Abstract
Widespread adoption of primary human papillomavirus (HPV)‐based screening has encouraged the search for a triage test which retains high sensitivity for the detection of cervical cancer and precancer, but increases specificity to avoid overtreatment. Methylation analysis of FAM19A4 and miR124‐2 genes has shown promise for the triage of high‐risk (hr) HPV‐positive women. In our study, we assessed the consistency of FAM19A4/miR124‐2 methylation analysis in the detection of cervical cancer in a series of 519 invasive cervical carcinomas (n = 314 cervical scrapes, n = 205 tissue specimens) from over 25 countries, using a quantitative methylation‐specific PCR (qMSP)‐based assay (QIAsure Methylation Test®). Positivity rates stratified per histotype, FIGO stage, hrHPV status, hrHPV genotype, sample type and geographical region were calculated. In total, 510 of the 519 cervical carcinomas (98.3%; 95% CI: 96.7–99.2) tested FAM19A4/miR124‐2 methylation‐positive. Test positivity was consistent across the different subgroups based on cervical cancer histotype, FIGO stage, hrHPV status, hrHPV genotype, sample type and geographical region. In conclusion, FAM19A4/miR124‐2 methylation analysis detects nearly all cervical carcinomas, including rare histotypes and hrHPV‐negative carcinomas. These results indicate that a negative FAM19A4/miR124‐2 methylation assay result is likely to rule out the presence of cervical cancer.
Keywords: DNA hypermethylation, human genome methylation, human papillomavirus, biomarker, cervical carcinoma, cervical screening
Short abstract
What's new?
Methylation analysis of host cell genes is a promising strategy for the triage of women who test positive for high‐risk human papillomavirus (hrHPV). Its ability to consistently detect cervical cancer, however, warrants further evaluation. In this retrospective cross‐sectional study of more than 500 cervical cancer cases worldwide, methylation analysis using FAM19A4 and miR124‐2 genes successfully detected the vast majority of cervical carcinomas. Detection by FAM19A4/miR124‐2 methylation analysis was consistent regardless of multiple factors, including hrHPV status and genotype, cancer histotype, sample type, and geographical region. The findings suggest that a negative FAM19A4/miR124‐2 methylation test result is likely to rule out cervical cancer.
Introduction
Cytology‐based cervical screening has reduced the incidence of and mortality from cervical cancer.1 However, with an estimated 570,000 new cases in 2018, cervical cancer still is the fourth most common cancer in women worldwide, and accounts for 7.5% of all female cancer deaths, with the majority occurring in low‐ and middle‐income settings.2 To facilitate a further reduction in cervical cancer incidence, an increasing number of countries are converting from cytology‐based screening to high‐risk human papillomavirus (hrHPV)‐based screening,3 which provides better protection against cervical cancer and precancer.4 Yet, given the 3–5% lower specificity of hrHPV testing compared to cytology, objective molecular triage tests are needed to identify hrHPV‐positive women with clinically relevant disease.4, 5
Since changes in host cell DNA methylation after a transforming hrHPV infection are central in cervical carcinogenesis, DNA methylation analysis has emerged as a promising and objective triage tool for hrHPV‐positive women.6 A number of studies have shown that host cell DNA methylation levels in cervical scrapes increase with underlying cervical disease severity and are highest in cervical cancer.7, 8, 9, 10, 11, 12 Accordingly, methylation biomarkers, including family with sequence similarity 19 (chemokine (C–C)‐motif)‐like) member A4 (FAM19A4) and microRNA 124‐2 (miR124‐2) genes, have been proposed as objective molecular triage tools for hrHPV‐positive women that allow the identification of women with cervical cancer or cervical intraepithelial neoplasia (CIN) lesions with a cancer‐like methylation profile that have a high risk of progression to cancer.6, 13, 14, 15, 16 Furthermore, recent longitudinal studies showed that hrHPV‐positive women negative for FAM19A4/miR124‐2 methylation have a similar risk of CIN3+, and lower risk of cervical cancer, at 14 years from the result as compared to hrHPV‐positive, cytology‐negative women.17, 18 Collectively, these data support the perspective of utilizing FAM19A4/miR124‐2 methylation analysis as an indicator of clinically significant cervical disease.
Demonstration of performance consistency of FAM19A4/miR124‐2 methylation analysis to detect cervical cancer is clearly important if this approach is to be used routinely as a triage test. So far, studies have largely been performed on populations from the Netherlands and encouraging clinical performance has been demonstrated at the level of CIN3+. However, these studies included relatively small sample series of invasive cervical cancers, comprising mainly squamous cell carcinomas.8, 13, 14 The aim of the present study was to determine test positivity rates of the FAM19A4/miR124‐2 methylation assay in a global series of cervical carcinomas, with a particular interest in histotypes that have not been studied in marked numbers up till now, including adenocarcinomas, adenosquamous carcinomas and rare types of cervical cancer such as hrHPV‐negative, neuroendocrine and clear cell carcinomas.
Methods
Clinical specimens
A multicenter, retrospective study was designed within the VALID‐SCREEN framework, a project funded by the EU Horizon 2020 program (project ID 666800), to determine the FAM19A4/miR124‐2 methylation positivity rate in cervical specimens of women with invasive cervical cancer. In total, 541 DNA extracts derived from cervical scrapes and cervical tissue samples (fresh frozen biopsies or formalin‐fixed, paraffin‐embedded biopsies) from screening or gynecologic outpatient populations from over 25 countries across five continents, were retrieved with DNA isolation performed according to local protocols. Data on the hrHPV status, hrHPV genotype, FIGO stage and pathology diagnosis of the cervical cancer specimens were obtained from the parent institutes. All participating institutes used clinically validated hrHPV DNA assays19 to determine hrHPV status and hrHPV genotype (if applicable). In samples testing hrHPV‐negative with a clinically validated hrHPV assay, additional HPV testing was performed with SPF10‐LiPA25 (Version‐1; Labo Biomedical Products, Rijswijk, The Netherlands) given this assay's high analytical sensitivity for detection of HPV.20 The cervical cancer histotype was classified according the WHO classification.21 Clinical cancer staging was done using the FIGO stage version operative at time of cervical cancer diagnosis.22 The work in this study with human‐derived material was conducted under national and international rules and legislation, as well as European standards of research ethics, as it is expressed in the applicable legislation/regulations (The Declaration of Helsinki; informed consent for participation of human subjects in medical and scientific research) and guidelines for Good Clinical Practice. The study was approved by the local ethics committees.
qMSP‐based methylation analysis
Aliquots of isolated DNA were shipped to the pathology department of Amsterdam UMC, Vrije Universiteit Amsterdam, The Netherlands. DNA was subjected to bisulphite treatment using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) as described previously.23 Bisulfite‐converted DNA was used as template for DNA methylation analysis for FAM19A4 and miR124‐2 genes using the QIAsure Methylation Test® (QIAGEN, Hilden, Germany) and was performed on the Rotorgene PCR‐platform (QIAGEN). The housekeeping gene β‐actin (ACTB) was used as a reference to assure successful bisulfite conversion, sample quality and normalization. A total of 22 (4.1%) samples were excluded from further analysis due to invalid FAM19A4/miR124‐2 methylation results, resulting in a final case series of 519 cervical carcinomas. The ΔΔCt values for FAM19A4 and miR124‐2 were calculated according to the manufacturer's instructions.
Data and statistical analysis
Cervical scrapes were scored positive for methylation based on preset ΔΔCt value thresholds according to the manufacturer's instructions. Thresholds for FAM19A4/miR124‐2 methylation analysis on cervical tissue samples were defined in a training set according to a predefined CIN3+ specificity of 70%. The 95% (exact) CI's were determined for the proportions of methylation‐positive samples. The proportions of the methylation‐positive samples per subcategory of histotype, FIGO stage, hrHPV status, hrHPV genotype, sample type and geographical region were compared using the Fishers’ exact test. Median age within groups was compared using the Mann–Whitney U test. For hrHPV genotyping, infections were categorized using hierarchical attribution, which categorizes hrHPV genotypes based on the genotype prevalence in our total series of cervical cancer. In our study population, HPV16 followed by HPV18 were the most prevalent genotypes, resulting in the following categories: (1) HPV16‐positive, (2) HPV16‐negative, HPV18‐positive and (3) HPV16/18‐negative, non‐16/18 hrHPVpositive. In cancers with multiple hrHPV infections, the more prevalent genotype was attributed to the case (e.g., an HPV16 and 18‐positive carcinoma was placed in Category 1). The interquartile range (IQR) was defined as the difference between the first and third quartiles. Analyses were performed in R (version 3.2.5, Vienna, Austria), SPSS statistics (version 22, IBM Corp, Armonk, NY) and Excel.
Results
Case series
A flowchart of the study is presented in Figure 1. The 519 invasive cervical carcinoma cases with a valid methylation result in our study comprised squamous cell carcinomas (n = 318), adenocarcinomas (n = 123), adenosquamous cell carcinomas (n = 42), other rare histotypes (n = 32) and histotype not specified (n = 4). The group of rare cervical cancer histotypes consisted of clear cell carcinomas (n = 14), neuroendocrine carcinomas (n = 13), adenocarcinoma mixed with neuroendocrine carcinoma (n = 1), mucinous adenocarcinoma gastric type (n = 2), mucinous adenocarcinoma, NOS (n = 1) and serous adenocarcinoma (n = 1). The median age of the women was 46.0 years (IQR; 38–56).
Figure 1.
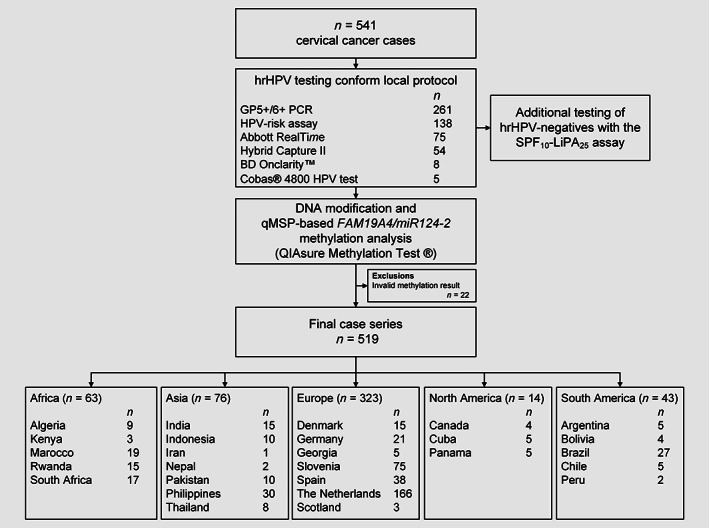
Study flowchart. Abbreviations: hrHPV, high‐risk human papillomavirus; qMSP, quantitative methylation‐specific PCR.
High‐risk HPV
Overall, 489 out of the 519 cancer cases were positive for hrHPV DNA, resulting in a hrHPV positivity rate of 94.2% (489/519; 95% CI: 91.9–96.1). Genotyping results were available for 429 out of the 489 hrHPV‐positive cancers. Based on hierarchical attribution, 255 cervical carcinoma were positive for HPV16 (59.4%), 92 for HPV18 (21.4%), and 82 for non‐16/18 hrHPV genotypes (19.1%). Of the initial 30 hrHPV‐negative cases, 10 tested positive with the additional SPF10‐LiPA25 assay, including 2 HPV16, 2 HPV18 and 6 non‐16/18 hrHPV types. One case was excluded from retesting with the SPF10‐LiPA25 assay due to insufficient residual sample, leaving 19 (3.7%) samples that remained negative for hrHPV DNA.
FAM19A4/miR124‐2 methylation analysis
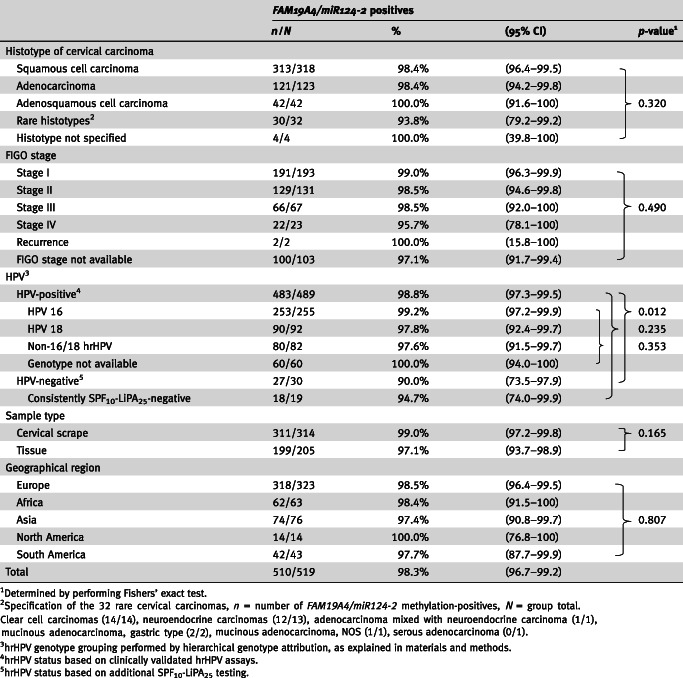
In total, 510 out of the 519 cancer cases tested positive for FAM19A4/miR124‐2 methylation, which yielded a methylation positivity rate of 98.3% (95% CI: 96.7–99.2). Table 1 shows the methylation positivity rates stratified for histotype, FIGO stage, hrHPV status, hrHPV genotype, sample type and geographical region. When considering results from clinically validated hrHPV assays only, a lower proportion of hrHPV‐negative samples tested methylation‐positive as compared to hrHPV‐positive samples: 90.0% (27/30, 95% CI: 73.5–97.9) vs. 98.8% (483/489; 95% CI: 97.3–99.5), p = 0.012. However, this observation did not hold when the analysis was restricted to consistently hrHPV‐negative cases based on additional SPF10‐LiPA25 testing with a methylation assay positivity rate of 94.7% (18/19; 95% CI: 74.0–99.9), p = 0.235. There was no difference in methylation positivity rates between hrHPV genotype, histotype, FIGO stage, sample type tested or geographical region (Table 1). In addition, no difference in median age between women with methylation‐positive (46 years, IQR 38–56) and methylation‐negative carcinomas (55 years, IQR 31–57) was found (p = 0.841).
Table 1.
FAM19A4/miR124‐2 positivity rates stratified per histotype, FIGO stage, hrHPV status, hrHPV genotype, sample type and geographical region
|
Considering the relative contribution of each marker, overall 410 cases (79.0%; 95% CI: 75.2–82.4) tested positive for both FAM19A4 and miR124‐2 genes, 72 (13.9%; 95% CI: 11.0–17.1) tested single FAM19A4 methylation‐positive and 28 (5.4%; 95% CI: 3.6–7.7) tested single miR124‐2 methylation‐positive. This frequency distribution was similar in subgroups based on hrHPV genotype, FIGO stage, sample type tested and geographical region. A higher proportion of samples was observed to be single miR124‐2 methylation‐positive in the consistently hrHPV‐negative cases as compared to the hrHPV‐positive cases (63.2% vs. 2.9%, p < 0.0001) and in rare histotypes as compared to the other histotypes merged as a group (28.1% vs. 3.9%, p < 0.0001).
Discussion
In our study, we evaluated FAM19A4/miR124‐2 methylation analysis in a large, worldwide series of cervical cancer, and demonstrated a positivity rate of over 98%. To the best of our knowledge, our study is the largest assessment of host cell DNA methylation in invasive cervical cancer to date. We showed that FAM19A4/miR124‐2 methylation is common to cervical cancer worldwide, including rare histotypes and hrHPV‐negative carcinomas. These results indicate that the FAM19A4/miR124‐2 methylation assay has a high sensitivity for cervical cancer and that a negative test result is likely to rule out the presence of cervical cancer.
Our data add a series of over 500 cases and show that aberrant FAM19A4/miR124‐2 methylation is rather universal to cervical carcinogenesis given that nearly all invasive cervical cancers were methylation‐positive for these human genes. This finding is in line with earlier studies using research tests on the same or different host cell genes but with lower numbers of included samples. In a Dutch series of 79 cervical carcinomas, CADM1/MAL/miR124‐2 methylation analysis identified 100% of the cervical cancers and in a series of 22 women, FAM19A4 detected all cervical carcinomas.8, 13 In addition to this, FAM19A4 detected 93.4% (57/61) of the cervical cancer cases in a Chinese cohort.15 Furthermore, a four‐gene methylation marker panel consisting out of the genes JAM3, EPB41L3, TERT and C13ORF18 detected 94.2% (65/69) of cervical carcinomas.10
The major strengths of our study are its large size, the inclusion of cases from over 25 countries and the use of a standardized FAM19A4/miR124‐2 methylation assay, with high intralaboratory and interlaboratory agreement as demonstrated in a recent international study.24 The present study indicates that the predefined methylation marker cut‐offs for test positivity appear independent of the population.
The current series of cervical cancers represented the most common histotypes, that is, squamous cell carcinomas and adeno(squamous)carcinomas, various rare histotypes including clear cell carcinomas and neuroendocrine carcinomas, and hrHPV‐negative cervical carcinomas. It should however be noted that this case series was selected for cases with sufficient DNA for methylation analysis and enriched for adeno(squamous)carcinomas, rare histotypes and hrHPV‐negative cervical carcinomas. Accordingly, this case series does not accurately represent the cervical cancer histotype distribution in the general population.
Although persistent hrHPV infection has been identified as the key causative agent for the development of cervical cancer,25 a (small) proportion of cervical carcinomas test hrHPV‐negative, as also confirmed in our series with very sensitive SPF10‐LiPA25 testing. It has been shown that these cases can be reclassified by surgical screening as either malignant tumors of other origin or finally as true hrHPV‐negative cervical cancers.26 Though rare, these cancer cases represent a challenge using hrHPV‐based screening methods. Since we showed that the FAM19A4/miR124‐2 methylation positivity rate was also high in this subgroup of hrHPV‐negative carcinomas, combining the FAM19A4/miR124‐2 methylation assay with HPV screening could exclude prevalent cancer with high reassurance. In addition, DNA methylation analysis on cervical scrapes might have the potential to detect noncervix gynecological cancers, like endometrial and ovarian carcinomas.8, 27 Our data indicate that also most cervical cancer cases with a rare histotype test FAM19A4/miR124‐2 methylation‐positive. The complementarity between FAM19A4/miR124‐2 methylation analysis was particularly noticeable in this subgroup, as well as in hrHPV‐negative carcinomas, allowing the combination of the two markers to consistently detect cervical cancer.
A limitation of our study, however, is that the number of rare cervical cancer histotypes remains relatively low despite the inclusion of samples from five continents, and hence needs to be expanded in further research for other histotypes than clear cell carcinomas and neuroendocrine carcinomas. In addition, the statistical analysis between subgroups should be interpreted with care, since comparisons were made with very small groups of FAM19A4/miR124‐2 methylation negative cancers. Another limitation of our study is that we were unable to evaluate additional H&E slides to assess whether a negative hrHPV test and/or a negative methylation test was caused by a very low or absent tumor fraction in the tested sample. Furthermore, given the retrospective design, not all data such as hrHPV genotype, cytology of the cervical scrapes or FIGO stage were available for all samples. It was noted, however, that all scrapes with known normal cytology (7 out of 7) and 99.0% of known FIGO stage I samples (191 out of 193) tested FAM19A4/miR124‐2 methylation‐positive. These findings are consistent with previous data of DNA hypermethylation being an early event in cervical cancer,15, 28 and support the potential of the methylation test for use in population‐based screening where most cervical cancers are early‐stage cancers.
The findings of our study highlight the value of FAM19A4/miR124‐2 methylation analysis as test with a very high sensitivity for cervical cancer, which is an essential characteristic of a triage test. An earlier study has shown that this translates into a low 14‐year cancer risk among hrHPV‐positive, methylation‐negative women (1.7%),17 and recently Dick et al. reported that a negative FAM19A4/miR124‐2 methylation triage test provides a similar long‐term CIN3+ risk compared to a negative cytology triage test among hrHPV‐positive women of 30 years and older.18 Altogether, these data indicate that further prospective studies are warranted to confirm the value of the FAM19A4/miR124‐2 methylation test as a safe triage alternative in hrHPV‐based screening programs. As FAM19A4/miR124‐2 methylation analysis is applicable to either a physician‐taken cervical scrape or a self‐collected specimen,29 objective, full molecular screening can be envisioned.
Overall, an approach combining hrHPV screening with methylation biomarkers, with or without supporting cytology, could allow for a more personalized screening with an improved balance between detection of treatment requiring disease and overtreatment, compared to the current implemented or proposed primary hrHPV‐based screening models. The combination of the FAM19A4/miR124‐2 methylation assay with hrHPV testing could provide marked reassurance for the subset of cases that are missed by hrHPV testing, especially when the high sensitivity of methylation analysis for adenocarcinoma and hrHPV‐negative cancers will also apply to adenocarcinoma in situ (AIS) and other non‐HPV‐related (glandular) high‐grade precursors.8, 30 Furthermore, with increasing uptake of prophylactic HPV vaccines, FAM19A4/miR124‐2 methylation analysis might be a good assay to rule out cervical cancers whether caused by hrHPV infection prior to vaccination, hrHPV genotypes not covered by the current prophylactic vaccines or characterized by the absence of hrHPV.
In conclusion, our study shows that methylation analysis of FAM19A4 and miR124‐2 genes detects nearly all cervical carcinomas, independent of histotype, FIGO stage, hrHPV status, hrHPV genotype, sample type and geographical region. These results indicate that a negative FAM19A4/miR124‐2 methylation assay result is likely to rule out the presence of cervical cancer.
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Ethics approval
The work in our study with human‐derived material was conducted under national and international rules and legislation, as well as European standards of research ethics, as it is expressed in the applicable legislation/regulations (The Declaration of Helsinki; informed consent for participation of human subjects in medical and scientific research) and guidelines for Good Clinical Practice. The study was approved by the local ethics committees.
Acknowledgements
This work is dedicated to our colleague Prof. Dr P.J.F. Snijders, who passed away on May 27, 2018. We thank W. Verlaat, S. Doorn, A. van Splunter and M. Bogaarts for excellent technical assistance. Our study was funded by the SME Instrument in the Horizon 2020 Work Program of the European Commission (Valid‐screen 666800). Personal funding: MP and AO were supported by the COHEAHR Network (grant no. 603019), which was funded by the 7th Framework Programme of DG Research and Innovation of the European Commission.
Author contributions: Project management: Vink FJ, Floore AN, Hesselink AT, Heideman DAM. Data collection: Clifford GM, Poljak M, Oštrbenk A, Petry KU, Rothe B, Bonde J, Pedersen H, de Sanjosé S, Torres M, del Pino M, Quint WGV, Cuschieri K, Alcañiz Boada E, van Trommel N, Bleeker MCG. Data analysis: Vink FJ, Lissenberg‐Witte B. Interpretation of data: Vink FJ, Meijer CJLM, Steenbergen RDM, Bleeker MCG, Heideman DAM. Writing of manuscript: Vink FJ, Meijer CJLM, Steenbergen RDM, Bleeker MCG, Heideman DAM. Review and Final approval of the manuscript: all authors.
Conflict of interest: DH, CM and RS are minority shareholders of Self‐screen B.V., a spin‐off company of VU University Medical Center (currently known as Amsterdam UMC, Vrije Universiteit Amsterdam). Self‐screen B.V. holds patents related to the work (i.e., high‐risk HPV test and methylation markers for cervical screening) and has developed and manufactured the methylation assay, which is licensed to QIAGEN (QIAsure Methylation Test®). CM has received speaker's fee from GSK, QIAGEN and SPMSD/Merck, and served occasionally on the scientific advisory board (expert meeting) of GSK, QIAGEN and SPMSD/Merck. CM has a very small number of shares of MDxHealth and QIAGEN and holds minority stock in Self‐Screen B.V. Until April 2016, he was minority shareholder of Diassay B.V. CM is part‐time director of Self‐screen B.V. since September 2017. AO has received reimbursement of travel expenses for attending conferences and honoraria for speaking from Abbott Molecular, QIAGEN and Seegene. KP has received speaker's fee from Mikrogen. JB is the PI of studies funded in part by BD Diagnostics, Agena Bioscience, Genomica SAU, LifeRiver Biotech and QIAGEN. He has received honoraria for lectures from BD Diagnostics, Roche Molecular Systems, QIAGEN and Genomica SAU. JB is an appointed member of the National Danish Cervical Screening Committee by the Danish Health Authority, and member of the Regional cervical screening steering committee of the Capital Region of Denmark. MDP has received occasionally speaker's fee from Roche diagnostics. MT's institution has received sponsorship grants from Merck & Co, Seegene, Hologic and GSR. WQ is shareholder of LBP, subcontractor of Self‐screen B.V. as the producer of the QIAsure Methylation Test. KC's and EAB's institution has received research funding or and/or associated consumables to support research from the following commercial entities in the last 3 years: Cepheid, Genomica, LifeRiver, Euroimmun, GeneFirst, Self‐screen B.V., QIAGEN and Hologic. AF and AH are employed by Self‐screen B.V. DH has been on the speakers bureau of QIAGEN and serves occasionally on the scientific advisory boards of Pfizer and Bristol‐Myers Squibb. All other authors declare that they have no conflicts of interest. The authors are solely responsible for final content of the manuscript and interpretation.
Data availability
The data that support the findings of our study are available from the corresponding author upon reasonable request.
References
- 1. Arbyn M, Raifu AO, Weiderpass E, et al. Trends of cervical cancer mortality in the member states of the European Union. Eur J Cancer 2009;45:2640–8. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 3. Cuschieri K, Ronco G, Lorincz A, et al. Eurogin roadmap 2017: triage strategies for the management of HPV‐positive women in cervical screening programs. Int J Cancer 2018;143:735–45. [DOI] [PubMed] [Google Scholar]
- 4. Ronco G, Dillner J, Elfstrom KM, et al. Efficacy of HPV‐based screening for prevention of invasive cervical cancer: follow‐up of four European randomised controlled trials. Lancet 2014;383:524–32. [DOI] [PubMed] [Google Scholar]
- 5. Arbyn M, Ronco G, Anttila A, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine 2012;30(Suppl 5):F88–99. [DOI] [PubMed] [Google Scholar]
- 6. Steenbergen RD, Snijders PJ, Heideman DA, et al. Clinical implications of (epi)genetic changes in HPV‐induced cervical precancerous lesions. Nat Rev Cancer 2014;14:395–405. [DOI] [PubMed] [Google Scholar]
- 7. Bierkens M, Hesselink AT, Meijer CJ, et al. CADM1 and MAL promoter methylation levels in hrHPV‐positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer 2013;133:1293–9. [DOI] [PubMed] [Google Scholar]
- 8. De Strooper LM, van Zummeren M, Steenbergen RD, et al. CADM1, MAL and miR124‐2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol 2014;67:1067–71. [DOI] [PubMed] [Google Scholar]
- 9. Cook DA, Krajden M, Brentnall AR, et al. Evaluation of a validated methylation triage signature for human papillomavirus positive women in the HPV FOCAL cervical cancer screening trial. Int J Cancer 2019;144:2587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eijsink JJ, Lendvai A, Deregowski V, et al. A four‐gene methylation marker panel as triage test in high‐risk human papillomavirus positive patients. Int J Cancer 2012;130:1861–9. [DOI] [PubMed] [Google Scholar]
- 11. Schmitz M, Eichelkraut K, Schmidt D, et al. Performance of a DNA methylation marker panel using liquid‐based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer 2018;18:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boers A, Wang R, van Leeuwen RW, et al. Discovery of new methylation markers to improve screening for cervical intraepithelial neoplasia grade 2/3. Clin Epigenetics 2016;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Strooper LM, Meijer CJ, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila) 2014;7:1251–7. [DOI] [PubMed] [Google Scholar]
- 14. Luttmer R, De Strooper LM, Berkhof J, et al. Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre)cancer in high‐risk HPV‐positive women of a gynecologic outpatient population (COMETH study). Int J Cancer 2016;138:992–1002. [DOI] [PubMed] [Google Scholar]
- 15. Bu Q, Wang S, Ma J, et al. The clinical significance of FAM19A4 methylation in high‐risk HPV‐positive cervical samples for the detection of cervical (pre)cancer in Chinese women. BMC Cancer 2018;18:1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilting SM, van Boerdonk RA, Henken FE, et al. Methylation‐mediated silencing and tumour suppressive function of hsa‐miR‐124 in cervical cancer. Mol Cancer 2010;9:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Strooper LMA, Berkhof J, Steenbergen RDM, et al. Cervical cancer risk in HPV‐positive women after a negative FAM19A4/mir124‐2 methylation test: a post hoc analysis in the POBASCAM trial with 14 year follow‐up. Int J Cancer 2018;143:1541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dick S, Kremer WW, De Strooper LMA, et al. Long‐term CIN3+ risk of HPV positive women after triage with FAM19A4/miR124‐2 methylation analysis. Gynecol Oncol 2019;154:368–73. [DOI] [PubMed] [Google Scholar]
- 19. Arbyn M, Snijders PJ, Meijer CJ, et al. Which high‐risk HPV assays fulfil criteria for use in primary cervical cancer screening? Clin Microbiol Infect 2015;21:817–26. [DOI] [PubMed] [Google Scholar]
- 20. Kleter B, van Doorn LJ, Schrauwen L, et al. Development and clinical evaluation of a highly sensitive PCR‐reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol 1999;37:2508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kurman RJ, Carcangiu ML, Herrington CS, et al. WHO classification of tumours of female reproductive organs, volume 6. WHO classification of tumours, 4th edn France: International Agency for Research on Cancer, 2014. 307. [Google Scholar]
- 22. Bhatla N, Denny L. FIGO cancer report 2018. Int J Gynaecol Obstet 2018;143(Suppl 2):2–3. [DOI] [PubMed] [Google Scholar]
- 23. Overmeer RM, Henken FE, Bierkens M, et al. Repression of MAL tumour suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol 2009;219:327–36. [DOI] [PubMed] [Google Scholar]
- 24. Floore A, Hesselink A, Ostrbenk A, et al. Intra‐ and inter‐laboratory agreement of the FAM19A4/mir124‐2 methylation test: results from an international study. J Clin Lab Anal 2019;33:e22854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999;189:12–9. [DOI] [PubMed] [Google Scholar]
- 26. Petry KU, Liebrich C, Luyten A, et al. Surgical staging identified false HPV‐negative cases in a large series of invasive cervical cancers. Papillomavirus Res 2017;4:85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang CC, Wang HC, Liao YP, et al. The feasibility of detecting endometrial and ovarian cancer using DNA methylation biomarkers in cervical scrapings. J Gynecol Oncol 2018;29:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verlaat W, Van Leeuwen RW, Novianti PW, et al. Host‐cell DNA methylation patterns during high‐risk HPV‐induced carcinogenesis reveal a heterogeneous nature of cervical pre‐cancer. Epigenetics 2018;13:769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luttmer R, De Strooper LM, Steenbergen RD, et al. Management of high‐risk HPV‐positive women for detection of cervical (pre)cancer. Expert Rev Mol Diagn 2016;16:961–74. [DOI] [PubMed] [Google Scholar]
- 30. van der Meide WF, Snellenberg S, Meijer CJ, et al. Promoter methylation analysis of WNT/beta‐catenin signaling pathway regulators to detect adenocarcinoma or its precursor lesion of the cervix. Gynecol Oncol 2011;123:116–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of our study are available from the corresponding author upon reasonable request.


