Abstract
Hepatitis B virus (HBV) RNA in serum is a novel biomarker that reflects cccDNA activity. We investigated whether HBV RNA can predict serological response to peginterferon (PEG‐IFN) treatment. Serum HBV RNA levels were retrospectively measured at weeks 0, 12, 24 and 52 of therapy and after treatment discontinuation (week 78) in 266 HBeAg‐positive chronic HBV patients who had participated in a global randomized controlled trial (HBV99‐01 study). Patients received 52 weeks PEG‐IFN monotherapy (n = 136) or PEG‐IFN and lamivudine (n = 130). The primary end point was HBeAg loss 24 weeks after PEG‐IFN discontinuation. At baseline, the mean serum level of HBV RNA was 6.8 (SD 1.2) log c/mL. HBV RNA levels declined to 4.7 (1.7) log c/mL after one year of PEG‐IFN therapy alone and to 3.3 (1.2)log c/mL after combination therapy. From week 12 onward, HBV RNA level was significantly lower in patients who achieved HBeAg loss at the end of follow‐up as compared to those who did not, regardless of treatment allocation (week 12:4.4 vs 5.1 log c/mL, P = .01; week 24:3.7 vs 4.9 log c/mL, P < .001). The performance of a multivariable model based on HBV RNA level was comparable at week 12 (AUC 0.68) and 24 (AUC 0.72) of therapy. HBV RNA level above 5.5 log c/mL at week 12 showed negative predictive values of 93/67/90/64% for HBV genotypes A/B/C/D for the prediction of HBeAg loss. In conclusion, HBV RNA in serum declines profoundly during PEG‐IFN treatment. Early on‐treatment HBV RNA level may be used to predict nonresponse.
Keywords: chronic hepatitis B infection, functional cure, peginterferon treatment, serum marker, treatment response
Abbreviations
- ALT
alanine aminotransferase
- BCP
basal core promoter
- cccDNA
covalently closed circular DNA
- CHB
chronic hepatitis B infection
- CI
confidence interval
- CR
combined response
- ETV
entecavir
- HBcAg
hepatitis B core antigen
- HBeAg
hepatitis B e antigen
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- LLD
lower limit of detection
- LTFU
long‐term follow‐up
- MITT
modified intention‐to‐treat
- NA(s)
nucleos(t)ide analogue(s)
- PC
precore
- PCR
polymerase chain reaction
- PEG‐IFN
peginterferon
- qHBsAg
quantitative hepatitis B surface antigen
- RACE
rapid amplification of cDNA ends
- RBV
ribavirin
- SD
standard deviation
- ULN
upper limit of normal
1. INTRODUCTION
Decades after the development of the first vaccine against hepatitis B virus (HBV), still almost one million individuals annually die due to acute or chronic HBV infection despite the available treatment strategies.1 Eight FDA‐approved drugs are currently available, including six nucleos(t)ide analogues (NA), pegylated interferon (PEG‐IFN) alfa‐2a and alfa‐2b. PEG‐IFN has anti‐proliferative, immunomodulatory and antiviral effects, while NA directly inhibits the HBV polymerase.2 Although PEG‐IFN treatment has a higher burden of side effects than NA treatment and is contraindicated in liver cirrhosis, its finite course and higher serological response rates makes it an interesting treatment option.3
However, one of the major limitations of currently available treatment options for chronic HBV infection is that they do not silence or eliminate the main HBV replication template, the covalently closed circular DNA (cccDNA).4 Persistence of active cccDNA during or after treatment limits the achievement of a functional cure. In the development of new therapeutic strategies, it therefore seems important to design agents that are able to interfere with cccDNA, but in order to reflect on the efficacy of these future agents, monitoring cccDNA activity is needed. Biomarkers reflecting cccDNA activity that can be obtained in a noninvasive way could play an important role.5 Studying these biomarkers in PEG‐IFN treatment is needed because PEG‐IFN is increasingly used in clinical studies of new therapeutic compounds, such as entry inhibitors.
HBV RNA in serum is a novel serum biomarker that can be measured using PCR techniques. Its nature has not been fully researched, but it most likely comprises encapsidated pregenomic RNA (pgRNA).6, 7 Pregenomic RNA is a more than whole‐genome‐length transcript that is derived from the cccDNA directly and is transcribed by reverse transcriptase to form relaxed circular DNA (rcDNA), eventually leading to replenishment of the cccDNA pool and formation of HBV DNA containing particles. Therefore, HBV RNA in serum is assumed to reflect cccDNA activity, which was indeed observed in recent studies.8, 9, 10
During treatment with nucleos(t)ide (NA), early on‐treatment levels of HBV RNA were found to be a an early predictor for HBeAg loss and were superior to serum levels of HBV DNA or hepatitis B surface antigen (HBsAg) in this context.11, 12 In a different, small population, a relation with HBsAg loss was observed.13 Fewer data are available on PEG‐IFN treatment, but HBV RNA levels showed distinct dynamics in patients achieving HBeAg seroconversion.7
However, the studies describing these dynamics were based on uncontrolled populations, used different molecular methods and study different treatment end points. Therefore, we have studied HBV RNA levels and dynamics before and during PEG‐IFN treatment in a well‐defined, multi‐ethnic population of HBeAg‐positive individuals who were previously treated in a randomized controlled trial.
2. PATIENTS AND METHODS
2.1. Treatment regimen and study population
Serum levels of full‐length polyadenylated HBV RNA were measured in available serum samples of 266 patients with HBeAg‐positive chronic HBV infection who were treated within a previously conducted global randomized controlled trial (99‐01 study). Detailed inclusion criteria have been described elsewhere.11, 14 In short, patients with active disease who had not received treatment ≥6 months prior to screening, did not have a coinfection and did not have a contraindication for PEG‐IFN treatment were included. Patients received 52 weeks of PEG‐IFNalpha‐2b and lamivudine (LAM) 100 mg or PEG‐IFN alpha alone (1:1 randomization). The dosage of PEG‐IFN was 100 µg/wk from baseline through week 32 and was then decreased to 50 µg/wk to prevent early treatment discontinuation due to side effects. After treatment discontinuation, all patients were followed for an additional 26 weeks.
2.2. End points
Levels of HBV RNA were measured at baseline, week 12, 24, end of treatment (EOT, week 52) and end of follow‐up (EOF, week 78). The primary end point was the association of serum HBV RNA levels and kinetics with loss of HBeAg or loss of HBsAg at the end of follow‐up. We additionally aimed to validate a recently proposed HBV RNA cut‐off to identify those patients not achieving loss of HBeAg.11
2.3. Serum HBV RNA quantification
Levels of polyadenylated HBV RNA were measured at a central laboratory (University Hospital Leipzig, Germany) from serum samples stored at −20°. For HBV RNA quantification, we used a rapid amplification of complimentary DNA (cDNA)‐ends (RACE)‐based real‐time polymerase chain reaction (PCR) technique that has been previously described. Quantification of HBV RNA was performed using specific primers (including HBV RNA RT primer 5′‐ACC ACG CTA TCG CTA CTC AC (t17)GWA GCT C) designed according to van Bömmel et al.15 The assay's lower limit of detection (LOD) for HBV RNA was 800 (2.9 log10) copies/mL (c/mL), with a corresponding linear range of 800 to 106 copies/mL. HBV RNA levels below the LOD were set to 450 c/mL for statistical analysis.
2.4. Other laboratory measurements
Routine biochemical and haematological tests were performed at each individual site. Serum ALT levels were standardized by calculating the value times for the ULN per centre. Virological tests were performed at one central laboratory (Erasmus Medical Center). HBV DNA was measured using a in‐house developed TaqMan‐based PCR assay (Roche Diagnostics; lower limit of detection of 400 copies/mL). HBV DNA results in copies/mL were converted into IU/mL using a conversion factor of 5.8 copies per IU. Serum qHBsAg levels were measured using the Architect HBsAg assay (Abbott Laboratories; range 0.05‐250 IU/mL). Serum HBeAg levels were quantified using the Cobas Elecsys HBeAg assay (Roche Diagnostics, measurement range 0.2‐100 IU/mL). HBV genotype analysis was performed using the INNO‐LiPA HBV genotype assay (Fujirebio Europe). The presence of PC and BCP mutants was assessed using the INNO‐LiPA HBV PreCore assay (Fujirebio Belgium), which detects precore (PC) mutations at nucleotide position 1896 and basal core promoter (BCP) mutations at nucleotide positions 1762 and 1764. Results were classified into four groups: wild type (WT, only WT virus detectable), PC (only PC or both PC and WT detectable), BCP (either or both BCP mutations detected, with or without WT) or as PC + BCP when both types of mutants were found.
2.5. Statistical analysis
SPSS version 25.0 (SPSS Inc) was used to perform statistical analyses. Skewed laboratory values were log‐transformed prior to analyses and were expressed as mean (standard deviation [SD]). Associations between variables were tested using Student's t test, chi‐squared test, Pearson correlation or their nonparametric equivalents when appropriate. Subgroup analysis for mean HBV RNA levels at baseline was performed using ANOVA with Bonferroni correction for intergroup comparison. We performed logistic regression analysis to determine factors associated with response. The factors we included in univariable analysis were age, sex, HBV genotype, BCP and PC variants, presence of cirrhosis and treatment history. Factors that were found to be related (P < .20) were analysed in multivariable analysis. The performances of the retrieved prediction models were tested with receiver operating characteristic (ROC) curve analysis. All analyses were performed two‐sided at the 0.05 level of significance.
3. RESULTS
3.1. Association of host and viral factors with HBV RNA levels at baseline
Patient characteristics are shown in Table 1. All major HBV genotypes were represented. At baseline, the mean serum level of HBV RNA was 6.8 (SD 1.2) log c/mL. In one patient, HBV RNA was below LLD (28‐year‐old female patient, HBV genotype D, BCP mutant, HBV DNA 5.1 log IU/mL).HBV RNA level significantly correlated to HBV DNA (r = 0.66, P < .001), qHBsAg (r = 0.47, P < .001), qHBeAg (r = 0.29, P < .001) and ALT (r = 0.25 (P < .001). In multivariable linear regression analysis adjusting for ALT and presence of mutations, HBV RNA level at baseline was lowest in patients infected with HBV genotype C and highest in HBV genotype B (A/B/C/D: 6.6/7.2/ 6.2/ 7.0 log c/mL; C vs D P = .002, B vs C P = .007, other P = n.s.). BCP mutation was associated with lower HBV RNA level (wild type/BCP only/PC only/BCP + PC: 6.9/6.1/7.0/7.2 c/mL; P < .002 for BCP mutation vs wild type).
Table 1.
Patient characteristics
| Characteristics | All patients (n = 266) | PEG‐IFN mono (n = 136) | PEG‐IFN + LAM (n = 130) |
|---|---|---|---|
| Demography | |||
| Age, years | 35 (13) | 36 (14) | 34 (12) |
| Male, n (%) | 20 (78) | 107 (79) | 100 (77) |
| Race, n (%) | |||
| Caucasian | 196 (74) | 101 (74) | 95 (73) |
| Asian | 53 (20) | 29 (21) | 24 (19) |
| Other | 17 (6) | 6 (5) | 11 (8) |
| HBV genotype, n (%) | |||
| A | 90 (34) | 47 (35) | 43 (33) |
| B | 23 (9) | 12 (9) | 11 (9) |
| C | 39 (15) | 21 (15) | 18 (14) |
| D | 103 (38) | 51 (38) | 52 (40) |
| Other | 11 (4) | 5 (4) | 6 (4) |
| BCP and PC variants, n (%) | |||
| Wildtype virus | 75 (28) | 40 (29) | 35 (27) |
| PC mutation | 56 (21) | 25 (18) | 31 (24) |
| BCP mutation | 47 (18) | 20 (15) | 27 (21) |
| PC & BCP mutation | 35 (13) | 19 (14) | 16 (12) |
| Cirrhosis, n (%) | 24 (9) | 11 (8) | 13 (10) |
| Treatment history | |||
| Previous NA | 33 (12) | 16 (12) | 17 (13) |
| Previous (PEG‐)IFN | 55 (21) | 28 (21) | 27 (21) |
| Laboratory results | |||
| HBV RNA § | 6.8 (1.2) | 6.9 (1.2) | 6.7 (1.2) |
| HBV DNA ‡ | 8.3 (1.0) | 8.3 (1.0) | 8.3 (1.0) |
| qHBsAg ‡ | 4.4 (0.6) | 4.4 (0.6) | 4.4 (0.6) |
| qHBeAg ‡ | 2.4 (0.8) | 2.4 (0.8) | 2.4 (0.8) |
| ALT (×ULN) † | 4.3 (3.5) | 4.3 (3.1) | 4.4 (3.9) |
Continuous variables are expressed as mean (SD), categorical variables as n (%).
Abbreviations: ALT, alanine aminotransferase; BCP, basal core promoter; HBV, hepatitis B virus; LLQ, lower limit of detection; NA, nucleos(t)ide analogue; (PEG‐)IFN, (peg)interferon; PC, precore; qHBeAg, quantitative hepatitis B e antigen; qHBsAg, quantitative hepatitis B surface antigen; SD, standard deviation; ULN, upper limit of normal.
Multiples of upper limit of the normal range.
Logarithmic scale, IU/mL.
Logarithmic scale, c/mL.
3.2. HBV RNA levels during and after PEG‐IFN treatment
In the overall population, HBV RNA levels showed a decline during PEG‐IFN based treatment (Figure 1). Mean declines in HBV RNA level were −2.0/−2.3/−2.9 log c/mL at weeks 12, 24 and 52 (EOT).The mean HBV RNA levels were 4.0 (1.6) log c/mLat EOT and 5.0 (1.9) c/mL at EOF. HBV RNA was undetectable in 54% patients at EOT and in 34% of patients at EOF.HBV RNA early on‐treatment decline differed by treatment regimen (Figure 2A). A significantly stronger on‐treatment decline was observed in patients treated with PEG‐IFN and LAM when compared to PEG‐IFN monotherapy (week 12: −5.5 vs −4.2 log c/mL, P < .001, week 24: −5.0 vs −3.8, P < .001 and week 52: −4.7 vs −3.3, P < .001).At EOF, however, HBV RNA levels were similar for both treatment arms (4.9 vs 5.0 log c/mL, P = .76; decline: −2.0 vs −1.8 log c/mL, P = .71).
Figure 1.
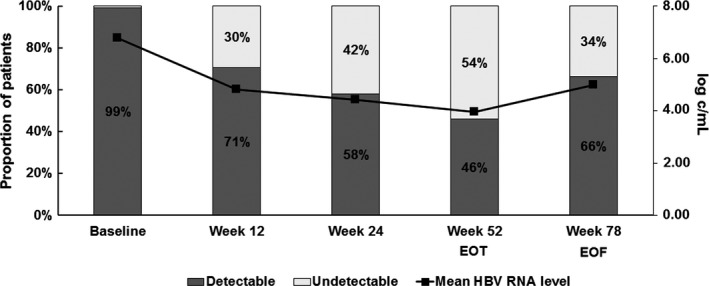
HBV RNA dynamics and detectability in the total study population (N = 266). The line represents mean level of HBV RNA (log c/mL); bars represent the proportion of patients with either detectable or undetectable HBV RNA
Figure 2.

Early on‐treatment HBV RNA dynamics according to therapy allocation and treatment response. Lines represent mean level of HBV RNA (log c/mL), according to therapy allocation (A), treatment response (B) or both (C,D). Grey lines in panels c and d represent individual HBV RNA dynamics in patients with HBsAg loss
HBV RNA decline from baseline to EOF (week 78) was stronger in patients infected with HBV genotype B than in those with genotype C (P = .04), but did not significantly differ across the other genotypes. By EOF, HBV RNA levels no longer differed between patients with wildtype virus at baseline and patients with PC and/or BCP mutations (overall P = .513), but there was a trend towards lower HBV RNA levels in patients infected with wildtype virus when compared to nonwildtype virus (4.7 vs 5.3 log c/mL, P = .09).
3.3. HBV RNA level in relation to response to PEG‐IFN treatment
HBeAg loss was achieved in 49 of 136 (36%) patients treated with PEG‐IFN monotherapy and 46 of 130 (35%) patients treated with PEG‐IFN and LAM (P = 1.00). HBsAg became negative in 9 of 136 (7%) and 9 of 130 (7%) patients, respectively (P = 1.00). Figure 2B‐D show early on‐treatment HBV RNA dynamics according to HBeAg loss, HBsAg loss and therapy allocation.
3.3.1. HBV RNA in relation to HBeAg loss
At baseline, mean HBV RNA levels did not differ between patients with or without HBeAg loss at the end of follow‐up (6.8 vs 6.6 log c/mL, P = .22; Figure 2). From week 12 onward, HBV RNA level was significantly lower in patients who achieved HBeAg loss at EOF, (Figure 3A, week 12:4.4 vs 5.1 log c/mL, P = .01; week 24:3.7 vs 4.9 log c/mL, P < .001). Also, the rates of HBV RNA undetectability were higher in patients with HBeAg loss from week 24 of treatment through EOF (Figure 3B). HBV RNA level was independently associated with HBeAg loss, at all time points from week 12 onward (week 12: OR 0.75, CI‐95% 0.59‐0.97, P = .02, adjusted for ALT, HBV genotype and mutations). When therapy allocation was introduced into the multivariable model (because it was not associated with HBeAg loss in univariable analysis), HBV RNA level remained significantly associated with HBeAg loss (OR 0.55, CI‐95% 0.44‐0.68, P < .001). In multivariable AUROC analysis again adjusting for ALT, HBV genotype and mutations, HBV RNA level at week 24 had the highest diagnostic accuracy when compared to baseline and week 12 (Figure 3C, AUC 0.75, CI‐95% 0.67‐0.84, P < .001).
Figure 3.
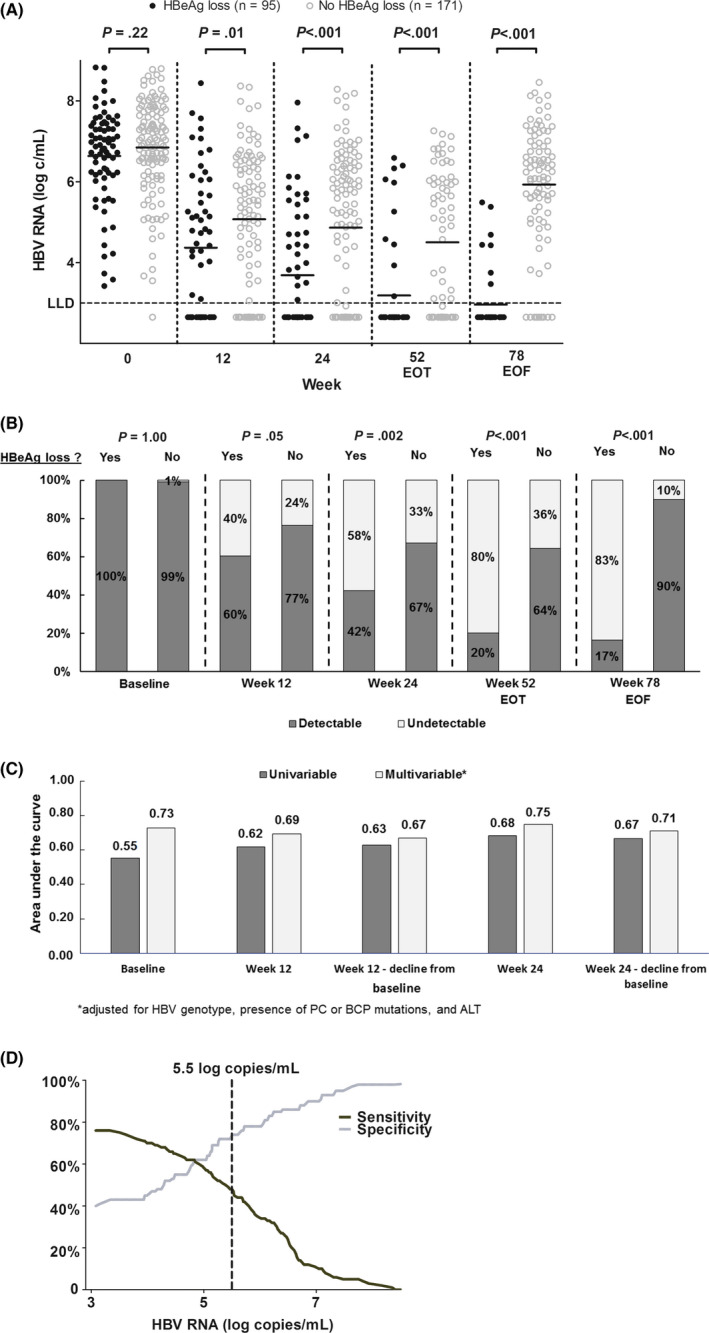
A, Individual HBV RNA measurements according to HBeAg loss at the end of follow‐up. Dots represent individual HBV RNA measurements according to HBeAg loss at the end of follow‐up, with lines representing the mean level of HBV RNA (log c/mL). The red dotted line indicates a previously proposed HBV DNA cut‐off for the early identification of nonresponse (10). B, HBV RNA detectability according to HBeAg loss at the end of follow‐up. Bars represent the proportion of patients with detectable or undetectable HBV RNA levels according to HBeAg loss at the end of follow‐up. C, Univariable and multivariable diagnostic accuracy of early on‐treatment HBV RNA for the prediction of HBeAg loss at the end of follow‐up. Bars represent the area under the curve (AUC) for HBV RNA at baseline, week 12 and week 24 for the prediction of HBeAg loss at the end of follow‐up. Bars in dark grey represent univariable analysis; bars in light grey represent multivariable analysis adjusting for HBV genotype, ALT level, and presence of BCP or PC mutations. D, HBV RNA cut‐off for the prediction of HBeAg loss at the end of follow‐up. The dark line and grey line represent sensitivity and specificity of HBV RNA level for the prediction of HBeAg loss at the end of follow‐up
The accuracy at this particular time point was AUC 0.68 after removing HBV RNA as a variable; the AUC was 0.79 when substituting HBV RNA by qHBsAg, the AUC was 0.78 when combining HBV RNA and qHBsAg, and the AUC was 0.80 when combining qHBsAg with HBV DNA. For the prediction of HBeAg nonresponse, we aimed to identify an HBV RNA cut‐off at week 12 as it was the earliest time point independently associated with response. When applying the HBV RNA cut‐off of 5.5 log c/mLat week 12, which was proposed in a recent study,11 the negative predictive values (NPV) for the prediction of nonresponse at the end of follow‐up were only 76% and 73% for the total study population and the PEG‐IFN monotherapy group, respectively. The performance of the cut‐off was genotype‐dependent, as NPVs for the main HBV genotypes A/B/C/D were 93/67/90/64%. Due to the high variance of HBV RNA results, which is illustrated in Figure 3A, no alternative cut‐off level for the prediction of nonresponse could be constructed at this time point or at week 24, when aiming for a prediction rule with high specificity for all HBV genotypes (Figure 3D). Also in subgroup analyses for genotypes B and D, no alternative cut‐off level met this requirement (data not shown).
3.3.2. HBV RNA in relation to HBsAg loss
At baseline, mean HBV RNA levels did not differ between patients with or without HBsAg loss at the end of follow‐up (6.5 vs 6.8 log c/mL, P = .40). At week 12, a trend was observed towards lower HBV RNA levels in patients with HBsAg loss (3.8 vs 4.9 log c/mL, P = .11) and more HBV RNA decline from baseline (−3.2 vs −2.0 log c/mL, P = .06). Patients with HBsAg loss at the end of follow‐up had a significantly lower HBV RNA level at week 24 than patients without HBsAg loss (2.8 vs 4.5 log c/mL, P < .001). Also within the group of 95 patients who achieved HBeAg loss, HBV RNA at week 24 was lower in those patients who also achieved subsequent HBsAg loss than in those who did not (2.8 vs 3.8 log c/mL, P = .001). The proportion of patients with undetectable HBV RNA at week 24 was threefold higher in patients in the overall cohort with vs without HBsAg loss (Figure 4A, 89% vs 39%, P = .005). However, vice versa, the probability of HBsAg loss at the end of follow‐up in patients with undetectable HBV RNA at week 24 was only 11%. In univariable AUROC analysis, the highest diagnostic accuracy of HBV RNA level for the prediction of HBsAg loss was found at week 24 (AUC 0.77, CI‐95% 0.67 −0.87, P = .006, Figure 4B). Because HBV RNA level at week 24 was available for 11 of 18 patients only, multivariable regression analysis could not be performed for this end point.
Figure 4.
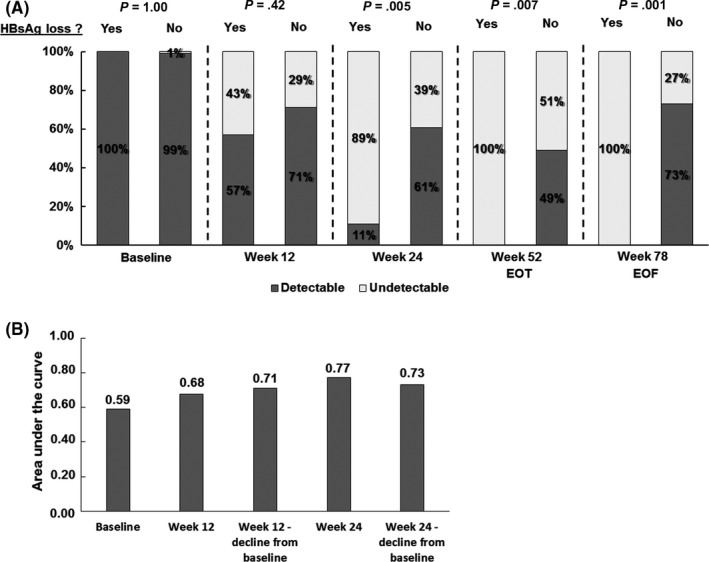
A, HBV RNA detectability according to HBsAg loss at the end of follow‐up. Bars represent the proportion of patients with detectable or undetectable HBV RNA levels according to HBsAg loss at the end of follow‐up. B, Univariable diagnostic accuracy of early on‐treatment HBV RNA for the prediction of HBeAg loss at the end of follow‐up. Bars represent the area under the curve (AUC) for HBV RNA at baseline, week 12 and week 24 for the prediction of HBeAg loss at the end of follow‐up. Because of the low number of patients with HBsAg loss, multivariable regression analysis could not be performed for this end point
3.4. HBV RNA dynamics in relation to dynamics of other biomarkers
Figure 5A shows the dynamics of HBV RNA in relation to HBV DNA, qHBsAg and qHBeAg, according to different definitions of treatment response. All four biomarkers showed a significantly stronger decline—most apparent at week 24—for patients with an HBeAg loss when compared to patients without. Among those patients who lost both HBeAg and HBsAg, week 24 HBV RNA (−3.8 vs 2.8 log c/mL, P < .001), HBV DNA (5.9 vs 3.2, P < .002) and qHBsAg levels (−3.9 vs −0.2, P < .001) were all significantly lower than in those with HBeAg loss only. Figure 5B shows univariable AUCs for HBV DNA, qHBsAg, qHBeAg and ALT in relation to AUC of early on‐treatment HBV RNA level.
Figure 5.
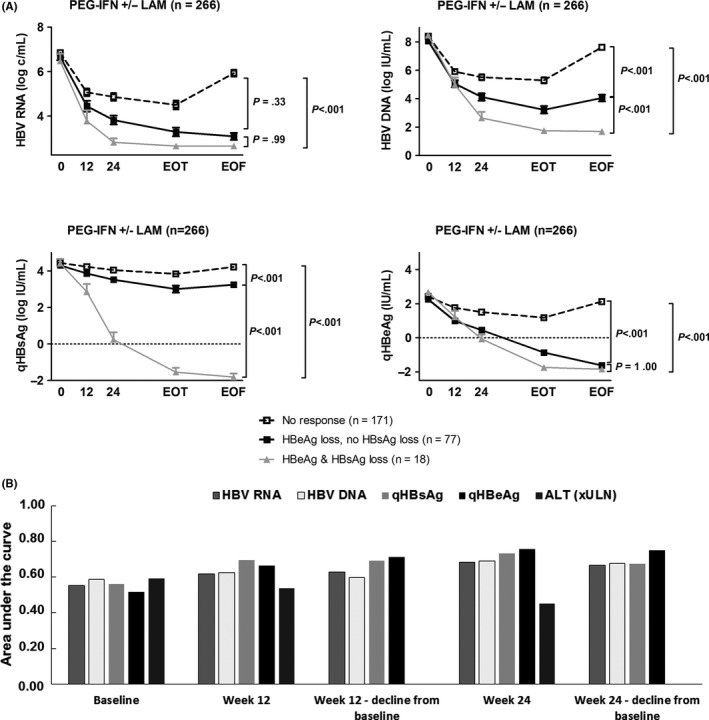
A, HBV RNA, HBV DNA, qHBsAg and qHBeAg dynamics according to HBeAg loss and HBsAg loss. Boxes represent biomarker dynamics for HBV RNA (upper left panel), HBV DNA (upper right panel), qHBsAg (lower left panel) and qHBeAg (lower right panel). Dotted lines represent patients without HBeAg loss, black lines represent patients with HBeAg loss but without HBsAg loss, and grey lines represent patients with HBsAg loss. B, Univariable diagnostic accuracy of early on‐treatment levels of various serum markers for the prediction of HBeAg loss at the end of follow‐up. Bars represent the area under the curve (AUC) at baseline, week 12 and week 24 for the prediction of HBeAg loss at the end of follow‐up, for HBV RNA, HBV DNA, qHBsAg, qHBeAg and ALT
4. DISCUSSION
In this study, we investigated HBV RNA dynamics during and after PEG‐IFN based treatment in a large multi‐ethnic cohort of 266 HBeAg‐positive patients previously treated in a randomized controlled trial. We observed that HBV RNA is a sensitive marker for HBeAg loss and HBsAg loss but has low specificity. Furthermore, we could show that HBV RNA levels showed a stronger decrease in those HBeAg‐positive patients who subsequently achieved HBeAg and HBsAg loss as compared to those who only lost HBeAg (P = .001). The performance of a previously proposed HBV RNA cut‐off of 5.5 log at week 12 for the identification of nonresponse did hold for HBV genotypes A and C.
HBV RNA in serum is particularly interesting as a biomarker because, at least in HBeAg‐positive patients, it represents most likely a direct transcript of the HBV cccDNA. HBV RNA may therefore be used to monitor the effect of currently available or future therapeutic agents, but also to improve the knowledge on the HBV life cycle. Recent reports indeed suggested that HBV RNA in serum is associated with response to NA therapy and PEG‐IFN therapy, and HBV RNA has already been used to illustrate the effects of new compounds.7, 11, 15, 16, 17, 18 A recent review highlighted the importance of studying this marker in multi‐ethnic populations.19
In our study, we could show that HBV RNA kinetics are not only associated with HBeAg loss, but also with HBsAg loss, which represents the ultimate goal of currently available and future treatments. Although a statistical prediction model for HBsAg loss was not possible owing to the small sample size in our study, we believe that the potential of HBV RNA kinetics for the prediction of a functional cure should be assessed in PEG‐IFN treated patient cohorts that include more patients who achieved HBsAg loss. HBV RNA monitoring should also be researched within the context of novel treatment approaches which target the HBV life cycle or cccDNA.
In our data set, we found that by the end of treatment, HBV RNA was undetectable in 54% of all patients. In line with earlier studies, initial HBV RNA decline was stronger in PEG‐IFN combination therapy than in PEG‐IFN monotherapy, but no more difference in HBV RNA level was observed 6 months after treatment discontinuation.7, 20 The degree of HBV RNA decline in the PEG‐IFN and LAM combination group was comparable to the previously reported decline in a much smaller population treated with PEG‐IFN in combination with adefovir, but it was weaker than reported in a study comparable to ours, in which patients were also treated with PEG‐IFN alone or in combination with LAM.7, 20 This difference may be explained by the higher rate of HBeAg loss in the prior study, by the greater proportion of genotype B and D patients in our population, or by a different prevalence of wildtype HBV. The correlations between HBV RNA and qHBeAg, and between HBV RNA and qHBsAg before treatment were also lower in our study, which could imply that patients may be in a different stage of infection.
We were able to confirm that HBV RNA level was significantly lower in patients with HBeAg loss than in patients without HBeAg loss from week 12 of treatment onward. Recently, a negative predictive value (NPV) of 93% was found for an HBV RNA cut‐off of 5.5 log c/mL to identify nonresponders in a population of individuals mainly infected with genotype C.20 In our study, we could confirm an NPV > 90% by using this cut‐off for individuals with genotypes A and C. The NPV was lower in patients with genotypes B and D. Accordingly, our group recently reported that among other factors, HBV genotype strongly influences HBV RNA levels and also correlations of HBV RNA level to other serum markers, such as qHBsAg and qHBeAg.21 This may explain why a universal HBV RNA cut‐off did not allow the prediction of serological nonresponse with a desirable NVP > 90% in all HBV genotypes.
Construction of a clinical decision rule regardless of HBV genotype was hampered by a large variance in HBV RNA results, which was observed across all HBV genotypes irrespective of PC and BCP mutations, and in both patients with and without a response (data not shown). HBV RNA levels during PEG‐IFN based treatments may therefore be influenced by other host or viral factors that have not been identified yet. One theory could be that PEG‐IFN treatment inhibits HBeAg, HBV RNA and HBsAg formation in a different way, which has not been described to date. The exact mode of action of PEG‐IFN has not completely been revealed yet, but effects include direct inhibition of transcription, epigenetic modifications of cccDNA, and immunomodulating effects.22, 23, 24, 25, 26, 27 Novel treatments for HBV infections that are currently being developed mostly target the HBV replication cycle, and therefore, under those treatments HBV RNA and other HBV biomarkers may show a clearer association to subsequent response.
To summarize, we showed that HBV RNA in serum declines significantly during PEG‐IFN based treatment. HBV RNA level is a sensitive predictor for PEG‐IFN induced HBeAg loss and possibly also for HBsAg loss, and may therefore be used to predict nonresponse in genotypes A and C. HBV RNA, however, was not a specific biomarker, as profound declines in HBV RNA level were also observed in patients without a response. Studies involving multiple HBV biomarkers, especially within the context of novel treatment approaches which target the HBV life cycle or cccDNA, are warranted to define more applications for this novel biomarker. As PEG‐IFN is increasingly used in clinical trials examining new therapeutic compounds, our findings may also be of particular value for these studies.
CONFLICT OF INTERESTS
FvB has been in speaker's bureau and advisory boards for Gilead Sciences, Bristol‐Myers Squibb, Roche Pharma, Abbvie, MSA and has received research grants from Roche Pharmaceuticals, Gilead Sciences and Bristol‐Myers Squibb. AB has been in consulting or in advisory boards for Gilead Sciences and Bristol‐Myers Squibb and has received research grants from Roche, Gilead Sciences, Fujirebio and Janssen. TB received grants and personal fees from AbbVie, Bristol‐Myers Squibb, Gilead Sciences, Janssen, Bayer, Vertex, Tibotec, Intercept, Merck Sharp & Dohme and Roche. HLAJ received grants from AbbVie, Bristol Myers Squibb, Gilead Sciences, Innogenetics, Janssen, Medimmune, Medtronic, Merck and Roche and is consultant for AbbVie, Benitec, Bristol Myers Squibb, Gilead Sciences, Janssen, Medimmune, Merck, Roche and Arbutus. The other authors have nothing to disclose.
AUTHOR CONTRIBUTIONS
MJHvC and FvB contributed to study coordination and design, data collection, data analysis, writing of manuscript, and approval of final version. MP, JF and DD involved in laboratory work, data analysis, critical review of the manuscript and approval of final version. HLAJ and TB contributed to study coordination and design, data collection, critical review of the manuscript, and approval of final version. AB and AJvV involved in data collection, critical review of the manuscript and approval of final version. BEH involved in study design, statistical analysis, critical review of the manuscript and approval of final version. All authors had access to the study data and have reviewed and approved the final manuscript.
ACKNOWLEDGEMENTS
The authors would like to thank Anthonie Groothuismink and Buddy Roovers from the Department of Gastroenterology and Hepatology, Erasmus MC University Medical Center (Rotterdam) for retrieval of serum samples.
van Campenhout MJH, van Bömmel F, Pfefferkorn M, et al. Serum hepatitis B virus RNA predicts response to peginterferon treatment in HBeAg‐positive chronic hepatitis B. J Viral Hepat. 2020;27:610–619. 10.1111/jvh.13272
This current study was a retrospective study and was therefore not registered at clinicaltrials.gov.
Funding information
This study was initiated and supported by the Foundation for Liver Research, Rotterdam, the Netherlands. Financial support was provided by F. Hoffmann‐La Roche Ltd., Basel, Switzerland. For the original studies, financial support, study medication and drug supply were provided by F. Hoffmann‐La Roche Ltd. (Basel, Switzerland), Bristol Myers Squibb (BMS, New York, United States), with additional financial support provided by the Virgo consortium, funded by the Dutch government project number FES0908, and by the Netherlands Genomics Initiative (NGI) project number 050‐060‐452. The funding sources did not have any influence on study design, data collection, analysis and interpretation of the data, writing of the report nor the decision to submit for publication.
REFERENCES
* Author names with asterisks designate shared co‐first authorship
- 1. World Health Organization . Hepatitis B. World Health Organization Fact Sheet 204 (Updated July 2017). 2017.
- 2. De Clercq E, Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29(3):695‐747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liver. EAftSot . EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67(2):370‐398. [DOI] [PubMed] [Google Scholar]
- 4. Lucifora J, Protzer U. Attacking hepatitis B virus cccDNA–The holy grail to hepatitis B cure. J Hepatol. 2016;64(1 Suppl):S41‐S48. [DOI] [PubMed] [Google Scholar]
- 5. Su TH, Kao JH. Unmet needs in clinical and basic hepatitis B virus research. J Infect Dis. 2017;216(suppl_8):S750‐S756. [DOI] [PubMed] [Google Scholar]
- 6. Wang J, Shen T, Huang X, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol. 2016;65(4):700‐710. [DOI] [PubMed] [Google Scholar]
- 7. Jansen L, Kootstra NA, van Dort KA, Takkenberg RB, Reesink HW, Zaaijer HL. Hepatitis B virus pregenomic RNA Is present in virions in plasma and is associated with a response to pegylated interferon alfa‐2a and nucleos(t)ide analogues. J Infect Dis. 2016;213(2):224‐232. [DOI] [PubMed] [Google Scholar]
- 8. Wang J, Du M, Huang H, et al. Reply to: "Serum HBV pgRNA as a clinical marker for cccDNA activity": Consistent loss of serum HBV RNA might predict the "para‐functional cure" of chronic hepatitis B. J Hepatol. 2017;66(2):462‐463. [DOI] [PubMed] [Google Scholar]
- 9. Giersch K, Allweiss L, Volz T, Dandri M, Lutgehetmann M. Serum HBV pgRNA as a clinical marker for cccDNA activity. J Hepatol. 2017;66(2):460‐462. [DOI] [PubMed] [Google Scholar]
- 10. Wang J, Niu J, Jiang J, Lu F. Serum HBV RNA can reflect the activity of intrahepatic cccDNA. Hepatol Int. 2017;11(1):S6‐S7. [Google Scholar]
- 11. van Bommel F, van Bommel A, Krauel A, et al. Serum HBV RNA as a predictor of peginterferon alfa‐2a response in patients with HBeAg‐positive chronic hepatitis B. J Infect Dis. 2018;218(7):1066‐1074. [DOI] [PubMed] [Google Scholar]
- 12. Ahmad Shah P, Rajab I, Choudhry S, et al. The application of novel HBV pgRNA assay to predict HBeAg clearance on long‐term nucleos (t)ide analogues. J Hepatol. 2019;70(1):e262‐e263. [Google Scholar]
- 13. Carey I, Gersch J, Moigboi C, et al. HBV DNA relapse after stopping nucleoside analogue therapy in patients with HBsAg loss: Detectable pre‐genomic HBV RNA is a better predictor of relapse than ultra‐sensitive HBsAg‐'implications for HBV cure’. J Hepatol. 2019;70(1):e254. [Google Scholar]
- 14. Janssen HL, van Zonneveld M, Senturk H, et al. Pegylated interferon alfa‐2b alone or in combination with lamivudine for HBeAg‐positive chronic hepatitis B: a randomised trial. Lancet. 2005;365(9454):123‐129. [DOI] [PubMed] [Google Scholar]
- 15. van Bommel F, Bartens A, Mysickova A, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61(1):66‐76. [DOI] [PubMed] [Google Scholar]
- 16. Lam AM, Ren S, Espiritu C, et al. Hepatitis B virus capsid assembly modulators, but not nucleoside analogs, inhibit the production of extracellular pregenomic RNA and spliced RNA variants. Antimicrob Agents Chemother. 2017;61(8):e00680‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Agarwal K, Gane EJ, Cheng W, et al. HBcrAg, HBV‐RNA declines in A phase 2a study evaluating the multi‐dose activity of ARB‐1467 in HBeAg‐positive and negative virally suppressed subjects with hepatitis B. Hepatology. 2017;66:22A‐23A. [Google Scholar]
- 18. Jia W, Zhu MQ, Zhang JM. Serum hepatitis B virus RNA levels as a predictor of HBeAg seroconversion during treatment with peginterferon alfa‐2a in hepatitis B e antigen‐positive patients. Hepatol Int. 2017;11(1):S702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu S, Zhou B, Valdes JD, Sun J, Guo H. Serum hepatitis B virus RNA: a new potential biomarker for chronic hepatitis B virus infection. Hepatology. 2019;69(4):1816‐1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Bömmel F*, van Bömmel A*, Krauel A, et al. Serum HBV RNA is an early predictor of HBeAg seroconversion in patients with chronic Hepatitis B (CHB) treated with pegylated interferon alfa‐2a (40KD). Hepatology 2015;62((Van Bömmel F.; Krauel A.; Deichsel D.; Berg T.; Boehm S.) Clinic of Gastroenterology and Rheumatology, University Hospital Leipzig, Leipzig, Germany):336A. [Google Scholar]
- 21. van Campenhout MJH, van Bommel F, Pfefferkorn M, et al. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology. 2018;68(3):839‐847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Belloni L, Allweiss L, Guerrieri F, et al. IFN‐alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest. 2012;122(2):529‐537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uprichard SL, Wieland SF, Althage A, Chisari FV. Transcriptional and posttranscriptional control of hepatitis B virus gene expression. Proc Natl Acad Sci USA. 2003;100(3):1310‐1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pasquetto V, Wieland SF, Uprichard SL, Tripodi M, Chisari FV. Cytokine‐sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J Virol. 2002;76(11):5646‐5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Biron CA. Interferons alpha and beta as immune regulators–a new look. Immunity. 2001;14(6):661‐664. [DOI] [PubMed] [Google Scholar]
- 26. Wieland SF, Guidotti LG, Chisari FV. Intrahepatic induction of alpha/beta interferon eliminates viral RNA‐containing capsids in hepatitis B virus transgenic mice. J Virol. 2000;74(9):4165‐4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rang A, Gunther S, Will H. Effect of interferon alpha on hepatitis B virus replication and gene expression in transiently transfected human hepatoma cells. J Hepatol. 1999;31(5):791‐799. [DOI] [PubMed] [Google Scholar]


