Summary
Compared to higher latitudes, tropical heterotrophic bacteria may be less responsive to warming because of strong bottom‐up control. In order to separate both drivers, we determined the growth responses of bacterial physiological groups to temperature after adding dissolved organic matter (DOM) from mangroves, seagrasses and glucose to natural seawater from the Great Barrier Reef. Low (LNA) and high (HNA) nucleic acid content, membrane‐intact (Live) and membrane‐damaged (Dead) plus actively respiring (CTC+) cells were monitored for 4 days. Specific growth rates of the whole community were significantly higher (1.9 day‐1) in the mangrove treatment relative to the rest (0.2–0.4 day‐1) at in situ temperature and their temperature dependence, estimated as activation energy, was also consistently higher. Strong bottom‐up control was suggested in the other treatments. Cell size depended more on DOM than temperature. Mangrove DOM resulted in significantly higher contributions of Live, HNA and CTC+ cells to total abundance, while the seagrass leachate reduced Live cells below 50%. Warming significantly decreased Live and CTC+ cells contributions in most treatments. Our results suggest that only in the presence of highly labile compounds, such as mangroves DOM, can we anticipate increases in heterotrophic bacteria biomass in response to warming in tropical regions.
Introduction
Tropical marine ecosystems naturally have warm surface temperatures year‐round. In the open ocean, high temperatures promote stratification, preventing the input of new nutrients, such that most low‐latitude waters show the oligotrophic characteristics of the typical tropical structure (TTS, Herbland and Voituriez, 1979), i.e. a warm, nutrient‐depleted upper mixed layer separated from the cooler waters underneath by a marked pycnocline (Bock et al., 2018). These conditions favour the prevalence of small plankton at the expense of larger organisms (Platt et al., 1983; Agawin et al., 2000). Prokaryotes, both autotrophic (cyanobacteria from the genera Synechococcus and Prochlorococcus) and heterotrophic (bacteria and archaea), are responsible for a large share of carbon biomass and fluxes in tropical regions (Flombaum et al., 2013; Richardson, 2019). In coastal waters, temperatures are generally further elevated compared with open waters, which combined with relatively constant supplies of nutrients and solar radiation (Nittrouer et al., 1995), may moderately increase primary production. Similar to other tropical areas, picophytoplankton (principally cyanobacteria) dominate the Great Barrier Reef (GBR) shelf waters in Australia (Crosbie and Furnas, 2001), accounting for 37%–99% of pelagic primary production (Furnas and Mitchell, 1987), and are ultimately the largest source of autochthonous DOM. However, besides being locally produced, DOM can also be advected. The most important allochthonous DOM sources in the GBR are mangroves and seagrasses (Maher and Eyre, 2010; Alongi and Mukhopadhyay, 2015). Changes in the relative importance of phytoplankton and these macrophytes as DOM sources (Stepanauskas et al., 2000; Sawstrom et al., 2016) will impact both the quantity and quality of carbon flowing through the microbial food web (Lønborg et al., 2018). Although temperatures are already elevated in the GBR, they will continue to rise (IPCC, 2013). Processing of DOM by heterotrophic prokaryotes is affected by temperature as shown in temperate and polar regions (e.g. Apple et al., 2006; Kirchman et al., 2009; Huete‐Stauffer et al., 2015). Yet, the combined impact of different DOM sources and temperature on tropical heterotrophic bacterioplankton has been rarely investigated.
The characteristics of heterotrophic prokaryotes related to phylogeny (i.e. relative nucleic acid content) and physiology (i.e. membrane integrity, respiration) were collectively termed as ‘physiological structure’ by del Giorgio and Gasol (2008). Flow cytometry has become the standard technique to assess the physiological structure of marine planktonic microbes across different environments (Gasol and Morán, 2015). Separated by their relative nucleic acid content and with a broadly distinct taxonomic composition (Schattenhofer et al., 2011; Vila‐Costa et al., 2012; Song et al., 2019), the universally distributed groups of high and low nucleic acid content (HNA and LNA) bacteria (Gasol et al., 1999; Bouvier et al., 2007) respond differently to most environmental factors, with LNA cells being generally less active in substrate incorporation and characterized by lower growth rates than HNA cells in natural conditions (e.g. Lebaron et al., 2001; Morán et al., 2007; Wang et al., 2009), especially in oligotrophic environments (Longnecker et al. 2005). Another useful distinction is the status of the bacterial membrane, either intact or damaged, which has been typically considered as characteristic of healthy (Live) and unhealthy (Dead) cells respectively (Grégori et al., 2001). The abundance of Dead cells rapidly increased in response to stressors such as high temperatures, elevated UV light or substrate limitation (Falcioni et al., 2008). Among the stains that target the electron transport chain, 5‐cyano‐2,3‐ditolyl tetrazolium chloride (CTC) has been extensively used to identify the actively respiring cells. Within the Live cell group, CTC positive (CTC+) cells represent the most active members of the community, frequently in both production and respiration (Longnecker et al., 2005; Gasol and Arístegui, 2007; Morán and Calvo‐Díaz, 2009). Most of the studies looking at the response of heterotrophic prokaryotes to environmental drivers have done so at the bulk level (i.e. total abundances), with only a few considering the physiological structure from an individual perspective (Gasol et al., 2009; Morán and Calvo‐Díaz, 2009; Morán et al., 2011; Huete‐Stauffer et al., 2015). However, rarely have the above‐mentioned physiological groups been jointly assessed in tropical ecosystems (Silva et al., 2019).
Recently, an extensive survey across the subtropical and tropical ocean has suggested that low latitude heterotrophic bacteria and archaea have a limited capability to respond positively to higher temperatures, because of either strong limitation by available substrate (i.e. bottom‐up control) or high mortality rates caused by protistan grazers or viruses (i.e. top‐down control, Morán et al., 2017). However, controlled laboratory experiments are needed to tease apart the specific effect of the different controls in these under‐sampled, yet globally important, low‐latitude regions. In order to advance our understanding of the role of bottom‐up and temperature controls, we assessed here the short‐term dynamics of LNA and HNA, Live and Dead and CTC+ cells in incubations of surface coastal water from the GBR at a site off Cape Cleveland (19.2°S, 147.1°E), Australia. These incubations included a control without DOM addition and others that were enriched with realistic increases in DOM (i.e. 40 μmol C L−1, the approximate seasonal build‐up of DOC, Lønborg et al., 2017) derived from mangrove and seagrass leaves, as well as glucose as an easily degradable compound for comparison. Experimental incubations were performed at three different temperatures (in situ plus 3°C below and above the ambient value in order to encompass 6°C without causing strong disturbance) aimed at comprehensively assessing the response of planktonic heterotrophic bacteria specific growth rates and carrying capacities at the individual level to allochthonous DOM and warming. The experimental response to temperature was described by the activation energies (E) of the metabolic theory of ecology (MTE) central equation (Brown et al., 2004), as in previous work (Huete‐Stauffer et al., 2015; Lønborg et al., 2016; Morán et al., 2018). Our hypotheses were (i) that heterotrophic bacteria growth would be enhanced by temperature in DOM‐rich conditions and (ii) that the contribution of the different physiological groups would change coherently with temperature and DOM. This study complements previous work by Baltar et al. (2017) and Lønborg et al. (2019) assessing, respectively, the proportion of cell‐free extracellular enzymatic activity and microbial carbon cycling in the same experiments.
Results
Physiological structure and cell size of heterotrophic bacteria
Total heterotrophic bacterial abundance (i.e. the sum of LNA and HNA cells) consistently increased from 1.2 to 1.4 × 105 cells mL−1 at day 0 until day 3, albeit with differences between treatments (Fig. 1), resulting in estimated specific growth rates at in situ temperature that ranged from 0.2 (Glucose) to 1.9 day‐1 (Mangrove). Total carrying capacity (i.e. the maximum abundance of LNA + HNA cells) was one order of magnitude higher in the Mangrove (3.0 × 106 cells mL−1) than in the other treatments (2.3–3.4 × 105 cells mL−1). Responses to DOM treatments were largely due to changes in HNA cells, while LNA cells were less responsive (Control and Glucose) or even decreased steadily during the course of the incubation (Mangrove and Seagrass; Table 1). Consequently, %HNA values increased with time in all DOM treatments and temperatures (Supporting Information Fig. S1) from the initial 35%–39% to mean values of 56% (Control), 53% (Glucose), 90% (Mangrove) and 67% (Seagrass) at in situ temperature (Fig. 2). Carrying capacities of LNA cells varied between 104 and 105 cells mL−1 while those of HNA ranged from 105 to 106 (1.1–29.6 × 105 cells mL−1; Table 1). The slight decline in heterotrophic bacteria abundance in the last day of all experiments (Fig. 1) was not caused by increased viral mortality since the abundance of viruses did not change much during the incubations (Supporting Information Fig. S2).
Figure 1.
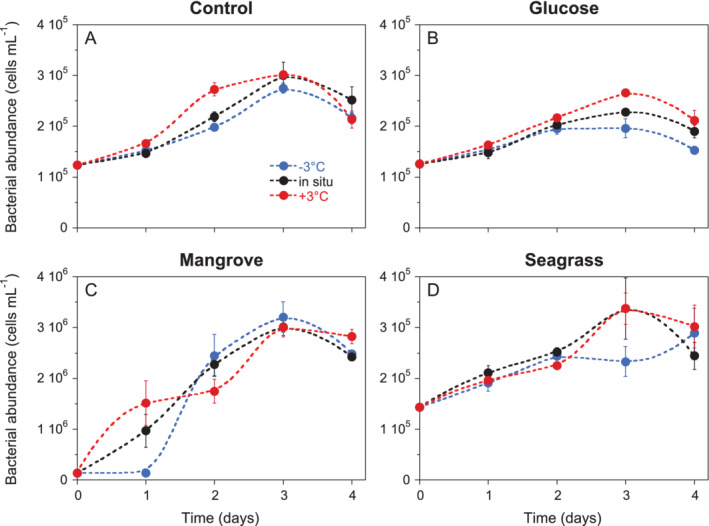
Dynamics of mean total [sum of low (LNA) and high (HNA) nucleic acid content bacteria] bacterial abundance during the 4 days incubations in the (A) Control, (B) Glucose, (C) Mangrove and (D) Seagrass treatments. Error bars represent standard deviations of 3 replicates. Dashed smooth fitting joins treatment points for clarity. Specific growth rate calculations included at least 3 sampling points (i.e. from day 0 to day 2) except in the Mangrove treatment at in situ and +3°C temperatures. Control at −3°C and in situ and Seagrass at in situ and +3°C temperatures included also day 3 while Seagrass at −3°C included the 4 days.
Table 1.
Mean ± SE specific growth rates (μ, day‐1) and mean ± SD carrying capacities (K, cells mL−1) in the experiments conducted at in situ temperature; n = 3.
| Treatment | Live | LNA | HNA | CTC+ | |
|---|---|---|---|---|---|
| Control | μ | 0.5 ± 0.0 | 0.1 ± 0.0 | 0.5 ± 0.0 | 1.0 ± 0.5 |
| K | 2.3 ± 0.3 × 105 | 1.1 ± 0.1 × 105 | 1.9 ± 0.1 × 105 | 0.3 ± 0.1 × 105 | |
| Glucose | μ | 0.3 ± 0.1 | 0.2 ± 0.1 | 0.5 ± 0.1 | 1.3 ± 0.4 |
| K | 1.5 ± 0.0 × 105 | 1.0 ± 0.1 × 105 | 1.3 ± 0.0 × 105 | 0.7 ± 0.0 × 105 | |
| Mangrove | μ | 1.7 ± 0.0 | −0.5 ± 0.2 | 2.9 ± 0.0 | 5.6 ± 0.0 |
| K | 17.2 ± 1.4 × 105 | 1.1 ± 1.0 × 105 | 29.6 ± 1.5 × 105 | 19.0 ± 2.1 × 105 | |
| Seagrass | μ | 0.3 ± 0.1 | −0.1 ± 0.1 | 0.5 ± 0.1 | 1.3 ± 0.5 |
| K | 1.5 ± 0.2 × 105 | 0.9 ± 0.1 × 105 a | 2.5 ± 0.5 × 105 | 0.6 ± 0.0 × 105 | |
The highest abundance was the initial.
Figure 2.
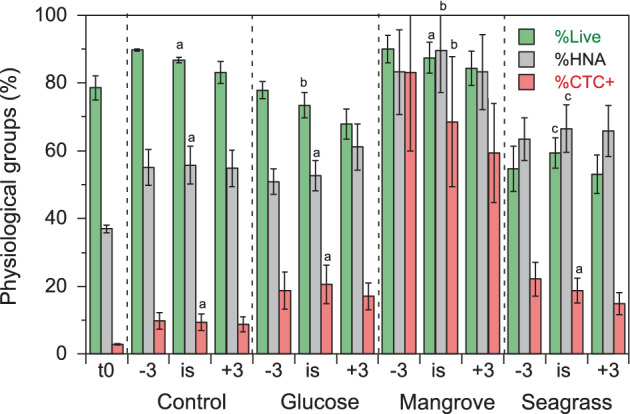
Mean percent contributions of Live, HNA and CTC+ bacteria in the different DOM and temperature treatments (n = 12) plus at the beginning of the experiments (t0, n = 3). −3, 3°C below in situ temperature; is, in situ temperature; +3, 3°C above in situ temperature. Different letters represent significant differences between DOM treatments at in situ temperature (ANOVA, P < 0.05, Tukey–Kramer HSD). Error bars represent standard deviations.
Cell sizes measured as biovolume increased noticeably over the course of the incubations except in the Control treatment (Fig. 3). In the Mangrove treatment, the cell size at in situ temperature averaged for the 4 days (n = 12) was 0.134 μm3, 1.8‐ to 3.2‐fold larger than the mean values in the other three treatments (Control: 0.042 μm3, Glucose: 0.068 μm3, Seagrass: 0.073 μm3). These differences were significant regardless of the temperature (one‐way ANOVA, P < 0.001, Tukey–Kramer HSD, Mangrove> Glucose = Seagrass > Control). Maxima were found between day 1 and day 2 for all treatments but glucose, in which cells increased in size until day 4 (Fig. 3).
Figure 3.
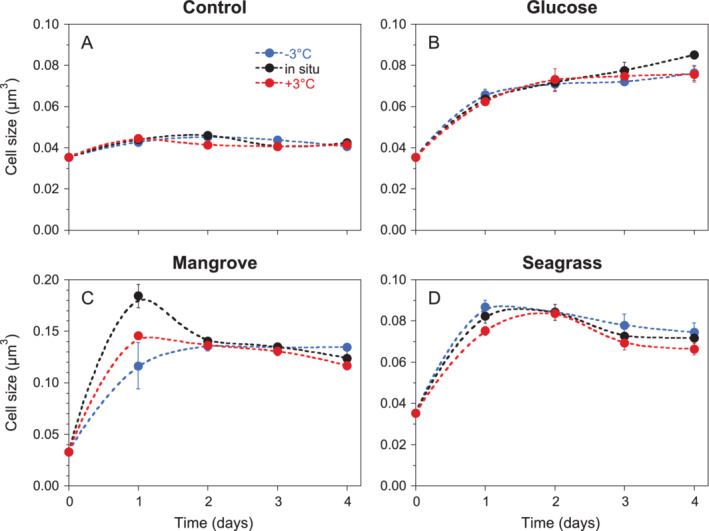
Dynamics of mean bacterial cell size during the 4 days incubations in the (A) Control, (B) Glucose, (C) Mangrove and (D) Seagrass treatments. Error bars and dashed lines are shown as in Fig. 1. Notice the different Y‐axis scale in C.
Both Live (initial abundance 5.9–7.0 × 104 cells mL−1) and Dead (6.1–17.1 × 103 cells mL−1) absolute cell abundance increased with time, although the patterns differed substantially among DOM treatments, affecting the relative contribution of Live cells to the total (%Live). %Live remained rather stable in the Control treatment (86%–90% except at −3°C), while it clearly decreased in the other experiments (Supporting Information Fig. S3). In Glucose and Seagrass, %Live increased again slightly in the last 2 days. Overall, the most conspicuous mortality was observed in Seagrass, where almost half of the population was made up by Dead cells at day 2, followed by Glucose (averaged %Live, Fig. 2; one‐way ANOVA, P < 0.001, Tukey–Kramer HSD, Control = Mangrove > Glucose > Seagrass). The specific growth rates of Live cells lay in between those for LNA and HNA bacteria, while their corresponding carrying capacities were lower than the total (i.e. LNA + HNA), indicating the presence of many cells with damaged membranes (Table 1).
The abundance of CTC+ cells at the beginning of the experiments was as expected low (3.3–4.5 × 103 cells mL−1), contributing little to the total community (%CTC+ ranged between 2.7% and 3.4%). Actively respiring cells increased in all treatments with higher specific growth rates than any other physiological group (1.0–5.6 day‐1 at in situ temperature, paired t‐test with total bacterial abundance specific growth rates, P = 0.0005, n = 12), thus resulting in higher contributions with time (Supporting Information Fig. S4). Averaged %CTC+ during the incubation ranged from 9% to 10% in the Control to most of the cells actively respiring in the Mangrove treatment (59%–83%; Fig. 2; one‐way ANOVA, P < 0.001, Tukey–Kramer HSD, Control < Glucose = Seagrass << Mangrove). Carrying capacities of CTC+ cells varied accordingly, from 0.3 to 19.0 × 105 cells mL−1 at in situ temperature (Table 1).
Both the specific growth rates and carrying capacities were significantly higher in the Mangrove treatment when pooling the values of the most active physiological groups (i.e. Live, HNA and CTC+ cells) at the three temperatures (one‐way ANOVA P = 0.031 and P < 0.001, respectively, Tukey–Kramer HSD, Mangrove > Control = Glucose = Seagrass, n = 36).
Temperature responses
The activation energy of the specific growth rate of the total assemblage ranged from 0.28 to 0.66 eV (Fig. 4). Except for the slightly negative values of Live cells in Glucose, CTC+ cells in Seagrass and the strong ones of LNA cells in Mangrove and Seagrass (see below), the activation energies of all physiological groups were positive, ranging from 0.08 to 0.72 eV (Supporting Information Fig. S5A). Strictly speaking, an activation energy cannot be below 0, so the negative values simply mean a decrease rather than an increase in specific growth rates with warming. With regard to LNA cells, their negative specific growth rates in Mangrove and Seagrass became more negative with warming yielding equivalent E values of ca. −1 eV (see a more detailed discussion in the study by Morán et al., 2018). Similar to the previous consideration, Dead cells cannot really have an activation energy, but we compared their apparent E values in order to assess their dynamics in response to temperature. The absolute abundance of Dead cells also increased markedly with temperature, except in the Seagrass treatment. Interestingly, the activation energies of LNA and Dead cells were significantly correlated when pooling the data from the four DOM treatments (r = 0.95, P = 0.05, n = 4), suggesting that unhealthy cells were made up mostly by the LNA group.
Figure 4.
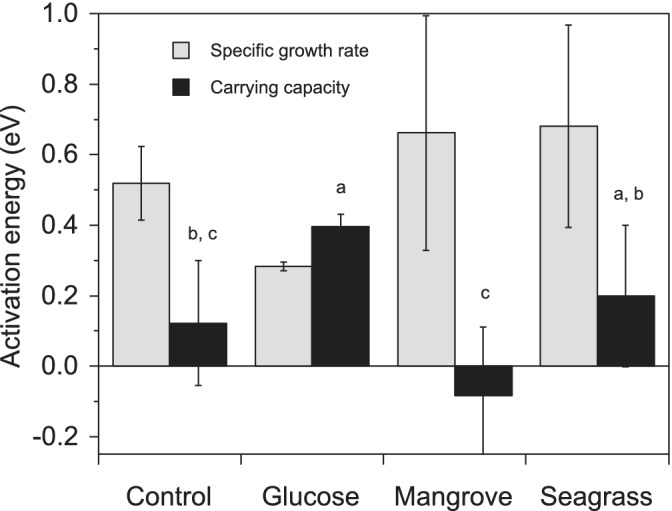
Activation energies of the specific growth rates and carrying capacities of the total bacterial community (sum of LNA and HNA cells). Error bars represent standard errors of the estimates. Different letters represent significant differences between DOM treatments (ANOVA, P < 0.05, Tukey–Kramer HSD).
The carrying capacities of Live, LNA, HNA and CTC+ cells at in situ temperature ranged over 2 orders of magnitude from 104 to 106 cells mL−1 (Table 1). Contrary to the overall positive (with the exception of LNA cells in the Mangrove and Seagrass treatments) temperature response of specific growth rates (Supporting Information Fig. S5A), a varied range of responses was found for the activation energy of carrying capacities (see Fig. 4 for total bacteria): generally positive values (i.e. carrying capacity increased with temperature) were found for total, LNA and HNA cells while those of CTC+ cells were consistently negative (Supporting Information Fig. S5B).
Table 2 summarizes the percent change with increasing temperature of the mean contribution of Live, HNA and CTC+ cells during the incubations already shown in Fig. 2. Both %Live and %CTC+ decreased with experimental warming in all treatments with values ranging from −0.2 to −3.9% °C−1 while %HNA changes did not show any consistent response.
Table 2.
Percent change (± SE) per °C of warming in the mean contribution of Live, HNA and CTC+ cells in the different DOM treatments (n = 3).
| Treatment | %Live (°C) | %HNA (°C) | %CTC+ (°C) |
|---|---|---|---|
| Control | −1.11 ± 0.06% | −0.04 ± 0.16%a | −0.17 ± 0.04% |
| Glucose | −1.66 ± 0.11% | 1.69 ± 0.65%a | −0.29 ± 0.52%a |
| Mangrove | −0.95 ± 0.04% | −0.01 ± 1.23%a | −3.94 ± 0.52% |
| Seagrass | −0.26 ± 1.03%a | 0.41 ± 0.37%a | −1.21 ± 0.06% |
Values were not significantly different from 0.
Differences among DOM treatments were only significant for %CTC+ (ANCOVA. Mangrove < Control = Glucose = Seagrass, P = 0.002–0.006).
Discussion
The response of tropical heterotrophic bacterioplankton to combined DOM additions and warming had not been attempted before this experiment (Lønborg et al., 2019; Baltar et al., 2017). Moreover, the physiological structure sensu del Giorgio and Gasol (2008) of tropical and subtropical assemblages has remained largely unexplored. A few individual reports on the distribution of LNA and HNA bacteria (Andrade et al., 2003; Andrade et al., 2007; Girault et al., 2015; Bock et al., 2018) and on Live and Dead cells (Gasol et al., 2009; Baltar et al., 2012) were recently complemented by a study on the annual variability in the abundance and specific growth rates of the same physiological groups assessed here at an equivalent northern latitude in the coastal Red Sea (Silva et al., 2019). Although experiments using sudden modifications in environmental conditions and monitored for a few days strongly limit our capability to assess future ecosystem responses, they provide a first and valuable step to set up possible directions of change.
Effects of DOM sources
Even if broad flow cytometric groups tell us little about the actual taxonomic diversity of heterotrophic bacteria in our experiments (Schiaffino et al., 2013; García et al., 2015), the corresponding cytograms (data not shown) strongly suggest that different groups of large, copiotrophic bacteria were selected depending on the amended DOM treatment. Our results show that there is a seed bank of pelagic bacteria able to rapidly exploit allochthonous DOM originated from adjacent coastal ecosystems (Alonso‐Sáez et al., 2015; Baltar et al., 2015).
The two clusters of bacteria differentiated by their relative nucleic acid content (LNA and HNA) that occur in virtually every aquatic ecosystem (Gasol et al., 1999; Bouvier et al., 2007; Wang et al., 2009) were also consistently observed in our tropical water experiments. The relative abundance of HNA cells (%HNA) in the inoculum and at the beginning of experiments ranged from 35% to 39%, a typical value for tropical, oligotrophic waters (Mary et al., 2006; Andrade et al., 2007; Gasol et al., 2009; Baltar et al., 2012; Bock et al., 2018). HNA always grew faster than LNA cells, resulting in increases of %HNA (Supporting Information Fig. S1). While LNA grew in the Control and Glucose treatments, though at lower rates than in the Red Sea (0.3–1.1 day‐1; Silva et al., 2019), their abundance decreased steadily in the Mangrove and Seagrass additions. It had been suggested that LNA could outperform HNA bacteria in nutrient‐limited waters in the Celtic Sea (Zubkov et al., 2001), but in our experiments, resource limitation (Lønborg et al., 2017) was alleviated by dilution in the Control treatment plus by the addition of DOM in the remaining experiments. However, this potentially higher nutrient availability did not result in marked increases in specific growth rates, except for the actively respiring cells in the Mangrove treatment. Except in the Control, there were new HNA groups (sometimes very large cells) appearing over the course of the incubation, increasing the mean size of the bacterial community in those treatments (Fig. 3). This led to noticeable increases in biomass, especially marked in the Mangrove treatment. Large increases in the mean size of HNA cells have also been observed in experiments conducted elsewhere (Longnecker et al., 2010; Calleja et al., 2018). Our results therefore suggest that DOM leaking from mangrove leaves might represent a highly labile, palatable substrate for coastal bacterioplankton in the tropics (Kristensen et al., 2008). The sustained increase in the mean size of bacteria in the Glucose treatment (Fig. 3B) could in turn be explained by the reported accumulation of an otherwise easy‐to‐degrade substrate (Thingstad et al., 2005) which, if not used for cell division (Fig. 1B), still has the potential to be transferred up other trophic groups of plankton (Jones et al., 2018). In any case, our results highlight the importance of accounting for cell size when investigating differences in bacterial biomass. For instance, bacterial biomass (see values in the study by Lønborg et al., 2019) in the Glucose treatment was higher than in the Control due to larger cell size (Fig. 3), not due to concomitant changes in bacterial abundance, which overall was lower in the former (Fig. 1). Larger individuals exacerbated the difference in biomass relative to abundance also in the Seagrass treatment.
The responses of Live and Dead cells in our experiments were surprisingly varied. Except in the Mangrove treatment, Live cells specific growth rates were substantially lower than in the tropical coastal waters of the central Red Sea (1–2 day‐1; Silva et al., 2019), indicating strong bottom‐up limitation in the GBR pelagic ecosystem, not overcome by DOM addition and bacterial dilution. The effect of temperature but especially the DOM treatment greatly affected the ‘healthiness’ (i.e. considered here as synonymous of the contribution of membrane‐intact or Live cells to the total pool of cells) of heterotrophic bacteria. Even in the treatment with higher bacterial μ values (Table 1), there was always a fraction of Dead cells, but it was in the Seagrass treatment in which a substantial portion of cells, occasionally most of them (i.e. >50%), were Dead. This finding could have been caused by the presence of damaging secondary compounds (e.g. antibiotics) in the Seagrass treatment (Puglisi et al., 2007). Indeed, the species Halodule uninervis was recently confirmed to produce antibiotics (Supriadi et al., 2016). The amount of labile dissolved compounds consumed in our experiments or bioavailable DOC (BDOC) was the lowest in the Seagrass experiment regardless of the temperature (Lønborg et al., 2019), with almost half the value of the Control (8 vs 14 μmol C L−1 d−1 at in situ temperature, Fig. 5), strongly indicating that this DOC addition did not translate into bacterial uptake over the 4 days incubations. Probably seagrass DOM was detrimental for many pelagic bacteria (which ultimately increased the Dead pool), while the rest of them (the Live ones) could indeed take advantage of the extra DOC and process it.
Of the physiological groups analysed here, the CTC+ cells are arguably responsible for much of the bacterial productivity of marine ecosystems (del Giorgio and Scarborough, 1995; Sherr et al., 1999; Morán and Calvo‐Díaz, 2009). In this regard, a highly significant correlation was found between the mean %CTC+ values shown in Fig. 2 and the corresponding bacterial growth efficiencies reported by Lønborg et al. (2019) in the different DOM and temperature treatments (r = 0.99, P < <0.001, n = 12). Contrary to LNA, HNA and Live, CTC+ cells grew at much higher rates in our experiments (overall range 0.90–5.59 day‐1) than in the coastal Red Sea with natural DOM (0.28–1.85 day‐1; Silva et al., 2019). The highest values recorded in temperate waters (up to more than 2 day‐1) were coincident with phytoplankton blooms or higher substrate availability (Morán et al., 2011; Huete‐Stauffer et al., 2015). The increase in CTC+ specific growth rate values in the Mangrove treatment was even more dramatic than for the other bacterial groups. It is clear that the Mangrove treatment triggered a response not observed in the other DOM additions or in the Control experiment. Limitation by other nutrients, likely P (Lønborg et al., 2019), precluded significant increases in bacterial biomass in the Control, Glucose and Seagrass treatments even if the amount of DOM consumed changed almost sixfold. Figure 5 summarizes this relationship, showing that over that large range of BDOC values (Lønborg et al., 2019), specific growth rates did not vary much and clustered around relatively low values (0.2–0.4 day‐1). A similar relationship was found between BDOC and total bacterial biomass produced in the incubations, with the Mangrove treatment standing out in the responses to DOM addition.
Figure 5.
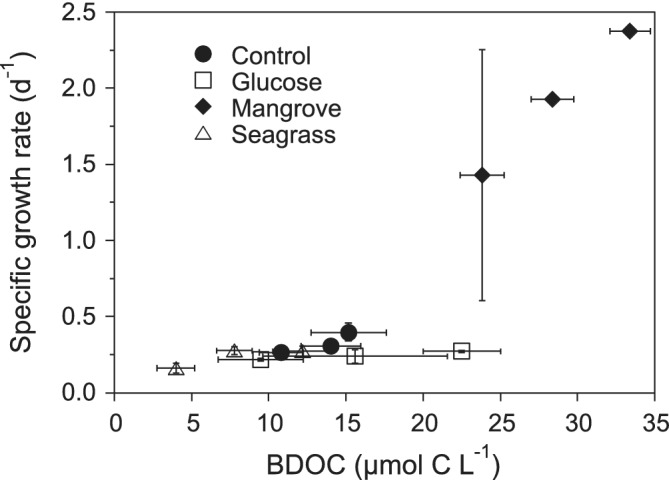
Scatterplot of the mean specific growth rates of the total bacterial community versus BDOC in the different DOM treatments and temperatures. Increasing BDOC values within each treatment match exactly increasing incubation temperatures. Error bars represent standard errors of three replicates.
Responses to temperature
The activation energies were used to describe the temperature response of the different physiological groups as in the study by Huete‐Stauffer et al. (2015). Simply put, a higher E value of the specific growth rate of heterotrophic bacteria when comparing physiological groups or DOM sources means that the response of that specific growth rate to temperature was more marked than the rest. Yet, a higher E value can also mean that the DOM source is more complex and less prone to bacterial uptake (Sierra, 2012; Lønborg et al., 2016). However, although it may require more energy to be processed, that extra energy was indeed provided by the mixture of DOM compounds being added, especially in the Mangrove and Seagrass treatments (Benner et al., 1986; Liu et al., 2018). In agreement with the view that a higher activation energy involves higher processing costs, the higher E values of Mangrove and Seagrass were also associated with a higher production of extracellular enzymes and higher E values of their corresponding activities (Baltar et al., 2017; Lønborg et al., 2019).
All treatments had a higher amount of DOM per bacterium compared to environmental conditions, which would partially alleviate bottom‐up control of bacteria while top‐down control from protistan grazers (reduced by pre‐filtration) and viruses (Supporting Information Fig. S2) was disregardable during the incubation. Given those conditions, control by temperature of the specific growth rates should become more prevalent (Morán et al., 2017). Indeed, for the total community, none of the activation energies were statistically different from 0.65 eV (t‐tests, P > 0.05), the value predicted for heterotrophs by the MTE (Brown et al., 2004; Allen et al., 2005), with virtually identical E values in the Mangrove and Seagrass treatments (0.65 and 0.67 eV, respectively, Fig. 4). In spite of that, the E of the different groups were variable (Supporting Information Fig. S5A), including four negative E values (i.e. decrease in growth rates with warming) if we exclude the E of Dead cells. The negative E for LNA cells deserves further discussion as argued below. With an overall mean value of 0.40 eV, E values were not significantly different among treatments. Surprisingly, bacteria exposed to extra 40 μmol L−1 glucose did not result in higher growth (Fig. 1) or showed a stronger response to temperature (i.e. higher E value) than the Control community (Fig. 4), strongly indicating that glucose alone was not sufficient for the dominant tropical bacteria to build up biomass, as previously shown in incubations in P‐deficient NW Mediterranean waters (Thingstad et al., 1998). It must be noted that the addition of glucose also created a disequilibrium of the C:N:P ratio of DOM. However, this can also be seen as a consequence that this molecule needs much lower energy to be processed by the microbial community (Lønborg et al. 2016 and references therein), contrary to the complex matrix of DOM compounds including humic substances added in the Seagrass and Mangrove treatments. Accordingly, extracellular enzymatic activities in the Glucose treatment, although higher than in the Control, were lower than in Seagrass and Mangrove and they did not show a marked response to temperature either (Baltar et al. 2017; Lønborg et al. 2019). On the contrary, DOM from the Seagrass and Mangrove treatments tended to increase the temperature sensitivity of the entire community although differences were not significant due to the low number of samples (Fig. 4). In any case, as previously argued, the ca. 0.15 eV extra activation energy in the Mangrove treatment would have been provided by highly labile DOM compounds, according to the notably higher amount of biodegradable DOC (Fig. 5). It is also worth mentioning that the Mangrove and Seagrass leachates provided also inorganic nutrients (Table 1 in the study by Lønborg et al., 2019). The activation energy of CTC+ cells specific growth rates was substantially higher in the Mangrove treatment (Supporting Information Fig. S5A), in agreement with their high contribution to total numbers (Fig. 2 and Supporting Information Fig. S4). Interestingly, we found that warming enhanced the loss rate of LNA cells in the two treatments showing negative growth (i.e. decreased rather than increased abundances) of this group, with virtually the same E value in Mangrove and Seagrass (Supporting Information Fig. S5A). The same response has been recently reported for temperate phytoplankton under nutrient limitation (Morán et al., 2018), suggesting that warming would exacerbate any metabolic and ecological processes resulting in either growth or decay of the various bacterial groups initially present.
While the specific growth rates generally (100% for total bacteria, 83% for the most active physiological groups, Figs. 4 and S5A) increased with temperature as expected (i.e. they showed significantly, positive values for the corresponding activation energies, Gillooly et al., 2001; Brown et al., 2004), the carrying capacities showed very different patterns, with the decreasing K values predicted by the MTE (Savage et al., 2004) only found in 25% of the experiments for total bacteria (Mangrove treatment, Fig. 4) and in 58% for live, HNA and CTC+ cells (Supporting Information Fig. S5B). Excluding the Mangrove treatment with maximum abundances exceeding 3 × 106 cells mL−1 (Table 1), total bacterioplankton carrying capacities were around one‐third of the canonical million of cells per mL. The abundance of heterotrophic bacteria in tropical waters may be well below this value (Silva et al., 2019; Al‐Otaibi et al., 2020). However, the 1 × 106 cells mL−1 value was reached in summer and fall in predator‐free incubations (Silva et al., 2019) of unamended Red Sea coastal water equivalent to our Control treatment, strongly suggesting that nutrient limitation was stronger in the GBR. The expectation of decreasing K with temperature is generally fulfilled for photosynthetic organisms (Bernhardt et al., 2018) but seems far from general for heterotrophic bacteria (Huete‐Stauffer et al., 2015; Bernhardt et al., 2018). The negative relationship between the carrying capacity of total bacteria and temperature was only found in the Mangrove treatment (Fig. 4, although not in all physiological groups, Supporting Information Fig. S5B), the only DOM addition consistently showing nutrient‐sufficient conditions for bacterial growth. Furthermore, the activation energy of the carrying capacity of CTC+ cells, usually driving heterotrophic bacterial production (Morán and Calvo‐Díaz, 2009), was markedly negative in that treatment (Supporting Information Fig. S5B), suggesting that no generalizations about the temperature response of the maximum abundances of physiological or phylogenetic groups of heterotrophic bacteria (Arandia‐Gorostidi et al., 2017) are feasible.
The same as for carrying capacities, which failed to show consistent decreases or increases with warming along the different treatments (Fig. 4 and Supporting Information Fig. S5B), cell size responses to temperature were inconclusive. Contrary to recent reports in temperate waters (Daufresne et al., 2009; Morán et al., 2015; Huete‐Stauffer et al., 2016), cell size did not significantly shrink with temperature in our incubations with tropical communities (Fig. 2), except in the Seagrass treatment, in which biovolume decreased 1.4% °C−1. We believe this is connected with the selection of different bacterial taxa after DOM addition (Baltar et al., 2015), which translated into larger cells of probably different phylogenetic groups in the Glucose and especially the Mangrove treatments (Fig. 3).
Regarding the bacterial physiological structure in warmer conditions (Table 2), the contribution of Live cells decreased significantly with warming in all DOM treatments except in the Seagrass treatment, likely because a large fraction of cells were already damaged by the suggested antibiotic property of this leachate. These short‐term experiments should be treated with caution, but it is surprising the agreement in the widespread loss of the healthy (i.e. Live) and more active (i.e. CTC+) cells with temperature. Falcioni et al. (2008) had also observed a decrease in the relative contribution of Live and CTC+ cells with higher temperatures in the Mediterranean. Contrary to Girault et al. (2015), no clear evidence of preference of HNA cells for higher temperatures (except though not significant in Glucose; Supporting Information Fig. S1B) was found in our experiments conducted with tropical water.
Advected DOM from nearby seagrass meadows and mangrove forests supplement natural concentrations in the GBR (Lønborg et al., 2017). We found that increased availability of autochthonous (by dilution) and allochthonous (by addition) DOM did not result in significant increases in the specific growth rates or standing stocks of the physiological groups examined, except in the mangrove extract, suggesting that there were other limiting nutrients or vitamins not provided by these coastal sources. In parallel, our results point out that warming affected negatively the health condition of natural bacterioplankton assemblages in all DOM sources. In conclusion, the expected effects of temperature on the specific growth rates (i.e. increasing) and carrying capacities (i.e. decreasing) of the most active physiological groups (Live, HNA and actively respiring cells) of tropical coastal bacteria in the south Pacific were only documented in the experiments with mangrove derived DOM. Even if the experiments were designed to expose heterotrophic bacteria to enhanced concentrations of a varied repertoire of substrates, our results are supportive of the hypothesis that strong bottom‐up control in low‐latitude waters hamper expectations of increased biomass of planktonic microbes in the coming decades; in other words, it seems that food will matter more than warming in the tropical south Pacific.
Experimental procedures
Experimental design
A detailed description of the study area and experimental setup is provided in the study by Lønborg et al. (2019). Briefly, surface seawater for preparing the DOM additions (with mean initial DOC concentration of 83 μmol L−1) and the bacterial inoculum was collected in October 2016 from a site of ca. 25 m depth off Cape Cleveland, Australia (19° 13′ 06” S, 147° 08′ 21″ E) and processed at the Australian Institute of Marine Science (AIMS). The expected oligotrophic conditions were found, with 0.20 μg chlorophyll a L−1 and dissolved inorganic nitrogen and phosphate concentrations of 0.25 and 0.08 μmol L−1 respectively. The four treatments consisted of natural seawater without any addition (Control) and with ca. 40 μmol C L−1 additions of glucose (Glucose), leachates from the leaves of Rhizophora stylosa mangrove (Mangrove) and Halodule uninervis seagrass (Seagrass). These mangrove and seagrass species are common in the coasts of the GBR. The bacterial inoculum consisted of seawater filtered through precombusted Whatman GF/C filters (nominal pore size 1.2 μm) in order to remove virtually all protistan grazers (the same filtration in the Red Sea removed on average 92% of the initial heterotrophic nanoflagellates present, E. I. Sabbagh et al., pers. comm.), which was then added to the treatment seawater with a 1/10 dilution, thus increasing the DOM to heterotrophic bacteria ratio. Each type of experimental water was thereafter distributed into 45 glass bottles (nine replicates per sampling day including time 0) of 500 ml. The bottles were divided into three groups and incubated for 4 days in the dark at the AIMS's National Sea Simulator (SeaSim) with an accuracy of ±0.1°C at in situ temperature (27.7°C, measured at the time of sampling), at 3°C below (24.7°C) and above (30.7°C) the in situ value. By incubating the sample at 3°C below and above, we aimed to make sure that we did not expose our samples to thermal stress. In order to enhance the independence among replicates, three replicate bottles from each treatment and temperature (36) were sacrificed every day for sampling the physiological structure of heterotrophic bacteria and other ancillary variables described in detail by Baltar et al. (2017) and Lønborg et al. (2019). Bioavailable DOC (BDOC) was calculated as the difference between the initial concentration and the minimum value recorded over the 4 days incubation.
Physiological groups of heterotrophic bacteria
Samples of ca. 20 ml were taken from each experimental bottle and analysed following the protocols described in detail by Gasol and Morán (2015), within 1 h for live protocols and within 1 week after the end of the experiments for fixed samples. All groups were analysed with a FACSVerse flow cytometer (BD Sciences). The healthiness of the community was assessed by separating the Live (i.e. membrane‐intact) from the Dead (i.e. membrane‐damaged) cells, which were analysed fresh in 0.4 ml subsamples after double staining with SYBR Green I (Molecular Probes) and propidium iodine (PI, Sigma) (Grégori et al., 2001). Dead cells were distinguished by a higher red fluorescence signal due to PI. About 0.4 ml samples for LNA and HNA content bacteria were previously fixed with 0.5% glutaraldehyde final concentration (25% EM‐grade, Merck), flash frozen in liquid nitrogen and stored at −80°C until analysis. After staining with SYBR Green I (0.1% final concentration), LNA and HNA cells were clearly separated in green fluorescence vs. right angle light scatter or side scatter (SSC) plots. Prochlorococcus cyanobacteria were present in the inoculum but after pre‐filtering, their abundance in the experimental incubations never exceeded 4.5 103 cells mL−1 and they could be easily separated from HNA cells. The SSC signal standardized to that of 1 μm fluorescent latex beads (Molecular Probes, ref. F‐13081) was used to estimate a mean cell diameter using the empirical calibration of Calvo‐Díaz and Morán (2006). This calibration was done using filtered samples through different pore sizes to create a regression between relative SSC and mean diameter (i.e. the estimated pore size retaining 50% of the initial abundance). Assuming spherical shape, cell volume was converted into cellular carbon content using the relationship of Gundersen et al. (2001). Heterotrophic bacteria with an active electron transport chain (i.e. those able to reduce 5‐cyano‐2,3‐ditolyl tetrazolium chloride or CTC+) were identified in red fluorescence and green fluorescence versus SSC cytograms of 0.25 ml fresh samples incubated with 5 mmol L−1 CTC for 1.5 h. The actual abundance of Live, Dead, LNA, HNA and CTC+ bacteria was calculated after gravimetric calibrating of the FACSVerse flow rate for every batch of samples analysed. The percent contribution of Live cells to the total abundance (%Live) was calculated as: Live/(Live + Dead); that of HNA cells (%HNA) as: HNA/(LNA + HNA) and finally, that of CTC+ cells (%CTC+) as: CTC+/(LNA + HNA).
Specific growth rates and carrying capacities
The specific growth rates (μ, day‐1) of the different physiological groups were estimated as the slope of linear regressions of ln‐transformed abundances vs. time for the exponential phase of growth (up to 2–3 days for most of the treatments before evident changes in slope, see, for instance, Fig. 1 for total bacterial abundance). Although the concept of carrying capacity (K) applies to pure cultures, we estimated the K of the different bacterial groups as the highest abundances recorded over the 4 days incubation. We assumed negligible effects of protistan grazers (by initial pre‐filtration of the inoculum) and viruses (due to the short incubation period, see below). In case of discrepancy when estimating the respective μ and K values, we consistently applied the same period for the three replicates per temperature and treatment.
Temperature responses
The temperature sensitivity of the specific growth rates was described by the activation energy (E, in eV), so that a higher response of a given treatment to a temperature increase (i.e. 1°C) means that it will also have a higher activation energy. E was initially developed for individual enzymes and substrates, but it is commonly applied for natural communities in the framework of the metabolic theory of ecology. Specific growth rates are expected to increase exponentially with temperature according to E under nutrient replete conditions (Brown et al., 2004). The temperature response has also been extensively described by the Q10 temperature coefficient. A specific growth rate Q10 value of 2 (i.e. the specific growth rate doubles with an increase of 10°C) would be roughly equivalent to a E value of 0.51 eV at 20°C. For each treatment and group, E was calculated using the slope of ln‐transformed values of the specific growth rates vs. temperature as 1/kT, with T as absolute temperature in Kelvin and k the Boltzmann's constant (8.62x10– 5 eV K−1). An example of this calculation is shown in the study by Huete‐Stauffer et al. (2015). For simplicity, and although activation energies refer properly to rates rather than stocks, we used the same approach for estimating the temperature response of carrying capacity (K‐TR, Huete‐Stauffer et al., 2015). Hence, a positive K‐TR value means that carrying capacity increased with increasing temperature. For other variables such as the percent contribution of HNA, Live or CTC+ cells to total numbers (%HNA, %Live and %CTC+) or cell size we decided to use the actual incubation temperature in °C rather than in K.
Viruses' concentrations
Unfiltered subsamples of 1 ml were collected in sterile 2 ml Eppendorf tubes and fixed with 0.5% glutaraldehyde final concentration (25% EM‐grade, Merck) for 15 min at 4°C, after which samples were flash frozen in liquid nitrogen and stored at −80°C until analysis within a month by flow cytometry. Viral abundance was determined using a FACSVerse flow cytometer (BD Sciences), according to Brussaard (2004). Samples were diluted (20–80 times) in TE buffer (Tris 10 mM, EDTA 1 mM, pH 8.0), stained with SYBR Green I (Molecular Probes) to a final concentration of 10−4 of the commercial stock solution. Viral samples were incubated at 80°C in the dark for 10 min. The trigger was set for green fluorescence and the data were analysed using Flowing Software 2.5.1 (freeware; http://flowingsoftware.btk.fi).
Statistical analyses
Ordinary least squares (model I) linear regressions for estimating the specific growth rates of Live, Dead (see discussion), LNA, HNA, total (i.e. the sum of LNA and HNA) and CTC+ cells were done with all replicates for the period of exponential growth identified for each DOM and temperature treatment. Model I linear regressions were also used for estimating the activation energies (E) of the specific growth rates and carrying capacities of the various physiological groups, as previously explained. Differences between the E values in the different DOM treatments were assessed by analysis of covariance (ANCOVA). For other variables such as the absolute and relative abundance of the different physiological groups, differences between DOM treatments were determined with one‐way ANOVAs and Tukey–Kramer HSD post hoc tests. General relationships between the variables were represented by Pearson's correlation coefficients. All statistical analyses were done with JMP software package.
Supporting information
Fig. S1 Contribution of HNA bacteria to total abundance (%) during the 4 days incubations in the A Control, B Glucose, C Mangrove and D Seagrass treatments. Error bars represent standard deviations of 3 replicates. Dashed smooth fitting joins treatment points for clarity.
Fig. S2. Dynamics of viruses during the 4 days incubations in the A Control, B Glucose, C Mangrove and D Seagrass treatments. Error bars and dashed lines as in Fig. S1.
Fig. S3. Contribution of Live bacteria to total abundance (%) during the 4 days incubations in the A Control, B Glucose, C Mangrove and D Seagrass treatments. Error bars and dashed lines as in Fig. S1.
Fig. S4. Percent contribution of CTC+ bacteria to total abundance (%) during the 4 days incubations in the A Control, B Glucose, C Mangrove and D Seagrass treatments. Error bars and dashed lines as in Fig. S1.
Fig. S5. Activation energies of A the specific growth rates of Live, Dead, LNA, HNA and CTC+ bacteria and B their respective carrying capacities (K‐TR, see the text for details). Error bars represent standard errors.
Acknowledgements
The authors are thankful to the SeaSim team at AIMS for their help and support when setting up the incubations. The study was co‐financed by the AIMS Visiting Fellowship Program as part of the Capability Development Fund (CDF), awarded to X.A.G.M. X.A.G.M. was also partly supported by his KAUST baseline funding. F.B. was supported by a University of Otago Research Grant. Thanks are due for the financial support to CESAM (UID/AMB/50017 ‐ POCI‐01‐0145‐FEDER‐007638), to FCT/MCTES through national funds (PIDDAC), and the co‐funding by the FEDER, within the PT2020 Partnership Agreement and Compete 2020. C.C. was funded by the Foundation for Science and Technology (SFRH/BPD/117746/2016).
References
- Agawin, N.S.R. , Duarte, C.M. , and Agustí, S. (2000) Nutrient and temperature control of the contribution of picoplankton to phytoplankton biomass and production. Limnol Oceanogr 45: 591–600. [Google Scholar]
- Allen, A.P. , Gillooly, J.F. , and Brown, J.H. (2005) Linking the global carbon cycle to individual metabolism. Funct Ecol 19: 202–213. [Google Scholar]
- Alongi, D.M. , and Mukhopadhyay, S.K. (2015) Contribution of mangroves to coastal carbon cycling in low latitude seas. Agric Meteorol 213: 266–272. [Google Scholar]
- Alonso‐Sáez, L. , Díaz‐Pérez, L. , and Morán, X.A.G. (2015) The hidden seasonality of the rare biosphere in coastal marine bacterioplankton. Environ Microbiol 17: 3766–3780. [DOI] [PubMed] [Google Scholar]
- Al‐Otaibi, N. , Huete‐Stauffer, T.M. , Calleja, M.L. , Irigoien, X. , and Moran, X.A.G. (2020) Seasonal variability and vertical distribution of autotrophic and heterotrophic picoplankton in the Central Red Sea. PeerJ 8: e8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, L. , Gonzalez, A.M. , Araujo, F.V. , and Paranhos, R. (2003) Flow cytometry assessment of bacterioplankton in tropical marine environments. J Microbiol Meth 55: 841–850. [DOI] [PubMed] [Google Scholar]
- Andrade, L. , Gonzalez, A.M. , Rezende, C.E. , Suzuki, M. , Valentin, J.L. , and Paranhos, R. (2007) Distribution of HNA and LNA bacterial groups in the Southwest Atlantic Ocean. Braz J Microbiol 38: 330–336. [Google Scholar]
- Apple, J.K. , del Giorgi, P.A. , and Kemp, W.M. (2006) Temperature regulation of bacterial production, respiration, and growth efficiency in a temperate salt‐marsh estuary. Aquat Microb Ecol 43: 243–254. [Google Scholar]
- Arandia‐Gorostidi, N. , Huete‐Stauffer, T.M. , Alonso‐Sáez, L. , and Morán, X.A.G. (2017) Testing the metabolic theory of ecology with marine bacteria: different temperature sensitivity of major phylogenetic groups during the spring phytoplankton bloom. Environ Microbiol 19: 4493–4505. [DOI] [PubMed] [Google Scholar]
- Baltar, F. , Morán, X.A.G. , and Lønborg, C. (2017) Warming and organic matter sources impact the proportion of dissolved to total activities in marine extracellular enzymatic rates. Biogeochemistry 133: 307–316. [Google Scholar]
- Baltar, F. , Arístegui, J. , Gasol, J.M. , and Herndl, G.J. (2012) Microbial functioning and community structure variability in the mesopelagic and epipelagic waters of the subtropical Northeast Atlantic Ocean. Appl Environ Microbiol 78: 3309–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltar, F. , Palovaara, J. , Vila‐Costa, M. , Salazar, G. , Calvo, E. , Pelejero, C. , et al (2015) Response of rare, common and abundant bacterioplankton to anthropogenic perturbations in a Mediterranean coastal site. FEMS Microbiol Ecol 91: fiv058. [DOI] [PubMed] [Google Scholar]
- Benner, R. , Peele, E.R. , and Hodson, R.E. (1986) Microbial utilization of dissolved organic‐matter from leaves of the red mangrove, Rhizophora‐Mangle, in the Fresh Creek estuary, Bahamas. Estuar Coast Shelf Sci 23: 607–619. [Google Scholar]
- Bernhardt, J.R. , Sunday, J.M. , and O'Connor, M.I. (2018) Metabolic theory and the temperature‐size rule explain the temperature dependence of population carrying capacity. Am Nat 192: 687–697. [DOI] [PubMed] [Google Scholar]
- Bock, N. , Van Wambeke, F. , Dion, M. , and Duhamel, S. (2018) Microbial community structure in the western tropical South Pacific. Biogeosciences 15: 3909–3925. [Google Scholar]
- Bouvier, T. , del Giorgio, P.A. , and Gasol, J.M. (2007) A comparative study of the cytometric characteristics of high and low nucleic‐acid bacterioplankton cells from different aquatic ecosystems. Environ Microbiol 9: 2050–2066. [DOI] [PubMed] [Google Scholar]
- Brown, J.H. , Gillooly, J.F. , Allen, A.P. , Savage, V.M. , and West, G.B. (2004) Toward a metabolic theory of ecology. Ecology 85: 1771–1789. [Google Scholar]
- Brussaard, C.P.D. (2004) Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol 70: 1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja, M.L., Ansari, M.I., Røstad, A., Silva, L., Kaartvedt, S., Irigoien, X., and Morán, X.A.G. (2018) The Mesopelagic Scattering Layer: A Hotspot for Heterotrophic Prokaryotes in the Red Sea Twilight Zone. Front Mar Sci 5 10.3389/fmars.2018.00259 [DOI] [Google Scholar]
- Calvo‐Díaz, A. , and Morán, X.A.G. (2006) Seasonal dynamics of picoplankton in shelf waters of the southern Bay of Biscay. Aquat Microb Ecol 42: 159–174. [Google Scholar]
- Crosbie, N.D. , and Furnas, M.J. (2001) Net growth rates of picocyanobacteria and nano/microphytoplankton inhabiting shelf waters of the central (17°S) and southern (20°S) great barrier reef. Aquat Microb Ecol 24: 209–224. [Google Scholar]
- Daufresne, M. , Lengfellner, K. , and Sommer, U. (2009) Global warming benefits the small in aquatic ecosystems. Proc Natl Acad Sci USA 106: 12788–12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Giorgio, P.A. , and Scarborough, G. (1995) Increase in the proportion of metabolically active bacteria along gradients of enrichment in freshwater and marine plankton: implications for the estimates of bacterial growth and production rates. J Plankton Res 17: 1905–1924. [Google Scholar]
- del Giorgio, P.A. , and Gasol, J.M. (2008) Physiological structure and single‐cell activity in marine bacterioplankton In Microbial Ecology of the Oceans, Kirchman D.L. (ed). Hoboken: John Wiley & Sons, pp. 243–298. [Google Scholar]
- Falcioni, T. , Papa, S. , and Gasol, J.M. (2008) Evaluating the flow‐cytometric nucleic acid double‐staining protocol in realistic situations of planktonic bacterial death. Appl Environ Microb 74: 1767–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flombaum, P. , Gallegos, J.L. , Gordillo, R.A. , Rincón, J. , Zabala, L.L. , Jiao, N. , et al (2013) Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus . P Natl Acad Sci USA 110: 9824–9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnas, M.J. , and Mitchell, A.W. (1987) Phytoplankton dynamics in the central great‐barrier‐Reef.2. Primary production. Cont Shelf Res 7: 1049–1062. [Google Scholar]
- García, F.C. , Alonso‐Sáez, L. , Morán, X.A.G. , and López‐Urrutia, A. (2015) Seasonality in molecular and cytometric diversity of marine bacterioplankton: the re‐shuffling of bacterial taxa by vertical mixing. Environ Microbiol 17: 4133–4142. [DOI] [PubMed] [Google Scholar]
- Gasol, J.M. , and Arístegui, J. (2007) Cytometric evidence reconciling the toxicity and usefulness of CTC as a marker of bacterial activity. Aquat Microb Ecol 46: 71–83. [Google Scholar]
- Gasol, J.M. , and Morán, X.A.G. (2015) Flow cytometric determination of microbial abundances and its use to obtain indices of community structure and relative activity In Hydrocarbon and Lipid Microbiology Protocols, McGenity T.J., Timmis K.N., and Nogales B. (eds). Berlin, Germany: Springer, pp. 159–187. [Google Scholar]
- Gasol, J.M. , Zweifel, U.L. , Peters, F. , Fuhrman, J.A. , and Hagström, Å. (1999) Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl Environ Microbiol 65: 4475–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasol, J.M. , Alonso‐Saez, L. , Vaque, D. , Baltar, F. , Calleja, M.L. , Duarte, C.M. , and Aristegui, J. (2009) Mesopelagic prokaryotic bulk and single‐cell heterotrophic activity and community composition in the NW Africa‐Canary Islands coastal‐transition zone. Prog Oceanogr 83: 189–196. [Google Scholar]
- Gillooly, J.F. , Brown, J.H. , West, G.B. , Savage, V.M. , and Charnov, E.L. (2001) Effects of size and temperature on metabolic rate. Science 293: 2248–2251. [DOI] [PubMed] [Google Scholar]
- Girault, M. , Arakawa, H. , Barani, A. , Ceccaldi, H.J. , Hashihama, F. , and Gregori, G. (2015) Heterotrophic prokaryote distribution along a 2300 km transect in the North Pacific subtropical gyre during a strong La Nina conditions: relationship between distribution and hydrological conditions. Biogeosciences 12: 3607–3621. [Google Scholar]
- Grégori, G. , Citterio, S. , Ghiani, A. , Labra, M. , Sgorbati, S. , Brown, S. , and Denis, M. (2001) Resolution of viable and membrane‐compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl Environ Microb 67: 4662–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen, K. , Orcutt, K.M. , Purdie, D.M. , Michaels, A.F. , and Knap, A.H. (2001) Particulate organic carbon mass distribution at the Bermuda Atlantic time‐series study (BATS) site. Deep Sea Res II 48: 1697–1718. [Google Scholar]
- Herbland, A. , and Voituriez, B. (1979) Hydrological structure analysis for estimating the primary production in the tropical Atlantic Ocean. J Mar Res 34: 87–101. [Google Scholar]
- Huete‐Stauffer, T.M. , Arandia‐Gorostidi, N. , Díaz‐Pérez, L. , and Morán, X.A.G. (2015) Temperature dependences of growth rates and carrying capacities of marine bacteria depart from metabolic theoretical predictions. FEMS Microbiol Ecol 91: fiv111. [DOI] [PubMed] [Google Scholar]
- Huete‐Stauffer, T.M. , Arandia‐Gorostidi, N. , Alonso‐Sáez, L. , and Morán, X.A.G. (2016) Experimental warming decreases the average size and nucleic acid content of marine bacterial communities. Front Microbiol 7: 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC . (2013) Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press. [Google Scholar]
- Jones, R.I. , Kankaala, P. , Nykanen, H. , Peura, S. , Rask, M. , and Vesala, S. (2018) Whole‐Lake sugar addition demonstrates trophic transfer of dissolved organic carbon to top consumers. Ecosystems 21: 495–506. [Google Scholar]
- Kirchman, D.L. , Morán, X.A.G. , and Ducklow, H. (2009) Microbial growth in the polar oceans ‐ role of temperature and potential impact of climate change. Nat Rev Microbiol 7: 451–459. [DOI] [PubMed] [Google Scholar]
- Kristensen, E. , Bouillon, S. , Dittmar, T. , and Marchand, C. (2008) Organic carbon dynamics in mangrove ecosystems: a review. Aquat Bot 89: 201–219. [Google Scholar]
- Lebaron, P. , Servais, P. , Agogue, H. , Courties, C. , and Joux, F. (2001) Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl Environ Microb 67: 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.L. , Jiang, Z.J. , Zhou, C.Y. , Wu, Y.C. , Arbi, I. , Zhang, J.P. , et al (2018) Leaching of dissolved organic matter from seagrass leaf litter and its biogeochemical implications. Acta Oceanol Sin 37: 84–90. [Google Scholar]
- Lønborg, C. , Baltar, F. , Carreira, C. , and Morán, X.A.G. (2019) Dissolved organic carbon source influences tropical heterotrophic bacterioplankton response to experimental warming. Front Microbiol 10: 2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lønborg, C. , Álvarez‐Salgado, X.A. , Duggan, S. , and Carreira, C. (2018) Organic matter bioavailability in tropical coastal waters: the great barrier reef. Limnol Oceanogr 63: 1015–1035. [Google Scholar]
- Lønborg, C. , Doyle, J. , Furnas, M. , Menendez, P. , Benthuysen, J. , and Carreira, C. (2017) Seasonal organic matter dynamics in the great barrier reef lagoon: contribution of carbohydrates and proteins. Cont Shelf Res 38: 95–105. [Google Scholar]
- Lønborg, C. , Cuevas, L.A. , Reinthaler, T. , Herndl, G.J. , Gasol, J.M. , Morán, X.A.G. , et al (2016) Depth dependent relationships between temperature and ocean heterotrophic prokaryotic production. Front Mar Sci 3: 90. [Google Scholar]
- Longnecker, K. , Sherr, B.F. , and Sherr, E.B. (2005) Activity and phylogenetic diversity of bacterial cells with high and low nucleic acid content and electron transport system activity in an upwelling ecosystem. Appl Environ Microb 71: 7737–7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker, K., Wilson, M.J., Sherr, E.B., and Sherr, B.F. (2010) Effect of top‐down control on cell‐specific activity and diversity of active marine bacterioplankton. Aquat Microb Ecol 58: 153–165. 10.3354/ame01366 [DOI] [Google Scholar]
- Maher, D.T. , and Eyre, B.D. (2010) Benthic fluxes of dissolved organic carbon in three temperate Australian estuaries: implications for global estimates of benthic DOC fluxes. J Geophys Res 115: G04039. [Google Scholar]
- Mary, I. , Heywood, J.L. , Fuchs, B.M. , Amann, R. , Tarran, G.A. , Burkill, P.H. , and Zubkov, M.V. (2006) SAR11 dominance among metabolically active low nucleic acid bacterioplankton in surface waters along an Atlantic meridional transect. Aquat Microb Ecol 45: 107–113. [Google Scholar]
- Morán, X.A.G. , and Calvo‐Díaz, A. (2009) Single‐cell vs. bulk activity properties of coastal bacterioplankton over an annual cycle in a temperate ecosystem. FEMS Microbiol Ecol 67: 43–56. [DOI] [PubMed] [Google Scholar]
- Morán, X.A.G. , Ducklow, H.W. , and Erickson, M. (2011) Single‐cell physiological structure and growth rates of heterotrophic bacteria in a temperate estuary (Waquoit Bay, Massachusetts). Limnol Oceanogr 2011: 37–48. [Google Scholar]
- Morán, X.A.G. , Bode, A. , Suárez, L.Á. , and Nogueira, E. (2007) Assessing the relevance of nucleic acid content as an indicator of marine bacterial activity. Aquat Microb Ecol 46: 141–152. [Google Scholar]
- Morán, X.A.G. , Calvo‐Díaz, A. , Arandia‐Gorostidi, N. , and Huete‐Stauffer, T.M. (2018) Temperature sensitivities of microbial plankton net growth rates are seasonally coherent and linked to nutrient availability. Environ Microbiol 20: 3798–3810. [DOI] [PubMed] [Google Scholar]
- Morán, X.A.G. , Gasol, J.M. , Pernice, M.C. , Mangot, J.F. , Massana, R. , Lara, E. , et al (2017) Temperature regulation of marine heterotrophic prokaryotes increases latitudinally as a breach between bottom‐up and top‐down controls. Glob Chang Biol 23: 3956–3964. [DOI] [PubMed] [Google Scholar]
- Morán, X.A.G. , Alonso‐Sáez, L. , Nogueira, E. , Ducklow, H.W. , Gonzalez, N. , López‐Urrutia, Á. , et al (2015) More, smaller bacteria in response to ocean's warming? Proc Roy Soc B‐Biol Sci 282: 20150371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nittrouer, C.A. , Brunskill, G.J. , and Figueiredo, A.G. (1995) Importance of tropical coastal environments. Geo‐Mar Lett 15: 121–126. [Google Scholar]
- Platt, T. , Subba Rao, D.W. , and Irwin, B. (1983) Photosynthesis of picoplankton in the oligotrophic ocean. Nature 301: 702–704. [Google Scholar]
- Puglisi, M.P. , Engel, S. , Jensen, P.R. , and Fenical, W. (2007) Antimicrobial activities of extracts from indo‐Pacific marine plants against marine pathogens and saprophytes. Mar Biol 150: 531–540. [Google Scholar]
- Richardson, T.L. (2019) Mechanisms and pathways of small‐phytoplankton export from the surface ocean. Ann Rev Mar Sci 11: 57–74. [DOI] [PubMed] [Google Scholar]
- Savage, V.M. , Gillooly, J.F. , Brown, J.H. , West, G.B. , and Charnov, E.L. (2004) Effects of body size and temperature on population growth. Am Nat 163: 429–441. [DOI] [PubMed] [Google Scholar]
- Sawstrom, C. , Hyndes, G.A. , Eyre, B.D. , Huggett, M.J. , Fraser, M.W. , Lavery, P.S. , et al (2016) Coastal connectivity and spatial subsidy from a microbial perspective. Ecol Evol 6: 6662–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattenhofer, M. , Wulf, J. , Kostadinov, I. , Glockner, F.O. , Zubkov, M.V. , and Fuchs, B.M. (2011) Phylogenetic characterisation of picoplanktonic populations with high and low nucleic acid content in the North Atlantic Ocean. Syst Appl Microbiol 34: 470–475. [DOI] [PubMed] [Google Scholar]
- Schiaffino, M.R. , Gasol, J.M. , Izaguirre, I. , and Unrein, F. (2013) Picoplankton abundance and cytometric group diversity along a trophic and latitudinal lake gradient. Aquat Microb Ecol 68: 231–250. [Google Scholar]
- Sherr, B.F. , del Giorgio, P. , and Sherr, E.B. (1999) Estimating abundance and single‐cell characteristics of respiring bacteria via the redox dye CTC. Aquat Microb Ecol 18: 117–131. [Google Scholar]
- Sierra, C.A. (2012) Temperature sensitivity of organic matter decomposition in the Arrhenius equation: some theoretical considerations. Biogeochemistry 108: 1–15. [Google Scholar]
- Silva, L. , Calleja, M.L. , Huete‐Stauffer, T.M. , Ivetic, S. , Ansari, M.I. , Viegas, M. , and Morán, X.A.G. (2019) Low abundances but high growth rates of coastal heterotrophic Bacteria in the Red Sea. Front Microbiol 9: 3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y.H. , Wang, Y.F. , Mao, G.N. , Gao, G.H. , and Wang, Y.Y. (2019) Impact of planktonic low nucleic acid‐content bacteria to bacterial community structure and associated ecological functions in a shallow lake. Sci Total Environ 658: 868–878. [DOI] [PubMed] [Google Scholar]
- Stepanauskas, R. , Farjalla, V.F. , Tranvik, L.J. , Svensson, J.M. , Esteves, F.A. , and Graneli, W. (2000) Bioavailability and sources of DOC and DON in macrophyte stands of a tropical coastal lake. Hydrobiologia 436: 241–248. [Google Scholar]
- Supriadi, A. , Baehaki, A. , and Pratama, M.C. (2016) Antibacterial activity of methanol extract from seagrass of Halodule uninervis in the coastal of Lampung. Pharm Lett 8: 77–79. [Google Scholar]
- Thingstad, T.F. , Zweifel, U.L. , and Rassoulzadegan, F. (1998) P limitation of heterotrophic bacteria and phytoplankton in the Northwest Mediterranean. Limnol Oceanogr 43: 88–94. [Google Scholar]
- Thingstad, T.F. , Øvreås, L. , Egge, J.K. , Løvdal, T. , and Heldal, M. (2005) Use of non‐limiting substrates to increase size; a generic strategy to simultaneously optimize uptake and minimize predation in pelagic osmotrophs? Ecol Lett 8: 675–682. [Google Scholar]
- Vila‐Costa, M. , Gasol, J.M. , Sharma, S. , and Moran, M.A. (2012) Community analysis of high‐ and low‐nucleic acid‐containing bacteria in NW Mediterranean coastal waters using 16S rDNA pyrosequencing. Environ Microbiol 14: 1390–1402. [DOI] [PubMed] [Google Scholar]
- Wang, Y.Y. , Hammes, F. , Boon, N. , Chami, M. , and Egli, T. (2009) Isolation and characterization of low nucleic acid (LNA)‐content bacteria. ISME J 3: 889–902. [DOI] [PubMed] [Google Scholar]
- Zubkov, M.V. , Fuchs, B.M. , Burkill, P.H. , and Amann, R. (2001) Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl Environ Microbiol 67: 5210–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Contribution of HNA bacteria to total abundance (%) during the 4 days incubations in the A Control, B Glucose, C Mangrove and D Seagrass treatments. Error bars represent standard deviations of 3 replicates. Dashed smooth fitting joins treatment points for clarity.
Fig. S2. Dynamics of viruses during the 4 days incubations in the A Control, B Glucose, C Mangrove and D Seagrass treatments. Error bars and dashed lines as in Fig. S1.
Fig. S3. Contribution of Live bacteria to total abundance (%) during the 4 days incubations in the A Control, B Glucose, C Mangrove and D Seagrass treatments. Error bars and dashed lines as in Fig. S1.
Fig. S4. Percent contribution of CTC+ bacteria to total abundance (%) during the 4 days incubations in the A Control, B Glucose, C Mangrove and D Seagrass treatments. Error bars and dashed lines as in Fig. S1.
Fig. S5. Activation energies of A the specific growth rates of Live, Dead, LNA, HNA and CTC+ bacteria and B their respective carrying capacities (K‐TR, see the text for details). Error bars represent standard errors.


