The outbreak of coronavirus disease 2019 (COVID-19) has spread across the world and was characterized as a pandemic. To protect medical laboratory personnel from infection, most laboratories inactivate the virus causing COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in clinical samples before testing. However, the effect of inactivation on the detection results remains unknown. Here, we used a digital PCR assay to determine the absolute SARS-CoV-2 RNA copy number in 63 nasopharyngeal swab samples and assess the effect of inactivation methods on viral RNA copy number.
KEYWORDS: COVID-19, inactivation, digital PCR, copy number
ABSTRACT
The outbreak of coronavirus disease 2019 (COVID-19) has spread across the world and was characterized as a pandemic. To protect medical laboratory personnel from infection, most laboratories inactivate the virus causing COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in clinical samples before testing. However, the effect of inactivation on the detection results remains unknown. Here, we used a digital PCR assay to determine the absolute SARS-CoV-2 RNA copy number in 63 nasopharyngeal swab samples and assess the effect of inactivation methods on viral RNA copy number. Viral inactivation was performed by three different methods: (i) incubation with the TRIzol LS reagent for 10 min at room temperature, (ii) heating in a water bath at 56°C for 30 min, and (iii) high-temperature treatment, including autoclaving at 121°C for 20 min, boiling at 100°C for 20 min, and heating at 80°C for 20 min. Compared to the amount of RNA in the original sample, TRIzol treatment destroyed 47.54% of the nucleocapsid protein (N) gene and 39.85% of open reading frame (ORF) 1ab. For samples treated at 56°C for 30 min, the copy number of the N gene and ORF 1ab was reduced by 48.55% and 56.40%, respectively. The viral RNA copy number dropped by 50 to 66% after heating at 80°C for 20 min. Nearly no viral RNA was detected after autoclaving at 121°C or boiling at 100°C for 20 min. These results indicate that inactivation reduced the quantity of detectable viral RNA and may cause false-negative results, especially in weakly positive cases. Thus, use of the TRIzol reagent rather than heat inactivation is recommended for sample inactivation, as the TRIzol reagent had the least effect on the RNA copy number among the tested methods.
INTRODUCTION
An outbreak of the respiratory illness coronavirus disease 2019 (COVID-19) has affected people in almost all countries and territories around the world (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/). On 12 March 2020, the World Health Organization (WHO) upgraded the status of the COVID-19 outbreak from epidemic to pandemic (www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic). COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is a coronavirus that has not been previously identified in humans (1).
Reverse transcription-quantitative PCR (RT-qPCR) is currently regarded as the gold standard for the diagnosis of SARS-CoV-2 infection (2). In China, nucleic acid assays of SARS-CoV-2 are required to be conducted in biosafety level 2 laboratories with personal biocontainment level 3 protection, and the U.S. Centers for Disease Control and Prevention (U.S. CDC) recommends that specimen processing be performed in a certified class II biological safety cabinet following biosafety level 2 or higher guidelines [www.who.int/publications-detail/laboratory-biosafety-guidance-related-to-coronavirus-disease-2019-(covid-19)]. However, the lack of protective materials has forced laboratories to adopt viral inactivation measures, such as through the use of the chemical substance the TRIzol reagent and heating at 56°C for 30 min, to allow downstream analysis of SARS-CoV-2 to be conducted outside high-containment laboratories. It has been estimated that in real COVID-19 cases, the false-negative rate (FNR) for one-time testing was 30% to 50% (3). Several factors may be related to the high FNR, such as the type of sample, the sampling procedure, the sample inactivation method, the sample transport condition, and the laboratory practice standard. The effect of sample inactivation on the test results remains unknown.
Digital PCR (dPCR) allows precise detection and quantification of the amount of nucleic acids (4). In dPCR, the sample is partitioned into numerous reaction chambers, such that most of the chambers contain zero or one template molecule. The amount of the template molecules in the sample can thus be quantified from the fraction of positive reactions after PCR amplification (5). dPCR has many potential advantages over quantitative PCR, including the capability to obtain absolute quantification without external references and the tolerance of PCR inhibitors (6). dPCR is increasingly used in clinical virology for the study of human-pathogenic viruses (7–12).
Here, we used a dPCR assay to determine the absolute SARS-CoV-2 RNA copy number in nasopharyngeal swab samples and assess the influence of sample inactivation on virus RNA copy number.
MATERIALS AND METHODS
Sample collection.
Nasopharyngeal swab samples were collected from suspected COVID-19 cases at fever clinics and their close contacts, placed in 1 to 3 ml virus transfer medium (VTM), and transferred to the Xi’an Center for Disease Control and Prevention (Xi’an CDC). All samples were tested using a commercial SARS-CoV-2 RNA detection kit (lot number ZC-HX-201-2; BioGerm Medical Biotechnology Co., Ltd., Shanghai, China) at the Xi’an CDC according to the manufacturer’s instructions. This RT-qPCR kit detects opening reading frame (ORF) 1ab and the nucleocapsid protein (N) gene simultaneously, as recommended by the China Center for Disease Control and Prevention (China CDC; http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html). The samples from 56 confirmed COVID-19 cases and 7 asymptomatic ones were chosen as the original group for this study.
Sample inactivation.
Viral inactivation was performed on each sample by three different methods: (i) incubation with the TRIzol LS reagent (Life Technologies Corp., Carlsbad, CA) for 10 min at room temperature (the Tri group), (ii) heating in a water bath at 56°C for 30 min (the 56°C 30 min group), and (iii) high-temperature treatment, including autoclaving at 121°C for 20 min (the 121°C 20 min group), boiling at 100°C for 20 min (the 100°C 20 min group), and heating at 80°C for 20 min (the 80°C 20 min group). In the high-temperature treatment group, only 16 out of the 61 samples were treated at 80°C, another 31 samples were treated at 100°C, and the remaining 14 samples were treated at 121°C. The detailed procedure was as follows: for the Tri group, 50 μl of sample was added to 200 μl of the TRIzol LS reagent (1:4) and the mixture was incubated for 10 min at room temperature; for the 56°C 30 min group, 300 μl of sample in a closed vial was put into a 56°C water bath for 30 min; and for the high-temperature treatment, 300 μl of sample in a closed vial was autoclaved at 121°C for 20 min, boiled at 100°C for 20 min, or heated at 80°C for 20 min.
RNA extraction.
The viral RNA in 200 μl of the original and treated samples was extracted using magnetic beads prefilled from RNA/DNA extraction kits (lot number T014; Tianlong Science and Technology Co., Xi’an, China) on a nucleic acid extractor (model NP968-S; Tianlong Science and Technology Co., Xi’an, China) according to the manufacturer’s instructions. The extracted RNA was eluted with 80 μl elution buffer.
Digital PCR detection.
dPCR was performed with a SARS-CoV-2 nucleic acid detection kit (digital PCR method) (TargetingOne, Beijing, China) and a TD-1 Droplet Digital PCR system (TargetingOne; licensed in China under registration numbers 20170025, 20190065, and 20192220517) following the manufacturer’s instructions. Briefly, 30 μl dPCR mixture was prepared with 15 μl RNA and 15 μl RT-PCR mix. Then, 30 μl dPCR mixture and 180 μl oil were loaded onto the droplet generation chip to produce droplets on a drop maker (TargetingOne). The droplets were thermally cycled using a protocol of 55°C for 15 min and 95°C for 10 min, followed by 40 cycles of 94°C for 30 s and 57°C for 1 min, and the temperature ramp rate was set to 1.5°C/s on a T100 thermal cycler (Bio-Rad Laboratories, Inc., Singapore). Finally, the droplets were detected on a chip reader (TargetingOne). dPCR was performed in duplicate.
Statistical analysis.
The Pearson correlation was used to assess the linear relationship between ORF 1ab and the N gene. As the samples were diluted in TRIzol at 1:4, we normalized the data for the Tri group by multiplying by the dilution factor before performing statistical analysis. A paired-sample t test was used to compare each treatment group with the original group. Samples with a viral load of less than 10 copies/test or more than 40,000 copies/test were excluded from the analysis to ensure reproducibility. The measurements for the 56°C 30 min and high-temperature treatment groups were logarithmically converted before comparison. The median for each group was calculated and used to determine the reduction in viral RNA copy number. SPSS (version 18.0) software was used for statistical analysis (two-tailed α = 0.05).
Ethical approval.
This study was approved by the Ethics Checking Committee of the Xi’an CDC (XACDC approval number ECC 01-2020), and because the samples were initially sent for COVID-19 case confirmation and epidemic control and only deidentified data were collected for this study, no written informed consent was needed.
RESULTS
Case information.
According to Chinese guidelines for the diagnosis of COVID-19 and on the basis of a combination of epidemiological history, clinical features, and radiographic and laboratory results, 56 cases were diagnosed with COVID-19 and 7 were considered asymptomatic carriers. Among them, 39 were male and 24 were female, and their ages ranged from 13 to 93 years (median ± interquartile range [IQR], 50 ± 29 years). The time of sample collection from the 56 cases was 2 to 26 days after illness onset (mean ± standard deviation [SD], 7.5 ± 8 days).
Detection of SARS-CoV-2 ORF 1ab and N gene in the original samples using RT-qPCR and dPCR.
We first tested the 63 original samples by RT-qPCR and dPCR. As shown in Fig. 1A, the results of RT-qPCR were expressed in cycle threshold (CT) values. The CT value of ORF 1ab in the original samples ranged from 11.9 to 38.2 (mean ± SD, 26.92 ± 4.99), and that of the N gene ranged from 11.54 to 35.32 (mean ± SD, 25.73 ± 4.93). Among the 63 specimens, dPCR found 61 to be positive and 2 to be negative, which was not consistent with the RT-qPCR results. The relative ratio of the absolute copy number of ORF 1ab to that of the N gene is listed in Fig. 1B, and ORF 1ab had 21.58% fewer copies than the N gene.
FIG 1.
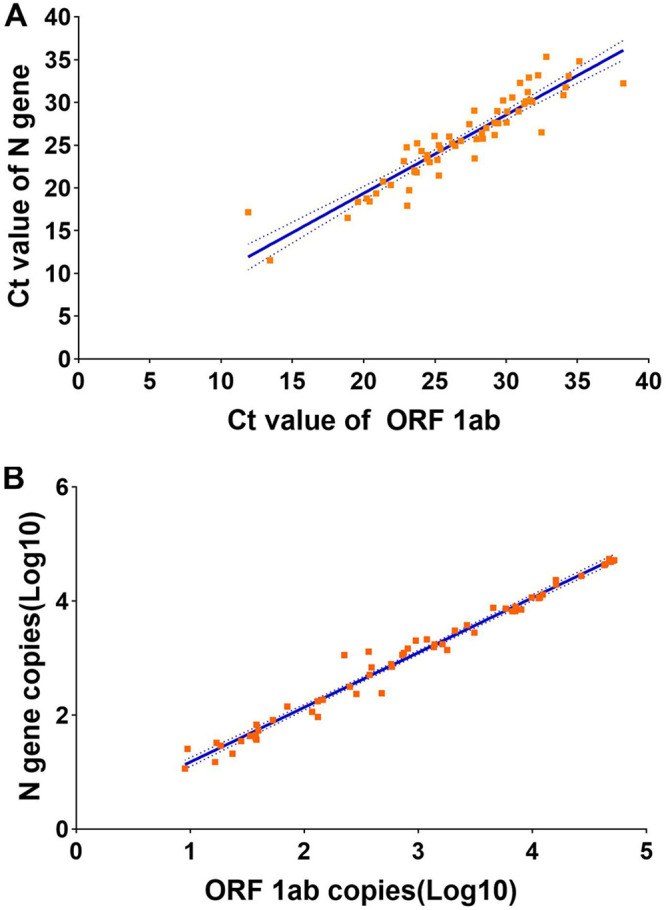
Detection of SARS-CoV-2 ORF 1ab and N gene in the original samples using RT-qPCR (A) and dPCR (B). The results of RT-qPCR were expressed in cycle threshold (CT) values. The solid lines show the linear regression of the N gene copy number against the ORF 1ab copy number. The dashed lines indicate the 95% confidence intervals of the regression line.
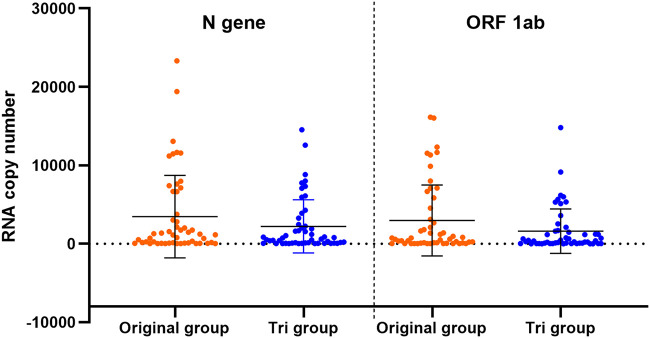
Comparison of RNA copy number between the Tri and original groups.
To investigate the influence of TRIzol LS reagent treatment on the RNA copy number, we compared the RNA copy number in the samples from the Tri and the original groups (Fig. 2). Fifty-one out of the 61 samples had viral loads of between 10 and 40,000 copies/test and were included in the analysis. Compared with the copy numbers in the original group, the N gene copy number was reduced by 47.54% (t = 2.783, P = 0.008) and the ORF 1ab copy number was reduced by 39.85% (t = 3.232, P = 0.002) in the Tri group.
FIG 2.
Comparison of RNA copy numbers (N gene and ORF 1ab) between the original group and the TRIzol treatment group (Tri). Data are shown as the mean ± SEM.
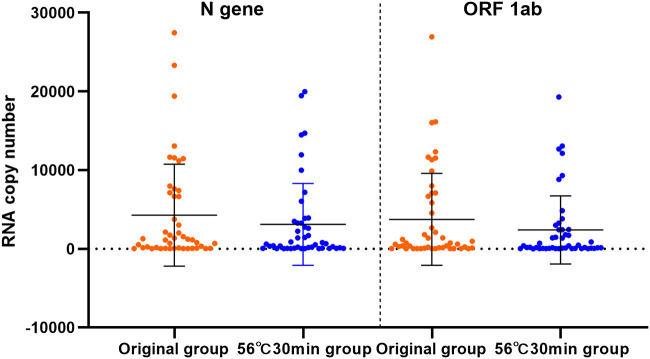
Comparison of RNA copy number between the 56°C 30 min group and the original group.
As the heating of samples at 56°C for 30 min is commonly used to inactivate SARS-CoV-2 in clinical practice, we compared the RNA copy number in the samples from the 56°C 30 min group and in those from the original group to study the effect of this treatment (Fig. 3). Forty-six out of 61 samples had viral loads of between 10 and 40,000 copies/test and were included in the analysis. In the 56°C 30 min group, the N gene and ORF 1ab copy numbers were reduced by 48.55% (t = 2.305, P = 0.026) and 56.40% (t = 2.931, P = 0.005), respectively, compared to those in the original group.
FIG 3.
Comparison of RNA copy numbers (N gene and ORF 1ab) between the original group and the 56°C 30 min treatment group. Data are shown as the mean ± SEM.
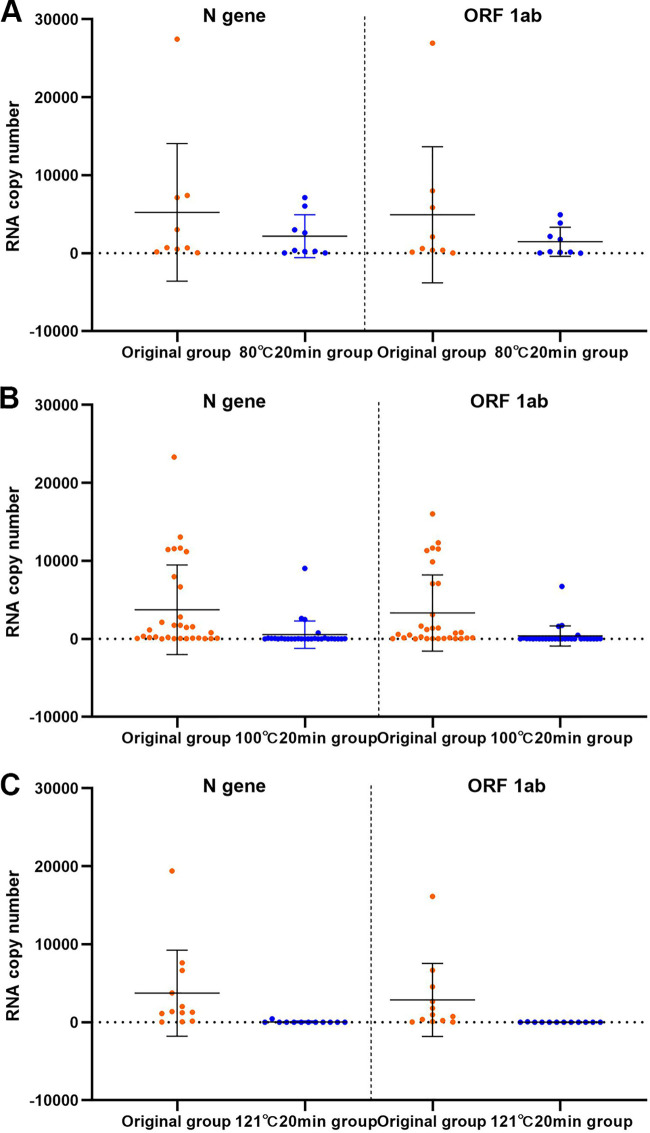
Comparison of RNA copy number between the high-temperature-treatment groups and the original group.
We also treated the samples with higher temperatures to examine the effect of high-temperature inactivation. Three temperatures were used to treat the samples for 20 min: 80°C, 100°C, and 121°C. In the 80°C 20 min group, 9 out of 16 samples had viral loads between 10 and 40,000 copies/test and were included in the analysis. Relative to the copy numbers in the original group, the N gene and ORF 1ab copy numbers in the 80°C 20 min group were reduced by 49.96% (t = 3.226, P = 0.012) and 65.96% (t = 4.504, P = 0.002), respectively (Fig. 4A).
FIG 4.
Comparison of RNA copy numbers (N gene and ORF 1ab) between the 80°C 20 min (A), 100°C 20 min (B), and 121°C 20 min (C) treatment groups and the original group. Data are shown as the mean ± SEM.
Autoclaving of the samples at 121°C or boiling at 100°C nearly destroyed all the viral particles, and almost no viral RNA was detected. Here, we plotted all the data points in Fig. 4B and C (31 samples and 14 samples for the 100°C 20 min group and the 121°C 20 min group, respectively).
DISCUSSION
The unexpected COVID-19 outbreak started at the end of 2019, and SARS-CoV-2 has been identified to be the pathogen. This virus mainly spreads through droplets and contact, and it is important to protect medical workers, including laboratory personnel, from infection. Facing a large number of suspected cases, laboratory assays were needed, but the scarcity of personal biocontainment equipment forced the use of inactivated specimens. Although viral nucleic acid detection by RT-qPCR is currently the gold standard for COVID-19 diagnosis, the reliability of this method has been questioned, as false-negative results have been reported (3, 13). We hypothesized that inactivation may contribute to the false-negative results. How inactivation affects the viral RNA copy number needs to be studied.
In this study, we evaluated three inactivation methods, including treatment with the TRIzol reagent, treatment at 56°C for 30 min, and high-temperature treatment (including autoclaving at 121°C for 20 min, boiling at 100°C for 20 min, and heating at 80°C for 20 min). The results showed that inactivation had a great impact on the amount of detectable RNA, suggesting that inactivation may cause false-negative results, especially in weakly positive cases. In addition, our results indicated that treatment with the TRIzol reagent had the least effect on the RNA copy number. TRIzol is commonly used in labs for RNA or DNA extraction and also can inactivate viruses. Mixing samples with TRIzol at a ratio of 1:3 or 1:4 at room temperature for 10 min can render suspensions of alphaviruses (western equine encephalomyelitis [WEE], eastern equine encephalomyelitis [EEE], and Venezuelan equine encephalomyelitis [VEE] viruses), flaviviruses (West Nile [WNV] and dengue [DEN] viruses), a bunyavirus (Rift Valley fever [RVF] virus), and filoviruses (Ebola and Marburg viruses) noninfectious to cultured cells, and this method is effective in inactivating Middle East respiratory syndrome coronavirus (MERS-CoV), which belongs to the same family as SARS-CoV-2 (14, 15). Therefore, we suggest using TRIzol to inactivate the virus before testing, if necessary.
The China CDC recommends that the primers and probes for detecting SARS-CoV-2 be designed to be specific for open reading frame (ORF) 1ab and the nucleocapsid protein (N) gene (http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html). We noticed that the copy number of the N gene was higher than that of ORF 1ab in any given sample. In the RT-qPCR assay, the CT value for ORF 1ab was 1.2 times larger than that of the N gene, and in dPCR, the copy number of the N gene was approximately 20% higher than that of ORF 1ab. This phenomenon has been reported for the severe acute respiratory syndrome coronavirus (SARS-CoV). A previous study found that the SARS-CoV nucleocapsid region is the ideal amplification target and exhibits the highest detection sensitivity. The N gene-specific primer set could detect as little as 1 copy of viral RNA and was more sensitive than primer sets specific for the spike and membrane genes (16). The authors of that study suggested that primer sets for detecting the SARS-CoV N region should be the first choice for highly sensitive detection.
dPCR is an important tool for nucleic acid analysis. The advantages of dPCR include absolute quantification without a calibration curve, high precision, and high accuracy even in the presence of inhibitors (5). dPCR and reverse transcription-dPCR have been used to determine the copy numbers of DNA and RNA viruses, such as human immunodeficiency virus (HIV), cytomegalovirus, hepatitis B virus, JC polyomavirus, human papillomavirus, human T-cell lymphotropic virus, and human rhinoviruses, in a variety of clinical specimens (7–12, 17, 18). Here, we employed dPCR to determine the RNA copy numbers of SARS-CoV-2 in the original nasopharyngeal swab samples and the inactivated samples and to evaluate the influence of different inactivation methods on the viral copy number. The targets of the dPCR detection kit include the ORF 1ab, the N gene, and a reference gene. According to the manufacturer’s instructions, the limit of blank (LoB), the limit of detection (LoD), and the limit of quantitation (LoQ) are 0 copy/reaction mixture, 5 copies/reaction mixture, and 50 copies/reaction mixture, respectively. The copy numbers of the ORF 1ab and the N gene in 2 out of the 63 samples were lower than the LoD, so the results for these two samples were classified as negative. This result was not consistent with the RT-qPCR results. Unfortunately, the cause of the false-negative results cannot be confirmed at this time, and we will investigate it in the future.
This study had some limitations. First, it is impossible to have the same RNA extraction rate for different samples. We used the same machine and the same lot of reagents and strictly followed the manufacturer’s instructions to minimize differences between the groups. Second, because of the limited biosafety biocontainment level, we did not isolate the virus to test the viral titer. Third, only one inactivation reagent (TRIzol) was evaluated in this study, so other reagents, such as AVL buffer, should be included in future studies. Fourth, our results suggest that the dPCR platform for SARS-CoV-2 should be further improved for a wider dynamic range.
In summary, we investigated the influence of clinical sample inactivation on the SARS-CoV-2 RNA copy number via dPCR. Inactivation of viruses reduced the amount of detectable viral RNA and may cause false-negative results. Nevertheless, TRIzol is suggested to be used for sample inactivation, as it had the least effect on the RNA copy number among the tested inactivation methods.
ACKNOWLEDGMENTS
This work was funded by emergency prevention and control projects for COVID-19 of the Xi’an Science and Technology Bureau (grant 20200002YX002-2) and the Science Technology+Action Plan-Medical Research Project of the Xi’an Science and Technology Bureau (grant 2019115713YX012SF050).
We gratefully acknowledge the medical workers for their arduous work in the treatment of COVID-19 patients and collecting the specimens and the Xi'an CDC staff for the epidemiological survey.
We declare no competing financial interest.
REFERENCES
- 1.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for Covid-19. 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon L. 2020. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Kang H, Liu X, Tong Z. 2020. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J Med Virol 92:538–539. doi: 10.1002/jmv.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B, Kinzler KW. 1999. Digital PCR. Proc Natl Acad Sci U S A 96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiss MM, Ortoleva-Donnelly L, Beer NR, Warner J, Bailey CG, Colston BW, Rothberg JM, Link DR, Leamon JH. 2008. High-throughput quantitative polymerase chain reaction in picoliter droplets. Anal Chem 80:8975–8981. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basu AS. 2017. Digital assays part I: partitioning statistics and digital PCR. SLAS Technol 22:369–386. doi: 10.1177/2472630317705680. [DOI] [PubMed] [Google Scholar]
- 7.Huang JT, Liu YJ, Wang J, Xu ZG, Yang Y, Shen F, Liu XH, Zhou X, Liu SM. 2015. Next generation digital PCR measurement of hepatitis B virus copy number in formalin-fixed paraffin-embedded hepatocellular carcinoma tissue. Clin Chem 61:290–296. doi: 10.1373/clinchem.2014.230227. [DOI] [PubMed] [Google Scholar]
- 8.Giovannelli I, Ciccone N, Vaggelli G, Malva ND, Torricelli F, Rossolini GM, Giannecchini S. 2016. Utility of droplet digital PCR for the quantitative detection of polyomavirus JC in clinical samples. J Clin Virol 82:70–75. doi: 10.1016/j.jcv.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Jeannot E, Becette V, Campitelli M, Calmejane MA, Lappartient E, Ruff E, Saada S, Holmes A, Bellet D, Sastre-Garau X. 2016. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. J Pathol Clin Res 2:201–209. doi: 10.1002/cjp2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunetto GS, Massoud R, Leibovitch EC, Caruso B, Johnson K, Ohayon J, Fenton K, Cortese I, Jacobson S. 2014. Digital droplet PCR (ddPCR) for the precise quantification of human T-lymphotropic virus 1 proviral loads in peripheral blood and cerebrospinal fluid of HAM/TSP patients and identification of viral mutations. J Neurovirol 20:341–351. doi: 10.1007/s13365-014-0249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lui YL, Tan EL. 2014. Droplet digital PCR as a useful tool for the quantitative detection of enterovirus 71. J Virol Methods 207:200–203. doi: 10.1016/j.jviromet.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR. 2012. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J Virol Methods 186:68–72. doi: 10.1016/j.jviromet.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders D, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25(3):pii=2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blow JA, Dohm DJ, Negley DL, Mores CN. 2004. Virus inactivation by nucleic acid extraction reagents. J Virol Methods 119:195–198. doi: 10.1016/j.jviromet.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Kumar M, Mazur S, Ork BL, Postnikova E, Hensley LE, Jahrling PB, Johnson R, Holbrook MR. 2015. Inactivation and safety testing of Middle East respiratory syndrome coronavirus. J Virol Methods 223:13–18. doi: 10.1016/j.jviromet.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang JL, Lin HT, Wang YM, Yeh YC, Peck K, Lin BL, Liu HW, Chen A, Lin CS. 2005. Rapid and sensitive detection of multiple genes from the SARS-coronavirus using quantitative RT-PCR with dual systems. J Med Virol 77:151–158. doi: 10.1002/jmv.20432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayden RT, Gu Z, Ingersoll J, Abdul-Ali D, Shi L, Pounds S, Caliendo AM. 2013. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J Clin Microbiol 51:540–546. doi: 10.1128/JCM.02620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sedlak RH, Nguyen T, Palileo I, Jerome KR, Kuypers J. 2017. Superiority of digital reverse transcription-PCR (RT-PCR) over real-time RT-PCR for quantitation of highly divergent human rhinoviruses. J Clin Microbiol 55:442–449. doi: 10.1128/JCM.01970-16. [DOI] [PMC free article] [PubMed] [Google Scholar]