of the other three assays. Combining both inpatient and outpatient sensitivity rates in convalescent-phase serum samples collected at least 15 days post-symptom onset or post-initial positive RT-PCR result, the overall sensitivity of the Abbott, Epitope, Euroimmun, and Ortho-Clinical IgG assays was 92.9% (78/84), 88.1% (74/84), 97.6% (82/84), and 98.8% (83/84), respectively, with overlapping 95% confidence intervals suggesting that these differences are insignificant (Table 4). At the individual patient level, among the 37 patients with at least one serum sample collected 15 days post-symptom onset (n = 14) or following the first positive SARS-CoV-2 molecular test (n = 23), the sensitivity of the Abbott, Epitope, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG assays was 97.3% (36/37), 73% (27/37), 94.6% (35/37), and 97.3% (36/37), respectively (Table 4). Notably, the 95% confidence intervals for the Abbott and Ortho-Clinical assays did not overlap the Epitope ELISA. Although all four assays are qualitative in design, the mean S/Co ratios for the Epitope, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG assays were nearly double in convalescent-phase sera from hospitalized patients versus convalescent-phase sera from outpatients—S/Co 3.8 versus 1.7 for Epitope, S/Co 8.5 versus 4.0 for Euroimmun, and S/Co 15.7 versus 9.0 for Ortho-Clinical (Fig. 2). The difference in mean S/Co ratios in convalescent-phase samples from inpatients and outpatients (S/Co 5.5 versus 4.3) for the Abbott anti-SARS-CoV-2 IgG assay was not as high.
TABLE 4.
Anti-SARS-CoV-2 IgG assay sensitivity in convalescent-phase sera and in individual patients tested ≥15 days post-symptom onset or first positive SARS-CoV-2 RT-PCR result
| Assay | Serum samples (n = 84)a
|
Individuals patients (n = 37)b
|
||
|---|---|---|---|---|
| % Positive (no.) | 95% CI | % Positive (no.)c | 95% CI | |
| Abbott | 92.9% (78/84) | 85%–97% | 97.3% (36/37) | 85%–100% |
| Epitope | 88.1% (74/84) | 79.3%–93.6% | 73% (27/37) | 56.9%–84.8% |
| Euroimmun | 97.6% (82/84) | 91.2%–99.9% | 94.6% (35/37) | 81.4%–99.4% |
| Ortho-Clinical | 98.8% (83/84) | 92.9%–100% | 97.3% (36/37) | 85%–100% |
The 84 serum samples included 61 samples collected at least 15 days post-symptom onset from inpatients and 23 samples collected 20 days or more post-initial positive SARS-CoV-2 RT-PCR result in outpatients (see Table 3).
The 37 individual patients included all 23 outpatients with sera collected 20 days or more post-initial positive SARS-CoV-2 RT-PCR result and 14 inpatients who had a serum sample collected 15 days or more post-symptom onset. Patients were counted as positive if at least one sample collected 15 days or more post-onset or first RT-PCR was positive.
One patient was negative by the Euroimmun and Ortho-Clinical assays, and a second patient was negative by both the Epitope and Abbott assays. One and two patients, respectively, were indeterminate by the Euroimmun and Epitope assays but positive by all other assays.
FIG 2.
Signal to cutoff (index value) distribution among SARS-CoV-2 RT-PCR-confirmed patients and controls. (A and B) Signal to cutoff (S/Co) ratios are shown relative to days post-symptom onset for inpatients (A) or days post-first SARS-CoV-2 RT-PCR-positive result in outpatients (B). (C) Signal intensities are also shown for healthy control samples collected in 2018 and for a cross-reactivity panel. Gray shaded bars represent the S/Co range for positive results by the four assays (range, 1.0 to 1.4 S/Co). AB, Abbott Laboratories; EDI, Epitope Diagnostics, Inc.; EI, Euroimmun; OCD, Ortho-Clinical Diagnostics.
Specificity of the anti-SARS-CoV-2 IgG assays.
Among 149 serum samples collected from healthy adults in 2018, the Abbott, Epitope, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG assays were negative in 100% (149/149), 100% (149/149), 99.3% (148/149), and 99.3% (148/149) of samples, respectively (Table 3). Using a set of 105 serum samples collected in late January to early February 2020, which were positive for antibodies to or nucleic acid from other respiratory viral or bacterial pathogens, the Abbott, Epitope, Euroimmun, and Ortho-Clinical IgG assays were negative in 99% (104/105), 99% (104/105; 3 indeterminate), 96.2% (101/105; 1 indeterminate), and 100% (105/105) of these samples, respectively. When combined with the healthy donor results, overall specificity was 99.6% (95% CI, 97.6% to 100%), 99.6% (95% CI, 97.6% to 100%), 98.0% (95% CI, 95.3% to 99.3%), and 99.6% (95% CI, 97.6% to 100%), respectively (Table 3). For this analysis, indeterminate results yielded by the Epitope and Euroimmun anti-SARS-CoV-2 IgG ELISAs were considered negative; however, if samples with indeterminate results were grouped as positive, the overall specificity of these two assays decreases to 98% (249/254; 95% CI, 95.3% to 99.3%) and 97.2% (247/254; 95% CI, 94.3% to 98.8%), respectively. The Abbott CMIA was positive (S/Co, 1.77) in a single serum sample positive for IgM and IgG antibodies against C. pneumoniae, the Epitope ELISA was positive in one serum sample with antibodies to C. pneumoniae (S/Co, 1.67), and the Euroimmun ELISA was positive in one anti-C. pneumoniae IgG-positive serum sample (S/Co, 2.09), one serum sample (S/Co, 1.91) from an RSV RT-PCR positive patient, and two serum samples (S/Co, 2.89, 2.98) from patients positive for Streptococcus pneumoniae antigen in urine (Fig. 2; data not shown). Notably, none of the samples found positive by one assay were found positive by any of the other three assays, suggesting that these are all false-positive results. Using a prevalence rate of 5%, our reported convalescent sensitivity in individual patients, and the overall specificity values, the positive predictive values of the Abbott, Epitope, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG immunoassays are 92.8% (95% CI, 81.6% to 97.5%), 90.6% (95% CI, 79.4% to 98.1%), 71.2% (95% CI, 59.3% to 80.8%), and 92.5% (95% CI, 81.6% to 97.5%), respectively.
DISCUSSION
This study presents a head-to-head comparison of four high-throughput, commercially available anti-SARS-CoV-2 IgG serologic tests from Abbott Laboratories, Epitope Diagnostics, Inc., Euroimmun, and Ortho-Clinical Diagnostics, using serially collected acute- and convalescent-phase sera from both hospitalized patients and outpatients with RT-PCR-confirmed COVID-19. We show that less than 20% (range, 0% to 18.2%) of serum samples collected within 7 days of symptom onset or first positive SARS-CoV-2 RT-PCR resulted as positive by any of the four evaluated methods. This is consistent with other studies evaluating the Abbott, Epitope, and Euroimmun anti-SARS-CoV-2 IgG assays (6–9). Seroconversion to anti-SARS-CoV-2 IgG positive in most individuals occurs toward the end of week 2 postinfection, and our data continue to underscore the importance of not relying solely on such testing to diagnose infection in acutely symptomatic patients (10, 11). As with other infections however, detecting seroconversion between acute- and convalescent-phase samples, collected 7 to 14 days apart, may be helpful to diagnose recent COVID-19 infection in certain challenging scenarios. Sensitivity increased significantly in sera collected 15 days or more post-symptom onset, and with the exception of the Epitope anti-SARS-CoV-2 IgG ELISA, all three remaining assays were positive in 92% to 97% of convalescent-phase samples. At the individual patient level, the Abbott, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG immunoassays were positive in 94% to 97% of patients tested during the convalescent phase. Although neither of these tests achieved 100% sensitivity, with one to two COVID-19 confirmed outpatients remaining seronegative or indeterminate at day 22 to 27 post-initial diagnosis, our findings indicate that the majority of infected individuals develop an immune response to SARS-CoV-2, irrespective of disease severity or the viral antigen used in the immunoassay.
The notable exception to this is the Epitope Diagnostics anti-SARS-CoV-2 IgG ELISA, which although positive in all serum samples collected from hospitalized patients with more than 15 days of symptoms, yielded negative results in 10 (45%) of the 23 outpatients tested during the convalescent phase. Of note, use of the manufacturer-established interpretive thresholds in this group resulted in 8 (35%) of the 23 outpatients remaining negative (data not shown). The modified Epitope IgG threshold used in our study maximized assay specificity to 99.6% (253/254), compared to manufacturer thresholds, which were associated with a specificity of 98% (250/254; data not shown). Additional serial serum samples to determine whether seroconversion by this assay occurred at a later time point were unavailable. The overall sensitivity of the Epitope IgG assay in convalescent-phase sera was 88% in our study, similar to the 90% sensitivity reported by a recent preprint manuscript in sera collected 20 days or more post-symptom onset (9). The Epitope IgG assay is based on a recombinant SARS-CoV-2 nucleocapsid protein, considered the most abundant coronavirus protein. Although prior studies suggest later seroconversion of anti-nucleocapsid-based assays, we did not observe a notable delay with the Abbott IgG CMIA, which also targets antibodies to the nucleocapsid, and the median time to seroconversion between all assays was similar (12 to 14 days) (10, 12). Collectively, these data suggest lower sensitivity of the Epitope IgG ELISA, which is potentially limited to mildly ill patients, although a definitive reason for this remains unclear.
The specificity of the four evaluated assays was high, above 97%, for all assays using sera from both healthy donors collected prior to the pandemic and samples positive for antibodies to or nucleic acid from other viral or bacterial pathogens. While we identified positive results for three of these assays in a number of samples positive for antibodies to bacterial pathogens, it is unlikely that they are causing the anti-SARS-CoV-2 IgG positivity. Although it is possible that these samples, collected in January and February of 2020, were from patients previously infected with SARS-CoV-2, none of them were positive by more than one anti-SARS-CoV-2 IgG assay, suggesting that the observed reactivity was nonspecific. The specificity of the Abbott anti-SARS-CoV-2 IgG assay was recently reported to be 99.4% and 99.9% by two separate studies, consistent with our findings (6, 8). Two prior published studies evaluated the Euroimmun anti-SARS-CoV-2 IgG ELISA and showed specificity ranges of approximately 95% (145/153) to 96% (195/203), which is lower than what we report here (97% to 98%) and may be attributed to differences in the populations tested, interlab performance, or lot-to-lot test kit variability (7, 8). The specificity of the Epitope anti-SARS-CoV-2 IgG ELISA was reported to range from 85% to 90% by the aforementioned preprint study, which is significantly lower than our overall finding of 99.6% (9). The specificity differences may be due to multiple factors, including our use of an automated ELISA platform to perform the testing, which may provide higher processing consistency, and our use of a higher index value threshold for positive results (≥1.21) compared to those recommended by the manufacturer (≥1.01). Finally, to our knowledge, this is the first study to independently report on the specificity of the Ortho-Clinical anti-SARS-CoV-2 IgG immunoassay, which we found to be 99.6%.
Although we report high specificity for the four assays evaluated here, the overall accuracy of these tests will be impacted significantly by the prevalence of the disease in the population tested. The current prevalence of SARS-CoV-2 in the United States and in other countries remains largely unknown due to the high rates of asymptomatic infection and varies from region to region. For assay comparison purposes, the FDA has chosen a 5% prevalence benchmark with which to evaluate the positive and negative predictive values (PPV and NPV, respectively) for EUA assays. Based on this prevalence rate, for our reported overall specificity and sensitivity in individual convalescent patients, we found that while the PPVs did not significantly differ between the Abbott, Epitope, and Ortho-Clinical anti-SARS-CoV-2 IgG immunoassays (90% to 92%), the Euroimmun assay was associated with a notably lower PPV (71%), despite a high overall specificity of 98%. In an effort to improve on the PPV given the still low prevalence of SARS-CoV-2 in many regions of the United States, the FDA has suggested that laboratories consider confirming initial antibody-positive results with a second serologic assay based on an alternative viral target antigen. Confirmatory testing of initially antibody-positive samples is not an uncommon practice to improve upon the overall diagnostic accuracy of serologic tests, and such an algorithmic approach will likely be useful, particularly for assays with lower specificity profiles.
There are a number of limitations associated with this study. First, the number of unique COVID-19 patients included in the inpatient and outpatient groups was low. However, aside from the Epitope anti-SARS-CoV-2 IgG ELISA, differences in sensitivity between these two groups were not notable for the other three assays. Second, the specificity of these assays was not evaluated using samples which were known to be positive for antibodies to the commonly circulating human coronaviruses (i.e., 229E, NL63, OC43, and HKU1). Prior seroprevalence studies show that 65% to 75% of children have antibodies to at least one of the commonly circulating coronaviruses, and over 90% of adults over the age of 50 years are seropositive for antibodies to all four common coronaviruses (13, 14). Collectively, based on these seroprevalence findings and the overall low false-positivity rates identified in this study, we anticipate that although false-positive reactions due to antibodies against other common coronaviruses can occur by these assays, the rate appears to be low. Additionally, samples in the cross-reactivity panel were collected during January and early February of 2020, thus preventing us from definitively ruling out prior infection with SARS-CoV-2 and potentially negatively biasing our overall specificity rate. However, applying the FDA-suggested approach of confirming positive samples with an assay based on an alternative SARS-CoV-2 antigen, all samples with positive results in this panel would be considered falsely positive. Third, due to a lack of reporting, the timing of serum collection for outpatients was compared to the first positive SARS-CoV-2 RT-PCR result, which may have led to an overestimation of anti-SARS-CoV-2 IgG assay sensitivity compared to the true date of infection. Similarly, symptom onset in hospitalized patients was extrapolated based on the date reported by the patient or assumed by the provider, which may be subject to recall inaccuracies. Finally, correlation of results from these assays relative to a neutralizing antibody (NAb) test was not performed. Anti-SARS-CoV-2 NAbs primarily develop against the spike protein, specifically to the S1 receptor-binding domain, and initial studies indicate that NAb titers correlate better to spike-based assays than to nucleocapsid assays (10, 15). While one study has shown high correlation of NAbs with the Euroimmun IgG assay, as NAb assays become increasingly accessible, more detailed correlation studies across assays will be possible (7).
In conclusion, we show that the Abbott, Epitope, Euroimmun, and Ortho-Clinical anti-SARS-CoV-2 IgG immunoassays perform similarly with respect to sensitivity in COVID-19 hospitalized patients and, with the exception of the Epitope assay, also in individuals with milder forms of the infection. The Abbott and Ortho-Clinical anti-SARS-CoV-2 IgG immunoassays provided the highest overall specificity, at over 99%. Given that one of the primary uses of serologic assays (at this time) is for the purpose of monitoring local and regional seroprevalence, and that SARS-CoV-2 prevalence rates remain largely unknown, it will be important to use immunoassay(s) with the highest specificity in an effort to maximize the PPV of results. With the unprecedented influx of commercially available test kits for detection of antibodies against SARS-CoV-2, it remains imperative that laboratories thoroughly evaluate these assays for accuracy prior to implementation.
ACKNOWLEDGMENTS
We acknowledge all of the patients who consented to participate in this study and the mobile phlebotomy unit at Mayo Clinic for their assistance in collecting acute- and convalescent-phase sera from outpatients confirmed positive for COVID-19. We also thank all of the front-line workers both at the bedside and at the bench for their incredible efforts during this global pandemic.
REFERENCES
- 1.Tang YW, Schmitz JE, Persing DH, Stratton CW. 2020. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J Clin Microbiol 58:e00512-20. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Theel ES, Slev P, Wheeler S, Couturier MR, Wong SJ, Kadkhoda K. 2020. The role of antibody testing for SARS-CoV-2: is there one? J Clin Microbiol doi: 10.1128/JCM.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.IDSA. 2020. IDSA COVID-19 antibody testing primer. https://www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-covid-19-antibody-testing-primer.pdf. Accessed 19 May 2020.
- 4.Patel R, Babady E, Theel ES, Storch GA, Pinsky BA, St George K, Smith TC, Bertuzzi S. 2020. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: value of diagnostic testing for SARS-CoV-2/COVID-19. mBio 11:e00722-20. doi: 10.1128/mBio.00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodino KG, Espy MJ, Buckwalter SP, Walchak RC, Germer JJ, Fernholz E, Boerger A, Schuetz AN, Yao JD, Binnicker MJ. 2020. Evaluation of saline, phosphate buffered saline and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J Clin Microbiol 58:e00590-20. doi: 10.1128/JCM.00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan A, Pepper G, Wener MH, Fink SL, Morishima C, Chaudhary A, Jerome KR, Mathias PC, Greninger AL. 2020. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken C, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. 2020. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis 26. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang MS, Hock KG, Logsdon NM, Hayes JE, Gronowski AM, Anderson NW, Farnsworth CW. 2020. Clinical performance of two SARS-CoV-2 serologic assays. Clin Chem doi: 10.1093/clinchem/hvaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, Rathore U, Goldgof GM, Whitty C, Woo JM, Gallman AE, Miller TE, Levine AG, Nguyen DN, Bapat SP, Balcerek J, Bylsma S, Lyons AM, Li S, Wong A-y, Gillis-Buck EM, Steinhart ZB, Lee Y, Apathy R, Lipke MJ, Smith JA, Zheng T, Boothby IC, Isaza E, Chan J, Acenas DD, Lee J, Macrae TA, Kyaw TS, Wu D, Ng DL, Gu W, York VA, Eskandarian HA, Callaway PC, Warrier L, Moreno ME, Levan J, Torres L, Farrington L, Loudermilk R, Koshal K, Zorn KC, Garcia-Beltran WF, Yang D, et al. 2020. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv doi: 10.1101/2020.04.25.20074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H, Wei X, Cai P, Ma WL. 2020. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cagliani R, Forni D, Clerici M, Sironi M. 2020. Computational inference of selection underlying the evolution of the novel coronavirus, SARS-CoV-2. J Virol 94. doi: 10.1128/JVI.00411-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorse GJ, Patel GB, Vitale JN, O’Connor TZ. 2010. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin Vaccine Immunol 17:1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Principi N, Bosis S, Esposito S. 2010. Effects of coronavirus infections in children. Emerg Infect Dis 16:183–188. doi: 10.3201/eid1602.090469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, Wei P, Ge J, Gou M, Li X, Sun L, Cao T, Wang P, Zhou C, Zhang R, Liang P, Guo H, Wang X, Qin CF, Chen F, Dong C. 2020. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 52:1–7. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]