Abstract
The study of island community assembly has been fertile ground for developing and testing theoretical ideas in ecology and evolution. The ecoevolutionary trajectory of lineages after colonization has been a particular interest, as this is a key component of understanding community assembly. In this system, existing ideas, such as the taxon cycle, posit that lineages pass through a regular sequence of ecoevolutionary changes after colonization, with lineages shifting toward reduced dispersal ability, increased ecological specialization, and declines in abundance. However, these predictions have historically been difficult to test. Here, we integrate phylogenomics, population genomics, and X‐ray microtomography/3D morphometrics, to test hypotheses for whether the ecomorphological diversity of trap‐jaw ants (Strumigenys) in the Fijian archipelago is assembled primarily through colonization or postcolonization radiation, and whether species show ecological shifts toward niche specialization, toward upland habitats, and decline in abundance after colonization. We infer that most Fijian endemic Strumigenys evolved in situ from a single colonization and have diversified to fill a large fraction of global morphospace occupied by the genus. Within this adaptive radiation, lineages trend to different degrees toward high elevation, reduced dispersal ability, and demographic decline, and we find no evidence of repeated colonization that displaces the initial radiation. Overall these results are only partially consistent with taxon cycle and associated ideas, while highlighting the potential role of priority effects in assembling island communities.
Keywords: 3D geometric morphometrics, community assembly, formicidae, phylogenomics, population genomics, taxon cycle
On remote islands, ecological communities assemble through transoceanic colonization and the subsequent evolution of lineages after colonization (Macarthur and Wilson 1967; Warren et al. 2015; Gillespie 2016). Their well‐defined boundaries make them attractive model systems for studying the interplay of ecological processes, such as dispersal and competition, and evolutionary processes such as adaptive shifts and speciation. Relevant ecoevolutionary theory ranges from the more stochastic perspectives, in which the central processes of dispersal and evolution are essentially unpredictable and lead to different outcomes for lineages (Hubbell 2001; Grant and Grant 2002; Fukami 2015), to more deterministic perspectives, in which lineages and communities assemble through regular pathways that are predictable and repeated (Losos et al. 1998; Gillespie 2004).
While such theorizing is nearly as old as biogeography, it has traditionally been difficult to test theoretical predictions and make strong links between observable static patterns and inferred dynamical processes (Warren et al. 2015). However, emerging data‐rich methods, including phylogenomics, community‐level population genomics, 3D imaging, and computational inference, give us a much broader window into the past and new opportunities to test longstanding ideas in biogeography (Economo and Sarnat 2012; Gillespie 2016; Cotoras et al. 2018).
Here, we examine a series of hypotheses about the community assembly of trap‐jaw ants (Strumigenys) in Fiji, a remote archipelago in eastern Melanesia. Strumigenys is the most species‐rich ant genus on this archipelago, which harbors a highly endemic ant fauna (Sarnat and Economo 2012) with several documented in situ radiations (Lucky and Sarnat 2010; Sarnat and Moreau 2011; Darwell et al. 2020). This Melanesian ant system was influential in the early development of island biogeography theory. In the 1950s, E.O. Wilson formulated two influential (but very different) ideas about how island communities assemble, the taxon cycle, and later his equilibrium theory of island biogeography. The taxon cycle hypothesis (Wilson 1959, 1961; Ricklefs and Bermingham 2002) has a more deterministic aspect. Inspired by patterns he observed through taxonomic work on Melanesian ants, Wilson (1959, 1961) proposed that lineages pass through phases of range expansion and contraction, associated with evolutionary niche shifts into and out of marginal habitats. On remote islands, lineages arrive at disturbed lowland habitats and subsequently evolve on trajectories toward the upland interior, eventually exhibiting ecomorphological specialization and a decline in both abundance and ecological dominance (Wilson 1959, 1961). Wilson (1959, 1961) envisioned new “expanding” lineages occasionally arriving and outcompeting the species derived from past colonizations, possibly even pushing them extinct. The taxon cycle has drawn interest by those analyzing various systems (e.g., Ricklefs and Cox 1972; Ricklefs and Bermingham 1999; Ricklefs and Bermingham 2002; Cook et al. 2008; Jønsson et al. 2014), as well as criticism (e.g., Pregill and Olson 1981; Losos 1992), but remains viewed (at least by some) as still a theoretically relevant idea in island biogeography (Ricklefs and Bermingham 2002; Steinbauer 2016; Whittaker et al. 2017). Recently, there has been renewed interest in testing its predictions using the original system that inspired it: Melanesian ants (Economo and Sarnat 2012; Economo et al. 2015; Matos‐Maraví et al. 2018a; Matos‐Maraví et al. 2018b; Darwell et al. 2020).
Both Wilson's original articles and subsequent works have touched on many different issues under the framework of the taxon cycle, making it sometimes unclear what exactly is being tested. However, most formulations of the taxon cycle include components that are more general themes discussed in the biogeographic literature. Rather than seek to test the taxon cycle as a singular hypothesis, we frame our study around these more general themes.
First, there is a question of whether the ecological diversity of the island community is assembled primarily through dispersal assembly or in situ radiation. The latter can occur in isolated islands when colonization is very infrequent (Whittaker 1998; Heaney 2000; Valente et al. 2020), where arriving lineages may take advantage of ecological opportunities (Schluter 2000). This can also occur, however, when dispersal rates are habitat dependent (Sukumaran and Knowles 2018), leading certain habitats to be colonized first on the island, followed by evolution of lineages into other “empty” niches or habitats. The particular habitats that are most connected may vary by taxon (Gillespie et al. 2012). For example, migrating birds may more frequently deposit plant propagules at high elevations in Hawaii, leading to greater colonization rates of high elevation forests (Carlquist 1967). In the case of Melanesian ants, it is likely that coastal, high‐disturbance habitats have higher connectivity, because rafting is thought to be a primary long‐distance dispersal mechanism of ants (Wilson 1959, 1961). Adaptation to such habitats facilitates range expansion of a lineage, while lineages adapted to high‐elevation interior habitats do not disperse long distances and colonize similar habitats in remote islands. Evidence for this habitat‐dependent dispersal has been found for Indo‐Pacific ants including the genus Pheidole (Economo et al. 2015), the genus Camponotus (Clouse et al. 2015), the Prenolepis‐genus group (Matos‐Maraví et al. 2018a), and Odontomachus (Matos‐Maraví et al. 2018b). Quantitative approaches are now available to test this statistically (Sukumaran et al. 2015; Matzke 2016; Sukumaran and Knowles 2018).
The fact that dispersal is habitat dependent, however, does not in itself mean that niches can shift after colonization and assemble the community. If niches are conserved after colonization, low elevation species should be more related to mainland low elevation species than to high elevation species in the same archipelago. In the taxon cycle hypothesis (Wilson 1959, 1961; Ricklefs and Bermingham 2002), niche shifts and niche specializations occur, as the lineage penetrates from the marginal habitats into the interior. Thus, disturbed coastal habitats are the “door” through which lineages first pass and (may) subsequently radiate to assemble the island community. The evidence here is a bit more fragmentary. Building on an inventory of the Fijian ants (Sarnat and Economo 2012), Economo and Sarnat (2012) showed that level of endemism is correlated to habitat affinity and elevation across the entire fauna, with the most endemic ants in Fiji being restricted to more pristine habitats and higher elevations. This is the pattern one would expect from directional niche shifts after colonization, but without phylogenetic information it is far from sufficient to show that they did occur (e.g., Losos 1992). Both Sarnat and Moreau (2011) and Economo and Sarnat (2012) used phylogenetic methods to show that high‐elevation, morphologically specialized lineages of the ant genus Pheidole evolved in situ from more generalized ancestors, even though they resemble spinescent forms found around the Indo‐Pacific (Sarnat et al. 2017). While this is a convincing example of niche shift, the extent to which it represents a special case restricted to Pheidole or a general pattern observed among Fiji's many other endemic ant radiations is unclear.
Another theme in both the taxon cycle and island biogeography in general is that after colonization, lineages undergo demographic decline and loss of dispersal ability (Wilson 1959, 1961; MacArthur and Wilson 1967; Gillespie 2002, Gillespie et al. 2012; Burns 2019). This could be because of an increase in ecological specialization leading to reduced competitive ability and increased vulnerability (Ricklefs and Bermingham 1999; Ricklefs and Bermingham 2002). This prediction has traditionally been difficult to test, and one weakness of many historical taxon cycle analyses is that lower dispersal ability for later‐stage endemic species was often inferred from their narrow ranges, which is somewhat circular without independent estimates of dispersal ability (e.g., from genetic data) (Dexter 2010). Modern population genomic methods offer a route to examining differences in dispersal ability and demography independent of geographic distribution per se. Although a recent study on spiders showed that species with high abundance and generalized ecologies can persist for millions of years after colonization (Gillespie et al. 2017), this prediction has not been tested directly in ants.
A related question is whether lineages evolve to acquire a consistent set of morphological changes that are expected to be associated with postcolonization evolution—the evolution of a morphological “island syndrome” of traits (Adler and Levins 1994; Novosolov et al. 2013; Burns 2019). An alternative is adaptive radiation, which is the diversification of lineages that take advantage of ecological opportunities (Schluter 2000). This can fill morphospace occupied by continental groups, and in some cases exceeding it. For example, rather than evolve toward a consistent set of traits, Fijian Pheidole have evolved to fill much of the global morphospace (Darwell et al. 2020) including the evolution of specialized traits such as extreme spinescence (Sarnat et al. 2017).
Finally, there is a question of whether after a lineage establishes on an island, the new arrivals can invade and penetrate the island's interior, eventually replacing the original inhabitants. Thus, we could see evidence of repeated colonization, with species derived from earlier colonizations more specialized and less abundant, a key prediction of the taxon cycle. One alternative to this would be a strong priority effect associated with ecological release. Ecological release is the expansion of species’ released niche when it arrives in a new environment with less competitors, and has been considered as an important process to shape community assembly of island fauna such as birds, lizards, spiders, and snails (Losos et al. 1998; Chiba 2004; Gillespie 2004; Reding et al. 2009). The first colonizing lineage seizes ecological opportunity and undergoes niche shift, radiation, and evolutionary community assembly—after which the community is resistant to subsequent colonists (Vannette and Fukami 2014; Fukami 2015). New arrivals may colonize the lowland disturbed habitats or deposit a lineage on the island but never manage to penetrate and usurp the existing community that has filled a diverse set of niches.
Here, we examine these dynamics using a combination of field inventory, phylogenomics, population genomics, X‐ray imaging, linear and 3D geometric morphometrics to unravel the assembly of a community of Strumigenys miniature trap‐jaw ants in Fiji, where it is the most speciose ant genus. The hypothesis that long‐distance ant dispersal is habitat dependent is best addressed on a regional or continental scale and has been tested for other ant groups in the region (Economo et al. 2015; Matos‐Maraví et al. 2018a, Matos‐Maraví et al. 2018b), and is beyond the scope of this study. We focus instead on the other themes listed above using a combined phylogenomic and population genomic analysis.
Specifically, we (i) use phylogenomics to test whether the Fijian Strumigenys community evolved primarily in situ within the archipelago or through dispersal assembly, (ii) use phenomic methods to test whether ecomorphological changes are consistent with evolution of an island syndrome of traits, or adaptive radiation into a broad range of ecomorphologies. Furthermore, we use population genomic approach to (iii) test whether niche shifts into interior and upland habitats are associated with an increase in geographic population structure, an indication of a decline in dispersal ability and connectivity, and (iv) we use demographic inference to test whether endemics are undergoing more demographic decline relative to newer arrivals, and whether evolution into more upland habitats and more specialized niches is associated with demographic decline. Finally, we examine (v) whether repeated colonizations lead to “takeovers” of the local community, would come in the form that multiple colonizations have occurred, and that in more than one colonization we see niche shift and ecomorphological radiation. If priority effects are strong, we would see most ecomorphological diversification descended from the initial colonizing lineage, with no evidence for niche shift and cladogenesis of more recent colonizations.
Material and Methods
THE STUDY SYSTEM AND TAXON SAMPLING
Many remote Pacific islands are thought to lack native ants entirely (Wilson and Taylor 1967), but Fiji supports a rich and distinctive ant fauna, including 45 genera and 188 known species, of which over 68% are endemic to Fiji (Sarnat 2006, 2008; Sarnat and Economo 2012). Strumigenys (Hymenoptera: Formicidae) is the third most speciose ant genus on earth with more than 1000 described species across tropical and subtropical forest (Bolton 2000), and it is also the most speciose ant genus in Fiji (23 species, with 20 endemics, Sarnat et al. 2019). Strumigenys is a pantropical, hyperdiverse clade of leaf‐litter predators (Wilson 1953; Brown and Wilson 1959). Strumigenys are known to vary along several important morphological axes including body size and relative mandible length. Relative mandible length (i.e., relative to head length), is thought to vary with hunting strategy and microhabitat (Brown and Wilson 1959; Bolton 2000). Species with long mandibles tend to be more epigaeic (aboveground active), and use a powerful trap‐jaw mechanism as a weapon for active hunting. In contrast, short‐mandibled species tend to be more hypogaeic (underground active)—they sit and wait until prey comes within close proximity of the mandibles, sometimes using chemical lures, then grip and hold onto their prey item until they can sting and immobilize it (Wilson 1953; Deyrup and Cover 2009). In a global revision of the genus, Bolton (2000) assigned Fijian species to six different morphological species groups, all of which are widespread in the Indo‐Pacific. These species groups provide some a priori expectations about community assembly of the archipelago: if morphological species groups are monophyletic, this would imply that the archipelago has been colonized a minimum of six times. Thus, dispersal would be a key mechanism assembling morphological diversity on the archipelago. To put the Fijian Strumigenys in phylogenetic context, we sampled 42 Strumigenys species (387 specimens in total; see Supporting information Table S1 for details), including 17 from Fiji and 25 non‐Fijian outgroups. We have another ongoing effort to reconstruct the global phylogeny of the genus with a dataset of 462 species representing all main geographic regions, thus, we chose these outgroups to be both representatives of global diversity and include closest relatives of the Fijian fauna on our broader phylogeny. Unfortunately, six rare Fijian species were not able to be included in the molecular study due to the lack of available material. However, we sequenced representative species from all the morphological groups in Fiji (Sarnat et al. 2019), and the missing species are closely allied with those taxa that were included. Thus, while we cannot completely rule out that these species represent distant lineages or independent colonizations of the island, we view it as highly unlikely.
SEQUENCING, RAD ASSEMBLY, AND GENOTYPING
The genomic DNA of each specimen was extracted using a nondestructive method following Tin et al. (2014). RAD library preparation was performed as in Tin et al. (2015) by using a Biomek FXP Laboratory Automation Workstation (Beckman Coulter). Equimolar concentrations of libraries with different barcodes were pooled, and a gel extraction was performed to select 200—400 bp DNA. Finally, the libraries were sequenced single end with read length of 55 bp on an Illumina HiSeq 2500 platform in DNA Sequencing Section (SQC) at the Okinawa Institute of Science and Technology Graduate University. Samples were demultiplexed, separated by individual, and filtered by quality using Trimmomatic (Bolger et al. 2014).
To assemble the dataset for phylogenomic analysis, we first assembled a dataset of a maximum of 10 specimens per species (the number of specimens range from 1 to 10, see Supporting information Table S2 for detail), by selecting the specimens with the highest data coverage after demultiplexing. We then de novo assembled the loci from raw RAD data using ipyrad (Eaton and Overcast 2018) pipeline. We largely followed the default setting for the assembling parameters, except we deleted the parameter 8 (“restriction_overhang”) in the parameter file since our samples were already demultiplexed. More specifically, we first filtered the reads with more than five ambiguous sites (Phred quality score <20). We then clustered the filtered reads within each sample using 85% sequence similarity. After the consensus sequences were called within each sample, we removed the potential paralogs by filtering out the consensus loci with more than two alleles. Finally, we clustered the loci across samples at 85% similarity and included all loci shared by at least four samples. The final assembled loci were exported in ipyrad formats *.loci (see ipyrad document for details). Because the 3’ edge of loci were not well aligned and might introduce false SNP calling during the ipyrad assembly, we trimmed the last 5 bp of each final aligned locus from the *.loci file, and then concatenated all loci into one supermatrix using custom python script.
To assemble datasets for population genomics analyses, we first selected eight Fijian Strumigenys species with enough specimens (at least three specimens from the same geographic population) sampled across the Fiji archipelago (two nonendemic species, including one Pacific‐widespread native species Strumigenys godeffroyi, and one exotic species Strumigenys rogeri, and six Fijian endemic species: Strumigenys trauma, Strumigenys basiliska, Strumigenys chernovi, Strumigenys ekasura, Strumigenys nidifex, and Strumigenys sulcata; Supporting information Table S3 to S10). We then applied separate ipyrad de novo assembly analyses on each species for genotyping (the minimum locus coverage across specimens = 70%). After assembly, we used VCFTools version 0.1.14, (Danecek et al. 2011) to remove individuals with average loci coverage <70%. SNPs with minimum minor allele frequency (MAF < 0.05) were also filtered out. Finally, for the downstream population structure inferences, we randomly selected one SNP per locus to reduce the effect of linkage disequilibrium.
PHYLOGENOMIC ANALYSIS
We included loci that had a high percentage of missing data across specimens, as previous work on RAD‐seq phylogenomics showed that loci missing across many or even most taxa can collectively contain a great deal of information about internal nodes in the phylogeny (Eaton et al. 2017). Our own informal testing on this and other datasets support this conclusion. Thus, we included even loci with low coverage across specimens. The final concatenated matrix for the phylogenomic analysis contained 330 413 loci and 13 993 656 bps with average 2.8 million bps present on the specimen level and 5.6 million bps presented per species (calculated from a consensus sequence within each species). To infer the history and the origins of Fijian Strumigenys, we reconstructed a maximum likelihood phylogeny in ExaML version 3.0.17 (Kozlov et al. 2015). All the starting trees were generated using RAxML version 8.2 (Stamatakis 2014), and we used a general GTR substitution model and a gamma distribution (GTR + G) of among site rate variation as selected by PartitionFinder 2 (Lanfear et al. 2017). We estimated the topology support by generating 100 bootstrap replicates using RAxML and then using ExaML to infer one ML tree per replicate.
We also inferenced an SVDquartets based species tree on Fijian Strumigenys using tetrad version 0.7.20, a program that is implemented in ipyrad (Chifman and Kubatko 2014; Eaton et al. 2017). We ran tetrad on the same assembled dataset that include 330, 288 unlinked SNPs. We inferred all 19 720 001 possible quartets for the 41 taxa, and then conducted 100 nonparametric bootstraps replicates to assess the support.
We estimated the divergence dates of Fijian endemic Strumigenys using BEAST version 2.4.4 (Bouckaert et al. 2014) with molecular evolution model set to GTR + G as suggested by PartitionFinder 2. To select the most appropriate clock model for our data, we applied the stepping‐stone sampling method implemented in BEAST version 2.4.4 to test whether the evolutionary rate of the taxa in our dataset is consistent (strict clock model vs. relaxed model). The result strongly suggests the relaxed model for our data (mean marginal likelihood: relaxed log normal model = −271 661.46, and strict model = −273 532.38). We then used the same stepping‐stone sampling on the two tree priors and selected Yule model as our tree prior (mean marginal likelihood: Yule = −271 661.46, Birth‐Death model = −272 948.05). We conducted the stepping‐stone sampling analyses with 50 steps and 200 000 generations within each step. The posterior distributions were sampled every 1000 generations and the convergence was accessed by examining the estimated marginal log likelihood values. For data calibration, we used the root divergence time of Strumigenys from Ward et al. 2015 as the minimum and maximum age constraint in our uniform distribution. We fixed the tree topology by using the phylogeny obtained from ExaML as the starting tree. We conducted two independent runs for 1 × 108 generations each, sampling every 10 000 generations. We assessed the convergence by examining whether the ESS of all parameters were greater than 200 in Tracer version 1.6 (Drummond and Rambaut 2007). We generated the maximum credibility clades trees using TreeAnnotator with the first 10% of posterior trees discarded as burnin (with burnin determined by the trace).
MORPHOMETRICS ANALYSES
We used both linear morphometrics and 3D geometric morphometrics to quantify the ecomorphological diversification of Fijian Strumigenys. We provide an overview here, but detailed methods can be found in Supporting information Appendix S1. The linear morphometrics were based on a series of measurements across the ant body, including head length, head width, scape length, eye length, Weber's length, and pronotum height following Bolton (2000), for each of the 14 Fijian Strumigenys species with total 281 specimens. To compare the level of morphological divergence in Fijian Strumigenys to that of the genus as a whole, we assembled a dataset of morphological measurements for 1056 non‐Fijian Strumigenys species (mainly using Bolton (2000) but adding our own measurements for some species, detail can be found in the Supporting information Appendix S1), and used it to characterize a global morphospace.
To quantify morphological shape variation in 3D using geometric morphometrics, we scanned 14 Strumigenys species using X‐ray microcomputed tomography (X‐ray micro‐CT) and applied a system of landmarks to the head and mesosoma (Supporting information Appendix S1) which were analyzed with the package geomorph. We estimated a phylomorphospace for both linear and 3D geometric morphometrics datatypes, and used phylomorphospace function in the R package phytools (Revell 2012) to evaluate the diversification of Fijian Strumigenys in morphospace. We reconstructed ancestral states for body size and elevational affinity of Fijian Strumigenys by using fastAnc function in the R package phytools (Revell 2012).
POPULATION GENOMICS ANALYSES
To test whether species decline in abundance and dispersal ability after colonization, we examined the degree of geographic population structure and the population demography. Specifically, we applied a population genomics approach to the eight Strumigenys species with sufficient sampling for analysis. These species include both nonendemic species and those from different clades within the Fijian endemics.
Population structure inference
We used three approaches to study the population structure of the eight Strumigenys species to assess the degree of genetic clustering of each species across different islands. We first examined genetic structure by conducting PCA on SNP data using the Python package scikit‐allele version 1.2.0 (Miles and Harding 2016). Second, we used sNMF version 1.2 (Frichot et al. 2014) to test the most likely number of genetic clusters within each species (k) and assign individuals to populations. We tested each k value (range from 1 to 10) with 20 replicated runs, and then examined their cross‐entropy criterion of each k. We visualized the sNMF results in R and determined the optimal k as the k value that minimizing the cross‐entropy criterion. Third, we used the Neighbor‐Net algorithm (Bryant and Moulton 2004) implemented in the program SplitsTree version 4.14.4 (Huson and Bryant 2006) to visualize population structure by generating phylogenetic networks. Genetic differentiation of populations in each species (F ST) were estimated at both per‐SNP and genome‐average level using Python package scikit‐allele. Calculation of F ST requires the designation of subpopulations. We generally used specimens collected from the same island as a single subpopulation, as we did not detect any within‐island structure for most species. For S. sulcata and S. trauma, specimens collected from western and eastern Viti Levu (the largest island) showed differentiation, and thus, were treated as separate subpopulations in the population structure analyses (in total, six populations for S. basiliska, four populations for S. chernovi, three populations for S. ekasura, five populations for S. godeffroyi, four populations for S. nidifex, four populations for S. rogeri, four populations for S. sulcata, four populations for S. trauma were selected; see Supporting information Appendix Table S3 to S10 for detail). We used Welch's two‐sample t test to investigate whether the F ST estimates between the species of the two clades were significantly different from each other.
Population demographic analyses
To test the different historical demographic syndromes (population expansion/contraction) of Fijian Strumigenys populations, we analyzed one dimensional site frequency spectra (1D‐SFS) using δaδi version 1.7.0 (Gutenkunst et al. 2009). We examined the following four alternative 1D models that differ in assumptions related to historical population change: (1) constant population size model, (2) instantaneous size change, (3) exponential size change, and (4) instantaneous size decline followed by exponential growth. The populations of each species were defined the same way as calculating F ST in the previous section. Parameter optimizations for each model were performed using the optimization routine procedure in Portik et al. (2017). We then used the Godambe Information Matrix (GIM, Coffman et al. 2015) to compare maximum likelihood models across the four demographic scenarios. For the best‐fit model, we estimated standard errors (SE) for each parameter using the GIM on 200 bootstrap data sets that randomly sampled from the original frequency spectrum using the built‐in sampling method in δaδi. We then calculated 95% confident intervals for each parameter estimate using the point estimate ±1.96 SE. We focused on the ratio of historical and current population size as an indicator of demographic growth or decline.
To cross‐validate the demographic results from δaδi, we also used fastsimcoal version 2.6 (Excoffier et al. 2013) to simulate a simple exponential population expansion or contraction model for each species. We set the prior distribution for this model as follows: time (TG) of instantaneous population size change ∼ uniform (U) (1 k, 1 million) generations ago, current effective population size (NCUR) ∼ U (100, 100K), and ancient effective population size (NANC) ∼U (100, 100K). We conducted 1 million coalescent simulations of this population expansion or contraction model with the following options: ‐ N 100 0000 (number of coalescent simulations), ‐L 40 [number of expectation‐maximization [M] cycles], ‐M 0.001 (minimum relative difference in parameter values for the stopping criterion), and ‐c 10 (threshold for observed SFS entry count, pooling all entries with less than 10 SNPs). For each species, we performed 100 independent fastsimcoal runs to determine the parameter estimates at maximum likelihood (Excoffier et al. 2013).
Results
IN SITU RADIATION OF FIJIAN STRUMIGENYS
The tree topology inferred with maximum likelihood method showed strong support (bootstrap 100%) for the monophyly of a single Fijian endemic long‐mandibled Strumigenys clade (Clade a in Fig. 1). The SVDQuartets based species tree matched the ML tree topology for all the endemic Fijian Strumigenys species (Supporting information Fig. S1). The monophyly indicates that the high endemism of long‐mandibled Strumigenys in Fiji is due to in situ radiation of a single colonist lineage, not independent colonizations of multiple lineages. The divergence dating analysis revealed the 14 extant species of long‐mandibled Strumigenys descended from a single lineage that colonized Fiji in the Miocene and split into two archipelago‐wide radiations (clade b and clade c in Fig. 2A).
Figure 1.
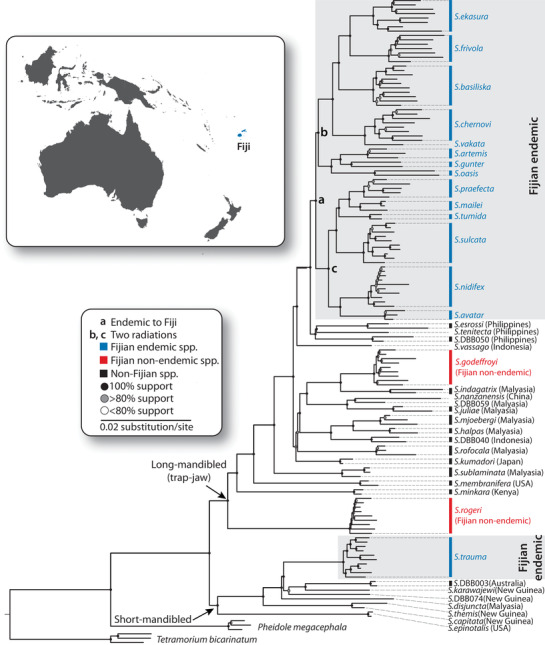
Maximum likelihood phylogeny of the ant genus Strumigenys in Fiji based on 330 413 Radseq loci. Nodes with 100% support values are represented by closed circles. Grey circles indicated the support value between 80 and 100%, while open circles indicated the nodes with less than 80% support.
Figure 2.
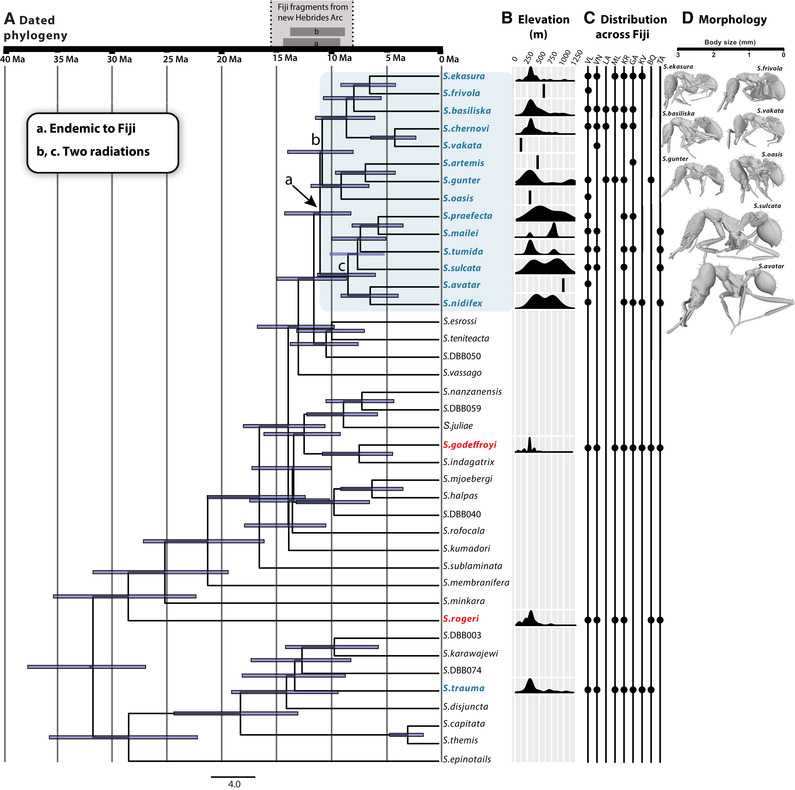
Radiation and diversification of Strumigenys in Fiji. (A) Dated chronogram inferred by Bayesian method based on 267 715 RAD loci showing the evolutionary radiations of Strumigenys in Fiji archipelago. (B) The elevation distribution (loess‐smoothed) indicated that in general, the species in small size clade tend to live in low elevation, while large Strumigenys species were often found in low and high elevation. (C) Species distribution across the whole archipelago. VL, Viti Levu; VN, Vanua Levu; LA, Ovalau; ML, Moala; KR, Koro; GA, Gau; KV, Kadavu; BQ, Beqa; TA, Taveuni. (D) 3D‐rendered models of Strumigenys species in the two clades to show the body size differences.
NICHE SPECIALIZATION AND ECOMORPHOLOGICAL DIVERSIFICATION
Within the endemic radiation, some species were only found in the lowland forest (e.g., Strumigenys ekasura, Strumigenys frivola, Strumigenys basiliska, Strumigenys chernovi, Strumigenys vakara, Strumigenys gunter, and Strumigenys oasis), while other species were more often found in high‐elevation forest (e.g., Strumigenys praefecta, Strumigenys mailei, Strumigenys tumida, Strumigenys sulcata, Strumigenys avatar, and Strumigenys nidifex; Fig. 2B). Species collected at high elevations were much larger than species collected at low elevations (Fig. 2D).
The distribution of the Fijian Strumigenys species in 2D morphospace indicates a considerable amount of morphological divergence between the two endemic clades (Fig. 3A, B). The first principal component (body size, variance explained = 91.3%) separated all endemic long‐mandibled Fijian Strumigenys into two major groups: large Strumigenys group with positive PC1 values and small Strumigenys group with negative PC1 values (Fig. 3A, B). Body size divergence in Strumigenys reflected phylogeny, with all the species in one endemic clade exhibiting small size (“small‐bodied clade”) and species in another endemic clade showing large size (“large‐bodied clade”). PC2 (eye size, variance explained = 3.98%) further separated ant species within each clade based on their eye size and other characteristics (Fig. 3A). Moreover, PC3 (mandible length, variance explained = 2.63%) revealed some variations in mandible length and other characteristics among species in the small‐bodied Strumigenys clade, while the mandible size in the large‐bodied Strumigenys clade was more conserved (Fig. 3B). The ancestral state reconstruction of body size showed that larger body‐sized form evolved with Fiji from more typical ancestors (Fig. 3C). The results indicate that the ecomorphological diversification of Fijian endemic Strumigenys evolved in situ within the archipelago. Moreover, we found that the Fijian Strumigenys lineages filled a large fraction of the entire global Strumigenys morphospace (Fig. 3D).
Figure 3.
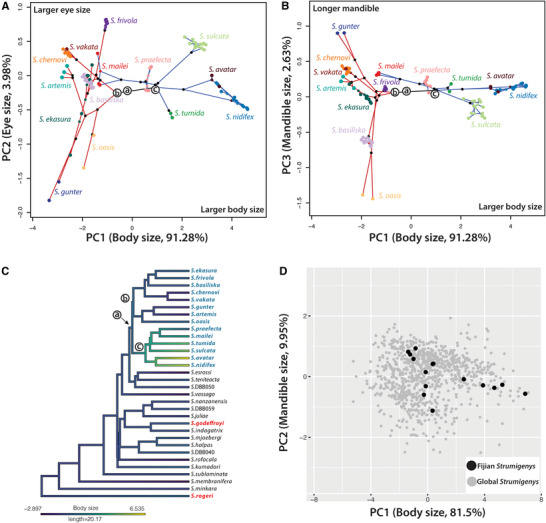
2D‐Phylomorphospace of morphological divergence in Fijian Strumigenys based on the morphological traits measurements. (A) PC1 (body size) versus PC2 (eye size). (B) PC1 (body size) versus PC3 (mandible length). The positive value indicates larger size. The dots in the same color indicate specimens from the same species. The red branch indicates large Strumigenys clade (negative PC1) and the blue branch represents the small Strumigenys clade (positive PC1). The key ancestral nodes are indicated using letters a, b, and c. (C) Ancestral state reconstruction of body size in long‐mandibled Strumigenys species, showing that the larger body sized Strumigenys species evolve in situ in Fiji. (D) The Fijian species (black dots) overlaid onto global Strumigenys morphospace which includes 1506 non‐Fijian Strumigenys species (grey dots).
In Fig. 4, 3D geometric morphometrics also revealed variation among species in head, mesosoma, and mandible shape. The small‐ and large‐bodied Strumigenys clades were consistently separated in each morphospace plot. For the head, PC1 (43% of variance) described the width of the posterior head, especially the breadth of the clypeus, and PC2 (20% of variance) described the depth of the posterior head (Fig. 4B). For the mesosoma, PC1 (39% of variance) described the relative size of the pronotum, and PC2 (18% of variance) described an apparent trade‐off in overall elongation of the pronotum versus propodeum (Fig. 4C). Lastly, the mandible varied from thick and triangular to thin and elongate (PC1; 63% of variance), with “teeth” along the same versus different axes (PC2; 16% of variance) (Fig. 4D).
Figure 4.
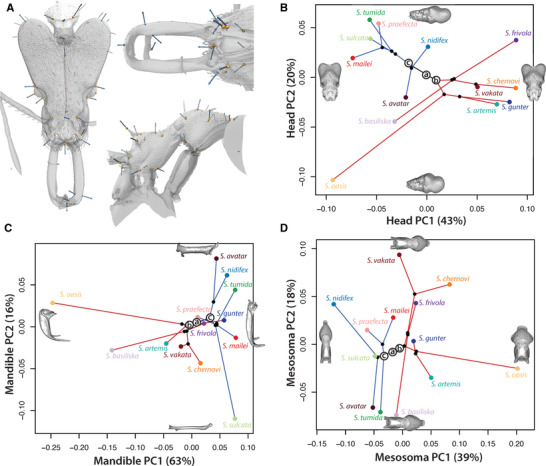
3D geometric morphometric analysis of morphological divergence in Fijian Strumigenys. (A) Landmarks placement on head, mandible, and mesosoma. (B‐D) Phylomorphospace of morphological divergence in head shape, mandible shape, and mesosoma shape. (B) For the head, PC1 (43% of variance) described species with a narrow anterior region of the head, especially the clypeus, and PC2 (20% of variance) described species with large eyes and a narrow posterior head. (C) The mandible varied from thick and triangular to thin and elongate (PC1; 63% of variance), with “teeth” along the same versus different axes (PC2; 16% of variance). Lastly, (D) for the mesosoma, PC1 (39% of variance) described species with a broad mesosoma and relatively enlarged pronotum, and PC2 (18% of variance) described species with a relatively enlarged mesonotum. The dots in the same color indicate specimens from the same species. The red branch indicates large Strumigenys clade and the blue branch represents the small Strumigenys clade.
POPULATION GENOMICS ANALYSES
Population structure
Inferred population structures of the selected Strumigenys species were largely consistent across methods, with no clear population structure in the two nonendemic species and clear population structure in endemic species (Fig. 5). More specifically, there were no clear geographic clustering patterns in the PCA plot for the two nonendemic species indicating low or no within‐archipelago differentiation (Supporting information Fig. S3). In contrast, PCA on SNP genotypes showed that all Fijian endemic Strumigenys species grouped geographically with individuals collected from the same locality (Supporting information Fig. S3). In sNMF, the best‐supported K value of the two nonendemic species S. rogeri and S. godeffroyi is 1, indicating no population structure among all the specimens. The optimum K values as well as the population assignments on the six endemic species were consistent with the clustering results from the PCA (Supporting information Fig. S4). Based on the Neighbor‐Net algorithm conducted on our SNP data, SplitsTree produced a tree for each species (Fig. 5) that was congruent with the genetic clusters inferred by sNMF and PCA.
Figure 5.
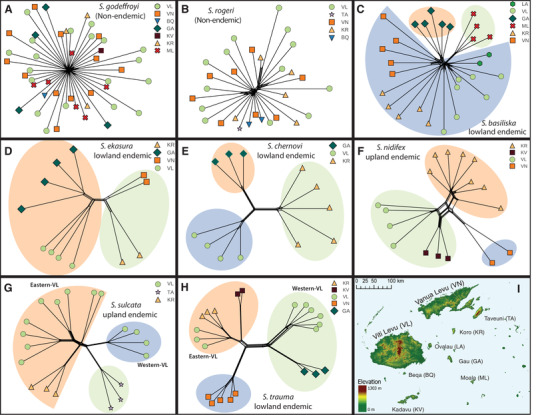
Neighbor‐net tree of eight Strumigenys species in Fiji inferred in SplitsTree to characterize genetic differentiation among different islands. No clear population structure was found in the two nonendemic species (A, B). All the endemic species exhibited clear population structures (C‐H). (I) The map of the Fiji archipelago. Colored nodes at the terminal of tree correspond to the sample from different islands. The colored shade indicates the genetic clusters inferred from sNMF.
The F ST values among all populations of different species vary from nearly 0 to 0.3 indicating the variable levels of geographic population structure across species (Fig. 6A). For example, the F ST values of S. godeffroyi and S. rogeri are very small suggesting little genetic differentiation among island populations in nonendemic species, while the F ST values in S. sulcata are around 0.3 indicating that populations from different islands were notably divergent from each other. It is worth noting that the three species from the small‐bodied clade (S. basiliska, S. chernovi, and S. ekasura) showed significantly lower genetic differentiation compared to the two species from the large‐bodied clade (S. nidifex and S. sulcata, P < 0.001, Welch's two‐sample t test).
Figure 6.
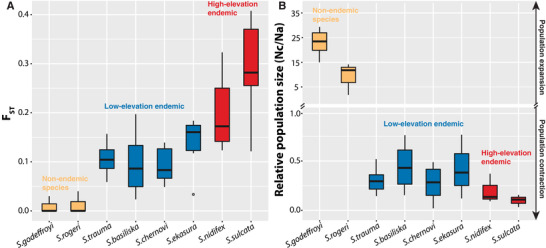
Average F ST and relative population size change of eight Strumigenys species in Fiji. (A) The average F ST for pairwise comparisons of all populations from different islands for each species. (B) The relative population size change of all populations for each species inferred from the best demographic model in dadi. Relative population size is represented by the ratio of current population size (Nc) to ancestral population size (Na). The box encloses the 25–75th percentiles of the values, the whisker extends to 1.5 times the interquartile range.
Population demographic history
The best demographic models inferred by δaδi indicate dramatic population expansion in the two nonendemic species with their population size increasing 22 and 10 times instantaneously after colonization (S. godeffroyi and S. rogeri, respectively, Supporting information Table S12, Fig. 6B). On the contrary, we detected exponential population contraction for all the endemic species with an average population decline of 0.25 times to their ancestral population size, Na (Supporting information Table S12, Fig. 6B). The population contraction of the one endemic species outside of the main radiation (S. trauma) was not significant (confidence interval including 0, Supporting information Table S12). Among the five other endemic species included in the analysis, the decline of population size in upland endemics (S. nidifex, and S. sulcata) was greater compared to that of the lowland endemic species (S. basiliska, S. ekasura, and S. chernovi) (0.15 Na compared to 0.36 Na; Fig. 6B, Supporting information Fig. S5).
Our fastsimcoal2 analysis produced similar results as the δaδi analysis: population contraction in all six endemic species, and population expansion in the two nonendemic species. Detailed results can be found in Supporting information Table S13.
Discussion
Understanding the processes assembling island biota has long been a fascination of ecological and evolutionary science. Modern methods of phylogenomics, geometric morphometrics, and population genomics applied to communities of species offer powerful new tools to test ideas about community assembly (Andújar et al. 2015; Sarnat et al. 2017; Cotoras et al. 2018; Maestri et al. 2018). Our multifaceted analyses found that the ecological and evolutionary assembly of Fijian Strumigenys community is partially consistent with a priori expectations based on island biogeography theory, and existing ideas in the system such as the taxon cycle.
COLONIZATION AND ADAPTIVE RADIATION IN FIJI
Our phylogenomic analysis of the Fijian Strumigenys reveals that all the endemic long‐mandibled species are derived from a single colonization from Asia 9–14 Ma followed by in situ cladogenesis. The colonization date (9–14 Ma) is also consistent with other studies of Fijian ant radiations (Lucky and Sarnat 2010; Sarnat and Moreau 2011). This was in contrast to the a priori expectation based on the fact that long‐mandibled species belong to different morphological species groups present in the Indo‐Pacific region. The only other endemic species, S. trauma, belongs to a widespread short‐mandibled group spread across the Indo‐Pacific.
Within the in situ radiation of Fijian Strumigenys, we found evidence of ecological niche diversification, with one endemic clade found in lower‐elevation habitats and the other found primarily in higher‐elevation habitats. However, because these two sister clades are derived from the same colonization event, it is difficult to determine the direction of niche shifts and reconstruct the ecological status of the original colonist. Ancestral state reconstruction inferred an intermediate ancestral elevation, although models of continuous trait evolution inherently assume evolution is undirected so ancestors are typically inferred to be within the variation of extant taxa (Supporting information Fig. S2). A dynamic, such as the taxon cycle, whereby all lineages shift in the same direction away from an ancestor will result in misleading inferences. So while we can reject dispersal assembly as a mechanism of community assembly of ecomorphological variation, it is clear that not all species shift into more highland/interior as predicted by the taxon cycle, rather there is diversification in elevational affinity.
In total, the morphological analysis is consistent with an adaptive radiation of Strumigenys in Fiji, rather than an evolution trend toward specific island syndromes. The phenotypic diversification was dramatic, with a relatively small number of Fijian species filling much of the global morphospace (Fig. 3D). The species in the clade associated with upland habitat have undergone a marked increase in body size relative to their ancestors (Fig. 3C), and S. nidifex is among the largest Strumigenys species in the world. In addition to body size divergence, the 3D geometric morphometric analysis also indicates morphological divergence in head, mandible, and mesosoma shapes among species and between those two clades (Fig. 4). Relative mandible size is notable as an important ecological trait correlated with more epigaiec or hypogaiec lifestyles (Wilson 1953; Brown and Wilson 1959). It general is highly conserved among species groups (Bolton 2000), and clades (D. Booher, unpubl data). Thus, the rapid shortening in mandible size and shift to hypogaeic lifestyle is a remarkable change within this small clade, and evidence for ecological opportunity promoting ecomorphological diversification after colonization.
DECLINES IN DISPERSAL ABILITY AND POPULATION SIZE
Our population genomic analysis largely supports the prediction that lineages show reduced dispersal ability and undergo demographic decline as they become affiliated with interior and higher‐elevation habitats (Wilson 1959, 1961). First, species from the upland clade both show greater genetic differentiation among their geographic subpopulations compared to the species from the lowland. For example, the average pairwise F ST in S. sulcata is about 0.3, while the average pairwise F ST in S. basiliska is about 0.1. This suggests that the upland species have reduced interisland dispersal ability and cannot maintain gene flow between their geographic populations. Moreover, the upland species also showed stronger population contraction compared to the lowland species. The strong link between morphological and ecological niche specialization—together with the decline in dispersal ability and population growth—supports the later phase of taxon cycles which suggests the shift requires adaptation and reduces commonness (Economo and Sarnat 2012; Economo et al. 2015). Future studies are needed to identify the morphological and geographic mechanisms behind this reduction of dispersal ability.
In contrast, the two nonendemic species showed very low genetic differentiation and high population growth. Both species are widely distributed across Southeast Asia and Oceania, and their strong dispersal ability should permit frequent gene flow among island populations. This is in part because in the lowland habitats they inhabit and likely to have a high connectivity, facilitating the oceanic dispersal among islands and reducing the level of genetic differentiation (Economo and Sarnat 2012; Economo et al. 2015).
ECOLOGICAL RELEASE AND THE ROLE OF PRIORITY EFFECTS
We examined whether there is any evidence that later arrivals are competitively superior and usurp the early colonist lineages, the key component of the taxon cycle (Wilson 1959, 1961 . This prediction would have been supported if we observed multiple independent colonization origins of Fijian endemic Strumigenys lineages with ecomorphological radiations. However, our results did not support this prediction since ecomorphological divergence of most Fijian endemic Strumigenys were descended from the initial colonization. On the contrary, the postcolonization ecomorphological diversification may suggest an important role of ecological release and priority effect in shaping community assembly of Strumigenys in Fiji.
Our phylogenomic analysis of Fijian Strumigenys revealed that larger body size of Fijian endemic Strumigenys ants resulted from the ecological release of its ancestor upon colonizing the archipelago. When the ancestral Strumigenys lineage arrived in the Fiji, it quickly expanded its range and ecological niche. Some descendent lineages evolved larger body size as a result of ecological release, enabling them to catch larger prey that was previously inaccessible expand their ecological dimensions (e.g., reaching higher elevation). Moreover, we also observed divergence of relative eye and mandible size as well as variations among head, mandible, and mesosoma shape in the Fijian Strumigenys, suggesting they have filled multiple available microhabitats (niches). For example, species with small eye size, such as S. oasis, are typically subterranean ants that live underground. This postcolonization ecomorphological radiation in the context of ecological release can produce a strong positive feedback that promotes evolution‐mediated priority effects: the radiation and evolution of early arrivals has strong influence on the community assembly (Vannette and Fukami 2014; Fukami 2015; De Meester et al. 2016).
It is worth noting that another potential reason why we did not find multiple radiations of Fijian Strumigenys could be because the colonization event was so rare since the degree of evolution‐mediated priority effect largely depends on the race between evolution of initial colonists and the immigration of preadapted species (De Meester et al. 2016). For example, thanks to human‐mediated invasions, the two nonendemic Strumigenys species are experiencing significant population expansion across the whole archipelago. It is unclear whether the nonendemic species represent the vanguard of new radiations.
Conclusions
Modern methods in genomics, imaging, and computation are finally allowing us to gain traction on testing ideas that have been around for decades. Using multiple lines of investigation, this study advances our knowledge of community assembly by unraveling the evolutionary history of Fijian Strumigenys ants. We find that the remote island community was primarily assembled through in situ diversification rather than dispersal assembly. The ecomorphological diversification was intense, filling much of the global morphospace and different ecological and elevational niches on the island. These niche shifts were also associated with loss of dispersal ability and demographic decline. However, although there is evidence of niche shifts, it is not clear whether they were unidirectional toward upland habitats or occurring in different directions as species diversified. It is clear that even old lineages can remain associated with lower elevation habitats even as others trend upwards, thus, there is not a single pathway of changes that all species follow. Overall, our findings for the Fijian Strumigenys echoes in large part patterns observed in the second most species‐rich genus in Fiji, Pheidole (Sarnat and Moreau 2011; Economo and Sarnat 2012; Darwell et al. 2020), and only partially support the presence of taxon cycles. It remains to be seen whether these cases reflect general patterns across the Fijian and broader Indo‐Pacific ant faunas, but the integration of phylogenomic, phenomic, and population genomic tools offers a powerful approach to evaluating longstanding ideas in ecology and evolution.
Associate Editor: I. Sanmartín
Handling Editor: M. R. Servedio
Supporting information
Figure S1. SVDQuartets based species tree of Fijian Strumigenys inferred by using 330,288 unlinked SNPs and 19,720,001 possible quartets.
Figure S2. Ancestral state reconstruction of the mean elevation in long‐mandibled Strumigenys species.
Figure S3. Principal components analyses (PCA) on SNP genotypes for eight Strumigenys species in Fiji to characterize genetic di_erentiation among di_erent islands.
Figure S4. Estimation of population structure of eight Strumigenys species in Fiji based on sNMF analysis on SNP genotype data from RADseq.
Figure S5. Inferred demographic model for eight Strumigenys species in Fiji.
Table S1. Strumigenys specimens information.
Supplementry Material
AUTHOR CONTRIBUTIONS
CL, EPE, and ASM conceived of the study. CL, EMS, EPE, and DB performed the specimen work. ASM performed the DNA sequencing, CL and FHG performed the CT scanning, NRF and CL performed the morphometrics analysis, CL and CD analyzed the molecular data. CL, EPE, ASM, and YK interpreted the data. CL and EPE wrote the article, with input from all coauthors.
ACKNOWLEDGMENTS
We wish to thank the Okinawa Institute of Science and Technology Graduate University (OIST) DNA Sequencing Section, Scientific Computing and Support Section, and Imaging Section for assistance with sequencing, HPC, and accessing the microCT scanner, respectively. We thank Issac Overcast for the suggestions on running ipyrad pipeline. We thank Mandy Tin, Jo Ann Tan, Julia Janicki, John Deyrup, Kenneth Dudley, and Masako Ogasawara for assisting in different aspects of the research. We thank Neal Evenhuis, Dan Bickel, Akanisi Caganitoba, Dave Olson, Sunil Prasad, and many Fijian landowners and local experts for their roles in the NSF Fiji Terrestrial Arthropod Survey project which collected many of the specimens used in this study. We also thank Richard Corlett and Nate Sanders for their comments on an early version of this manuscript. We appreciate the valuable comments from the Editor and two anonymous reviewers that improved the manuscript. This work was supported by subsidy funding to OIST, and JSPS Kakenhi grants (No. 17K15180 to E. P. Economo, No. 17K15178 to N. R. Friedman, No. 18K14768 to F. H. Garcia, No. 16J00372 to C. Liu). E. P. Economo was additionally funded by a grant from the Japan Ministry of Environment (Environment Research and Technology Development Fund no. 4‐1904).
DATA ARCHIVING
All the sequence data have been submitted to DNA Data Bank of Japan (DDBJ) under the bioproject PRJDB9763. The Dryad doi for our data is https://doi.org/10.5061/dryad.k3j9kd54m.
LITERATURE CITED
- Adler, G. H. , and Levins R.. 1994. The island syndrome in rodent populations. Q. Rev. Biol. 69:473–489. [DOI] [PubMed] [Google Scholar]
- Andújar, C. , Arribas P., Ruzicka F., Crampton‐Platt A., Timmermans M. J. T. N., and Vogler A. P.. 2015. Phylogenetic community ecology of soil biodiversity using mitochondrial metagenomics. Mol. Ecol. 24:3603–3617. [DOI] [PubMed] [Google Scholar]
- Balke, M. , Wewalka G., Alarie Y., and Ribera I.. 2007. Molecular phylogeny of pacific island colymbetinae: radiation of New Caledonian and Fijian species (Coleoptera, Dytiscidae). Zool. Scr. 36:173–200. [Google Scholar]
- Bolger, A. M. , Lohse M., and Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, B. 2000. The ant tribe Dacetini. American Entomological Institute, Gainesville, FL. [Google Scholar]
- Bouckaert, R. , Heled J., Kühnert D., Vaughan T., Wu C. H., Xie D., Suchard M. A., Rambaut A., and Drummond A. J.. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W. L. , and Wilson E. O.. 1959. The evolution of the Dacetine ants. Q. Rev. Biol. 34:278–294. [Google Scholar]
- Bryant, D. , and Moulton V.. 2004. Neighbor‐net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255–265. [DOI] [PubMed] [Google Scholar]
- Burns, K. C. 2019. Evolution in isolation: the search for an island syndrome in plants. Cambridge Univ. Press, Cambridge, U.K. [Google Scholar]
- Carlquist, S. 1967. The biota of long‐distance dispersal. V. plant dispersal to Pacific islands. Bull. Torrey Bot. Club 94:129–162. [Google Scholar]
- Chiba, S. 2004. Ecological and morphological patterns in communities of land snails of the genus Mandarina from the Bonin islands. J. Evol. Biol. 17:131–143. [DOI] [PubMed] [Google Scholar]
- Chifman, J. , and Kubatko L.. 2014. Quartet inference from SNP data under the coalescent model. Bioinformatics 30:3317–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse, R. M. , Janda M., Blanchard B., Sharma P., Hoffmann B. D., Andersen A. N., Czekanski‐Moir J. E., Krushelnycky P., Rabeling C., Wilson E. O., et al. 2015. Molecular phylogeny of Indo‐Pacific carpenter ants (Hymenoptera: Formicidae, Camponotus) reveals waves of dispersal and colonization from diverse source areas. Cladistics 31:424–437. [DOI] [PubMed] [Google Scholar]
- Coffman, A. J. , Gutenkunst R. N., Hsieh P. H., and Gravel S.. 2015. Computationally efficient composite likelihood statistics for demographic inference. Mol. Biol. Evol. 33:591–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, B. D. , Pringle C. M., and Hughes J. M.. 2008. Molecular evidence for sequential colonization and taxon cycling in freshwater decapod shrimps on a Caribbean island. Mol. Ecol. 17:1066–1075. [DOI] [PubMed] [Google Scholar]
- Cotoras, D. D. , Bi K., Brewer M. S., Lindberg D. R., Prost S., and Gillespie R. G.. 2018. Co‐occurrence of ecologically similar species of Hawaiian spiders reveals critical early phase of adaptive radiation. BMC Evol. Biol. 18:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek, P. , Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwell, C. T. , Fischer G., Sarnat E. M., Friedman N. R., Liu C., Baiao G., Mikheyev A. S., and Economo E. P.. 2020. Genomic and phenomic analysis of island ant community assembly. Mol. Ecol. 00:1–17. [DOI] [PubMed] [Google Scholar]
- De Meester, L. , Vanoverbeke J., Kilsdonk L. J., and Urban M. C.. 2016. Evolving perspectives on monopolization and priority effects. Trends Ecol. Evol. 31:136–146. [DOI] [PubMed] [Google Scholar]
- Dexter, K. G. 2010. The influence of dispersal on macroecological patterns of lesser Antillean birds. J. Biogeogr. 37:2137–2147. [Google Scholar]
- Deyrup, M. , and Cover S.. 2009. Dacetine ants in southeastern North America (Hymenoptera: Formicidae). Southeast. Nat. 8:191–212. [Google Scholar]
- Drummond, A. J. , and Rambaut A.. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton, D. A. R. , and Overcast I.. 2018. ipyrad: interactive assembly and analysis of RADseq data sets. [DOI] [PubMed]
- Eaton, D. A. R. , and Ree R. H.. 2013. Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae). Syst. Biol. 62:689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton, D. A. R. , Elizabeth L. S., Park B., and Donoghue M. J.. 2017. Misconceptions on missing data in RAD‐seq phylogenetics with a deep‐scale example from flowering plants. Syst. Biol. 66:399–412. [DOI] [PubMed] [Google Scholar]
- Economo, E. P. , and Sarnat E. M.. 2012. Revisiting the ants of Melanesia and the taxon cycle: historical and human‐mediated invasions of a tropical archipelago. Am. Nat. 180:E1–E16. [DOI] [PubMed] [Google Scholar]
- Economo, E. P. , Sarnat E. M., Janda M., Clouse R., Klimov P. B., Fischer G., Blanchard B. D., Ramirez L. N., Andersen A. N., Berman M., et al. 2015. Breaking out of biogeographical modules: range expansion and taxon cycles in the hyperdiverse ant genus Pheidole . J. Biogeogr. 42:2289–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier, L. , Dupanloup I., Huerta‐Sánchez E., Sousa V. C., and Foll M.. 2013. Robust demographic inference from genomic and SNP data. PLoS Genet 9:e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frichot, E. , Mathieu F., Trouillon T., Bouchard G., and François O.. 2014. Fast and efficient estimation of individual ancestry coefficients. Genetics 196:973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami, T. 2015. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 46:1–23. [Google Scholar]
- Gillespie, R. 2004. Community assembly through adaptive radiation in Hawaiian spiders. Science 303:356–359. [DOI] [PubMed] [Google Scholar]
- Gillespie, R. G. 2002. Biogeography of spiders on remote oceanicislands of the Pacific: archipelagoes as stepping stones? J. Biogeogr. 29:655–662. [Google Scholar]
- Gillespie, R. G. 2016. Island time and the interplay between ecology and evolution in species diversification. Evol. Appl. 9:53–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, R. G. , Baldwin B. G., Waters J. M., Fraser C., Nikula R., and Roderick G. K.. 2012. Long‐distance dispersal—a framework for hypothesis testing. Trends Ecol. Evol. 27:47–56. [DOI] [PubMed] [Google Scholar]
- Gillespie, R. G. , Brewer M. S., and Roderick G. K.. 2017. Ancient biogeography of generalist predators on remote oceanic islands. J. Biogeogr. 44:1098–1109. [Google Scholar]
- Grant, P. R. , and Grant B. R.. 2002. Unpredictable evolution in a 30‐year study of Darwin's finches. Science 296:707–711. [DOI] [PubMed] [Google Scholar]
- Gutenkunst, R. N. , Hernandez R. D., Williamson S. H., and Bustamante C. D.. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5:e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney, L. 2000. Dynamic disequilibrium: a long‐term, large‐scale perspective on the equilibrium model of island biogeography. Glob. Ecol. Biogeogr. 9:59–74. [Google Scholar]
- Hubbell, S. P. 2001. The unified neutral theory of biodiversity and biogeography. Princeton Univ. Press, Princeton, NJ. [DOI] [PubMed] [Google Scholar]
- Huson, D. H. , and Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267. [DOI] [PubMed] [Google Scholar]
- Jønsson, K. A. , Irestedt M., Christidis L., Clegg S. M., Holt B. G., and Fjeldså J.. 2014. Evidence of taxon cycles in an Indo‐Pacific passerine bird radiation (Aves: Pachycephala). Proc. R. Soc. B 281:20131727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov, A. M. , Aberer A. J., and Stamatakis A.. 2015. ExaML version 3: a tool for phylogenomic analyses on supercomputers. Bioinformatics 31:2577–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear, R. , Frandsen P. B., Wright A. M., Senfeld T., and Calcott B.. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34:772–773. [DOI] [PubMed] [Google Scholar]
- Losos, J. B. 1992. A critical comparison of the taxon‐cycle and character‐displacement models for size evolution of Anolis lizards in the Lesser Antilles. Copeia 2:279–288. [Google Scholar]
- Losos, J. B. , Jackman T. R., Larson A., Queiroz K., and Rodríguez‐Schettino L.. 1998. Contingency and determinism in replicated adaptive radiations of island lizards. Science 279:2115–2118. [DOI] [PubMed] [Google Scholar]
- Lucky, A. , and Sarnat E. M.. 2010. Biogeography and diversification of the Pacific ant genus Lordomyrma Emery. J. Biogeogr. 37:624–634. [Google Scholar]
- Macarthur, R. H. , and Wilson E. O.. 1967. The theory of island biogeography. Princeton Univ. Press, Princeton, NJ. [Google Scholar]
- Maestri, R. , Monteiro L. R., Fornel R., de Freitas T. R. O., and Patterson B. D.. 2018. Geometric morphometrics meets metacommunity ecology: environment and lineage distribution affects spatial variation in shape. Ecography 41:90–100. [Google Scholar]
- Matos‐Maraví, P. , Clouse R. M., Sarnat E. M., Economo E. P., LaPolla J. S., Borovanska M., Rabeling C., Czekanski‐Moir J., Latumahina F., Wilson E. O., et al. 2018a. An ant genus‐group (Prenolepis) illuminates the biogeography and drivers of insect diversification in the Indo‐Pacific. Mol. Phylogenetics Evol. 123:16–25. [DOI] [PubMed] [Google Scholar]
- Matos‐Maraví, P. , Matzke N. J., Larabee F. J., Clouse R. M., Wheeler W. C., Sorger D. M., Suarez A. V., and Janda M.. 2018b. Taxon cycle predictions supported by model‐based inference in Indo‐Pacific trap‐jaw ants (Hymenoptera: Formicidae: Odontomachus). Mol. Ecol. 27:4090–4107. [DOI] [PubMed] [Google Scholar]
- Matzke, N. J. 2016. Trait‐dependent dispersal models for phylogenetic biogeography, in the R package BioGeoBEARS. Integr. Comp. Biol. 56:E330. [Google Scholar]
- Miles, A. , and Harding N.. 2016. scikit‐allel: a Python package for exploring and analysing genetic variation data. Available from https://github.com/cggh/scikit-allel
- Novosolov, M. , Raia P., and Meiri S.. 2013. The island syndrome in lizards. Glob. Ecol. Biogeogr. 22:184–191. [Google Scholar]
- Portik, D. M. , Leaché A. D., Rivera D., Barej M. F., Burger M., Hirschfeld M., Rödel M. O., Blackburn D. C., and Fujita M. K.. 2017. Evaluating mechanisms of diversification in a Guineo‐Congolian tropical forest frog using demographic model selection. Mol. Ecol. 26:5245–5263. [DOI] [PubMed] [Google Scholar]
- Pregill, G. K. , and Olson S. L.. 1981. Zoogeography of West Indian vertebrates in relation to Pleistocene climatic cycles. Annu. Rev. Ecol. Evol. Syst. 12:75–98. [Google Scholar]
- Reding, D. M. , Foster J. T., James H. F., Pratt H. D., and Fleischer R. C.. 2009. Convergent evolution of ‘creepers’ in the Hawaiian honeycreeper radiation. Biol. Lett. 5:221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell, L. J. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3:217–223. [Google Scholar]
- Ricklefs, R. E. , and Bermingham E.. 1999. Taxon cycles in the Lesser Antillean avifauna. Ostrich 70:49–59. [Google Scholar]
- Ricklefs, R. E. , and Bermingham E.. 2002. The concept of the taxon cycle in biogeography. Glob. Ecol. Biogeogr. 11:353–361. [Google Scholar]
- Ricklefs, R. E. , and Cox G. W.. 1972. Taxon cycles in the west Indian avifauna. Am. Nat. 106:195–219. [Google Scholar]
- Roughgarden, J. D. , and Pacala S.. 1989. Taxon cycle among Anolis lizard populations: review of evidence Pp. 403—432 in Otte D. and Endler J. A., eds. Speciation and adaptation. National Academy of Science, Philadelphia, PA. [Google Scholar]
- Sarnat, E. M. 2006. Lordomyrma (Hymenoptera: Formicidae) of the Fiji islands. Occas. Pap. Bernice P. Bishop Mus. 90:9–42. [Google Scholar]
- Sarnat, E. M. 2008. A taxonomic revision of the Pheidole roosevelti‐group (Hymenoptera: Formicidae) in Fiji. Zootaxa 1767:1–36. [Google Scholar]
- Sarnat, E. M. , and Economo E. P.. 2012. Ants of Fiji. Univ. Cal. Publ. Entomol. 132:1–398. [Google Scholar]
- Sarnat, E. M. , and Moreau C. S.. 2011. Biogeography and morphological evolution in a Pacific island ant radiation. Mol. Ecol. 20:114–130. [DOI] [PubMed] [Google Scholar]
- Sarnat, E. M. , Friedman N. R., Fischer G., Lecroq‐Bennet B., and Economo E. P.. 2017. Rise of the spiny ants: diversification, ecology and function of extreme traits in the hyperdiverse genus Pheidole (Hymenoptera: Formicidae ). Biol. J. Linnean Soc. 122:514–538. [Google Scholar]
- Sarnat, E. M. , Hita Garcia F., Dudley K., Liu C., Fischer G., and Economo E. P.. 2019. Ready species one: exploring the use of augmented reality to enhance systematic biology with a revision of Fijian Strumigenys (Hymenoptera: Formicidae). Insect Syst. Divers. 3:1–43. [Google Scholar]
- Schluter, D. 2000. The ecology of adaptive radiation. Oxford Univ. Press, Oxford, U.K. [Google Scholar]
- Stamatakis, A. 2014. RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbauer, M. J. 2016. A generalization of the taxon cycle. J. Biogeogr. 44:1110–1112. [Google Scholar]
- Sukumaran, J. , and Knowles L. L.. 2018. Trait‐dependent biogeography: (re)integrating biology into probabilistic historical biogeographical models. Trends Ecol. Evol. 33:390–398. [DOI] [PubMed] [Google Scholar]
- Sukumaran, J. , Economo E. P., and Knowles L. L.. 2015. Machine learning biogeographic processes from biotic patterns: a new trait‐dependent dispersal and diversification model with model choice by simulation‐trained discriminant analysis. Syst. Biol. 65:525–545. [DOI] [PubMed] [Google Scholar]
- Tin, M. M. Y. , Economo E. P., and Mikheyev A. S.. 2014. Sequencing degraded DNA from non‐destructively sampled museum specimens for RAD‐tagging and low‐coverage shotgun phylogenetics. PLoS One 9:e96793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tin, M. M. Y. , Rheindt F. E., Cros E., and Mikheyev A. S.. 2015. Degenerate adaptor sequences for detecting PCR duplicates in reduced representation sequencing data improve genotype calling accuracy. Mol. Ecol. Resour. 15:329–336. [DOI] [PubMed] [Google Scholar]
- Vannette, R. L. , and Fukami T.. 2014. Historical contingency in species interactions: towards niche‐based predictions. Ecol. Lett. 17:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente, L. , Phillimore A. B., Melo M., Warren B. H., Clegg S. M., Havenstein K., Tiedemann R., Illera J. C., Thébaud C., Aschenbach T., et al. 2020. A simple dynamic model explains the diversity of island birds worldwide. Nature 579:92–96. [DOI] [PubMed] [Google Scholar]
- Ward, P. S. , Brady S. G., Fisher B. L., and Schultz T. R.. 2015. The evolution of myrmicine ants: phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae). Syst. Entomol. 40:61–81. [Google Scholar]
- Warren, B. H. , Simberloff D., Ricklefs R. E., Aguilée R., Condamine F. L., Gravel D., Morlon H., Mouquet N., Rosindell J., Casquet J., et al. 2015. Islands as model systems in ecology and evolution: prospects fifty years after MacArthur‐Wilson. Ecol. Lett. 18:200–217. [DOI] [PubMed] [Google Scholar]
- Whittaker, R. J. 1998. Island biogeography: ecology, evolution, and conservation. Oxford Univ. Press, Oxford, U.K. [Google Scholar]
- Whittaker, R. J. , Fernández‐Palacios J. M., Matthews T. J., Borregaard M. K., and Triantis K. A.. 2017. Island biogeography: taking the long view of nature's laboratories. Science 357:1–7. [DOI] [PubMed] [Google Scholar]
- Wilson, E. O. 1953. The ecology of some north American Dacetine ants. Ann. Entomol. Soc. Am. 46:479–495. [Google Scholar]
- Wilson, E. O. 1959. Adaptive shift and dispersal in a tropical ant fauna. Evolution 13:122–144. [Google Scholar]
- Wilson, E. O. 1961. The nature of the taxon cycle in the melanesian ant fauna. Am. Nat. 95:169–193. [Google Scholar]
- Wilson, E. O. , and Taylor R. W.. 1967. The ants of Polynesia (Hymenoptera, Formicidae). Entomology Department, Bernice P. Bishop Museum, Honolulu, HI. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. SVDQuartets based species tree of Fijian Strumigenys inferred by using 330,288 unlinked SNPs and 19,720,001 possible quartets.
Figure S2. Ancestral state reconstruction of the mean elevation in long‐mandibled Strumigenys species.
Figure S3. Principal components analyses (PCA) on SNP genotypes for eight Strumigenys species in Fiji to characterize genetic di_erentiation among di_erent islands.
Figure S4. Estimation of population structure of eight Strumigenys species in Fiji based on sNMF analysis on SNP genotype data from RADseq.
Figure S5. Inferred demographic model for eight Strumigenys species in Fiji.
Table S1. Strumigenys specimens information.
Supplementry Material


