Abstract
Growing plants with modified cell wall compositions is a promising strategy to improve resistance to pathogens, increase biomass digestibility, and tune other important properties. In order to alter biomass architecture, a detailed knowledge of cell wall structure and biosynthesis is a prerequisite. We report here a glycan array‐based assay for the high‐throughput identification and characterization of plant cell wall biosynthetic glycosyltransferases (GTs). We demonstrate that different heterologously expressed galactosyl‐, fucosyl‐, and xylosyltransferases can transfer azido‐functionalized sugar nucleotide donors to selected synthetic plant cell wall oligosaccharides on the array and that the transferred monosaccharides can be visualized “on chip” by a 1,3‐dipolar cycloaddition reaction with an alkynyl‐modified dye. The opportunity to simultaneously screen thousands of combinations of putative GTs, nucleotide sugar donors, and oligosaccharide acceptors will dramatically accelerate plant cell wall biosynthesis research.
Keywords: carbohydrates, glycan array, glycosyltransferases, plant cell wall, sugar nucleotides
Express and screen: A high‐throughput assay based on the application of azido‐functionalized sugar nucleotide donors on a synthetic glycan array for screening heterologously expressed plant glycosyltransferases was developed. The opportunity to express and screen large numbers of glycosyltransferases instead of rationally selected candidates will markedly accelerate the elucidation of plant cell wall biosynthetic pathways.
Introduction
As the global population increases, the demands for food, energy, and materials will continue to grow dramatically, resulting in a clear need for increased crop productivity and improved utilization of biomass‐derived bioenergy and sustainable bio‐based products and materials. Plant biomass, which is comprised of carbohydrate‐rich plant cell walls, represents the most abundant renewable resource on Earth. Despite their ubiquity, major gaps in our knowledge on the structure, function, and synthesis of the cognate building blocks of plant cell walls remain. While the last few years have seen significant advances in our understanding of how plant cell walls are both constructed and decomposed, we are still far away from tailoring plant cell wall composition and architecture to our needs.1 To identify potentially beneficial traits that could be introduced into modern breeding varieties as well as enhance the economic viability of lignocellulosic biomass as a renewable resource, a detailed understanding of plant cell wall architecture and biosynthetic pathways that are involved in its construction are required.
The main components of plant cell walls include a variety of glycans, proteins, and phenolic polymers.2 In these cell walls, cellulose microfibrils are cross‐linked by a group of highly complex and heterogeneous polysaccharides, the hemicelluloses and pectins. In plants, synthesis of cellulose occurs at the plasma membrane, while the remainder of cell wall glycans are made in the Golgi through glycosyltransferase (GT)‐catalyzed additions of monosaccharide residues from an activated nucleotide sugar donor onto an acceptor, typically a saccharide, protein, or small molecule.3 The genome of the model plant Arabidopsis thaliana encodes more than 561 GTs (nearly 2 % of total genes) distributed across 42 sequence‐based families, identified thus far, in the Carbohydrate‐Active enZYme (CAZy) database,3 and only a handful have been biochemically validated. For example, despite their enormous importance for cell wall biosynthesis and structural properties, to date only 22 of the more than 100 GT activities theoretically required for plant cell wall glycan synthesis across all species have been confirmed via in vitro assays, largely due to the historic difficulties associated with biochemical characterization of enzymes involved in glycan synthesis.4
The ability of a putative GT to transfer a certain sugar nucleotide to an acceptor substrate is most commonly evaluated using MS,5 HPLC,6 or less accurate radioactivity‐based approaches.7 However, as every reaction has to be performed and analyzed individually, screening the overwhelmingly large number of possible combinations of GTs, donor substrates, and acceptor substrates becomes very difficult and impractical.
Glycan microarrays have become immensely powerful tools for the high‐throughput analysis of carbohydrate‐protein interactions,8 but have not been widely applied for screening carbohydrate‐active enzymes such as GTs. Determining the substrate specificities of GTs on glycan arrays is challenging, as enzymes do not permanently bind to the immobilized acceptor substrates and cannot directly be detected on the array. One option is to use chemically functionalized sugar nucleotide donors that enable a direct detection of the acceptor after transfer of the modified glycosyl residue without the need for radiolabeled donors.9 This format allows for the use of a standard glycan array platform with maximum throughput and sensitivity, suitable for many different applications. It remains unclear if such unnatural donor substrates will be accepted by all classes of GTs. However, small modifications of sugar nucleotide donors including alkynyl‐ and azido‐modifications are usually tolerated well by GTs, as observed in numerous metabolic glycan engineering studies, not only in mammals and bacteria,10 but also in plants.11
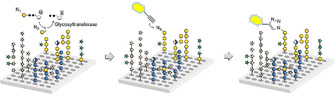
We have recently developed a glycan array equipped with 88 synthetic plant oligosaccharides to determine the binding epitopes of cell wall glycan‐directed antibodies.12 These oligosaccharides represent fragments of natural hemicellulose, hydroxyproline‐rich glycoproteins, and pectic polysaccharides, and include arabinoxylan‐,13 type I and type II arabinogalactan‐,14 xyloglucan‐,15 and mixed‐linkage glucan‐16 related structures.17 This array is being continuously expanded with newly synthesized oligosaccharides to increase the covered chemical space. In combination with chemically modified nucleotide donors, the synthetic plant glycan array provides a powerful platform for developing a high‐throughput screening method for the identification and characterization of new plant GTs. Here we report that incubation of this array with putative GTs and azido‐ or amino‐functionalized nucleotide sugars followed by visualization of transferred monosaccharides by reaction with a functionalized dye allows the simultaneous screening of thousands of individual combinations of enzyme, donor, and acceptor (Scheme 1).
Scheme 1.
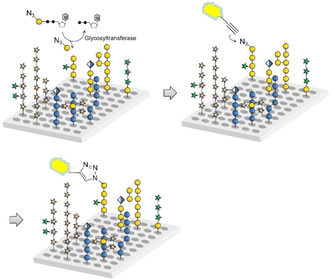
Glycan array‐based assay for the identification and characterization of plant GTs. The array is incubated with a chemically modified nucleotide sugar donor and a putative GT, followed by visualization of any transferred monosaccharide by an “on chip” reaction with an alkynyl‐functionalized dye.
Results and Discussion
Plant cell wall biosynthetic GTs are primarily transmembrane proteins that for research purposes are commonly expressed in eukaryotic systems such as yeast, tobacco leaves, or mammalian cell lines.4 The ability of these eukaryotic expression systems to perform post‐translational modifications is often required for successful production and sufficient yield of active enzymes. A particularly powerful method is the expression of putative GTs in a soluble form (truncated to remove their transmembrane domain and with an NH2‐terminal secretion signal) in eukaryotic HEK293 cells.18 HEK293 cell cultures have been proven to be a highly successful system for robust expression of functional plant glycosyl‐ and O‐acetyltransferases.5, 19 All enzymes studied in this work were produced in this expression system.
We prepared 6‐ and 4‐azido‐functionalized (“clickable”) uridine diphosphate (UDP) galactose donors 1 and 2 and 4‐azido‐xylose donor 3 by coupling the respective per‐acetylated sugar 1‐phosphates with cycloSal‐activated uridine monophosphate (Figure 1), a strategy that had proven successful for the preparation of a range of natural and non‐natural sugar nucleotides.20 The required sugar 1‐phosphates were prepared following literature reports or in analogy.21 UDP‐Gal derivatives 1 and 2 were subsequently reduced by hydrogenolysis to afford the corresponding amino‐functionalized derivatives 5 and 6 in moderate purity (see the Supporting Information).
Figure 1.
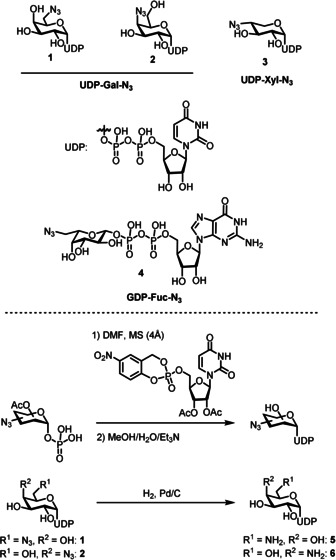
Sugar nucleotide donors used in this study and their syntheses.
To test the ability of heterologously expressed plant GTs to transfer functionalized UDP sugar derivatives to acceptor substrates on our synthetic plant glycan array, the array was incubated with UDP‐N3‐Gal derivatives 1 or 2 and the well‐characterized plant GT GALS1 from the model plant Arabidopsis thaliana that transfers UDP‐Gal to growing β‐1,4‐galactan sidechains in the pectic polysaccharide rhamnogalacturonan I (RG‐I) (Figure 2).22
Figure 2.
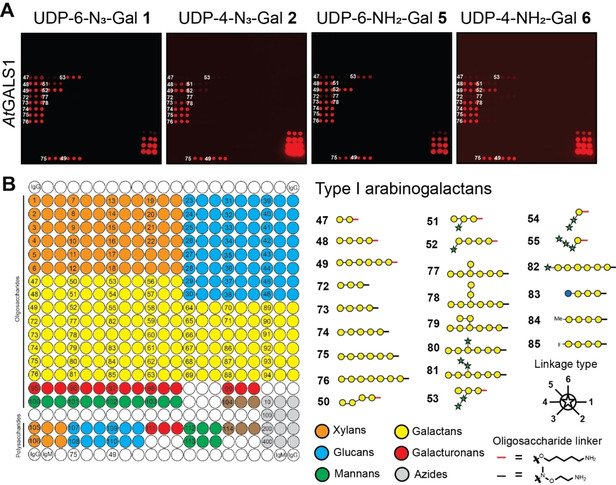
A) AtGALS1‐catalyzed transfer of azido‐ (1 and 2) and amino‐functionalized (5 and 6) UDP‐Gal derivatives to selected oligosaccharides on the array. B) Printing pattern for the synthetic plant glycan array. Azide controls were printed in the bottom right corner of the array in 10, 100, 200, and 400 μm concentrations. Subset of β‐1,4‐linked galactan acceptor substrates is shown.
Any transferred galactose was subsequently visualized “on‐chip” with an alkynyl functionalized dye in a copper‐catalyzed 1,3‐dipolar cyloaddition.23 As expected, the reaction occurred exclusively with β‐1,4‐linked galactan oligosaccharides. Interestingly, the enzyme discriminated between different substitution patterns on the oligosaccharide acceptors. We observed that α‐arabinofuranosyl substitutions in the 3‐position of some galactose residues (acceptors 51–53) were accepted by the enzyme, while β‐galactosyl and α‐arabinofuranosyl substitutions in the 6‐position were not (acceptors 77–81). Both UDP‐N3‐Gal derivatives were accepted as donor substrates, but 1 was transferred more extensively, probably because transfer of 2 is chain terminating due to the presence of N3 at the 4‐position, while 1 can be transferred multiple times to the same acceptor oligosaccharide (see Supplementary Figure 2). Note that the arrays were scanned with different photomultiplier gains. The overall efficiency of donor transfer can be estimated using the azide‐controls printed in the bottom right corner of the array. Amino‐functionalized UDP‐Gal derivatives 5 and 6 were also transferred to the same oligosaccharides on the array and were visualized using an NHS‐azide crosslinker before reaction with the alkynyl‐functionalized dye. However, transfer was diminished compared to UDP‐N3‐Gal derivatives 1 and 2.
Next, we analyzed related enzymes, such as PtGALS1 from Populus and the close AtGALS1 ortholog AtGALS2.24 Both enzymes transferred the UDP‐N3‐Gal‐donors 1 and 2, and only very slight differences were observed in the pattern of recognized acceptors compared to AtGALS1 (Figure 3 A). Surprising results were obtained when the array was incubated with UDP‐6‐N3‐Gal 1 and AtGalT31A, which was previously implicated in the synthesis of β‐1,6‐linked galactans in arabinogalactan proteins (AGPs).25 We found this GT to galactosylate substituted and unsubstituted β‐1,3‐galactan oligosaccharides rather than purely β‐1,6‐linked galactan oligosaccharides, indicating β‐1,3‐galactosylation rather than β‐1,6‐galactosylation activity. Similar to GALS1, galactosylation was dependent on the individual substitution pattern of the acceptor substrates. We have also assayed three fucosyltransferases (AtFUT4, AtFUT6, and AtFUT7) from Arabidopsis using GDP‐6‐N3‐Fuc derivative 4 and observed fucosylation of essentially all oligosaccharides containing arabinofuranose residues α‐1,3‐linked to galactose. AtFUT4 and AtFUT6 have previously been reported to fucosylate arabinose in AGPs based on preliminary enzyme assays and the analysis of knockout mutants.26 The biochemical function of AtFUT7 was previously unknown, and these data indicate that it shares similar acceptor substrate specificity with AtFUT4 and AtFUT6, indicating it is likely a previously undiscovered member of the AGP biosynthesis pathway.
Figure 3.
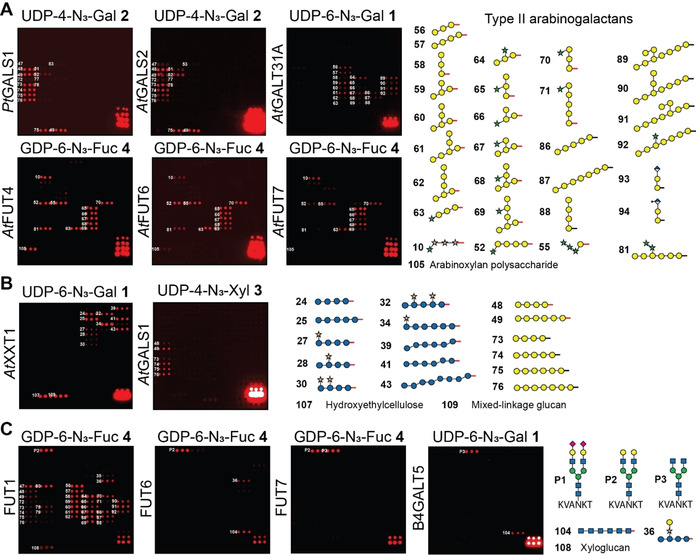
A) Glycan array‐based characterization of plant GTs PtGALS1, AtGALS2, AtGalT31A, and fucosyltransferases AtFUT4, AtFUT6, and AtFUT7. B) Glycan array‐based characterization of xylosyltransferase AtXXT1. AtXXT1 and AtGALS1 show loose donor substrate specificity and accept functionalized UDP‐Gal and UDP‐Xyl donors, respectively. C) Selected human galactosyl‐ and fucosyltransferases recognize not only their natural acceptor substrates, but also fungal and plant cell wall oligosaccharides. Subset of acceptor glycans is shown. The full set of printed glycans is presented in Supplementary Figure 1 (see the Supporting Information).
When we tested the Arabidopsis thaliana AtXXT1, an α‐1,6‐xylosyltransferase involved in adding xylose to the β‐1,4‐glucan backbone in xyloglucan,27 for its ability to transfer UDP‐4‐N3‐Xyl derivative 3 to suitable acceptor substrates on the array, we did not observe any enzymatic activity. Apparently, AtXXT1 does not accept the azido‐modification in 3. Instead, AtXXT1 was able to transfer UDP‐6‐N3‐Gal derivative 1 to a number of unsubstituted and xylose‐substituted glucan oligosaccharides (Figure 3 B, see also Supplementary Figure 3). AtXXT1 was found to prefer unsubstituted and lowly substituted glucans over highly substituted acceptors. Interestingly, not only purely β‐1,4‐linked, but also mixed linkage glucans (β‐1,3‐β‐1,4‐linked) were recognized by AtXXT1 when reacted with UDP‐Gal‐N3 derivative 1, as long as longer stretches of β‐1,4‐linked Glc residues were present. Also, polymeric hydroxylethylcellulose and natural mixed‐linkage glucan served as acceptor glycans. We were surprised to see that, while the xylosyltransferase AtXXT1 was unable to transfer UDP‐Xyl‐N3 derivative 3, the galactosyltransferase AtGALS1 did transfer UDP‐Xyl‐N3 derivative 3, albeit to a much lesser extent than UDP‐Gal‐N3. Thus, due to the natural donor and acceptor substrate promiscuity of these GTs, we have generated unnatural xylogalactan oligosaccharides and galacto‐mixed linkage glucan (β‐1,3‐β‐1,4‐linked) oligo‐ and polysaccharides on the array.
To evaluate the possibility of assaying also mammalian GTs using this glycan array platform,18 we added three peptides containing mammalian‐type N‐glycans (P1–P3)28 to the array (Figure 3 C). These peptides were purified from egg yolk and enzymatically trimmed. We incubated the array with UDP‐Gal‐N3 1 and 2 and GDP‐Fuc‐N3 4 and the different GTs. HsFUT1 is a galactoside α‐1,2‐fucosyltransferase that is involved in ABO blood‐group antigen synthesis, primarily on red blood cells. Surprisingly, this mammalian GT glycosylated a number of plant oligosaccharide acceptors to a similar extent as the mammalian N‐glycan substrate. Besides galactose terminated N‐glycan P2, a large number of galactose‐containing plant oligosaccharides, including galactan and xyloglucan structures, were recognized. On the other hand, α‐1,3‐fucosyltransferases HsFUT6 and HsFUT7, which are responsible for the synthesis of sialyl Lewis X oligosaccharides in human cells, fucosylated non‐mammalian glycans only to a limited extent or not at all. While HsFUT6 recognized galactose terminated N‐glycan P2, HsFUT7 fucosylated both P2 and de‐galactosylated N‐glycan P3. In addition, HsFUT6 accepted chitin 104, galactosylated xyloglucan 36, and mixed‐linkage glucans 38 and 41. None of the tested mammalian fucosyltransferases accepted α‐2,6‐sialylated glycan P1. Human galactosyltransferase B4GALT5, which normally extends Glc‐ceramide to form lactosylceramide in animal cells, galactosylated N‐acetylglucosamine‐terminated N‐glycan P3 and recognized chitin oligosaccharide 104 to a lesser extent. The observed tolerance of some GTs for non‐natural substrates may be explored to enzymatically modify natural polysaccharides such as xyloglucan, mixed‐linkage glucan, and chitin to produce new unnatural classes of polysaccharides with tailor‐made physico‐chemical properties suited for applications in the materials sciences.
Conclusion
In conclusion, we have established a high‐throughput assay for the identification and characterization of plant cell wall biosynthetic GTs based on the use of functionalized sugar nucleotide donors on glycan microarrays that are equipped with synthetic cell wall oligosaccharides. Utilizing glycan microarray technology, the interactions of more than 100 different acceptor oligosaccharides with several enzymes and sugar nucleotide donors were investigated simultaneously on a single glass slide. Current efforts are directed at the synthesis of further donor and acceptor substrates to enable comprehensive screens for new plant GT activities. Advances in plant cell wall biosynthesis research will set the stage for production of tailor‐made plants with improved properties, including crop resistance to pathogens, biomass digestibility, material strength, and the shelf life of fruits and vegetables.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We gratefully acknowledge support from the Max Planck Society and the German Research Foundation (DFG, Emmy Noether program PF850/1‐1 to F.P.). This work has also been supported by the Center for Bioenergy Innovation (Oak Ridge National Laboratory), a US Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science and NIH grants P41GM103390, R01GM130915 and P01GM107012. M.H.C. is grateful to the Villum Foundation for support of the PLANET Project (grant no. 9283). H.O. and D.V.S. thank the ERC ETN Marie‐Curie project GlyCoCan for financial support. Work on Arabidopsis galactosyltransferases was supported by DE‐SC0015662. We thank Dr. Martina Delbianco for providing oligosaccharide 104.
C. Ruprecht, M. P. Bartetzko, D. Senf, A. Lakhina, P. J. Smith, M. J. Soto, H. Oh, J.-Y. Yang, D. Chapla, D. Varon Silva, M. H. Clausen, M. G. Hahn, K. W. Moremen, B. R. Urbanowicz, F. Pfrengle, Angew. Chem. Int. Ed. 2020, 59, 12493.
In memory of Professor Rolf Huisgen
Contributor Information
Prof. Dr. Breeanna R. Urbanowicz, Email: breeanna@uga.edu.
Prof. Dr. Fabian Pfrengle, Email: fabian.pfrengle@boku.ac.at.
References
- 1. Doblin M. S., Johnson K. L., Humphries J., Newbigin E. J., Bacic A., Curr. Opin. Biotechnol. 2014, 26, 108–114. [DOI] [PubMed] [Google Scholar]
- 2. Albersheim P., Darvill A., Roberts K., Sederoff R., Staehelin A., Plant Cell Walls: From Chemistry to Biology, Garland Science, New York, 2011, pp. 365–409. [Google Scholar]
- 3. Coutinho P. M., Deleury E., Davies G. J., Henrissat B., J. Mol. Biol. 2003, 328, 307–317. [DOI] [PubMed] [Google Scholar]
- 4. Amos R. A., Mohnen D., Front. Plant Sci. 2019, 10, 00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Urbanowicz B. R., Peña M. J., Moniz H. A., Moremen K. W., York W. S., Plant J. 2014, 80, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Urbanowicz B. R., Bharadwaj V. S., Alahuhta M., Pena M. J., Lunin V. V., Bomble Y. J., Wang S., Yang J. Y., Tuomivaara S. T., Himmel M. E., Moremen K. W., York W. S., Crowley M. F., Plant J. 2017, 91, 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wagner G. K., Pesnot T., ChemBioChem 2010, 11, 1939–1949. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Rillahan C. D., Paulson J. C., Annu. Rev. Biochem. 2011, 80, 797–823; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Geissner A., Seeberger P. H., Annu. Rev. Anal. Chem. 2016, 9, 223–247; For a glycan array-based method to profile glycosyl hydrolases, see: [DOI] [PubMed] [Google Scholar]
- 8c. van Munster J. M., Thomas B., Riese M., Davis A. L., Gray C. J., Archer D. B., Flitsch S. L., Sci. Rep. 2017, 7, 43117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a. Blixt O., Allin K., Bohorov O., Liu X. F., Andersson-Sand H., Hoffmann J., Razi N., Glycoconjugate J. 2008, 25, 59–68; For mass spectrometry-based methods, see: [DOI] [PubMed] [Google Scholar]
- 9b. Ban L., Pettit N., Li L., Stuparu A. D., Cai L., Chen W., Guan W., Han W., Wang P. G., Mrksich M., Nat. Chem. Biol. 2012, 8, 769–773; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9c. Beloqui A., Calvo J., Serna S., Yan S., Wilson I. B. H., Martin-Lomas M., Reichardt N. C., Angew. Chem. Int. Ed. 2013, 52, 7477–7481; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 7625–7629; For a method using radiolabeled donors, see: [Google Scholar]
- 9d. Serna S., Hokke C. H., Weissenborn M., Flitsch S., Martin-Lomas M., Reichardt N.-C., ChemBioChem 2013, 14, 862–869; For a method using product detection by lectins, see: [DOI] [PubMed] [Google Scholar]
- 9e. Yan S., Serna S., Reichardt N.-C., Paschinger K., Wilson I. B. H., J. Biol. Chem. 2013, 288, 21015–21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.
- 10a. Prescher J. A., Dube D. H., Bertozzi C. R., Nature 2004, 430, 873–877; [DOI] [PubMed] [Google Scholar]
- 10b. Sminia T. J., Zuilhof H., Wennekes T., Carb. Res. 2016, 435, 121–141; For chemo-enzymatic cell surface glycan modifications using azido-modified donors, see: [DOI] [PubMed] [Google Scholar]
- 10c. Briard J. G., Jiang H., Moremen K. W., Macauley M. S., Wu P., Nat. Commun. 2018, 9, 880; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10d. Mbua N. E., Li X., Flanagan-Steet H. R., Meng L., Aoki K., Moremen K. W., Wolfert M. A., Steet R., Boons G.-J., Angew. Chem. Int. Ed. 2013, 52, 13012–13015; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 13250–13253. [Google Scholar]
- 11.
- 11a. Zhu Y., Wu J., Chen X., Angew. Chem. Int. Ed. 2016, 55, 9301–9305; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 9447–9451; [Google Scholar]
- 11b. McClosky D. D., Wang B., Chen G., Anderson C. T., Phytochemistry 2016, 123, 16–24; [DOI] [PubMed] [Google Scholar]
- 11c. Dumont A., Malleron A., Awwad M., Dukan S., Vauzeilles B., Angew. Chem. Int. Ed. 2012, 51, 3143–3146; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 3197–3200; [Google Scholar]
- 11d. Dumont M., Lehner A., Vauzeilles B., Malassis J., Marchant A., Smyth K., Linclau B., Baron A., Pons J. M., Anderson C. T., Schapman D., Galas L., Mollet J. C., Lerouge P., Plant J. 2016, 85, 437–447; [DOI] [PubMed] [Google Scholar]
- 11e. Anderson C. T., Wallace I. S., Somerville C. R., Proc. Natl. Acad. Sci. USA 2012, 109, 1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Ruprecht C., Bartetzko M. P., Senf D., Dallabernadina P., Boos I., Andersen M. C. F., Kotake T., Knox J. P., Hahn M. G., Clausen M. H., Pfrengle F., Plant Physiol. 2017, 175, 1094–1104; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b. Ruprecht C., Geissner A., Seeberger P. H., Pfrengle F., Carb. Res. 2019, 481, 31–35. [DOI] [PubMed] [Google Scholar]
- 13.
- 13a. Schmidt D., Schuhmacher F., Geissner A., Seeberger P. H., Pfrengle F., Chem. Eur. J. 2015, 21, 5709–5713; [DOI] [PubMed] [Google Scholar]
- 13b. Senf D., Ruprecht C., de Kruijff G. H. M., Simonetti S. O., Schuhmacher F., Seeberger P. H., Pfrengle F., Chem. Eur. J. 2017, 23, 3197–3205. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Bartetzko M. P., Schuhmacher F., Hahm H. S., Seeberger P. H., Pfrengle F., Org. Lett. 2015, 17, 4344–4347; [DOI] [PubMed] [Google Scholar]
- 14b. Bartetzko M. P., Schuhmacher F., Seeberger P. H., Pfrengle F., J. Org. Chem. 2017, 82, 1842–1850; [DOI] [PubMed] [Google Scholar]
- 14c. Andersen M. C. F., Kracun S. K., Rydahl M. G., Willats W. G. T., Clausen M. H., Chem. Eur. J. 2016, 22, 11543–11548; [DOI] [PubMed] [Google Scholar]
- 14d. Andersen M. C. F., Boos I., Kinnaert C., Awan S. I., Pedersen H. L., Kracun S. K., Lanz G., Rydahl M. G., Kjaerulff L., Hakansson M., Kimbung R., Logan D. T., Gotfredsen C. H., Willats W. G. T., Clausen M. H., Org. Biomol. Chem. 2018, 16, 1157–1162; [DOI] [PubMed] [Google Scholar]
- 14e. Andersen M. C. F., Boos I., Ruprecht C., Willats W. G. T., Pfrengle F., Clausen M. H., J. Org. Chem. 2017, 82, 12066–12084. [DOI] [PubMed] [Google Scholar]
- 15.
- 15a. Dallabernardina P., Schuhmacher F., Seeberger P. H., Pfrengle F., Org. Biomol. Chem. 2016, 14, 309–313; [DOI] [PubMed] [Google Scholar]
- 15b. Dallabernardina P., Ruprecht C., Smith P. J., Hahn M. G., Urbanowicz B. R., Pfrengle F., Org. Biomol. Chem. 2017, 15, 9996–10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dallabernardina P., Schuhmacher F., Seeberger P. H., Pfrengle F., Chem. Eur. J. 2017, 23, 3191–3196. [DOI] [PubMed] [Google Scholar]
- 17. Bartetzko M. P., Pfrengle F., ChemBioChem 2019, 20, 877–885. [DOI] [PubMed] [Google Scholar]
- 18. Moremen K. W., Ramiah A., Stuart M., Steel J., Meng L., Forouhar F., Moniz H. A., Gahlay G., Gao Z. W., Chapla D., Wang S., Yang J. Y., Prabhakar P. K., Johnson R., dela Rosa M., Geisler C., Nairn A. V., Seetharaman J., Wu S. C., Tong L., Gilbert H. J., LaBaer J., Jarvis D. L., Nat. Chem. Biol. 2018, 14, 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.
- 19a. Ruprecht C., Dallabernardina P., Smith P. J., Urbanowicz B. R., Pfrengle F., ChemBioChem 2018, 19, 793–798; [DOI] [PubMed] [Google Scholar]
- 19b. Amos R. A., Pattathil S., Yang J. Y., Atmodjo M. A., Urbanowicz B. R., Moremen K. W., Mohnen D., J. Biol. Chem. 2018, 293, 19047–19063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.
- 20a. Wolf S., Zismann T., Lunau N., Meier C., Chem. Eur. J. 2009, 15, 7656–7664; [DOI] [PubMed] [Google Scholar]
- 20b. Wendicke S., Warnecke S., Meier C., Angew. Chem. Int. Ed. 2008, 47, 1500–1502; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2008, 120, 1523–1525. [Google Scholar]
- 21.
- 21a. Takaya K., Nagahori N., Kurogochi M., Furuike T., Miura N., Monde K., Lee Y. C., Nishimura S.-I., J. Med. Chem. 2005, 48, 6054–6065; [DOI] [PubMed] [Google Scholar]
- 21b. Beahm B. J., Dehnert K. W., Derr N. L., Kuhn J., Eberhart J. K., Spillmann D., Amacher S. L., Bertozzi C. R., Angew. Chem. Int. Ed. 2014, 53, 3347–3352; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 3415–3420. [Google Scholar]
- 22.
- 22a. Liwanag A. J. M., Ebert B., Verhertbruggen Y., Rennie E. A., Rautengarten C., Oikawa A., Andersen M. C. F., Clausen M. H., Scheller H. V., Plant Cell 2012, 24, 5024–5036; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22b. Laursen T., Stonebloom S. H., Pidatala V. R., Birdseye D. S., Clausen M. H., Mortimer J. C., Scheller H. V., Plant J. 2018, 94, 340–351. [DOI] [PubMed] [Google Scholar]
- 23.
- 23a. Rillahan C. D., Schwartz E., Rademacher C., McBride R., Rangarajan J., Fokin V. V., Paulson J. C., ACS Chem. Biol. 2013, 8, 1417–1422; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23b. Rostovtsev V. V., Green L. G., Fokin V. V., Sharpless K. B., Angew. Chem. Int. Ed. 2002, 41, 2596–2599; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2002, 114, 2708–2711; [Google Scholar]
- 23c. Huisgen R., Proc. Chem. Soc. 1961, 357–396; [Google Scholar]
- 23d. Breugst M., Reissig H.-U., Angew. Chem. Int. Ed. 2020, 10.1002/anie.202003115; [DOI] [Google Scholar]; Angew. Chem. 2020, 10.1002/ange.202003115. [DOI] [Google Scholar]
- 24. Ebert B., Birdseye D., Liwanag A. J. M., Laursen T., Rennie E. A., Guo X., Catena M., Rautengarten C., Stonebloom S. H., Gluza P., Pidatala V. R., Andersen M. C. F., Cheetamun R., Mortimer J. C., Heazlewood J. L., Bacic A., Clausen M. H., Willats W. G. T., Scheller H. V., Plant Cell Physiol. 2018, 59, 2624–2636. [DOI] [PubMed] [Google Scholar]
- 25. Geshi N., Johansen J. N., Dilokpimol A., Rolland A., Belcram K., Verger S., Kotake T., Tsumuraya Y., Kaneko S., Tryfona T., Dupree P., Scheller H. V., Höfte H., Mouille G., Plant J. 2013, 76, 128–137. [DOI] [PubMed] [Google Scholar]
- 26. Liang Y., Basu D., Pattathil S., Xu W.-l., Venetos A., Martin S. L., Faik A., Hahn M. G., Showalter A. M., J. Exp. Bot. 2013, 64, 5537–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Faik A., Price N. J., Raikhel N. V., Keegstra K., Proc. Natl. Acad. Sci. USA 2002, 99, 7797–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu L., Prudden A. R., Bosman G. P., Boons G.-J., Carb. Res. 2017, 452, 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


