Abstract
Purpose
Pseudocontinuous arterial spin labeling (pCASL) allows for noninvasive measurement of regional cerebral blood flow (CBF), which has the potential to serve as biomarker for neurodegenerative and cardiovascular diseases. This work aimed to implement and validate pCASL on the dedicated MRI system within the population‐based Rotterdam Study, which was installed in 2005 and for which software and hardware configurations have remained fixed.
Methods
Imaging was performed on two 1.5T MRI systems (General Electric); (I) the Rotterdam Study system, and (II) a hospital‐based system with a product pCASL sequence. An in‐house implementation of pCASL was created on scanner I. A flow phantom and three healthy volunteers (<27 years) were scanned on both systems for validation purposes. The data of the first 30 participants (86 ± 4 years) of the Rotterdam Study undergoing pCASL scans on scanner I only were analyzed with and without partial volume correction for gray matter.
Results
The validation study showed a difference in blood flow velocity, sensitivity, and spatial coefficient of variation of the perfusion‐weighted signal between the two scanners, which was accounted for during post‐processing. Gray matter CBF for the Rotterdam Study participants was 52.4 ± 8.2 ml/100 g/min, uncorrected for partial volume effects of gray matter. In this elderly cohort, partial volume correction for gray matter had a variable effect on measured CBF in a range of cortical and sub‐cortical regions of interest.
Conclusion
Regional CBF measurements are now included to investigate novel biomarkers in the Rotterdam Study. This work highlights that when it is not feasible to purchase a novel ASL sequence, an in‐house implementation is valuable.
Keywords: arterial spin labeling, cerebral blood flow, flow phantom, population imaging
1. INTRODUCTION
Measurement of regional cerebral blood flow (CBF) with pseudocontinuous arterial spin labeling (pCASL) is a cost‐effective and safe option for repeated assessment of regional cerebral perfusion without the need to administer contrast media. This technique allows for quantification of CBF in ml/100 g/min and is, therefore, especially attractive for large and longitudinal population‐based studies in which healthy participants undergo MRI scans at multiple time points along the course of the study period.
The Rotterdam Study 1 is a prospective population‐based cohort study ongoing since 1990 in the city of Rotterdam in The Netherlands, which targets cardiovascular, endocrine, hepatic, neurological, ophthalmic, psychiatric, dermatological, otolaryngological, locomotor, and respiratory diseases. The Rotterdam Scan Study 2 is a substudy and focuses specifically on (repeat) brain MRI examinations to study neurodegenerative and cerebrovascular disease and was officially embedded in the core protocol of the Rotterdam Study in 2005, when a dedicated MRI system was installed in the Rotterdam Study Research Centre. To maintain consistency in image acquisition, the MRI hardware and software have not changed since.
Evidence for regional CBF as a biomarker in healthy aging, 3 , 4 cardiovascular disease, 5 and neurodegeneration 6 is steadily building in the literature. However, the original MRI protocol of the Rotterdam Scan Study only contained a phase contrast assessment of blood flow through major brain feeding arteries, assessing total CBF. The reason for this is that, despite ASL first being introduced in the early 1990s, the adoption of ASL as one of the standard sequences available on MRI systems is only a recent development due to the 2015 consensus paper. 7
Here we present the process of implementing and validating pCASL on the MRI system within the Rotterdam Study Research Centre. The validation of this pCASL sequence was done by CBF measurements of a novel perfusion phantom and healthy volunteers and comparing these to CBF measurements with a novel product pCASL sequence on one of the 1.5T MRI systems at the main site of the Erasmus Medical Centre in Rotterdam. In addition, the first 30 ASL datasets were assessed for their ability to measure regional CBF, corrected for partial volume estimates of gray matter, in the eldest cohort of participants of the Rotterdam Study.
We present this work to illustrate how local, multidisciplinary collaborations between MR physicists, engineers, and radiologists can result in successful implementation of a novel imaging technique. The need for ASL in the Rotterdam Study was given by a radiologist and epidemiologist and the process of programming and compiling the ASL sequence was done by physicists. During the optimization of the image analysis pipeline, the role of the engineer was to show that the sequence was working as intended (introduction of the phantom experiment) and communicate between the physics and radiologists teams in optimizing the processing pipeline. The final implementation was done via close collaboration between the MR Physics and Population‐Based Imaging groups at the department of Radiology and Nuclear Medicine of the Erasmus MC. This work shows that porting a novel MRI sequence to an older MRI system is valuable and, therefore, may be beneficial for sites that do not have the option to purchase novel ASL sequences for their MRI systems and software versions.
2. METHODS
All imaging was performed on 1.5T MRI systems (General Electric Healthcare, Milwaukee, WI, USA). Experiments were conducted under approval of the institutional ethics committee, in compliance with the declaration of Helsinki.
2.1. Implementation and validation of ASL in population based imaging
Imaging was performed on two different 1.5T MRI systems; (a) the population imaging scanner, a Signa EXCITE with software version 11 (released November 2003) and an 8‐channel head coil; (b) an Optima MR450w 1.5T with software version DV26 (released June 2017) and a 24‐channel head coil, which includes the General Electric product version of pCASL. 8 The implementation of pCASL at scanner I was done in‐house, converting the product pCASL sequence from software version DV26 to 11. This was done by mimicking the product sequence as much as possible hereby keeping up with the theoretical background of the implementation of the product sequence. 9 , 10 This was a debugging process that required downgrading novel functions and input structures from DV26, ensuring that the sequence could be compiled for software version 11. This particular part of porting back the sequence is highly dependent on both software versions and required close collaboration with GE. The final step in coding the in‐house pulse sequence was to fine‐tune the unbalancing of the radiofrequency (RF) gradients used to generate the label and control images. This process ensures that the effects of phase shifts between RF pulses in the pseudocontinuous labeling caused by magnetic field inhomogeneities at the labeling plane are minimalized, ensuring that the labeling efficiency of the pCASL scan was as high as possible. 10 , 11 Please note that we cannot publish any code of the pulse sequence, since this is GE proprietary information. However, via the GE research collaboration portal, we are open to collaborations in which this experience is shared.
Acquisition parameters were the same for both pCASL implementations; 3D spiral gradient echo read‐out with background suppression, 512 points per arm, eight arms, reconstruction diameter = 200 mm, reconstruction matrix 128 × 128 × 30, slice thickness 4 mm, post label delay (PLD) 1.525 s, label duration 1.45 s, number of excitations = 3 , echo time = 0.011 s, repetition time = 4.6 s. Each pCASL acquisition included a proton density weighted image for normalization (the same acquisition parameters, but no labeling applied 9 ).
2.2. Phantom experiment
A perfusion phantom (beta test version of QASPER, Gold Standard Phantoms, London, UK), which allows for setting variable continuous flow rates, was used. Measurements were done on both MRIs systems at two different days, 2 weeks apart. Per session, 12 pCASL scans were made in random order for different flow rates (200‐475 ml/min, steps of 25 ml/min). Voxelwise perfusion‐weighted images (ΔM/M 0) were calculated for each pCASL scan. 9 Median values were calculated over a circular region of interest (ROI), which was placed over the perfusion region of the phantom via visual inspection. A scaling factor between the two sequences was calculated by dividing the ROI median ΔM/M 0,DV26 by the ROI median ΔM/M 0,11.
2.3. Healthy, young volunteer experiment
Three young healthy volunteers (all female, <27 years) underwent pCASL scanning on both MR systems on the same day. Perfusion‐weighted pCASL images before quantification were compared by calculating the signal‐to‐noise ratio (SNR) of ΔM as the mean perfusion‐weighted signal in the GM divided by the standard deviation of a circular ROI (radius of 10 voxels) well outside the brain 12 and by calculating the spatial coefficient of variation (CoV) within the whole brain GM ROI. 13 The whole brain GM mask was obtained via segmentation of a T1 – weighted structural scan (fast in FSL), which was linearly registered to the pCASL image (flirt in FSL) and only included voxels with partial volume estimates for GM (PVEGM) > 70%. CBF maps were quantified with oxford_asl (FSL, version 6.0.1, Oxford, UK), 7 taking into account the scaling factor between the two implementations resulting from the phantom experiment. In addition, 16 cortical and sub‐cortical ROIs from the Harvard‐Oxford atlas (thresholded at partial volume estimates > 25%) were nonlinearly transformed from MNI standard space to the participant’s pCASL space and used to calculate the absolute differences in ROI CBF calculated between scanner I and scanner II.
2.4. Application of ASL imaging in the Rotterdam scan study
Thirty elderly participants (20 females, mean age 86 ± 4 years) of the RS‐I cohort in the Rotterdam Study (study entry in 1989‐1993) underwent scanning on scanner I only (Sep‐Nov 2018). SNR, CoV, and ROI average CBF were calculated in the same manner as for the young volunteers. To show the potential and advantage of being able to investigate regional CBF, a comparison was made of CBF calculated with and without PVE correction to assess the effect of local GM volume (BASIL within FSL 14 ).
3. RESULTS
3.1. Implementation and validation of pCASL
The flow phantom results illustrate a difference in flow sensitivity of ΔM/M 0 between the two pCASL implementations, with the pCASL on scanner I being more sensitive to increasing flow velocities (Figure 1). The scaling factor between the two pCASL implementations has an exponential relationship, with an asymptote at 30.9 (Figure 1B).
FIGURE 1.
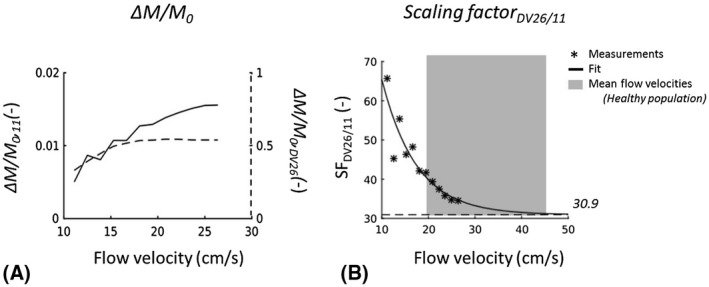
A, ΔM/M 0 plotted against the flow velocity set for the flow phantom for the pCASL implementation at software version 11 (solid line, left y‐axis) and for the implementation at DV26 (dashed line, right y‐axis). B, Scaling factor (SF) between ΔM/M 0 within perfusion ROI of the phantom collected with pCASL sequences at DV26 and at 11. SF is dependent on the flow velocity of the perfusate at the location of labeling, with a fitted asymptote of 30.9 (close to GE’s scaling factor of 32, used in image recon at DV26). The gray box indicates the range of mean flow velocities in the internal carotid and vertebral arteries in a healthy population (N = 180, 20‐79 years), measured with Doppler sonography 17
For the healthy, young volunteers the SNR of the perfusion‐weighted signal directly resulting from image reconstruction was smaller and CoV was larger for scanner I than for scanner II (Table 1 and Figure 2). When taking the scaling factor and additional smoothing into account, group averaged CBFGM (uncorrected for PVE of GM) for scanner I and II in the healthy volunteers were similar: 63.3 ± 1.8 ml/100 g/min and 62.2 ± 2.0 ml/100 g/min, respectively. Figure 3 shows the differences in CBF between the two scanners for the whole brain GM and 16 cortical and sub‐cortical ROIs calculated for all three volunteers. Note that, except for the brainstem, ROI differences in CBF for the two scanners are on the order of ±6 ml/100 g/min.
TABLE 1.
SNR for perfusion‐weighted (ΔM) signal within the whole brain GM ROI (thresholded at PVEGM > 70%)
| Participants | SNR ΔM GM | CoV ΔM GM |
|---|---|---|
| Young (N = 3) | ||
| Scanner I | 8.9 ± 0.4 | 30.3 ± 0.3 % |
| Scanner II | 18.6 ± 1.4 | 25.1 ± 0.9 % |
| Elderly (N = 29) | ||
| Scanner I | 5.9 ± 1.1 | 36.9 ± 2.2% |
FIGURE 2.
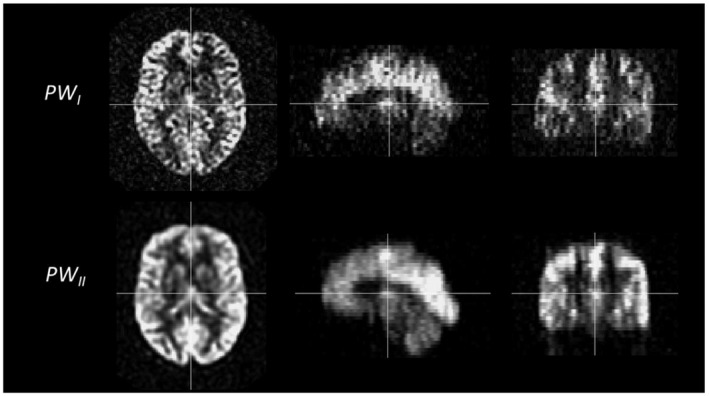
Example of perfusion weighted (ΔM) images of one young volunteer. Note the difference in smoothing occurring during the reconstruction of the images. On scanner I with software version 11 (top row), no smoothing is implemented during reconstruction. On scanner II with software version DV26 (bottom row), smoothing occurs during reconstruction of the images
FIGURE 3.
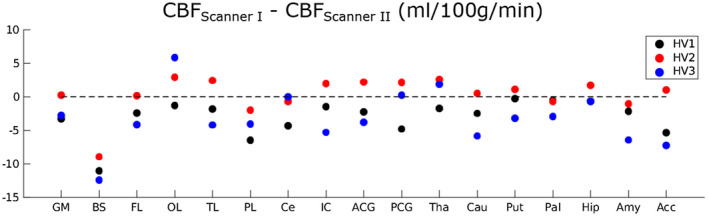
Absolute differences in regional CBF estimates between scanner I and scanner II in healthy volunteers. Note that these differences are calculated in ml/100 g/min. The dotted line is there as a reference at 0, that is, represents no difference in ROI average CBF between scanner I and scanner II. GM = gray matter, BS = brainstem, FL = frontal lobe, OL = occipital lobe, TL = temporal lobe, PL = parietal lobe, Ce = cerebellum, Ins = insular cortex, ACC = anterior cingulate cortex, PCC = posterior cingulate cortex, Tha = thalamus, Cau = caudate nucleus, Put = putamen, Pal = pallidum, Hip = hippocampus, Am = amygdala, Ac = Accumbens
3.2. Application of ASL imaging in the Rotterdam scan study
The dataset of one elderly participant was excluded due to severe head motion during the pCASL acquisition. Group averaged SNR of the perfusion‐weighted signal was lower for the elderly participants than for the younger, while spatial CoV of the perfusion‐weighted signal for the whole brain GM was higher than for the young volunteers. Both values are reported in Table 1. Group averaged whole brain GM CBF for the 29 remaining elderly participants was increased when correcting for PVEGM: 52.4 ± 8.2 ml/100 g/min vs 63.0 ± 8.9 ml/100 g/min, uncorrected and corrected for PVEGM, respectively (paired t test, P < .001). Regional investigation showed that the increase in GM CBF with partial volume correction varies across the brain, ranging from no significant change in CBF in brainstem (paired t test, P = 0.353) and cerebellum (paired t test, P = 0.345) to a little over 50% in the caudate nucleus (Figure 4).
FIGURE 4.
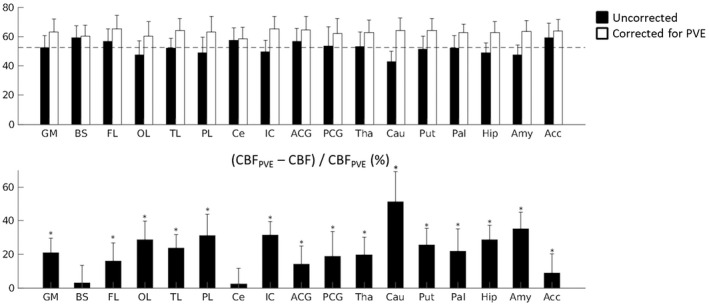
Top: Group averaged (N = 29) regional CBF values for elderly participants of the Rotterdam Scan Study, uncorrected and corrected for partial volume estimates of gray matter (GM). The error bars indicate the standard deviation across the group. The dotted line indicates the group averaged, uncorrected value for whole brain grey matter CBF. Bottom: The percentage in CBF per ROI after partial volume correction. GM = gray matter, BS = brainstem, FL = frontal lobe, OL = occipital lobe, TL = temporal lobe, PL = parietal lobe, Ce = cerebellum, Ins = insular cortex, ACC = anterior cingulate cortex, PCC = posterior cingulate cortex, Tha = thalamus, Cau = caudate nucleus, Put = putamen, Pal = pallidum, Hip = hippocampus, Am = amygdala, Ac = Accumbens. *Significantly larger than 0, P < .001. (Bonferroni‐corrected P‐value = .0029)
4. DISCUSSION
This work shows the in‐house implementation and validation of pCASL on a population imaging MRI system, with an outdated software platform. The first 30 pCASL datasets acquired in the Rotterdam Study highlight the potential of using pCASL for monitoring regional cerebral perfusion in population imaging, even on older software platforms.
The flow phantom scans led to the inclusion of a scaling factor in the quantification of CBF in the pCASL scans of the population‐based scanner. The results of the phantom experiment indicate that labeling efficiency decreases at lower blood flow velocities (<30 cm/s) in our in‐house implementation of pCASL. This is likely due to the difference in gradient coils between the two scanners. The asymptote of the scaling factor between the two different implementations (Figure 1B) suggests that, in the healthy population, a constant scaling factor is sufficient to correct for this difference in labeling efficiency. However, care should be taken in participants of the Rotterdam Study with blood flow velocities through the major cerebral arteries (<30 cm/s) in the labeling plane of the pCASL sequence. Within the Rotterdam Scan Study protocol phase contrast measurements of the flow velocities through the internal carotid and basilar arteries are measured. Therefore, the non‐constant scaling factor as measured with the flow phantom is being incorporated in the CBF analysis of the complete cohort.
The use of a flow phantom is highly recommended when comparing pCASL implementations, because it allows for an objective manner to test the reproducibility of the CBF measurements between two different scanners. However, should a flow phantom not be available, it is important to note that, in healthy volunteers, cerebral perfusion is variable due to several physiological parameters, including blood caffeine levels, blood pressure, hormone levels, etc. In absence of a flow phantom, it is important to compare sequences/scanners in light of previously reported reproducibility of ASL‐based measurements of CBF. 15
There are different reconstruction engines implemented in the software versions of scanner I and scanner II, which results in different inherent scaling of the perfusion‐weighted signal compared with the M0 image as well as different smoothing of images directly resulting from the reconstruction. The reconstruction engine as implemented in scanner II (DV26), does not allow for the reconstruction of the raw ΔM images and therefore it was not feasible to investigate ΔM images without smoothing for scanner II. The differences in smoothing between the scanners leads to the higher spatial CoV in the perfusion‐weighted images of scanner I, clearly seen in the healthy volunteers (Figure 2), which is the reason for including additional spatial smoothing in the post processing of these images. The higher SNR for the perfusion‐weighted signal on scanner II is likely caused by both the scaling and smoothing differences during reconstruction. However, note that the SNR for ΔM for young volunteers at scanner I is already in line with the results for 3D acquisitions as reported by Vidorreta et al 12 (and using the latest reconstruction engine clearly improves this value). Rather than changing these aspects of the reconstruction engines, which can lead to revoking CE/FDA marking of the system, we opted for adapting the post‐processing pipeline of the pCASL data for scanner I.
Limited differences were found between regional CBF measured with pCASL on scanner I and scanner II when taking the smoothing and scaling differences between the two software versions into account in the processing pipeline of perfusion‐weighted images from scanner I. The variability of CBF measured with ASL at different scanners is known to fluctuate in the order of >17%, as reported previously. 15 We therefore deemed the ±6 ml/100 g/min (<10% of the whole brain GM average) an acceptable range of fluctuation for the validation of this exam. The exception to this is the region of the brainstem, with a lower average CBF in scanner I. From previous work it is known that ASL requires optimisation for accurate brainstem CBF measurement. 16 Our validation study highlights this aspect and indicates that it may not be feasible to make inferences about CBF in the brainstem within the Rotterdam Scan study. Note that extending the number of young healthy volunteers could have aided a more thorough comparison in regional CBF measurements between scanner I and scanner II. However, the combination of the phantom and young volunteer results gave us confidence that the pCASL implementation on scanner I was functioning appropriately and application in the Rotterdam Scan Study followed.
The added value of measuring regional CBF within the Rotterdam Scan Study is illustrated by the first datasets of the elderly cohort. The regional PVEGM correction for CBF (Figure 4) illustrates the capability of assessing the effect of local GM volume on CBF measurement and highlights that the underestimation of underlying GM CBF due to partial volume effects is dependent on the ROI. The latter can be explained by the aspect that for subcortical gray matter structures (such as the caudate nucleus) the voxel size within the ASL sequence allows for limited voxels solely consisting of GM. The possibility of taking these regional variations in GM volume into account for the CBF measurements will allow for disentangling the effect of changes in measured CBF due to a true change in GM perfusion or to GM atrophy. 14 Note that this is valuable information in the context of the longitudinal Rotterdam Study and the incorporation of the current ASL processing pipeline within the analysis of the complete dataset of this cohort is now in progress. Future work with this data set, therefore, includes investigating the effect of aging and GM atrophy on regional CBF measurements.
A limitation of this work is the PLD of 1525 ms, which we chose based on the T1 of blood at 1.5T (1350 ms), the knowledge that the majority of the participants being scanned in the Rotterdam Scan Study are healthy, and to keep the imaging protocol consistent across all cohorts. Although this PLD led to SNR and CoV values comparable with the literature for the young volunteers, the lower SNR and higher CoV for the elderly participants may be improved with a longer PLD, for example, 2000 ms as recommended by the ASL White Paper. 7 This is based on the assumptions that the longer PLD will lead to less macrovascular artifacts within the perfusion‐weighted images, because it would accommodate the expected longer arterial arrival times occurring in healthy aging 7 and that this affects the SNR and CoV in a stronger positive manner than the decrease in signal of the labeled blood because of T1 relaxation during the prolonged PLD. In summary, this work illustrates the successful application of an in‐house implementation of pCASL in the Rotterdam Scan Study, including validation of this sequence with a novel flow phantom. The regional CBF data that are now acquired within the study population will be used in the future to investigate novel biomarkers of neurodegenerative and cardiovascular disease in the Rotterdam Study.
CONFLICT OF INTEREST
Mika Vogel is an employee of General Electric Global Research.
ACKNOWLEDGMENTS
Dr. Warnert has received funding from the Dutch Research Council (project “Food for thought”, veni 016.196.121). Dr. Ikram has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (project: ORACLE, grant agreement No: 678543).
Warnert EAH, Steketee RME, Vernooij MW, et al. Implementation and validation of ASL perfusion measurements for population imaging. Magn Reson Med. 2020;84:2048–2054. 10.1002/mrm.28271
REFERENCES
- 1. Ikram MA, Brusselle GGO, Murad SD, et al. The Rotterdam study: 2018 update on objectives, design and main results. Eur J Epidemiol. 2017;32:807–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ikram MA, van der Lugt A, Niessen WJ, et al. The Rotterdam scan study: Design update 2016 and main findings. Eur J Epidemiol. 2015;30:1299–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang N, Gordon ML, Ma Y, et al. The age‐related perfusion pattern measured with arterial spin labeling MRI in healthy subjects. Front Aging Neurosci. 2018;10:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Vis JB, Peng S‐L, Chen XI, et al. Arterial‐spin‐labeling (ASL) perfusion MRI predicts cognitive function in elderly individuals: A 4‐year longitudinal study. J Magn Reson Imaging. 2018;48:449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Warnert EAH, Rodrigues JCL, Burchell AE, et al. Is high blood pressure self‐protection for the brain? Circ Res. 2016;119:e140‐e151. [DOI] [PubMed] [Google Scholar]
- 6. Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the national institute on Aging‐Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011;7:280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alsop DC, Detre JA, Golay X, et al. Recommended implementation of arterial spin‐labeled Perfusion mri for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med. 2015;73:102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maleki N, Dai W, Alsop DC. Optimization of background suppression for arterial spin labeling perfusion imaging. Magn Reson Mater Physics, Biol Med. 2012;25:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dai W, Shankaranarayanan A, Alsop DC. Volumetric measurement of perfusion and arterial transit delay using hadamard encoded continuous arterial spin labeling. Magn Reson Med. 2013;69:1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dai W, Garcia D, de Bazelaire C, Alsop DC. Continuous flow‐driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn Reson Med. 2008;60:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao L, Vidorreta M, Soman S, Detre JA, Alsop DC. Improving the robustness of pseudo‐continuous arterial spin labeling to off‐resonance and pulsatile flow velocity. Magn Reson Med. 2017;78:1342–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vidorreta M, Wang ZE, Rodríguez I, Pastor MA, Detre JA, Fernández‐Seara MA. Comparison of 2D and 3D single‐shot ASL perfusion fMRI sequences. Neuroimage. 2012;66C:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mutsaerts HJMM, Petr J, Václavů L, et al. The spatial coefficient of variation in arterial spin labeling cerebral blood flow images. J Cereb Blood Flow Metab. 2017;37:3184–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chappell MA, Groves AR, MacIntosh BJ, Donahue MJ, Jezzard P, Woolrich MW. Partial volume correction of multiple inversion time arterial spin labeling MRI data. Magn Reson Med. 2011;65:1173–1183. [DOI] [PubMed] [Google Scholar]
- 15. Gevers S, van Osch MJ, Bokkers RPH, et al. Intra‐ and multicenter reproducibility of pulsed, continuous and pseudo‐continuous arterial spin labeling methods for measuring cerebral perfusion. J Cereb Blood Flow Metab. 2011;31:1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Warnert EAH, Harris AD, Murphy K, et al. Measuring tissue perfusion in the human brainstem using multi‐inversion time pulsed arterial spin labelling. Proc Intl Soc Mag Reson Med. 2013;21:2182. [Google Scholar]
- 17. Albayrak R, Degırmencı B, Acar M, Haktanır A, Colbay M, Yaman M. Doppler sonography evaluation of flow velocity and volume of the extracranial internal carotid and vertebral arteries in healthy adults. J Clin Ultrasound. 2007;35:27–33. [DOI] [PubMed] [Google Scholar]


