Abstract
Two CoII 4L4 tetrahedral cages prepared from similar building blocks showed contrasting host–guest properties. One cage did not bind guests, whereas the second encapsulated a series of anions, due to electronic and geometric effects. When the building blocks of both cages were present during self‐assembly, a library of five CoII LA x LB 4−x cages was formed in a statistical ratio in the absence of guests. Upon incorporation of anions able to interact preferentially with some library members, the products obtained were redistributed in favor of the best anion binders. To quantify the magnitudes of these templation effects, ESI‐MS was used to gauge the effect of each template upon library redistribution.
Keywords: host-guest systems, mass spectrometry, metal-organic Cages, self-assembly, supramolecular library
A statistical library of dynamic tetrahedral cages was formed by self‐assembly of building blocks of similar sizes. ESI‐MS was used to probe the subtle effects of anion binding on the dynamic library formed. Upon the addition of anions able to bind within the library members, an increase or decrease of the proportion of each congener of the library was observed, allowing for information on relative binding affinities to be gained via MS.
Molecular recognition is a fundamental process within biological systems. The conformational restructuring which a system undergoes upon the introduction of additional components1 is a crucial aspect of processes such as drug–protein interactions.2 Synthetic supramolecular systems can be engineered to mimic their biological counterparts,3 where the reconfiguration of species is enabled by the dynamic nature of the interactions between building blocks.4 Coordination cages are capable of reorganization upon stimuli such as light,5 pH changes,6 or guest templation7 and thus represent attractive reorganizing systems to study.
The outcome of self‐sorting8 in dynamic libraries can be influenced by the addition of new components to libraries,9 which interact preferentially with certain library members.10 Mass spectrometry has proven to be a useful technique to quantify templation effects on these systems,11 which could allow for better understanding of the binding processes that induce molecular reconfiguration.
The FeII 4L4 tetrahedral cages prepared from trianilines A or B were reported not to encapsulate guest molecules, due to their small cavity sizes.12 We anticipated that preparing analogous cages with CoII instead of FeII might yield structures with slightly larger cavities, thus enabling guest uptake, as CoII cages had previously been shown to encapsulate larger guests than their FeII analogs.13 Tetrahedral cages 1 and 2 (Figure 1) were thus synthesized by the reaction of the corresponding trianiline A or B (4 equiv), 2‐formylpyridine (12 equiv) and Co(NTf2)2 (4 equiv). The 1H NMR spectra of 1 and 2 were consistent with species of overall T symmetry (Figures S1 and S4), while ESI‐MS confirmed CoII 4L4 stoichiometry (Figures S2 and S5). Single crystals of 1(ClO4)8 and 2(ClO4)8 suitable for X‐ray diffraction were obtained by slow vapor diffusion of either Et2O or EtOAc into solutions of 1 or 2 in CH3CN. The crystal structures (Figures 1, S10, S11) confirm the formation of the tetrahedral cages. The average CoII–CoII distances for 1 and 2 were both found to be 12.0 Å, which is slightly larger than that of the FeII analogs (11.9 and 11.8 Å respectively).12 While cage 1 was found to crystallize without a guest bound inside its cavity, the crystal structure of 2 showed an encapsulated ClO4 − anion.
Figure 1.
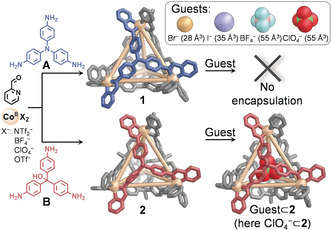
A and B self‐assembled with 2‐formylpyridine and CoII to form 1 and 2, respectively. Three‐dimensional views are constructed from the X‐ray crystal structures of 1 and ClO4 −⊂2 (CoII: orange, LA: blue, LB: red; orange lines indicate the closest metal–metal separations, to illustrate geometry). The insert gives the list of guests encapsulated by 2 but not 1 along with their volumes (Å3).
Br−, I−, BF4 −, and ClO4 − anions, as well as acetonitrile, were found to bind within 2 (see Supporting Information section 4.2). Internal guest binding was indicated by the appearance of a new set of 1H NMR signals upon addition of these anions, consistent with the formation of a host–guest complex in slow exchange on the NMR time scale.14 The relative binding affinities depended on guest size, with smaller species binding more strongly within 2, in the order Br− > I− ≫ BF4 − ≥ ClO4 − ≫ CH3CN.15 As the amount of anion X− added increased, the concentration of host‐guest complex (X−⊂2) reached a plateau at a concentration below 1:1 binding (Figures S31, S32). The analysis described in section 5 in the Supporting Information allowed us to conclude that anion encapsulation was competing with anion‐TBA+ association (Figures S29, S30) and with the encapsulation of CH3CN, in geared equilibrium processes. These three association phenomena were considered together (Supporting Information section 5.3), producing the following association constants in CD3CN: K Br‐=16000±1000 m −1, K I‐=4800±400 m −1, K ‐=580±50 m −1, K ‐=1100±100 m −1, K =0.0072±0.0004 m −1.
No binding was observed within 1 when the anions studied were added following cage formation (Figure S18) or when cage 1 was formed in the presence of the anions (Figures S19 and S20). Given that 2 encapsulates a variety of anions, we were surprised that host 1 did not encapsulate any of these guests. Calculations of the cages’ internal voids (Figure S15) indicated that the cavities of 1 and 2 were of nearly equal volumes (59 Å3 and 56 Å3, respectively). The cavity in 1 was expected to accommodate Br− and I−, with occupancies of 48 % and 59 %, respectively (Table S2).16 Although BF4 −⊂1 and ClO4 −⊂1 are predicted to have ≈95 % occupancy, the X‐ray structure of ClO4 −⊂2 demonstrated that such high occupancy is possible. Guest binding may be hindered by the lesser degree of pyramidalization in nitrogen‐centered LA than in hydroxymethyl‐centered LB (Supporting Information Section 3.4). In addition, we hypothesize that the p orbitals on four facial nitrogen atoms contribute electron density to the microenvironment within the cavity, thus destabilizing the binding of anionic guests. This hypothesis is supported by molecular orbital and electrostatic potential calculations (Supporting Information section 3.6).
Given the similarities in size and geometry between tritopic amines A and B, and the product complexes 1 and 2, we investigated the formation of heteroleptic libraries of CoII LA x LB 4−x cages in the presence of guest molecules. The reaction of A and B (1 equiv each), 2‐formylpyridine (6 equiv) and Co(NTf2)2 (2 equiv) in CH3CN led to the formation of a library of cages (Lib ) as confirmed by ESI‐MS and 1H NMR (Figures 2 a and b, S33 and S34). Slow diffusion of Et2O into the solution of Lib in the presence of ClO4 − produced crystals suitable for diffraction studies (Supporting Information section 3.3). Single‐crystal X‐ray diffraction afforded a structure displaying disorder that was best modeled whereby each face of each cage had a 50 % probability of incorporating a residue of A, and 50 % B.
Figure 2.
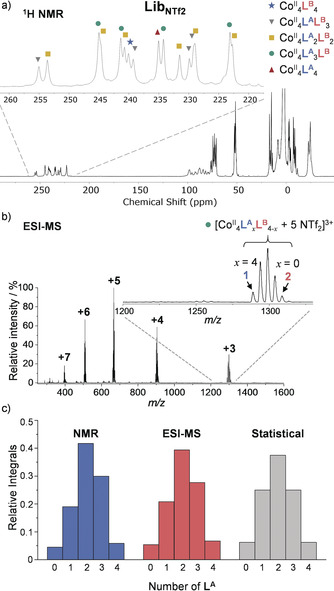
a) 1H NMR spectrum of Lib with each of the imine signals attributed to one of the cage species in the insert. b) Low‐resolution ESI‐MS of Lib . The +3 region of the mass spectra is expanded, showing the peak clusters corresponding to cages in the absence of a template. c) Normalized integrals of Lib obtained by NMR and ESI‐MS compared to the statistical binomial distribution.
When the reaction was carried with sub‐stoichiometric amounts of 2‐formylpyridine (3 equiv instead of 6 equiv), a different library Lib′ was formed, where structures of composition CoII 4 LA x LB 4−x with 2≤x≤4 were expressed predominantly (Figure S37). Three sets of imine signals, containing six, four, and one resonance, respectively, were observed in the 1H NMR spectrum of Lib′ (Figure S38). Based upon symmetry, these sets of signals were assigned to CoII 4 LA 2 LB 2, CoII 4 LA 3 LB and CoII 4 LA 4, respectively. Each of the 16 signals observed in the imine region of the 1H NMR spectrum of Lib were thus attributed to one of the CoII LA x LB 4−x cage congeners (Figures 2 a and S39). Monitoring the formation of Lib over time, suggested that the final equilibrated library developed from a more complex mixture of species present upon mixing. Heating at 70 °C for 18 h was required to approach equilibrium (Supporting Information section 6.3).
The distribution of species within Lib was examined with 1H NMR and ESI‐MS (Supporting Information section 7.2). Since similar ESI response factors were observed for 1 and 2 (Figure S48), we hypothesized that the heteroleptic CoII LA x LB 4−x cages might have similar response factors as well. The integral of the signal for each species might thus reflect its concentration, relative to the others present in solution. No significant difference between the ratio of species in Lib was observed with either analytical method (NMR or ESI‐MS), validating the use of ESI‐MS spectra to determine the relative concentrations of cage species in solution. In addition, the collection of CoII 4 LA x LB (4−x) species was observed in an approximately 1:4:6:4:1 ratio both by NMR and ESI‐MS, close to the expected binomial distribution (Figure 2 c). The ratio of species was also found to be time‐independent in the ESI‐MS traces (Supporting Information Section 8).
Four libraries of mixed cages (LibX, where X=Br−, I−, BF4 − or ClO4 −) were prepared by the addition of the tetrabutylammonium salt of the corresponding anion X− (2 equiv) to Lib (Figure 5). The overlap between some signals and the low intensity of the 1H NMR signals corresponding to the host–guest complexes limited our ability to deconvolute them (Figure S42 and S45), therefore precluding quantification of the species present in solution. Thus, detailed ESI‐MS studies were undertaken.
Figure 5.

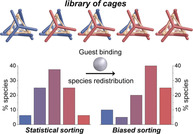
Formation of a library of CoII LA x LB (4−x), 0≤x≤4. The library expressed a near‐statistical distribution of cage species in the absence of a guest. A bias towards structures incorporating more LB was observed when guests were added.
The ESI‐MS spectra of the libraries in the presence of template anions (LibX) displayed clusters of peaks corresponding to individual CoII 4 LA x LB (4−x) cages associated with zero, one or more X− (Figures 3 and S43, S44, S46, S47), indicative of X− associating either externally or internally with the cages. The influence of encapsulating anion X− on the library constitution could be deciphered by analyzing the distribution of the library members X−⊂CoII 4 LA x LB 4−x, which was obtained by subtracting the distributions of library members having externally associated X− (Supporting Information Section 7.3). The distributions thus observed deviated strongly from the near‐binomial distribution observed for Lib , with anion templation favoring the incorporation of LB into the structures. The greatest deviations were observed for the smaller anions (Figure 4 a), corresponding to the trends observed for anion binding affinity within 2: Br− > I− ≫ BF4 − > ClO4 −.
Figure 3.
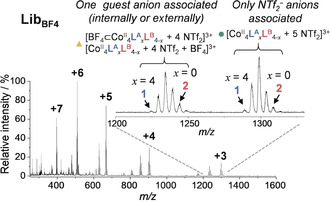
Low‐resolution ESI‐MS of Lib obtained after addition of TBABF4 (2 equiv) to Lib . The +3 region of the mass spectra is expanded, showing the peak clusters corresponding to cages with no BF4 − (green circle) and cages with one BF4 − associated (yellow triangle).
Figure 4.
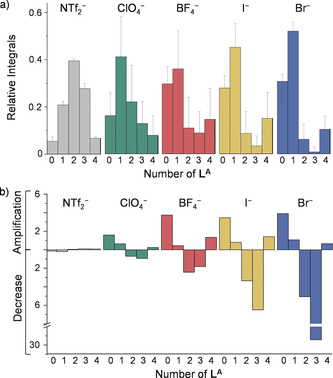
a) Normalized ESI‐MS integrals of libraries with no guest (NTf2 −, gray) and after addition of guests (Br−, I−, BF4 − and ClO4 − in blue, yellow, red, and green, respectively). Each experiment was repeated three times and averaged across all charge states to obtain standard deviations, of which only positive values are shown as error bars for clarity. b) Factor of amplification or decrease of each congener generated in the libraries relative to the binomial distribution baseline with no amplification.
Cage 2 was not observed to be the major species present in solution, with CoII LALB 3 species being favoured statistically (Figure 4 a). We calculated the deviation of the proportions of the generated cages from the expected binomial distribution for each of the five libraries (Figure 4 b and Supporting Information Section 7.3). While no significant changes were observed for Lib , greater perturbations were observed for the libraries in the presence of templating anions. The amplitude of changes was observed to correlate with anion binding affinities. Cage 2 was amplified the most, with amplification factors of 160 % for Lib , 370 % for Lib , 350 % for LibI and 390 % for LibBr compared to Lib . CoII LA 2 LB 2 and CoII LA 3 LB were clearly disfavoured, with decreases of up to 29‐fold observed in the case of LibBr. However, cage 1, which incorporates only A residues, also exhibited amplification as a result of the anion templation of other cages in solution. Cage 1 was not observed to encapsulate any of the anions present in the libraries tested (Figures S18–S20). Thus, we suggest the high proportion of 1 to be due to it serving as a ‘sink’ for residual A that were not incorporated into the anion binding cages (Figure 5).17
The magnitudes of the deviations of the libraries of CoII 4 LA x LB 4−x complexes (Figure 4 a) from the statistically‐expected values were used to calculate relative Gibbs free energies for each library member (Supporting Information section 7).18 This analysis was based on the observation that mass spectrometric response factors were similar for cages 1 and 2, and that as a consequence, the values of the integrals reflected the quantity of each species in solution. These energies were plotted as a function of the number of LA incorporated for each anion used (Figure 6). Cage 2 was the most stable structure in all libraries, as reflected in the greater degree of amplification of this species. This observation suggests the differences in relative energies of a few kJ mol−1 not to be enough to lead to the exclusive formation of cage 2. The energetic cost was raised for each LA incorporated due to the poorer fit for anions within cavities surrounded by more A residues, as discussed above. The library member CoII LA 3 LB, which should be affected by the presence of a template to a greater extent, had the least favorable relative energy in most cases. Anions which bound most strongly within 2 perturbed the libraries from a binomial distribution to a greater degree.
Figure 6.
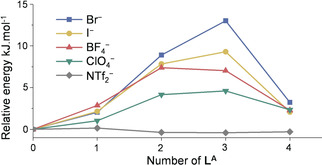
Relative energies (kJ mol−1) of each CoII 4 LA x LB 4−x cage within the libraries. Each experiment was repeated three times and averaged across all charge states.
A fine‐grained study of guest binding within complex heteroleptic supramolecular libraries was carried out. Cage 2, the strongest anion binder, was amplified to a greater degree than any other species, while CoII LA 3 LB was the least favored structure. Large anions that fit most poorly within the cage cavities resulted in lower stabilization of library members, and thus less species redistribution. ESI‐MS hence allowed quantification of the degree to which guest binding favored structures that incorporated more of the ligand that is best able to accommodate the guest. Related ESI‐MS methods, used alongside other quantitative techniques such as NMR, may thus enable quantification of molecular interactions and reconfigurations in increasingly complex systems.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
This project has received funding from the European Union's Horizon 2020 research and innovation program under Marie Sklodowska‐Curie grant agreement No 642192 and was supported by the UK Engineering and Physical Sciences Research Council (EPSRC, EP/P027067/1) as well as the Marie Curie Incoming International Fellowship Scheme of the 7th EU Framework Program. The authors thank Diamond Light Source (UK) for synchrotron beamtime on I19 (MT6791), the Department of Chemistry NMR facility at the University of Cambridge, the Australian Research Council, the EPSRC UK National Mass Spectrometry Facility at Swansea University and the UK National Crystallography Service at Southampton. J.D.T. acknowledges the Rashkind Family Endowment and the Chenery Endowment from Randolph‐Macon College. The authors thank Jack Davies for his contribution to the processing of ESI‐MS data.
M. Kieffer, R. A. Bilbeisi, J. D. Thoburn, J. K. Clegg, J. R. Nitschke, Angew. Chem. Int. Ed. 2020, 59, 11369.
References
- 1. Hochgürtel M., Kroth H., Piecha D., Hofmann M. W., Nicolau C., Krause S., Schaaf O., Sonnenmoser G., Eliseev A. V., Proc. Natl. Acad. Sci. USA 2002, 99, 3382–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scott D. E., Bayly A. R., Abell C., Skidmore J., Nat. Rev. Drug Discovery 2016, 15, 533. [DOI] [PubMed] [Google Scholar]
- 3.
- 3a. Wiester M. J., Ulmann P. A., Mirkin C. A., Angew. Chem. Int. Ed. 2011, 50, 114; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011, 123, 118; [Google Scholar]
- 3b. Mondal M., Hirsch A. K. H., Chem. Soc. Rev. 2015, 44, 2455–2488; [DOI] [PubMed] [Google Scholar]
- 3c. Zamora-Olivares D., Kaoud T. S., Dalby K. N., Anslyn E. V., J. Am. Chem. Soc. 2013, 135, 14814–14820; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3d. Yang G., Zheng W., Tao G., Wu L., Zhou Q.-F., Kochovski Z., Ji T., Chen H., Li X., Lu Y., Ding H.-m., Yang H.-B., Chen G., Jiang M., ACS Nano 2019, 13, 13474–13485; [DOI] [PubMed] [Google Scholar]
- 3e. Ulrich S., Dumy P., Chem. Commun. 2014, 50, 5810–5825; [DOI] [PubMed] [Google Scholar]
- 3f. Punt P. M., Clever G. H., Chem. Sci. 2019, 10, 2513–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Rowan S. J., Cantrill S. J., Cousins G. R. L., Sanders J. K. M., Stoddart J. F., Angew. Chem. Int. Ed. 2002, 41, 898–952; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2002, 114, 938–993; [Google Scholar]
- 4b. Komáromy D., Stuart M. C. A., Monreal Santiago G., Tezcan M., Krasnikov V. V., Otto S., J. Am. Chem. Soc. 2017, 139, 6234–6241; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4c. Lehn J.-M., Chem. Soc. Rev. 2007, 36, 151–160; [DOI] [PubMed] [Google Scholar]
- 4d. Wang L., Cheng L., Li G., Liu K., Zhang Z., Li P., Dong S., Yu W., Huang F., Yan X., J. Am. Chem. Soc. 2020, 142, 2051–2058. [DOI] [PubMed] [Google Scholar]
- 5.
- 5a. Li R.-J., Holstein J. J., Hiller W. G., Andréasson J., Clever G. H., J. Am. Chem. Soc. 2019, 141, 2097–2103; [DOI] [PubMed] [Google Scholar]
- 5b. Oldknow S., Martir D. R., Pritchard V. E., Blitz M. A., Fishwick C. W. G., Zysman-Colman E., Hardie M. J., Chem. Sci. 2018, 9, 8150–8159; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5c. Burke M. J., Nichol G. S., Lusby P. J., J. Am. Chem. Soc. 2016, 138, 9308–9315; [DOI] [PubMed] [Google Scholar]
- 5d. Samanta D., Mukherjee P. S., J. Am. Chem. Soc. 2014, 136, 17006–17009. [DOI] [PubMed] [Google Scholar]
- 6.
- 6a. Kurihara K., Yazaki K., Akita M., Yoshizawa M., Angew. Chem. Int. Ed. 2017, 56, 11360–11364; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 11518–11522; [Google Scholar]
- 6b. Liu Y., Shi B., Wang H., Shangguan L., Li Z., Zhang M., Huang F., Macromol. Rapid Commun. 2018, 39, 1800655; [DOI] [PubMed] [Google Scholar]
- 6c. Jansze S. M., Cecot G., Severin K., Chem. Sci. 2018, 9, 4253–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.
- 7a. Paul R. L., Bell Z. R., Jeffery J. C., McCleverty J. A., Ward M. D., Proc. Natl. Acad. Sci. USA 2002, 99, 4883–4888; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7b. Custelcean R., Chem. Soc. Rev. 2014, 43, 1813–1824; [DOI] [PubMed] [Google Scholar]
- 7c. Chifotides H. T., Dunbar K. R., Acc. Chem. Res. 2013, 46, 894–906; [DOI] [PubMed] [Google Scholar]
- 7d. Wang B., Zang Z., Wang H., Dou W., Tang X., Liu W., Shao Y., Ma J., Li Y., Zhou J., Angew. Chem. Int. Ed. 2013, 52, 3756–3759; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 3844–3847; [Google Scholar]
- 7e. Mirtschin S., Slabon-Turski A., Scopelliti R., Velders A. H., Severin K., J. Am. Chem. Soc. 2010, 132, 14004–14005; [DOI] [PubMed] [Google Scholar]
- 7f. Kumazawa K., Yamanoi Y., Yoshizawa M., Kusukawa T., Fujita M., Angew. Chem. Int. Ed. 2004, 43, 5936–5940; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2004, 116, 6062–6066; [Google Scholar]
- 7g. Sekiya R., Fukuda M., Kuroda R., J. Am. Chem. Soc. 2012, 134, 10987–10997; [DOI] [PubMed] [Google Scholar]
- 7h. Hong C. M., Kaphan D. M., Bergman R. G., Raymond K. N., Toste F. D., J. Am. Chem. Soc. 2017, 139, 8013–8021. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. He Z., Jiang W., Schalley C. A., Chem. Soc. Rev. 2015, 44, 779–789; [DOI] [PubMed] [Google Scholar]
- 8b. Safont-Sempere M. M., Fernández G., Würthner F., Chem. Rev. 2011, 111, 5784–5814; [DOI] [PubMed] [Google Scholar]
- 8c. Zheng Y.-R., Yang H.-B., Northrop B. H., Ghosh K., Stang P. J., Inorg. Chem. 2008, 47, 4706–4711; [DOI] [PubMed] [Google Scholar]
- 8d. Kołodziejski M., Stefankiewicz A. R., Lehn J.-M., Chem. Sci. 2019, 10, 1836–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a. Beaudoin D., Rominger F., Mastalerz M., Angew. Chem. Int. Ed. 2017, 56, 1244–1248; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 1264–1268; [Google Scholar]
- 9b. Black S. P., Wood D. M., Schwarz F. B., Ronson T. K., Holstein J. J., Stefankiewicz A. R., Schalley C. A., Sanders J. K. M., Nitschke J. R., Chem. Sci. 2016, 7, 2614–2620; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9c. Hsu C.-W., Miljanić O. Š., Chem. Commun. 2016, 52, 12357–12359; [DOI] [PubMed] [Google Scholar]
- 9d. Stefankiewicz A. R., Sanders J. K. M., Chem. Commun. 2013, 49, 5820–5822; [DOI] [PubMed] [Google Scholar]
- 9e. Makiguchi W., Tanabe J., Yamada H., Iida H., Taura D., Ousaka N., Yashima E., Nat. Commun. 2015, 6, 7236; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9f. Badetti E., Carmo dos Santos N. A., Scaramuzzo F. A., Bravin C., Wurst K., Licini G., Zonta C., RSC Adv. 2018, 8, 19494–19498; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9g. Wiley C. A., Holloway L. R., Miller T. F., Lyon Y., Julian R. R., Hooley R. J., Inorg. Chem. 2016, 55, 9805–9815; [DOI] [PubMed] [Google Scholar]
- 9h. Men G., Lehn J.-M., Chem. Sci. 2019, 10, 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.
- 10a. Miljanić O. Š., Chem 2017, 2, 502–524; [Google Scholar]
- 10b. Atcher J., Moure A., Bujons J., Alfonso I., Chem. Eur. J. 2015, 21, 6869–6878; [DOI] [PubMed] [Google Scholar]
- 10c. Valdivielso A. M., Puig-Castellví F., Atcher J., Solà J., Tauler R., Alfonso I., Chem. Eur. J. 2017, 23, 10789–10799; [DOI] [PubMed] [Google Scholar]
- 10d. Blanco-Gómez A., Rama T., Domarco O., Neira I., Blanco V., Quintela J. M., García M. D., Peinador C., Dalton Trans. 2017, 46, 15671–15675; [DOI] [PubMed] [Google Scholar]
- 10e. Septavaux J., Germain G., Leclaire J., Acc. Chem. Res. 2017, 50, 1692–1701; [DOI] [PubMed] [Google Scholar]
- 10f. Ziegler M., Miranda J. J., Andersen U. N., Johnson D. W., Leary J. A., Raymond K. N., Angew. Chem. Int. Ed. 2001, 40, 733–736; [PubMed] [Google Scholar]; Angew. Chem. 2001, 113, 755–758; [Google Scholar]
- 10g. Severin K., Chem. Eur. J. 2004, 10, 2565–2580; [DOI] [PubMed] [Google Scholar]
- 10h. Halina N., Jaroslaw P., Rafal G., Robert M., Dominik T., Barbara P., Andrzej B., Anna P., Jean-Francois M., Johann G., Marc Le B., Comb. Chem. High Throughput Screening 2006, 9, 753–770. [Google Scholar]
- 11.
- 11a. Bravin C., Badetti E., Puttreddy R., Pan F., Rissanen K., Licini G., Zonta C., Chem. Eur. J. 2018, 24, 2936–2943; [DOI] [PubMed] [Google Scholar]
- 11b. Warzok U., Marianski M., Hoffmann W., Turunen L., Rissanen K., Pagel K., Schalley C. A., Chem. Sci. 2018, 9, 8343–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bilbeisi R. A., Clegg J. K., Elgrishi N., de Hatten X., Devillard M., Breiner B., Mal P., Nitschke J. R., J. Am. Chem. Soc. 2012, 134, 5110–5119. [DOI] [PubMed] [Google Scholar]
- 13. Ronson T. K., Giri C., Kodiah Beyeh N., Minkkinen A., Topić F., Holstein J. J., Rissanen K., Nitschke J. R., Chem. Eur. J. 2013, 19, 3374–3382. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Rizzuto F. J., Carpenter J. P., Nitschke J. R., J. Am. Chem. Soc. 2019, 141, 9087–9095; [DOI] [PubMed] [Google Scholar]
- 14b. Rizzuto F. J., Wu W.-Y., Ronson T. K., Nitschke J. R., Angew. Chem. Int. Ed. 2016, 55, 7958–7962; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 8090–8094. [Google Scholar]
- 15. Clegg J. K., Cremers J., Hogben A. J., Breiner B., Smulders M. M. J., Thoburn J. D., Nitschke J. R., Chem. Sci. 2013, 4, 68–76. [Google Scholar]
- 16. Mecozzi S., Rebek J., Chem. Eur. J. 1998, 4, 1016–1022. [Google Scholar]
- 17. Osypenko A., Dhers S., Lehn J.-M., J. Am. Chem. Soc. 2019, 141, 12724–12737. [DOI] [PubMed] [Google Scholar]
- 18. Kieffer M., Pilgrim B. S., Ronson T. K., Roberts D. A., Aleksanyan M., Nitschke J. R., J. Am. Chem. Soc. 2016, 138, 6813–6821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


