Abstract
Poly[adenosine diphosphate (ADP) ribose]polymerase (PARP) has multifaceted roles in the maintenance of genomic integrity, deoxyribonucleic acid (DNA) repair and replication, and the maintenance of immune-system homeostasis. PARP inhibitors are an attractive oncologic therapy, causing direct cancer cell cytotoxicity by propagating DNA damage and indirectly, by various mechanisms of immunostimulation, including activation of the cGAS/STING pathway, paracrine stimulation of dendritic cells, increased T-cell infiltration, and upregulation of death-ligand receptors to increase susceptibility to natural-killer-cell killing. However, these immunostimulatory effects are counterbalanced by PARPi-mediated upregulation of programmed cell-death-ligand 1 (PD-L1), which leads to immunosuppression. Combining PARP inhibition with immune-checkpoint blockade seeks to exploit the immune stimulatory effects of PARP inhibition while negating the immunosuppressive effects of PD-L1 upregulation.
Keywords: gynecologic cancer, immune-checkpoint inhibition, immune modulation, ovarian cancer, PARP inhibitor, PD-L1, STING
Introduction
Poly[adenosine diphosphate (ADP) ribose]polymerase (PARP) inhibitors (PARPi) and immune-checkpoint inhibitors have revolutionized the treatment paradigm for many cancer types. This is particularly notable in gynecologic cancers, with multiple US Food and Drug Administration (FDA) approvals for PARPi in recent years across all lines of treatment in ovarian cancer. Here, we review the mechanisms of PARP inhibition, the connection between deoxyribonucleic acid (DNA) damage, PARP, and immunogenicity, and the rationale for combined PARP inhibition and immune-checkpoint blockade (ICB) within the context of gynecologic cancers.
The PARP superfamily is composed of 17 proteins with varying catalytic abilities and functions. Though the majority of PARP proteins generate mono(ADP ribose; MAR) modifications,1 a few (PARP1, PARP2, and PARP5A/B) generate poly(ADP ribose; PAR) moieties. As the majority of available data focuses on PARP1 and PARP2, this review will also focus on these PARP proteins. PARP1 and PARP2 share a common Trp-Gly-Arg (WGR) domain, which interacts with DNA and regulates catalytic activity.2 PARP1 differs in having zinc-finger domains, a BRCA-C-terminus (BRCT) domain, and a WGR domain that is activated by DNA breaks, irrespective of the presence of phosphorylation groups. PARP2 has only a short N-terminal domain and a WGR domain that preferentially binds phosphorylated DNA breaks. These variances may underlie some differences in these protein’s functions, though globally, both PARP1 and PARP2 play roles in gene expression, cell signaling, and genome integrity. Overall, PARP1 is thus far considered to contribute the majority of known PARP function.
To date, the PARP inhibitors olaparib, rucaparib, and niraparib are FDA approved for the treatment of epithelial ovarian cancer (EOC). All three drugs potently inhibit PARP1 and PARP2, with nanomolar half-maximal inhibitory concentration values.3 They are generally considered of comparable clinical efficacy, though harbor some differences in toxicity profiles.4 Currently, there are no approved indications for PARPi in the treatment of other gynecologic malignancies, though PARPi are under active study in endometrial and cervical cancers.
DNA repair and the role of PARP
High-fidelity repair of DNA damage is critical for cell survival. If DNA damage or replicative errors are unrepaired and propagated forward, the accumulation of progressive genomic instability can lead to cell death. PARPi are used in the treatment of gynecologic cancers, particularly in the context of existing DNA damage-repair deficiencies, for this purpose.
In response to particular types of DNA damage, PARP1 is recruited to and binds the sites of damage.5 DNA binding triggers a conformational change and PARP1 activation, leading to the addition of PAR moieties to itself and other proteins. PAR chains act as binding sites, recruiting and activating proteins involved in the DNA repair process upon PAR binding. This displaces PARP1 from DNA and catalyzes the repair process. In parallel, PARP1 will stall and protect replication forks, allowing time for DNA repair.
PARP1 is crucial to several repair pathways.5,6 Single-strand breaks (SSBs) in DNA, generated spontaneously or through base modifications, are rapidly recognized by PARP1. Upon binding, PARP1 recruits target proteins including the scaffold protein XRCC1, DNA ligase-3 and DNA polymerase β, catalyzing SSB repair.5,7 Base excision repair (BER), in which SSBs are generated in the process of an endonuclease removing a damaged base, therefore may also rely on PARP1,8 though data is conflicting as to whether PARP1 is essential for BER.8–10 Additionally, PARP1 may function in the nucleotide excision repair (NER) pathway, which removes stretches of damaged single-strand DNA (ssDNA), fills in the resultant gap, and ligates the repaired strand.5,11
Double-strand breaks (DSBs), generated by DNA-damaging agents or from the collapse of replication forks, rely on PARP1 in multiple roles in several repair pathways. Firstly, PARP1 recognizes, and is recruited to, sites of DSBs, where PARylation recruits additional repair proteins, including ataxia–telangiectasia mutated (ATM) and the nuclease MRE,11 as part of the MRE11/RAD50/NBS1 (MRN) complex.12 DSBs can be repaired through homologous recombination (HR) or non-homologous end joining (NHEJ), a branchpoint influenced by the balance between functional BRCA1 and 53BP1. Evidence suggests that PARP2 limits 53BP1 accumulation, thereby promoting end-resection and HR.13 In high-fidelity HR repair, BRCA1 is recruited by PARP1 to sites of DSBs and helps to stabilize end-resected DNA.14 Alternatively, DSBs can be repaired through the less-fidelitous NHEJ pathway.15,16 In both classical and alternative NHEJ, PARP1 remains important for the appropriate recruitment of NHEJ-associated factors. NHEJ is particularly error prone compared with HR, due to the lack of a sister chromatid template and resultant insertions and deletions.15
PARP1 additionally plays a vital role in DNA synthesis by engaging with DNA replication-associated proteins and stabilizing replication forks in the setting of replication stress.17 Importantly, PARP1 interaction with the DNA helicase RECQ1 slows and reverses replication forks, preventing inappropriate fork movement into unrepaired DNA lesions, which would cause replication fork collapse and formation of DSBs.17,18 Loading of RAD51 to damaged replication forks is regulated by both PARP1 and PARP2.10
PARP1 impacts chromatin structure to better facilitate DNA accessibility and allow DNA damage repair. After recruitment to sites of DNA damage, PARP1 PARylates histones, which promote nucleosome disassociation and recruit chromatin remodelers to further induce chromatin relaxation so damaged DNA is more accessible for repair.19,20
Therefore, PARPi can be highly deleterious due to effects at multiple points in the DNA repair and synthesis process. The negative effects of PARPi are amplified in the context of a cancer cell that may have underlying DNA repair- or cell-cycle-associated alterations. For example, high grade serous ovarian carcinoma (HGSOC), the most common subtype of EOC, is typified by high copy number variation, uncontrolled cell proliferation (most commonly due to p53 loss of function), and defective homologous recombination, all contributing to a high baseline degree of genomic instability. Therefore, HGSOC is exquisitely predisposed to additional perturbation in the DNA repair process, such as by PARPi.
This forms the basis for synthetic lethality by PARPi in BRCA1/2-mutated cancers.5,21 PARP inhibition leads to the accumulation of unrepaired SSBs, which, in the setting of replication, are processed into DSBs that, due to the lack of a functional BRCA1/2, cannot be repaired through HR. DSBs are additionally unable to be repaired through alternative NHEJ, despite BRCA-deficient cells relying on polymerase-θ-mediated alternative NHEJ, as this process also relies on PARP. PARPi also disrupt the carefully controlled action of the PARP1-recruited MRE11 nuclease at the sites of replication-fork restart, which requires PARP1 and BRCA2 to appropriately disengage. In the setting of PARP inhibition and BRCA2 deficiency, the nuclease remains engaged on the DNA strand, leading to uncontrolled strand degradation, replication fork collapse, and DSBs that cannot be repaired. Lastly, PARP inhibition traps PARP on DNA, generating a DNA-protein complex that stalls replication forks; the process of replication-fork restart requires BRCA1/2 and functional PARP.
The immune system: role of PARP and effects of PARP deficiency or inhibition
It is now well recognized that the interface between the immune system and cancer is a dynamic, complex process. The immune system is engaged throughout tumorigenesis, including recognition of malignant cells and inflammation, immune exhaustion and pruning, and immune surveillance. It is therefore important to understand what roles PARP plays in the functioning immune system and what effects PARP deficiency and/or inhibition may have on specific immune cells.
Effects on T-cells
T-cell development is highly regulated, occurring through several steps of maturation and involving a complex system of transcription factors and cytokines. T-cells play numerous roles in cancer development, shaped by the signaling milieu in the tumor microenvironment.22
PARP2 appears to be involved early in T-cell maturation, specifically in the development of CD4/CD8 double-positive thymocytes.23 PARP2 deficiency was associated with reduced total and double-positive populations of thymocytes; this was not seen with PARP1 deficiency.23,24 In double-positive thymocytes, PARP2 is theorized to be critical for repair of DNA strand breaks generated in the process of T-cell-receptor-α rearrangement.24 Without PARP2, unrepaired DNA breaks initiate a pro-apoptotic cascade.
In a murine model of dually PARP1/2-deficient T-cells, total populations of CD4 single-positive and CD8 single-positive T-cells were reduced, with greater reduction in the CD8 lineage. These results were seen only in dual PARP1/2 deficiency and not in singular PARP1 or singular PARP2 deficiency, with concurrently elevated markers of DNA damage and apoptosis suggesting that reduced T-cell populations were due to accumulation of genomic instability precipitating cell death and not solely a block in maturation.23 In the same murine model of background PARP1 deficiency and selective PARP2 deficiency in T-cells, implanted breast cancer cells grew larger and more rapidly compared with single PARP1- or single PARP2-deficient cells;25 intratumoral CD4 and CD8 T-cell infiltration was decreased, likely related to dual PARP1/2-deficiency-related lymphocyte cell death.
Expression of Foxp3 marks the differentiation of CD4 single-positive T-cells in regulatory T-cells (Tregs) and imparts the immunosuppressive capability of Tregs. The expression of Foxp3 is regulated by PARP1, which acts post-translationally to PARylate Foxp3, marking it for ubiquitination by the Stub1 E3 ubiquitin ligase and subsequent degradation.26,27 The role of PARP1 in indirectly modulating Foxp3 transcription via Smad3 binding at the Foxp3 enhancer is debated.26,28 Overall, in the setting of PARP1 deficiency, the population of CD4/Foxp3-positive Tregs increased, due to persistence of Foxp3.26 Consequently, expression of genes downstream of Foxp3 was increased, including of CD25, CTLA-4, and interleukin 10 (IL-10). Though one study noted that the increase in expression was associated with greater suppressive function of Tregs on peripheral blood mononuclear cells,26 this may not wholly reflect a tumor microenvironment. For example, the role of secreted IL-10 has been shown to be immunostimulatory, rather than suppressive, in different tumor contexts.29–31
CD4 T-cell differentiation is driven by differential gene expression regulated by the NFAT (nuclear factor of activated T-cells) family of transcription factors.32 NFAT activity is itself modulated by PARP1, whereby PARP1 binds and PARylates NFAT, increasing its DNA binding ability and regulating its nuclear import and export.33,34 It is important to note that this activity of PARP1 occurred secondary to T-cell stimulation and not due to the presence of DNA damage.34 Therefore, PARP1 directly impacts T-cell differentiation. PARP1 deficiency in T-cells resulted in reduced expression of cytokines reliant on NFAT, including IL-2 and IL-4, suggesting further downstream effects on immune-cell differentiation.33 Furthermore, PARP1 deficiency and/or inhibition may bias CD4 T-cell differentiation to a Th1 phenotype rather than a Th2 phenotype,35–37 though conflicting data may underscore context-specific differences. In a model of airway inflammation, olaparib treatment yielded increases in the Th1-associated cytokine interferon-γ (IFNγ) and expression of T-bet, a Th1-associated T-box transcription factor, while suppressing expression of the Th2-associated cytokines IL-4, IL-5, IL6-, IL-13, and M-CSF,36 suggesting a skew toward a Th1 phenotype. Conversely, in a model of inflammatory arthritis, PARP inhibition was associated with reduced expression of Th1-associated cytokines TNFα and IFNγ and partially inhibited Th1-cell clonal expansion.38
Furthermore, PARP1 modulates transforming growth factor β (TGFβ)-receptor expression on CD4 T-cells. At least for TGFβ-receptor 2, this appears to be through direct binding of PARP on the tgfbr2 promoter to affect its transcription.28 Interestingly, PARP1 deficiency was associated with higher expression of TGFβ receptors, but inhibition of PARP1 enzymatic activity was associated only with increased TGFβ-receptor-1 expression, suggesting differential regulation. PARP inhibition also predisposed T-cells to greater sensitivity to TGFβ, and PARP1 deficiency with concurrent TGFβ treatment was associated with an increased Th17 population, which requires TGFβ for differentiation,28 suggesting that PARP1 plays this additional role in T-cell differentiation.
In addition to affecting T-cell differentiation, PARP1 and PARP2 affect T-cell function. In a murine model of background PARP1 deficiency with selective PARP2 deficiency in T-cells, the populations of activated CD4 and CD8 T-cells secreting IL-2 and IFNγ in response to viral inoculation were diminished.23 Dual PARP1/2 deficient models had a more dramatic reduction compared with models of singular PARP1 or singular PARP2 deficiency, suggesting additive roles in effector T-cell function. Furthermore, in the same murine model, CD4 and CD8 T-cells infiltrating implanted breast cancer tumors had reduced expression of genes associated with chemotaxis, T-cell activation, and T-cell-mediated cytotoxicity.25 Notably, gene expression was not changed in either PARP1 or PARP2 deficiency.
In vivo models of BRCA1-deficient ovarian cancer demonstrated that PARP inhibition using olaparib significantly increased the number of effector CD4 and CD8 T-cells intratumorally and peripherally, demonstrating the global effects of PARP inhibition.39 Moreover, olaparib-treated CD8 T-cells had reduced expression of the immune-checkpoint receptors PD-1, Tim-3, and Lag-3, associated with T-cell inhibition and exhaustion, and produced significantly higher levels of TNFα and IFNγ. Interestingly, intratumoral CD4/Foxp3 Tregs were not increased following olaparib treatment, counter to expectations in considering PARP1 modulation of Foxp3 expression. These results suggest overall that PARP inhibition was associated with an activated effector T-cell response with alteration in immune-checkpoint receptor expression that could predispose to response to immune-checkpoint blockade (ICB).
Effects on B cells
There is increasing evidence for a role of B-cells in malignancy, including both pro- and antitumor functions depending on the tumor microenvironment.40,41 B-cells play important roles in generating antibodies, but also modulate immunity independently of antibody generation, via interactions with effector cells and antigen-presenting cells.
V(D)J gene recombination is critical for the appropriate generation of immunoglobulins, occurring in the pre-B-cell stage. The generation and pairing of VLJL and VHDJH generate immunoglobulin M (IgM) in immature B-cells. Later on, mature B-cells undergo class-switching recombination, altering the immunoglobulin isotype, for example to IgG. Both the V(D)J and class-switching recombination processes generate DSBs which are repaired through the PARP1-mediated NHEJ pathway, thus giving rise to the question of whether PARPi may impact humoral immunity. In steady-state conditions without introduction of an antigen stimulus, serum IgM and IgG levels were comparable between PARP1/2-proficient, singular PARP1 deficient, singular PARP2 deficient, and dual PARP1/2 deficient mice.42 Therefore, despite the role of PARP in NHEJ, PARP1/2 did not appear to be critical for V(D)J recombination nor class switching. Interestingly, dual PARP1/2 deficiency in B-cells did not impact Ig V(D)J recombination, baseline serum levels of IgM and IgG, or antibody responses to T-cell-dependent antigens,23,42 but led to reduced serum IgG levels in response to T-cell-independent antigens.42
PARP plays a role in maintaining B-cell homeostasis, most notably in mediating the differentiation of transitional B-cells into follicular B-ells. Bone marrow B-cell progenitors and peripheral mature B-cells were preserved in a genetically engineered mouse model of dual PARP1/2 deficiency; however, peripheral transitional and follicular B-cell populations were significantly depleted.42 This suggests a crucial role of PARP at that level of B-cell differentiation. Notably, dual PARP1/2-deficient B-cells accumulated DNA damage and apoptosed at a faster rate compared with control, leading to B-cell lymphopenia, and underscoring the important role of PARP1/2 in maintaining genomic stability, even in immune cells.
PARP may additionally play a role in the expression of Bcl-6, a transcription factor essential for the generation of germinal centers and high-affinity antibodies. PARP1 binds the first intron of Bcl-6 and suppresses its transcription. PARP inhibition and PARP knockdown in vitro induced expression of Bcl-6, corroborating the inhibitory role of PARP1 in B-cell differentiation into germinal-center B-cells.43 It remains unknown at this time how B-cell homeostasis and immunoglobulin responses are affected by PARP-inhibition treatment in solid malignancies.
Effects on dendritic cells
Dendritic cells are pivotal antigen-presenting cells with the ability to activate and induce differentiation of T-cells,44,45 conferring an antitumor microenvironment.
PARP1 has been shown in several contexts to be important for the recruitment of dendritic cells to sites of inflammation, possibly through regulation of VCAM-1 expression.46–48 In contrast, in a murine model of PARP1 deficiency and selective PARP2 deficiency in T-cells, intratumoral infiltration by CD11b dendritic cells was higher compared with settings of PARP1 deficiency, PARP2 deficiency, or control.25 Whether PARP is critical to the function of dendritic cells remains unclear, as some studies suggest that PARPi impaired the maturation and antigen presenting function of DCs,47,48 while other studies did not.46
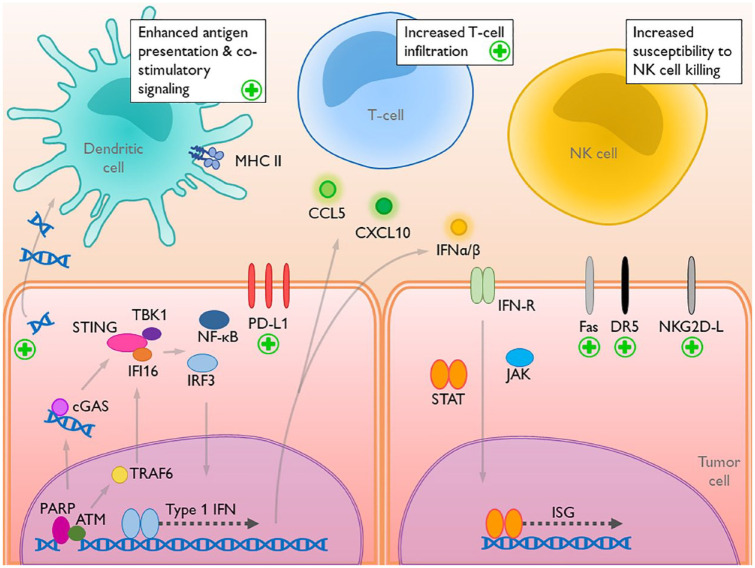
There is evidence for an indirect role of PARP1 in activating dendritic cells (Figure 1). The synthetic lethality of PARP1 in an HR-deficient setting generates DNA damage and genomic instability, leading to micronuclei and cytosolic DNA. Cytosolic DNA activates the cyclic guanosine monophosphate (GMP)–adenine monophosphate synthase (cGAS)/stimulator of interferon genes (STING) pathway within the tumor cell, but is also exocytosed to act in a paracrine fashion, activating the cGAS/STING pathway in neighboring dendritic cells.49,50 In a BRCA1-deficient model of triple-negative breast cancer, olaparib was associated with significantly upregulated levels of tank-binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3), markers of activated cGAS/STING.49 However, olaparib treatment of dendritic cells alone did not induce cGAS/STING pathway activation, implying that the action of PARPi on dendritic cells was indirect. Ultimately, the in vivo dendritic cell population increased and demonstrated increased antigen presentation and recruitment of CD8 T-cells. These findings were replicated in a BRCA1-deficient model of ovarian cancer.39 In response to treatment with olaparib, tumor-associated dendritic cells were increased in number, with upregulated cell-surface costimulatory CD80 and CD86 and antigen-presenting major histocompatibility complex class II. Additionally, co-culturing olaparib-treated ovarian cancer cells with naïve dendritic cells led to increased levels of TBK1, IRF3, CXCL10, and IFNβ, indicating cGAS/STING activation and downstream gene expression, confirming the paracrine effect of PARPi on dendritic cells. Dendritic cells activated by cytosolic DNA stimulated CD4 T-cells to generate Th1-type cytokines, and induced formation of cytotoxic CD4 and CD8 T-cells.51
Figure 1.
The interaction between DNA damage, intratumoral alterations, and activation of the immune cells.
Plus signs denote where PARP inhibition acts to influence the tumor–immune interaction. PARP inhibition increases DNA damage, generating cytosolic DNA that activates cGAS/STING intratumorally and in dendritic cells. PARP inhibition upregulates costimulatory CD80/CD86 and MHC class II on dendritic cells, enhancing antigen presentation and T-cell interactions. PD-L1 is upregulated following PARP inhibition, due to several mechanisms. Increased expression of T-cell chemokines increase tumor infiltration of activated, effector T-cells. Increased expression of cell-death receptor ligands and NKG2D ligands increase tumor-cell sensitivity to NK-cell killing.
ATM, ataxia–telangiectasia mutated; CCL5, C-C motif ligand 5; cGAS, cyclic guanosine monophosphate–adenosine monophosphate synthase; CXCL10, C-X-C motif chemokine 10; DNA, deoxyribonucleic acid; DR5, death receptor 5; IFI16, gamma-interferon inducible protein IFI-16; IFN, interferon; IFN-R, interferon receptor; ISG, IFN-stimulated gene; JAK, Janus kinase; MHC II, major histocompatibility complex class II; NF-kB, nuclear-factor-kappa light-chain enhancer of activated B cells; NK, natural killer; NKG2D-L, natural-killer group 2 member D ligand; PARP, poly(ADP ribose)polymerase; PD-L1, programmed cell-death ligand 1; STAT, signal transducer and activator of transcription proteins; STING, stimulator of interferon genes; TBK1, tank-binding kinase 1; TRAF6, tumor necrosis factor receptor-associated factor 6.
Effects on macrophages
Macrophage phenotype is influenced by exposure to specific antigens and cytokines.52 Stimulation by Th1-associated cytokines promote a pro-inflammatory M1 phenotype, while stimulation by Th2-associated cytokines promote an anti-inflammatory M2 phenotype. However, macrophage polarization and functions exist on a continuum between M1 and M2 and therefore, in turn, may be tumoricidal or tumorigenic.52,53
PARP1 may modulate macrophage phenotype polarization through its regulation of high-mobility group box protein 1 (HMGB1), an inflammatory mediator with macrophage-differentiating effects. In the setting of lipopolysaccharide stimulation, PARP1 PARylates HMGB1, facilitating its acetylation and subsequent displacement from chromatin, inducing migration of HMGB1 from the nucleus to the cytoplasm.54 This is paralleled by HMGB1 cytosolic translocation due to PARP1 activation in the setting of alkylating DNA damage.55 Cytosolic HMGB1 can be exocytosed through a lysosomal pathway and secreted into the extracellular space as a damage-associated molecular pattern (DAMP) with cytokine and chemokine functions. Though HMGB1 can signal polarization into an M1 phenotype via interaction with the receptor for advanced glycation products (RAGE),56 it can also direct M2 polarization through interaction with C1q complement.57 Under oxidative stress, PARP1 appears to protect M1 macrophages from cell death.58 In a pancreatic cancer model, PARP1 deficiency was associated with significantly fewer tumor-infiltrating macrophages, thought to be related to the concomitant decrease in vascular endothelial growth-factor receptor (VEGFR) expression.59,60 Ultimately, the effects of PARP and PARPi on macrophages will be directed by the immediate microenvironment and signaling milieu.
The cancer cell: role of PARP and effects of PARP inhibition
DNA damage and the immune response
Maintenance of genomic stability requires careful coordination of DNA damage repair, DNA synthesis, and cell-cycle regulation. Under conditions of genomic stress, double-stranded DNA is released into the cytoplasm,61 initiating a cell-intrinsic innate immune response through the well-characterized cGAS/STING pathway.
The numerous roles of PARP in maintaining genomic stability result in significant deleterious effects of PARPi on the genome. Expectedly, PARP inhibition is associated with increased levels of cytosolic DNA,62 which are detected by cGAS, leading to production of the secondary messenger cyclic GMP. cGMP activates STING, prompting the recruitment of TBK1 and IFI16, activation of the IRF3 and NF-κB transcription factors, nuclear translocation of IRF3 and NF-κB, and expression of several genes that mediate an innate immune response. This includes expression of type 1 interferons and T-cell-recruiting chemokines (CCL5, CXCL10), leading to higher percentages of tumor-infiltrating T-cells.49,62–66 The specific phenotypes of recruited T-cells are an area of active investigation, as there are conflicting data regarding increased levels of CD4 T-cells, CD8 T-cells, and CD4/Foxp3 Tregs, with PARPi alone.39,49,62,64,67 Notably, downstream markers of cGAS/STING activation were greater in the setting of BRCA deficiency (i.e. greater genomic instability) compared with BRCA-proficient cells.49,68 Secreted IFN can act in an autocrine or paracrine manner, stimulating the JAK/STAT (Janus kinase/signal transducer and activator of transcription proteins) pathway and expression of interferon-stimulated genes.68
Additionally, PARP1’s role in sensing DNA damage mediates a non-canonical pathway of STING activation.69 Upon binding DSBs, PARP1 recruits and activates ATM, which subsequently activates the ubiquitin ligase TRAF6. Translocation of TRAF6 to the cytosol and association with IFI16 and p53 results in STING activation. This non-canonical pathway, independent of cGAS, preferentially generates the pro-inflammatory transcription factor NF-κB, and to a lesser extent, generates activated IRF3.69–72
Evasion of NK-cell-mediated cancer cell death
NK cells possess potent cytolytic abilities, mediated by direct cytotoxicity through release of perforin and granzyme, direct interaction with target cells via TNF-related apoptosis-inducing ligand (TRAIL) and Fas ligand, or indirectly through secretion of IFNγ and TNFα to stimulate apoptotic pathways in target cells.73,74 NK cells play crucial roles in the antitumor immune response, as they are able to kill tumor cells without prior antigen exposure.
In tumor cells, similar to in immune-system cells, PARP1 is required for HMGB1 localization to the cytoplasm. However, in the tumor-cell context, PARP1 activation occurred via stimulation by TRAIL; subsequent HMGB1 cytoplasmic localization promoted an autophagic response, protecting the tumor cell from TRAIL-induced caspase-8-mediated apoptosis.75 Suppression of the PARP1/HMGB1 pathway via PARP inhibition and PARP deficiency reversed this resistance and sensitized cancer cells to TRAIL-mediated cell death.75 This suggests that antitumor effects of PARP inhibition may include enhanced tumor-cell sensitivity to NK-cell-mediated TRAIL activation and apoptosis.
PARPi-modulated susceptibility to NK-cell-mediated cytotoxicity may also occur through upregulation of death receptors on tumor cells. Olaparib and veliparib upregulated transcription and protein expression of the death receptors Fas and death receptor 5, sensitizing several cell lines, including ovarian cancer, lung cancer, and leukemic cells, to TRAIL-induced cell death.76 The effects of PARPi appeared to be specific to inhibition of PARP1 and PARP2 and not of the other PARP isoforms. These findings were recapitulated in prostate cancer cells, wherein NK cell killing was greater in cells pretreated with olaparib compared with control.77 These findings demonstrate that PARPi-induced antitumor effects are at least partially mediated by NK cells.
Furthermore, cancer-cell-surface expression of NKG2D ligands, which interact with NKG2D on NK cells to effect tumor-cell death, is downregulated by PARP1. In vitro and in vivo AML models demonstrated PARP1-mediated suppression of NKG2D ligands, and PARP inhibition was sufficient to suppress leukemogenesis.78 Interestingly, in addition to immune cells, cancer cells can also express the NKG2D receptor, co-opting it for autonomous stimulation and oncogenic signaling. In ovarian cancer, NKG2D expression was associated with increased cancer cell self-renewal capacity and tumor spheroid formation,79 though it is unclear how PARP may regulate autonomous NKG2D signaling.
Regulation of immune-checkpoint ligands
Olaparib, talazoparib, rucaparib, and PARP1 knockdown resulted in higher levels of cancer cell PD-L1 expression in breast cancer cells in vitro and in vivo, an effect seen regardless of BRCA proficiency or deficiency.67 There are several possible mechanisms of PD-L1 upregulation. Transcription factor NF-kB, generated in response to activated IFI16/STING/TBK1 in human papillomavirus (HPV)-positive cervical cancer, binds the PD-L1 promoter, upregulating transcription.80 However, knockdown of IRF3, another crucial transcription factor in the cGAS/STING pathway, was sufficient to abrogate upregulation of PD-L1 in response to PARPi,64 suggesting multiple mechanisms of PD-L1 regulation. JAK1/2 activation in tumor cells is also sufficient to induce PD-L1,81 and therefore it is possible that PD-L1 is upregulated due to autocrine or paracrine JAK/STAT signaling downstream of a cGAS/STING/type 1 IFN response. IFN-y was sufficient to induce PD-L1 expression in a non-small-cell lung cancer cell line,63 and therefore PD-L1 may be upregulated in response to IFNγ secretion by T-cells or NK cells recruited following the cGAS/STING/type 1 interferon response. PD-L1 expression is additionally modulated by glycogen synthase kinase 3β (GSK3β), which induces phosphorylation-dependent degradation of PD-L1.82 Inactivation of GSK3β was associated with stabilized expression of PD-L1. PARP inhibition generates inactivated GSK3β, thereby preventing PD-L1 degradation,67 though the exact mechanism linking PARP and GSK3β is not yet known.
Rationale for combined immune-checkpoint blockade and PARP inhibition
Preclinical and correlative data provide strong support for the combination of PARP inhibition and ICB.
PARP inhibition has widespread effects on cells in both innate and adaptive immune responses (Figure 1). DNA damage and genomic instability generated by PARPi activates the cytosolic DNA sensing cGAS/STING pathway, culminating in a type 1 interferon response with several immunogenic effects. T-cell-associated chemokines increase T-cell recruitment and tumoral infiltration. PARP inhibition, via its effect on NFAT, may bias CD4 T-cell differentiation into pro-inflammatory Th1 cells, and may promote pro-inflammatory Th17-cell differentiation via its effects on the TGFβ receptor. PARPi-treated CD8 T-cells downregulated the immune-checkpoint receptors PD-1, Tim-3, and Lag-3, suggesting decreased propensity for T-cell inhibition and exhaustion, despite PARPi-associated increases in tumor-cell expression of PD-L1. Additionally, tumoral cytosolic DNA can act in a paracrine manner to activate the cGAS/STING pathway in dendritic cells, leading to increased antigen-presenting ability. Furthermore, PARP inhibition increases the sensitivity of tumor cells to NK-cell-mediated TRAIL-induced apoptosis. Taken together, PARPi may stimulate an immunogenic tumor microenvironment.
Disruption of immune-checkpoint interactions between tumor cells and T-cells has been the primary focus of immuno-oncologic development. In view of the numerous mechanisms of PARPi-associated PD-L1 upregulation, ICB is a logical pairing. In a murine model of small-cell lung cancer, PARP inhibition significantly increased PD-L1 expression, and dual inhibition with olaparib and an anti-PD-L1 agent induced significant tumor regression, greater than either agent alone.64 Correlative tumor analysis demonstrated markedly increased populations of CD3 T-cells and cytotoxic CD8 T-cells, and decreased populations of CD4/Foxp3 Tregs and exhausted PD-1/Tim-3 CD8 T-cells. The cGAS/STING pathway was directly implicated, as cGAS knockdown abolished the antitumor effect of combination olaparib/anti-PD-L1 therapy. In a BRCA1-deficient ovarian cancer model, combined olaparib and anti-PD-1 therapy significantly extended the survival of tumor-bearing mice compared with olaparib alone.39 In a breast cancer model, olaparib upregulated PD-L1 in both BRCA-deficient and -proficient cells, and combined olaparib and anti-PD-L1 treatment resulted in T-cell-mediated tumor cell death that was greater than olaparib alone.67 A preclinical study of niraparib and pembrolizumab in BRCA-mutated breast cancer, BRCA-mutated ovarian cancer, and BRCA wild-type skin-cancer cells demonstrated that niraparib induced T-cell infiltration regardless of BRCA status.66 Combination niraparib/pembrolizumab yielded better antitumor activity than either agent alone. There is early evidence that dual inhibition may reprogram the tumor microenvironment in a durable fashion. One mouse achieved a complete response with niraparib/pembrolizumab and growth of a second implanted tumor was prevented, even in the absence of active treatment, indicating an enduring antitumor response.66 This study also reported that niraparib/pembrolizumab treatment inhibited tumor growth in a sarcoma xenograft previously refractory to anti-PD-1 treatment. This suggests that the addition of PARPi was able to induce an inflammatory immune response sufficient to overcome prior ICB resistance.
Perspectives for gynecologic cancers
In theory, the combination of PARPi and ICB may provide benefit in two specific situations: to induce a greater or more durable response in settings of PARPi sensitivity or HR deficiency, or to gain antitumor effect in the setting of PARPi resistance or HR proficiency.
Combination PARPi + ICB in HR deficiency
In the setting of HR deficiency, and therefore assumed PARPi sensitivity, the combination of PARPi with ICB would presumably capitalize on the synthetic lethality of PARPi, a potentially higher neoantigen load, and the immunogenic effects of both. For example, HGSOC accounts for the majority of epithelial ovarian cancer (EOC). Within HGSOC, approximately 50% are HR deficient,83,84 harboring germline (~14%) and somatic (6%) BRCA1/2 mutations, BRCA1 promoter methylation (10%), and alterations in other HR-associated genes, such as RAD51C. In trials of PARPi as a single agent and in combinations in frontline maintenance, recurrence monotherapy, and recurrence maintenance settings, almost all of which were restricted to HGSOC, the subgroup of HR-deficient EOC has consistently achieved greater benefit than HR-proficient subgroups, underscoring the significant effect of synthetic lethality.85–90 Moreover, HR-deficient HGSOC exhibits higher neoantigen loads, increased CD3 and CD8 T-cell infiltration, a higher CD8:CD4 T-cell ratio, and higher PD-L1 expression compared with HR-proficient HGSOC, emphasizing the inherent immunogenicity of tumors with defective DNA repair.91 The phase II MEDIOLA trial [ClinicalTrials.gov identifier: NCT02734004] evaluating the combination of olaparib and durvalumab in 32 patients with germline BRCA1/2-mutated platinum-sensitive recurrent EOC found an overall response rate (ORR) of 71.9%, 28-week disease control rate (DCR) of 65.9%, median progression-free survival (PFS) of 11.1 months, and median duration of response of 10.2 months.92 There were seven patients (21.8%) with complete responses (CRs) and median overall survival (OS) was not yet reached at time of data presentation. In comparison, in the randomized phase III SOLO3 trial comparing olaparib alone with physician’s choice non-platinum chemotherapy in patients with germline BRCA1/2-mutated platinum-sensitive recurrent EOC, treatment with olaparib yielded an ORR of 72.2%, including 14 patients (9.3%) achieving a CR, and median PFS of 13.4 months, by blinded independent central review.93 Of olaparib-treated patients with a partial response (PR) or CR, median duration of response was 9.4 months. Data immaturity precluded OS estimation. Acknowledging the limitations of cross-trial comparisons, the MEDIOLA and SOLO3 trials appear to have similar response rates, median PFS estimates, and durations of response. This prompts the questions of whether and to what extent preclinical evidence of combination PARPi and ICB is borne out in clinical practice, and whether HR deficiency is the best context in which to visualize possible benefits of combined PARPi and ICB. Ongoing clinical trials (FIRST trial of niraparib/dostarlimab [ClinicalTrials.gov identifier: NCT03602859], ATHENA trial of rucaparib/nivolumab [ClinicalTrials.gov identifier: NCT03522246]) in the maintenance setting after first-line therapy will hopefully address whether addition of ICB is superior to PARPi alone in HR-deficient ovarian cancer.
This has bearing on other gynecologic malignancies in which subsets of disease are also HR deficient. For example, in endometrial cancer, alterations in HR-related genes as detected by next-generation sequencing was found in 20–30% of cases, occurring primarily in non-endometrioid, p53-mutant endometrial cancers,94,95 likely reflecting the copy-number-high/serous-like molecular subgroup.96 In one study, 46% of non-endometrioid endometrial cancer specimens were HR deficient by functional assay.94 Loss or deficiency of MRE11, part of the MRN complex crucial for end resection in HR, sensitized an endometrial carcinoma cell line to talazoparib.97 Furthermore, PTEN-deficient endometrial cancer cells predispose to PARPi sensitivity in vitro, theorized to be due to transcriptional downregulation of RAD51.98–101 Case reports describe PARPi inducing clinical responses in endometrial carcinomas, one case involving a patient with PTEN-deficient disease102 and another involving a patient with BRCA-mutated endometrial carcinoma;103 however, to date there are no published clinical trial data of PARPi in HR-deficient endometrial carcinoma. Nonetheless, trials of combination PARPi and ICB in endometrial carcinoma are ongoing, including combinations of olaparib and durvalumab [ClinicalTrials.gov identifier: NCT03951415], rucaparib and nivolumab [ClinicalTrials.gov identifier: NCT03572478], and niraparib and TSR-042 (dostarlimab, anti-PD-1) [ClinicalTrials.gov identifier: NCT03016338]. The combination of avelumab and talazoparib [ClinicalTrials.gov identifier: NCT02912572] is being investigated in microsatellite-stable (MSS) endometrial carcinoma, a population also typified by genomic stability.
HPV-positive cervical cancer, accounting for nearly all cervical cancer cases, may also be deficient in effective HR. While the prevailing understanding of HPV-induced carcinogenesis involves E6- and E7-mediated cell-cycle dysregulation through their well-documented effects on p53 and Rb, evidence is emerging for virally mediated HR deficiency and genomic instability.104–106 This may explain in part the platinum and radiation sensitivity seen clinically. High-risk HPV E6 and E7 are thought to impair HR by initiating HR in the wrong phase of the cell cycle (i.e. in G1), when a sister chromatid template is not present, thereby preventing completion of HR despite upregulation of HR-related genes105,107 and by impairing the correct localization of RAD51 to DSBs, further hindering HR. There may be additional mechanisms contributing to ineffective HR and/or genomic instability, including decreased levels of RAD52,108 hypermethylation-related downregulation of RAD51,109 impaired recruitment of downstream repair factors due to E7-mediated interaction with the E3 ubiquitin ligase RNF168,110 and suppression of NHEJ and shunting of DSB repair toward the more error-prone microhomology-mediated end joining.111 Interestingly, in vitro analysis of nine patient-derived cervical-cancer cell lines, eight of which were HPV 16- or HPV 18-positive, found that none met the criteria of HR deficiency, defined using log2-ratios and allele frequencies to generate a loss of heterozygosity score in a method used previously in EOC trials.112 Despite this, olaparib inhibited tumor growth and induced apoptosis in three of the cell lines and suppressed xenograft tumor growth from a sensitive cell line. Therefore, a subset of cervical cancer may be amenable to synthetic lethality using PARPi; adding ICB may take further advantage of PD-L1 expression and/or amplification.113,114 Though there are trials of PARPi combined with chemotherapy and/or radiotherapy in cervical cancer, there are thus far no published clinical trials of PARPi monotherapy nor, to our knowledge, ongoing trials of PARPi and ICB.
Combination PARPi + ICB in HR proficiency
The ability of combination PARPi and ICB to inhibit tumor growth regardless of BRCA status (i.e. regardless of HR repair status) suggests that this combination should not be limited only to states of HR deficiency. Indeed, the benefit of PARPi/ICB regardless of HR status is exemplified in the phase I/II TOPACIO/KEYNOTE-162 trial of combined niraparib and pembrolizumab.115 In the pooled platinum-resistant EOC cohort of 62 patients, response rates were similar regardless of tumor BRCA or HR status. The majority of patients with EOC had BRCA wild-type (79%) or HR-proficient (53%) disease. An ORR of 19% was seen in subgroup analysis in both the BRCA wild-type and HR-proficient groups, similar to the ORR of 18% in the BRCA-mutated group, and ORR of 14% in the HR-deficient group. Intriguingly, five of eight patients achieving a PR or CR lasting longer than 6 months had tumors that were BRCA wild type. Correlative work, which included immunogenomic profiling and highly multiplexed single-cell imaging on tumor samples from patients enrolled the study, identified two determinants of response; mutational signature 3 reflecting defective HR, and positive immune score as a surrogate of interferon-primed exhausted CD8 + T-cells in the tumor microenvironment.116 Absence of both features yielded no responses, while presence of one or both features captured all objective responses. Single-cell spatial analysis revealed prominent interactions of exhausted CD8 + T-cells with PD-L1 + macrophages and/or PD-L1 + tumor cells as mechanistic determinants of response. Of note, two extreme responders showed differential clustering of exhausted CD8 + T-cells either with PD-L1 + macrophages in the first patient, or with cancer cells harboring genomic PD-L1 and PD-L2 amplification in the second patient.
Similarly, a phase II trial of olaparib and durvalumab [ClinicalTrials.gov identifier: NCT02484404] in 35 patients with predominantly platinum-resistant (86%), BRCA wild-type (77%) recurrent EOC yielded an ORR of 14%.117 However, a subset of patients experienced a durable benefit, as evidenced by a clinical benefit rate (PR + SD ⩾6 months) of 34%, including 10 platinum resistant patients, of whom three patients attained a PR with a median duration on study of 17.2 months, and of whom seven patients achieved a median of 7.3 months’ disease stabilization. Correlative studies of paired pre- and on-treatment tissue and blood specimens indicated that the combination of olaparib and durvalumab promoted an immune-inflamed environment, with increased tumoral IFNγ and CXCL9/CXCL10 expression and increased systemic IFNγ and TNFα production. Increased systemic IFNγ was associated with clinical benefit and improved PFS (HR 0.37, p = 0.023). Treatment significantly increased tumor-infiltrating lymphocytes (TILs) and increased PD-L1 expression compared with pre-treatment specimens. Interestingly, PARPi may not induce an effect through the STING pathway. Only 4 of 14 patients with available samples had increased expression of STING following treatment, none of whom experienced a response. However, in patients without increased STING expression but who achieved clinical benefit, the type 1 interferon downstream inflammatory chemokines CCL4 and CCL5 were still increased, suggesting an alternative pathway of activation. Additionally, increased post-treatment levels of VEGF3R3 were associated with worse PFS, suggesting that activation of the VEGF/VEGFR pathway may arise to compensate for treatment-related immunostimulatory changes. Accepting the limitations of cross-trial comparisons, response rates in this and the TOPACIO trial are in line with responses to PARPi monotherapy in similar populations.88,118
Conclusion
Preclinical studies suggest a strong mechanistic rationale for pairing PARPi and ICB, specifically capitalizing on PARPi-associated PD-L1 upregulation, and preliminary evidence of clinical activity has been demonstrated in early-phase trials. Though most extensively studied in EOC, evidence for HR deficiency in endometrial and cervical cancers highlights additional opportunities to study PARPi and ICB combinations. Several phase III studies investigating combination PARPi/ICB are ongoing, including the phase III ATHENA trial of rucaparib/nivolumab [ClinicalTrials.gov identifier: NCT03522246] and the phase III FIRST trial of niraparib/dostarlimab (TSR-042) [ClinicalTrials.gov identifier: NCT03602859], both in the frontline treatment of EOC and unrestricted for HR status. These studies will also evaluate whether the benefit of adding ICB to PARPi is confined to HR-deficient or HR-proficient ovarian cancers or both. Importantly, these trials will compare combination PARPi/ICB against monotherapy of either or both agents, as a critical question in the pursuit of novel combinations is whether, and to what extent, combination therapy improves upon the benefit seen with monotherapy. Moving forward, it will be imperative to understand how PARPi/ICB alters the tumor microenvironment and immune milieu in a clinical setting, whether there might be any clinically relevant difference in activity between HR-deficient or -proficient cancers, and what resistance mechanisms may arise from combination treatment.
Footnotes
Conflict of interest statement: EKL declares no conflicting interests.
PAK reports participation in advisory boards from GSK/Tesaro, Merck, AstraZeneca, and Bayer, outside the submitted work.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Elizabeth K. Lee  https://orcid.org/0000-0001-7533-7853
https://orcid.org/0000-0001-7533-7853
Panagiotis A. Konstantinopoulos  https://orcid.org/0000-0002-1032-1479
https://orcid.org/0000-0002-1032-1479
Contributor Information
Elizabeth K. Lee, Department of Medical Oncology, Division of Gynecologic Oncology, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02115, USA
Panagiotis A. Konstantinopoulos, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA.
References
- 1. Vyas S, Matic I, Uchima L, et al. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun 2014; 5: 4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Langelier MF, Eisemann T, Riccio AA, et al. PARP family enzymes: regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr Opin Struct Biol 2018; 53: 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antolin AA, Ameratunga M, Banerji U, et al. The kinase polypharmacology landscape of clinical PARP inhibitors. Sci Rep 2020; 10: 2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. LaFargue CJ, Molin GZD, Sood AK, et al. Exploring and comparing adverse events between PARP inhibitors. Lancet Oncol 2019; 20: e15–e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 2017; 18: 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Konstantinopoulos PA, Matulonis UA. PARP inhibitors in ovarian cancer: a trailblazing and transformative journey. Clin Cancer Res 2018; 24: 4062–4065. [DOI] [PubMed] [Google Scholar]
- 7. El-Khamisy SF, Masutani M, Suzuki H, et al. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res 2003; 31: 5526–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dantzer F, Schreiber V, Niedergang C, et al. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie 1999; 81: 69–75. [DOI] [PubMed] [Google Scholar]
- 9. Vodenicharov MD. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res 2000; 28: 3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ronson GE, Piberger AL, Higgs MR, et al. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat Commun 2018; 9: 746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pines A, Vrouwe MG, Marteijn JA, et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol 2012; 199: 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haince JF, McDonald D, Rodrigue A, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 2008; 283: 1197–1208. [DOI] [PubMed] [Google Scholar]
- 13. Fouquin A, Guirouilh-Barbat J, Lopez B, et al. PARP2 controls double-strand break repair pathway choice by limiting 53BP1 accumulation at DNA damage sites and promoting end-resection. Nucleic Acids Res 2017; 45: 12325–12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 2013; 23: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet 2013; 47: 433–455. [DOI] [PubMed] [Google Scholar]
- 16. Ceccaldi R, Liu JC, Amunugama R, et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 2015; 518: 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ray Chaudhuri A, Hashimoto Y, Herrador R, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol 2012; 19: 417–423. [DOI] [PubMed] [Google Scholar]
- 18. Berti M, Chaudhuri AR, Thangavel S, et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol 2013; 20: 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Messner S, Altmeyer M, Zhao H, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res 2010; 38: 6350–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poirier GG, De Murcia G, Jongstra-Bilen J, et al. Poly (ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A 1982; 79: 3423–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D’Andrea AD. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair (Amst) 2018; 71: 172–176. [DOI] [PubMed] [Google Scholar]
- 22. Thommen DS, Schumacher TN. T cell dysfunction in cancer. Cancer Cell 2018; 33: 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Navarro J, Gozalbo-López B, Méndez AC, et al. PARP-1/PARP-2 double deficiency in mouse T cells results in faulty immune responses and T lymphomas. Nat Publ Gr 2017; 7: 41962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yélamos J, Monreal Y, Saenz L, et al. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J 2006; 25: 4350–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moreno-Lama L, Galindo-Campos MA, Martínez C, et al. Coordinated signals from PARP-1 and PARP-2 are required to establish a proper T cell immune response to breast tumors in mice. Oncogene 2020; 39: 2835–2843. [DOI] [PubMed] [Google Scholar]
- 26. Luo X, Nie J, Wang S, et al. Poly(ADP-ribosyl)ation of FOXP3 protein mediated by PARP-1 protein regulates the function of regulatory T cells. J Biol Chem 2015; 290: 28675–28682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang P, Maruyama T, Konkel JE, et al. PARP-1 controls immunosuppressive function of regulatory T cells by destabilizing Foxp3. PLoS One 2013; 8: e71590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang P, Nakatsukasa H, Tu E, et al. PARP-1 regulates expression of TGF-β receptors in T cells. Blood 2013; 122: 2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li L, Ma Y, Liu S, et al. Interleukin 10 promotes immune response by increasing the survival of activated CD8+ T cells in human papillomavirus 16-infected cervical cancer. Tumor Biol 2016; 37: 16093–16101. [DOI] [PubMed] [Google Scholar]
- 30. Zheng LM, Ojcius DM, Garaud F, et al. Interleukin-10 inhibits tumor metastasis through an NK cell-dependent mechanism. J Exp Med 1996; 184: 579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berman RM, Suzuki T, Tahara H, et al. Systemic administration of cellular IL-10 induces an effective, specific, and long-lived immune response against established tumors in mice. J Immunol 1996; 157: 231–238. [PubMed] [Google Scholar]
- 32. Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol 2005; 5: 472–484. [DOI] [PubMed] [Google Scholar]
- 33. Olabisi OA, Soto-Nieves N, Nieves E, et al. Regulation of transcription factor NFAT by ADP-ribosylation. Mol Cell Biol 2008; 28: 2860–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valdor R, Schreiber V, Saenz L, et al. Regulation of NFAT by poly(ADP-ribose) polymerase activity in T cells. Mol Immunol 2008; 45: 1863–1871. [DOI] [PubMed] [Google Scholar]
- 35. Saenz L, Lozano JJ, Valdor R, et al. Transcriptional regulation by Poly(ADP-ribose) polymerase-1 during T cell activation. BMC Genomics 2008; 9: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ghonim MA, Pyakurel K, Ibba SV, et al. PARP inhibition by olaparib or gene knockout blocks asthma-like manifestation in mice by modulating CD4+ T cell function. J Transl Med 2015; 13: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sambucci M, Laudisi F, Novelli F, et al. Effects of PARP-1 deficiency on Th1 and Th2 cell differentiation. Sci World J 2013; 2013: 375024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gonzalez-Rey E, Martínez-Romero R, O’Valle F, et al. Therapeutic effect of a poly(ADP-ribose) polymerase-1 inhibitor on experimental arthritis by downregulating inflammation and Th1 response. PLoS One 2007; 2: e1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ding L, Kim HJ, Wang Q, et al. PARP inhibition elicits STING-dependent antitumor immunity in brca1-deficient ovarian cancer. Cell Rep 2018; 25: 2972–2980.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Largeot A, Pagano G, Gonder S, et al. The B-side of cancer immunity: the underrated tune. Cells 2019; 8: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsou P, Katayama H, Ostrin EJ, et al. The emerging role of B cells in tumor immunity. Cancer Res 2016; 76: 5591–5601. [DOI] [PubMed] [Google Scholar]
- 42. Galindo-Campos MA, Bedora-Faure M, Farrés J, et al. Coordinated signals from the DNA repair enzymes PARP-1 and PARP-2 promotes B-cell development and function. Cell Death Differ 2019; 26: 2667–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ambrose HE, Papadopoulou V, Beswick RW, et al. Poly-(ADP-ribose) polymerase-1 (Parp-1) binds in a sequence-specific manner at the Bcl-6 locus and contributes to the regulation of Bcl-6 transcription. Oncogene 2007; 26: 6244–6252. [DOI] [PubMed] [Google Scholar]
- 44. Patente TA, Pinho MP, Oliveira AA, et al. Human dendritic cells: their heterogeneity and clinical application potential in cancer immunotherapy. Front Immunol 2019; 10: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tran Janco JM, Lamichhane P, Karyampudi L, et al. Tumor-infiltrating dendritic cells in cancer pathogenesis. J Immunol 2015; 194: 2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Echeverri Tirado LC, Ghonim MA, Wang J, et al. PARP-1 is critical for recruitment of dendritic cells to the lung in a mouse model of asthma but dispensable for their differentiation and function. Mediators Inflamm 2019; 2019: 1656484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cavone L, Aldinucci A, Ballerini C, et al. PARP-1 inhibition prevents CNS migration of dendritic cells during EAE, suppressing the encephalitogenic response and relapse severity. Mult Scler J 2011; 17: 794–807. [DOI] [PubMed] [Google Scholar]
- 48. Wang JQ, Tang Y, Li QS, et al. PARG regulates the proliferation and differentiation of DCs and T cells via PARP/NF-κB in tumour metastases of colon carcinoma. Oncol Rep 2019; 41: 2657–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pantelidou C, Sonzogni O, De Oliveria Taveira M, et al. PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov 2019; 9: 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mouw KW, Goldberg MS, Konstantinopoulos PA, et al. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov 2017; 7: 675–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kis-Toth K, Szanto A, Thai TH, et al. Cytosolic DNA-activated human dendritic cells are potent activators of the adaptive immune response. J Immunol 2011; 187: 1222–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 2014; 14: 392–404. [DOI] [PubMed] [Google Scholar]
- 53. Krishnan V, Schaar B, Tallapragada S, et al. Tumor associated macrophages in gynecologic cancers. Gynecol Oncol 2018; 149: 205–213. [DOI] [PubMed] [Google Scholar]
- 54. Yang Z, Li L, Chen L, et al. PARP-1 mediates LPS-induced HMGB1 release by macrophages through regulation of HMGB1 acetylation. J Immunol 2014; 193: 6114–6123. [DOI] [PubMed] [Google Scholar]
- 55. Ditsworth D, Zong WX, Thompson CB. Activation of poly(ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J Biol Chem 2007; 282: 17845–17854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu T, Xiang A, Peng T, et al. HMGB1–C1q complexes regulate macrophage function by switching between leukotriene and specialized proresolving mediator biosynthesis. Proc Natl Acad Sci U S A 2019; 116: 23254–23263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Son M, Porat A, He M, et al. C1q and HMGB1 reciprocally regulate human macrophage polarization. Blood 2016; 128: 2218–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tokarz P, Płoszaj T, Regdon Z, et al. PARP1-LSD1 functional interplay controls transcription of SOD2 that protects human pro-inflammatory macrophages from death under an oxidative condition. Free Radic Biol Med 2019; 131: 218–224. [DOI] [PubMed] [Google Scholar]
- 59. Martínez-Bosch N, Iglesias M, Munné-Collado J, et al. PARP-1 genetic ablation in Ela-myc mice unveils novel roles for PARP-1 in pancreatic cancer. J Pathol 2014; 234: 214–227. [DOI] [PubMed] [Google Scholar]
- 60. Li C, Liu B, Dai Z, et al. Knockdown of VEGF receptor-1 (VEGFR-1) impairs macrophage infiltration, angiogenesis and growth of clear cell renal cell carcinoma (CRCC). Cancer Biol Ther 2011; 12: 872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dhanwani R, Takahashi M, Sharma S. Cytosolic sensing of immuno-stimulatory DNA, the enemy within. Curr Opin Immunol 2018; 50: 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shen J, Zhao W, Ju Z, et al. PARPi Triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res 2019; 79: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chabanon RM, Lord CJ, Postel-Vinay S. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J Clin Invest 2019; 129: 1211–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sen T, Rodriguez BL, Chen L, et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov 2019; 9: 646–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang J, Wang L, Cong Z, et al. The PARP1 inhibitor BMN 673 exhibits immunoregulatory effects in a Brca1−/– murine model of ovarian cancer. Biochem Biophys Res Commun 2015; 463: 551–556. [DOI] [PubMed] [Google Scholar]
- 66. Wang Z, Sun K, Xiao Y, et al. Niraparib activates interferon signaling and potentiates anti-PD-1 antibody efficacy in tumor models. Sci Rep 2019; 9: 1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jiao S, Xia W, Yamaguchi H, et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res 2017; 23: 3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reisländer T, Lombardi EP, Groelly FJ, et al. BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat Commun 2019; 10: 3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dunphy G, Flannery SM, Almine JF, et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol Cell 2018; 71: 745–760.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Miyamoto S. Nuclear initiated NF-κB signaling: NEMO and ATM take center stage. Cell Res 2011; 21: 116–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bakhoum SF, Ngo B, Laughney Ashley M, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018; 553: 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu ZH, Shi Y, Tibbetts RS, et al. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 2006; 311: 1141–1146. [DOI] [PubMed] [Google Scholar]
- 73. Souza-Fonseca-Guimaraes F, Cursons J, Huntington ND. The emergence of natural killer cells as a major target in cancer immunotherapy. Trends Immunol 2019; 40: 142–158. [DOI] [PubMed] [Google Scholar]
- 74. Barrow AD, Colonna M. Tailoring natural killer cell immunotherapy to the tumour microenvironment. Semin Immunol 2017; 31: 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang M, Liu L, Xie M, et al. Poly-ADP-ribosylation of HMGB1 regulates TNFSF10/TRAIL resistance through autophagy. Autophagy 2015; 11: 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Meng XW, Koh BD, Zhang JS, et al. Poly(ADP-ribose) polymerase inhibitors sensitize cancer cells to death receptor-mediated apoptosis by enhancing death receptor expression. J Biol Chem 2014; 289: 20543–20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fenerty KE, Padget M, Wolfson B, et al. Immunotherapy utilizing the combination of natural killer- and antibody dependent cellular cytotoxicity (ADCC)-mediating agents with poly (ADP-ribose) polymerase (PARP) inhibition 11 medical and health sciences 1112 oncology and carcinogenesis 11 medical. J Immunother Cancer 2018; 6: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Paczulla AM, Rothfelder K, Raffel S, et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature 2019; 572: 254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cai X, Caballero-Benitez A, Gewe MM, et al. Control of tumor initiation by NKG2D naturally expressed on ovarian cancer cells 1,2. Neoplasia 2017; 19: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cai H, Yan L, Liu N, et al. IFI16 promotes cervical cancer progression by upregulating PD-L1 in immunomicroenvironment through STING-TBK1-NF-kB pathway. Biomed Pharmacother 2020; 123: 109790. [DOI] [PubMed] [Google Scholar]
- 81. Bellucci R, Martin A, Bommarito D, et al. Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Oncoimmunology 2015; 4: e1008824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li CW, Lim SO, Xia W, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun 2016; 7: 12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Konstantinopoulos PA, Ceccaldi R, Shapiro GI, et al. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov 2015; 5: 1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bell D, Berchuck A, Birrer M, et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. González-Martín A, Pothuri B, Vergote I, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2019; 381: 2391–2402. [DOI] [PubMed] [Google Scholar]
- 86. Coleman RL, Fleming GF, Brady MF, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med 2019; 381: 2403–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ray-Coquard I, Pautier P, Pignata S, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019; 381: 2416–2428. [DOI] [PubMed] [Google Scholar]
- 88. Moore KN, Secord AA, Geller MA, et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol 2019; 20: 636–648. [DOI] [PubMed] [Google Scholar]
- 89. Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017; 390: 1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Oza AM, Tinker AV, Oaknin A, et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: integrated analysis of data from study 10 and ARIEL2. Gynecol Oncol 2017; 147: 267–275. [DOI] [PubMed] [Google Scholar]
- 91. Strickland KC, Howitt BE, Shukla SA, et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016; 7: 13587–13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Drew Y, Kaufman B, Banerjee S, et al. Phase II study of olaparib + durvalumab (MEDIOLA): updated results in germline BRCA-mutated platinum-sensitive relapsed (PSR) ovarian cancer (OC). Ann Oncol 2019; 30: v485–v486. [Google Scholar]
- 93. Penson RT, Villalobos Valencia R, Cibula D, et al. Olaparib versus nonplatinum chemotherapy in patients with platinum-sensitive relapsed ovarian cancer and a germline BRCA1/2 mutation (SOLO3): a randomized phase III trial. J Clin Oncol 2020; 38: 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. De Jonge MM, Auguste A, Van Wijk LM, et al. Frequent homologous recombination deficiency in high-grade endometrial carcinomas. Clin Cancer Res 2019; 25: 1087–1097. [DOI] [PubMed] [Google Scholar]
- 95. Heeke AL, Pishvaian MJ, Lynce F, et al. Prevalence of homologous recombination–related gene mutations across multiple cancer types. JCO Precis Oncol 2018; 2: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cancer Genome Atlas Research Network; Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013; 497: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Koppensteiner R, Samartzis EP, Noske A, et al. Effect of MRE11 loss on PARP-inhibitor sensitivity in endometrial cancer in vitro. PLoS One 2014; 9: 100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dedes KJ, Wetterskog D, Mendes-Pereira AM, et al. PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med 2010; 2: 53–75. [DOI] [PubMed] [Google Scholar]
- 99. Mendes-Pereira AM, Martin SA, Brough R, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med 2009; 1: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Shen WH, Balajee AS, Wang J, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 2007; 128: 157–170. [DOI] [PubMed] [Google Scholar]
- 101. Philip CA, Laskov I, Beauchamp MC, et al. Inhibition of PI3K-AKT-mTOR pathway sensitizes endometrial cancer cell lines to PARP inhibitors. BMC Cancer 2017; 17: 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Forster MD, Dedes KJ, Sandhu S, et al. Treatment with olaparib in a patient with PTEN-deficient endometrioid endometrial cancer. Nat Rev Clin Oncol 2011; 8: 302–306. [DOI] [PubMed] [Google Scholar]
- 103. Gockley AA, Kolin DL, Awtrey CS, et al. Durable response in a woman with recurrent low-grade endometrioid endometrial cancer and a germline BRCA2 mutation treated with a PARP inhibitor. Gynecol Oncol 2018; 150: 219–226. [DOI] [PubMed] [Google Scholar]
- 104. Wallace NA. Catching HPV in the homologous recombination cookie jar. Trends Microbiol 2020; 28: 191–201. [DOI] [PubMed] [Google Scholar]
- 105. Wallace NA, Khanal S, Robinson KL, et al. High-risk alphapapillomavirus oncogenes impair the homologous recombination pathway. J Virol 2017; 91: e01084-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liu Q, Ma L, Jones T, et al. Subjugation of TGFb signaling by human papilloma virus in head and neck squamous cell carcinoma shifts DNA repair from homologous recombination to alternative end joining. Clin Cancer Res 2018; 24: 6001–6014. [DOI] [PubMed] [Google Scholar]
- 107. Roszik J, Ring KL, Wani KM, et al. Gene expression analysis identifies novel targets for cervical cancer therapy. Front Immunol 2018; 9: 2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Shi TY, Yang G, Tu XY, et al. RAD52 variants predict platinum resistance and prognosis of cervical cancer. PLoS One 2012; 7: e50461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Rieke DT, Ochsenreither S, Klinghammer K, et al. Methylation of RAD51B, XRCC3 and other homologous recombination genes is associated with expression of immune checkpoints and an inflammatory signature in squamous cell carcinoma of the head and neck, lung and cervix. Oncotarget 2016; 7: 75379–75393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Sitz J, Blanchet SA, Gameiro SF, et al. Human papillomavirus E7 oncoprotein targets RNF168 to hijack the host DNA damage response. Proc Natl Acad Sci U S A 2019; 116: 19552–19562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Leeman JE, Li Y, Bell A, et al. Human papillomavirus 16 promotes microhomology-mediated end-joining. Proc Natl Acad Sci U S A 2019; 116: 21573–21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bianchi A, Lopez S, Altwerger G, et al. PARP-1 activity (PAR) determines the sensitivity of cervical cancer to olaparib. Gynecol Oncol 2019; 155: 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Burk RD, Chen Z, Saller C, et al. Integrated genomic and molecular characterization of cervical cancer. Nature 2017; 543: 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Mezache L, Paniccia B, Nyinawabera A, et al. Enhanced expression of PD L1 in cervical intraepithelial neoplasia and cervical cancers. Mod Pathol 2015; 28: 1594–1602. [DOI] [PubMed] [Google Scholar]
- 115. Konstantinopoulos PA, Waggoner S, Vidal GA, et al. Single-arm phases 1 and 2 trial of niraparib in combination with pembrolizumab in patients with recurrent platinum-resistant ovarian carcinoma. JAMA Oncol 2019; 5: 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Färkkilä A, Gulhan DC, Casado J, et al. Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat Commun 2020; 11: 1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Lampert EJ, Zimmer AS, Padget MR, et al. Combination of PARP inhibitor olaparib, and PD-L1 inhibitor durvalumab, in recurrent ovarian cancer: a proof-of-concept phase 2 study. Clin Cancer Res. Epub ahead of print 12 May 2020. DOI: 10.1158/1078-0432.CCR-20-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Vanderstichele A, Van Nieuwenhuysen E, Han S, et al. Randomized phase II CLIO study on olaparib monotherapy versus chemotherapy in platinum-resistant ovarian cancer. J Clin Oncol 2019; 37(Suppl. 15): 5507. [Google Scholar]