Summary
Bacteria in nature often encounter non‐antibiotic antibacterials (NAAs), such as disinfectants and heavy metals, and they can evolve resistance via mechanisms that are also involved in antibiotic resistance. Understanding whether susceptibility to different types of antibacterials is non‐randomly associated across natural and clinical bacteria is therefore important for predicting the spread of resistance, yet there is no consensus about the extent of such associations or underlying mechanisms. We tested for associations between susceptibility phenotypes of 93 natural and clinical Escherichia coli isolates to various NAAs and antibiotics. Across all compound combinations, we detected a small number of non‐random associations, including a trio of positive associations among chloramphenicol, triclosan and benzalkonium chloride. We investigated genetic mechanisms that can explain such associations using genomic information, genetic knockouts and experimental evolution. This revealed some mutations that are selected for by experimental exposure to one compound and confer cross‐resistance to other compounds. Surprisingly, these interactions were asymmetric: selection for chloramphenicol resistance conferred cross‐resistance to triclosan and benzalkonium chloride, but selection for triclosan resistance did not confer cross‐resistance to other compounds. These results identify genetic changes involved in variable cross‐resistance across antibiotics and NAAs, potentially contributing to associations in natural and clinical bacteria.
Introduction
Antibiotic resistance is a growing problem worldwide. A major driver of resistance is exposure of bacteria to antibiotics (Andersson and Hughes, 2012), but in nature bacteria also encounter other types of antibacterial compounds, such as disinfectants, preservatives and heavy metals. Laboratory experiments show bacteria can evolve resistance to these non‐antibiotic antibacterials (NAAs) (McMurry et al., 1998b; Langsrud et al., 2004). Some environmental, human and animal isolates also show relatively high resistance to NAAs (Jeanthon and Prieur, 1990; Copitch et al., 2010; Deus et al., 2017; Pidot et al., 2018). NAA resistance can be conferred by mechanisms that also confer resistance to antibiotics, such as increased expression of nonspecific efflux pumps or compound‐degrading enzymes (Levy, 2002; Stepanauskas et al., 2005). Even when they are encoded by different mechanisms, antibiotic‐ and metal‐resistance genes can co‐occur in the same bacteria (Baker‐Austin et al., 2006; Sandegren et al., 2012; Pal et al., 2015), especially in bacteria associated with humans or human activities (Li et al., 2017). This means exposure of bacteria to NAAs potentially contributes to the spread of antibiotic resistance. However, the evidence of whether NAA resistance and antibiotic resistance co‐occur at higher than expected frequencies in natural populations, and whether adaptation to NAAs of natural bacteria simultaneously increases their resistance to clinically relevant antibiotics, is not conclusive. For example, some studies found co‐occurrence of reduced susceptibility to antibiotics and biocides such as triclosan among natural isolates (Copitch et al., 2010; Middleton and Salierno, 2013), while others did not (Lear et al., 2002; Cameron et al., 2019). Similarly, upon experimental challenge with biocides, bacteria have been observed to sometimes acquire cross‐resistance to antibiotics (Braoudaki and Hilton, 2004a, 2004b; Pycke et al., 2010), but in other cases not to (Suller, 2000; Ledder et al., 2006).
It is especially important to know more about the relationship between susceptibility to antibiotics and to heavy metals and disinfectants. Both of these compound classes are used in various industries and consumer products. Once released into the environment, they resist degradation and persist in water and soil (Wales and Davies, 2015; Tronsmo et al., 2016). Metal pollutants can come from various sources, such as zinc and cadmium from metal production processes, coal combustion, battery recycling and agriculture (Bradl, 2005). Disinfectants and preservatives, such as the polychloro phenoxy phenol triclosan and the quaternary ammonium compound benzalkonium chloride, are used in a wide range of consumer products, such as soap, toothpaste and deodorants (Bhargava and Leonard, 1996; Merchel Piovesan Pereira and Tagkopoulos, 2019), from where they can enter the environment. Derivative molecules of such compounds can be detected in human bodily fluids, waste water, lakes and rivers (Carey and McNamara, 2014; Zhang et al., 2015; Azzouz et al., 2016). Similarly, detergents such as SDS are very widely used in cleaning and hygiene products (Bondi et al., 2015), but also in food production (Byelashov et al., 2008) and laboratory procedures (Swank and Munkres, 1971). As for other NAAs, evolution of resistance against heavy metals and disinfectants has been described and associations with antibiotic resistance have been observed, including in environmental isolates (McMurry et al., 1998b; Langsrud et al., 2004; Braoudaki and Hilton, 2004a, 2004b; Stepanauskas et al., 2005; Dickinson et al., 2019). However, there is little consensus over the extent and importance of the interplay between antibiotics and these types of NAAs in terms of resistance evolution. Moreover, the relative contributions of different genetic mechanisms that can drive such associations are not fully understood. For example, correlated resistance to different types of antibacterials can result from the same allele (a resistance gene or mutation) affecting multiple susceptibility phenotypes simultaneously [cross‐resistance (Baker‐Austin et al., 2006); e.g. via efflux pumps (Levy, 2002; Poole, 2002)], or from co‐occurrence of multiple resistance alleles in the same organism [co‐resistance (Baker‐Austin et al., 2006); e.g. plasmids carrying multiple resistance genes (Sandegren et al., 2012; Pal et al., 2015)].
Here, we ask the following questions: (i) Are antibiotic and NAA (here, heavy metals and disinfectants/detergents) susceptibility non‐randomly associated (phenotypically and genotypically) across natural and clinical isolates of Escherichia coli? (ii) Can we explain non‐random associations between susceptibility toward different classes of antimicrobials by looking at carriage of particular resistance genes, or individual mutations that affect multiple resistance phenotypes simultaneously? (iii) In cases where resistance to one type of antibacterial causes altered resistance to another, are these effects reciprocal? For example, if exposure to an NAA results in cross‐resistance to an antibiotic, does exposure to the antibiotic result in the same type of cross‐resistance? This is important for understanding how separate exposure to each type of compound in environmental or clinical settings may contribute to the spread of resistance to both of them. Among antibiotics, cross‐resistance observed in resistant laboratory isolates is sometimes reciprocal (selection with either compound generates cross‐resistance to both) and sometimes asymmetric (the cross‐resistance phenotype depends on which compound bacteria were exposed to first) (Imamovic and Sommer, 2013; Lázár et al., 2014), but this remains unclear for cross‐resistance across antibiotics and NAAs.
To quantify associations among susceptibility phenotypes toward different NAAs, we tested 93 natural and clinical isolates for their susceptibility to five NAAs (zinc, cadmium, benzalkonium chloride, triclosan and SDS). We did this with E. coli, a widespread species and key pathogen (Kaper et al., 2004) for which antibiotic resistance is a growing challenge (Baquero et al., 2008; Allen et al., 2010), and can evolve via conserved mechanisms also relevant in other species (Carattoli, 2009; Du et al., 2014). We mapped these results against existing antibiotic susceptibility data for the same isolates, to test whether susceptibility to NAAs was associated with antibiotic susceptibility, correcting for phylogenetic relatedness among the isolates. Where relevant, we interpreted our data in the context of epidemiological cut‐off values (ECOFF), where sensitivity/resistance categorization is based on the species‐wide distribution of minimal inhibitory concentrations (MICs) (Kahlmeter et al., 2003; Morrissey et al., 2014). To investigate possible genetic mechanisms driving observed associations, we (i) analysed data on the metal and biocide resistance gene (MBRG) and antibiotic resistance gene (ARG) contents of the isolates, testing for non‐random associations between individual resistance genes and resistance phenotypes, and (ii) used genetic knockouts and experimentally evolved resistant mutants to screen for genes that affect multiple phenotypes simultaneously. We found a small number of non‐random associations between antibiotic‐ and NAA‐susceptibility phenotypes across natural and clinical isolates. Observed associations were mostly positive. We found that the same gene could affect susceptibility to both antibiotics and NAAs, and that mutants isolated after experimental challenge with one type of compound were sometimes cross‐resistant to others, but such interactions were asymmetric across compounds.
Results and discussion
Positive association between susceptibility to benzalkonium chloride and triclosan, but not zinc, cadmium or SDS
We quantified the susceptibility phenotypes of natural and clinical E. coli isolates against five different NAAs (benzalkonium chloride, cadmium, SDS, triclosan, zinc), before testing for non‐random associations between each pair of susceptibility phenotypes (each pair of NAA compounds) across the isolates (lower part of Fig. 1). This showed natural and clinical isolates with relatively high triclosan resistance also had relatively high resistance to benzalkonium chloride (tested using phylogenetic generalized least squares, PGLS, that accounts for phylogenetic relatedness among the isolates and with correction for multiple testing, see Section 4; beta = 0.371, t 2.256,91 = 4.089, corrected p < 0.01; Fig. 1). For all other pairs of NAAs we tested, we found no statistically significant associations across natural and clinical isolates. Despite this, observed associations were on average positive (mean ± SD of t‐values of regression coefficient without significant associations: 0.542 ± 1.109; Fig. 1). Therefore, we do not rule out other associations that are weaker but would only be detected with a much larger sample size, or across other combinations of compounds and strains. We found significant evidence that phylogenetically closely related isolates had more similar susceptibility levels only for benzalkonium chloride (λ = 0.466, corrected p < 0.001; Fig. 2). Compared with concentrations reported elsewhere as being likely to select for resistance, 12 isolates (12.9%) had an IC90 (the concentration required to inhibit growth by 90%; see Section 4) for cadmium above the concentration of 1 mM considered by Khan et al. (2015) as indicative for cadmium resistance. For zinc, the IC90s we estimated were comparable with MICs of human and avian isolates of E. coli reported previously [mean ± SD IC90 for zinc in this study: 272.526 ± 63.232 μg/ml; mean ± SD MIC reported by Deus et al. (2017)): 348.69 ± 148.470 μg/ml]. For triclosan and benzalkonium chloride, all isolates had IC90s below reported ECOFF concentrations (4 and 128 μg/ml respectively). For SDS, Kramer et al. (1984) considered Enterobacteriaceae as SDS‐resistant if they could grow in 5% SDS. According to this criterion, 21 of our isolates (22.6%) can be considered SDS‐resistant, because they were not inhibited by 4% SDS, the highest concentration tested.
Figure 1.

Associations between susceptibility to different antibacterials across natural and clinical Escherichia coli isolates.
The matrix shows the strength of association (t‐statistic) for individual compound combinations, obtained with the pgls function of R's caper package (see Section 4). Susceptibility here is taken as the IC90 for each compound (see Section 4). Thus, a strong association means isolates that were relatively resistant to one compound were also relatively resistant to the other.
Significant combinations are indicated with black asterisks (significance codes: ‘**’ <0.01; ‘*’ <0.05, p‐values corrected for multiple testing by sequential Bonferroni correction). Benzalk.chl. is benzalkonium chloride.
Figure 2.
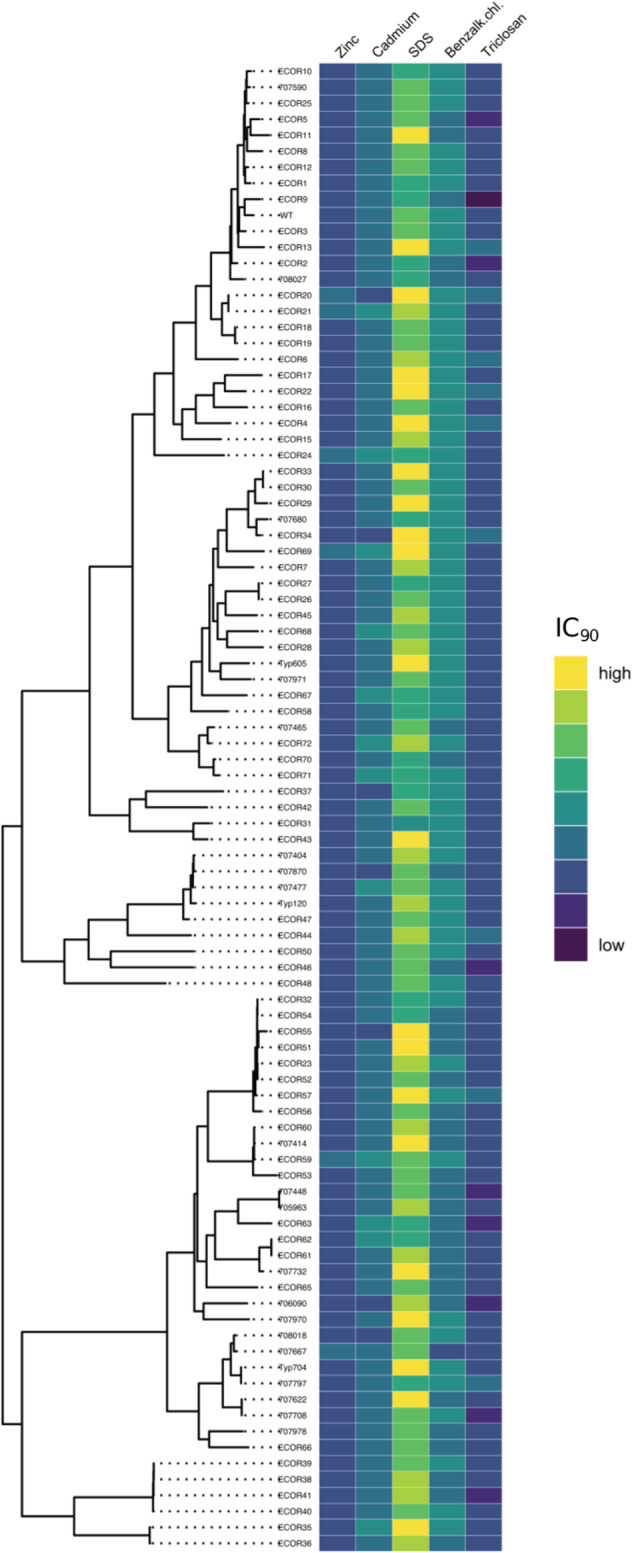
Susceptibility profiles of natural and clinical isolates toward the five tested NAAs, grouped according to core‐genome phylogeny.
In the heat map, blue shades indicate an isolate with a relatively low IC90 to the range of tested concentrations, yellow shades indicate a relatively high IC90.
The core‐genome phylogenetic tree for natural and clinical isolates at left was adapted from (Allen et al., 2017). Benzalk.chl. is benzalkonium chloride.
Range of tested concentrations for each compound (twofold broth dilution): zinc 8192–16 μg/ml; cadmium 20–0.0390625 mmol/l; SDS 4–0.0078125 mg/ml; benzalkonium chloride 256–0.5 μg/ml; triclosan 4–0.0078125 μg/ml.
Links between chloramphenicol, triclosan and benzalkonium chloride susceptibility
Having observed a significant positive association across our isolates for susceptibility to the two NAAs benzalkonium chloride and triclosan, we tested for associations between susceptibility to NAAs and to antibiotics. For this, we obtained data for susceptibility to nine antibiotics from a previous study with the same isolates (Allen et al., 2017). We found susceptibility to benzalkonium chloride and triclosan was also positively associated with chloramphenicol susceptibility (chloramphenicol–benzalkonium chloride association by PGLS: beta = 0.307, t 2.254,91 = 4.484, corrected p < 0.01; chloramphenicol‐triclosan association by PGLS: beta = 0.285, t 2.261,91 = 4.031, corrected p < 0.01). Triclosan resistance was also positively associated with resistance to another antibiotic, tigecycline (PGLS, beta = 0.316, t 2.277,91 = 3.837, corrected p < 0.05). One isolate (ECOR9, Fig. S1) was highly susceptible to each of the compounds for which we observed positive associations. Exclusion of this isolate did not render the observed associations nonsignificant (triclosan–tigecycline p new < 0.05; benzalkonium chloride–chloramphenicol p new < 0.001; benzalkonium chloride–triclosan p new < 0.001; triclosan–chloramphenicol p new < 0.01). The strongest negative associations we observed, such as between ciprofloxacin and cadmium susceptibility or chloramphenicol and SDS susceptibility, were not significant. In summary, the only significant associations we observed across isolates for resistance to NAAs and antibiotics were positive, in particular the triad of benzalkonium chloride, triclosan and chloramphenicol, and this was not driven by a single isolate being highly susceptible to all of them.
Known metal and biocide resistance genes predict some phenotypes
We next tested if the observed non‐random associations between NAA and antibiotic susceptibility phenotypes across our isolates were linked to carriage of similar sets of dedicated resistance genes. We found the presence/absence of some individual MBRGs (identified from genomic data using BacMet; see Section 4) was associated with susceptibility to individual antibacterials (Fig. S2). Isolates with the gene sitD, encoding an iron/manganese transport system inner membrane protein, had a lower IC90 for benzalkonium chloride (PGLS: beta = −0.936, t 4.668,90 = −4.751, corrected p < 0.01). Two related genes (sitA and sitBC) were also positively associated with susceptibility to benzalkonium chloride, and all three genes (sitA, sitBC, sitD) showed a similar association with triclosan susceptibility, although these associations were not significant after accounting for multiple testing (Fig. S2). FetA (ybbL), the ATP binding subunit of a putative ABC family iron transporter, was associated with decreased cefotaxime and ciprofloxacin susceptibility (fetA‐cefotaxime association by PGLS: beta = 3.487, t 4.814,90 = 4.169, corrected p < 0.05; fetA‐ciprofloxacin association by PGLS: beta = 5.204, t 4.546,90 = 7.285, corrected p < 0.001). It has been previously reported that fetA and fetB, the second component of the iron transport system, play a role in resistance to oxidative stress (Nicolaou et al., 2013) and antibiotics (Conley et al., 2019).
We found isolates with similar overall MBRG profiles (a pair of isolates have similar profiles when they have a small Euclidean distance across all MBRGs in terms of the presence and absence of each MBRG) did not have similar profiles in terms of NAA susceptibility (partial Mantel test: r = 0.003, p > 0.05) or antibiotic susceptibility (partial Mantel test: r = 0.031, p > 0.05). Similarly, isolates with a similar number of MBRGs did not have more similar NAA susceptibility profiles (partial Mantel test: r = 0.023, p > 0.05), antibiotic susceptibility profiles (partial Mantel test: r = −0.013, p > 0.05) or lower average susceptibility (mean IC90) to NAAs (PGLS, beta = −1.411, t = −2.222, corrected p > 0.05) or antibiotics (PGLS, beta = −0.010, t = −0.023, p > 0.05). Finally, we also tested whether NAA susceptibility profiles were linked to ARG content of the isolates (identified from genomic data using ResFinder; see Section 4) but found no strong association (partial Mantel test: r = −0.089, p > 0.05). Despite this, Allen et al. (2019) found the ARG profiles of the same isolates were predictive of their antibiotic resistance profiles, and that in some cases ARGs occur on the same contigs as plasmid‐associated sequences (replicons). Thus, known MBRG content of these isolates appeared to be less closely linked to their overall susceptibility phenotypes (across antibiotics and NAAs) than the link between ARG content and the susceptibility phenotypes. As for ARGs, we found some instances where MBRGs occurred on the same contigs as plasmid replicons. We found six such instances, each in a different isolate (although we note that there may be more such cases that are undetected by this approach). These included four IncF‐plasmid replicons, one IncI‐plasmid replicon and one IncQ‐plasmid replicon, and various MBRGs associated with arsenic, mercury, manganese and quaternary ammonium compound (qac) resistance.
In summary, despite some individual associations between known MBRGs and phenotypic variation, known MBRG content did not explain overall variation of NAA susceptibility phenotypes, or the non‐random associations we observed for susceptibility to benzalkonium chloride, chloramphenicol and triclosan.
acrA and acrB affect resistance to benzalkonium chloride, chloramphenicol and triclosan
One possible mechanism by which the observed positive associations between antibiotic and NAA susceptibility across natural and clinical isolates can emerge is by individual genes or alleles affecting multiple resistance phenotypes simultaneously. Because our natural and clinical isolates vary at many different loci, we cannot infer the effects of individual genes on resistance phenotypes here. We therefore next asked, as proof‐of‐principle, whether genetic changes at various loci in an isogenic strain background would alter multiple phenotypes simultaneously. When we tested single‐gene deletion mutants of several candidate genes (Table S1), we found deletion of rfaY, acrA and acrB increased susceptibility to benzalkonium chloride (Fig. 3). Deletion of rfaY, which is a lipopolysaccharide core heptose (II) kinase, did not increase susceptibility to the other tested antibacterials. However, deletion of acrA and acrB, which are subunits of the AcrA‐AcrB‐TolX multidrug efflux complex (Okusu et al., 1996; Levy, 2002; Tikhonova and Zgurskaya, 2004), also increased susceptibility to both chloramphenicol and triclosan (Fig. 3), though the effect for triclosan was moderate and not significant after correction for multiple testing. After testing an additional 10 antibacterials, we found deletion of acrA and acrB also strongly affected susceptibility to the surfactant SDS (Fig. S3), and moderately increased resistance to zinc. This shows genetic variation at a single locus, such as acrA or acrB, can affect multiple susceptibility phenotypes simultaneously. Analysis of the genetic sequences of all 93 isolates revealed they all carried a copy of both acrA and acrB, evidenced by high bit scores S′ for all isolates (S′ for acrA for all isolates between 686 and 700, S′ for acrB for all isolates between 1983 and 2011; S′ is a normalized version of the raw alignment score S, with higher S′ indicating better alignment (Madden, 2002); see Section 4). This suggests the same effect we observed for single‐gene knockouts of these genes is unlikely to explain the observed associations across natural and clinical isolates for benzalkonium chloride, triclosan and chloramphenicol susceptibility. Nevertheless, we cannot exclude other types of variation affecting such genes, such as regulatory mutations that increase or decrease expression levels.
Figure 3.
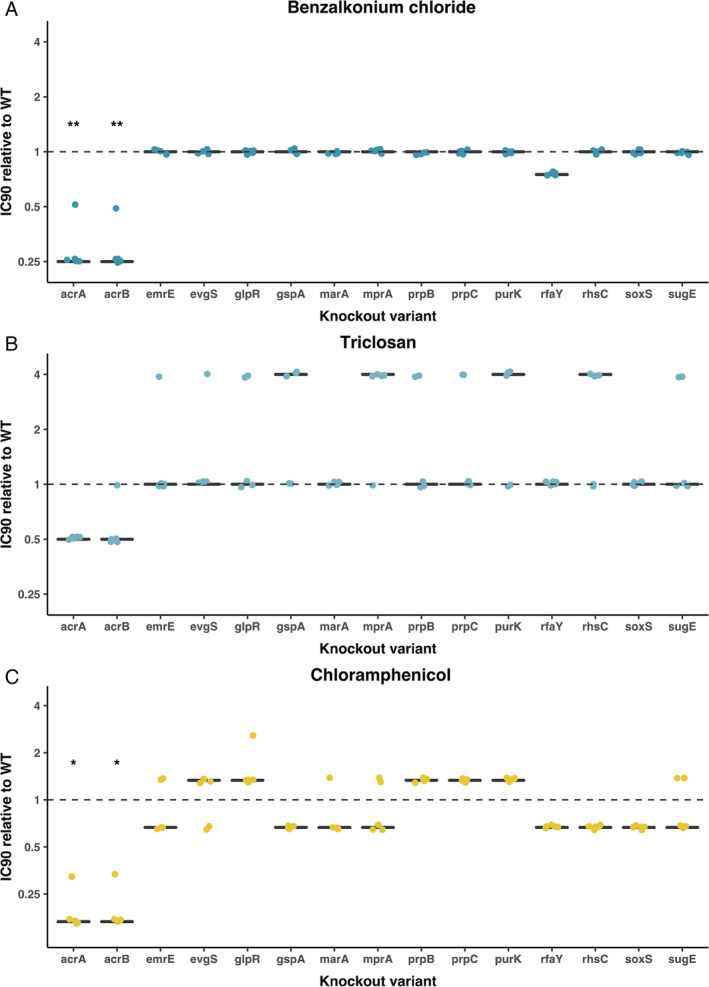
Antibacterial susceptibility of knockout variants.
Each panel shows the IC90 of 15 knockout variants from the Keio knockout collection relative to the wild‐type Escherichia coli BW25113, tested against one of three compounds [(A) benzalkonium chloride; (B) triclosan; (C) chloramphenicol)].
Each black line gives the median of five independent replicates; individual replicates are shown as coloured dots.
Significant differences from the ancestral score (1.0) are indicated with black asterisks (significance codes: ‘**’ <0.01; ‘*’ <0.05, p‐values corrected for multiple testing by sequential Bonferroni correction). [Color figure can be viewed at wileyonlinelibrary.com]
Asymmetric cross‐resistance in isolated resistant mutants
We next asked whether genetic changes that occur upon experimental challenge with one of the three compounds for which we observed non‐random associations (triclosan, benzalkonium chloride and chloramphenicol) would confer reduced susceptibility to all three. To answer this question, we isolated mutants of E. coli K‐12 MG1655 selected for resistance to benzalkonium chloride, chloramphenicol or triclosan (see Section 4). We found that all five benzalkonium chloride‐resistant mutants had marginally reduced benzalkonium chloride susceptibility compared with the wild type, but there was no consistent change in susceptibility to the other compounds tested (Fig. 4A). The decrease in benzalkonium chloride susceptibility was only consistent across replicate assays for two mutants, BKC_05 and BKC_06 (Fig. 4A). Genome sequencing revealed these two mutants were the only ones in which we detected mutations (Fig. 5A, Table S3). One (BKC_05) harboured a nucleotide change in msbA, a gene encoding for a lipid ABC transporter permease. The other (BKC_06) had a mutation in rpoC, which encodes an RNA polymerase subunit.
Figure 4.
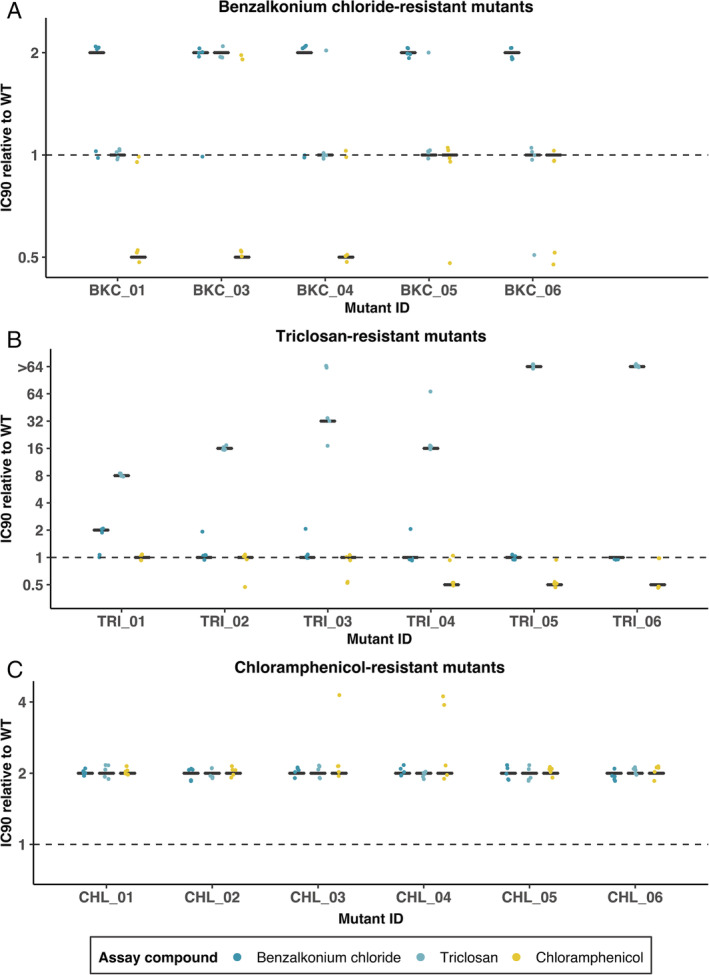
Antibacterial susceptibility of resistant mutants isolated after experimental challenge with (A) benzalkonium chloride, (B) triclosan or (C) chloramphenicol.
Each panel shows the IC90 of five or six independently isolated mutants (x‐axis) relative to the wild‐type Escherichia coli K‐12 MG1655.
For each mutant, the three black lines show susceptibility to all three compounds (see legend).
Each black line gives the median of five independent replicates; individual replicates are shown as coloured dots. Benzalkonium chloride‐ and triclosan‐resistant mutants (panels A and B) were generally only resistant to the antibacterial compound they were selected for resistance against, chloramphenicol‐resistant mutants (panel C) showed increased resistance to all three antibacterials.
Figure 5.
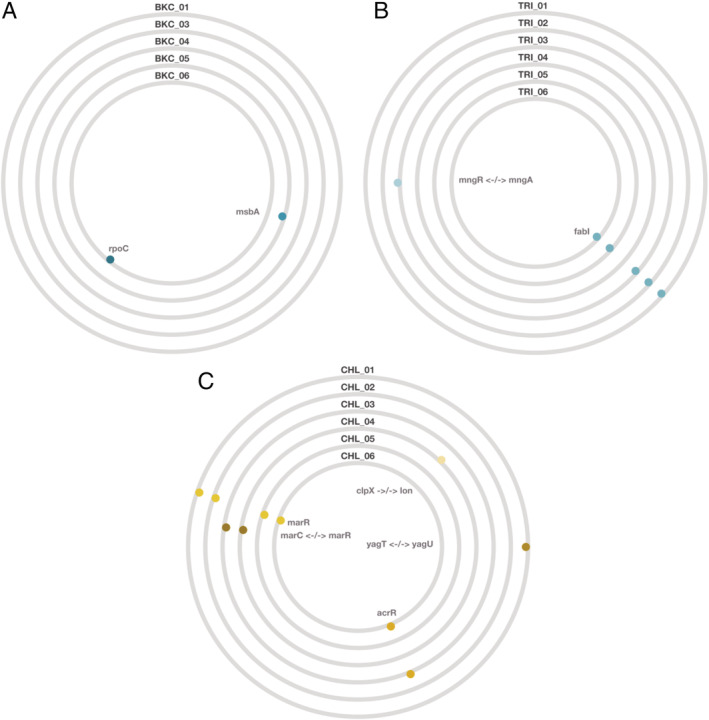
Mutations detected in resistant mutants isolated after experimental challenge with (A) benzalkonium chloride, (B) triclosan or (C) chloramphenicol.
Each BioCircos plot (panel) shows the mutations detected in the five to six resistant mutants for each compound. Each circle is one resistant mutant.
Affected genes are shown as coloured dots, with different genes in different colours, and gene names inside the circles; arrows indicate intergenic mutations. Further details are given in Table S3.
All six triclosan‐resistant mutants had greatly increased triclosan resistance relative to the wild type but no consistent change in resistance to the other tested compounds (Fig. 4B). Five of six triclosan‐resistant mutants had nucleotide changes in fabI (Fig. 5B, Table S3), which encodes the known triclosan target FabI, an enoyl‐acyl carrier protein reductase (McMurry et al., 1998b). Despite the observed increase in triclosan resistance, no mutation was detected in TRI_04; this might be explained by other mechanisms of resistance, such as a transient increase in expression of efflux pump‐encoding genes (Fernández and Hancock, 2012). Thus, five of six triclosan‐resistant mutants acquired a specialized resistance mechanism (mutations in fabI), consistent with the past work showing high‐level triclosan resistance evolving via the same pathway (McMurry et al., 1998b; Pycke et al., 2010). Note that experimental challenge with triclosan has in other scenarios and species been associated with generalized resistance mechanisms such as efflux (Chuanchuen et al., 2001; Birošová and Mikulášová, 2009), conferring cross resistance to antibiotics including chloramphenicol (Braoudaki and Hilton, 2004b), particularly after selection at lower triclosan concentrations (Pycke et al., 2010).
Unlike for triclosan or benzalkonium chloride, mutants selected for chloramphenicol resistance were consistently more resistant to all three tested antibacterials relative to the wild type (Fig. 4C). All six of these mutants had mutations in or near genes associated with expression of multidrug efflux pumps (Fig. 5C, Table S3). This included four mutants with changes in marR, two with changes in acrR and two with intergenic mutations between marC and marR (some mutants had more than one mutation). MarR (‘Multiple antibiotic resistance’) is the repressor of the marRAB operon, which is normally not transcribed. Induction of the mar phenotype (marRAB transcription) can be caused by a variety of antimicrobial agents or by mutations in marR (Seoane and Levy, 1995). The mar phenotype is linked to increased expression of the multidrug efflux pump acrAB (Alekshun and Levy, 1997). AcrR in turn is the repressor of acrAB (Lázár et al., 2014), and deleterious mutations in acrR are known to result in increased expression of acrA (Wang et al., 2001). AcrAB multidrug efflux pump has broad substrate specificity and can, as we have seen in our knockout experiments, affect multiple resistance phenotypes (Levy, 2002). Consistent with this, past work showed that increased acrAB expression, either directly or through upregulation or downregulation of genes upstream of acrAB in the regulatory network, can confer cross‐resistance to NAAs (McMurry et al., 1998a; Webber et al., 2015).
The observed asymmetry in cross‐resistance (that chloramphenicol‐resistant mutants were also resistant to benzalkonium chloride and triclosan but not vice versa) can be explained by the different types of genetic changes in mutants selected for resistance to different compounds. In triclosan‐selected mutants, we observed a specialized resistance mechanism: mutations in fabI that conferred relatively large increases in triclosan resistance and were therefore more likely to be sampled in our mutant screen with triclosan than the generalized resistance mutations in acrA and marR that conferred moderately increased resistance to all three compounds. This asymmetry is important for predicting how resistance evolves upon exposure to such compounds. It suggests exposure to chloramphenicol or other compounds that promote generalized resistance mechanisms can confer cross‐resistance to disinfectants such as triclosan and benzalkonium chloride (McMurry et al., 1998a). However, this pattern is not necessarily reciprocal, in that exposure to triclosan can instead promote specialized resistance mechanisms that do not affect antibiotic resistance. Such asymmetry can also occur across different combinations of antibiotics: Lázár et al. (2014) detected asymmetric cross‐resistance for some antibiotic combinations, and Imamovic and Sommer (2013) observed asymmetric collateral sensitivities for some combinations of 23 tested antibiotics. Across disinfectants, Braoudaki and Hilton (2004b) showed adaptation to benzalkonium chloride and to chlorhexidine resulted in asymmetric cross resistance patterns. Our results show that such asymmetric patterns of cross‐resistance/collateral effects are also important across different classes of antibacterials (here, antibiotics and disinfectants).
The positive associations we detected across benzalkonium chloride, triclosan and chloramphenicol in natural and clinical isolates more closely resembled the cross‐resistant phenotypes of our chloramphenicol‐resistant laboratory mutants than benzalkonium chloride‐ or triclosan‐resistant laboratory mutants. It is therefore possible that a similar generalized resistance mechanism as we observed in laboratory mutants, in particular mutations affecting marR, could underlie these associations in natural and clinical isolates. Consistent with this, mutations in acrR and marR have been widely reported in connection with individual resistance phenotypes in natural and clinical isolates of E. coli from various sources (Maneewannakul and Levy, 1996; Oethinger et al., 1998; Wang et al., 2001; Sáenz et al., 2004; Webber et al., 2005). Similarly, among triclosan‐resistant isolates, mutations affecting FabI have been reported in multiple species (Fan et al., 2002; Chen et al., 2009). Thus, the same types of mutations we observed during experimental evolution may also play a role in resistance in nature, including asymmetric patterns of cross‐resistance as we observed here.
With only 93 isolates and applying conservative criteria (correction for phylogenetic relatedness and for multiple testing), our experimental design restricts us to detecting relatively strong associations and only in a single species. There may be other associations that would be detected by testing different species, antibacterials or larger numbers of isolates. Additionally, we cannot exclude that some of the positive associations we detected resulted instead from carriage of multiple dedicated resistance genes, as observed elsewhere across NAAs and antibiotics (Poirel et al., 2000; Riaño et al., 2006; Pal et al., 2015; Li et al., 2017), and for the isolates we studied across some pairs of antibiotics (Allen et al., 2017). We therefore expect that, in general, both explanations (generalized resistance mechanisms and carriage of multiple resistance genes) play a role in explaining resistance phenotypes in natural environments. We also note that the positive associations we detected represented only a small fraction of the compound combinations we tested. Furthermore, for benzalkonium chloride and triclosan, no isolates were above reported ECOFF values. Therefore, while our data help predict how exposure to one compound may influence susceptibility to others, this does not necessarily result in resistance above clinical breakpoints and ECOFF values.
Analogous positive associations between triclosan and antibiotic susceptibility, including chloramphenicol, have been observed for faecal coliforms isolated from near to waste water treatment plants (Middleton and Salierno, 2013) and a wide range of human and animal isolates of Salmonella enterica (Copitch et al., 2010). By contrast, other studies found no such associations (Cottell et al., 2009; Cameron et al., 2019). How can we explain this variability of associations between susceptibility to different antibacterials in nature? Our finding that cross‐resistance of laboratory isolates after exposure to different compounds can be strongly asymmetric (e.g. chloramphenicol vs triclosan), and this can be explained by the phenotypic effects of the different resistance mutations involved, suggests such associations are at least partly predictable from information about the types of genetic changes involved in resistance to different compounds and concentration. A limitation of such predictions is that, as noted above, associations in natural populations can be influenced by horizontal acquisition of dedicated resistance genes, as well as chromosomal mutation as in our experimentally evolved isolates. A key area for future work is therefore to collect more information from clinically relevant bacteria about the conditions (e.g. compounds or concentrations) that select for generalized vs specialized resistance mechanisms against NAAs.
Conclusion
We found associations between susceptibility to antibiotics and NAAs in natural and clinical isolates for a small number of compound combinations. Genome‐wide variation among natural and clinical isolates makes it inherently challenging to attribute observed associations to individual loci in such data sets. However, using genetic knockouts and experimental evolution of an isogenic strain in vitro, we were able to show that exposure to one antibacterial compound sometimes results in acquisition of generalized resistance mechanisms, such as changes in efflux pump expression, that confer resistance to multiple antibacterials with unrelated mechanisms of action. Crucially, observed patterns of cross‐resistance were asymmetric, in that exposure to chloramphenicol conferred cross‐resistance to other compounds including triclosan but not vice versa. We identified the genetic changes involved in these generalized resistance phenotypes, as well as more specialized ones emerging upon exposure to triclosan in our experiment. A key implication of these findings is that the emergence of cross‐resistance across different antibacterials in nature will depend on which of them, and in which concentrations, bacteria are exposed to. This suggests predicting how exposure to antibiotics or NAAs in nature will influence the spread of multiresistance phenotypes requires information about whether cross‐resistance/collateral effects are reciprocal or asymmetric. More generally, our results improve our quantitative understanding of associations between resistance phenotypes across different types of antibacterials in nature and provide new insights into possible mechanisms explaining the observed variability of these associations.
Experimental procedures
Organisms and growth conditions
The isolates we analysed included 69 isolates from the E. coli Reference (ECOR) Collection, 23 clinical isolates from patients with urinary tract infections obtained from the University Hospital Basel, Switzerland, in 2016, and K‐12 MG1655 (93 isolates total). We stored all isolates in 25% glycerol at −80°C. We performed routine culturing in lysogeny broth (LB) at an incubation temperature of 37°C. We purchased amoxicillin (product number A8523), benzalkonium chloride (product number 12060), cadmium chloride hydrate (product number 208299), chloramphenicol (product number 23275), ciprofloxacin (product number 17850), gentamicin (product number 48760), polymyxin B (product number 5291), rifampicin (product number 3501), SDS (product number L4509), triclosan (product number PHR1338), trimethoprim (product number 92131) and zinc chloride (product number 208086) from Sigma‐Aldrich (Merck KGaA, Germany) and cefotaxime (product number 219380) and tigecycline (product number 610225) from Merck (Merck KGaA, Germany). We prepared stock solutions at the outset of the experiments and filter sterilized (Filtropur S 0.2, Sarstedt, Germany) and stored them according to the manufacturer's instructions [stock solutions: amoxicillin 25 mg/ml in sterile distilled water (dH2O); benzalkonium chloride: 100 mg/ml in dH2O; cadmium chloride hydrate: 100 mg/ml in dH2O; chloramphenicol 50 mg/ml in 70% ethanol; ciprofloxacin 20 mg/ml in 0.1 M HCl; gentamicin 50 mg/ml in dH2O; polymyxin B 20 mg/ml in dH2O; rifampicin 50 mg/ml in methanol; SDS: 4% in LB; tigecycline 25 mg/ml in DMSO; triclosan: 4 mg/ml in 70% ethanol; trimethoprim 25 mg/ml in DMSO; zinc chloride: 200 mg/ml in dH2O].
Measuring susceptibility of natural and clinical isolates to NAAs
We defined the 90% inhibitory concentration (IC90) as the lowest concentration tested above which bacterial growth did not exceed 10% of growth of the same isolate in the absence of antibacterials. We estimated the IC90 for each isolate for five NAAs (benzalkonium chloride, cadmium, SDS, triclosan and zinc) by measuring their growth in liquid culture at 10 different concentrations. We transferred independent LB‐overnight cultures [cultured in flat‐based 96‐well microplates (Sarstedt, Germany)] into microplates filled with 10 concentrations of test compound and plain LB as control (twofold broth dilution, concentration ranges: benzalkonium chloride 256–0.5 μg/ml; cadmium 20–0.0390625 mmol/l; SDS 4–0.0078125 mg/ml; triclosan 4–0.0078125 μg/ml; zinc 8192–16 μg/ml) using a pin replicator (1/100 dilution, 1.5 μl in 150 μl). We selected these concentration ranges using previously defined resistance benchmarks such as ECOFFs (Morrissey et al., 2014) or values reported in the literature (Bednorz et al., 2013; Khan et al., 2015). Because triclosan was dissolved in ethanol, an additional control microplate was filled with LB and the amount of ethanol corresponding to the amount in the microplate with the highest triclosan concentration, to control for ethanol exposure of isolates. We incubated the microplates overnight at 37°C and quantified bacterial growth by measuring optical density at 600 nm (OD600) with a microplate reader (Infinite® 200 PRO, Tecan Trading AG, Switzerland) at the beginning and end of the experiment (0 and 24 h). We corrected the obtained values for the optical density of the media. We performed every assay in triplicate. We conducted the assays for different test compounds on different days.
We tested for associations between susceptibility phenotypes to different NAAs using PGLS (pgls function) in the caper package (version 1.0.1) of R [R version 3.5.3 (2019‐03‐11)]. This function fits a linear model while correcting for phylogenetic relatedness between the isolates, which is known to violate the assumption of non‐independence (Felsenstein, 1985). We calculated phylogenetic relatedness as the patristic distance (the sum of branch lengths between two strains), where branch length is the expected number of nucleotide substitutions per site. We took the IC90 for a relevant NAA or antibiotic as the susceptibility phenotype, and for each compound the IC90 concentrations were converted to ranks (each of the n concentrations tested was assigned a rank from 1 to n; if no concentration was sufficient to suppress growth by 90% we assigned a value of n + 1). This accounted for the fact that the concentration range varied across assay compounds and linearized the scale. We used the t‐value returned by pgls to estimate the strength of the association between susceptibility to different antibacterials. We imported the phylogenetic tree used by Allen et al. (2017), which is based on 1424 core genes shared by all isolates, with the ape package (version 5.3). We also tested separately whether individual susceptibility phenotypes were more similar among isolates with higher phylogenetic relatedness, using the fitDiscrete function of R's geiger package (version 2.0.6) to fit Pagel's λ, using the equal rates model for the evolution of the discrete parameter. We adjusted all p‐values for multiple testing using the Holm–Bonferroni method (sequential Bonferroni).
Analysis of metal and biocide resistance gene and antibiotic resistance gene data
We compared the susceptibility phenotypes measured above for natural and clinical isolates with data on the MBRG and ARG content for the same isolates obtained previously (Allen et al., 2019). In brief, MBRGs were identified from genomic data using the antibacterial biocide and metal resistance genes database BacMet (www.bacmet.biomedicine.gu.se/index.html) by using Blastx, and ARGs were identified from genomic data using the ARGs database ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/). When multiple resistance genes were found in the same operon, they were merged to single entries. That is, we treated unique haplotypes as single events, rather than counting resistance genes that always co‐occur on the same operon as separate alleles, which would inflate the weight of larger/multi‐ARG operons.
To look for cases where MBRGs co‐occurred with plasmid replicons, we identified contigs (fragments of the genome de novo assembled from raw reads) that contained plasmid replicons and MBRGs using the data available in previous papers (Allen et al., 2017, 2019). Note this approach does not represent a systematic screen for co‐occurrence because of the significant possibility that plasmids can be represented by multiple contigs in the draft genomes.
We tested for correlations between MBRG or ARG profiles and susceptibility to NAAs and antibiotics with a partial Mantel test, applying the permutation method from Harmon and Glor (2010) and correcting for phylogenetic relatedness of the isolates. This tests whether isolates with similar susceptibility profiles across different antibacterial compounds also have similar sets of resistance genes. We also tested for an association between the presence/absence of each individual MBRG across natural and clinical isolates with variation of susceptibility to each antibacterial compound across the same isolates, using the pgls function (see above), adjusting all p‐values for multiple testing using the Holm–Bonferroni method (sequential Bonferroni). We also used the pgls function to determine whether isolates with a greater number of MBRGs had lower average antibacterial susceptibility.
Knockout experiment
We used existing literature to identify candidate genes representing a diverse range of functions or potentially involved in resistance to NAAs and antibiotics (Table S1). We used single‐gene knockouts from the Keio Knockout Collection (Baba et al., 2006) to test for a change in the IC90 of these knockout variants relative to the ancestral strain of the knockout collection, E. coli BW25113. We measured IC90 here as described above for benzalkonium chloride, triclosan and chloramphenicol (five concentrations of each compound plus antibacterial‐free; five replicates in every combination). We conducted an additional experiment with only two knockouts, acrA and acrB (plus the wild type) to determine the IC90 toward 10 further antibacterials (3 NAAs, 7 antibiotics), because these genes showed strong effects in the first experiment and we wanted to test whether they also influenced other susceptibility phenotypes.
For analysis of the susceptibility testing of Keio knockout variants, we used a t‐test to compare susceptibility of each knockout variant with the wild type for every assay compound, adjusting p‐values to account for multiple testing using the Holm–Bonferroni method (sequential Bonferroni).
Having observed that acrA and acrB affected multiple phenotypes in E. coli BW25113, we aimed to determine whether these genes were present in the natural and clinical isolates used above. To do this, we downloaded protein sequences for both genes from the Comprehensive Antibiotic Resistance Database CARD (https://card.mcmaster.ca) and used NCBI's tblastn program to compare the protein query sequences to the nucleotide sequences of our isolates. We used a bit‐score cut‐off of 670 and 1900 for acrA and acrB, respectively, to determine if each isolate contained a copy of these genes, and only considered hits with E values <10−6 as significant.
Isolation of resistant mutants
To test whether a single allele that confers a beneficial resistance phenotype against one compound would alter susceptibility to others, and to identify the genetic changes involved, we isolated mutants of E. coli K‐12 MG1655 with resistance to benzalkonium chloride, triclosan and chloramphenicol. We used these three antibacterials as selection compounds here because of the observed significant associations across the isolates tested above.
We started the experiment by picking independent colonies from one overnight agar plate (one colony per selection compound), incubating them for 2 h in 5 ml LB (37°C, 180 rpm), then diluting the cultures 1/10 in fresh LB (1 ml bacterial culture in 9 ml). We then inoculated 45 microplate wells per selection compound with diluted bacterial culture (5 μl bacterial culture in 245 μl LB) and incubated overnight at 37°C. For every selection compound, we prepared 45 LB agar plates supplemented with three different concentrations of selection compound (15 agar plates per concentration per selection compound). We chose concentrations 1.5‐, 2‐ or 4‐times higher than the previously determined IC90s of the wild‐type strain in liquid. We plated all 250 μl of each overnight culture onto a supplemented agar plate and incubated overnight at 37°C. We then picked six mutant colonies for each compound. We prioritized colonies from plates with higher concentrations, and re‐streaked each colony on a fresh agar plate supplemented with the same concentration of antibacterial, before making a liquid culture and cryopreserving it in 25% glycerol at −80°C. This resulted in a total of 17 resistant mutants (5 or 6 per selection compound, for more details see Table S2). Mutant generation for the different selection compounds was done on different days.
We tested the susceptibility phenotype (IC90) of the isolated resistant mutants (n = 17) and the ancestral strain E. coli K‐12 MG1655 against the three antibacterials benzalkonium chloride, chloramphenicol and triclosan as described above. In brief, we inoculated a 96‐well microplate with five independent colonies per isolate. After overnight incubation at 37°C, we used this microplate to inoculate 25 microplates (150 μl LB, overnight incubation at 37°C). We filled 25 microplates with eight concentrations of the assessed antibacterials [3 antibacterials × 8 concentrations + 1 control (LB medium) = 25 environments], each environment split over 2 microplates. We quantified bacterial growth by measuring OD600 with a microplate reader at the beginning and end of the experiment (0 and 24 h), correcting obtained values for the optical density of the media. We conducted the assays for all test compounds on the same day.
Genetic analysis of resistant mutants
We used the QIAGEN Genomic‐tip 20/G (Cat. No. 10223) according to the manufacturer's instructions for genomic DNA (gDNA) extraction. Isolated resistant mutants were streaked (from glycerol stock) on selective agar plates before inoculation of non‐selective liquid medium with a single colony; one of the mutants isolated as benzalkonium chloride‐resistant (BKC_02) did not grow on selective agar in this step and was excluded from genetic sequencing and subsequent resistance phenotyping. Overnight cultures grown in plain LB were centrifuged at 5000g at room temperature for 10 min. After removal of the supernatant, we stored cell pellets at −20°C until further processing. We quantified the obtained gDNA using the Quant‐iTTM dsDNA BR (Broad Range) Assay Kit (Thermo Fisher Scientific) in the Qubit™ Fluorometer (Thermo Fisher Scientific). We used a Nanodrop (Thermo Fisher Scientific) to control the purity of gDNA (ratios A260 /A280). We sequenced all isolated resistant mutants and the ancestral strain (E. coli K‐12 MG1655) at the Functional Genomic Center, Zurich, Switzerland, using the Illumina Hiseq 4000 platform after library preparation with the Nextera XT DNA Library Prep Kit (Illumina).
We trimmed and quality‐filtered all sequences with trimmomatic (Bolger et al., 2014). We mapped the sequence of the ancestral strain of our resistance isolation experiment against the reference sequence of E. coli K‐12 MG1655 (NCBI accession number: U00096) using breseq 0.33.1 (Deatherage and Barrick, 2014). We used gdtools implemented in breseq to integrate the identified mutations into the reference genome. We then mapped all sequences of the evolved isolates for variant calling against the refined reference genome using breseq. The sequencing data have been deposited in the European Nucleotide Archive under the study accession number PRJEB35347 (https://www.ebi.ac.uk/ena).
We used R's BioCircos package to visualize genetic data. The BioCircos package relies on the JavaScript library BioCircos.js (Cui et al., 2016).
Supporting information
Appendix S1: Supplementary Data
Acknowledgement
A.H. acknowledges Swiss National Science Foundation project 31003A_165803.
References
- Alekshun, M.N. , and Levy, S.B. (1997) Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother 41: 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, H.K. , Donato, J. , Wang, H.H. , Cloud‐Hansen, K.A. , Davies, J. , and Handelsman, J. (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8: 251–259. [DOI] [PubMed] [Google Scholar]
- Allen, R.C. , Pfrunder‐Cardozo, K.R. , Meinel, D. , Egli, A. , and Hall, A.R. (2017) Associations among antibiotic and phage resistance phenotypes in natural and clinical Escherichia coli isolates. MBio 8: e01341‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, R.C. , Angst, D.C. , and Hall, A.R. (2019) Resistance gene carriage predicts growth of natural and clinical Escherichia coli isolates in the absence of antibiotics. Appl Environ Microbiol 85 AEM.02111‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, D.I. , and Hughes, D. (2012) Evolution of antibiotic resistance at non‐lethal drug concentrations. Drug Resist Updat 15: 162–172. [DOI] [PubMed] [Google Scholar]
- Azzouz, A. , Rascón, A.J. , and Ballesteros, E. (2016) Simultaneous determination of parabens, alkylphenols, phenylphenols, bisphenol a and triclosan in human urine, blood and breast milk by continuous solid‐phase extraction and gas chromatography‐mass spectrometry. J Pharm Biomed Anal 119: 16–26. [DOI] [PubMed] [Google Scholar]
- Baba, T. , Ara, T. , Hasegawa, M. , Takai, Y. , Okumura, Y. , Baba, M. , et al (2006) Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker‐Austin, C. , Wright, M.S. , Stepanauskas, R. , and McArthur, J.V. (2006) Co‐selection of antibiotic and metal resistance. Trends Microbiol 14: 176–182. [DOI] [PubMed] [Google Scholar]
- Baquero, F. , Martínez, J.L. , and Cantón, R. (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19: 260–265. [DOI] [PubMed] [Google Scholar]
- Bednorz, C. , Oelgeschläger, K. , Kinnemann, B. , Hartmann, S. , Neumann, K. , Pieper, R. , et al (2013) The broader context of antibiotic resistance: zinc feed supplementation of piglets increases the proportion of multi‐resistant Escherichia coli in vivo. Int J Med Microbiol 303: 396–403. [DOI] [PubMed] [Google Scholar]
- Bhargava, H.N. , and Leonard, P.A. (1996) Triclosan: applications and safety. Am J Infect Control 24: 209–218. [DOI] [PubMed] [Google Scholar]
- Birošová, L. , and Mikulášová, M. (2009) Development of triclosan and antibiotic resistance in Salmonella enterica serovar Typhimurium. J Med Microbiol 58: 436–441. [DOI] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. , and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondi, C.A.M. , Marks, J.L. , Wroblewski, L.B. , Raatikainen, H.S. , Lenox, S.R. , and Gebhardt, K.E. (2015) Human and environmental toxicity of sodium lauryl sulfate (SLS): evidence for safe use in household cleaning products. Environ Health Insights 9: 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradl, H.B. (2005) Chapter 1 Sources and origins of heavy metals. Interface Sci Technol 6: 1–27. [Google Scholar]
- Braoudaki, M. , and Hilton, A.C. (2004a) Adaptive resistance to biocides in salmonella enterica and Escherichia coli O157 and cross‐resistance to antimicrobial agents. J Clin Microbiol 42: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braoudaki, M. , and Hilton, A.C. (2004b) Low level of cross‐resistance between triclosan and antibiotics in Escherichia coli K‐12 and E. coli O55 compared to E. coli O157. FEMS Microbiol Lett 235: 305–309. [DOI] [PubMed] [Google Scholar]
- Byelashov, O.A. , Kendall, P.A. , Belk, K.E. , Scanga, J.A. , and Sofos, J.N. (2008) Control of Listeria monocytogenes on vacuum‐packaged frankfurters sprayed with lactic acid alone or in combination with sodium lauryl sulfate. J Food Prot 71: 728–734. [DOI] [PubMed] [Google Scholar]
- Cameron, A. , Barbieri, R. , Read, R. , Church, D. , Adator, E.H. , Zaheer, R. , and McAllister, T.A. (2019) Functional screening for triclosan resistance in a wastewater metagenome and isolates of Escherichia coli and enterococcus spp. from a large Canadian healthcare region. PLoS One 14: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli, A. (2009) Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53: 2227–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey, D.E. , and McNamara, P.J. (2014) The impact of triclosan on the spread of antibiotic resistance in the environment. Front Microbiol 5: 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Pi, B. , Zhou, H. , Yu, N. , and Li, L. (2009) Triclosan resistance in clinical isolates of Acinetobacter baumannii. J Med Microbiol 58: 1086–1091. [DOI] [PubMed] [Google Scholar]
- Chuanchuen, R. , Beinlich, K. , Hoang, T.T. , Becher, A. , Karkhoff‐schweizer, R.R. , Schweizer, H.P. , et al (2001) Cross‐resistance between Triclosan and antibiotics in. Society 45: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley, Z.C. , Carlson‐Banning, K.M. , Carter, A.G. , De La Cova, A. , Song, Y. , and Zechiedrich, L. (2019) Sugar and iron: toward understanding the antibacterial effect of ciclopirox in Escherichia coli . PLoS One 14: e0210547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copitch, J.L. , Whitehead, R.N. , and Webber, M.A. (2010) Prevalence of decreased susceptibility to triclosan in Salmonella enterica isolates from animals and humans and association with multiple drug resistance. Int J Antimicrob Agents 36: 247–251. [DOI] [PubMed] [Google Scholar]
- Cottell, A. , Denyer, S.P. , Hanlon, G.W. , Ochs, D. , and Maillard, J.Y. (2009) Triclosan‐tolerant bacteria: changes in susceptibility to antibiotics. J Hosp Infect 72: 71–76. [DOI] [PubMed] [Google Scholar]
- Cui, Y. , Chen, X. , Luo, H. , Fan, Z. , Luo, J. , He, S. , et al (2016) BioCircos.Js: an interactive Circos JavaScript library for biological data visualization on web applications. Bioinformatics 32: 1740–1742. [DOI] [PubMed] [Google Scholar]
- Deatherage, D.E. , and Barrick, J.E. (2014) Identification of mutations in laboratory‐evolved microbes from next‐generation sequencing data using breseq. Methods Mol Biol 1151: 165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deus, D. , Krischek, C. , Pfeifer, Y. , Sharifi, A.R. , Fiegen, U. , Reich, F. , et al (2017) Comparative analysis of the susceptibility to biocides and heavy metals of extended‐spectrum β‐lactamase‐producing Escherichia coli isolates of human and avian origin, Germany. Diagn Microbiol Infect Dis 88: 88–92. [DOI] [PubMed] [Google Scholar]
- Dickinson, A.W. , Power, A. , Hansen, M.G. , Brandt, K.K. , Piliposian, G. , Appleby, P. , et al (2019) Heavy metal pollution and co‐selection for antibiotic resistance: a microbial palaeontology approach. Environ Int 132: 105117. [DOI] [PubMed] [Google Scholar]
- Du, D. , Wang, Z. , James, N.R. , Voss, J.E. , Klimont, E. , Ohene‐Agyei, T. , et al (2014) Structure of the AcrAB‐TolC multidrug efflux pump. Nature 509: 512–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, F. , Yan, K. , Wallis, N.G. , Reed, S. , Moore, T.D. , Rittenhouse, S.F. , et al (2002) Defining and combating the mechanisms of triclosan resistance in clinical isolates of Staphylococcus aureus . Antimicrob Agents Chemother 46: 3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985) Phylogenies and the comparative method. Am Nat 125: 1–15. [Google Scholar]
- Fernández, L. , and Hancock, R.E.W. (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25: 661–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon, L.J. , and Glor, R.E. (2010) Poor statistical performance of the mantel test in phylogenetic comparative analyses. Evolution (NY) 64: 2173–2178. [DOI] [PubMed] [Google Scholar]
- Imamovic, L. , and Sommer, M.O.A. (2013) Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci Transl Med 5: 204ra132. [DOI] [PubMed] [Google Scholar]
- Jeanthon, C. , and Prieur, D. (1990) Resistance to heavy metals of heterotrophic bacteria isolated from the deep‐sea hydrothermal vent polychaete, Alvinella pompejana. Prog Oceanogr 24: 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlmeter, G. , Brown, D.F.J. , Goldstein, F.W. , MacGowan, A.P. , Mouton, J.W. , Österlund, A. , et al (2003) European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. J Antimicrob Chemother 52: 145–148. [DOI] [PubMed] [Google Scholar]
- Kaper, J.B. , Nataro, J.P. , and Mobley, H.L.T. (2004) Pathogenic Escherichia coli . Nat Rev Microbiol 2: 123–140. [DOI] [PubMed] [Google Scholar]
- Khan, Z. , Nisar, M.A. , Hussain, S.Z. , Arshad, M.N. , and Rehman, A. (2015) Cadmium resistance mechanism in Escherichia coli P4 and its potential use to bioremediate environmental cadmium. Appl Microbiol Biotechnol 99: 10745–10757. [DOI] [PubMed] [Google Scholar]
- Kramer, V.C. , Nickerson, K.W. , Hamlett, N.V. , and O'Hara, C. (1984) Prevalence of extreme detergent resistance among the Enterobacteriaceae. Can J Microbiol 30: 711–713. [DOI] [PubMed] [Google Scholar]
- Langsrud, S. , Sundheim, G. , and Holck, A.L. (2004) Cross‐resistance to antibiotics of Escherichia coli adapted to benzalkonium chloride or exposed to stress‐inducers. J Appl Microbiol 96: 201–208. [DOI] [PubMed] [Google Scholar]
- Lázár, V. , Nagy, I. , Spohn, R. , Csörgo', B. , Györkei, Á. , Nyerges, Á. , et al (2014) Genome‐wide analysis captures the determinants of the antibiotic cross‐resistance interaction network. Nat Commun 5: 4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear, J.C. , Maillard, J.Y. , Dettmar, P.W. , Goddard, P.A. , and Russell, A.D. (2002) Chloroxylenol‐ and triclosan‐tolerant bacteria from industrial sources. J Ind Microbiol Biotechnol 29: 238–242. [DOI] [PubMed] [Google Scholar]
- Ledder, R.G. , Gilbert, P. , Willis, C. , and McBain, A.J. (2006) Effects of chronic triclosan exposure upon the antimicrobial susceptibility of 40 ex‐situ environmental and human isolates. J Appl Microbiol 100: 1132–1140. [DOI] [PubMed] [Google Scholar]
- Levy, S.B. (2002) Active efflux, a common mechanism for biocide and antibiotic resistance. J Appl Microbiol 92: 65S–71S. [PubMed] [Google Scholar]
- Li, L.G. , Xia, Y. , and Zhang, T. (2017) Co‐occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J 11: 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden, T. (2002) Chapter 16: The BLAST Sequence Analysis Tool. In The NCBI Handbook [Internet], pp. 1–15.
- Maneewannakul, K. , and Levy, S.B. (1996) Identification of mar mutants among quinolone‐resistant clinical isolates of Escherichia coli . Antimicrob Agents Chemother 40: 1695–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry, L.M. , Oethinger, M. , and Levy, S.B. (1998a) Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli . FEMS Microbiol Lett 166: 305–309. [DOI] [PubMed] [Google Scholar]
- McMurry, L.M. , Oethinger, M. , and Levy, S.B. (1998b) Triclosan targets lipid synthesis. Nature 394: 531–532. [DOI] [PubMed] [Google Scholar]
- Merchel Piovesan Pereira, B. , and Tagkopoulos, I. (2019) Benzalkonium chlorides: uses, regulatory status, and microbial resistance. Appl Environ Microbiol 85: e00377–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton, J.H. , and Salierno, J.D. (2013) Antibiotic resistance in triclosan tolerant fecal coliforms isolated from surface waters near wastewater treatment plant outflows (Morris County, NJ, USA). Ecotoxicol Environ Saf 88: 79–88. [DOI] [PubMed] [Google Scholar]
- Morrissey, I. , Oggioni, M.R. , Knight, D. , Curiao, T. , Coque, T. , Kalkanci, A. , et al (2014) Evaluation of epidemiological cut‐off values indicates that biocide resistant subpopulations are uncommon in natural isolates of clinically‐relevant microorganisms. PLoS One 9: e86669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou, S.A. , Fast, A.G. , Nakamaru‐Ogiso, E. , and Papoutsakis, E.T. (2013) Overexpression of fetA (ybbL) and fetB (ybbM), encoding an iron exporter, enhances resistance to oxidative stress in Escherichia coli . Appl Environ Microbiol 79: 7210–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oethinger, M. , Podglajen, I. , Kern, W.V. , and Levy, S.B. (1998) Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli . Antimicrob Agents Chemother 42: 2089–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okusu, H. , Ma, D. , and Nikaido, H. (1996) AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple‐antibiotic‐resistance (mar) mutants. J Bacteriol 178: 306–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, C. , Bengtsson‐Palme, J. , Kristiansson, E. , and Larsson, D.G.J. (2015) Co‐occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co‐selection potential. BMC Genomics 16: 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidot, S.J. , Gao, W. , Buultjens, A.H. , Monk, I.R. , Guerillot, R. , Carter, G.P. , et al (2018) Increasing tolerance of hospital Enterococcus faecium to handwash alcohols. Sci Transl Med 10: 6115. [DOI] [PubMed] [Google Scholar]
- Poirel, L. , Le Thomas, I. , Naas, T. , Karim, A. , and Nordmann, P. (2000) Biochemical sequence analyses of GES‐1, a novel class a extended‐ spectrum β‐lactamase, and the class 1 integron In52 from Klebsiella pneumoniae . Antimicrob Agents Chemother 44: 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, K. (2002) Mechanisms of bacterial biocide and antibiotic resistance. J Appl Microbiol 92: 55S–64S. [PubMed] [Google Scholar]
- Pycke, B.F.G. , Crabbé, A. , Verstraete, W. , and Leys, N. (2010) Characterization of triclosan‐resistant mutants reveals multiple antimicrobial resistance mechanisms in rhodospirillum rubrum S1H. Appl Environ Microbiol 76: 3116–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaño, I. , Moreno, M.A. , Teshager, T. , Sáenz, Y. , Domínguez, L. , and Torres, C. (2006) Detection and characterization of extended‐spectrum β‐lactamases in Salmonella enterica strains of healthy food animals in Spain. J Antimicrob Chemother 58: 844–847. [DOI] [PubMed] [Google Scholar]
- Sáenz, Y. , Briñas, L. , Domínguez, E. , Ruiz, J. , Zarazaga, M. , Vila, J. , and Torres, C. (2004) Mechanisms of resistance in multiple‐antibiotic‐resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother 48: 3996–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandegren, L. , Linkevicius, M. , Lytsy, B. , Melhus, Å. , and Andersson, D.I. (2012) Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae‐specific plasmid associated with a major nosocomial outbreak. J Antimicrob Chemother 67: 74–83. [DOI] [PubMed] [Google Scholar]
- Seoane, A.S. , and Levy, S.B. (1995) Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli . J Bacteriol 177: 3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanauskas, R. , Glenn, T.C. , Jagoe, C.H. , Tuckfield, R.C. , Lindell, A.H. , and McArthur, J.V. (2005) Elevated microbial tolerance to metals and antibiotics in metal‐contaminated industrial environments. Environ Sci Technol 39: 3671–3678. [DOI] [PubMed] [Google Scholar]
- Suller, M.T.E. (2000) Triclosan and antibiotic resistance in Staphylococcus aureus . J Antimicrob Chemother 46: 11–18. [DOI] [PubMed] [Google Scholar]
- Swank, R.T. , and Munkres, K.D. (1971) Molecular weight analysis of oligopeptides by electrophoresis in polyacrylamide gel with sodium dodecyl sulfate. Anal Biochem 39: 462–477. [DOI] [PubMed] [Google Scholar]
- Tikhonova, E.B. , and Zgurskaya, H.I. (2004) AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem 279: 32116–32124. [DOI] [PubMed] [Google Scholar]
- Tronsmo, A. , Gjøen, T. , Sørum, H. , and Yazdankhah, S. (2016) Antimicrobial resistance due to the use of biocides and heavy metals: a literature review, Norwegian Committee for Food Safety (VKM).
- Wales, A.D. , and Davies, R.H. (2015) Co‐selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 4: 567–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Dzink‐Fox, J.L. , Chen, M. , and Levy, S.B. (2001) Genetic characterization of highly fluoroquinolone‐resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother 45: 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, M.A. , Talukder, A. , and Piddock, L.J.V. (2005) Contribution of mutation at amino acid 45 of AcrR to acrB expression and ciprofloxacin resistance in clinical and veterinary Escherichia coli isolates. Antimicrob Agents Chemother 49: 4390–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, M.A. , Whitehead, R.N. , Mount, M. , Loman, N.J. , Pallen, M.J. , and Piddock, L.J.V. (2015) Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemother 70: 2241–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Cui, F. , Zeng, G.M. , Jiang, M. , Yang, Z.Z. , Yu, Z.G. , et al (2015) Quaternary ammonium compounds (QACs): a review on occurrence, fate and toxicity in the environment. Sci Total Environ 518–519: 352–362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supplementary Data


