Abstract
Background:
Hidradenitis suppurativa (HS) is a chronic, follicular, and inflammatory skin disease with multifactorial pathogenesis, and its definite molecule mechanism is still not fully elucidated.
Objective:
The purpose of this research was to identify crucial differentially expressed genes (DEGs) and biological processes and pathways involved in HS through bioinformatics analysis.
Methods:
A total of 30 tissues from patients with HS, 17 lesional and 13 healthy, were obtained from the GSE72702 data set of the Gene Expression Omnibus database. The DEGs were sorted by GEO2R and analyzed by Gene ontology and Kyoto Encyclopedia of Genes and Genomes analysis. MCODE analysis was visualized using Cytoscape.
Results:
Of the 723 identified DEGs, 364 were upregulated and 359 were downregulated. Upregulated DEGs were significantly enriched in immune response, inflammatory response, tumor necrosis factor signaling pathway, and so on and downregulated DEGs were enriched in the positive regulation of the peroxisome proliferators-activated receptor pathways, the regulation of lipolysis in adipocytes, and so on. The most significant DEGs group of 35 genes were screened.
Conclusion:
The internal biological information in HS can be revealed by bioinformatic methods, providing direction for further research and potential therapeutic targets.
Keywords: hidradenitis suppurativa, bioinformatics, differentially expressed gene, pathway
Introduction
Hidradenitis suppurativa (HS), also called acne inversa (AI), is a chronic, inflammatory, and recurrent follicular disease that primarily affects axillary, inguinal, and anogenital regions.1 The estimated incidence rate of the disease ranges between 1% and 4%.2 Young adults aged between 20 and 40 years have the highest incidence rates (4%), and the incidence rates are lower in individuals aged 50 years.3 Hidradenitis suppurativa is a painful and distressing disorder, is associated with depression, and significantly reduces the quality of life of the affected individuals.4 A broad range of comorbidities, such as metabolic syndrome, Crohn’s disease, and SpA, have been observed in patients with HS. Most importantly, HS is associated with substantially increased mortality (mean incidence ratio of 1.35; 95% CI: 1.15-1.59) particularly due to cardiovascular events (mean incidence ratio of 1.95; 95% CI: 1.42-2.67).5 Some advancements have been made in the elucidation of the pathological mechanism involved in the development of HS. Genetic and environmental factors, lifestyle, hormonal status, and microbiota induce immune activation around terminal hair follicles and the hyperkeratosis of the infundibulum.6 These changes result in follicular plugging and stasis. Bacterial propagation on the intertriginous skin usually acts as a booster for immune activation. The resident and migrated cells of the innate immune system (such as macrophages and granulocytes) and adaptive immune system (such as type1 T helper [TH1] cells and TH17 cells) secrete pro-inflammatory cytokines (such as tumor necrosis factor [TNF], interleukin [IL]-1β, and IL-17) to activate tissue cells, further enhancing immune cell infiltration and inflammation.
The pharmacological management of HS is generally aimed at producing anti-inflammatory effects. The intralesional application of glucocorticoid triamcinolone quickly alleviates pain.7 Traditional immunosuppressive drugs, such as cyclosporine, have good efficacy only in a small case series.8 In the recent years, evidence based on randomized controlled trials (RCTs) supports the efficacy of anti-TNF antibodies (such as adalimumab and infliximab).9-11 Open case series and small exploratory RCTs indicate that the targeting of other cytokines, such as IL-1, and those involved in the IL-12/IL-23 pathways, might be useful in the management of HS.12-14 Thus, finding molecular targets, such as genes, signaling pathways, and proteins, are highly necessary for improving the treatment outcome of HS.
In the present study, we aim to identify crucial genes, biological processes (BPs), pathways for putative molecular targets through bioinformatics analysis. The GEO2R online tool was used. Then, Gene ontology analysis (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, protein–protein interaction (PPI) network, and MCODE analysis were performed.
Materials and Methods
Microarray Data Information
We selected a gene expression profile in which the messenger RNA (mRNA) expression levels of lesional and clinically healthy skin of patients with HS significantly differ. We obtained the gene expression profile of GSE72702 between the lesional and clinically healthy skin of patients with HS from NCBI-GEO, a free public database of microarray profiles. The microarray data of GSE72702 was on account of GPL13158 Platforms ([HT_HG-U133_Plus_PM] Affymetrix HT HG-U133+ PM Array Plate) and included 17 lesional issues and 13 nonlesional tissues of patients with HS.
Data Processing of Differentially Expressed Genes
Differentially expressed genes (DEGs) between HS and healthy HS specimens were identified using GEO2R online tools with |logFC| >2, and the adjusted P value was <.05.15 The DEGs with log FC of <0 were considered downregulated genes, whereas the DEGs with log FC of >0 were considered upregulated genes.16
Gene Ontology and Pathway Enrichment Analysis
Gene ontology analysis was used for the unification of biology, collecting defined, structured, and controlled vocabulary for gene annotation, which mainly includes the following 3 categories: molecular function (MF), BP, and cellular component (CC).17 The KEGG is a collection of databases dealing with genomes, diseases, biological pathways, drugs, and chemical materials.18 DAVID, an online bioinformatic tool, facilitates the identification of the functions of a large number of genes and proteins.19 We used DAVID to visualize the DEG enrichment of BP, MF, CC, and pathways.
Protein–Protein Interaction Network and MCODE Analysis
Protein–protein interaction information can be evaluated by an online tool called STRING (Search Tool for the Retrieval of Interacting Genes). In this study, the STRING application in Cytoscape was used in examining the potential correlations among these DEGs (confidence score = 0.4). The application was also used in checking the modules of the PPI network and screening the most significant DEG group (degree cutoff = 2; maximum depth = 100; k-core = 2; and node score cutoff = 0.2).
Results
Identification of DEGs in HS
A total of 723 genes were differentially expressed in lesional skin tissues compared to nonlesional skin tissues, 364 genes were upregulated and 359 were downregulated. We listed only the top 10 upregulated DEGs and the top 10 downregulated DEGs in Table 1.
Table 1.
Top 10 Upregulated Genes and Top 10 Downregulated DEGs Comparing the Lesional Skin Tissues to Nonlesional Skin Tissues of Patients With HS.
| Expression | Gene name | Adjust P value | LogFC |
|---|---|---|---|
| Upregulated | S100A7A | 8.37E-05 | 4.85 |
| SERPINB4 | 1.02E-03 | 4.27 | |
| DEFB4A | 3.81E-04 | 3.81 | |
| S100A9 | 4.17E-05 | 3.54 | |
| ADAMDEC1 | 2.03E-04 | 3.38 | |
| SERPINB3 | 1.14E-04 | 3.21 | |
| IGKC | 1.74E-03 | 3.13 | |
| TCN1 | 2.59E-03 | 3.12 | |
| AKR1B10 | 1.27E-05 | 3.07 | |
| TDO2 | 9.05E-04 | 3.05 | |
| Downregulated | PIP | 4.66E-06 | −5.42 |
| DCD | 1.70E-04 | −5.31 | |
| SCGB2A2 | 2.76E-04 | −4.94 | |
| ADIPOQ | 2.24E-06 | −4.51 | |
| THRSP | 3.24E-03 | −3.8 | |
| TSPAN8 | 1.16E-05 | −3.79 | |
| SCGB1D2 | 1.02E-04 | −3.79 | |
| WIF1 | 2.24E-06 | −3.69 | |
| PLIN1 | 2.80E-07 | −3.44 | |
| BTC | 2.74E-04 | −3.05 |
Abbreviations: DEGs, differentially expressed genes; HS, hidradenitis suppurativa.
Gene Ontology and KEGG Pathway Analysis in HS
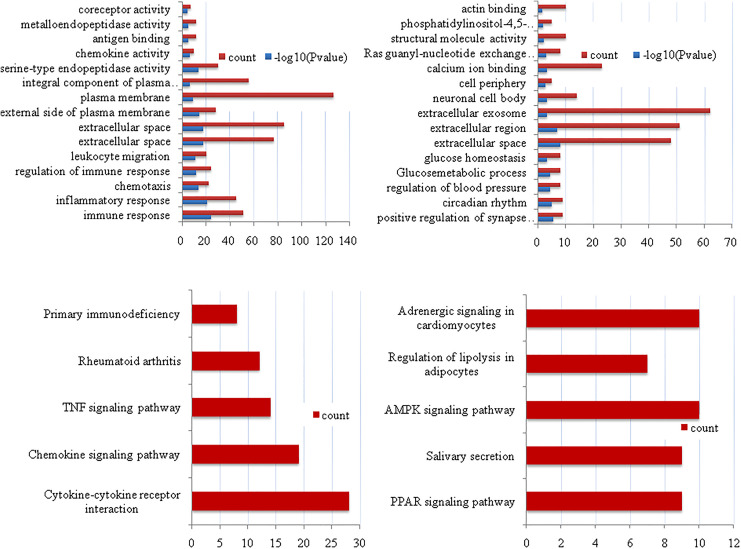
All 723 DEGs were analyzed using the DAVID software. The results (Table 2 and Figure 1) indicated that (1) for BP, upregulated DEGs were particularly enriched in the regulation of immune and inflammatory responses, chemotaxis, regulation of immune response, and leukocyte migration. The downregulated DEGs were enriched in the positive regulation of synapse assembly, circadian rhythm, extracellular space, Ras guanyl-nucleotide exchange factor activity, structural molecule activity, and phosphatidylinositol-4,5-bisphosphate 3-kinase activity. (2) For MF, upregulated DEGs were enriched in serine-type endopeptidase activity, chemokine activity, antigen binding, metalloendopeptidase activity, antigen binding, metalloendopeptidase activity, and coreceptor activity, and downregulated DEGs were enriched in calcium ion binding, ras guanyl-nucleotide exchange factor activity, structural molecule activity, phosphatidylinositol-4,5-bisphosphate 3-kinase activity, and actin binding. (3) For CC, upregulated DEGs were enriched in extracellular space, extracellular space, external side of plasma membrane, plasma membrane, and the integral component of the plasma membrane, and downregulated DEGs were enriched in extracellular space, extracellular region, extracellular exosome, neuronal cell body, and cell periphery.
Table 2.
Gene Ontology Analysis of Differentially Expressed Genes in Hidradenitis Suppurativa.
| Expression | Category | Term | P value | Count | FDR |
|---|---|---|---|---|---|
| Upregulated | BP | Immune response | 8.18E-25 | 51 | 1.40E-21 |
| BP | Inflammatory response | 1.65E-21 | 45 | 2.83E-18 | |
| BP | Chemotaxis | 3.32E-14 | 22 | 5.68E-11 | |
| BP | Regulation of immune response | 1.08E-12 | 24 | 1.84E-09 | |
| BP | Leukocyte migration | 3.63E-12 | 20 | 6.22E-09 | |
| CC | Extracellular space | 1.24E-18 | 77 | 1.61E-15 | |
| CC | Extracellular space | 1.53E-18 | 85 | 1.99E-15 | |
| CC | External side of plasma membrane | 4.50E-15 | 28 | 5.92E-15 | |
| CC | Plasma membrane | 1.34E-09 | 127 | 1.75E-06 | |
| CC | Integral component of plasma membrane | 1.81E-07 | 56 | 2.36E-04 | |
| MF | Serine-type endopeptidase activity | 1.74E-14 | 30 | 2.49E-11 | |
| MF | Chemokine activity | 3.67E-07 | 10 | 5.24E-04 | |
| MF | Antigen binding | 5.12E-06 | 12 | 0.007306 | |
| MF | Metalloendopeptidase activity | 1.26E-05 | 12 | 0.017925 | |
| MF | Coreceptor activity | 2.06E-05 | 7 | 0.029385 | |
| Downregulated | BP | Positive regulation of synapse assembly | 2.69E-06 | 9 | 0.004484 |
| BP | Circadian rhythm | 1.15E-05 | 9 | 0.019073 | |
| BP | Regulation of blood pressure | 2.69E-06 | 9 | 0.063886 | |
| BP | Glucose metabolic process | 1.15E-05 | 8 | 0.077889 | |
| BP | Glucose homeostasis | 3.84E-05 | 8 | 1.017447 | |
| CC | Extracellular space | 4.68E-05 | 8 | 1.30E-05 | |
| CC | Extracellular region | 6.14E-04 | 48 | 1.68E-04 | |
| CC | Extracellular exosome | 1.00E-08 | 51 | 0.589427 | |
| CC | Neuronal cell body | 1.29E-07 | 62 | 0.840461 | |
| CC | Cell periphery | 4.54E-04 | 14 | 2.931534 | |
| MF | Calcium ion binding | 6.47E-04 | 5 | 0.671142 | |
| MF | Ras guanyl-nucleotide exchange factor activity | 0.002281 | 23 | 1.63037 | |
| MF | Structural molecule activity | 4.77E-04 | 8 | 10.84205 | |
| MF | Phosphatidylinositol-4,5-bisphosphate 3-kinase activity | 0.001164 | 10 | 14.48553 | |
| MF | Actin binding | 0.025202 | 10 | 21.00713 |
Abbreviations: BP, biological process; CC, cellular component; MF, molecular function.
Figure 1.
Gene ontology enrichment analysis and KEGG pathway analysis. (A) Upregulated DEGs; (B) downregulated DEGs; (C) upregulated DEGs; and (D) downregulated DEGs. DEGs indicates differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Kyoto Encyclopedia of Genes and Genomes analysis results are shown in Table 3 and Figure 1. The upregulated DEGs were particularly enriched in cytokine–cytokine receptor interaction, chemokine signaling pathway, TNF signaling pathway, rheumatoid arthritis, and primary immunodeficiency, and the downregulated DEGs were enriched in the peroxisome proliferators-activated receptor (PPAR) signaling pathway, salivary secretion, adenosine 5’-monophosphate (AMP)-activated protein kinase (AMPK) signaling pathway, the regulation of lipolysis in adipocytes, and adrenergic signaling in cardiomyocytes.
Table 3.
KEGG Pathway Analysis of DEGs in Hidradenitis Suppurativa.
| Expression | Pathway | Count | Gene name |
|---|---|---|---|
| Upregulated | Cytokine–cytokine receptor interaction | 28 | CXCL1, CCL2, CCR1, CXCL2, IL21R, CXCL9, CCL8, CXCL6, CCL5, IL7R, CXCL10, IL12RB1, CCL20, CXCR4, IL4R, CXCR6, CSF2RB, IL1B, IL2RG, IL13RA1, CD27, IL6, TNFRSF17, IL24, CCL18, TNFSF10, CCR5, CCR2 |
| Chemokine signaling pathway | 19 | CXCL1, CCL2, FGR, LYN, CCR1, CXCL2, CXCL9, CCL8, CXCL6, CCL5, CCL18, CXCL10, RAC2, CCR5, CCL20, CXCR4, CCR2, CXCR6, JAK3 | |
| TNF signaling pathway | 14 | CXCL1, IL6, CCL2, SOCS3, MMP9, CXCL2, BIRC3, CCL5, MMP3, CXCL10, CCL20, BCL3, IL1B, SELE | |
| Rheumatoid arthritis | 12 | ITGAL, IL6, CD86, CCL2, CCL20, CTLA4, IL1B, CXCL6, CCL5, MMP3, MMP1, CD28 | |
| Primary immunodeficiency | 8 | CIITA, PTPRC, CD3D, CD3E, LCK, IL2RG, CD79A, IL7R | |
| Downregulated | PPAR signaling pathway | 9 | LPL, SORBS1, PLIN1, FABP4, SCD5, FABP7, ACADL, ADIPOQ, ACSBG1 |
| Salivary secretion | 9 | ADCY2, ADRB1, PLCB4, NOS1, CHRM3, SLC12A2, AQP5, ADRA1A, ATP1A2 | |
| AMPK signaling pathway | 10 | LEP, IRS4, IRS2, CAB39 L, EEF2 K, ADRA1A, PRKAA2, SCD5, ADIPOQ, PPARGC1A | |
| Regulation of lipolysis in adipocytes | 7 | IRS4, IRS2, ADCY2, ADRB1, PLIN1, FABP4, NPY1R | |
| Adrenergic signaling in cardiomyocytes | 10 | TNNT2, AGTR1, ADCY2, ADRB1, PLCB4, PPP1R1A, PLN, ADRA1A, SCN7A, ATP1A2 |
Abbreviations: AMPK, adenosine 5’-monophosphate (AMP)-activated protein kinase; DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPAR, peroxisome proliferators-activated receptor.
Protein–Protein Interaction Network and Modular Analysis
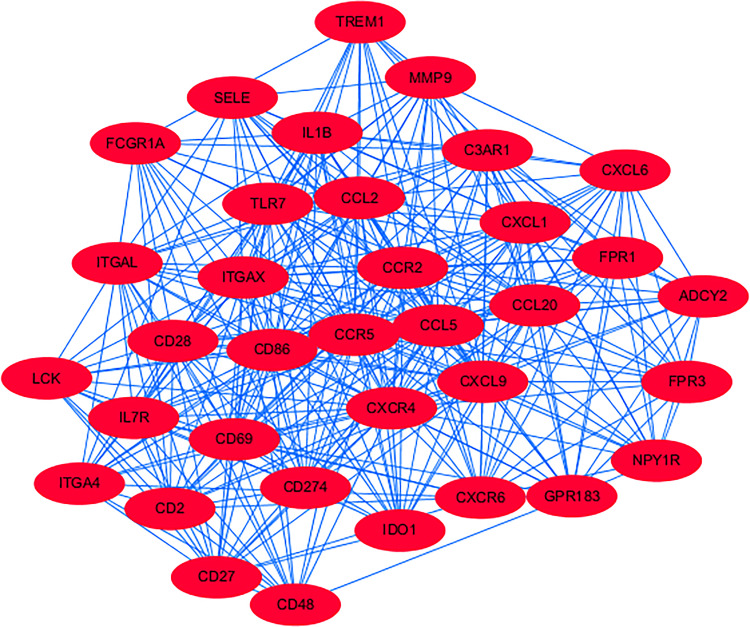
A total of 723 DEGs were analyzed for PPI. Then, we applied Cytoscape MCODE module for further analysis, and results showed that the most significant DEGs group shown in Figure 2 mainly includes 35 genes (High affinity immunoglobulin gamma Fc receptor I [FCGR1A], Indoleamine 2,3-dioxygenase 1 [IDO1], C-X-C Motif Chemokine Receptor 6 [CXCR6], IL1B, Integrin Subunit Alpha 4 [ITGA4], GPR183, and complement component 3a receptor 1 [C3AR1]).
Figure 2.
MCODE analysis via cytoscape software (degree cutoff = 2, node score cut off = 0.2, K-core = 2, and max, Depth = 100).
Discussion
Hidradenitis suppurativa is characterized by presence of recurrent abscesses and nodules and sinus tract formation and is mainly localized in body folds. The pathogenesis is poorly understood. Mutations in genes encoding the essential compounds of the transmembrane protease ү-secretase, including NCSTN, PSEN1, and PSENEN, have been identified in familial HS.20,21 Additionally, inflammatory pathways and immune dysregulation likely play a role. To acquire a better understanding of the molecular pathogenesis of HS, we investigated HS-related mRNA expression patterns through the analysis of GEO.
In this study, we identified 723 differentially expressed genes involved in HS by comparing the mRNA microarray in lesional skin tissues with that of non-lesional skin tissues. Based on the validation from GEO data sets, GO and KEGG using DAVID methods facilitate the discovery of key BPs and pathways involved in the HS.22 Then, 35 vital DEGs were screened by Cytoscape software using PPI network and MCODE analysis.
A total 13 upregulated DEGs were enriched in the TNF-signaling pathway. Tumor necrosis factor is a pro-inflammatory cytokine produced by various cells of different types, mainly macrophages and monocytes. In addition, TNF induces a wide range of chemokines (such as CXCL8, CXC11, chemokine ligand 20 [CCL20], and CCL2), which attract neutrophils, subsets of T cells, and monocytes in the skin.23 In addition, TNF activates endothelial cells, thereby strengthening the expression of adhesion molecules. Matusiak et al24 showed that TNF levels were significantly elevated in the blood samples of patients with Zee et al25 showed that TNF was elevated in inflamed HS skin lesions compared to that in healthy skin. The significance of TNF in HS is supported by patient response to anti-TNF therapy.9-11 The results of our study provide the evidence for the significance of TNF signaling pathway in the occurrence and development of HS, supporting again the efficacy of anti-TNF biologics.
Interleukin 1β, one of the crucial 35 DEGs discovered, plays a vital role in HS. Similar to TNF, IL-1β is a pro-inflammatory cytokine produced by many different types of cells, mainly monocytes and macrophages. Among skin cells, the highest levels of the receptor for IL-1β were found in fibroblasts with those attracting neutrophilic granulocytes (such as CXCL1 and CXCL6).26 Zee et al25 reported relatively high levels of IL-1β, which is found in HS skin. The result of our study might support that IL-1β is a potential vital target for various drugs, such as colchicine or biologics, and such as IL-1β receptor antagonists and anti-IL-1 monoclonal antibodies. However, this is a new pharmacological concept that needs further investigations.
Both TNF and IL-1β, as mentioned earlier, induce various immune cell-attracting chemokines and activate endothelial cells, thereby facilitating immune cell infiltration into the skin. This increase in immune cell infiltration contributes to the following inflammatory nodules and dermal abscesses. Interleukin 1β can promote the production of extracellullar matrix-degrading enzymes, such as matrix metalloproteinases (MMPs). The MMPs can facilitate the rupture of neighboring dilated hair follicles and promote final destruction of the skin with abscess and tunnel formation.27 Tsaousi et al28 showed that high lesional MMP levels were accompanied by elevated that positively correlated with TNF blood levels and disease severity assessed by the Sartorius score, especially with the number of regions with inflammatory nodules and abscesses. They recommend the use of MMP8 as a blood biomarker for HS disease activity assessment. Moreover, Bechara et al29 found that MMP1 was significantly higher in HS lesions compared non-lesional skin (P < .05). In our study, MMP9 is one of the 35 DEGs screened by the MCODE analysis. The definite correlation between MMP9 and the occurrence and development of HS warrants further research.
The most frequently reported condition associated with HS skin alteration, including metabolic syndrome. Increased prevalence of metabolic syndrome (in up to 50% of the patients), based on concomitant obesity, dyslipididemia, hyperglycemia, and hypertension, has been noted in patients with HS.30,31 Our study showed that the downregulated DEGs were enriched in the regulation of lipolysis in adipocytes. The definite pathogenesis between them also needs further research.
In addition to the genes and pathway mentioned earlier, our study also showed the other significant key genes (eg, CGR1A, IDO1, CXCR6, IL1B, ITGA4, GPR183, and C3AR1) and pathways (eg, PPAR signaling pathway, salivary secretion, and AMPK signaling pathway) which were rarely reported to be related to HS in the literature before. Therefore, the data in our study can provide useful information and direction for future study.
Conclusion
In conclusion, these hub genes may have various roles in the occurrence and development of the HS. Combined with bioinformatics analysis, the current study identified key genes and cellular pathways involved in the occurrence and development of HS. The present study may provide a basis for improving understanding of HS. However, the current findings are limited by the lack of experimental verification in vivo and in vitro. Therefore, future experimental studies that confirm the expression and functions of identified genes on the protein level, whether these alterations in the pathway activation above correlate with HS disease activity, and whether any of these molecules lend themselves to the development of targeted therapy for HS which may be a new area of future research.
Footnotes
Author Contributions: Yan Teng and Xiaohua Tao contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Yibin Fan  https://orcid.org/0000-0002-3660-2316
https://orcid.org/0000-0002-3660-2316
References
- 1. Saunte DML, Jemec GBE. Hidradenitis suppurativa: advances in diagnosis and treatment. JAMA. 2017;318(20);2019–2032. [DOI] [PubMed] [Google Scholar]
- 2. Revuz JE, Canoui-Poitrine F, Wolkenstein P, et al. Prevalence and factors associated with hidradenitis suppurativa: results from two case–control studies. J Am Acad Dermatol. 2008;59(4):596–601. [DOI] [PubMed] [Google Scholar]
- 3. Jemec GB, Heidenheim M, Nielsen NH. The prevalence of hidradenitis suppurativa and its potential precursor lesions. J Am Acad Dermatol. 1996;35(2 pt 1):191–194. [DOI] [PubMed] [Google Scholar]
- 4. Matusiak L, Bieniek A, Szepietowski JC. Hidradenitis suppurativa markedly decreases quality of life and professional activity. J Am Acad Dermatol. 2010;62(4):706–708. [DOI] [PubMed] [Google Scholar]
- 5. Egeberg A, Gislason GH, Hansen PR. Risk of major adverse cardiovascular events and all-cause mortality in patients with hidradenitis suppurativa. JAMA Dermatol. 2016;152(4):429–434. [DOI] [PubMed] [Google Scholar]
- 6. Sabat R, Jemec GBE, Matusiak Ł, Kimball AB, Prens E, Wolk K. Hidradenitis suppurativa. Nat Rev Dis Primers. 2020;6(1):18 doi:10.1038/s41572-020-0149 -1 [DOI] [PubMed] [Google Scholar]
- 7. Hunger RE, Laffitte E, Läuchli S, et al. Swiss practice recommendations for the management of hidradenitis suppurativa/acne inversa. Dermatology. 2017;233(2-3):113–119. [DOI] [PubMed] [Google Scholar]
- 8. von der Werth JM, Williams HC. The natural history of hidradenitis suppurativa. J Eur Acad Dermatol Venereol. 2000;14(5):389–392. [DOI] [PubMed] [Google Scholar]
- 9. Kimball AB, Okun MM, Williams DA, et al. Two phase 3 trials of adalimumab for hidradenitis suppurativa. N Engl J Med. 2016;375(5):422–434. [DOI] [PubMed] [Google Scholar]
- 10. Kimball AB, Kerdel F, Adams D, et al. Adalimumab for the treatment of moderate to severe hidradenitis suppurativa: a parallel randomized trial. Ann Intern Med. 2012;157(12):846–855. [DOI] [PubMed] [Google Scholar]
- 11. Sullivan TP, Welsh E, Kerdel FA, Burdick AE, Kirsner RS. Infliximab for hidradenitis suppurativa. Br J Dermatol. 2003;149(5):1046–1049. [DOI] [PubMed] [Google Scholar]
- 12. Tzanetakou V, Kanni T, Giatrakou S, et al. Safety and efficacy of anakinra in severe hidradenitis suppurativa: a randomized clinical trial. JAMA Dermatol. 2016;152(1):52–59. [DOI] [PubMed] [Google Scholar]
- 13. Maarouf M, Clark AK, Lee DE, Shi VY. Targeted treatments for hidradenitis suppurativa: a review of the current literature and ongoing clinical trials. J Dermatol Treat. 2018;29(5):441–449. [DOI] [PubMed] [Google Scholar]
- 14. Kanni T, Argyropoulou M, Spyridopoulos T, et al. MABp1 targeting IL-1α for moderate to severe hidradenitis suppurativa not eligible for adalimumab: a randomized study. J Invest Dermatol. 2018;138(4):795–801. [DOI] [PubMed] [Google Scholar]
- 15. Meltzer DPS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. [DOI] [PubMed] [Google Scholar]
- 16. Davis S, Meltzer PS. GEOquery: a bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. [DOI] [PubMed] [Google Scholar]
- 17. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang d W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 20. Wang B, Yang W, Wen W, et al. Gamma-secretase gene mutations in familial acne inversa. Science. 2010;330(6007):1065. [DOI] [PubMed] [Google Scholar]
- 21. Pink AE, Simpson MA, Brice GW, et al. PSENEN and NCSTN mutations in familial hidradenitis suppurativa (acne inversa). J Invest Dermatol. 2011;131(7):1568–1570. [DOI] [PubMed] [Google Scholar]
- 22. Liu XD, Xie DF, Wang YL, Guan H, Huang RX, Zhou PK. Integrated analysis of lncRNA-mRNA co-expression networks in the α-particle induced carcinogenesis of human branchial epithelial cells. Int J Radiat Biol. 2019;95(2):144–155. [DOI] [PubMed] [Google Scholar]
- 23. Wolk K, Haugen HS, Xu W, et al. IL-22 and IL-20 are key mediators of the epidermal alterations in psoriasis while IL-17 and IFN-gamma are not. J Mol Med. 2009;87(5):523–536. [DOI] [PubMed] [Google Scholar]
- 24. Matusiak L, Bieniek A, Szepietowski JC. Increased serum tumour necrosis factor in hidradenitis suppurativa patients: is there a basis for treatment with anti-tumour necrosis factor- agents? Acta Dermato Venereologica. 2009;89(6):601–603. [DOI] [PubMed] [Google Scholar]
- 25. Zee HHVD, Ruiter LD, Broecke DGVD, Dik WA, Laman JD, Prens EP. Elevated levels of tumour necrosis factor (TNF)-, interleukin (IL)-1β and IL-10 in hidradenitis suppurativa skin: a rationale for targeting TNF- and IL-1β. Br J Dermatol. 2011;164(6):1292–1298. [DOI] [PubMed] [Google Scholar]
- 26. Witte Handel E, Wolk K, Tsaousi A, et al. The IL-1 pathway is hyperactive in hidradenitis suppurativa and contributes to skin infiltration and destruction. J Invest Dermatol. 2019;139(6):1294–1305. [DOI] [PubMed] [Google Scholar]
- 27. Witte Handel E, Wolk K, Tsaousi A, et al. The IL-1 pathway is hyperactive in hidradenitis suppurativa and contributes to skin infiltration and destruction. J Invest Dermatol. 2019;139(6):1294–1305. [DOI] [PubMed] [Google Scholar]
- 28. Tsaousi A, Witte E, Witte K, et al. MMP8 is increased in lesions and blood of acne inversa patients: a potential link to skin destruction and metabolic alterations. Mediators Inflamm. 2016;2016:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bechara FG, Sand M, Skrygan M, Kreuter A, Altmeyer P, Gambichler T. Acne inversa: evaluating antimicrobial peptides and proteins. Ann Dermatol. 2012;24(4):393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sabat R, Chanwangpong A, Schneider-Burrus S, et al. Increased prevalence of metabolic syndrome in patients with acne inversa. PLoS One. 2012;7(3):e31810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gold DA, Reeder VJ, Mahan MG, Hamzavi IH. The prevalence of metabolic syndrome in patients with hidradenitis suppurativa. J Am Acad Dermatol. 2014;70(4):699–703. [DOI] [PubMed] [Google Scholar]