Abstract
Kinesin family member 11 (KIF11) is a plus end‐directed kinesin indispensable for the formation of the bipolar spindle in metaphase, where it objects to the action of minus end‐directed molecular motors. Here, we hypothesize that KIF11 might be a therapeutic target of breast cancer and regulated by miR‐30a. Cell Counting Kit 8 assays were used to investigate cell proliferation. Invasion assays were used to survey the motility of cells. Kaplan‐Meier and Cox proportional analyses were employed for this outcome study. The prognostic significance and performance of KIF11 were validated on 17 worldwide independent microarray datasets and two The Cancer Genome Atlas‐Breast Invasive Carcinoma sets. microRNA was predicted targeting KIF11 through sequence alignment in microRNA.org and confirmed by coexpression analysis in human breast cancer samples. Dual‐luciferase reporter assays were employed to validate the interaction between miR‐30a and KIF11 further. Higher KIF11 mRNA levels and lower miR‐30a were significantly associated with poor survival of breast cancer patients. Inhibition of KIF11 by small‐hairpin RNA significantly reduced the proliferation and invasion capabilities of the breast cancer cells. Meanwhile, downregulation of KIF11 could enhance the cytotoxicity of adriamycin in breast cancer cell lines MCF‐7 and MDA‐MB‐231. A population study also validated that chemotherapy and radiotherapy significantly improved survival in early‐stage breast cancer patients with low KIF11 expression levels. Further bioinformatics analysis demonstrated that miR‐30a could interact with KIF11 and validated by dual‐luciferase reporter assays. Therefore, KIF11 is a potential therapeutic target of breast cancer. miR‐30a could specifically interact with KIF11 and suppress its expression in breast cancer.
Keywords: breast cancer, kinesin family member 11, microRNA‐30a, prognostic biomarker, therapeutic target
1. INTRODUCTION
Breast cancer is one kind of the most common malignant cancers among females in the world, with approximately 1 000 000 new cases each year. 1 , 2 It is also the second‐leading cause of death among women, accounts for 15% of all cancer deaths. 3 Multiple oncogenes, tumor suppressor genes, sex steroid hormones, and their receptors are involved in the genesis and development of breast cancer. Breast cancer is a heterogeneous tumor. There are a variety of subtypes with different biological behaviors and clinicopathologic features that can result in obviously different prognoses, which can be divided into four major molecular subtypes: luminal A (LumA), luminal B (LumB), triple‐negative/basal‐like, and human epidermal growth factor receptor 2 (HER2) type. 4 , 5 , 6 , 7 , 8 This classification of breast cancers has been used for selecting the appropriate therapeutic method. Currently, personalized precision medicine is an emerging field, however, underdeveloped in breast cancer. More targets and corresponding inhibitors need to be explored to improve treatment efficacy and to reduce adverse side effects.
The protein of the kinesin family could function as molecular nanomotors. It converts the free energy of nucleotide hydrolysis in coordinating the mechanical movement of microtubules. 9 , 10 As a member of the kinesin family, KIF11 is a microtubule‐dependent motor protein encoded by the KIF11 gene located at 10q24.1, with a primary function in mitotic spindle formation. 11 KIF11 is still an essential element for maintaining proper spindle dynamics and preserving spindle bipolarity in cell division. It has a catalytic motor/ATPase domain that mediates its interaction with ATP and microtubules. KIF11 utilizes the energy released by ATP hydrolysis to move forward along microtubules. It facilitates spindle assembly by forming a homotetramer. The homotetramer can cross‐link and push apart antiparallel microtubules. 12 , 13 In the previous study, the KIF11 has been implicated in tumourigenesis. It overexpresses in blast crisis chronic myeloid leukemia, activation in mouse B‐cell leukemia, and triggering of genomic instability in transgenic mice. 14 , 15 , 16 KIF11 has also been identified as a molecule involved in pancreatic cancer, non–muscle invasive bladder urothelial carcinoma, non–small cell lung cancer, and glioblastoma. 17 , 18 , 19 , 20 These studies suggest that KIF11 may be involved in the pathogenesis of multiple kinds of cancer. Because of its participating in dividing cells, KIF11 is an essential anticancer target with the trait to avoid the deficiencies of traditional anti‐mitosis drugs. 21 , 22 Drug candidates like ispinesib inhibit KIF11 and cause mitotic arrest, then apoptosis. The research and development of ligand are continually driven partly by the observation of deactivating mutations in the drug binding region, and lack of successful monotherapies based on KIF11 inhibition. Although in the course of our research, one study has discussed the function of KIF11 in breast cancer, 23 whether KIF11 is a potential therapeutic target for breast cancer remains unclarified currently, and the transcriptional regulation on KIF11 also needs to be elucidated.
As an essential transcriptional regulator, the differential expression pattern of microRNAs (miRNAs) in health and disease, therapeutic response, and resistance has resulted in its application as robust biomarkers. 24 Gene regulation by miRNAs and reciprocal regulation of miRNAs have now been studied for over 15 years and extensively reviewed. 25 In general, one miRNA could target multiple genes. Meanwhile, one messenger RNA (mRNA) can be targeted by multiple miRNAs, which highlighted the complexity of miRNA biology. 26 Previous studies showed that the outcome of cancer is closely related to the variable expression and the specific expression signatures of miRNA in cancer tissues. 27 In particular, there is some existing evidence that miRNAs are tightly linked to the development of human breast cancer. 28 , 29 , 30 , 31 miRNAs are attractive candidates as upstream regulators of breast tumor progression and metastasis by regulating entire sets of genes. miRNA signature can subclassify breast cancer 32 and can even determine new subtypes, as recently reported. 33 miR‐30a has been validated as a tumor suppressor via targeting multiple genes in diverse cancer. 34 , 35 , 36 , 37 Here, we predicted target miRNAs of KIF11 using both predicting KIF11‐related miRNAs in microRNA.org and correlation analysis for KIF11 and miRNAs in the GSE22220 dataset, screening out target‐KIF11 miRNAs. Favorably, miR‐30a was one of five miRNAs that could bind to the 3'‐untranslated region (3'‐UTR) of KIF11 mRNA. Thus, miR‐30a may be involved in the regulation of KIF11 in cancer progression.
In the present study, we investigated the role of KIF11 in the tumorigenesis and treatment of breast cancer and the possible role of miR‐30a in the regulation of KIF11 in this process.
2. MATERIALS AND METHODS
2.1. Cell culture and chemicals
Human breast cancer cells, MCF‐7 and MDA‐MB‐231, were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were incubated in Roswell Park Memorial Institute‐1640 medium supplying with 10% fetal bovine serum (Gibco, Carlsbad, CA) and l‐glutamine (Invitrogen, Carlsbad, CA) at 37°C in a humidified atmosphere containing 5% CO2. The passage time of all cell lines was less than 3 months.
2.2. The quantitative reverse transcription‐polymerase chain reaction analysis
A detailed protocol of quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis could be found in our previous publication. 38 Quantitative RT‐PCR for mRNA was detected using an ABI 7500 real‐time PCR system and Absolute qPCR SYBR Green Mix (Applied Biosystems, Foster City, CA). The primer sequences used for KIF11 mRNA detection were 5′‐GATGGACGTAAGGCAGCTCA‐3′ (forward) and 5′‐TGTGGTGTCGTACCTGTTGG‐3′ (reverse). C t values for KIF11 mRNA were normalized to β‐actin mRNA, which was used as internal controls. The method was applied to calculate the relative expression of mRNA.
2.3. KIF11 small‐hairpin RNA plasmids and transient transfectants construction
pGPU6/GFP/Neo was used to express small‐hairpin RNA (shRNA). pGPU6/GFP/Neo‐ KIF11‐Homo vectors were purchased from GenePharma (Shanghai). The target sequence of pGPU6/GFP/Neo‐KIF11‐404 was GCGGAAAGCTAGCGCCCATTC, and the target sequence of pGPU6/GFP/Neo‐KIF11‐1152 was GCTCGGGAAGCTGGAAATATA. MCF‐7 cells at 2 × 105/well and MDA‐MB‐231 cells at 3 × 105/well in a six‐well plates were transduced with the lipofection shRNAs and selected with 600 µg/mL G418 (BBL Life Science, Shanghai).
2.4. miR‐30a plasmids construction, transient transfection, and luciferase assay
To construct a plasmid containing the KIF11 3′‐UTR fused to the 3′‐end of the luciferase reporter, sequences containing the predicted miR‐30a target sites were synthesized and ligated into the pGLO‐control vector (Promega, Madison, WI). KIF11 3′‐UTR was amplified with the primers 5′‐AAACTAGCGGCCGCAATTTATATTCTTTTGTTTACAT‐3′ (forward) and 5′‐CTAGATGTAAACAAAAGAATATAAATTGCGGCCGCTAGTTT‐3′ (reverse) and was subcloned into a pGLO control vector with the restriction endonuclease XbaI site (italic font) to generate pGLO‐KIF11‐3′‐UTR. The 3′‐UTR of KIF11 containing two putative miR‐30a‐binding sites was amplified and cloned into a pmirGLO‐control vector separately. In the mutated fragment, three mutational bases were introduced into the predicted miR‐30a target sites. Subsequently, cells were plated into 24‐well plates, cotransfected with the constructed plasmids, the plasmids with either miR‐30a or miR‐NC; were purchased from Vigene Bioscience (Rockville, MD). Collected cells and measured their luciferase activity after 48 hours using the Dual‐Luciferase Reporter Assay Kit (Promega, Madison, WI). The results are shown as the relative luciferase activity of the firefly/renilla ratio. All the transient transfections were performed using Lipofectamine 2000 (Invitrogen).
2.5. Cell proliferation analysis and drug treatment
Cell Counting Kit 8 (CCK8) was employed to determine the viable cells in cytotoxicity and proliferation assays. According to the manufacturer's instructions, we seeded 2500 cells per well for proliferation assay and 5000 cells/well for cytotoxicity test in a 96‐well plates. The incubation times for proliferation and cytotoxicity were 72 and 48 hours, respectively. Total 10 µL of the CCK8 reagent (Bimake, Houston, TX) directly added to each well, after incubating for 4 hours, reading the optical density (OD) at 450 nm with a BioTeK Synergy H1 plate reader (Winooski, VT). The value of OD 450 nm represents the number of viable cells.
2.6. Invasion assays
Details of the invasion assay were described in a previous publication. 39 About 50 000 cells were seeded on the Matrigel (BD Biosciences, San Jose, CA) insert of the 24‐well chambers. After incubation for 72 hours in 5% CO2 at 37°C, the top of the Matrigel inserts were wiped with a cotton‐tipped swab to remove cells that had not migrated through the membrane. The cells on the lower surface of the membrane were stained with crystal violet solution and counted. Each experiment was performed three times.
2.7. Western blot analysis
Denatured total protein was extracted from breast cancer cells after transfected by sh‐NC or sh‐KIF11. Proteins separated by 10% sodium dodecyl sulphate‐polyacrylamide gel electrophoresis were transferred to polyvinylidene difluoride membranes. The membranes were blocked with 3% bovine serum albumin or 5% nonfat powdered milk in TBST for 2 hours at room temperature, then incubated with primary antibodies overnight at 4°C. The primary antibodies used were against KIF11 (Proteintech, Wuhan, China), E‐cadherin, extracellular‐signal‐regulated kinase (Erk), phospho Erk (pErk), protein kinase B (Akt), phosphorylated Akt (pAkt), (all from Cell Signaling Technology, Danvers, MA), and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) (Sungene, Tianjin, China). After washing with TBST three times, the membranes were incubated with secondary antibodies conjugated with horseradish peroxidase‐conjugated goat anti‐rabbit IgG or goat anti‐mouse IgG (ImmunoWay) for 1 hour at room temperature. After washing with TBST three times, the protein bands were measured by an Enhanced Chemiluminescence Kit (Beyotime) through a Clinx Science Instruments (Shanghai). The intensity of the specific bands was measured by Image J software.
2.8. Worldwide gene expression datasets
A total of 17 published microarray datasets containing survival information of breast cancer patients was downloaded from the Array Express database (www.ebi.ac.uk/arrayexpress) including GSE7390, 40 GSE1456, 41 GSE2034, 42 GSE4922, 43 GSE10885, 44 GSE24450, 45 GSE25066, 46 GSE53031, 47 GSE58812, 48 GSE22220, 49 GSE3143, 50 GSE3494, 51 GSE11121, 52 GSE12276, 53 GSE22226, 54 GSE6532, 55 and NKI 56 ; two of The Cancer Genome Atlas‐Breast Invasive Carcinoma (TCGA‐BRCA) was downloaded from TCGA (https://www.cbioportal.org/). Datasets without prognostic outcome information were excluded. The clinical relevance and prognostic significance of KIF11 in breast cancer were evaluated on the above datasets. Detailed information of the microarray datasets is summarized in Table S1.
The disease‐free survival (DFS) period was defined as the time from initial surgery until tumor recurrence, including local relapse and distant metastasis. The overall survival (OS) period was calculated as the time from initial surgery to the date when the patient was last seen. To normalize the mRNA expression levels among the above datasets, we restratified the scores of KIF11 and other related genes into four grades (Q1, Q2, Q3, and Q4) based on the percentile for each independently downloaded dataset. For Cox analysis, less than the median was regarded as KIF11‐low (Q1 + Q2), while greater than or equal to the median was regarded as KIF11‐high (Q3 + Q4). The demographic distribution of KIF11 is presented in Table S2.
2.9. Gene set enrichment analysis
To evaluate the correlations between KIF11 expression and cancer‐related pathways, we conducted gene set enrichment analysis (GSEA) using the above microarray dataset GSE1456. The detailed protocol of GSEA was available on the Broad Institute GSEA website (www.broad.mit.edu/gsea) or from related references. 57 Briefly, datasets and phenotype label files were created and loaded into GSEA software (v2.0.13). The gene sets were downloaded from the Gene Expression Omnibus (http://www.cancergenome.nih.gov/geo/). The phenotype label was KIF11 expression. We set the number of permutations to 1000.
2.10. Target prediction and functional analysis of miRNA
The presumed target of KIF11‐related miRNA, especially the most significant hsa‐miR‐30a, we searched in microRNA.org (http://www.microrna.org/microrna/home.do). The above breast cancer microarray dataset GSE22220 was used to assess the role of miR‐30a and KIF11 in breast cancer progression and prognosis.
2.11. Data management and statistical analysis
All data were analyzed using the SAS statistical software, version 9.2 (SAS Institute, Cary, NC), unless otherwise noted. The Student t test and one‐way analysis of variance were used for continuous data analyses, and the Pearson χ 2 test was used for categorical data analyses. We used Kaplan‐Meier survival analysis to draw the proportion of the population that was OS or DFS by follow‐up time in months. We calculated hazard ratios (HR) with 95% confidence intervals (CI) using Cox proportional hazards regression analysis to survey the association of KIF11 expression levels with patient survival. Two‐sided P values less than .05 were considered statistically significant. Missing data were coded and excluded from the analysis.
3. RESULTS
3.1. Inhibition of KIF11 causes growth blockage and invasion decrease in breast cancer cells
To explore the role of KIF11 in the development of breast cancer, we analyzed mRNA expression of KIF11 in both cell lines and the GSE70947 dataset. Analysis results showed that mRNA expression levels of KIF11 were significantly higher in tumor than in normal control (Figure 1A). To verify whether inhibition of KIF11 could alleviate the development of breast cancer in vitro, the shRNA was used to downregulate the expression of KIF11 in estrogen receptor (ER)‐positive breast cancer cell line MCF‐7 and ER‐negative breast cancer cell line MDA‐MB‐231. sh‐NC plasmids were employed as control. The qPCR results indicated that sh‐KIF11 reduced the mRNA level of KIF11 in both MCF‐7 and MDA‐MB‐231 (Figure 1B). After knocking down KIF11 in MCF‐7 and MDA‐MB‐231, expression levels of KIF11, E‐cadherin, Akt, p‐Akt, Erk, and p‐Erk levels were measured. The levels of Akt, p‐Akt, and Erk did not change significantly after KIF11 reduction, whereas p‐Erk levels slightly decreased in both MCF‐7 and MDA‐MB‐231, then E‐cadherin slightly increased in MCF‐7 but not expressed in MDA‐MB‐231 (Figure 1C). Inhibition of KIF11 significantly delayed cell growth of both MCF‐7 and MDA‐MB‐231 (Figure 1D,E). The invasion assay also showed that sh‐KIF11 could reduce invasive cells from 132.0 ± 12.4 per field (sh‐NC) to 92.5 ± 16.7 per field (sh‐KIF11‐404) and 83.5 ± 20.5 per field (sh‐KIF11‐1152) in MCF‐7 cells (P < .01), and from 118 ± 26.4 per field (sh‐NC) to 55.0 ± 20.4 per field (sh‐KIF11‐1152) in MDA‐MB‐231 cells (P < .05) (Figure 1F). Expression levels of MMP were significantly reduced (Figure 1G). These findings suggested that inhibition of KIF11 could significantly inhibit the growth and invasive ability of both MCF‐7 (ER‐positive) and MDA‐MB‐231 (ER‐negative) cell lines.
Figure 1.
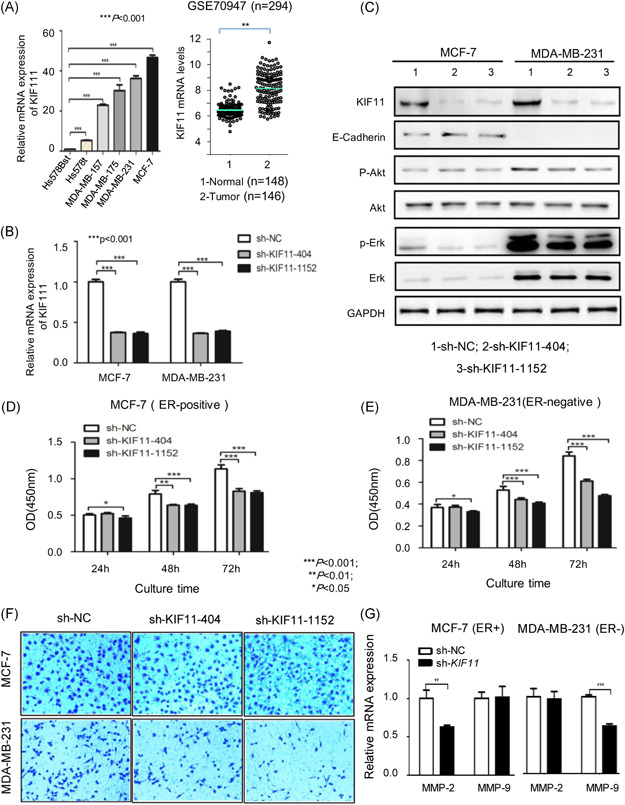
Inhibition of KIF11 causes growth blockage, and invasion decrease in breast cancer cells. A, Expression of KIF11 in cell lines and GSE70947 set. B, Relative mRNA expression of KIF11 in MCF‐7 and MDA‐MB‐231 cells following transfection with sh‐KIF11, sh‐NC as control. C, The expression levels of KIF11, E‐cadherin, Akt, p‐Akt, Erk, and p‐Erk in transfectants were visualized by Western blot. D, The cell growth curve for MCF‐7 transfectants was measured with CCK8 assay. E, The cell growth curve for MDA‐MB‐231 transfectants was measured with CCK8 assay. F, The invasion chamber detected invasion ability in breast cancer cells. G, Expression levels of MMP were detected by qPCR. Shown were the representative results (mean ± standard deviation) of three independent experiments. Akt, protein kinase B; CCK8, Cell Counting Kit 8; ER, estrogen receptor; Erk, extracellular‐signal‐regulated kinase; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; KIF11, kinesin family member 11; MMP, matrix metalloproteinase; mRNA, messenger RNA; OD, optical density; p‐Akt, phosphorylated Akt; p‐Erk, phospho Erk; qPCR, quantitative polymerase chain reaction; sh‐NC, short hairpin negative control. *P < .05, **P < .01, ***P < .001 [Color figure can be viewed at wileyonlinelibrary.com]
3.2. KIF11 is associated with poor differentiation and aggressive phenotypes of breast cancer
The expression data of KIF11 could be obtained from all collected datasets to investigate the clinical relevance. Analysis results showed that mRNA expression levels of KIF11 were significantly associated with the younger patient, lower ER levels, bigger tumor size, lymph node, and higher grade of breast cancer in none‐TCGA (Figure 2A) and TCGA datasets (Figure 2B). In TCGA‐BRCA‐set 2, mRNA expression levels of KIF11 were significantly higher in aggressive molecular subtypes such as triple‐negative breast cancer (TNBC) than in normal‐like or LumA breast cancer (Figure 2C). The genes were coexpressed with KIF11 in TCGA‐BRCA‐set 2, which including cyclin‐dependent kinase, abnormal spindle microtubule assembly, epithelial cell transforming, and mitotic checkpoint serine/threonine kinase were shown (Figure 2D). A further GSEA analysis indicated that high‐expression of KIF11 significantly enriched in the gene signatures related to poor prognosis. The normalized enrichment score (NES) was 2.32 (Figure 2E). The correlation between KIF11 and poor prognosis was further verified. Besides, more NES related to poor differentiation, metastasis, and so forth were indicated in breast cancer (Figure 2F). All of the above findings validated that mRNA levels of KIF11 were significantly associated with aggressive phenotypes in breast cancer.
Figure 2.
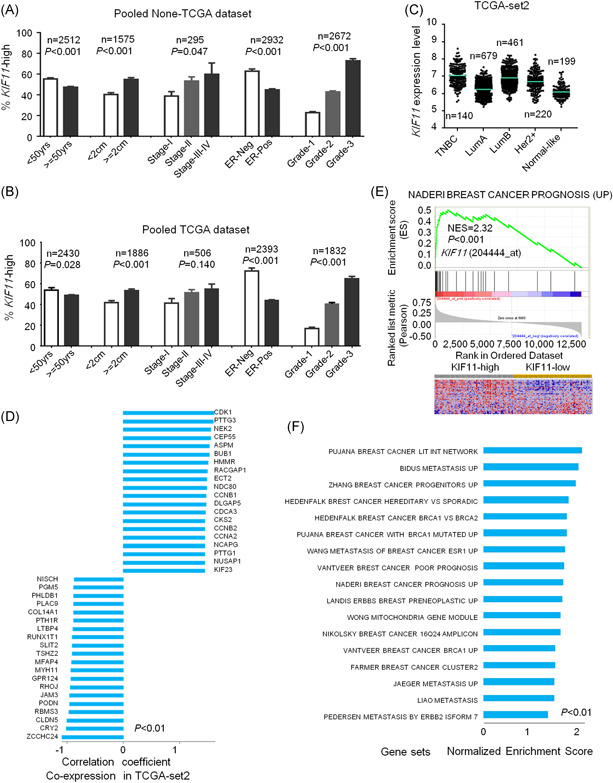
High expression of KIF11 is related to invasiveness in breast cancer in downloaded datasets. A, The mRNA levels of KIF11 and age, tumor size, ER status, and Elson grade of breast cancer in pooled dataset in non‐TCGA. Here, the KIF11‐high was defined as the KIF11 mRNA level equal to or larger than median mRNA levels in each dataset. B, The mRNA levels of KIF11 and age, tumor size, ER status, and Elson grade of breast cancer in pooled dataset in TCGA. C, The mRNA expression level of KIF11 in different molecular subtypes of breast cancer in TCGA‐BRCA‐set2. D, Genes were coexpressed with KIF11 in TCGA‐BRCA‐set2. E, Enriched gene signatures associated with poor prognosis were displayed. Heatmap depicts the expression levels of these genes. Red represents upregulation, and blue represents downregulation. F, NES of KIF11 shown in a related pathway in ordered datasets. ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; KIF11, kinesin family member 11; LumA, luminal A; LumB, luminal B; mRNA, messenger RNA; NES, normalized enrichment score; TCGA‐BRCA, The Cancer Genome Atlas‐Breast Invasive Carcinoma; TNBC, triple‐negative breast cancer [Color figure can be viewed at wileyonlinelibrary.com]
3.3. Prognostic significance of KIF11 for breast cancers
Since NES of KIF11 was associated with poor prognosis, poor differentiation, and metastasis of breast cancer, the expression of KIF11 could be related to poor outcomes in breast cancer. To address this hypothesis, we conducted a survival analysis on public microarray gene expression datasets. Here, we recategorized participants of each dataset into four subgroups (Q1, Q2, Q3, and Q4) according to the expression levels of KIF11. In Figure 3A, the mRNA levels of KIF11 significantly impacted poor OS (left) of breast cancer in none‐TCGA datasets. Following increased KIF11 levels, OS decreased in a dose‐dependent manner. The prognostic significance of KIF11 was further analyzed in TCGA datasets (Figure 3B). ER‐negative breast cancers (including the HER2‐positive and TNBC subtypes) have a poor prognosis. 58 Our results in Figure 2A,B, showed that KIF11 had higher expression levels in ER‐negative breast cancers. Further stratified Kaplan‐Meier analysis with the pooled data explored that KIF11 mRNA levels were significantly associated with poor OS (Figure 3C) and poor DFS (Figure 3D) in not only ER‐negative but also in ER‐positive breast cancers. It was confirmed that KIF11 expression significantly impacted the poor survival of breast cancer.
Figure 3.
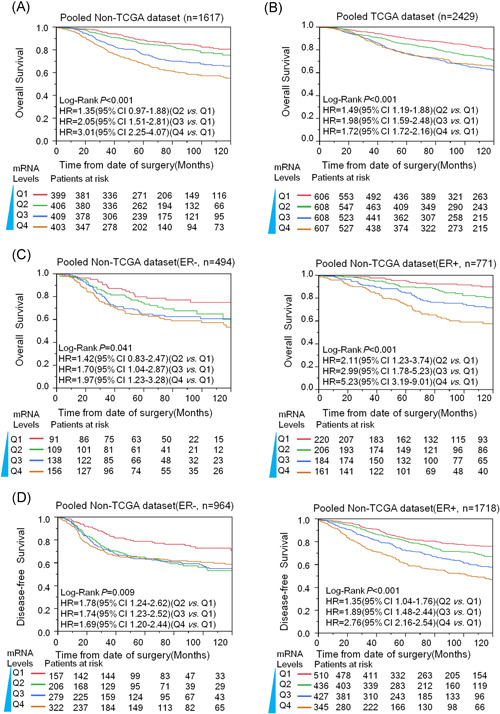
Kaplan‐Meier analysis was performed to investigate the KIF11 and outcome of breast cancer among microarray datasets. We pooled all eligible breast cancers after normalizing. A, The overall survival (OS) analysis results of KIF11 in pooled datasets in non‐TCGA. B, Analysis of KIF11 for the OS in pooled datasets in TCGA. C, The prognostic performance of KIF11 for the OS of both ER‐negative and ER‐positive breast cancers in non‐TCGA. D, Disease‐free survival results of KIF11 from pooled datasets with disease‐free survival information in non‐TCGA. CI, confidence interval; ER, estrogen receptor; HR, hazard ratio; KIF11, kinesin family member 11; mRNA, messenger RNA; TCGA, The Cancer Genome Atlas [Color figure can be viewed at wileyonlinelibrary.com]
Further survival analysis was conducted on every independent dataset by employing unique and multiple Cox proportional hazard analysis. The results are listed in Table 1. Q1 was the lowest expression subgroup, which was set as the relative point of reference. The HR of KIF11 for OS and DFS mostly increased along with KIF11 expression levels increased in none‐TCGA datasets. Particularly in higher KIF11 levels (Q4), the significance almost could be observed in all datasets. The overall pooled analysis indicated that the HR of higher KIF11 (Q4) for OS and DFS were 3.01 (95% CI: 2.25‐4.07) and 2.11 (95% CI: 1.78‐2.50), respectively. In TCGA datasets, the HR of KIF11 for OS also increased along with KIF11 expression levels increased, the HR of higher KIF11 (Q4) for OS was 1.72 (95% CI: 1.38‐2.16).
Table 1.
Uni‐ and multivariate analysis for KIF11 and survival in microarray datasets
| Overall survival | Disease‐free survival | ||||
|---|---|---|---|---|---|
| HR | Adjusted HR | HR | Adjusted HR | ||
| Dataset | (95% CI) | (95% CI) a | (95% CI) | (95% CI) a | |
| GSE7390 | |||||
| Q1 | Reference | Reference | Reference | Reference | |
| Q2 | 2.30 (0.97‐6.03) | 2.24 (0.94‐5.92) | 1.70 (0.94‐3.19) | 1.64 (0.90‐3.09) | |
| Q3 | 1.97 (0.80‐5.24) | 2.01 (0.78‐5.57) | 1.33 (0.71‐2.54) | 1.19 (0.60‐2.40) | |
| Q4 | 3.42 (1.52 ‐8.69)** | 2.98 (1.23‐7.99)* | 1.82 (0.99‐3.43) | 1.65 (0.84‐3.31) | |
| GSE1456 | |||||
| Q1 | Reference | Reference | Reference | Reference | |
| Q2 | 3.74 (1.14‐16.69)* | 4.45 (1.13‐29.38)* | 2.51 (0.70‐11.65) | 2.08 (0.57‐9.73) | |
| Q3 | 5.75 (1.89‐24.84)** | 5.01 (1.18‐34.83)* | 5.83 (1.92‐25.20)** | 3.32 (0.93‐15.86) | |
| Q4 | 4.72 (1.50‐20.70)** | 4.68 (1.02‐34.28)* | 6.42 (2.12‐27.73)** | 3.71 (0.99‐18.55) | |
| GSE2034 | |||||
| Q1 | N/A | N/A | Reference | Reference | |
| Q2 | 1.40 (0.77‐2.60) | 1.40 (0.77‐2.61) | |||
| Q3 | 1.82 (1.03‐3.33)* | 1.85 (1.04‐3.38)* | |||
| Q4 | 2.31 (1.31‐4.18)** | 2.36 (1.33‐4.30)** | |||
| GSE4922 | |||||
| Q1 | N/A | N/A | Reference | Reference | |
| Q2 | 1.67 (0.83‐3.52) | 1.61 (0.79‐3.39) | |||
| Q3 | 2.82 (1.47‐5.77)** | 2.30 (1.16‐4.83)* | |||
| Q4 | 3.48 (1.82‐7.09)** | 2.65 (1.28‐5.76)** | |||
| GSE25066 | |||||
| Q1 | N/A | N/A | Reference | Reference | |
| Q2 | 1.91 (1.08‐3.44)* | 1.31 (0.71‐2.44) | |||
| Q3 | 1.58 (0.93‐2.75) | 1.16 (0.66‐2.08) | |||
| Q4 | 1.52 (0.87‐2.71) | 1.10 (0.61‐2.03) | |||
| GSE10885 | |||||
| Q1 | Reference | Reference | Reference | Reference | |
| Q2 | 3.97 (1.18‐17.91)* | 4.54 (1.28‐21.27)* | 2.96 (1.15‐8.54)* | 3.31 (1.19‐10.67)* | |
| Q3 | 5.14 (1.64‐22.52)** | 4.53 (1.34‐20.82)* | 3.84 (1.60‐10.62)** | 3.91 (1.38‐12.99)** | |
| Q4 | 7.16 (2.35‐31.06)** | 5.91 (1.59‐29.05)** | 4.11 (1.70‐11.41)** | 3.49 (1.12‐12.58)* | |
| GSE22226 | |||||
| Q1 | Reference | Reference | Reference | Reference | |
| Q2 | 0.55 (0.32‐0.94)* | 0.57 (0.32‐1.01) | 1.66 (0.67‐4.47) | 1.52 (0.57‐4.48) | |
| Q3 | 0.55 (0.31‐0.95)* | 0.57 (0.32‐1.03) | 1.60 (0.61‐4.40) | 1.23 (0.43‐3.76) | |
| Q4 | 0.56 (0.32‐1.00)* | 0.57 (0.31‐1.06) | 1.84 (0.73‐5.01) | 1.52 (0.55‐4.57) | |
| GSE58812 | |||||
| Q1 | Reference | Reference | Reference | Reference | |
| Q2 | 0.56 (0.15‐1.84) | 0.54 (0.14‐1.80) | 1.63 (0.54‐5.41) | 1.56 (0.52‐5.16) | |
| Q3 | 1.54 (0.59‐4.23) | 1.92 (0.73‐5.32) | 1.98 (0.68‐6.44) | 2.43 (0.83‐7.98) | |
| Q4 | 1.11 (0.40‐3.17) | 1.26 (0.45‐3.60) | 1.83 (0.63‐5.96) | 2.06 (0.71‐6.72) | |
| GSE24550 | |||||
| Q1 | Reference | N/A | Reference | N/A | |
| Q2 | 0.75 (0.48‐1.18) | 0.98 (0.63‐1.52) | |||
| Q3 | 0.78 (0.49‐1.24) | 0.93 (0.58‐1.47) | |||
| Q4 | 0.85 (0.52‐1.37) | 0.96 (0.58‐1.57) | |||
| GSE53031 | |||||
| Q1 | N/A | N/A | Reference | Reference | |
| Q2 | 1.39 (0.83‐2.32) | 1.41 (0.83‐2.38) | |||
| Q3 | 1.15 (0.70‐1.92) | 1.12 (0.66‐1.92) | |||
| Q4 | 1.06 (0.64‐1.77) | 1.08 (0.61‐1.90) | |||
| GSE3494 | |||||
| Q1 | Reference | Reference | N/A | N/A | |
| Q2 | 1.62 (0.64‐4.39) | 1.58 (0.59‐4.64) | |||
| Q3 | 2.46 (1.05‐6.40)* | 2.24 (0.89‐6.40)* | |||
| Q4 | 3.96 (1.76‐10.07)** | 3.22 (1.25‐9.46) | |||
| GSE6532 | |||||
| Q1 | Reference | Reference | |||
| Q2 | 0.97 (0.64‐1.48) | 1.01 (0.52‐1.64) | |||
| Q3 | 0.36 (0.22‐0.56)** | 0.52 (0.30‐0.87)* | |||
| Q4 | 0.20 (0.12‐0.34)* | 0.39 (0.20‐0.72)* | |||
| GSE11121 | |||||
| Q1 | Reference | Reference | |||
| Q2 | 1.69 (0.66‐4.58) | 1.44 (0.56‐3.95) | |||
| Q3 | 1.49 (0.59‐4.06) | 1.13 (0.43‐3.21) | |||
| Q4 | 2.90 (1.24‐7.54)* | 1.95 (0.75‐5.53) | |||
| GSE12276 | |||||
| Q1 | Reference | N/A | |||
| Q2 | 1.34 (0.91‐1.99) | ||||
| Q3 | 1.71 (1.15‐2.55)** | ||||
| Q4 | 1.40 (0.94‐2.07) | ||||
| NKI set | |||||
| Q1 | Reference | Reference | Reference | Reference | |
| Q2 | 4.60 (2.09‐11.53)** | 3.12 (1.40‐7.92)** | 2.78 (1.55‐5.20)** | 2.35 (1.30‐4.41)** | |
| Q3 | 4.37 (1.96‐11.04)** | 2.38 (1.03‐6.19)* | 3.22 (1.81‐5.98)** | 2.41 (1.40‐4.22)** | |
| Q4 | 4.80 (2.17‐12.11)** | 1.83 (0.75‐4.96) | 2.95 (1.65‐5.52)** | 1.89 (0.98‐3.76) | |
| Pooled GEO dataset | |||||
| Q1 | Reference | Reference | Reference | Reference | |
| Q2 | 1.35 (0.97‐1.88) | 1.63 (1.05‐2.59)* | 1.41 (1.18‐1.69)** | 1.47 (1.15‐1.88)** | |
| Q3 | 2.05 (1.51‐2.81)** | 1.80 (1.16‐2.85)** | 1.69 (1.42‐2.01)** | 1.66 (1.31‐2.12)** | |
| Q4 | 3.01 (2.25‐4.07)** | 2.69 (1.75‐4.24)** | 2.11 (1.78‐2.50)** | 1.95 (1.53‐2.49)** | |
| TCGA‐BRCA‐set1 | |||||
| Q1 | Reference | Reference | Reference | Reference | |
| Q2 | 0.93 (0.50‐1.71) | 1.07 (0.57‐1.98) | 1.05 (0.49‐2.25) | 1.15 (0.53‐2.51) | |
| Q3 | 1.19 (0.66‐2.14) | 1.41 (0.76‐2.59) | 1.12 (0.52‐2.41) | 1.16 (0.52‐2.58) | |
| Q4 | 0.81 (0.43‐1.48) | 0.88 (0.43‐1.75) | 1.27 (0.63‐2.63) | 1.14 (0.52‐2.52) | |
| TCGA‐BRCA‐set2 | |||||
| Q1 | Reference | Reference | NA | NA | |
| Q2 | 1.62 (1.26‐2.08)** | 1.50 (1.16‐1.94)** | |||
| Q3 | 2.16 (1.70‐2.75)** | 1.75 (1.37‐2.25)** | |||
| Q4 | 1.92 (1.51‐2.46)** | 1.38 (1.07‐1.80)* | |||
| Pooled TCGA dataset | |||||
| Q1 | Reference | Reference | Reference | Reference | |
| Q2 | 1.49 (1.19‐1.88)** | 1.52 (1.18‐1.97)** | 1.05 (0.63‐2.84) | 1.14 (0.52‐2.48) | |
| Q3 | 1.98 (1.59‐2.48)** | 1.73 (1.35‐2.24)** | 1.12 (0.52‐2.41) | 1.22 (0.55‐2.60) | |
| Q4 | 1.72 (1.38‐2.16)** | 1.39 (1.07‐1.82)* | 1.27 (0.63‐2.63) | 1.15 (0.53‐2.56) | |
Note: Uni‐ and multivariate analysis were conducted to evaluate HR of KIF11.
Abbreviations: CI, confidence interval; ER, estrogen receptor; GEO, Gene Expression Omnibus; HR, hazard ratio; KIF11, kinesin family member 11; NA, not applicable; TCGA‐BRCA, The Cancer Genome Atlas‐Breast Invasive Carcinoma.
For multivariate analysis, HR was adjusted by age, ER status, Elston grade in the GSE7390, GSE4922, GSE25066, GSE10885, GSE2226, GSE53031, GSE3494, GSE3494, NKI set, TCGA‐BRCA‐set2, pooled GEO analysis, and in pooled TCGA analysis. In the GSE1456, HR was adjusted by Elston grade and ER; in the GSE2034, HR was adjusted by ER status; in the GSE58812 and TCGA‐BRCA‐set1, HR was adjusted by age and ER status; and in the GSE11121, HR was adjusted by grade.
Statistical significance, P < .05.
Statistical significance, P < .01.
3.4. Reduction of KIF11 sensitizes chemotherapy and radiotherapy in breast cancer
Chemotherapy is usually performed to patients with advanced stages of breast cancer (stage III or IV). For stage II breast cancer, the application of chemotherapy is determined by tumor size, grade, and other indicators. In these patients, a therapeutic biomarker would be beneficial for selecting chemotherapy. Our population study demonstrated that chemotherapy significantly improved the OS of stage II breast cancer patients with KIF11‐low expression in NKI dataset (logrank P = .004; HR = 0.23; 95% CI, 0.07‐0.63) rather than with the KIF11‐high expression (logrank P = .59; HR = 0.82; 95% CI, 0.38‐1.69) (Figure 4A). In TCGA‐BRCA‐set 2, radiotherapy significantly improved the OS of T2 breast cancer patients with KIF11‐low expression but not with the KIF11‐high expression (Figure 4B). Inhibition of KIF11 by shRNA could improve the efficacy of adriamycin on breast cancer cells (Figure 4C). These findings suggest that the silence of KIF11 can significantly enhance the effects of chemotherapy and radiotherapy in breast cancer. KIF11 can also be employed as a therapeutic target and can serve as a biomarker for selecting chemotherapy and radiotherapy in breast cancer treatment.
Figure 4.
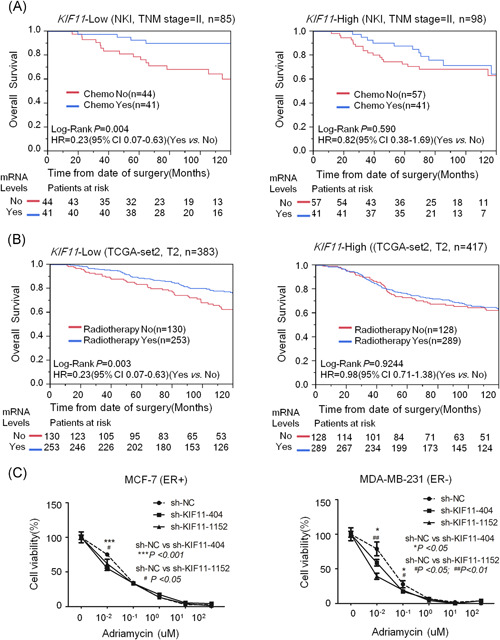
Reduction of KIF11 expression might enhance chemo‐ and radiotherapy efficiency in breast cancers. A, The stratification analysis was conducted on stage II breast cancer patients to compare the chemotherapy efficacy between KIF11‐high expression and KIF11‐low expression in the NKI set. The OS of breast cancer patients was shown. B, The stratification analysis was conducted on T2 breast cancer patients to compare the radiotherapy efficacy between KIF11‐high expression and KIF11‐low expression in TCGA‐BRCA‐set2. The OS of breast cancer patients was shown. C, Reduction of KIF11 expression might enhance the cytotoxicity of adriamycin in breast cancers. About 5000 cells (MCF‐7 or MDA‐MB‐231) per well were seeded on 96‐cell plates and transfected with sh‐NC, sh‐KIF11. After transfection, cancer cells were treated with 0, 0.01, 0.1, 1, 10, and 100 µM of adriamycin for 48 hours. Then, the cytotoxicity was measured by CCK8 assay. Shown were the representative results (mean ± standard deviation) of three independent experiments. CCK8, Cell Counting Kit 8; CI, confidence interval; ER, estrogen receptor; HR, hazard ratio; KIF11, kinesin family member 11; mRNA, messenger RNA; OS, overall survival; sh‐NC, short hairpin negative control; TCGA‐BRCA, The Cancer Genome Atlas‐Breast Invasive Carcinoma; TNM, tumor, node, metastasis [Color figure can be viewed at wileyonlinelibrary.com]
3.5. Screening of KIF11‐targeting miR‐30a
miRNA is an essential transcriptional regulator involved in the various cancerous process. Here, we screened out eligible target miRNAs using both predicting KIF11‐related miRNAs in microRNA.org and correlation analysis for KIF11 and miRNAs in the GSE22220 dataset (Figure 5A), to validate in cell culture study and to conduct clinical relevance analysis. Hsa‐miR‐30a expression was negatively correlated with KIF11 mRNA expression (Figure 5B). The sequence of hsa‐miR‐30a target KIF11 was shown in Figure 5C. miR‐30a transfection inhibited KIF11 mRNA and protein expression (Figure 5D, left), luciferase assay further verified inhibited effects of miR‐30a to KIF11 (Figure 5D, right). Analysis results based on the GSE22220 set showed that expression levels of hsa‐miR‐30a were significantly positively correlated with disease‐relapse‐free survival of breast cancer (Figure 5E). The prognostic performance of KIF11 and miR‐30a could be compared with age, tumor size, and grade. KIF11 and hsa‐miR‐30a had better prognostic capabilities than lymph node involvement (Figure 5F). The above findings suggest that KIF11 and miR‐30a could serve as a prognostic biomarker to predict poor outcome in breast cancers, and miR‐30a in breast cancer could suppress the expression of KIF11.
Figure 5.
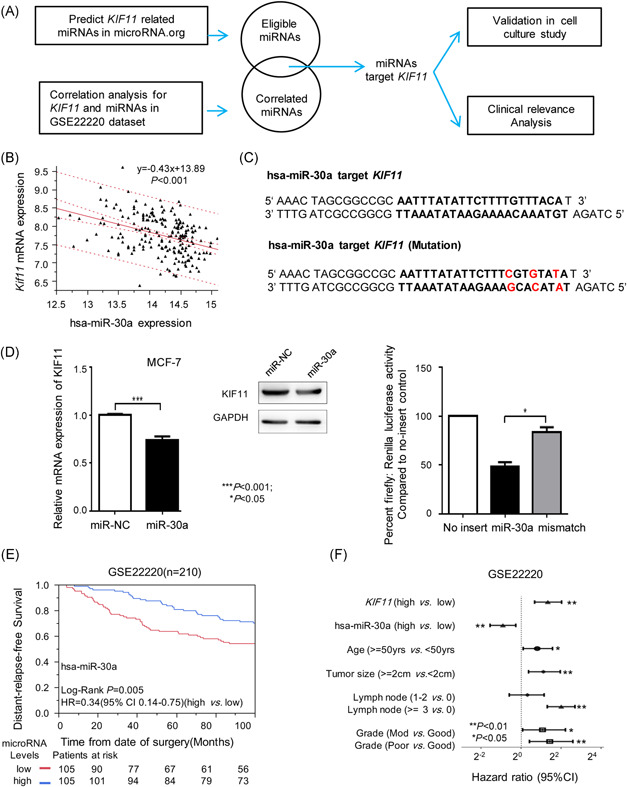
Correlated analysis of KIF11 and hsa‐miR‐30a. A, The flowchart of the analysis and function validation about KIF11‐related miRNAs. B, Linear regression analysis between KIF11 mRNA expression and hsa‐miR‐30a expression in the GSE22220 dataset. C, The sequence of hsa‐miR‐30a target KIF11 was shown. D, Relative mRNA expression and protein expression of KIF11 after miR‐30a transfection (left). Luciferase assay (right). E, Kaplan‐Meier distant‐relapse‐free survival analysis of hsa‐miR‐30a in the GSE22220. F, Comparison of prognostic performance of KIF11 and hsa‐miR‐30a with tumor size, lymph node involvement, and Elston grade in breast cancer. CI, confidence interval; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; HR, hazard ratio; miRNA, microRNAs; mRNA, messenger RNA; KIF11, kinesin family member 11. *P < .05, **P < .01, ***P < .001 [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
In this study, we demonstrated that KIF11 is highly expressed in breast cancer cell lines and closely related to poor differentiation and aggressive phenotypes in breast cancer patients. By GSEA analysis, we verified that KIF11 enrichment occurs in breast cancer. Our data showed that KIF11 mRNA levels were consistent with patient prognosis through pooled dataset analysis. Moreover, our data further indicate that high expression of KIF11 may compromise chemo‐ and radiotherapy efficiency.
Previous studies have identified the potential role of kinesins in the breast cancer cell. Elevated expression levels of kinesins KIF14, KIF4A, KIF20A, and KIF23, KIF2C, and KIFC1 have been reported in breast cancer cell lines, other kinesins KIF10, KIF18A, and KIF15 have been linked to prognostic indicators in breast cancer patients. 59 One study found that KIF26B is independently linked to the prognostic outcome in breast cancer. 60 KIF11 was found overexpressed in human pancreatic cancer samples and promoted cell proliferation in an ATPase activity‐dependent manner, leading to the accumulation of polyploid cells. 17 Another research showed that overexpression of KIF11 was related to poor differentiation of bladder cancer. 18 Inhibition of KIF11 with a highly specific small‐molecule inhibitor has been proven to delay the growth of commonly treatment‐resistant glioblastoma tumor cells and to hamper tumor initiation. 20 KIF11 increasingly expressed in high stage and malignant tumor cells. 23 KIF11 expression has also predicted treatment response with platinum chemotherapy in patients with advanced NSCLC. Here, downregulation of KIF11 suppresses cell proliferation, invasion, and chemoresistance in MCF‐7 and MDA‐MB‐231 cells.
Furthermore, our validation in vitro study and clinical relevance analysis suggest that miR‐30a could be a negative regulator of KIF11 in breast cancer development. miRNAs have proven to be a distinguished group of ribonucleotides that play a crucial role in human cancer. miRNA expression profiles have been proposed as a method to classify tumor stages and prognosis. 61 , 62 , 63 , 64 A recent review highlighted the predictive value of miRNAs in breast cancer patients before chemotherapy, radiotherapy, or surgical intervention. 65 Researchers have discussed the potential role of miRNAs in breast cancer management, particularly in improving current prognosis and achieving individualized cancer care. On one side, miRNAs can function as oncogenes via negative inhibition of tumor suppressor genes; on the other side, induction of tumor suppressor miRNAs may result in the prevention or treatment of breast tumors. Further investigation of the functional roles of miRNAs would help us in gathering more knowledge of carcinogenesis, tumor biomarkers, and therapeutic drug discovery. 66
Expression of KIF11 and miR‐30a is associated with the development and outcome of breast cancer. First, in vitro assays with KIF11 knockdown significantly inhibited cell proliferation and invasion. Second, the OS and DFS in breast cancer databases were significantly lower in high‐KIF11 breast cancer than in low‐KIF11 breast cancer. Third, the expression between KIF11 and miR‐30a shared a negative correlation (Figure 5B). Given these findings, we propose that KIF11 contributes to the development of breast cancer, and miR‐30a suppresses the KIF11 expression. Undoubtedly, extensive investigations are required to illuminate the elaborate mechanism of KIF11 in the development and regulation of breast cancer, and in‐depth studies are also needed to uncover the interactions between KIF11 and miR‐30a.
Taken together, we demonstrate a critical role of KIF11 in promoting invasion and predicting poor prognosis in breast cancer patients. The levels between KIF11 and miR‐30a present a significantly negative correlation in breast cancer databases. Our findings highlight that miR‐30a and KIF11 could be employed as promising prognostic biomarkers and therapeutic targets for breast cancers.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
This study was supported by an unrestricted starting package from The First Affiliated Hospital of Soochow University to Yiqiang Wang and a grant from the National Natural Science Foundation of China (81600076) to Dandan Lin.
Wang B, Yu J, Sun Z, et al. Kinesin family member 11 is a potential therapeutic target and is suppressed by microRNA‐30a in breast cancer. Molecular Carcinogenesis. 2020;59:908–922. 10.1002/mc.23203
Benfang Wang, Jianjiang Yu, and Zhenjiang Sun contributed equally to this study.
Contributor Information
Benfang Wang, Email: benfangwang@163.com.
Xiyong Liu, Email: xiyongliu@sacfamerica.org.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 2. McPherson K, Steel CM, Dixon JM. ABC of breast diseases. Breast cancer‐epidemiology, risk factors, and genetics. BMJ. 2000;321:624‐628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DeSantis CE, Siegel RL, Sauer AG, et al. Cancer statistics for African Americans, 2016: progress and opportunities in reducing racial disparities. CA Cancer J Clin. 2016;66:290‐308. [DOI] [PubMed] [Google Scholar]
- 4. Prat A, Lluch A, Albanell J, et al. Predicting response and survival in chemotherapy‐treated triple‐negative breast cancer. Br J Cancer. 2014;111:1532‐1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shin VY, Siu JM, Cheuk I, Ng EKO, Kwong A. Circulating cell‐free miRNAs as biomarker for triple‐negative breast cancer. Br J Cancer. 2015;112:1751‐1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woolston C. Breast cancer. Nature. 2015;527:S101. [DOI] [PubMed] [Google Scholar]
- 7. Yaffe MJ, Jong RA. Adjunctive ultrasonography in breast cancer screening. Lancet. 2016;387:313‐314. [DOI] [PubMed] [Google Scholar]
- 8. Braunstein LZ, Taghian AG, Niemierko A, et al. Breast‐cancer subtype, age, and lymph node status as predictors of local recurrence following breast‐conserving therapy. Breast Cancer Res Treat. 2017;161:173‐179. [DOI] [PubMed] [Google Scholar]
- 9. Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus‐end‐directed microtubule motor. Nature. 1992;359:540‐543. [DOI] [PubMed] [Google Scholar]
- 10. Mishima M, Pavicic V, Grüneberg U, Nigg EA, Glotzer M. Cell cycle regulation of central spindle assembly. Nature. 2004;430:908‐913. [DOI] [PubMed] [Google Scholar]
- 11. Kashina AS, Rogers GC, Scholey JM. The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim Biophys Acta. 1997;1357:257‐271. [DOI] [PubMed] [Google Scholar]
- 12. Valentine MT, Gilbert SP. To step or not to step? How biochemistry and mechanics influence processivity in kinesin and Eg5. Curr Opin Cell Biol. 2007;19:75‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114‐118. [DOI] [PubMed] [Google Scholar]
- 14. Hansen GM, Justice MJ. Activation of Hex and mEg5 by retroviral insertion may contribute to mouse B‐cell leukemia. Oncogene. 1999;18:6531‐6539. [DOI] [PubMed] [Google Scholar]
- 15. Nowicki MO, Pawlowski P, Fischer T, Hess G, Pawlowski T, Skorski T. Chronic myelogenous leukemia molecular signature. Oncogene. 2003;22:3952‐3963. [DOI] [PubMed] [Google Scholar]
- 16. Castillo A, Morse HC 3rd, Godfrey VL, Naeem R, Justice MJ. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138‐10147. [DOI] [PubMed] [Google Scholar]
- 17. Liu M, Wang X, Yang Y, et al. Ectopic expression of the microtubule‐dependent motor protein Eg5 promotes pancreatic tumourigenesis. J Pathol. 2010;221:221‐228. [DOI] [PubMed] [Google Scholar]
- 18. Ding S, Xing N, Lu J, et al. Overexpression of Eg5 predicts unfavorable prognosis in non‐muscle invasive bladder urothelial carcinoma. Int J Urol. 2011;18:432‐438. [DOI] [PubMed] [Google Scholar]
- 19. Saijo T, Ishii G, Ochiai A, et al. Eg5 expression is closely correlated with the response of advanced non‐small cell lung cancer to antimitotic agents combined with platinum chemotherapy. Lung Cancer. 2006;54:217‐225. [DOI] [PubMed] [Google Scholar]
- 20. Venere M, Horbinski C, Crish JF, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self‐renewal in glioblastoma. Sci Transl Med. 2015;7:304ra143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797‐806. [DOI] [PubMed] [Google Scholar]
- 22. Dumontet C, Jordan MA. Microtubule‐binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov. 2010;9:790‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou J, Chen WR, Yang LC, et al. KIF11 Functions as an oncogene and is associated with poor outcomes from breast cancer. Cancer Res Treat. 2019;51:1207‐1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith B, Agarwal P, Bhowmick NA. MicroRNA applications for prostate, ovarian and breast cancer in the era of precision medicine. Endocr Relat Cancer. 2017;24:R157‐R172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015;25:137‐147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Evans‐Knowell A, LaRue AC, Findlay VJ. MicroRNAs and their impact on breast cancer, the tumor microenvironment, and disparities. Adv Cancer Res. 2017;133:51‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA‐21 and microRNA‐205 overexpression in non‐small cell lung cancer by quantitative real‐time RT‐PCR. Clin Chem. 2008;54:1696‐1704. [DOI] [PubMed] [Google Scholar]
- 28. Shi M, Guo N. MicroRNA expression and its implications for the diagnosis and therapeutic strategies of breast cancer. Cancer Treat Rev. 2009;35:328‐334. [DOI] [PubMed] [Google Scholar]
- 29. Adams BD, Wali VB, Cheng CJ, et al. miR‐34a silences c‐SRC to attenuate tumor growth in triple‐negative breast cancer. Cancer Res. 2016;76:927‐939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodriguez‐Barrueco R, Nekritz EA, Bertucci F, et al. miR‐424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31:553‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhan MN, Yu XT, Tang J, et al. MicroRNA‐494 inhibits breast cancer progression by directly targeting PAK1. Cell Death Dis. 2017;8:e2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bhattacharyya M, Nath J, Bandyopadhyay S. MicroRNA signatures highlight new breast cancer subtypes. Gene. 2015;556:192‐198. [DOI] [PubMed] [Google Scholar]
- 34. Zhu J, Zeng Y, Li W, et al. CD73/NT5E is a target of miR‐30a‐5p and plays an important role in the pathogenesis of non‐small cell lung cancer. Mol Cancer. 2017;16:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu M, Huang F, Zhang D, et al. Heterochromatin protein HP1gamma promotes colorectal cancer progression and is regulated by miR‐30a . Cancer Res. 2015;75:4593‐4604. [DOI] [PubMed] [Google Scholar]
- 36. Liu X, Ji Q, Zhang C, et al. miR‐30a acts as a tumor suppressor by double‐targeting COX‐2 and BCL9 in H. pylori gastric cancer models. Sci Rep. 2017;7:7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L, Kang L, Zhao W, et al. miR‐30a‐5p suppresses breast tumor growth and metastasis through inhibition of LDHA‐mediated Warburg effect. Cancer Lett. 2017;400:89‐98. [DOI] [PubMed] [Google Scholar]
- 38. Zhu J, Zeng Y, Xu C, et al. Expression profile analysis of microRNAs and downregulated miR‐486‐5p and miR‐30a‐5p in non‐small cell lung cancer. Oncol Rep. 2015;34:1779‐1786. [DOI] [PubMed] [Google Scholar]
- 39. Liu X, Lai L, Wang X, et al. Ribonucleotide reductase small subunit M2B prognoses better survival in colorectal cancer. Cancer Res. 2011;71:3202‐3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76‐gene prognostic signature for node‐negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207‐3214. [DOI] [PubMed] [Google Scholar]
- 41. Smeds J, Miller LD, Bjöhle J, et al. Gene profile and response to treatment. Ann Oncol. 2005;16(suppl 2):ii195‐ii202. [DOI] [PubMed] [Google Scholar]
- 42. Wang Y, Klijn JG, Zhang Y, et al. Gene‐expression profiles to predict distant metastasis of lymph‐node‐negative primary breast cancer. Lancet. 2005;365:671‐679. [DOI] [PubMed] [Google Scholar]
- 43. Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292‐10301. [DOI] [PubMed] [Google Scholar]
- 44. Hennessy BT, Gonzalez‐Angulo AM, Stemke‐Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial‐to‐mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116‐4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Muranen TA, Greco D, Fagerholm R, et al. Breast tumors from CHEK2 1100delC‐mutation carriers: genomic landscape and clinical implications. Breast Cancer Res. 2011;13:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hatzis C. A genomic predictor of response and survival following taxane‐anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873‐1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Azim HA Jr, Brohee S, Peccatori FA, et al. Biology of breast cancer during pregnancy using genomic profiling. Endocr Relat Cancer. 2014;21:545‐554. [DOI] [PubMed] [Google Scholar]
- 48. Jézéquel P, Loussouarn D, Guérin‐Charbonnel C, et al. Gene‐expression molecular subtyping of triple‐negative breast cancer tumours: importance of immune response. Breast Cancer Res. 2015;17:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Buffa FM, Camps C, Winchester L, et al. microRNA‐associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635‐5645. [DOI] [PubMed] [Google Scholar]
- 50. Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353‐357. [DOI] [PubMed] [Google Scholar]
- 51. Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550‐13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schmidt M, Bohm D, von Torne C, et al. The humoral immune system has a key prognostic impact in node‐negative breast cancer. Cancer Res. 2008;68:5405‐5413. [DOI] [PubMed] [Google Scholar]
- 53. Bos PD, Zhang XHF, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Esserman LJ, Berry DA, Cheang MC, et al. Chemotherapy response and recurrence‐free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I‐SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res Treat. 2012;132:1049‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loi S, Haibe‐Kains B, Majjaj S, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor‐positive breast cancer. Proc Natl Acad Sci USA. 2010;107:10208‐10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van de Vijver MJ, He YD, van't Veer LJ, et al. A gene‐expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999‐2009. [DOI] [PubMed] [Google Scholar]
- 57. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492‐2502. [DOI] [PubMed] [Google Scholar]
- 59. Wolter P, Hanselmann S, Pattschull G, Schruf E, Gaubatz S. Central spindle proteins and mitotic kinesins are direct transcriptional targets of MuvB, B‐MYB and FOXM1 in breast cancer cell lines and are potential targets for therapy. Oncotarget. 2017;8:11160‐11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang Q, Zhao ZB, Wang G, et al. High expression of KIF26B in breast cancer associates with poor prognosis. PLoS One. 2013;8:e61640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van Schooneveld E, Wouters MC, Van der Auwera I, et al. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012;14:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hofsli E, Sjursen W, Prestvik WS, et al. Identification of serum microRNA profiles in colon cancer. Br J Cancer. 2013;108:1712‐1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kanaan Z, Roberts H, Eichenberger MR, et al. A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg. 2013;258:400‐408. [DOI] [PubMed] [Google Scholar]
- 64. Zheng G, Du L, Yang X, et al. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014;111:1985‐1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34‐49. [DOI] [PubMed] [Google Scholar]
- 66. Lowery AJ, Miller N, McNeill RE, Kerin MJ. MicroRNAs as prognostic indicators and therapeutic targets: potential effect on breast cancer management. Clin Cancer Res. 2008;14:360‐365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


