Abstract
Mitochondrial respiration using the oxygraph‐2k respirometer (Oroboros) is widely used to estimate mitochondrial capacity in human skeletal muscle. Here, we measured mitochondrial respiration variability, in a relatively large sample, and for the first time, using statistical simulations, we provide the sample size required to detect meaningful respiration changes following lifestyle intervention. Muscle biopsies were taken from healthy, young men from the Gene SMART cohort, at multiple time points. We utilized samples for each measurement with two technical repeats using two respirometer chambers (n = 160 pairs of same muscle after removal of low‐quality samples). We measured the Technical Error of measurement (TEM) and the coefficient of variation (CV) for each mitochondrial complex. There was a high correlation between measurements from the two chambers (R > 0.7 P < .001) for all complexes, but the TEM was large (7.9‐27 pmol s−1 mg−1; complex dependent), and the CV was >15% for all complexes. We performed statistical simulations of a range of effect sizes at 80% power and found that 75 participants (with duplicate measurements) are required to detect a 6% change in mitochondrial respiration after an intervention, while for interventions with 11% effect size, ~24 participants are sufficient. The high variability in respiration suggests that the typical sample sizes in exercise studies may not be sufficient to capture exercise‐induced changes.
Keywords: exercise, intervention, mitochondria, OXPHOS, oxygen consumption
Abbreviations
- ATP
adenosine triphosphate
- CI
complex I, electron input through CI
- CV
coefficient of variation
- CIP
oxidative phosphorylation (OXPHOS) capacity (P) through CI
- ETS
electron transport system
- CI+CII
convergent electron input through CI and CII CI+CIIE, capacity (E) through CI+CII
- CI+CIIP
complex II (CII) linked respiration
- E
ETS capacity
- ETS
measurement of electron transport system
- FCCP
p‐trifluoromethoxyphenylhydrazone
- Gene SMART
genes and skeletal muscle adaptive response to training
- Inv‐RCR
inverse of respiratory control ratio (CIL/CI+IIP)
- L
leak respiration
- LCR
leak control ratio (CIL/CI+IIE)
- OXPHOS
oxidative phosphorylation
- P
oxphos capacity
- PCR
phosphorylation control ratio (CI+IIP/CI+IIE)
- ROX
residual oxygen consumption
- SCR
substrate control ratio at constant P (CIP/CI+IIP)
- TCA
tricarboxylic acid
- TEM
technical error of measurement
1. INTRODUCTION
The mitochondrion is a membrane‐enclosed organelle found in eukaryotic cells. With its five specialized complexes, it produces adenosine triphosphate (ATP) and thus constitutes the powerhouse of the cell.1 ATP is produced during oxidative phosphorylation (OXPHOS) via the tricarboxylic acid (TCA) cycle inside the mitochondrial matrix, and via the electron transport system (ETS) along the inner mitochondrial membrane.2 Mitochondrial respiration measured by maximal oxygen consumption in skeletal muscle fibers, is currently the primary way of assessing mitochondrial capacity.3, 4 The most common method in human skeletal muscle uses fibers permeabilized by saponin.5, 6 Using a combination of different substrates, this technique is able to mimic the processes (ie, TCA cycle and ETS) occurring within the mitochondrion.
Regular exercise has a beneficial effect on mitochondrial function.7 Endurance exercise training improves mitochondrial respiration,8, 9, 10, 11, 12, 13 with high‐intensity exercise training leading to larger improvements (up to 40%)8, 10, 11, 12 than moderate continuous exercise training (up to 20%).14, 15 However, changes in mitochondrial respiration using permeabilized muscle fibers following exercise training have been assessed in relatively small sample sizes, typically within a range of n = 8‐15,16 and without assessing respiration changes in a control group.17 Mitochondrial respiration values, as well as improvements in mitochondrial respiration following exercise training are also quite variable between studies.3, 10, 14, 16, 18, 19 For example, Irving et al18 observed a ~1.5‐fold change in mitochondrial respiration after moderate intensity endurance training (n = 34), while Robach et al20 did not observe any change after a similar intervention (n = 17). Mitochondrial respiration capacity in human exercise intervention studies is not exclusive to skeletal muscle samples. Although such investigations are less common, different studies have measured respiration capacity in adipose tissue,21, 22 and blood cells (including lymphocytes and platelet).23, 24, 25, 26
Tests run in duplicates (or more) enable researchers to capture technical variability, and/or biological day‐to‐day variability within participants.17 The typical error of measurement (TEM), also called “within‐subject standard deviation,” provides an estimate of such variability.17 In the only study to date investigating the reliability of mitochondrial respiration measurement,3 the TEM between two fiber bundles from the same biopsy was 10.5 pmol s−1 mg−1 in the maximal oxidative phosphorylation (OXPHOS) and the coefficient of variations (CVs) were worryingly high (15.2% between two fiber bundles, 23.9% between legs, and 33.1% at different time points).3 More studies are required to confirm whether such variability is consistently high, but, more importantly, this technical variability needs to be put into perspective with typical mitochondrial respiration changes following interventions (eg, exercise training interventions). The qualifiers “high” and “low” variability only make sense when compared with the magnitude of the intervention‐induced changes, which will determine how likely we are to detect those changes.
Therefore, the aim of the present study was to investigate the reliability of mitochondrial respiration measurements in human vastus lateralis muscle using large number of duplicate fiber bundles (160 pairs) collected from a range of participants at different time points. We correlated mitochondrial respiration values between two chambers containing bundles of same muscle sample for complexes CIP, CI + CIIP and CI + CIIE, and calculated the TEM and the CV between experiments. Using the estimated TEM and CV, we performed simulations to determine the minimum number of participants required to detect meaningful mitochondrial respiration changes of various effect sizes following a hypothetical intervention, at 80% power.
2. MATERIALS AND METHODS
2.1. Participants
Participants were from the Genes and Skeletal Muscle Adaptive Response to Training (Gene SMART) cohort. The full study methodology has been previously described elsewhere.27 68 apparently healthy, Caucasian men (age = 31.4 ± 8.2 years old; BMI = 25.2 ± 3.2 kg/m2) participated in the study and signed a written informed consent. Participants were excluded if they had any preexisting heart condition, health issues, and/or preexisting injury that could potentially impair exercise capacity. The study was approved by the Human Ethics Research Committee at Victoria University (HRE13‐223).
2.2. Muscle biopsies
A controlled diet for 48 hours prior to the muscle biopsies was provided to the participants, according to the guidelines of the Australian National Health & Medical Research Council (NHMRC). Muscle biopsies were taken by an experienced medical doctor from the vastus lateralis muscle of the participants’ dominant leg, following local anesthesia (2 mL, 1% Lidocaine (Lignocaine)). The needle was inserted in the participant leg and manual suction was applied for muscle collection. Care was taken not to contaminate the muscle samples with local anesthetic during the biopsy. Approximately 2‐6 mg of muscle was then immediately placed in ice‐cold BIOPS for determination of mitochondrial respiration in two individual chambers (duplicates).
2.3. Mitochondrial respiration
Immediately after each biopsy (within max 30 minutes of collection), 2‐6 mg of muscle fibers were mechanically separated using pointed forceps under a binocular microscope in 2‐mL ice‐cold biopsy preservation solution on ice (BIOPS, 2.77 mM CaK2EGTA, 7.23 mM K2EGTA, 5.77 mM Na2ATP, 6.56 mM MgCl2•6H2O, 20 mM Taurine, 15 mM Na2Phosphocreatine, 20 mM Imidazole, 0.5 mM Dithiothreitol, and 50 mM K+‐MES at pH 7.1).6 Permeabilization of the plasma membrane occurred in the same solution with 50 μg/ml of saponin (Sigma‐Aldrich, St Louis, USA) for 30 minutes rotating on ice. This was followed by rinsing the muscle fibers for 3 × 7 minutes in mitochondrial respiration medium (MiR05, 0.5 mM EGTA, 3 mM MgCl2•6H2O, 60 mM K‐lactobionate, 20 mM Taurine, 10 mM KH2PO4, 20 mM Hepes, 110 mM sucrose, and 1 g/L bovine serum albumin at pH 7.1)6 on ice. Mitochondrial respiration was measured in duplicates in washed muscle fibers (between 1 and 3 mg wet weight of muscle fibers/chamber) in MiR05 at 37°C using the high‐resolution Oxygraph‐2k (Oroboros, Innsbruck,Austria), with additional substrates. Oxygen concentration (mM) and flux (pmol × s‐1 × mg−1) were recorded using DatLab software. Reoxygenation by direct syringe injection of O2 was necessary to maintain O2 levels between 275 and 450 mM and to avoid potential oxygen diffusion limitation. A substrate‐uncoupler‐inhibition tritation sequence was used. The following substrates were added (final concentration): malate (2 mM) and pyruvate (5 mM) were added to measure the LEAK respiration (L) through Complex I (CI) (CIL), followed by MgCl2 (3mM) and ADP (5 mM) to measure oxidative phosphorylation (OXPHOS) capacity (P) through CI (CIP), followed by succinate (10 mM) to measure P through CI + Complex II (CII) linked respiration (CI + CIIP).28 This respiration state represents the maximal respiratory capacity in the respirometer chamber.28 Cytochrome c (10 μM) was used to test the integrity of the outer mitochondrial membrane, in this step if the respiration increased >10% when cytochrome c was added, values from that chamber were removed due to a damaged membrane. A series of steps (steps of 0.5 μM) p‐trifluoromethoxyphenylhydrazone (FCCP) titrations followed for measurement of electron transport system (ETS) capacity (E) through CI + CII (CI + CIIE). Antimycin (3.75 μM) was added to block the activity of complex III and to measure the residual oxygen consumption (ROX) indicative of non‐mitochondrial oxygen consumption. Substrate and coupling control ratios were calculated from the different titration steps obtained from the protocol used.7 A background calibration for the Oroboros machine was performed every 3 months, and air calibrations were performed before each experiment. The results were pasted into the excel spreadsheet supplied by the manufacturer (Oroboros). If air calibrations presented more than 5% deviation in the results, membranes were changed, and new background calibration was done. Instrument backgrounds were performed in MiR05, and oxygen levels were at 450 nmol/mL. Highly variable graphs are indicative of poor quality, as shown on the O2K software, were removed.
2.4. Citrate synthase activity
Intrinsic changes in the mitochondria can be determined by quantitative measurements of specific markers. Such measurements can estimate the content of the mitochondria and are commonly used to normalize global measurements of mitochondrial function. The most commonly used measurement is the citrate synthase (CS) enzyme activity.29 Thus, we have normalized our results by CS activity.
Complete enzyme extractions, from small pieces of frozen tissues, were performed in an ice‐cold buffer (KH2PO4 & K2HPO4) using a TissueLyser II (Qiagen, Hilden, Germany). Protein concentration was assessed using the bicinchoninic acid assay. Total citrate synthase (CS) activity was measured (30°C, pH 7.5) using standard spectrophotometric assays. CS activity is presented in international units (IU).
2.5. Statistical analyses
We calculated three different metrics to show the reliability of mitochondrial respiration measurements. First, we calculated the correlation between duplicates from the two chambers, for each complex, using non‐parametric Spearman’s test to downweight the influence of outliers, and a stringent P value < .005 for significance. Then, we calculated the within‐subject standard deviation, also called typical (or technical) error of measurement (TEM)17:
where n is the number of pairs of duplicates and x is the respiration measurement.
We calculated the coefficient of variability (CV) estimated by:
, where µ is the mean respiration across all duplicates and all samples. While TEM is expressed in the units of mitochondrial respiration (pmol s−1 mg−1), CV is a percentage.
Lastly, we performed simulations based on the TEM for the CI + CIIP and CI + CIIE respiration values. We simulated increases of 1%‐50% in mitochondrial respiration in each participant after a hypothetical intervention and estimated the sample size (number of participants) required to detect this change at 80% power.
All analyses were performed using the R software.
3. RESULTS
3.1. Large technical error in mitochondrial respiration measurement
Two fiber bundles from the same muscle biopsy were run simultaneously in two chambers totaling 160 duplicate pairs after removal of results that indicated damaged membrane (ie, cyt‐c increased >10%). Respiration measurements were correlated between chambers (R ≥ 0.71‐0.75, P < .005 for all) (Figure 1). Yet when compared with correlations obtained for gene expression data (generally R > 0.9),30, 31 the correlation values obtained here are rather low.
Figure 1.
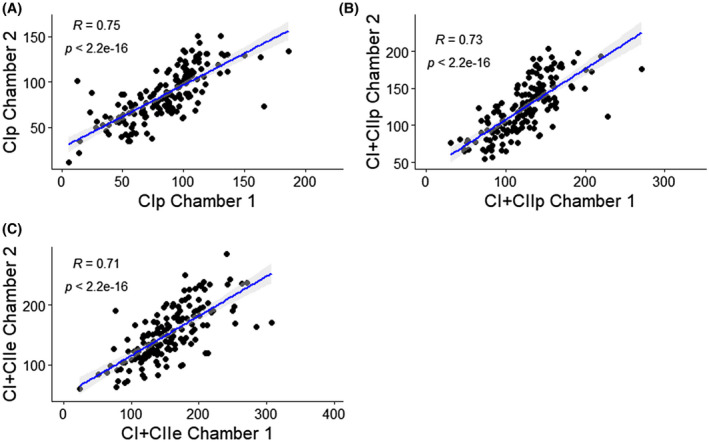
Spearman’s correlation between chambers after the addition of (A) oxidative phosphorylation (OXPHOS) capacity (P) through Complex I (CIP), (B), measure P through CI+Complex II (CII) linked respiration (CI+CIIP), (C) electron transport system (ETS) capacity (E) through CI+CII (CI+CIIE). All values are in pmol s−1 mg−1
The poor correlation between chambers was consistent with high TEM and CV estimates for all complexes (Table 1), as all complexes showed a CV ≥ 15%. When reporting the Flux Control Ratios (FCRs), to account for lab‐to‐lab variability,7 the TEM and CV estimates were also significantly elevated, with some reaching more than 100%.
Table 1.
Chamber‐specific respiration values and FCRs, typical error of measurement and coefficient of variation for each substrate
| Mean ± SD | Chamber 2 | TEM | CV (%) | |
|---|---|---|---|---|
| Chamber 1 | ||||
| CIP (pmol s−1 mg−1) | 82.9 ± 30.6 | 85.1 ± 28.7 | 14.9 | 17.5 |
| CI + CIIP (pmol s−1 mg−1) | 123.1 ± 39.0 | 123.0 ± 36.0 | 19.0 | 15.3 |
| CI + IIE (pmol s−1 mg−1) | 154.9 ± 48.8 | 150.6 ± 44.1 | 24.4 | 15.9 |
| LCR (CIL/CI + IIE) | 0.08 ± 0.08 | 0.09 ± 0.07 | 0.04 | 50.8 |
| PCR (CI + IIP/CI + IIE) | 0.80 ± 0.12 | 0.82 ± 0.11 | 0.07 | 9.0 |
| RCR (CI + IIP/CIL) | 11.1 ± 31.3 | 11.6 ± 16.7 | 22.8 | 193.7 |
| Inv_RCR (CIL/CI + IIP) | 0.11 ± 0.09 | 0.11 ± 0.08 | 0.05 | 48.6 |
| SCR (CIP/CI + IIP) | 0.67 ± 0.11 | 0.69 ± 0.08 | 0.07 | 10.8 |
Abbreviations: CI, complex I; CI, electron input through CI; CI+II, convergent electron input through CI and CII; CI+CII, complex I & II; E, ETS capacity; L, leak respiration; Inv‐RCR, inverse of respiratory control ratio (CIL/CI+IIP); LCR, leak control ratio (CIL/CI+IIE); P, oxphos capacity; PCR, phosphorylation control ratio (CI+IIP/CI+IIE); RCR, respiratory control ratio (CI+IIP/CIL); SCR, substrate control ratio at constant P (CIP/CI+IIP).
FCR were calculated from mass‐specific mitochondrial respiration measurements in permeabilized muscle fibers (vastus lateralis).
The poor correlation along with high TEM and CV (> 15%) estimates for all complexes was still observed when we normalized mitochondrial respiration with CS activity, (Table 2).
Table 2.
Chamber‐specific respiration values normalized by CS activity, typical error of measurement, and coefficient of variation for each substrate
| Mean ± SD | Chamber 2 | TEM | CV (%) | |
|---|---|---|---|---|
| Chamber 1 | ||||
| CIP*/CS** | 5.38 ± 1.92 | 5.60 ± 1.79 | 1.02 | 18.5 |
| CI+CIIP*/CS** | 8.13 ± 2.73 | 8.24 ± 2.52 | 1.25 | 15.2 |
| CI+IIE*/CS** | 10.35 ± 3.65 | 10.22 ± 3.32 | 1.57 | 15.3 |
Results based on 128 muscle samples.
(pmol s−1 mg−1);
(mIU × mg protein−1).
3.2. Simulations to estimate the sample size required to detect changes in mitochondrial respiration at 80% power
We performed simulations for both the CI + CIIP and CI + IIE respiration values since they are the most commonly reported respiration measurements in the literature, as well as the PCR and SCR ratios. We estimated the sample size required to detect true changes in mitochondrial respiration at 80% power. Since TEM and CV values for mitochondrial respiration and mitochondrial respiration/CS were similar we have not conducted simulations for the latter. However, we have attached the code for this calculation in the supplementary data.
For the coupled and uncoupled respiration, the minimum sample size required to observe a percentage increase at 80% power is shown on Figure 2. An intervention that increases mitochondrial respiration by 10% at the group level requires a minimum of 23 participants to detect changes for CI + CIIP and CI + CIIE at 80% power. Our results suggest that with the typical sample size in exercise training studies (n = 12), only changes of >15% in mitochondrial respiration following training would be detectable at 80% power.
Figure 2.
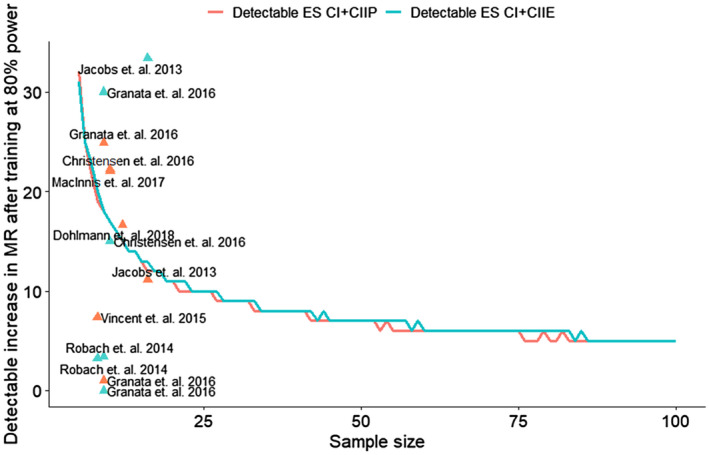
Minimum sample size required to detect increases in mitochondrial respiration (MR) after training at 80% power. A minimum of ~55 (CI+CIIP) and ~60 (CI+CIIE) pairs of duplicate samples are necessary to detect an increase of 6% in mitochondrial respiration at the group level, at 80% power. An intervention with ~20 samples/individuals would require a change of at least 11% in mitochondrial respiration to achieve 80% power for both CI+CIIP and CI+CIIE respiration. Experiments with less than 15 individuals would require a change of at least 13% to achieve 80% power. The triangles represent real effect sizes and sample sizes reported in different studies 8, 10, 11, 12, 14, 20, 35
For the PCR and SCR respiration ratios, the minimum sample size required to observe a percentage increase at 80% power is shown on Figure 3. An intervention that increases mitochondrial respiration by 10%, at the group level, requires a minimum of 11 participants to detect changes for PCR ratio and 22 participants for the SCR ratio at 80% power.
Figure 3.
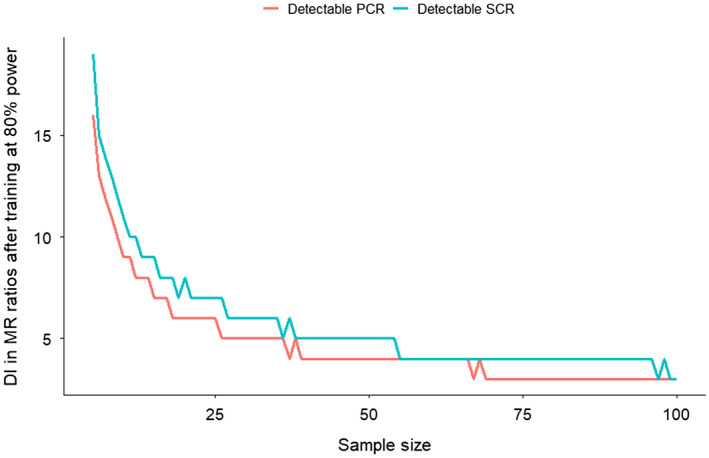
Minimum sample size required to detect increases (DI) in mitochondrial respiration (MR) ratios after training at 80% power. A minimum of ~26 (PCR) and ~38 (SCR) pairs of duplicate samples are necessary to detect an increase of 5% in mitochondrial respiration at the group level, at 80% power. An intervention with ~20 samples/individuals would require a change of at least 6% and 8% in mitochondrial respiration to achieve 80% power for PCR and SCR ratios, respectively. Experiments with less than 20 individuals would require a change of at least 7%‐10% to achieve 80% power. Due to inconsistencies in the literature in which ratios each study calculates we have not included data from published studies in our ratios graph
We have also simulated percentage increases after hypothetical exercise training intervention in a cohort of 20 individuals (Figure 4A). We observed that an increase of ~11% or more in mitochondrial respiration is necessary for changes to be detected at 80% statistical power if each participant had duplicate respiration measurements. While for ratios (Figure 4B), a minimum of ~6% increase for PCR phosphorylation control ratio (CI + IIP/CI + IIE) and ~7% increase for SCR substrate control ratio at constant P (CIP/CI + IIP) is required to be detected at 80% power, if each participant had duplicate respiration measurements.
Figure 4.
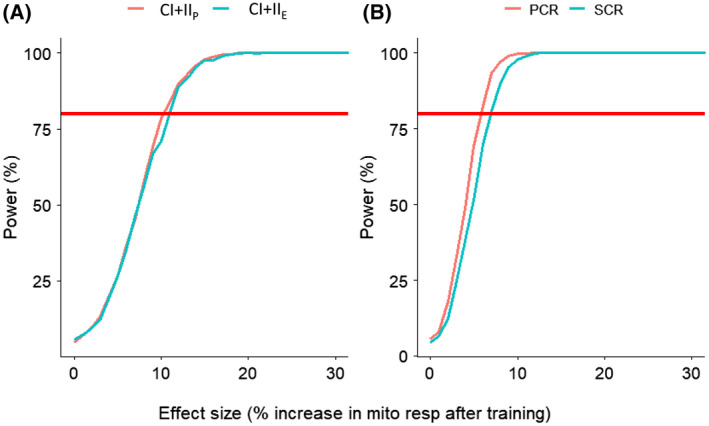
A, Power to detect percentage change in mitochondrial respiration (effect size) after a training intervention with n = 20 participants. A minimum of ~11% increase in mitochondrial respiration is needed to be detected at 80% power for both the CI+CIIP and CI+CIIE, if each participant had duplicate respiration measurements. B, Power to detect percentage change in mitochondrial respiration ratios (effect size) after a training intervention with n = 20 participants. A minimum of ~6% increase for PCR phosphorylation control ratio (CI+IIP/CI+IIE) and ~7% increase for SCR substrate control ratio at constant P (CIP/CI+IIP) is required to be detected at 80% power, if each participant had duplicate respiration measurements
4. DISCUSSION
In the present study, we reported the TEM and CV for measurements of mitochondrial respiration for CIp, CI + CIIP, and CI + CIIE, using the OROBOROS equipment, and in a large sample (n = 160 pairs of duplicate respiration measurements). We also performed statistical simulations to uncover the minimum number of participants required to detect an intervention‐induced change in mitochondrial respiration at 80% power. We found a very large variability in all measurements (CV > 15%), suggesting that this measurement may only be appropriate in studies using large sample sizes (n ≥ 55) or that detect large effect sizes (>15%). To account for between‐lab (lab‐by‐lab) variability, we have also computed the TEM and CV for mitochondrial respiration ratios including: LCR (L/E), PCR (P/E), RCR (P/L), Inv RCR (L/P), and SCR (constant P). Not surprisingly, the TEM and CV remained >9%, and the statistical simulations suggest a sample size of ≥ 26 is required to achieve 80% power. Mitochondrial respirations values were also normalized by CS activity, but no significant changes in TEM or CVs were observed. In other words, the type of training should be carefully selected to achieve effect sizes in mitochondrial respiration experiments if sample size is a limitation. For example, participants who did sprint interval training (SIT) (n = 9) presented >19% change in mitochondrial respiration after 4 weeks of training, while participants who did only moderate intensity (n = 9) or high intensity interval training (n = 11) did not show any changes in mitochondrial respiration after 4 weeks of training.10 Higher numbers of technical replicates (ie, number of chambers used for the same muscle—here we used two) could potentially lower the TEM, in which case the required sample size would be lower to detect a given effect size.
TEM includes variability due to machine calibration and human error, and is potentially specific to each research facility.17 However, Cardinale et al3 recently reported a similarly high CV (15.2%), suggesting that the high variability we observed occurs across research facilities and is intrinsic to the OROBOROS equipment. Permeabilization of muscle pieces involves taking a small piece of muscle (typically 2‐6 mg) and placing it in a dish with BIOPS solution; then, a technician uses two pairs of sharp forceps to separate individual fiber bundles.32 Since this process is complicated, it is recommended that the same person performs the procedure in a given study to avoid variability between technicians.33 The degree of fiber separation determines the amount of mitochondria present after the permeabilization, thus affecting the respiration measurements.33 It is plausible that technicians vary in their ability to efficiently separate fibers, and this could lead to higher respiration values and potentially higher variability as well. Thus, different technical staff/researchers conducting the experiment can explain why experiments are variable. The largest variability in measurements, recently reported by Cardinale et al3 was in experiments conducted by two different technicians working on the same piece of muscle (mean ± SD =31.3 ± 7.1 vs 26.3 ± 8.1 pmol s‐1 mg‐1, P = .12). In the present study, some of the experiments were conducted by one technician, and some by another technician (ie, the two technicians never handled the same piece of muscle), which might explain some of the variability we reported. Unfortunately, we are unable to calculate the variability due to technicians in this study as they worked on different muscle samples. It should also be noted that it is common to use creatine in the respiration chambers when working with permeabilized muscle. However, several papers have not used creatine in their experiments and have reported valid and replicable results.7, 8, 10, 12
To the best of our knowledge, the smallest meaningful change in mitochondrial respiration after training or other lifestyle interventions has never been reported, since this is dependent on the overall aim of each study.10 In the present study, we calculated the minimum number of participants required to detect a change in mitochondrial respiration at 80% power. Our results suggest that with the typical sample size in exercise training studies (n = 12), only changes of >18% in mitochondrial respiration following training would be detectable at 80% power. This means that it would be a challenge to observe true changes in mitochondrial capacity using the OROBOROS technology, since most studies do not report such large increases following exercise training (−9% to 20%).18, 19, 32, 34, 35, 36, 37, 38 We acknowledge that some of the studies have observed significant changes in mitochondrial respiration without reaching the effect sizes we presented here. This implies that although such studies were significant, the sample sizes were too low to detect the magnitude of changes they reported at 80% power. The simulations presented in this paper provide important information for planning experiments investigating mitochondrial capacity. Future studies can use this data and the code we provided (see Supplementary File 1) as a guide to determine the number of participants required to detect changes in mitochondrial respiration in their study. Alternatively, if the number of participants is a limitation, then a careful consideration of the exercise intervention duration is recommended, to trigger changes that are large enough to achieve 80% power.
In conclusion, we found very large variability in mitochondrial respiration measurements, reflected by TEM and CV, and calculated the required sample size necessary for studies aimed at detecting changes in mitochondrial respiration. We recommend that future studies utilizing this method in skeletal muscle would follow the guidelines we provided here to detect significant changes in mitochondrial capacity, following lifestyle interventions. Finally, it should be noted that mitochondrial respiration is only one measure of mitochondrial function. The use of integrative approaches,1, 16, 32 such as mitochondrial protein expression, mitochondrial content quantification, mitochondrial DNA sequencing, and appropriate statistical methods17 may allow discoveries in the complex and integrative nature of exercise adaptations16, 39 as well as strengthen results from mitochondrial respiration measurements.
CONFLICT OF INTEREST
The authors have no conflict of interest on the production of this manuscript.
AUTHOR CONTRIBUTIONS
M Jacques performed the experiments, analyzed the data, and drafted the manuscript; J Kuang performed the experiments and revised the manuscript; DJ Bishop revised the manuscript; X Yan participated in the research design and revised the manuscript; J Alvarez‐Romero performed the experiments; F Munson, I Papadimitriou; and A Garnham performed and collected the muscle biopsies and revised the manuscript; S Voisin performed the data analyses and assisted in drafting the manuscript; and N Eynon designed the research and assisted in drafting the manuscript.
Supporting information
ACKNOWLEDGMENT
This study was chiefly supported by Nir Eynon’s National Health and Medical Research Council, Australia Career Development Fellowship (NHMRC CDF# APP1140644), and Sarah Voisin’s NHMRC Early Career Fellowship (APP11577321). The authors would like to acknowledge Dr Lannie O’Keefe for providing the participant’s‐controlled diet according to the NHMRC guidelines.
Jacques M, Kuang J, Bishop DJ, et al. Mitochondrial respiration variability and simulations in human skeletal muscle: The Gene SMART study. The FASEB Journal. 2020;34:2978–2986. 10.1096/fj.201901997RR
Sarah Voisin and Nir Eynon are Co‐senior authorship.
REFERENCES
- 1. Bishop DJ, Granata C, Eynon N. Can we optimise the exercise training prescription to maximise improvements in mitochondria function and content? Biochim Biophys Acta – Gen Subj. 2014;1840:1266‐1275. [DOI] [PubMed] [Google Scholar]
- 2. Saraste, M . Oxidative phosphorylation at the fin de siecle. Science. 1999;283:1488‐1493 [DOI] [PubMed] [Google Scholar]
- 3. Cardinale DA, Gejl KD, Ortenblad N, Ekblom B, Blomstrand E, Larsen FJ. Reliability of maximal mitochondrial oxidative phosphorylation in permeabilized fibers from the vastus lateralis employing high‐resolution respirometry. Physiol Rep. 2018;6(4):e13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wibom R, Hultman E. ATP production rate in mitochondria isolated from microsamples of human muscle. Am J Physiol. 1990;259(2 Pt 1):E204‐E209. 10.1152/ajpendo.1990.259.2.E204. [DOI] [PubMed] [Google Scholar]
- 5. Kuznetsov AV, Veksler V, Gellerich FN, Saks V, Margreiter R, Kunz WS. Analysis of mitochondrial function in situ in permeabilized muscle fibers, tissues and cells. Nat Protoc. 2008;3:965‐976. 10.1038/nprot.2008.61. [DOI] [PubMed] [Google Scholar]
- 6. Pesta D, Gnaiger E. High‐resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol Biol. 2012;810:25‐58. 10.1007/978-1-61779-382-0_3. [DOI] [PubMed] [Google Scholar]
- 7. Granata C, Oliveira RSF, Little JP, Renner K, Bishop DJ. Mitochondrial adaptations to high‐volume exercise training are rapidly reversed after a reduction in training volume in human skeletal muscle. FASEB J. 2016;30:3413‐3423. [DOI] [PubMed] [Google Scholar]
- 8. Christensen PM, Jacobs RA, Bonne T, Flück D, Bangsbo J, Lundby C. A short period of high‐intensity interval training improves skeletal muscle mitochondrial function and pulmonary oxygen uptake kinetics. J Appl Physiol. 2016;120:1319‐1327. 10.1152/japplphysiol.00115.2015. [DOI] [PubMed] [Google Scholar]
- 9. Daussin FN, Zoll J, Dufour SP, et al. Effect of interval versus continuous training on cardiorespiratory and mitochondrial functions: relationship to aerobic performance improvements in sedentary subjects. Am J Physiol Regul Integr Comp Physiol. 2008;295:R264‐R272. [DOI] [PubMed] [Google Scholar]
- 10. Granata C, Oliveira RSF, Little JP, Renner K, Bishop DJ. Training intensity modulates changes in PGC‐1α and p53 protein content and mitochondrial respiration, but not markers of mitochondrial content in human skeletal muscle. FASEB J. 2016;30:959‐970. [DOI] [PubMed] [Google Scholar]
- 11. Jacobs RA, Flück D, Bonne TC, et al. Improvements in exercise performance with high‐intensity interval training coincide with an increase in skeletal muscle mitochondrial content and function. J Appl Physiol. 2013;115:785‐793. [DOI] [PubMed] [Google Scholar]
- 12. Vincent G, Lamon S, Gant N, et al. Changes in mitochondrial function and mitochondria associated protein expression in response to 2‐weeks of high intensity interval training. Front Physiol. 2015;6:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walsh B, Tonkonogi M, Sahlin K. Effect of endurance training on oxidative and antioxidative function in human permeabilized muscle fibres. Pflugers Arch. 2001;442:420‐425. 10.1007/s004240100538. [DOI] [PubMed] [Google Scholar]
- 14. MacInnis MJ, Zacharewicz E, Martin BJ, et al. Superior mitochondrial adaptations in human skeletal muscle after interval compared to continuous single‐leg cycling matched for total work. J. Physiol. 2017;595:2955‐2968. 10.1113/JP272570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Montero D, Cathomen A, Jacobs RA, et al. Haematological rather than skeletal muscle adaptations contribute to the increase in peak oxygen uptake induced by moderate endurance training. J. Physiol. 2015;593:4677‐4688. 10.1113/JP270250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Granata C, Jamnick NA, Bishop DJ. Training‐induced changes in mitochondrial content and respiratory function in human skeletal muscle. Sport Med. 2018;48:1809‐1828. [DOI] [PubMed] [Google Scholar]
- 17. Voisin S, Jacques M, Lucia A, Bishop DJ, Eynon N. Statistical considerations for exercise protocols aimed at measuring trainability. Exerc Sport Sci Rev. 2019;47:37‐45. 10.1249/JES.0000000000000176. [DOI] [PubMed] [Google Scholar]
- 18. Irving BA, Lanza IR, Henderson GC, Rao RR, Spiegelman BM, Sreekumaran NK. Combined training enhances skeletal muscle mitochondrial oxidative capacity independent of age. J Clin Endocrinol Metab. 2015;100:1654‐1663. 10.1210/jc.2014-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leckey JJ, Hoffman NJ, Parr EB, et al. High dietary fat intake increases fat oxidation and reduces skeletal muscle mitochondrial respiration in trained humans. FASEB J. 2018;32:2979‐2991. [DOI] [PubMed] [Google Scholar]
- 20. Robach P, Bonne T, Flück D, et al. Hypoxic training: effect on mitochondrial function and aerobic performance in hypoxia. Med Sci Sports Exerc. 2014;46:1936‐1945. [DOI] [PubMed] [Google Scholar]
- 21. Brandao CFC, de Carvalho FG, Souza AO, et al. Physical training, UCP1 expression, mitochondrial density, and coupling in adipose tissue from women with obesity. Scand J Med Sci Sports. 2019;29:1699‐1706. 10.1111/sms.13514. [DOI] [PubMed] [Google Scholar]
- 22. Goedecke JH, Mendham AE, Clamp L, et al. An exercise intervention to unravel the mechanisms underlying insulin resistance in a cohort of black South African women: protocol for a randomized controlled trial and baseline characteristics of participants. JMIR Res Protoc. 2018;7:e75 10.2196/resprot.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Lucas RD, Caputo F, Mendes de Souza K, et al. Increased platelet oxidative metabolism, blood oxidative stress and neopterin levels after ultra‐endurance exercise. J Sports Sci. 2014;32:22‐30. 10.1080/02640414.2013.797098. [DOI] [PubMed] [Google Scholar]
- 24. Gatterer H, Menz V, Salazar‐Martinez E, et al. Exercise performance, muscle oxygen extraction and blood cell mitochondrial respiration after repeated‐sprint and sprint interval training in hypoxia: a pilot study. J Sport Sci Med. 2018;17:339‐347. [PMC free article] [PubMed] [Google Scholar]
- 25. Hedges CP, Woodhead JST, Wang HW, et al. Peripheral blood mononuclear cells do not reflect skeletal muscle mitochondrial function or adaptation to high‐intensity interval training in healthy young men. J Appl Physiol. 2019;126:454‐461. 10.1152/japplphysiol.00777.2018. [DOI] [PubMed] [Google Scholar]
- 26. Tsai HH, Chang SC, Chou CH, Weng TP, Hsu CC, Wang JS. Exercise training alleviates hypoxia‐induced mitochondrial dysfunction in the lymphocytes of sedentary males. Sci Rep. 2016;6:35170 10.1038/srep35170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan X, Eynon N, Papadimitriou ID, et al. The gene SMART study: method, study design, and preliminary findings. BMC Genomics. 2017;18(Suppl 8):821 10.1186/s12864-017-4186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363‐2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larsen S, Nielsen J, Hansen CN, et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J Physiol. 2012;14:3349‐3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kitchen RR, Sabine VS, Sims AH, et al. Correcting for intra‐experiment variation in Illumina BeadChip data is necessary to generate robust gene‐expression profiles. BMC Genomics. 2010;11:134 10.1186/1471-2164-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McHale CM, Zhang L, Lan Q, et al. Global gene expression profiling of a population exposed to a range of benzene levels. Environ Health Perspect. 2011;119:628‐634. 10.1289/ehp.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boushel R, Gnaiger E, Schjerling P, Skovbro M, Kraunsøe R, Dela F. Patients with type 2 diabetes have normal mitochondrial function in skeletal muscle. Diabetologia. 2007;50:790‐796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larsen S, Kraunsøe R, Gram M, Gnaiger E, Helge JW, Dela F. The best approach: homogenization or manual permeabilization of human skeletal muscle fibers for respirometry? Anal Biochem. 2014;446:64‐68. [DOI] [PubMed] [Google Scholar]
- 34. Doerrier C, Garcia‐Souza LF, Krumschnabel G, Wohlfarter Y, Mészáros AT, Gnaiger E. High‐resolution fluorespirometry and oxphos protocols for human cells, permeabilized fibers from small biopsies of muscle, and isolated Mitochondria. 2018. [DOI] [PubMed]
- 35. Dohlmann TL, Hindsø M, Dela F, Helge JW, Larsen S. High‐intensity interval training changes mitochondrial respiratory capacity differently in adipose tissue and skeletal muscle. Physiol Rep. 2018;6:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Konopka AR, Laurin JL, Schoenberg HM, et al. Metformin inhibits mitochondrial adaptations to aerobic exercise training in older adults. Aging Cell. 2019;18(1):e12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larsen FJ, Schiffer TA, Ørtenblad N, et al. High‐intensity sprint training inhibits mitochondrial respiration through aconitase inactivation. FASEB J. 2016;30:417‐427. [DOI] [PubMed] [Google Scholar]
- 38. Porter C, Reidy PT, Bhattarai N, Sidossis LS, Rasmussen BB. Resistance exercise training alters mitochondrial function in human skeletal muscle. Med Sci Sports Exerc. 2015;47:1922‐1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whitham M, Febbraio MA. The ever‐expanding myokinome: discovery challenges and therapeutic implications. Nat Rev Drug Discov. 2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


