Summary
Receptor‐like protein kinases (RLKs) play key roles in regulating plant growth, development and stress adaptations. There are at least 610 RLKs (including receptor‐like cytoplasmic kinases) in Arabidopsis. The functions of the majority of RLKs have not yet been determined.
We previously generated promoter::GUS transgenic plants for all leucine‐rich repeat (LRR)‐RLKs in Arabidopsis and analyzed their expression patterns during various developmental stages. We found the expression of two LRR‐RLKs, MUSTACHES (MUS) and MUSTACHES‐LIKE (MUL), are overlapped in lateral root primordia. Independent mutants, mus‐3 mul‐1 and mus‐4 mul‐2, show a significantly decreased emerged lateral root phenotype.
Our analyses indicate that the defects of the double mutant occur mainly at stage I of lateral root development. Exogenous application of auxin can dramatically enhance the transcription of MUS, which is largely dependent on AUXIN RESPONSE FACTOR 7 (ARF7) and ARF19. MUS and MUL are inactive kinases in vitro but are phosphorylated in planta, possibly by an unknown kinase. The kinase activity of MUS is dispensable for its function in lateral root development. Many cell wall related genes are down regulated in mus‐3 mul‐1.
In conclusion, we identified MUS and MUL, two kinase‐inactive RLKs, in controlling the early development of lateral root primordia likely via regulating cell wall synthesis and remodeling.
Keywords: Arabidopsis, auxin, lateral root, MUSTACHES, MUSTACHES‐LIKE, RECEPTOR‐LIKE KINASE, root
Introduction
Plant growth, development, and adaptation to various stresses largely rely on cell‐to‐cell and cell‐to‐environment communications. Roots are important organs anchoring plants in soil and directly sensing signals from their surrounding environments. Roots also absorb water and nutrients from soil to provide fundamental compounds to plants for survival (Bellini et al., 2014). Dicotyledonous plants mainly possess a taproot system, consisting of a primary root and numerous lateral roots. Lateral roots, including their number, length and distribution, play a key role in determining the entire architecture of a root system, directly associating with its biological functions (Lynch, 1995). Initiation and development of lateral root are regulated by both internal and external signals (Malamy, 2005). Molecular mechanisms controlling lateral root development, however, are poorly understood.
In model plant Arabidopsis, transverse section of a primary root exhibits a pattern organized in concentric layers, consisting of epidermis, cortex, endodermis, pericycle, and vascular tissues from outside to inside (Dolan et al., 1993; Verbelen et al., 2006). The pericycle contains two different types of cells, phloem‐pole‐pericycle cells and xylem‐pole‐pericycle (XPP) cells (Mähönen et al., 2006; Parizot et al., 2008). Differentiated XPP cells have the ability to re‐initiate cell division, the first step of lateral root development, and these XPP cells are named lateral root founder cells (Casimiro et al., 2003). Lateral root organogenesis can be divided into four steps, lateral root positioning, lateral root initiation, lateral root primordium development and lateral root primordium emergence (Du & Scheres, 2018). Lateral root positioning determines the spatial arrangement of lateral roots along a primary root (Van Norman et al., 2013). Lateral roots initiate from regularly spaced lateral root founder cells, undergoing the first asymmetric cell division (Dubrovsky et al., 2000). Lateral root primordium development can be artificially classified into eight different stages, stages I–VIII. A new meristem is gradually established through lateral root primordium cell division, differentiation and growth (Malamy & Benfey, 1997; Benková & Bielach, 2010). As the lateral root primordia develops, it emerges from the primary root by crossing the overlying endodermis, cortex and epidermis (Kumpf et al., 2013; Vilches‐Barro & Maizel, 2015).
Cell‐to‐cell and cell‐to‐environment communications are critical during lateral root development. Receptor‐like protein kinases (RLKs) are a group of transmembrane proteins playing significant roles in these communications (Walker & Zhang, 1990). RLKs contain an extracellular domain mainly involved in perceiving small molecules from adjacent cells or from the surrounding environment, a single‐pass transmembrane domain anchoring the protein to the plasma membrane, and a cytoplasmic kinase domain responsible for transducing an extracellular signal to a cellular signaling cascade predominantly via protein phosphorylation and dephosphorylation (Shiu & Bleecker, 2001). There are at least 610 RLKs and receptor‐like cytoplasmic kinases (RLCKs) in the model plant Arabidopsis (Shiu & Bleecker, 2001). Based on the composition of the extracellular domain, Arabidopsis RLKs can be grouped into more than 21 subfamilies, in which leucine‐rich repeat (LRR)‐RLKs belong to the most abundant subfamily. There are at least 223 LRR‐RLKs in Arabidopsis (Shiu & Bleecker, 2001; Gou et al., 2010; Wu et al., 2016). Generally, RLKs play fundamental roles in regulating both plant development and defense (Li & Tax, 2013). Functional characterization of receptor kinases has become one of the frontiers in plant biology. Up to date, only a small fraction of RLKs has been functionally defined, and the majority of them still need to be functionally characterized.
It has been reported that several RLKs regulate lateral root growth, development, and adaptations to various adverse environments. For example, Arabidopsis CRINKLY4 (ACR4) is involved in controlling the division of lateral root founder cells during lateral root initiation and lateral root primordium development at stage I (Gifford et al., 2005; De Smet et al., 2008). HAESA (HAE), HAESA‐LIKE 2 (HSL2), and their peptide ligand INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) play important functions in controlling cell separation during lateral root emergence (Kumpf et al., 2013). Under heterogeneous nitrogen conditions, CEP RECEPTOR 1 (CEPR1) and CEPR2 can perceive C‐TERMINALLY ENCODED PEPTIDE 1 (CEP1), which can be transported through roots and shoots, and ultimately promote the growth of lateral roots where there are sufficient nitrogen sources, and dampen the growth under nutrient‐poor conditions (Tabata et al., 2014; Ohkubo et al., 2017). This strategy helps plants to coordinate regulation and adapt to the heterogeneity of the soil environment by controlling lateral root growth (Bisseling & Scheres, 2014; Dimitrov & Tax, 2018). When nitrogen is deficient, CLAVATA 3/EMBRYO SURROUNDING REGION (CLE) 1, CLE3, CLE4 and CLE7 can be synthesized and secreted in the pericycle layer, which are perceived by CLAVATA 1 (CLV1) in the adjacent phloem cells, inhibiting lateral root formation (Araya et al., 2014). ROOT MERISTEM GROWTH FACTOR 1 (RGF1) can reduce the lateral root numbers of Columbia‐0 (Col‐0). Lateral roots of a quadruple mutant rgi1,2,3,4 of the RGF1 receptors, RGF1 INSENSITIVES (RGIs), cannot be inhibited by RGF1 (Ou et al., 2016). In addition, TRACHEARY ELEMENT DIFFEENTIATION INHIBITORY FACTOR (TDIF) and its receptor PHLOEM INTERCALATED WITH XYLEM (PXY) interact with BRASSINOSTEROID‐INSENSITIVE2 (BIN2) in pericycle and regulate lateral root development by promoting auxin signal transduction (Okushima et al., 2007; Cho et al., 2014). Loss of function mutants in the PEPTIDE RAPID ALKALINIZATION FACTOR (RALF) 34 and its receptor THESEUS1 (THE1) show irregular asymmetric founder cell divisions during lateral root initiation (Murphy et al., 2016; Gonneau et al., 2018). In a recent study, it was found that peptide hormone TARGET OF LBD SIXTEEN2 (TOLS2) and its receptor RECEPTOR‐LIKE KINASE7 (RLK7) govern the number of lateral root founder cells, negatively regulating lateral root initiation and ensuring proper lateral root spacing (Toyokura et al., 2019).
Our laboratory is mainly interested in revealing the biological function of RLKs, especially LRR‐RLKs, in model plant Arabidopsis. From a previous study (Wu et al., 2016), we identified a receptor kinase, encoded by At1G75640, that is mainly expressed during lateral root initiation and development. This gene was previously named MUSTACHES (MUS) (Keerthisinghe et al., 2015). MUS protein was identified in stomata and regulates stomatal bilateral symmetry, although we failed to detect its expression in pMUS::GUS transgenic plants possibly due to the limitation of the GUS staining approach we used. Here we report a new role of MUS in regulating lateral root development. Our phylogenetic analysis indicated that MUS has a closely related protein which is encoded by At4G36180 and we subsequently named it as MUSTACHES‐LIKE (MUL). In vitro analysis indicated MUS and MUL are kinase‐inactive RLKs. However, MUS and MUL can be phosphorylated in planta, possibly by another undetermined kinase. Single loss‐of‐function mutants of MUS and MUL, including mus‐3, mus‐4, mul‐1 and mul‐2, show no lateral root developmental defects. However, two independent double mutants, mus‐3 mul‐1 and mus‐4 mul‐2, show significantly decreased emerged lateral root density. Further investigation using one of the two independent double mutants, mus‐3 mul‐1, revealed that the decreased emerged lateral root phenotype of the double mutant is not caused by defects in the initiation of lateral root primordia, but mainly by delayed lateral root primordium development at stage I. Interestingly, MUS was found to have a polar localization in primary and lateral root cells. Exogenous application of auxin can dramatically enhance the expression of MUS in the primary root differentiation zone and in the lateral root primordia. The upregulation of MUS by auxin is AUXIN RESPONSE FACTOR 7 (ARF7) and ARF19 dependent. RNA‐seq data showed that MUS and MUL affect lateral root development likely via regulating cell wall biosynthesis and remodeling. In summary, we found MUS and MUL are kinase‐inactive RLKs and play an important role during an early stage of lateral root development.
Materials and Methods
Plant materials and growth conditions
Wild‐type and mutants used in this study are all in Arabidopsis thaliana ecotype Col‐0 background. The T‐DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (ABRC), including mus‐3 (SALK_101029), mus‐4 (SALK_072166), mul‐1 (SALK_042322), mul‐2 (GK‐593C11), arf7‐1 arf19‐1 (CS24629). All plants were grown in glasshouses (16 h light and 8 h dark, 22°C). For seedling experiments, seeds were sterilized in 75% ethanol for 1 min and 1% sodium hypochlorite for 10 min, and washed in double distilled water (ddH2O) five times. The seeds were then vernalized at 4°C for 2 d. Seeds were germinated and the seedlings were grown on ½ Murashige and Skoog (MS) plates containing 1% sucrose and 0.9% agar in a growth chamber with 16 h light and 8 h dark at 22°C. For liquid culture, seeds in ½MS liquid medium were shaken gently at 120 rpm under the same growing condition.
Generation of double mutants and transgenic plants
Double mutants mus‐3 mul‐1 and mus‐4 mul‐2 were obtained by crossing single mutants mus‐3, mul‐1, mus‐4, mul‐2, correspondingly. The promoter and coding sequences of MUS, MUL and BAM2 were amplified with primers listed in the Supporting Information Table S1. The PCR products were cloned into an entry vector pDONR/Zeo using a BP reaction (Invitrogen, Carlsbad, CA, USA). The sequences from the obtained entry clones were recloned into an appropriate destination vector. For expression pattern analyses, the promoter sequences of MUS (2160 bases before translation initiation codon) and MUL (1276 bases before translation initiation codon) were cloned into the pBIB‐BASTA‐GUS‐GWR vector. For protein localization and complementation, genomic sequences driven by the native promoter of MUS or MUL were cloned into pBIB‐BASTA‐GWR‐YFP and pBIB‐BASTA‐GWR‐GFP, respectively. For phosphorylation analyses in planta, the coding sequences of MUS, MUL and BAM2 were cloned into pBIB‐BASTA‐UBQ10‐GWR‐FLAG. The arginine (R) at 867 aa of MUS or at 869 aa of MUL, the ATP binding sites, were mutated to a glutamic acid (E) and lysine (K) by PCR‐mediated mutagenesis. The mutated gene was also cloned into pBIB‐BASTA‐UBQ10‐GWR‐FLAG. All transgenic plants were generated by the floral dip method using Agrobacterium tumefaciens GV3101 strain. All the vectors were used to construct transgenic plants in this article as described previously (Gou et al., 2010).
Staining and microscopic analyses
Homozygous seedling lines of pMUS::GUS and pMUL::GUS were incubated in a GUS staining solution at 37°C for 6 h as described previously (Wu et al., 2016), and then were decolorized by a gradient of ethanol solutions. Whole plants were analyzed and photographed using a stereomicroscope (M165C; Leica, Weztlar, Germany). The primary roots and the lateral root primordia were cleared with two clearing solutions, solution made by mixing chloral hydrate, glycerol, water (4 : 1 : 1, w/w/w) for Fig. 1, and another clearing solution as reported previously (Malamy & Benfey, 1997). A microscope (DM6000B; Leica) was used for analyzing and photographing the root GUS staining. For protein localization of MUS, MUL, PIN1 and PIN3, the corresponding seedlings were stained in 100 ng µl−1 propidium iodide (PI) for 8 min. For stomatal phenotypes of Col‐0, mus‐3, mul‐1 and mus‐3 mul‐1, the corresponding seedlings were stained in 25 ng µl−1 SynaptoRed C2 (FM4‐64) (70021; Biotium, Fremont, CA, USA) for 30 min. Seedlings were photographed using a confocal fluorescence microscope (TCS SP8; Leica).
Fig. 1.
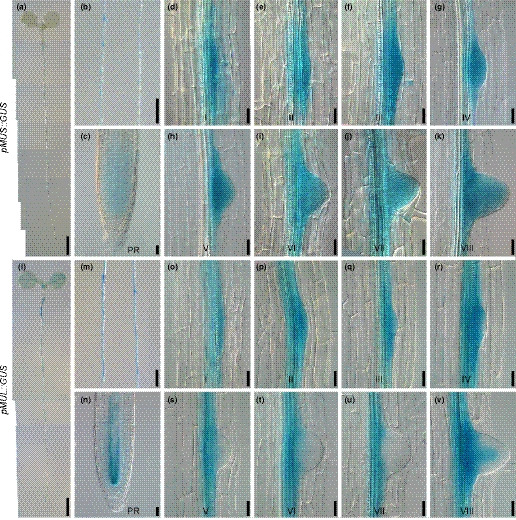
The expression patterns of MUS and MUL in roots. (a–c) MUS is mainly expressed in lateral roots and weakly expressed in the meristematic zone of the primary roots. (d–k) MUS is expressed in the lateral root primordia at all eight different developmental stages. (l–v) MUL is expressed in the vasculature of the seedlings and in the early stages of lateral root formation. The seedlings used for GUS staining were 5 d (a–c, l–n) or 8 d (d–k, o–v) after germination. PR represents primary root. I–VIII represent the eight development stages of lateral root primordia. Bars: (a, l) 2mm; (b, m) 1mm; (c–k, n–v) 25 μm.
RNA extraction and transcription analysis
Total RNA from Col‐0 and mus‐3 mul‐1 plants was extracted from 7‐d‐old seedling roots by using an RNeasy Plant Mini Kit (74903; Qiagen, Hilden, Germany) for RNA‐seq or total RNA pure Plant Kits (DP437; Tiangen; Beijing, China) for quantitative reverse transcription polymerase chain reaction (qRT‐PCR). The purified RNA was used for synthesizing the cDNA by using M‐MLV reverse transcriptase (C28025; Invitrogen, Carlsbad, CA, USA). The cDNA was used to test homozygous T‐DNA insertions of genes by RT‐PCR. Relative expression levels were obtained after normalized by ACTIN2. The primers used for RT‐PCR and qRT‐PCR are shown in Table S1.
Lateral root induction
Seedlings were grown on ½MS for 3 d after germination. The primary root tip was artificially bent to 90° with the root tip pointing towards the gravity. The roots were collected every 6 h and stained with GUS solution before analyzing under a microscope (DM6000B; Leica).
IAA treatment
The pMUS::GUS and pMUL::GUS transgenic plants were grown on ½MS medium for 8 d, then treated with 0.01% dimethyl sulfoxide (DMSO) or 1 µM indole‐3‐acetic acid (IAA) liquid medium for 12 h, stained with GUS solution for 2 h and photographed with a Leica microscope, as described earlier. Col‐0, arf7‐1 arf19‐1 seedlings were grown on 1/5 MS medium for 8 d, and treated in 1 µM IAA liquid medium for 3 h. Total RNA was extracted from the seedling roots and expression levels of MUS and MUL were detected by qRT‐PCR.
Protein induction, purification, phosphorylation assays in vitro
For in vitro induction and purification of proteins, the cDNA sequence coding of cytoplasmic domains (CDs) of MUS and the kinase domain (KD) of CLV1 were cloned into a pDEST15 vector with an in‐frame GST tag. Whereas the cDNA sequence coding for the cytoplasmic domain of MUL was cloned into pET‐15b with an in‐frame His tag. The resulting construct was transformed into Escherichia coli Rosetta for protein expression with isopropyl‐β‐d‐thiogalactoside (IPTG) induction. GST‐MUSCD, GST‐MUSCD R867E, GST‐MUSCD R867K, His‐MULCD, His‐MULCD R869E, His‐MULCD R869K, GST and GST‐CLV1KD were purified by using glutathione agarose beads (C600031; Sangon Biotech, Shanghai, China) in binding buffer (50 mM Tris‐HCl (pH 7.9), 100 mM sodium chloride (NaCl), 0.2% NP40, 10% glycerol, 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF)) at 4°C for 3 h.
Phosphorylation was assayed as described previously with modifications (Li et al., 2002). For in vitro phosphorylation assays, 1 µg of purified GST‐CLV1KD, GST‐MUSCD, GST‐MUSCD R867E, GST‐MUSCD R867K, His‐MULCD, His‐MULCD R869E, His‐MULCD R869K, GST, MBP substrate were incubated with gentle shaking in the kinase assay buffer 20 mM Tris‐HCl (pH 7.6), 1 mM DTT, 10 mM magnesium chloride (MgCl2), 1 mM manganese chloride (MnCl2), 0.1 mM ATP) containing 1 µl [γ‐32P]ATP at 30°C for 1 h. The proteins were mixed with 2 × SDS loading buffer and boiled for 5 min. The proteins were separated by 12% SDS‐PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis), quantitated by Coomassie brilliant blue and analyzed by autoradiography.
Total protein extraction and in planta phosphorylation assays
Ten‐day‐old transgenic seedlings from liquid culture were ground to powder and lysed in extraction buffer (10 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 10% glycerol, 0.5% Triton X‐100, complete protease inhibitor cocktail (04693132001; Roche, Basel, Swizerland) and phosphatase inhibitors). The supernatant was incubated with anti‐FLAG beads (A2220; Sigma, St Louis, MO, USA) and gently shaken at 4°C for 3 h. The beads were washed three times with the extraction buffer. Proteins bound to the beads were mixed with 1 × SDS loading buffer and boiled for 5 min. The proteins were then analyzed by immunoblotting with anti‐FLAG antibody (M20008L; Abmart, Shanghai, China) for quantification or with anti‐pThr antibody (9381S; Cell Signaling Technology, Boston, MA, USA) for phosphorylation analysis.
Results
MUS is mainly expressed in lateral root primordia
Within the last two decades, our laboratory has been mainly focusing on elucidating the biological functions of RLKs in Arabidopsis. We generated promoter::GUS transgenic plants for all 223 LRR‐RLKs and analyzed their detailed expression patterns (Wu et al., 2016). A number of tissue‐specifically expressed RLKs were identified during these assays. One of these RLKs, At1G75640, is predominantly expressed during lateral root development. This gene, previously reported as MUSTACHES (MUS), was found to be involved in stomatal development (Keerthisinghe et al., 2015). Our GUS staining analysis indicated that MUS is strongly expressed in the spots where lateral roots are formed and weakly expressed in the meristematic zone of primary roots (Fig. 1a–c). The expression of MUS, based on our GUS analysis, is beyond the detection levels in rosette leaves, inflorescences, flowers and siliques (Supporting Information Fig. S2a–d, see later). Detailed investigations were carried out in cleared seedlings by using chloral hydrate solution (chloral hydrate, glycerol, water (4 : 1 : 1, w/w/w)), revealing that MUS is expressed during all eight development stages of lateral root primordia (Fig. 1d–k).
The unique expression patterns of MUS suggest that MUS may be involved in lateral root development. We therefore set to examine the biological function of MUS in lateral root development. MUS is a member of the LRR‐RLK subfamily VII. This subfamily contains 10 members. Phylogenetic analysis based on full‐length amino acid sequences of subfamily VII members indicated that the closest paralog of MUS is At4G36180, which was subsequently named as MUSTACHES‐LIKE (MUL) (Fig. S1). Analysis of transgenic plants harboring pMUL::GUS showed that MUL is expressed not only in lateral roots but also in other tissues (Fig. 1l). Detailed analysis indicated that MUL is also expressed in young leaves, shoot apical meristems of 2‐wk‐old seedlings, pistils, the transmitting tract of pollen tubes, at the junction of siliques and petioles, in lateral root primordia stages I–IV, and also in the vascular tissue at stage VIII (Figs 1l–v, S2e–h). The overlapping expression patterns of MUS and MUL in lateral root development suggest MUS and MUL may play redundant roles during early stages of lateral root development.
Both mus‐3 mul‐1 and mus‐4 mul‐2 double mutants show a significantly reduced lateral root formation phenotype
To test whether MUS and MUL are truly involved in lateral root development, we obtained two independent T‐DNA insertion lines for both MUS and MUL, named mus‐3, mus‐4, mul‐1 and mul‐2, respectively (Fig. 2a). RT‐PCR analysis with primers that amplify full‐length transcripts of MUS and MUL confirmed that the two independent T‐DNA insertion lines for both genes are true null alleles (Fig. 2b). All of the single knockout mutants showed primary and lateral roots indistinguishable from those of wild type (Fig. S3a–d). These results suggest that MUS and MUL may play functionally redundant roles. To test this hypothesis, two independent double mutants, mus‐3 mul‐1 and mus‐4 mul‐2 were generated by genetic crosses. Interestingly, both mus‐3 mul‐1 and mus‐4 mul‐2 seedlings displayed slightly shortened primary roots and significantly less emerged lateral roots compared to wild‐type Col‐0 (Fig. 2c–f). The emerged lateral root densities (number of lateral roots/root length) of mus‐3 mul‐1 and mus‐4 mul‐2 were nearly half of those from Col‐0 (Fig. 2f). The aerial parts of the mutants showed no obvious differences from Col‐0 (Fig. S3e). Because mus‐3 mul‐1 and mus‐4 mul‐2 showed identical root defects, further analyses were mainly focused on mus‐3 mul‐1.
Fig. 2.

Two independent double mutants, mus‐3 mul‐1 and mus‐4 mul‐2, show a similar lateral root defective phenotype. (a) Diagrams showing the T‐DNA insertion sites of two independent alleles for MUS (mus‐3 and mus‐4) and MUL (mul‐1 and mul‐2). The insertion sites of the T‐DNA are indicated as triangles and the T‐DNA orientations are marked with arrows. (b) RT‐PCR results show no full length transcripts for both MUS and MUL in two independent double mutants. (c, d) Both mus‐3 mul‐1 and mus‐4 mul‐2 show significantly reduced number of emerged lateral roots than Col‐0. The seedlings used for the photograph were 9 d after germination. Bars, 1 cm. (e, f) Measurements of primary root length and emerged lateral root density of Col‐0, mus‐3 mul‐1 and mus‐4 mul‐2. Data shown represent average and SD (n > 30). Student's t‐tests were used to compare the measurements from mus‐3 mul‐1 and mus‐4 mul‐2 with those from Col‐0 (***, P < 0.001).
In order to further confirm the decreased lateral root density phenotype of mus‐3 mul‐1 was indeed due to the absence of both MUS and MUL, we constructed binary constructs pMUS::MUS‐YFP and pMUL::MUL‐YFP, and transferred them into mus‐3 mul‐1 respectively. More than 10 independent transgenic lines for each construct were subsequently generated. Transgenic plants with only one T‐DNA insertion event, which was determined via segregation analysis in T2, were selected and homozygous lines were subsequently obtained. Seeds of two randomly selected independent homozygous transgenic lines from each construct were planted on ½MS medium, with Col‐0 and mus‐3 mul‐1 as controls. The results indicated that two independent lines of pMUS::MUS‐YFP in mus‐3 mul‐1 or pMUL::MUL‐YFP in mus‐3 mul‐1 showed completely complemented primary and lateral root phenotypes, similar to wild type Col‐0 (Fig. 3a–c). Consistently, qRT‐PCR analysis indicated that, in all independent transgenic lines of pMUS::MUS‐YFP in mus‐3 mul‐1 or pMUL::MUL‐YFP in mus‐3 mul‐1, the expression of MUS or MUL was recovered to the level either similar or higher than Col‐0 (Fig. 3d,e). These results demonstrate that the lateral root defective phenotype of mus‐3 mul‐1 is indeed caused by the simultaneous knockouts of both MUS and MUL.
Fig. 3.

Expression of MUS or MUL driven by their respective native promoters can rescue the lateral root defects of mus‐3 mul‐1. (a) Root phenotypes of Col‐0, mus‐3 mul‐1, and two independent transgenic lines of pMUS::gMUS‐YFP and pMUL::gMUL‐YFP in mus‐3 mul‐1. The seedlings used for photographs and measurements were 9 d after germination. Bars, 1 cm. (b, c) Measurements of primary root length and emerged lateral root density of the seedlings, respectively. Data shown represent average and SD (n > 25). (d, e) qRT‐PCR results indicating the relative expression levels of MUS and MUL in Col‐0, mus‐3 mul‐1, pMUS::gMUS‐YFP in mus‐3 mul‐1, and pMUL::gMUL‐YFP in mus‐3 mul‐1. Data shown represent average and SD (n = 3). Student's t‐test was used to compare the measurements from wild‐type Col‐0, two independent transgenic lines of pMUS::gMUS‐YFP in mus‐3 mul‐1, and pMUL::gMUL‐YFP in mus‐3 mul‐1 with those from mus‐3 mul‐1 (***, P < 0.001).
MUS and MUL regulate lateral root primordia at an early developmental stage
To understand the detailed mechanisms of MUS and MUL in regulating lateral root development, we investigated the specific stages in which Col‐0 and mus‐3 mul‐1 show developmental differences. Decreased emerged lateral roots may be caused by defects at the initiation stage of lateral roots, lateral root primordium emergence, or developmental abnormalities in lateral root primordia. At early developmental stages, lateral root primordia are tiny and easily miscounted under a microscope. Auxin is known to accumulate at all developmental stages of lateral root primordia. To facilitate our analysis, we transformed pDR5::GUS into Col‐0 and mus‐3 mul‐1, and counted the primordia in both pDR5::GUS in Col‐0 and pDR5::GUS in mus‐3 mul‐1 transgenic seedlings after GUS staining. Our data suggested that the density of stage I lateral root primordia in mus‐3 mul‐1 was significantly greater than that of Col‐0, nearly twice that of Col‐0 (Fig. 4a). The density of stages II–VIII lateral root primordia in Col‐0 and mus‐3 mul‐1 showed no significant differences. The density of nonemerged lateral roots in mus‐3 mul‐1 was dramatically increased compared to that of wild type. The density of emerged lateral roots in mus‐3 mul‐1, however, was significantly decreased relative to Col‐0, although the densities of total lateral roots (emerged and nonemerged together) in mus‐3 mul‐1 and Col‐0 showed no significant difference (Fig. 4a). These results indicated that MUS and MUL play an important role mainly at stage I of lateral root primordium development.
Fig. 4.

Lateral root primordium development at stage I of mus‐3 mul‐1 is significantly delayed. (a) Comparison of lateral root density of Col‐0 and mus‐3 mul‐1 at various developmental stages. Nine‐day‐old seedlings were used for analysis. Data shown represent average and SD (n > 40). Student's t‐tests were used to compare the measurements of mus‐3 mul‐1 with those from Col‐0 (***, P < 0.001; ns, no significance at P > 0.05). (b) Diagram showing a classic approach which was used to induce lateral root initiation and formation. Bending the horizontally placed primary root tip 90° and allowing the root tip to grow in the direction of gravity can induce the initiation and formation of lateral roots at the convex side of the bending site, as indicated by an asterisk. (c) Measurements of the percentage of lateral root formation of Col‐0 and mus‐3 mul‐1 at various developmental stages (n > 50). Measurements were carried out every 6 h after the artificial induction shown in (b). (d) Number of primordium cells at stage I when the first periclinal division occurs in Col‐0 (n = 59) and mus‐3 mul‐1 (n = 50). Transgenic plants carrying pDR5::GUS in either Col‐0 or mus‐3 mul‐1 were used for the analyses as GUS staining can reveal the presence of lateral root primordia. NE, no emerged; E, emerged; Total, all lateral roots including lateral root primordia and emerged lateral roots. At least three independent biological replicates were carried out and consistent results were obtained. Results from one representative experiment are shown.
To further confirm the prominent role of MUS and MUL in lateral root primordium development, we designed a lateral root induction experiment based on an early observation that bending roots can induce lateral root formation at the convex surface due to the preferential accumulation of auxin (Ditengou et al., 2008; Moreno‐Risueno et al., 2010). Seedlings were grown on ½MS medium for 3 d after germination, and then placed horizontally with their primary root tips bending 90° towards the gravity (Fig. 4b). After different induction times, the roots were collected for further analyses. After 6 and 12 h of lateral root induction, the percentage of stage I lateral root primordia were almost the same between Col‐0 and mus‐3 mul‐1 at the bending site, indicating that the initiation of lateral root primordia is unaffected in mus‐3 mul‐1 (Fig. 4c). Interestingly, after 30 h, Col‐0 lateral root primordia were all developed at stage II and beyond, while 41% lateral root primordia in mus‐3 mul‐1 were still at stage I (Fig. 4c). After 48 h, 8.4% lateral root primordia in mus‐3 mul‐1 were still at stage I, whereas all the lateral root primordia in wild type were already developed at stage III and beyond (Fig. 4c). These results further confirmed that MUS and MUL regulate the development of the lateral root primordia at stage I.
The completion of lateral root initiation represents the beginning the stage I of lateral root primordium development, which contains four cells, two big and two small ones (Malamy & Benfey, 1997). After several anticlinal divisions, there are six or more cells at stage I. The center cells of stage I undergo a periclinal division and produce two layers of cells, symbolizing the beginning of stage II (Malamy & Benfey, 1997). In order to uncover what causes lateral roots to be retained at stage I in mus‐3 mul‐1, we counted cell numbers in Col‐0 and mus‐3 mul‐1 at stage I just after the first periclinal division starts. Statistical analysis showed that in Col‐0 the first periclinal division occurs when 5.7 cells are formed at stage I (Fig. 4d). However, in mus‐3 mul‐1 the first periclinal division occurs when 6.7 cells are formed at stage I (Fig. 4d). It was reported that cell width of the center founder cells affects early development of lateral root (Vermeer et al., 2014; Ramakrishna et al., 2019; Vilches Barro et al., 2019). We measured the width of lateral root founder cells in Col‐0 and mus‐3 mul‐1 before or after the first asymmetric division, using transgenic plants carrying pPIN3::PIN3‐GFP in Col‐0 and in mus‐3 mul‐1. We found no significant cell width difference in Col‐0 and mus‐3 mul‐1 (Fig. S4). These results indicated that the more time needed to finish stage I in mus‐3 mul‐1 is partially due to the fact that additional cell divisions are required in comparison with that of Col‐0.
MUS shows a polar localization in primary and lateral root cells
To understand the biological functions of MUS and MUL in regulating lateral root development, it is critical to know the protein localization of the two RLKs. Because transgenic plants carrying pMUS::MUS‐YFP and pMUL::MUL‐YFP in mus‐3 mul‐1 were able to complement the lateral root defects of mus‐3 mul‐1, the localization of MUS‐YFP and MUL‐YFP observed in the transgenic plants should represent its true protein localization. MUS can be detected in primary root meristem zone and in all eight developmental stages of lateral root primordia (Fig. 5a–k). Interestingly, MUS shows strong polar localization at anticlinal sides of the primary roots, the lateral root founder cells, and stage I lateral root primordia (Fig. 5a–o). We measured the mean fluorescence intensity of the MUS‐YFP signal at the anticlinal and periclinal sides of cells in primary roots and stage I lateral roots. The polarity index is then calculated by dividing the anticlinal side fluorescence intensity by periclinal side fluorescence intensity (Fig. 5p). The polarity index is about 2.7 in primary roots and 2.5 in lateral roots. Polar localization of MUS was confirmed in transgenic plants carrying 35S::MUS‐GFP. We found that MUS‐GFP was located at the root‐tip side of the cells in the primary roots (Fig. S5). Meanwhile, we analyzed protein localization of MUL in transgenic plants carrying pMUL::MUL‐YFP and found no obvious polar localization (Fig. 5q–a1). MUL‐YFP signal could be strongly observed in endodermis and weakly in pericycle and vascular cells of the primary root tips (Fig. 5q–s). Relatively weak MUL‐YFP signal could be detected in lateral root primordia (Fig. 5t–a1).
Fig. 5.

MUS and MUL proteins can be detected in both primary and lateral roots, and MUS shows polar localization in primary and lateral root cells. (a–c) MUS‐YFP from pMUS::gMUS‐YFP transgenic plants can be visualized in the primary root meristematic zone and shows clear polar localization under a confocal microscope. (d–k) MUS‐YFP can be detected at all eight developmental stages of lateral root primordia. (l–o) Different numbers of anticlinal cell divisions during early stages of lateral root development. MUS‐YFP is mainly localized at the cell transverse side of lateral root founder cells and stage I lateral root primordia. (p) Polarity index of MUS‐YFP in primary roots and stage I lateral root primordia (ratio of MUS intensity at anticlinal vs periclinal sides). Data shown represent average and SD (n > 40). (q–s) MUL‐YFP from pMUL::gMUL‐YFP transgenic plants can be detected in the endodermal cells of primary root without obvious polar localization under a confocal microscope. (t–a1) MUL‐YFP shows weak signals during all eight developmental stages of lateral root primordia. Nine‐day‐old seedlings were stained with 100 ng µl−1 PI for 8 min before confocal analyses. Bars: (b, c, r, s) 5 μm; (a, d–o, q, t–a1) 25 μm. PR represents primary root. I–VIII represent the eight development stages of lateral root primordia.
IAA treatment can induce the expression of MUS in primary and lateral roots
Auxin plays important roles in all stages of lateral root formation. Some auxin‐related mutants show similar phenotypes with mus‐3 mul‐1 in lateral root development, such as a double mutant of auxin response factors, arf7‐1 arf19‐1 (Fig. S6) (Okushima et al., 2007; Ito et al., 2016). We proposed that MUS may be regulated by auxin in regulating lateral root development. In order to examine the relationship between MUS and auxin, we treated pMUS::GUS transgenic plants with 1 µM IAA for 12 h and analyzed the GUS intensity by GUS staining. The MUS promoter activity was significantly enhanced after IAA treatment (Fig. S7). Detailed analysis showed that MUS promoter activity was enhanced mainly in the vascular tissues of primary roots and at early stages of lateral root primordia (Fig. 6a–l). However, based on GUS staining results, IAA treatment showed no obvious effects on MUL promoter activity in primary and lateral roots (Fig. 6m–x). We next performed qRT‐PCR analysis to further examine the expression levels of MUS and MUL in Col‐0 roots treated with 1 µM IAA for 3 h. The results showed that the expression level of MUS was increased more than eight‐fold, the expression level of MUL was actually also increased by about 60% (Fig. 6y,z). Previous studies indicated that high concentration of auxin could regulate expression levels of its downstream target genes via auxin‐mediated signaling pathways (Weijers et al., 2005; Ito et al., 2016). For example, auxin could up‐regulate two transcription factors, ARF7 and ARF19. ARF7 and ARF9 could directly regulate the expression of their target genes to affect the lateral root growth and development (Okushima et al., 2007). We examined the expression levels of MUS and MUL in arf7‐1 arf19‐1 double mutant with or without the IAA treatment. Interestingly, the up‐regulation of MUS and MUL by IAA was significantly decreased in arf7‐1 arf19‐1 compared to that in wild‐type (Fig. 6y,z). These data suggest that auxin enhances the expression levels of MUS and MUL largely dependent on the function of ARF7 and ARF19.
Fig. 6.

IAA significantly upregulates the expression of MUS in an ARF7 and ARF19 dependent manner. (a–l) GUS stained roots from pMUS::GUS transgenic seedlings showing the expression of MUS is enhanced in the vasculature of root differentiation zone and at the early developmental stages of lateral root primordia. The pMUS::GUS transgenic roots were treated either with 0.01% DMSO as controls (a–f) or with 1 µM IAA (g–l) for 12 h. (m–x) GUS stained roots from pMUL::GUS transgenic seedlings indicating the upregulation of MUL after IAA treatment cannot be obviously observed in primary and lateral roots. pMUL::GUS transgenic seedlings treated with 0.01% DMSO were used as controls (m–r) or 1 µM IAA (s–x) for 12 h. Bars: (a, b, g, h, m, n, s, t) 100 μm; (c–f, i–l, o–r, u–x) 25 μm. (y, z) The relative expression levels of MUS and MUL in Col‐0 and arf7‐1 arf19‐1 without or with 1 µM IAA treatment for 3 h. Total RNA extracted from roots was used for qRT‐PCR analyses. Data shown represent average and SD (n = 3). Student's t‐test was used to compare the expression levels of MUS and MUL after treated with DMSO or IAA (***, P < 0.001; **, P < 0.01; *, P < 0.05). At least three independent biological replicates were performed and similar results were obtained. The data shown represent the results from one of the representative experiments.
MUS and MUL are kinase‐inactive RLKs
MUS and MUL are two receptor kinases in the LRR VII subfamily. MUS contains 1140 amino acid residues, while MUL contains 1136 amino acid residues. Both proteins contain 25 LRRs in their extracellular domains. Generally, receptor kinases possess kinase activities which can phosphorylate downstream components to transmit extracellular signals. However, some receptor kinases show no kinase activity and they transmit signals mainly via protein–protein interactions (Castells & Casacuberta, 2007). Previous studies predicted that MUS is likely a kinase‐inactive RLK (Castells & Casacuberta, 2007; Keerthisinghe et al., 2015). To test the kinase activities of MUS and MUL, we carried out in vitro kinase assays. The cDNA sequences coding for the cytoplasmic domain of MUS and the kinase domain of CLV1 were cloned into a pDEST15 vector. The resulting constructs encode MUSCD and CLV1KD with an N‐terminal in‐frame fused GST tag, respectively. Whereas the cDNA sequence coding for the cytoplasmic domain of MUL was cloned into pET‐15b. The resulting construct encodes MULCD with an N‐terminal in‐frame fused 6 X His tag. GST‐MUSCD, His‐MULCD and GST‐CLV1KD were purified from E. coli and incubated in the kinase assay buffer supplemented with [γ‐32P]ATP. Kinase activity analysis via autoradiography showed that MUS and MUL did not exhibit any auto‐phosphorylation or transphosphorylation (towards the substrate MBP, myelin basic protein) activity in vitro. CLV1, as a positive control, however, showed strong auto‐phosphorylation and transphosphorylation activity towards MBP (Fig. 7a). In many kinase active RLKs, there is a conservative lysine (K) residue within their ATP binding motif. For example, K911 and K317 are these conservative lysine residues in the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) and co‐receptor BRI1‐ASSOCIATED RECEPTOR KINASE (BAK1), respectively (Li et al., 2002). In MUS and MUL, the corresponding residues are arginines (R), R867 for MUS and R869 for MUL. To test whether the natural occuring mutation from K to R resulted in the inactive kinases, we mutated R867 and R869 into K867 and K869, and glutamic acid (E) 867 and E869. We expected that MUSCD R867K and MULCD R869K could regain kinase activities if the mutation from K to R is the reason for these two proteins to lose their kinase activities, whereas MUSCD R867E and, MULCD R869E should not have kinase activities. We tested the kinase activities of these mutated proteins. Surprisingly, we found that all MUSCD R867E, MUSCD R867K, MULCD R869E, MULCD R869K still lacked kinase activities (Fig. 7a). These results showed that MUS and MUL are indeed kinase‐inactive RLKs in vitro. At the meantime, we detected the phosphorylation levels of MUS, MUL and BARELY ANY MERISTEM 2 (BAM2) in planta. We constructed transgenic seedlings carrying pUBQ10::MUS‐FLAG, pUBQ10::MUL‐FLAG, and pUBQ10::BAM2‐FLAG with the coding sequences of MUS, MUL and BAM2 being cloned into pBIB‐BASTA‐UBQ10‐GWR‐FLAG. MUS‐FLAG, MUL‐FLAG and BAM2‐FLAG were immunoprecipitated by anti‐FLAG beads. The phosphorylation levels of these proteins were analyzed by using an anti‐pThr antibody. Our results showed that all three fusion proteins were actually phosphorylated with the phosphorylation level of MUS much stronger than that of BAM2 in planta. MUL was also phosphorylated in planta and its phosphorylation level was slightly stronger than BAM2 but weaker than MUS (Fig. 7b).
Fig. 7.

MUS and MUL are kinase‐inactive receptor‐like kinases, and kinase activity is not essential for the function of MUS in regulating lateral root development. (a) The kinase activity assays indicate that MUSCD, MUSCD R867E, MUSCD R867K, MULCD, MULCD R869E and MULCD R869k cannot be auto‐phosphorylated in vitro. CLV1KD was used as a positive control. MBP (myelin basic protein) was used as substrate of kinases. [γ‐32P]ATP was added in the reaction buffer for the kinase activity assay. CBB, Coomassie brilliant blue. (b) The phosphorylation level of MUS is stronger than BAM2, a positive control, in planta. MUL can also be phosphorylated in planta. (c) MUSR867E, in which an arginine residue in the ATP binding site was mutated to a glutamic acid, can still be phosphorylated in planta. MUS‐FLAG, MUL‐FLAG, BAM2‐FLAG and MUSR867E‐FLAG were overexpressed and stable transgenic plants were obtained. (d, e) Mutation in the ATP binding site of MUS does not affect the biological function of MUS in lateral root development. Transgenic plants carrying pMUS::MUSR867E‐GFP can restore the developmental defects of the lateral roots of mus‐3 mul‐1. Seedlings were cultured on 1/5 MS medium for 8 d. Bars, 1 cm. (f, g) Measurements of primary root length and emerged lateral root numbers of Col‐0, mus‐3 mul‐1 and two independent transgenic lines #1, #2 of pMUS::MUSR867E‐GFP in mus‐3 mul‐1, respectively. Data shown represent average and SD (n > 35). Student's t‐test was used to compare measurements from Col‐0, two independent transgenic lines #1, #2 of pMUS::MUSR867E‐GFP in mus‐3 mul‐1 with those from mus‐3 mul‐1 (***, P < 0.001).
MUS and MUL have no auto‐phosphorylation activity in vitro, but are phosphorylated in planta, suggesting that there is another kinase in planta to phosphorylate MUS and MUL. We used MUS as an example to test whether the kinase activity of MUS is essential for its function, we detected the phosphorylation level of MUSR867E in planta and found that it was similar to that of MUS (Fig. 7c). We next transformed pMUS::MUSR867E‐GFP into mus‐3 mul‐1 and dozens of individual transgenic lines were obtained. In most of these transgenic lines, MUSR867E was able to complement the lateral root phenotypes of mus‐3 mul‐1. Two representative lines were analyzed in detail (Fig. 7d–g). These data suggest that MUS and MUL are kinase‐inactive RLKs and affect the early development of lateral root primordia. Our result indicated that MUS functions independently of its kinase activity. In other words, the kinase activity of MUS is dispensable for its biological function.
The expression levels of cell wall related genes are significantly down‐regulated in mus‐3 mul‐1
To investigate possible molecular mechanisms of MUS and MUL in regulating early lateral root development, we carried out an RNA‐seq analysis to compare the transcription profiles of mus‐3 mul‐1 with those of wild‐type Col‐0. Total RNA was extracted from 7‐d‐old seedling roots of mus‐3 mul‐1 and wild type Col‐0. The obtained transcripts were analyzed by RNA‐seq. Three biological replicates were performed. The volcano‐plot distribution map showed relative expression of genes that differ more than two‐fold between Col‐0 and mus‐3 mul‐1. There were 136 differentially expressed genes based on a two‐fold cutoff, including 39 up‐regulated genes and 97 down‐regulated genes in the double mutant relative to wild‐type (Fig. 8a,b; Table S2). Thirteen of these down‐regulated genes are classified as cell wall related and mapped by the expression clustering heatmap based on log2 (FPKM) (Fig. 8c). Nine of the 13 genes were further confirmed by qRT‐PCR analysis (Fig. 8d). These nine genes include At1G06520 (GPAT1), At2G24980 (EXTENSIN 6), At3G28300 (AT14A), At3G62820, At4G33810, At4G37780 (MYB87), At5G06640 (EXTENSIN 10), At5G26080, At5G44400 (ATBBE26). We also analyzed the transcriptional levels of other cell wall remodeling genes which were found to be reduced at least 1.5‐fold but less than two‐fold based on the RNA‐seq results, such as XYLOGLUCAN ENDOTRANSGLYCOSYLASE 6 (XTR6), EXPANSIN 1 (EXP1), EXP17 and POLYGALACTURONASE ABSCISSION ZONE Arabidopsis thaliana (PGAZAT). Our qRT‐PCR results indicated that the transcriptional levels of these four genes were all down‐regulated in mus‐3 mul‐1 compared to wild‐type (Fig. 8e) (Cai & Lashbrook, 2008; Ogawa et al., 2009; González‐Carranza et al., 2012; Lee & Kim, 2013). Cell wall biosynthesis and remodeling are important during early development of lateral root primordia. For example, during the lateral root initiation, the lateral root founder cells expand and the stage I lateral root primordia show a distinct bulge. During these processes, the cell walls of overlying endodermal cells begin to relax, and endodermal cells undergo morphological changes, which promotes lateral root primordium emergence and lateral root development (Péret et al., 2009; Vermeer et al., 2014; Ramakrishna et al., 2019). After multiple anticlinal divisions during stage I, new cell walls need to be synthesized. The RNA‐seq results suggest that MUS and MUL regulate early lateral root primordium development possibly via mediating cell wall synthesis and remodeling (Fig. 9).
Fig. 8.

A number of genes involved in cell wall biosynthesis and remodeling are significantly down‐regulated in mus‐3 mul‐1. (a) The volcano‐plot distribution map between Col‐0 and mus‐3 mul‐1. Red dots represent up‐regulated genes and blue dots represent down‐regulated genes, fold change > 2 or < −2, q‐value < 0.001. (b) The expression clustering heatmap of differentially expressed genes between Col‐0 and mus‐3 mul‐1. Red and blue represent the expression after the conversion log2 (FPKM). (c) The expression clustering heatmap of cell wall‐related genes between Col‐0 and mus‐3 mul‐1. These genes were selected from the data shown in (b). (d) The transcriptional levels of cell wall‐related genes are down‐regulated in mus‐3 mul‐1 compared to Col‐0. (e) The transcriptional levels of several cell wall remodeling genes that have been reported in the literature are decreased in mus‐3 mul‐1. Total RNA extracted from 7‐d‐old seedling roots of Col‐0 and mus‐3 mul‐1 was used for qRT‐PCR analyses. Three biological replicates for RNA sequencing were conducted for Col‐0 and mus‐3 mul‐1. Data shown represent average and SD (n = 3). Student's t‐test was carried out to test whether the data are significantly different between mus‐3 mul‐1 and Col‐0 (*** , P < 0.001; **, P < 0.01; *, P < 0.05).
Fig. 9.

A hypothetical model showing the roles of MUS and MUL in regulating lateral root development at stage I. Auxin can enhance the expression levels of MUS and MUL in primary and lateral roots. The regulation is largely dependent on ARF7 and ARF19. MUS and MUL, two kinase‐inactive RLKs, can be phosphorylated by another unknown kinase in planta and control lateral root development at an early development stage through cell wall synthesis and remodeling. Diagram represents the early development of lateral root primordia in Col‐0. Cell division is delayed due to impaired cell wall biosynthesis and remodeling at stage I of the lateral root development in mus‐3 mul‐1. In addition, more stage I cells are required for the double mutant before entering to stage II. These two defects lead to the retention of lateral root primordia at stage I in the double mutant.
Discussion
Here, we report our identification of an LRR‐RLK whose expression pattern appears to be very unique. This LRR‐RLK encodes a previously described protein named MUS. MUS protein was identified in stomata. The mus mutants show a stomatal bilateral asymmetry phenotype. Since our GUS staining data indicated that MUS is predominantly expressed in lateral root primordia and weakly expressed in primary roots (Fig. 1a–k), we proposed that MUS may also have a role in mediating lateral root development. To test this assumption, we isolated two independent T‐DNA insertional lines for MUS, named mus‐3 and mus‐4, and analyzed their possible defective phenotypes, especially in primary and lateral roots. Both alleles displayed a stomatal defect similar to what was reported previously (Keerthisinghe et al., 2015), but they did not show any lateral root defects (Figs S3, S9, see later). We believed that failing to identify lateral root defects could be caused by functional redundancy of MUS with its paralogs. Phylogenic analysis indicated that MUS is a member of LRR‐RLK subfamily VII which consists of 10 total members (Fig. S1). We named its most closely related protein as MUSTACHES‐LIKE (MUL). GUS staining analysis using transgenic plants carrying pMUL::GUS indicated that MUL promoter activity can be strongly detected in the vascular tissues of primary roots, stems, leaves, and areas where lateral root primordium initiates (Fig. 1l–v). Detailed protein localization analysis in transgenic plants carrying pMUS::MUS‐YFP and pMUL::MUL‐YFP further confirmed that these proteins are indeed localized in the lateral root primordia. The overlapped expression patterns in lateral root primordia further suggest that the two LRR‐RLKs may redundantly regulate lateral root growth and development. Our detailed genetic and cell biology analyses suggested that MUS and MUL regulate lateral root development at stage I.
Our results showed that MUS and MUL are kinase‐inactive receptor kinases, which can be phosphorylated by an unknown kinase in planta and transmit signals to regulate early lateral root development (Figs 7, 9). Our analyses confirmed that the kinase activity of MUS is dispensable for the function of MUS in lateral root development. In Arabidopsis thaliana, sequence analysis found that nearly 20% of the RLKs are not conserved in their catalytic kinase domains, which were predicted to be kinase‐inactive RLKs (Castells & Casacuberta, 2007). These kinase‐inactive RLKs are also important in signal transduction and may act as scaffolds by interacting with other kinases (Kroiher et al., 2001). For example, RECEPTOR DEAD KINASE1 (RDK1) is a kinase‐dead RLK, and its kinase activity is not required for ABA‐mediated early seedling development (Kumar et al., 2017). The Arabidopsis thaliana CRINKLY4 RELATED 2 (AtCRR2) is a homolog of ACR4 and has no catalytic kinase activity. However, AtCRR2 could be phosphorylated by ACR4 in vitro, which indicates that AtCRR2 might act as heterodimer with the kinase‐active ACR4 to regulate ACR4‐mediated seed coat signaling (Cao et al., 2005). STRUBBELIG (SUB) is also a kinase‐inactive receptor kinase which plays an indispensable role in regulating root epidermal patterning (Chevalier et al., 2005). To understand the detailed molecular mechanisms of MUS and MUL in regulating lateral root development will rely on the identification of an assumed kinase which can phosphorylate MUS and MUL in planta. Future genetic and proteomic approaches can be used to find such an active kinase.
Interestingly, MUS shows a polar localization in the primary and lateral root cells (Fig. 5a–o). This localization is similar to the polar distribution of an auxin efflux transporter PIN1, which has been reported to regulate lateral root development. PIN1 expression can be detected in the vascular organization of primary roots, the lateral root founder cells and lateral root primordia cells (Omelyanchuk et al., 2016). The polar localization of PIN1 determines the direction of auxin transport, leading to gradient distribution of auxin (Blilou et al., 2005). Accumulation of auxin in lateral root founder cells can promote the formation of lateral root primordia (Benková et al., 2003). Similar polar localization and similar biological function at lateral root development suggest that MUS and MUL may interact with PIN1. We therefore examined the protein level and localization of PIN1 in lateral root primordia of mus‐3 mul‐1. Our preliminary results indicated that the localization and protein level of PIN1 in lateral root primordia were not affected in mus‐3 mul‐1 (Fig. S8). However, exogenous application of auxin could significantly enhance the expression of MUS in the early stages of lateral root development, which is dependent on the functions of ARF7 and ARF19 (Fig. 6). These data suggest that MUS and MUL are downstream components of auxin signaling pathway to regulate lateral root development. Whether MUS and MUL interact with PIN1 in vivo needs to be investigated in the future. It is worth noting that another member in the LRR VII subfamily, INFLORESCENCE AND ROOT APICES RECEPTOR KINASE (IRK), also shows polar distribution in the endodermis of Arabidopsis primary roots (Campos et al., 2020). Whether additional members in this subfamily show polar localization is another interesting question to be explored.
RNA‐seq data and qRT‐PCR analyses showed the transcriptional levels of some cell wall synthesis and remodeling genes in mus‐3 mul‐1 were down‐regulated compared to Col‐0 (Fig. 8). These data suggest that MUS and MUL control early lateral root development likely via regulating cell wall synthesis and remodeling. It is possible that the cell division at stage I of mus‐3 mul‐1 is delayed due to the cell wall biosynthesis defects. MUS localization analysis suggests that MUS is mainly accumulated at newly formed cell plates after the first few anticlinal cell divisions at stage I of lateral root primordium development (Fig. 5l–o). The observation that MUS is mainly accumulated at the newly formed cell plate during stage I is consistent with the RNA‐seq results, both of which suggest that MUS and MUL may function in cell wall biosynthesis and remodeling at the early stages of lateral root development.
MUS and MUL may regulate microtubule arrangement. A previous report suggested that a loss‐of‐function mutant of MUS showed an altered stomatal bilateral symmetry phenotype due to defects in polarity growth and the radial arrangement of microtubules (Keerthisinghe et al., 2015). We reinvestigated this result with our mutants. FM4‐64 stained true leaves from 7‐d‐old seedlings were photographed using confocal fluorescence microscopy. Stomata of Col‐0 and mul were bilaterally symmetrical, while stomata of mus‐3 and mus‐3 mul‐1 display impaired bilateral symmetry (Fig. S9). These results are consistent with the observation from the previous report (Keerthisinghe et al., 2015). In addition, the polar growth of microtubules affects cytoskeleton remodeling of pericycle and endodermis cells, which is required for the asymmetric expansion of founder cells when lateral root initiation occurs (Vilches Barro et al., 2019). Thus, MUS and MUL may influence the early development of lateral root primordia at stage I partially through regulating the polar growth of microtubules. This issue also needs to be clarified in the future.
In summary, we proved MUS and MUL are two RLKs regulating early development of lateral root primordia. We propose that during lateral root primordium development, auxin can be accumulated in the center cells at stage I, which can upregulate the expression of MUS and MUL. As a result, cell division can be accelerated partially due to enhanced cell wall biosynthesis and remodeling. At the meantime, MUS and MUL also can promote the first periclinal cell division, allowing lateral root primordium to proceed into stage II (Fig. 9). Lateral roots are extremely critical for the entire architecture of the root system. Identification of RLKs in regulating lateral root initiation and development will help us to understand how environmental and developmental cures play significant roles in root system formation.
Data availability
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org/). Accession number: MUS (At1G75640), MUL (At4G36180), ARF7 (At5G20730), ARF19 (At1G19220), CLV1 (At1G75820), BAM2 (At3G49670), PGAZAT (At2G41850), XTR6 (At4G25810), EXP1 (At1G69530), EXP17 (At4G01630).
Author contributions
JL, FET and QX designed all experiments, analyzed the data and wrote the manuscript. QX performed most of the experiments and prepared the data. YW and HL, contributed to generate the transgenic plants. YO, JC and KH participated in discussions and provided some suggestions for the project. XG provided GST‐CLV1KD bacterial strain, and transgenic plant pBIB‐BASTA‐UBQ10::BAM2‐FLAG.
Supporting information
Fig. S1 Phylogenetic analysis of LRR‐RLKs from subfamily VII.
Fig. S2 The expression of MUS cannot be detected in the aerial parts of Arabidopsis seedlings, while the expression of MUL can be detected in the vasculature of the aerial parts.
Fig. S3 Single mutant mus‐3 or mul‐1 shows the root and above‐ground phenotypes similar to Col‐0.
Fig. S4 The width of lateral root founder cells in Col‐0 and mus‐3 mul‐1 showed no significant difference before or after the first asymmetric division.
Fig. S5 MUS shows a polar localization in primary roots.
Fig. S6 The arf7‐1 arf19‐1 double mutant shows a defective lateral root phenotype.
Fig. S7 Auxin can upregulate the expression level of MUS in roots.
Fig. S8 Protein levels and localization of an auxin efflux transporter PIN1 are similar in Col‐0 and mus‐3 mul‐1.
Fig. S9 MUS regulates stomatal bilateral symmetry and morphogenesis.
Table S1 Primers used in this study.
Table S2 Differentially expressed genes with fold change ≥ 2, P < 0.001 in roots of Col‐0 and mus‐3 mul‐1.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
The authors are grateful to the Arabidopsis Biological Resource Center for the T‐DNA insertion lines used in this study. The authors thank Guangqin Guo for the pPIN1::PIN1‐GFP and pPIN3::PIN3‐GFP transgenic plants. This study was supported by the National Natural Science Foundation of China (31720103902 and 31530005 to JL), Ministry of Agriculture of the People's Republic of China (2016ZX08009‐003‐002).
References
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wiren N, Takahashi H. 2014. CLE‐CLAVATA1 peptide‐receptor signaling module regulates the expansion of plant root systems in a nitrogen‐dependent manner. Proceedings of the National Academy of Sciences, USA 111: 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I. 2014. Adventitious roots and lateral roots: similarities and differences. Annual Review of Plant Biology 65: 639–666. [DOI] [PubMed] [Google Scholar]
- Benková E, Bielach A. 2010. Lateral root organogenesis – from cell to organ. Current Opinion in Plant Biology 13: 677–683. [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux‐dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bisseling T, Scheres B. 2014. Plant Science. Nutrient computation for root architecture. Science 346: 300–301. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Cai SQ, Lashbrook CC. 2008. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2 . Plant Physiology 146: 1305–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos R, Goff J, Rodriguez‐Furlan C, Van Norman JM. 2020. The Arabidopsis receptor kinase IRK is polarized and represses specific cell divisions in roots. Developmental Cell 52: 183–195 e184. [DOI] [PubMed] [Google Scholar]
- Cao X, Li K, Suh SG, Guo T, Becraft PW. 2005. Molecular analysis of the CRINKLY4 gene family in Arabidopsis thaliana . Planta 220: 645–657. [DOI] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. 2003. Dissecting Arabidopsis lateral root development. Trends in Plant Science 8: 165–171. [DOI] [PubMed] [Google Scholar]
- Castells E, Casacuberta JM. 2007. Signalling through kinase‐defective domains: the prevalence of atypical receptor‐like kinases in plants. Journal of Experimental Botany 58: 3503–3511. [DOI] [PubMed] [Google Scholar]
- Chevalier D, Batoux M, Fulton L, Pfister K, Yadav RK, Schellenberg M, Schneitz K. 2005. STRUBBELIG defines a receptor kinase‐mediated signaling pathway regulating organ development in Arabidopsis . Proceedings of the National Academy of Sciences, USA 102: 9074–9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ et al 2014. A secreted peptide acts on BIN2‐mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nature Cell Biology 16: 66–76. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R et al 2008. Receptor‐like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597. [DOI] [PubMed] [Google Scholar]
- Dimitrov I, Tax FE. 2018. Lateral root growth in Arabidopsis is controlled by short and long distance signaling through the LRR RLKs XIP1/CEPR1 and CEPR2. Plant Signaling & Behavior 13: e1489667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li X, Nitschke R et al 2008. Mechanical induction of lateral root initiation in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 105: 18818–18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Du Y, Scheres B. 2018. Lateral root formation and the multiple roles of auxin. Journal of Experimental Botany 69: 155–167. [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Doerner PW, Colon‐Carmona A, Rost TL. 2000. Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiology 124: 1648–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Robertson FC, Soares DC, Ingram GC. 2005. ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 17: 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Martin M, Doblas VG, Bacete L, Miart F, Sormani R, Hématy K, Renou J, Landrein B et al 2018. Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis . Current Biology 28: 2452–2458. [DOI] [PubMed] [Google Scholar]
- González‐Carranza ZH, Shahid AA, Zhang L, Liu Y, Ninsuwan U, Roberts JA. 2012. A novel approach to dissect the abscission process in Arabidopsis. Plant Physiology 160: 1342–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, He K, Yang H, Yuan T, Lin H, Clouse SD, Li J. 2010. Genome‐wide cloning and sequence analysis of leucine‐rich repeat receptor‐like protein kinase genes in Arabidopsis thaliana . BMC Genomics 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Fukaki H, Onoda M, Li L, Li C, Tasaka M, Furutani M. 2016. Auxin‐dependent compositional change in Mediator in ARF7‐ and ARF19‐mediated transcription. Proceedings of the National Academy of Sciences, USA 113: 6562–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthisinghe S, Nadeau JA, Lucas JR, Nakagawa T, Sack FD. 2015. The Arabidopsis leucine‐rich repeat receptor‐like kinase MUSTACHES enforces stomatal bilateral symmetry in Arabidopsis. The Plant Journal 81: 684–694. [DOI] [PubMed] [Google Scholar]
- Kroiher M, Miller MA, Steele RE. 2001. Deceiving appearances: signaling by "dead" and "fractured" receptor protein‐tyrosine kinases. BioEssays 23: 69–76. [DOI] [PubMed] [Google Scholar]
- Kumar D, Kumar R, Baek D, Hyun TK, Chung WS, Yun DJ, Kim JY. 2017. Arabidopsis thaliana RECEPTOR DEAD KINASE1 functions as a positive regulator in plant responses to ABA. Molecular Plant 10: 223–243. [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Sto IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB. 2013. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proceedings of the National Academy of Sciences, USA 110: 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim J. 2013. EXPANSINA17 up‐regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant and Cell Physiology 54: 1600–1611. [DOI] [PubMed] [Google Scholar]
- Li J, Tax FE. 2013. Receptor‐like kinases: key regulators of plant development and defense. Journal of Integrative Plant Biology 55: 1184–1187. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. 2002. BAK1, an Arabidopsis LRR receptor‐like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Lynch J. 1995. Root architecture and plant productivity. Plant Physiology 109: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. 2006. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98. [DOI] [PubMed] [Google Scholar]
- Malamy JE. 2005. Intrinsic and environmental response pathways that regulate root system architecture. Plant, Cell & Environment 28: 67–77. [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana . Development 124: 33–44. [DOI] [PubMed] [Google Scholar]
- Moreno‐Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Vu LD, Van den Broeck L, Lin Z, Ramakrishna P, van de Cotte B, Gaudinier A, Goh T, Slane D, Beeckman T et al 2016. RALFL34 regulates formative cell divisions in Arabidopsis pericycle during lateral root initiation. Journal of Experimental Botany 67: 4863–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Kay P, Wilson S, Swain SM. 2009. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis . Plant Cell 21: 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y, Tanaka M, Tabata R, Ogawa‐Ohnishi M, Matsubayashi Y. 2017. Shoot‐to‐root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nature Plants 3: 17029. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis . Plant Cell 19: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelyanchuk NA, Kovrizhnykh VV, Oshchepkova EA, Pasternak T, Palme K, Mironova VV. 2016. A detailed expression map of the PIN1 auxin transporter in Arabidopsis thaliana root. BMC Plant Biology 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Lu X, Zi Q, Xun Q, Zhang J, Wu Y, Shi H, Wei Z, Zhao B, Zhang X et al 2016. RGF1 INSENSITIVE 1 to 5, a group of LRR receptor‐like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana . Cell Research 26: 686–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, Boucheron‐Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig SR, Helariutta Y, Haseloff J et al 2008. Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiology 146: 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Larrieu A, Bennett MJ. 2009. Lateral root emergence: a difficult birth. Journal of Experimental Botany 60: 3637–3643. [DOI] [PubMed] [Google Scholar]
- Ramakrishna P, Ruiz Duarte P, Rance GA, Schubert M, Vordermaier V, Vu LD, Murphy E, Vilches Barro A, Swarup K, Moirangthem K et al 2019. EXPANSIN A1‐mediated radial swelling of pericycle cells positions anticlinal cell divisions during lateral root initiation. Proceedings of the National Academy of Sciences, USA 116: 8597–8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. 2001. Receptor‐like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences, USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y. 2014. Perception of root‐derived peptides by shoot LRR‐RKs mediates systemic N‐demand signaling. Science 346: 343–346. [DOI] [PubMed] [Google Scholar]
- Toyokura K, Goh T, Shinohara H, Shinoda A, Kondo Y, Okamoto Y, Uehara T, Fujimoto K, Okushima Y, Ikeyama Y et al 2019. Lateral inhibition by a peptide hormone‐receptor cascade during Arabidopsis lateral root founder cell formation. Developmental Cell 48: 64–75 e65. [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Xuan W, Beeckman T, Benfey PN. 2013. To branch or not to branch: the role of pre‐patterning in lateral root formation. Development 140: 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbelen JP, De Cnodder T, Le J, Vissenberg K, Baluska F. 2006. The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities: meristematic zone, transition zone, fast elongation zone and growth terminating zone. Plant Signaling & Behavior 1: 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JE, von Wangenheim D, Barberon M, Lee Y, Stelzer EH, Maizel A, Geldner N. 2014. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis . Science 343: 178–183. [DOI] [PubMed] [Google Scholar]
- Vilches Barro A, Stockle D, Thellmann M, Ruiz‐Duarte P, Bald L, Louveaux M, von Born P, Denninger P, Goh T, Fukaki H et al 2019. Cytoskeleton dynamics are necessary for early events of lateral root initiation in Arabidopsis . Current Biology 29: 2443–2454. [DOI] [PubMed] [Google Scholar]
- Vilches‐Barro A, Maizel A. 2015. Talking through walls: mechanisms of lateral root emergence in Arabidopsis thaliana . Current Opinion in Plant Biology 23: 31–38. [DOI] [PubMed] [Google Scholar]
- Walker JC, Zhang R. 1990. Relationship of a putative receptor protein kinase from maize to the S‐locus glycoproteins of Brassica . Nature 345: 743–746. [DOI] [PubMed] [Google Scholar]
- Weijers D, Benková E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jurgens G. 2005. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO Journal 24: 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Xun Q, Guo Y, Zhang J, Cheng K, Shi T, He K, Hou S, Gou X, Li J. 2016. Genome‐wide expression pattern analyses of the Arabidopsis Leucine‐rich repeat receptor‐like kinases. Molecular Plant 9: 289–300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phylogenetic analysis of LRR‐RLKs from subfamily VII.
Fig. S2 The expression of MUS cannot be detected in the aerial parts of Arabidopsis seedlings, while the expression of MUL can be detected in the vasculature of the aerial parts.
Fig. S3 Single mutant mus‐3 or mul‐1 shows the root and above‐ground phenotypes similar to Col‐0.
Fig. S4 The width of lateral root founder cells in Col‐0 and mus‐3 mul‐1 showed no significant difference before or after the first asymmetric division.
Fig. S5 MUS shows a polar localization in primary roots.
Fig. S6 The arf7‐1 arf19‐1 double mutant shows a defective lateral root phenotype.
Fig. S7 Auxin can upregulate the expression level of MUS in roots.
Fig. S8 Protein levels and localization of an auxin efflux transporter PIN1 are similar in Col‐0 and mus‐3 mul‐1.
Fig. S9 MUS regulates stomatal bilateral symmetry and morphogenesis.
Table S1 Primers used in this study.
Table S2 Differentially expressed genes with fold change ≥ 2, P < 0.001 in roots of Col‐0 and mus‐3 mul‐1.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org/). Accession number: MUS (At1G75640), MUL (At4G36180), ARF7 (At5G20730), ARF19 (At1G19220), CLV1 (At1G75820), BAM2 (At3G49670), PGAZAT (At2G41850), XTR6 (At4G25810), EXP1 (At1G69530), EXP17 (At4G01630).


