Molecular testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the gold standard for diagnosis of coronavirus disease 2019 (COVID-19), but the clinical performance of these tests is still poorly understood, particularly with regard to disease course, patient-specific factors, and viral shedding. From 10 March to 1 May 2020, NewYork-Presbyterian laboratories performed 27,377 SARS-CoV-2 molecular assays from 22,338 patients. Repeat testing was performed for 3,432 patients, of which 2,413 had initial negative and 802 had initial positive results.
KEYWORDS: COVID-19, SARS-CoV-2, sensitivity, laboratory utilization, negative predictive value
ABSTRACT
Molecular testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the gold standard for diagnosis of coronavirus disease 2019 (COVID-19), but the clinical performance of these tests is still poorly understood, particularly with regard to disease course, patient-specific factors, and viral shedding. From 10 March to 1 May 2020, NewYork-Presbyterian laboratories performed 27,377 SARS-CoV-2 molecular assays from 22,338 patients. Repeat testing was performed for 3,432 patients, of which 2,413 had initial negative and 802 had initial positive results. Repeat-tested patients were more likely to have severe disease and low viral loads. The negative predictive value of the first-day result among repeat-tested patients was 81.3% The clinical sensitivity of SARS-CoV-2 molecular assays was estimated between 58% and 96%, depending on the unknown number of false-negative results in single-tested patients. Conversion to negative was unlikely to occur before 15 to 20 days after initial testing or 20 to 30 days after the onset of symptoms, with 50% conversion occurring at 28 days after initial testing. Conversion from first-day negative to positive results increased linearly with each day of testing, reaching 25% probability in 20 days. Sixty patients fluctuated between positive and negative results over several weeks, suggesting that caution is needed when single-test results are acted upon. In summary, our study provides estimates of the clinical performance of SARS-CoV-2 molecular assays and suggests time frames for appropriate repeat testing, namely, 15 to 20 days after a positive test and the same day or next 2 days after a negative test for patients with high suspicion for COVID-19.
INTRODUCTION
Coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1, 2), is a global pandemic with mortality significantly higher than that of seasonal influenza (3). The rapid sequencing of SARS-CoV-2 genomes (1, 2) has allowed the development of multiple real-time reverse transcription-PCR (RT-PCR) assays that have become the gold standard to detect viral RNA and identify patients with COVID-19 as well as asymptomatic carriers. However, some patients with positive chest radiologic findings and symptoms suspicious of COVID-19 have been reported to test negative by SARS-CoV-2 RT-PCR and require multiple consecutive tests to convert to a positive result (4, 5). There is limited information on the clinical performance characteristics of the SARS-CoV-2 molecular tests in the clinical setting, in particular regarding the predictive value of a negative result in patients suspected of COVID-19 and the relationship between the course of the disease, viral shedding, and positivity of the various molecular assays. During the 2020 pandemic of COVID-19, we tested 27,377 samples from 22,338 patients with SARS-CoV-2 molecular assays performed at NewYork-Presbyterian (NYP) clinical laboratories. The assays used for this patient population included six platforms based on nucleic acid amplification to detect SARS-CoV-2-specific RNA sequences. Our goal was to determine the clinical performance characteristics of SARS-CoV-2 molecular assays for diagnosis and stratification of COVID-19 patients and to determine the dynamics of SARS-CoV-2 molecular assay results over time in a large data set from multiple hospital locations in the New York City area.
MATERIALS AND METHODS
Laboratory data set.
We extracted records of all the SARS-CoV-2 tests performed at NYP laboratories from our laboratory information system (Cerner Millennium; Cerner Corporation, North Kansas City, MO) using a custom Cerner Command Language query. We tested a total number of 22,338 patients between 10 March 2020 and 1 May 2020 for SARS-CoV-2 by molecular assays at NewYork-Presbyterian-affiliated hospitals and performed a total of 27,377 assays on nasopharyngeal (initially also on oropharyngeal) swab samples. The majority of the SARS-CoV-2 tests were RT-PCR assays performed with the high-throughput automated cobas 6800 (Roche Molecular Systems, Inc., Branchburg, NJ) platform (n = 19,195), which went live at the Columbia University Irving Medical Center-NYP laboratory on 15 March 2020 and at the Weill Cornell Medical Center-NYP laboratory on 30 March 2020. In-house-developed assays that received United States Food and Drug Administration emergency use authorization were performed using the using the Rotor-Gene Q (n = 1,795; Qiagen, Valencia, CA) and 7500 Fast (n = 89; Thermo Fisher Scientific, Waltham, MA) instruments beginning 10 March 2020. From 3 April 2020, additional platforms were introduced, including the Xpert Xpress SARS-CoV-2 test analyzed on the GeneXpert (n = 265) and Infinity (n = 5,954) platforms (Cepheid, Inc., Sunnyvale, CA), the ID NOW instrument (n = 53) (Abbott, Scarborough, ME), and the Panther Fusion instrument (n = 26) (Hologic Inc., San Diego, CA). Commercial molecular viral assays were validated and performed at NYP hospital laboratories following manufacturers’ recommendations.
The cobas SARS-CoV-2 RT-PCR assay amplifies two specific targets: target 1 is located in the ORF1ab nonstructural region that is unique to SARS-CoV-2, and target 2 is a conserved region of the structural protein envelope E gene common to all members of the Sarbecovirus subgenus of coronavirus, which include SARS-CoV-2 and SARS-CoV (6, 7). The cobas SARS-CoV-2 RT-PCR assay also includes an internal control for assay performance that has no homology to the coronaviruses. For Cepheid assays, target 1 is the SARS-CoV-2-specific N2 gene and target 2 is the pan-Sarbecovirus E gene. The Panther Fusion assay amplifies two targets in the ORF1ab region. Both in-house tests are variants of the Centers for Disease Control and Prevention SARS-CoV-2 assay and amplify two regions of the SARS-CoV-2 N gene (N1 and N2). The Abbott ID NOW uses isothermal nucleic acid amplification of a unique region of the RdRp (RNA-dependent RNA polymerase) region to detect SARS-CoV-2 RNA.
Results were reported as “detected,” “not detected,” “indeterminate,” and “invalid.” “Indeterminate” results were considered when target 1 was negative but target 2 was positive in the cobas and Xpert Xpress SARS-CoV-2 RT-PCR assays or when only one of the two N gene targets was positive. “Invalid” results were reported when the internal control failed to amplify.
Patients.
The NYP laboratory SARS-CoV-2 testing data set contains basic demographic information on each patient, including age, gender, race, and health care encounter type and location at the time of order. To better understand SARS-CoV2-2 test utilization and investigate the association of clinical information with the SARS-CoV-2 test results for a subset of patients, we merged our laboratory data set with the Columbia COVID-19 CARE clinical database. The COVID-19 CARE clinical database is an interdisciplinary database managed by the Columbia Division of Infectious Diseases and contains data on all patients tested for SARS-CoV-2 at the NewYork Presbyterian West Campus (Milstein Hospital, Allen Hospital, and Morgan Stanley Children’s Hospital). This interdisciplinary database contains the work of numerous divisions within the Departments of Medicine, Pediatrics, Neurology, and Obstetrics and Gynecology. The database is composed of a general primary instrument and multiple subspecialty instruments, resulting in over 720 curated fields per patient. Data were extracted from the medical record through manual review by a team of medical students and subspecialty fellows and attendings. Manually curated data were combined with structured data from the electronic medical records, pharmacy, and laboratory systems to complete the data set. As of 1 May 2020, 1,624 patients had undergone manual review (see Table S1 in the supplemental material).
Clinical factors and other patient characteristics are compared between SARS-CoV-2-positive and SARS-CoV-2-negative patients in Table S2 and between repeat-tested and single-tested patients in Table S3. This study was approved by the Institutional Review Boards of Columbia University and Weill Cornell Medicine.
Data analysis.
Categorical data are presented as frequencies and percentages. Continuous data are presented as means (standard deviations) or medians and ranges. Differences between groups were compared using chi-squared tests for categorical variables and linear analysis of variance (ANOVA) for continuous variables. Correction for multiple testing was performed using the false-discovery-rate method of Benjamini and Hochberg (8). Confidence intervals for clinical sensitivity and negative predictive values were calculated using the epiR R package. Times to event, determined using Kaplan-Meier curves, were used to investigate time to conversion from initially negative to positive and from initially positive to negative. All statistical analyses were performed using the R statistical language, version 3.6.3 (9).
RESULTS
A total of 18,906 assays were performed once per patient, and 8,471 assays were performed for 3,432 (15.4%) patients, with tests being repeated over a span of 1 to 49 (median = 8) days between assays. There were 2,630 patients with multiple SARS-CoV-2 molecular assays performed who had an initial negative, invalid, or indeterminate result and 802 patients who had an initial positive result.
Demographic and clinical characteristics.
Patients with any positive SARS-CoV-2 molecular assay result differed from those that were never positive for SARS-CoV-2 even with repeat testing (Table S2). Notably, SARS-CoV-2-positive patients were more likely to be male, older than 44 years, and of self-reported African-American or Hispanic/Latino ethnicity and less likely to be Asian or Caucasian (Table S2) (all P < 0.001). Not surprisingly, severe disease was more likely in SARS-CoV-2-positive patients, as indicated by a higher fatality rate (6.2% versus 3.8%; P = 0.048), presence of symptoms (91.8% versus 72.4%; P = 0.004), need for intubation (10.7% versus 7%; P = 0.026), and frequency of decompensation, as defined by an outcome of intubation, death, or discharge to hospice (13.4% versus 8.4%; P = 0.005).
To determine which factors were associated with repeat testing for SARS-CoV-2, we analyzed demographic and clinical features of repeat-tested compared to single-tested patients (Table S3). Our data show higher age (median = 59.9 versus 53.4; P < 0.001), higher frequency of male gender (52.2% versus 44.3%; P < 0.001), and different distribution of self-reported race and ethnicity in repeat-tested compared to single-tested patients, with African-Americans and Hispanics/Latinos being more likely to be repeat-tested (P < 0.001). At the time of the first test order, admitted patients were significantly more highly represented in the repeat-tested population in contrast to patients visiting the emergency department and outpatients (P < 0.001). Compared with single-tested patients, repeat-tested patients were 3.2 times more likely to be admitted to the intensive care unit (29.7% versus 9.4%; P < 0.001), 4.0 times more likely to be intubated (24.4% versus 6.1%; P < 0.001), 3.4 times more likely to decompensate (27.6% versus 8.1%; P < 0.001), and 1.7 times more likely to die during the observation period (8.0% versus 4.7%; P = 0.038).
SARS-CoV-2 test performance.
The characteristics of the SARS-CoV-2 tests performed for repeat-tested compared to single-tested patients are described in Table S4. The vast majority of tests were performed with the cobas 6800, and the use of the various assays was not significantly different between repeat- and single-tested patients, except for a small number of patients initially tested with the Thermo Fisher 7500 assay, which was more likely in repeat-tested patients (0.6% versus 0.2%; P < 0.001, standardized Pearson residuals for repeat-tested patients = 3.9).
Among all the repeat-tested patients, 23.4% were positive on the first test (26.7% when indeterminate results were included). When a negative test was repeated on the first day of testing, the positivity rate increased to 27.7% (29.7% with indeterminate results). Among patients that converted from negative to positive on the same day, the mean interval between sample collections was 10.8 ± 6.0 h. In only 3 cases were the same samples retested, of which two were initially negative by the Abbott ID NOW COVID-19 test and one was initially negative by the Cepheid Xpert Xpress assay. All other repeat tests were performed on a subsequently collected sample.
Overall positivity among repeat-tested patients over the course of the study period was 39.9%, in contrast to 49.0% for single-tested patients (P < 0.001). When indeterminate results were counted as positive, 42.9% of repeat-tested patients were positive over the course of the study period, in contrast to 50.0% of single-tested patients (P < 0.001).
Indeterminate results are generally considered presumptively positive and occur when the SARS-CoV-2-specific target is negative and the pan-Sarbecovirus target is positive. In our repeat-tested patients with an initial result of “indeterminate,” 53.9% ultimately had a result of “detected,” compared to 7.0% that remained indeterminate upon repeat testing and 39.1% that converted to a negative status during our study period, thus suggesting that it is acceptable to consider these patients positive. It is unlikely that the initially indeterminate results for patients that subsequently tested negative were false positives, as SARS-CoV is not in circulation; therefore, detection of the pan-Sarbecovirus locus is essentially specific to SARS-CoV-2. It is rather more likely that the patient presented with low viral loads, possibly in the recovery phase of a mild infection.
In contrast, a result of “invalid” reflects the failure to amplify the internal control and is likely related to poor sampling or inadequate RNA extraction usually due to high viscosity of the sample. In our study of repeat test patients, 52.3% ultimately became positive, 1.3% had a repeat-test result of “indeterminate,” and 46.0% had a repeat-test result of “not detected.” For the analysis of clinical sensitivity of SARS-CoV-2 molecular tests, we counted patients with invalid results as “negative,” as these results can be considered clinically false negatives in the sense that the patient may be infected and the test failed to yield a positive result. In practice, tests yielding invalid results should always be repeated, preferably with a new sample, as the results are unpredictable.
Initial negative, invalid, or indeterminate SARS-CoV-2 test results were much more frequent among repeat-tested patients (Table S4; P < 0.001), and patients without a positive initial result were more likely to be repeat-tested (21%) than initially positive patients (8%; P < 0.001).
A subset of the cobas 6800 tests (n = 5,343) had cycle threshold (CT) values available for analysis. The CT represents the PCR cycle, interpolated to two decimal digits, at which the fluorescent signal crosses a predefined threshold for positivity. The CT is inversely proportional to the viral load.
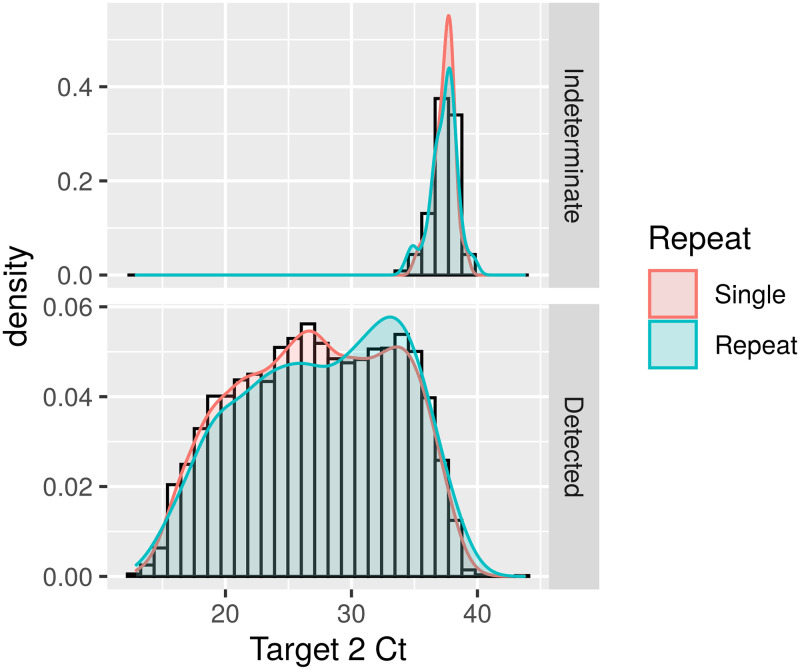
When positive and indeterminate results from repeat- and single-tested patients were compared, there were no differences in indeterminate CT values, which is expected, as by definition the indeterminate results represent high CT values. In contrast, the repeat-tested group (n = 795) had significantly higher target 2 CT values than single-tested patients (n = 4,548) (median = 29.1 versus 27.3; P < 0.001) and a frequency of target 2 CT values above 30 (45.8% versus 36.9%; P < 0.001), indicating lower viral loads in the repeat-tested samples (Fig. 1 and Table S4).
FIG 1.
Density distribution of cobas SARS-2-CoV-2 target 2 (E gene, pan-Sarbecovirus target) CT values in repeat-tested versus single-tested patients. (Top) CT values from results reported as “intermediate”; (bottom) CT values from results reported as “detected.”
Analysis of conversion rates of repeat-tested patients.
In this study, we classified repeat-tested patients in two groups according to their initial SARS-CoV-2 results: those that were initially positive, for whom repeat testing was most likely intended to ascertain recovery and noninfectiousness, and those that had an initial result of negative, indeterminate, or invalid, in whom persistent clinical suspicion for COVID-19 likely motivated ordering of the repeat test. Table S4 shows the different clinical characteristics between repeat- and single-tested patients.
For the time-dependent analysis of conversion rates, we considered “initially positive” patients with any “detected” or “indeterminate” SARS-CoV-2 result obtained during the first calendar day of testing rather than the first positive test, to reduce bias due to nasopharyngeal sampling inadequacy (Table 1). Conversely, patients without a result of “detected” or “indeterminate” on the first day were labeled as “initially negative.” Among the 2,413 initially negative repeat-tested patients, 18.6% became positive upon repeat testing on subsequent days (Table S5), indicating a negative predictive value of 81.3% (95% confidence interval [CI] = 79.7 to 82.8) in this repeat-tested population, with an average prevalence of 43% at the time of the study.
TABLE 1.
Number of SARS-CoV-2 molecular test results over the course of repeat testing, grouped by the highest test result on day 1
| Highest day 1 result | No. (%) with highest result (any day) |
||||
|---|---|---|---|---|---|
| Invalid (n = 1) | Not detected (n = 1,960) | Indeterminate (n = 100) | Detected (n = 1,371) | Totala (n = 3,432) | |
| Invalid | 0 (0.0) | 0 (0.0) | 0 (0.0) | 79 (5.8) | 79 (2.3) |
| Not detected | 1 (100) | 1,960 (100) | 38 (38.0) | 335 (24.4) | 2,334 (68.0) |
| Indeterminate | 0 (0.0) | 0 (0.0) | 62 (62.0) | 7 (0.5) | 69 (2.0) |
| Detected | 0 (0.0) | 0 (0.0) | 0 (0.0) | 950 (69.3) | 950 (27.7) |
P < 0.001, Pearson’s chi-squared test (adjusted for multiple comparisons).
In a separate analysis, we compared the results of the first test with the results from tests repeated on the same day (Table S5) or repeated any time after the first result (Table S6). Among the patients with repeat testing who had initial results of “invalid” (241), “not detected” (2,280), or “indeterminate” (114), 5.6% had “detected” results upon repeat testing on the first day of testing (Table S5). This increase in positivity rate most likely results from a false-negative initial test due to preanalytic factors, such as sample inadequacy, incorrect swabbing technique, or stochastic sampling bias from low viral loads in the patient nasopharynx. After the first day of testing, repeated testing after invalid, negative, or indeterminate results on the same day resulted in about 0.6% additional positive results per day, for a total of 17.0% positives that were missed by the first test.
Among the 1,371 repeat-tested patients with one or more SARS-CoV-2 results of “detected,” which can be assumed to be truly infected, only 58.2% had the result “detected” on the initial test (Table S5), and only 69.3% were reported as “detected” on the first day (Table 1). Considering “detected” and “indeterminate” as positive, 1,471 repeat-tested patients had one or more SARS-CoV-2-positive results over time; only 61.9% were positive on the initial test, and only 69.3% had a positive result on the first day (Table 1). These data provide an estimate of the clinical sensitivity of the assay in the repeat-tested population and establish a baseline to look at rates of conversion from negative to positive.
Table 2 shows the number of patients who had an initial result of “not detected” or "indeterminate" on day 1 and who converted to a SARS-CoV-2-positive status, grouped by time after the initial test, as well as the number of repeat tests in patients with a status of “detected” on day 1. Figure S1 shows the density and cumulative distributions per day after onset of symptoms (Fig. S1a and b) or after initial testing (Fig. S1c to f) of conversions from positive to negative (left) and from negative to positive (right) in the two groups of repeat-tested patients. For this analysis, positive status included “detected” and “indeterminate” and negative status included results of “not detected,” with “invalid” results being excluded. Among the initially positive patients, the unadjusted distributions show a peak of conversion to negative between 30 and 40 days after symptoms or around 20 days after initial testing. Less than 10% of the patients who converted to negative converted before 15 days after onset of symptoms or 10 days after initial testing (Fig. S1a and c, respectively). In general, there were no significant differences in the last CT values between patients who converted to negative and patients whose last result was positive (results not shown). However, those that became negative in less than 10 days had mean CT values significantly higher than those that converted after 10 days (target 1, 22.6 ± 6.4 versus 27.5 ± 6.5; target 2, 23.6 ± 7.0 versus 29.1 ± 7.7; both P < 0.001 [results not shown]). In contrast, among the patients with initially negative results who converted to positive, most conversions occurred 10 days or less after onset of symptoms and in the first 1 to 3 days after initial testing (Fig. S1b and d, respectively). Whereas the mean target 1 CT values of the first positive result in initially negative patients (26.5 ± 6.4) were comparable to the mean first CT values for single-tested patients (26.2 ± 5.5), the mean target 2 CT values (28.0 ± 7.4) were significantly higher than the mean first CT values of single-tested patients (27.3 ± 6.1; P < 0.001) (Table S4 and results not shown).
TABLE 2.
Distribution of repeat tests per day after a first-day result of “not detected” or “indeterminate” or after a first-day result of “detected”
| First-day result | Day of testing | No. (%a
) of repeat-test results |
|||
|---|---|---|---|---|---|
| Not detected | Indeterminate | Detected | Total | ||
| Not detected or indeterminate | 1 | 1,995 (97.4) | 54 (2.6) | 0 (0.0) | 2,049 |
| 2 | 556 (80.1) | 39 (5.6) | 99 (14.3) | 694 | |
| 3–6 | 601 (87.0) | 7 (1.0) | 83 (2.0) | 691 | |
| 7–9 | 327 (89.6) | 6 (1.6) | 32 (8.8) | 365 | |
| 10–15 | 469 (87.0) | 6 (1.1) | 64 (11.9) | 539 | |
| >16 | 565 (86.3) | 6 (0.9) | 84 (12.8) | 655 | |
| Detected | 1 | 0 (0.0) | 46 (5.8) | 754 (94.2) | 800 |
| 2 | 5 (7.9) | 2 (3.2) | 56 (88.9) | 63 | |
| 3–6 | 7 (11.3) | 1 (1.6) | 54 (87.1) | 62 | |
| 7–9 | 12 (16.4) | 6 (8.2) | 55 (75.3) | 73 | |
| 10–16 | 43 (20.2) | 19 (8.9) | 151 (70.9) | 213 | |
| 16–20 | 74 (40.9) | 16 (8.8) | 91 (50.3) | 181 | |
| 21–30 | 225 (57.7) | 35 (9.0) | 130 (33.3) | 390 | |
| >30 | 148 (69.5) | 13 (6.1) | 52 (24.4) | 213 | |
Percentages are the proportion of each result relative to the total tests for that day.
Since we cannot be certain about the conversion rates due to a significant proportion of repeat-test patients having insufficient testing performed to detect conversion (right-censoring), we used a Kaplan-Meier approach to estimate the conversion rate by day of testing (Fig. 2 and 3 and Fig. S1e to h) with the following assumptions. (i) When there were multiple tests performed per patient in one day, the results were aggregated to the highest result, i.e., “detected” > “indeterminate” > “not detected.” (ii) For initially positive patients, an event was defined as the highest result in a day of “not detected.” (iii) Conversely, an event was defined as conversion from a “not detected” or “invalid” result at day 1 to “detected” or “indeterminate” on subsequent days. (iv) If the last test result was unchanged relative to the first-day result, the patient was considered right-censored at that time. (v) Only the first day and either the censoring day or the event day were used for each patient, and indeterminate results were ignored.
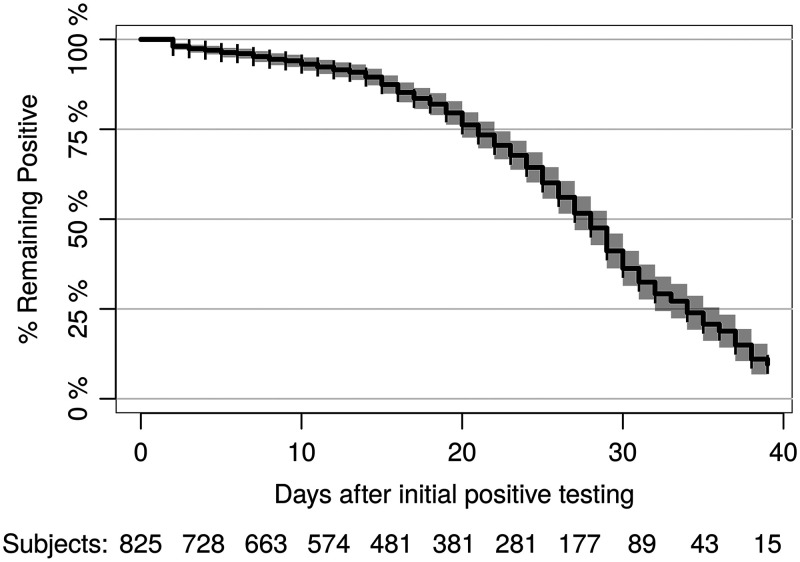
FIG 2.
Kaplan-Meier estimate of conversion from an initially positive SARS-CoV-2 status on day 1 to a subsequent negative result. The number of patients at risk is shown at the bottom for each time point after removal of censored patients, represented by vertical ticks in the curve.
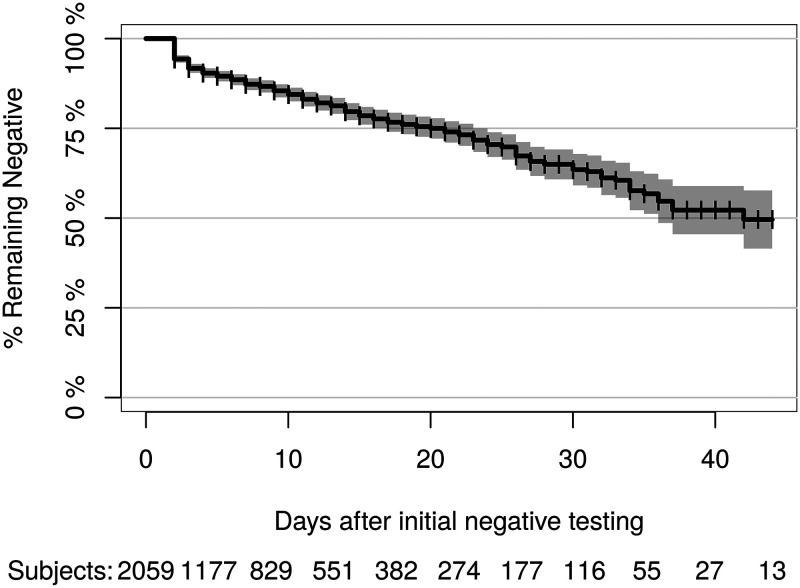
FIG 3.
Kaplan-Meier estimate of conversion rate from initially negative SARS-CoV-2 status on day 1 to a subsequent positive result.
The results show that the probability of converting from positive to negative in the initially positive, repeat-tested population is minimal until about 15 to 20 days after initial testing and reaches 50% at 28 days (95% CI = 27 to 29 days) (Fig. 2). In the initially negative repeat-tested population, the risk linearly increased every day, with a 25% probability of conversion to positive at 20 days (95% CI = 17 to 23 days) (Fig. 3).
Since lack of conversion could be due to death of the patient, which occurred in approximately 8% of the repeat-tested population, we performed a competing risk analysis with death as the alternative event, using the timereg R package (10), and did not find significant differences in either the positive-to-negative or the negative-to-positive Kaplan-Meier cumulative probability of conversion (results not shown).
Interestingly, we identified 11 patients that converted from an initial SARS-CoV-2-negative result to positive and back to negative over several days (Fig. S2A). This pattern may be expected in the course of infection, especially for patients with low viral loads near the limit of detection. However, we also observed some unusual patterns, such as patient 5, who repeatedly tested negative for 35 days, then tested indeterminate at day 36, negative at day 46, positive at day 48, and negative at day 49. We also identified 49 initially positive patients who converted to negative and later reverted to positive again (Fig. S2B). These cases likely represent persistence of viral RNA at low levels and demonstrate that previous repeat negative results cannot completely rule out a subsequent positive test.
DISCUSSION
Our results from repeat-tested patients can be used to estimate the clinical sensitivity of the SARS-CoV-2 molecular testing in the population of patients that were selected for repeat testing, because a subsequent positive result is almost definitive evidence that the patient was infected. In all likelihood, most repeat testing on initial negative patients was performed either to follow up a history of exposure, when the clinical profile did not fit the initial results, or when clinical presentation deteriorated after the initial result. Consistent with this hypothesis, repeat-tested patients were more likely to be older, male, and of non-Caucasian race than single-tested patients (Table S3), consistent with the demographics of COVID-19-positive patients (Table S2). Importantly, repeat-tested patients had worse outcomes, as demonstrated by higher rates of decompensation, intubation, and mortality. Interestingly, repeat-tested patients who converted from positive to negative tended to be younger and present as outpatients, compared to patients that remained positive (Table S7), but there were no significant differences in gender, ethnicity, race, or clinical outcomes. In contrast, repeat-tested patients that remained negative tended to be female, younger, and inpatients and to have longer admission duration and were more likely to be extubated than those who converted to positive (Table S8), suggesting that a significant number of repeat tests for SARS-CoV-2-negative patients were performed for inpatients admitted before the pandemic or for non-COVID-19 reasons. Indeed, repeat-tested patients admitted before 1 March 2020 were much more likely to have a persistently negative result (71.4%) than those admitted after 1 March 2020 (40.0%; P < 0.001).
In the absence of a more sensitive gold standard, the repeat-tested patients with an eventual positive result can be considered true positives, as the analytical specificity of molecular testing is very high (3, 6, 7, 11, 12). It may be tempting to add the 9,272 single-tested positive patients to the 1,371 repeat-tested positive patients to determine clinical sensitivity. However, we do not know how many of the “not detected” results for single-tested patients are false negatives, especially given the high frequency of asymptomatic or mildly symptomatic COVID-19 patients (13–15). Therefore, considering only the positive patients will inflate the estimated clinical sensitivity. Nevertheless, if we consider all negative results (repeated or not) to be true negative (i.e., a specificity of 100%), we can estimate the upper bound of the clinical sensitivity of a first initial result to be 94.6% (95% CI = 94.2 to 95.0%). If the test is repeated on the first testing day to account for nasopharyngeal sampling inadequacy, the upper boundary of the estimated sensitivity with these assumptions would be 96.0% (95% CI = 95.7 to 96.4%). A lower bound of clinical sensitivity can be estimated by considering the worst-case scenario that the same percentage of false negatives identified in repeat-tested patients would apply to the general population. With these assumptions, the clinical sensitivity of the SARS-CoV-2 assay can be estimated to be 57.9% (95% CI = 55.2 to 60.5) to 94.6% for a single initial test or 69.3% (95% CI = 66.8 to 71.7) to 96.0% when first-day repeat testing is considered.
The lower clinical sensitivity of the first-day results in the ultimately positive repeat-tested patients suggests several possibilities. (i) Viral shedding increases over time in a recently infected patient and will eventually cross the detection threshold in subsequent samples. This possibility is suggested by the lower CT values for target 1 and particularly target 2 for the repeat-tested patients (Table S4 and Fig. 1). (ii) A significant number of samples are improperly collected, and repeat testing increases the probability of detection; this is particularly likely in the first day of testing, when suspicion may be high but the initial test results are negative or inconclusive, as shown in Table S5. (iii) Initially tested patients were truly negative and acquired the infection nosocomially after admission.
There were no significant differences in mean testing interval in initially negative patients who converted to positive (6.1 ± 8.1 days) versus those that remained negative (6.5 ± 7.6 days) (Table S8). The mean interval between an initial negative test result and the first positive result in patients who converted in our study was 9.4 days (95% CI = 8.4 to 10.5 days; n = 335), which is longer than that reported by Ai et al., who showed a mean interval between initial negative and positive RT-PCR results of 5.1 ± 1.5 days, with a median of 4 days (n = 15) (4). We have insufficient data to calculate incubation time, as only 1.3% of the patients developed symptoms after the first test, with a median time between testing and symptoms of 1.3 days. The median interval between start of symptoms and the first test in our study of 4.8 days (95% CI = 4.5 to 5.4 days) is in line with the data from Lauer et al., who calculated a median incubation time of 5.1 days (95% CI = 4.5 to 5.8 days) in patients from China (16).
The third possibility, i.e., hospital-acquired infection explaining a low rate of initial true negative results, is less likely, because patients who converted from negative to positive were less likely to be inpatients, had shorter intervals between admission and the first test, and had shorter duration of admission than those who remained negative (Table S8); also, there were no significant differences between repeat-tested inpatients and outpatients in the average time interval between the initial negative test and the first positive test (results not shown). If there were a large number of inpatients acquiring the infection in the hospital, one would expect longer intervals between admission and positivity due to the cohort of patients already admitted before the pandemic, compared to patients recently admitted with COVID-19. While symptomatic rates were high in both cohorts, our data did not demonstrate a difference in severe clinical outcome between persistently negative patients and patients that converted from negative to positive (Table S8). These data suggest a pattern of repeated test ordering for uninfected inpatients with symptoms suggestive of COVID-19 but with a lower likelihood of conversion.
Our analysis of repeat-tested patients with an initial positive result, using the Kaplan-Meier estimator, indicates that conversion to a negative result is unlikely to occur until about 15 to 20 days after initial testing or 20 to 30 days after start of symptoms, when the odds ratio significantly increases (Fig. 2 and Fig. S1). Conversely, when a patient is suspected of COVID-19 but the initial test is negative, repeating the test steadily increases the probably of conversion to “detected” every day (Fig. 3). Repeat testing is especially likely to yield a positive result if the initial test is indeterminate or invalid. For indeterminate results, this likely reflects low levels of the virus in the sample or low viral shedding in the nasopharynx early or late in the course of the infection. For invalid results, this likely reflects sampling inadequacy, often due to excess mucous in the sample. Our finding of significantly higher CT values in repeat-tested patients than single-tested patients supports this hypothesis.
This study has some limitations. This is an observational study without selection bias for laboratory data, as all results were included in the analysis. However, clinical data were restricted to a subset of patients seen at Columbia University Irving Medical Center campuses. Nevertheless, rates of positivity and demographic variables captured in the laboratory data set were not significantly different between the other campuses, suggesting that the population in the clinical data set is generally representative of the New York City patients tested for COVID-19. Other limitations of the study of repeat-tested patients include the facts that only 15% of the total patients tested had repeat testing done and that ordering of repeat testing was at the discretion of the health care providers and not performed according to a standard protocol, although test ordering was mostly accomplished through standardized electronic medical record order sets.
In summary, our data suggest that patients with a high clinical suspicion or exposure setting suggestive of COVID-19 should be tested by a molecular SARS-CoV-2 assay and that it is appropriate to repeat the test the same day or on subsequent days if the results are initially negative and there is high suspicion or prevalence of COVID-19. Conversely, for patients with a positive SARS-CoV-2 molecular assay result, repeating the test before at least 15 days after the first test is unlikely to yield a negative result. Whether repeat positivity represents active infection or, more likely, detection of nonviable viral RNA is unknown. Further studies are needed to develop predictive models of the course and outcomes of COVID-19 using well-curated demographic, clinical, and laboratory data sets.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the contributions of all the dedicated medical technologists and laboratory technicians who performed testing at NYP laboratories, the efforts of the NYP Laboratory Information Team—in particular, Kelvin Espinal, Sarah Russell, Dennis Camp, Yingzhe Kuang, and Bulent Oral for designing queries and providing data extracts—and the volunteers who helped gather data from electronic medical records for the COVID-19 CARE database.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sector.
Lars F. Westblade has served on an advisory board for Roche Molecular Systems, Inc. All other authors have no conflicts.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, China Novel Coronavirus Investigating and Research Team, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. 2020. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. 2020. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. 2020. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfefferle S, Reucher S, Nörz D, Lütgehetmann M. 2020. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill 25:2000152. doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smithgall MC, Scherberkova I, Whittier S, Green DA. 2020. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the Rapid Detection of SARS-CoV-2. J Clin Virol 128:104428. doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 9.R Development Core Team. 2017. R: a language and environment for statistical computing.
- 10.Scheike TH, Zhang M-J. 2011. Analyzing competing risk data using the R timereg package. 1. J Stat Soft 38:i02. doi: 10.18637/jss.v038.i02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordes AK, Heim A. 2020. Rapid random access detection of the novel SARS-coronavirus-2 (SARS-CoV-2, previously 2019-nCoV) using an open access protocol for the Panther Fusion. J Clin Virol 125:104305. doi: 10.1016/j.jcv.2020.104305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhen W, Smith E, Manji R, Schron D, Berry GJ. 2020. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol doi: 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai C-C, Liu YH, Wang C-Y, Wang Y-H, Hsueh S-C, Yen M-Y, Ko W-C, Hsueh P-R. 2020. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J Microbiol Immunol Infect 53:404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Ji F, Wang L, Wang L, Hao J, Dai M, Liu Y, Pan X, Fu J, Li L, Yang G, Yang J, Yan X, Gu B. 2020. Early release—asymptomatic and human-to-human transmission of SARS-CoV-2 in a 2-family cluster, Xuzhou, China. Emerg Infect Dis 26. doi: 10.3201/eid2607.200718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Liu Y, Liu L, Wang X, Luo N, Li L. 2020. Clinical outcomes in 55 patients with severe acute respiratory syndrome coronavirus 2 who were asymptomatic at hospital admission in Shenzhen, China. J Infect Dis 221:1770–1774. doi: 10.1093/infdis/jiaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. 2020. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 172:577–682. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.