Abstract
Co-essentiality mapping has been useful to systematically cluster genes into biological pathways and identify gene functions1–3. Here, using the debiased sparse partial correlation (DSPC) method3, we construct a functional co-essentiality map for cellular metabolic processes across human cancer cell lines. This analysis reveals 35 modules associated with known metabolic pathways and further assigns metabolic functions to unknown genes. In particular, we discover C12orf49 as an essential regulator of cholesterol and fatty acid metabolism in mammalian cells. Mechanistically, C12orf49 localizes to the Golgi, binds site 1 protease (MBTPS1) and is necessary for the cleavage of its substrates, including SREBP transcription factors. This function depends on the evolutionarily conserved uncharacterized domain (DUF2054) and promotes cell proliferation under cholesterol depletion. Notably, c12orf49 depletion in zebrafish blocks dietary lipid clearance in vivo, phenocopying mbtps1 mutants. Finally, in an EHR-linked DNA biobank, C12orf49 is associated with hyperlipidemia through phenome analysis. Altogether, our findings reveal a conserved role for C12orf49 in cholesterol and lipid homeostasis and provide a platform to identify unknown components of other metabolic pathways.
While most components of metabolic pathways have been well-defined, a significant portion of metabolic reactions still has unidentified enzymes or regulatory components, even in lower organisms4–8. Co-essentiality mapping was previously used for systematic identification of large-scale relationships among individual components of gene sets1–3. Perturbation of enzymes or regulatory units involved in the same metabolic pathway should display similar effects on cellular fitness across cell lines, suggesting that correlation of essentiality profiles may provide the unique opportunity to identify unknown components associated with a particular metabolic function.
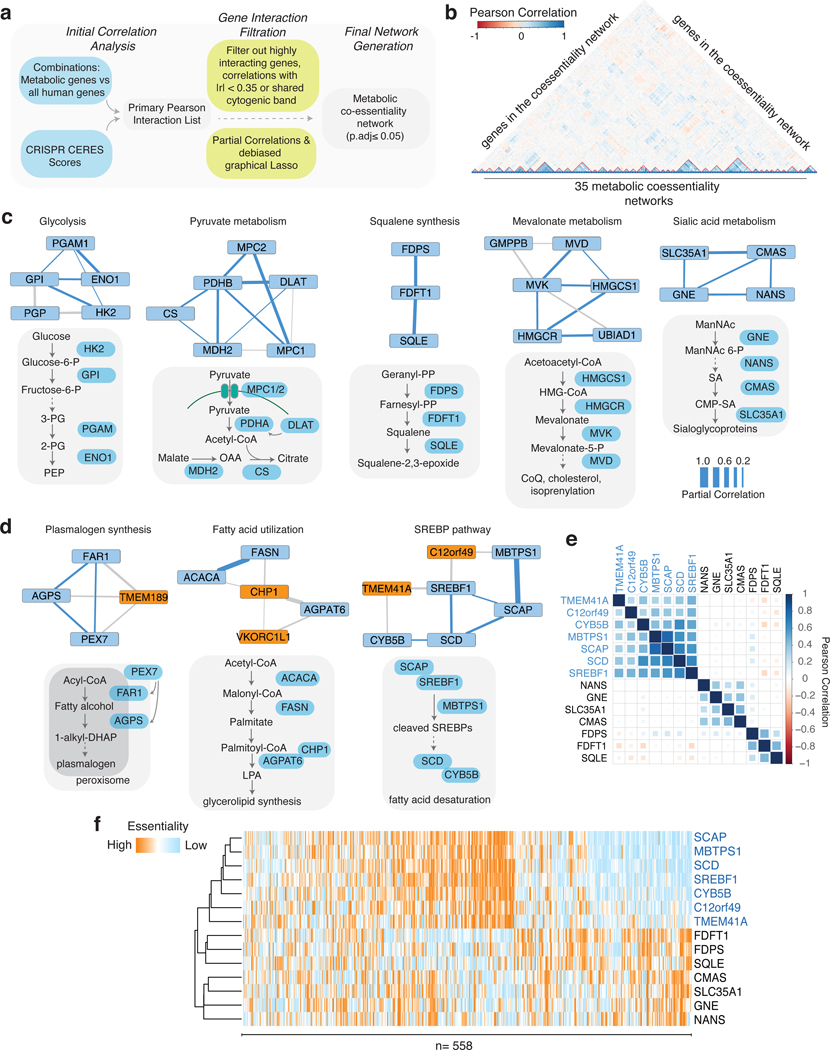
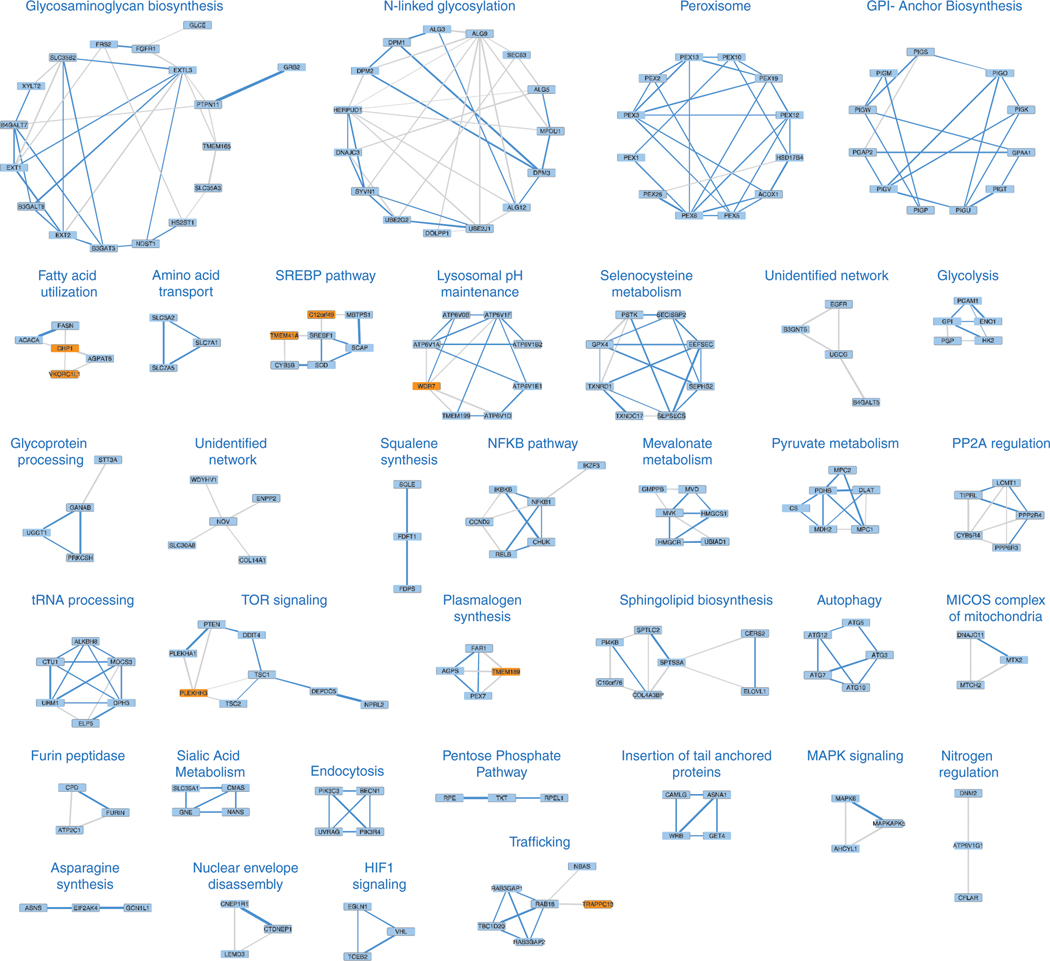
To generate a putative co-essentiality network for metabolic genes, we analyzed genetic perturbation datasets from the DepMap project collected from 558 cancer cell lines (Fig. 1a)9–11. Existing computational methods for constructing co-essentiality networks primarily rely on Pearson correlation, which is not suitable for distinguishing between direct and indirect gene associations and leads to false positive edges in the network (Extended Data Fig. 1a,b). However, gaussian graphical models (GGM) calculate partial correlation and offer unique advantage over commonly used Pearson correlation networks by automatically removing indirect associations among genes from the network, hence reducing false positives and producing a small number of high confidence set of putative interactions for follow-up validation12. We therefore applied debiased sparse partial correlation (DSPC), a GGM technique, to measure associations between the essentiality scores of genes from human cancer cell lines. In prior work13, we have successfully used DSPC to build networks among metabolites and identified new biological compounds. Of note, this method, while useful for generating high confidence lists, does not account for dependence among cell lines, a key strength of previously published work3,11. After removing networks with large numbers of components (i.e. electron transport chain), we focused on genes with a high Pearson correlation (|r|>0.35) with at least one of the 2,998 metabolism-related genes in the dataset. Our analysis of positively correlated genes revealed a set of 202 genes organized in 35 metabolic networks, 33 of which we can assign a metabolic function using literature searches and STRING database (Fig. 1b, Extended Data Fig. 2).
Figure 1, Genetic coessentiality analysis assigns metabolic functions to uncharacterized genes.
A. Scheme of the computational steps to generate the metabolic coessentiality network.
B. Heatmap depicting the partial correlation values of the essentialities of genes in the metabolic coessentiality networks.
C. Correlated essentialities of the genes encoding members of glycolysis, pyruvate metabolism, squalene synthesis, mevalonate and sialic acid metabolism. The thickness of the lines indicates the level of partial correlation.
D. Genetic coessentiality analysis assigns metabolic functions to uncharacterized genes. Orange and blue boxes show genes with unknown and known functions, respectively. The thickness of the lines is indicative of partial correlation.
E. Pearson correlation values of the essentiality scores of genes in indicated metabolic networks.
F. Unbiased clustering of fitness variation of indicated genes across 558 human cancer cell lines.
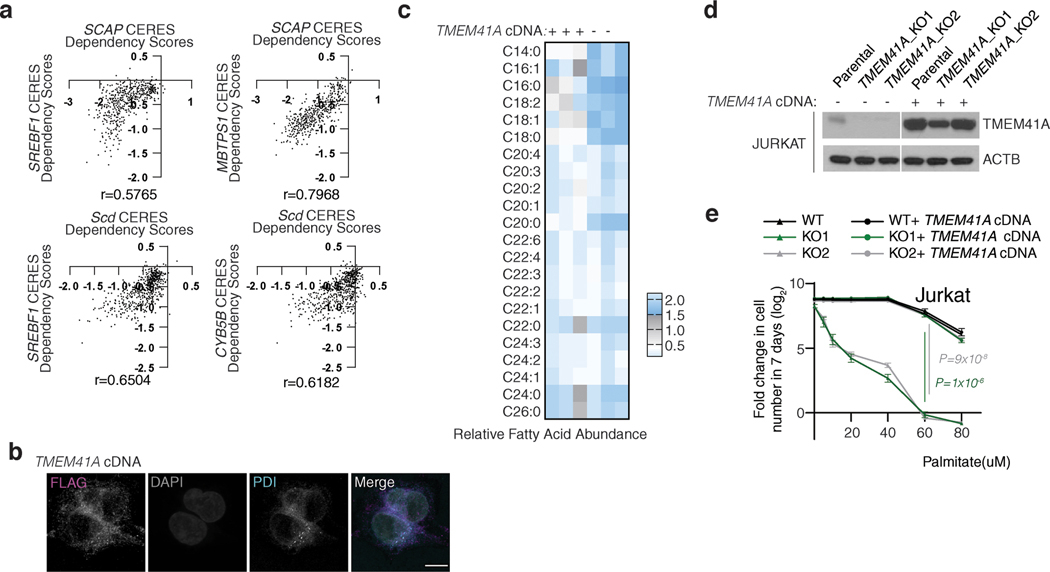
Among these networks are glycolysis (PGAM1, GPI, ENO1, HK2, PGP), squalene synthesis (FDPS, FDFT1, SQLE), sialic acid metabolism (SLC35A1, CMAS, GNE, NANS), plasmalogen synthesis (FAR1, AGPS, TMEM189, PEX7) and pyruvate utilization (MPC2, PDHB, DLAT, CS, MDH2, MPC1) but also networks that were not part of a known metabolic pathway, suggesting the presence of unidentified metabolic pathways (Fig. 1c). Our analysis also identified associations between genes of unknown function and those encoding components of well-characterized metabolic pathways. Interestingly, the functions of three of these genes have recently been discovered (Fig. 1d, Extended Data Fig. 2). UBIAD1, a prenyltransferase, has been shown to bind to HMGCR to promote its degradation at ER in the presence of sterols14. CHP1, which is associated with glycerolipid synthesis pathway in our analysis, binds to and is necessary for the function of the protein product of AGPAT6, the rate-limiting enzyme for glycerolipid synthesis15. Additionally, a recent study identified TMEM189, a gene associated with plasmalogen synthesis, as the elusive plasmanylethanolamine desaturase16. Interestingly, squalene and mevalonate synthesis clustered into different networks, consistent with additional functions of the branches of cholesterol metabolism. Indeed, while loss of HMG-CoA synthase would decrease all intermediates as well as cholesterol, loss of squalene synthase or downstream enzymes would decrease cholesterol but increase upstream intermediates, hence leading to different cellular outcomes17. Finally, several genes of unknown function, such as C12orf49 and TMEM41A, have correlated essentialities with those of genes encoding components of sterol regulatory element binding proteins (SREBP)-regulated lipid metabolism, raising the possibility that they may be involved in the regulation of SREBPs or their downstream targets (Fig. 1e,f; Extended Data Fig. 3a). Due to their strong correlation and unknown function, we focused our attention on these two genes.
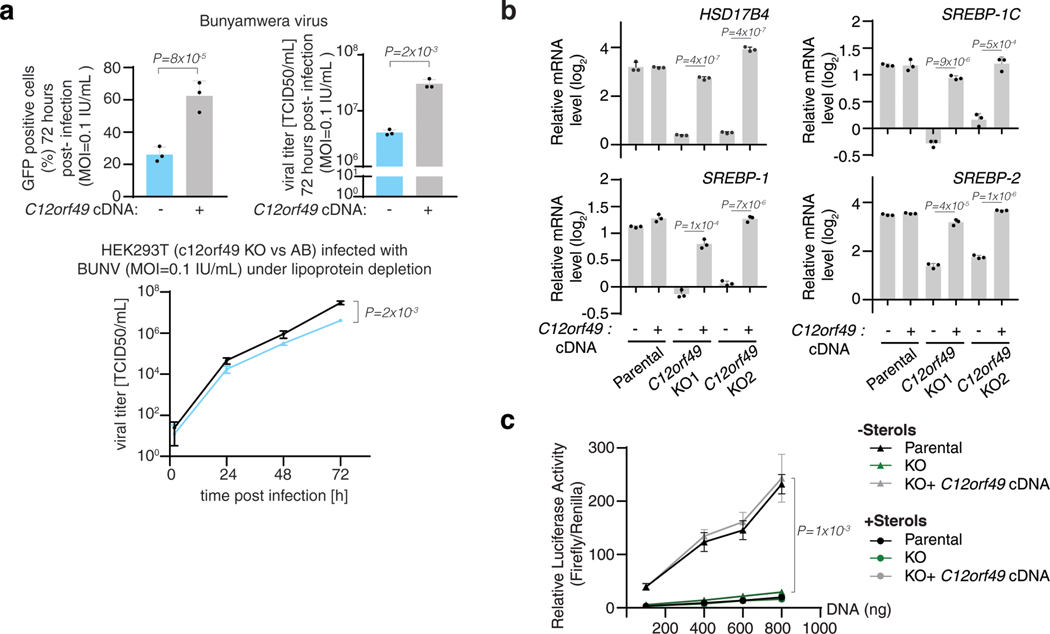
Sterol regulatory element binding proteins (SREBPs) are transcription factors that regulate transcription of genes encoding many enzymes in the cholesterol and fatty acid synthesis18. SREBPs are normally bound to endoplasmic reticulum (ER) membranes and are activated through a proteolytic cascade regulated by sterols19,20. Cleaved SREBPs localize to nucleus and induce expression of cholesterol synthesis genes enabling cells to survive under sterol depletion21,22. Given the strong coessentialities of C12orf49 and TMEM41A with the SREBP pathway, we hypothesized that these uncharacterized genes may be required for the activation of cholesterol synthesis and cell proliferation upon cholesterol deprivation. To address this possibility, we generated a small CRISPR library consisting of 103 sgRNAs targeting genes involved in SREBP maturation and lipid metabolism (3–8 sgRNA/gene) (Fig. 2a). Using this focused library, we performed negative selection screens for genes whose loss potentiates anti-proliferative effects of lipoprotein depletion. Among the scoring genes were MBTPS1 and SCAP, both of which are involved in SREBP processing23–25, but also C12orf49, a gene of unknown function that has not been previously linked to cholesterol metabolism (Fig. 2b, Extended Data Fig. 3b). Consistent with the screening results, depletion of C12orf49 strongly decreases proliferation of HEK293T, Jurkat and other cancer cell lines (U87 and MDA-MB-435) under cholesterol depletion, indicating a generalized role for C12orf49 in cholesterol homeostasis (Fig. 2c,d; Extended Data Fig. 3c,d). Importantly, expression of an sgRNA-resistant human C12orf49 cDNA in the null cells or free cholesterol addition completely restores proliferation under lipoprotein depletion (Extended Data Fig. 3e,f). None of the SREBPs scored likely due to highly complementary and redundant functions. Notably, TMEM41A was not a scoring gene in these screens, suggesting that it may function in other downstream processes regulated by SREBPs, such as lipid biosynthesis or saturation. Indeed, TMEM41A, similar to fatty acid synthesis enzymes, localizes to ER and its loss substantially impacts cellular lipid composition (Extended Data Fig. 4a-c). In individual assays, TMEM41A-null cells are more sensitive to the treatment of palmitate, which kills cells at high concentrations likely due to the dysregulation of the membrane saturation (Extended Data Fig. 4d,e). Altogether, these results identify C12orf49 and TMEM41A as major components of cholesterol and fatty acid metabolism.
Figure 2, C12orf49 is necessary for cholesterol synthesis and SREBP-induced gene expression in human cells.
A. Schematic for the focused CRISPR-Cas9 based genetic screen.
B. Differential sgRNA scores for the indicated genes. Blue bars indicate genes that are significantly and differentially essential under lipoprotein depletion. Boxes represent the median, and the first and third quartiles, and the whiskers represent the minimum and maximum of all data points. n=8 independent sgRNAs targeting each gene except for previously validated sgRNAs for ACSL3 (n=3) and ACSL4 (n=4)15.
C. Immunoblot of C12orf49 in the indicated cancer cell lines (left). Actin was used as the loading control. Fold change in cell number (log2) of Jurkat wild type and C12orf49_KO cells following 6-day growth under lipoprotein depletion with the indicated treatments (mean ± SD, n=3 biologically independent samples) (middle). Representative images of indicated cell lines under lipoprotein depletion at the end of the experiment (right).
D. Fold change in cell number (log2) of HEK293T wild type and C12orf49_KO cells following 6-day growth under lipoprotein depletion with the indicated treatments (mean ± SD, n=3 biologically independent samples).
E. Mass isotopologue analysis of cholesterol in Jurkat wild type and C12orf49_KO cells in the absence and presence of sterols after 48 hours of incubation with 13C-acetate (mean ± SD, n=3 biologically independent samples).
F. Fold change in mRNA levels (log2) of SREBP target genes in indicated Jurkat cell lines following 8h growth under lipoprotein depletion in the presence and absence of sterols (mean ± SD, n=3 biologically independent samples).
G. Immunoblots of SREBP target proteins in indicated Jurkat cell lines following 24h growth under lipoprotein depletion in the presence and absence of sterols. Actin was used as the loading control.
H. Immunoblots of mature SREBP1 and SREBP2 in indicated Jurkat cell lines following 24h growth under lipoprotein depletion in the presence and absence of sterols. Lamin B1 was used as the loading control.
I. Localization of SREBP1 in C12orf49-null HEK293T cells expressing control or C12orf49 cDNA under lipoprotein depletion in the presence or absence of sterols (Scale bar, 8 μm).
The experiments were repeated independently at least twice with similar results. Statistical significance was determined by two-tailed unpaired t-test.
We next sought to understand why cells require C12orf49 to proliferate under cholesterol depletion. To first determine whether C12orf49 is necessary for de novo cholesterol synthesis, we performed metabolite tracing experiments in Jurkat cells using [U-13C]-Acetate (Fig. 2e). While acetate contributes to cellular cholesterol under lipoprotein depletion, we observed significantly lower labeling in C12orf49-null cells, indicating a problem in the synthesis (Fig. 2e). Consistent with the requirement of sterols for viral infection26–28, C12orf49 loss also decreases Bunyamwera virus infectivity in mammalian cell lines and total viral titers (Extended Data Fig. 5a). As cholesterol synthesis pathway comprises over thirty successive steps that are transcriptionally regulated 22,29–32, we considered that a dysfunction in gene expression might lead to defective synthesis and reliance on extracellular cholesterol. Indeed, C12orf49-null cells fail to induce expression of cholesterol metabolism genes under sterol depletion (Fig. 2f,g). Furthermore, in line with the role of SREBPs in the transcription of cholesterol synthesis genes, loss of C12orf49 reduced mature (cleaved) SREBP protein levels and blocked nuclear translocation of SREBPs (Fig. 2h,i). Similarly, expression of other genes known to be induced by SREBPs, such as fatty acid synthase (FASN), low density lipoprotein receptor (LDLR), acetyl-coA carboxylase (ACC) and ATP citrate lyase (ACLY) did not change in C12orf49-null cells (Fig. 2f,g, Extended Data Fig. 5b). Finally, SREBPs fail to induce the transcription of the reporter luciferase under the control of sterol regulatory elements in C12orf49-null cells (Extended Data Fig. 5c). These results suggest that C12orf49, like SCAP and MBTPS1, is necessary for SREBP activation and subsequent regulation of its biosynthetic targets.
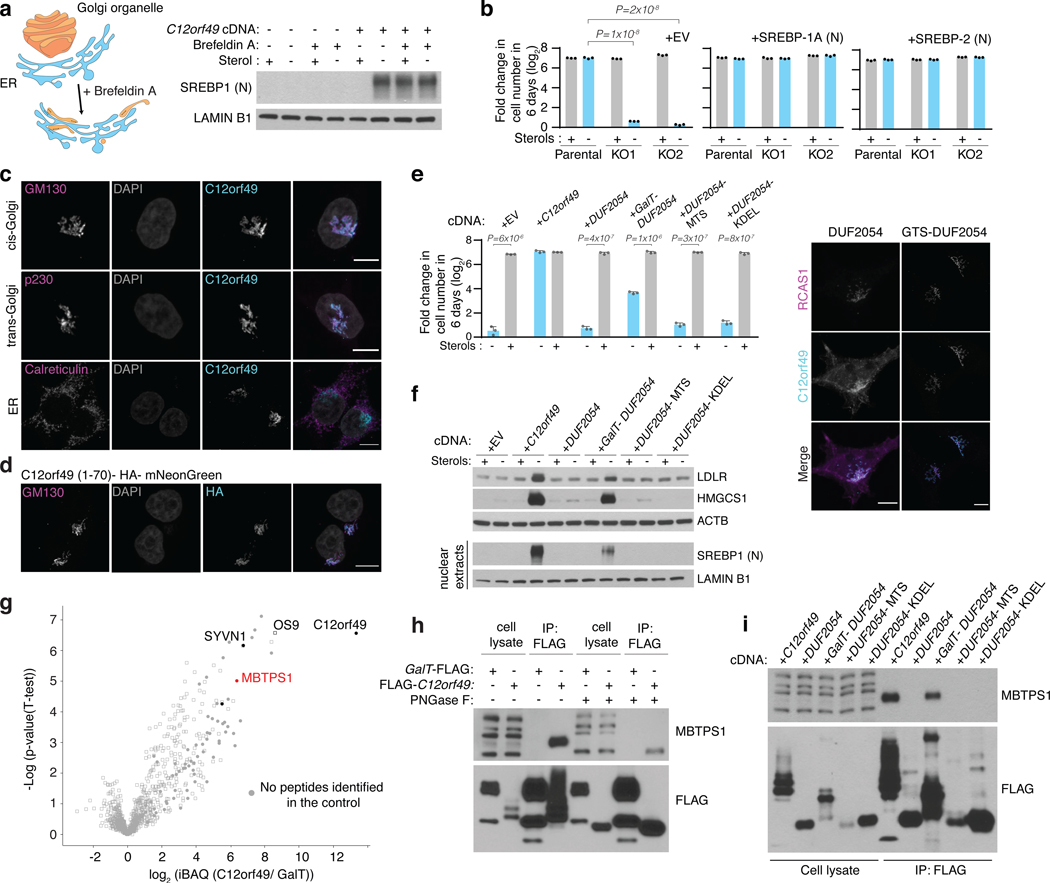
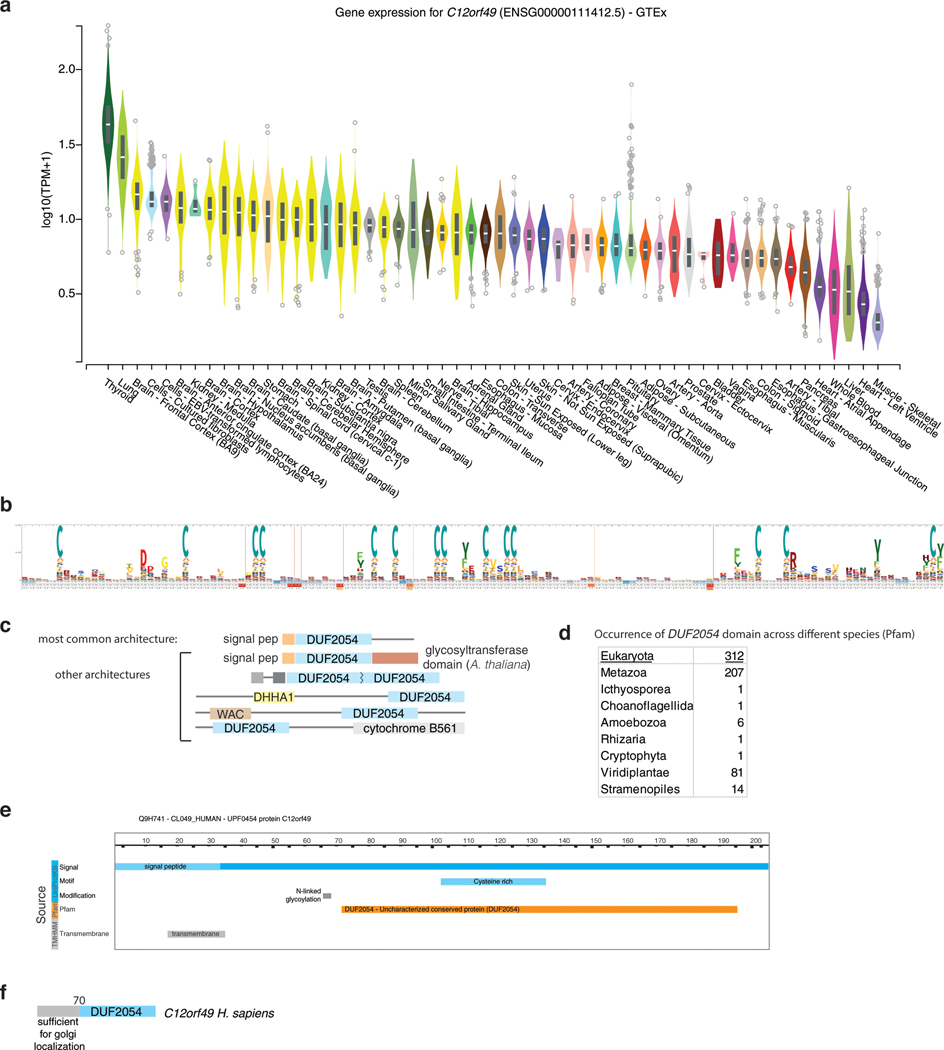
C12orf49 is ubiquitously expressed among different tissues (Extended Data Fig. 6a) and contains an uncharacterized conserved domain, DUF2054 (Extended Data Fig. 6b-e). Upon sterol depletion, SCAP, a chaperone protein, transports SREBP to the Golgi complex where it is subsequently cleaved by membrane bound transcription factor peptidase, Site 1 (MBTPS1, site-1-protease). The evidence that a primary role of C12orf49 may be in SREBP processing raised the question of where within this pathway C12orf49 functions. To address this, we treated cells with brefeldin A, which disassembles the Golgi compartments and redistributes them to the ER, eliminating the need for SREBP transport to the Golgi and allowing the cleavage of SREBP1 regardless of the presence of sterols33,34. Interestingly, brefeldin A treatment failed to induce SREBP cleavage in C12orf49-null cells, strongly suggesting that C12orf49 functions downstream of SCAP localization (Fig. 3a). Notably, overexpression of the mature SREBP isoforms completely eliminated the sensitivity of C12orf49-null cells, indicating that C12orf49 does not impact nuclear function of mature SREBP (Fig. 3b). Consistent with its role downstream of SCAP, C12orf49 mainly localizes to cis- and trans- Golgi (GM130 and p230, respectively) (Fig. 3c). While N-terminal region of C12orf49 provides the Golgi localization signal of the protein, this region is dispensable for SREBP activation (Fig. 3d). Instead, localizing the conserved DUF2054 domain to Golgi, but not to other organelles (ER and mitochondria), is sufficient to activate SREBP cleavage and signaling, as well as proliferation under lipoprotein depletion (Fig. 3e,f; Extended Data Fig. 6f).
Figure 3, C12orf49 is a Golgi localized protein and binds S1P to regulate cholesterol metabolism.
A. Scheme depicting the action of Brefeldin A which disassembles the Golgi compartments and redistributes them to the ER (left). Immunoblots of mature SREBP1 and SREBP2 in indicated Jurkat cells in the presence and absence of sterols or Brefeldin A (1 ug/ml) for 6 hours in the lipoprotein depleted serum (right). Lamin B1 was used as the loading control.
B. Fold change in cell number (log2) of Jurkat wild type and C12orf49_KO cells overexpressing a control or mature SREBP cDNA following 7-day growth under lipoprotein depleted serum in the absence or presence of sterols (mean ± SD, n=3 biologically independent samples).
C. Localization of C12orf49 to the Golgi. Wild type HEK293T cells expressing C12orf49 cDNA were processed for immunofluorescence analysis using antibodies against c12orf49, calreticulin (ER), p230 (trans-Golgi) and GM130 (cis-Golgi). White color indicates overlap. (Scale bar, 8 μm).
D. N-terminal region of C12orf49 is sufficient for Golgi localization. Wild type HEK293T cells expressing C12orf49(1–70)- HA-mNeonGreen cDNA were processed for immunofluorescence analysis using antibodies against HA and GM130 (Golgi). White color indicates overlap. (Scale bar, 8 μm)
E. Fold change in cell number (log2) of Jurkat C12orf49_KO cells overexpressing indicated cDNAs following 6-day growth under lipoprotein depletion serum with indicated sterol concentrations (mean ± SD, n=3 biologically independent samples) (left). Immunofluorescence analysis of overexpressed DUF2054 domain alone or tagged with the Golgi targeting sequence of B3GALT1 (amino acids 1–61) in HEK293T cells (right). White indicates overlap (Scale bar, 8 μm).
F. Immunoblots of SREBP1 and several SREBP target proteins of Jurkat C12orf49_KO cell lines expressing the indicated cDNAs following 24h growth under lipoprotein depletion in the presence and absence of sterols. Actin and Lamin B1 were used as the loading controls for whole cell and nuclear extracts, respectively.
G. iBAQ based mass spectrometric analysis identified proteins immunoprecipitated from HEK293T cells expressing FLAG-C12orf49 (n=6 biologically independent samples) or GalT-FLAG cDNA (n=2 biologically independent samples). Log2 transformed fold differences are indicated on x-axis. Selected proteins are marked to show proteins of particular interest. Filled circles indicates that a protein was not detectable in the control samples. For visualization, an unpaired two-tailed t-test was performed.
H. Immunoblot analysis of C12orf49 interaction partners. Glycosylated MBTPS1 co-immunoprecipitated with c12orf49. GalT- FLAG was used as a near-neighbor control immunoprecipitation.
I. Immunoblot analysis of c12orf49 immunoprecipitates in the HEK293T C12orf49_KO cells expressing the indicated cDNAs. DUF2054 was localized to mitochondria, ER or Golgi using specified targeted sequences.
The experiments were repeated independently at least twice with similar results. Statistical significance was determined by two-tailed unpaired t-test.
To begin to understand the precise mechanism by which C12orf49 regulates SREBP processing and cholesterol metabolism, we sought to identify candidate regulators of SREBP processing that interact with C12orf49. Mass spectrometric analyses of immunoprecipitates of C12orf49, as compared to a Golgi-localized control, revealed the presence of several proteins including OS9 and MBTPS1 (Fig. 3g, Extended Data Fig. 7a). MBTPS1 is a member of the subtilisin-like proprotein convertase family and originally made as an inactive precursor in the ER35. This inactive precursor undergoes a series of autocatalytic cleavage at 2 sites, creating active forms, which can be glycosylated33,36. In turn, active forms of site-1-protease catalyze the proteolytic cleavage of its substrates including SREBPs. In individual immunoprecipitation experiments, C12orf49 specifically immunoprecipitates with an N-glycosylated form of S1P, as shown by its sensitivity to PNGase F, a glycosidase that cleaves the asparagine linked glycosylation residues (Fig. 3h). This interaction requires the correct localization of the protein to the Golgi and the presence of DUF2054 domain, as forced localization of the protein to other organelles prevents the interaction (Fig. 3i). Notably, loss of C12orf49 impacts cleavage of S1P targets including GNPTAB37, CREB3L2 and CREB438, though at different levels (Extended Data Fig. 7b). Consistent with the dysfunction of the Golgi-ER recycling of SCAP in the absence of S1P activity 39, SCAP localizes to the Golgi even in the presence of sterols in the C12of49 knockouts. These experiments suggest that the Golgi-localized C12orf49 binds and regulates S1P function (Extended Data Fig. 7c).
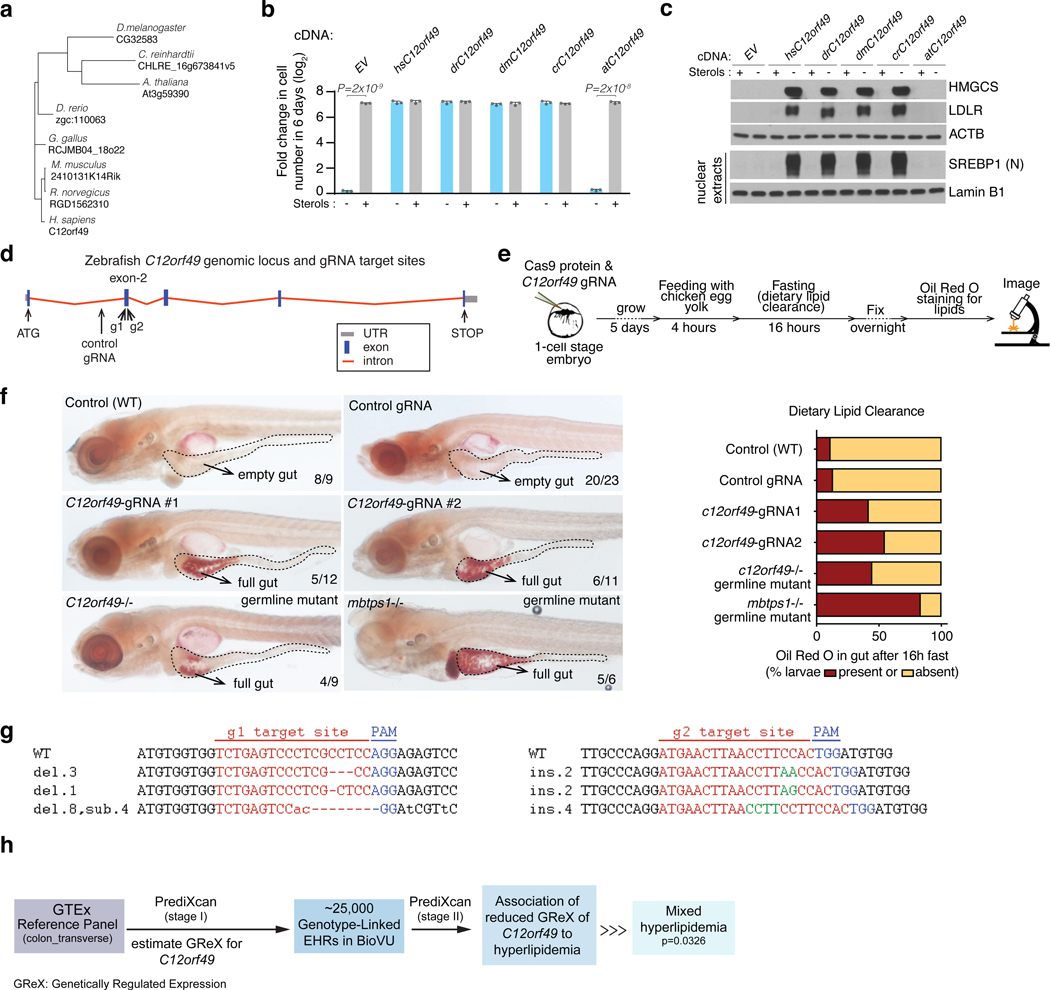
Because C12orf49 is conserved in the metazoa and in some plants, we next asked whether these homologs could replace C12orf49 in human cells, when expressed (Fig. 4a; Extended Data Fig. 8a). With the exception of the A.thaliana homolog, overexpression of any of the C12orf49 homologs rescued the sensitivity of Jurkat C12orf49-knockout cells to cholesterol depletion and restored SREBP activation (Fig. 4b,c). Notably, A. thaliana C12orf49 possesses a long C-terminus glycosyltransferase domain, raising the possibility that this protein may have evolved an additional role in plants (Extended Data Fig. 6c). Collectively, these results suggest that the functional relationship between C12orf49 and S1P is evolutionarily conserved.
Figure 4, C12orf49 function is conserved and essential for organismal lipid homeostasis.
A. Phylogenetic tree of C12orf49 in organisms.
B. Fold change in cell number (log2) of Jurkat C12orf49_KO cells overexpressing indicated C12orf49 cDNAs of different organisms following a 6-day growth under lipoprotein depletion in the presence or absence of sterols (mean ± SD, n=3 biologically independent samples). Statistical significance was determined by two-tailed unpaired t-test.
C. Immunoblots of SREBP1 (nuclear) and SREBP target proteins of Jurkat c12orf49_KO cell lines expressing the indicated cDNAs following 24h growth under lipoprotein depletion in the presence and absence of sterols. Actin and Lamin B1 were used as the loading controls for whole cell and nuclear extracts, respectively. The experiment was repeated independently twice with similar results.
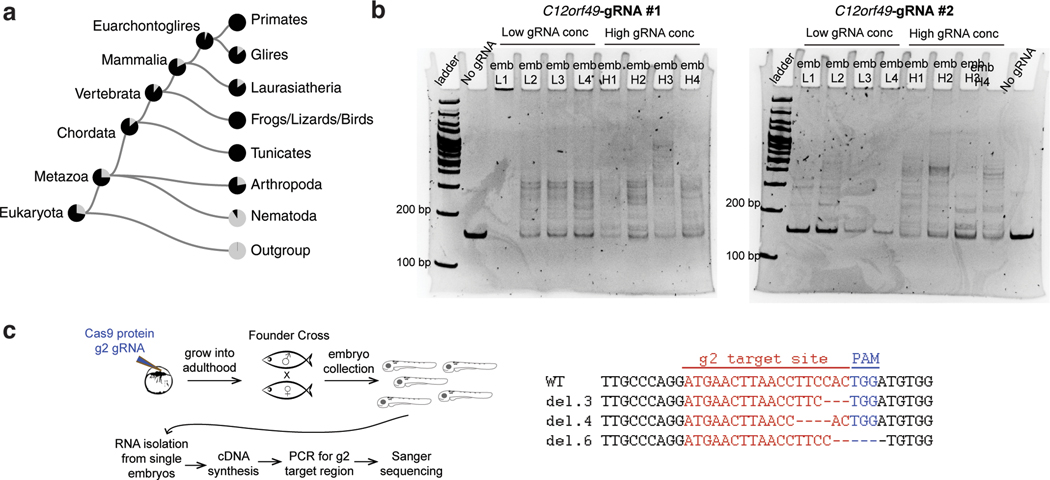
D. Schematic showing genomic locus of zebrafish c12orf49, g1 and g2 guide RNA target sites are marked by arrows.
E. Experimental strategy for feeding and dietary clearance assay.
F. Lipid absorption defects are marked by Oil Red O staining (full gut) in mutant larvae. Quantification shows similar defects in c12orf49 g1/g2 (trans-heterozygous germline mutant) and mbtps1hi1487/hi1487 germline mutants, as well as c12orf49-gRNA injected larvae (c12orf49 g1and c12orf49 g2). Number of larvae with represented phenotype is indicated on corresponding images. Gut is demarcated by dashed lines.
G. CRISPR-Cas9 generated mutations detected in c12orf49 g1 and c12orf49 g2 injected larvae. del: deletion, ins: insertion, sub: substitution. Number of base pair changes are indicated. Dashes indicate deletions, insertions are shown in green, substitutions in small-case letters.
H. Flow chart describing disease association study using PrediXcan method in BioVU biobank.
Significance is tested by logistic regression analysis (two-sided), n = 25,000. Multiple testing adjustment is done using Bonferroni correction. GTEx: Genotype-Tissue Expression, EHR: electronic health record.
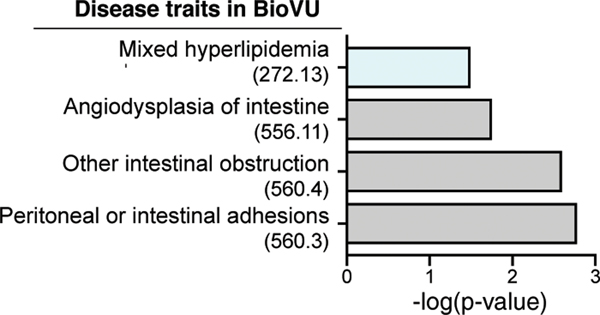
Building upon the conserved function and to further study C12orf49 in a more physiologically relevant context, we used zebrafish as a model organism. Since our biochemical data show that S1P is unable to cleave and activate SREBP in the absence of C12orf49, we postulated that zebrafish s1p-mutant (mbtps1hi1487 allele shown to block SREBP activation40) and c12orf49-mutant models would demonstrate comparable phenotypes in their lipid metabolism. Indeed, a dietary lipid clearance assay on a high-cholesterol diet revealed similar intestinal lipid absorption blockade in both s1phi1487 and c12orf49 mutants generated by CRISPR/Cas9 system (Fig. 4d-g; Extended Data Fig. 8b,c). While previous studies showed cranioskeletal malformations associated with mbtps1 mutations, c12orf49 mutants do not display these phenotypes, suggesting that mbtps1 targets may be affected to a different extent upon c12orf49 loss (Extended Data Fig. 7b) or alternative pathways exist to compensate for the loss in different tissues. Collectively, these results suggest that C12orf49, like S1P, may regulate lipid metabolism in vivo. To gain insight into C12orf49 function in human physiology, we also examined disease associations to reduced genetically regulated expression (GReX) of C12orf49 in the genotype-linked Electronic Health Records (EHR) of BioVU biobank41,42 using PrediXcan43 method. This analysis performed in ~25,000 BioVU subjects revealed a significant association of reduced C12orf49 GReX to mixed hyperlipidemia (p=0.0326) and other secondary intestinal phenotypes (Fig. 4h; Extended Data Fig. 9). These results collectively suggest that C12orf49 functions in organismal lipid homeostasis and may be associated with dysregulated lipid metabolism in humans.
Metabolic coessentiality network offers an alternative method to discover unknown components of cellular metabolism and functionally assign them to existing pathways. Using this method, here, we identify C12orf49 as an essential component of SREBP processing and cholesterol-sensing in mammalian cells. Precisely how C12orf49 contributes to the proteolysis of SREBPs is not known but our findings suggest that its interaction with S1P is likely involved in the regulation of cholesterol metabolism. Remarkably, C12orf49 is highly conserved, even in lower organisms. As a subset of these organisms does not have an SREBP ortholog yet harbor orthologs of C12orf49 and MBTPS1, the association between C12orf49 and S1P is likely relevant to cellular processes other than SREBP in these organisms. Interestingly, C12orf49 is associated with hyperlipidemia, so future line of work is needed to understand whether this protein may be implicated in human disease or have any clinical value. In conclusion, our work adds a new component to cellular cholesterol regulation and provides a platform to determine the function of other unknown metabolic components.
MATERIALS AND METHODS
Metabolic Coessentiality analysis
We adopted a three-step method to build putative interaction network among genes based on their co-essentiality scores. In step I, we removed genes which were strongly correlated with a large number of genes because pathway analysis literature suggest that few proteins have many interaction partners. To do this, we calculated a Pearson correlation network among all 17,638 genes with a threshold of |r|=0.25. Then we ranked the genes based on their degrees in this network and removed the top 10% from downstream analysis.
In steps II and III, we built partial correlation networks following the Correlation Analysis workflow proposed in Section 3.1 of previous work13. Since calculating partial correlation among essentiality scores of many genes using fewer cell lines is computationally intensive, this workflow builds on a useful property of Gaussian graphical models that was previously established44. This property ensures that genes in different connected components of the partial correlation network are marginally uncorrelated. Therefore, we can first construct a network by applying a threshold on Pearson correlation, and then estimate partial correlation networks separately for each of its connected components.
In step II of our analysis, we built such a Pearson correlation network with a threshold |r|=0.35. Since we are only interested in finding novel genes that interact with metabolic genes, we removed all the non-metabolic genes that are not connected to any metabolic genes in this network, using a curated metabolic gene set45–47. Of note, we curated this metabolic gene set by exhaustive analysis of every known human gene combined with searches of KEGG database and literature verifying the known or proposed metabolic function of each gene45. Focusing on positive Pearson correlations, this led to a network with 515 genes (275 metabolic genes, 240 non-metabolic genes) consisting of 55 components (component size varied between 3 and 20).
In step III, we calculated separate partial correlation matrices for each of these connected components and used statistically significant partial correlations (FDR < 0.05) to construct the putative interaction network. We used R function ‘pcor’ from library ‘ppcor’, and debiased graphical lasso48 implemented in the DSPC software13, as two different ways to calculate partial correlation networks. The debiased graphical lasso has an in-built regularization step and is particularly suitable when the number of genes in the network is high compared to the number of cell lines. Since the Pearson network components were reasonably small, the results of the two methods were qualitatively similar and we reported the output from ‘pcor’ in this paper. Finally, we removed interactions of genes in −/+1 cytogenic bands of each other in order to reduce false interactions as CRISPR-Cas9 genome editing was reported to induce large truncations49,50.
Cell lines
Cell lines HEK293T, Jurkat, MDA-MB-435, U-87 and BHK-21 were purchased from the ATCC. Cell lines were verified to be free of mycoplasma contamination and the identities of all were authenticated by STR profiling.
Antibodies, compounds and constructs
Custom antibody for c12orf49 and TMEM41A were designed and generated at YenZym Antibodies, using synthetic peptides with QEERAVRDRNLLQVHDHNQP (amino acids 37–56 of c12orf49) and ETSTANHIHSRKDT (amino acids 251–264 of TMEM41A). Other antibodies, compounds, supplies, equipment, software, experimental models and constructs are provided in the supplementary files.
Cell Culture Conditions
Jurkat were maintained in RPMI media (GIBCO) containing 2 mM glutamine, 10% fetal bovine serum, penicillin and streptomycin. HEK293T, U87M and MDA-MB-435 cells were maintained in DMEM media (GIBCO) containing 4.5g/L glucose, 4mM glutamine, 10% fetal bovine serum, penicillin and streptomycin. All cells were maintained in monolayer culture at 37ºC and 5% CO2.
Focused CRISPR-based genetic screen
The highly focused sgRNA library was designed by including representation of each gene within the SREBP module. For some of the genes, our sgRNAs have previously been published and validated15, we therefore used smaller number of sgRNAs for particular genes. Oligonucleotides for sgRNAs were synthesized by Integrated DNA Technologies and annealed before they were introduced in lentiCRISPR-v2 vector using a T4 DNA ligase kit (NEB), following manufacturer’s instructions. Ligation products were then transformed in NEB stable competent E. coli (NEB) and the resulting colonies were grown overnight at 32 °C and plasmids isolated by Miniprep (QIAGEN). This plasmid pool was used to generate a lentiviral library containing five sgRNAs per gene target. This viral supernatant was titred in each cell line by infecting target cells at increasing amounts of virus in the presence of polybrene (8 μg ml−1) and by determination of cell survival after 3 days of selection with puromycin. One million Jurkat cells were infected at a MOI of 1 before selection with puromycin for 3 days. An initial pool of one million cells was collected. Infected cells were then cultured for 14 population doublings in the lipoprotein depleted serum containing media in the presence or absence of cholesterol, after which one million cells were collected and their genomic DNA was extracted by a DNeasy Blood & Tissue kit (QIAGEN). For amplification of sgRNA inserts, we performed PCR using specific primers for each condition. PCR amplicons were then purified and sequenced on a MiSeq (Illumina). Sequencing reads were mapped and the abundance of each sgRNA was measured. sgRNA score is defined as the log2 fold change in the abundance between the initial and final population the sgRNA targeting a particular gene. Report of the guide scores and sequences of the guides are available in Supplementary Table 1.
Generation of knockout and cDNA overexpression cell lines
For knockout experiments of C12orf49, sgRNA (5′-TTTCAGGCTACGTTTGCGAG-3′) was cloned into lentiCRISPR-v1-GFP vector by T4 DNA ligase (NEB) after linearization with BsmBI. Vector was transfected into HEK293T cells with lentiviral packaging vectors VSV-G and Delta-VPR using XtremeGene transfection reagent (Roche). Media was changed 24 hr after transfection. The virus containing supernatant was collected at 48h and filtered through 0.45 uM filter before use. Jurkat cells were spin-infected at a MOI of 1 in 6-well tissue culture plates using 8 μg ml−1 of polybrene at 1,200g for 1.5 h. Virus was removed 24 hours after infection and single cell sorting was performed into 96 well plates using GFP. Separately, HEK293T cells were transfected with the same vector and single cell sorted similarly following selection by puromycin for 3 days. For overexpressions, gBlocks(IDT) containing the guide-resistant version of c12orf49 and other indicated cDNAs were cloned into the pMXs retroviral vector by linearizing with BamHI and NotI, followed by Gibson assembly. Epitope tags were added to the cDNAs when indicated. Overexpression plasmids were transfected with retroviral packaging plasmids Gag-pol and VSV-G into HEK293T cells. After transduction, cells were selected with blasticidin.
Immunoblotting
Cell pellets were washed twice with ice-cold PBS before lysis in SDS lysis buffer (10 mM Tris-HCl pH 6.8, 100mM NaCl, 1 mM EDTA, 1mM EGTA, 1% SDS) supplemented with protease inhibitors. Each cell lysate was sonicated thrice for 15s on ice with a 2 min interval between each sonication. Proteins from membranes and nuclei are isolated using the Cell Fractionation Kit (CST #9038). Protein concentrations of the samples were determined by a Pierce BCA Protein Assay Kit (Thermo Scientific) with bovine serum albumin as a protein standard. Samples were mixed with 5x SDS loading buffer and boiled for 5 min. Finally, samples were resolved on 8%, 12% or 16% SDS–PAGE gels and analyzed by immunoblotting. Immunoblot analysis of c12orf49 knockouts were performed following deglycosylation with PNGase F (New England Biolabs) under denaturing conditions, according to the manufacturer’s instructions.
For SREBP targets, 24 hours before extraction, Jurkat cells were washed three times with PBS and plated as triplicates (1 × 106 cells per replicate) in 6-well plates using RPMI medium supplemented with 10% LPDS supplemented with 50uM compactin and 50uM sodium mevalonate in the presence or absence of sterols (10 μg ml−1 cholesterol, 1 μg ml−1 25-hydroxycholesterol). For nuclear extracts, cells were also provided 25 μg ml−1 N-acetly-leucinal-leucinal-norleucinal for the last 3 hours. Rest of the immunoblotting was performed as described above. Immunoprecipitated proteins were equally split into different tubes and reactions were performed under denaturing conditions with the indicated deglycosylation enzyme according to the manufacturer’s manual.
Proliferation assays
Cell lines were cultured as triplicates in 96-well plates at 500 cells (suspension) or 200 cells (adherent) per well in a final volume of 0.2 ml RPMI-1640 medium (suspension) or DMEM media (adherent) supplemented with 10% lipoprotein depleted serum (Kalen) with indicated treatments. A duplicate plate was setup to determine initial luminescence on the day plates were set up, without any treatment. To measure luminescence, 40 μl of Cell Titer Glo reagent (Promega) was added in each well according to the manufacturer’s instructions and data was obtained using a SpectraMax M3 plate reader (Molecular Devices). Data are presented as relative fold change in luminescence of the final measurement to the initials. For proliferation assays under lipoprotein depletion luminescence was measured after 6 days of growth. In cholesterol rescue experiments, 100 μg ml−1 LDL (corresponding to total 50 μg ml−1 of cholesterol) or 10 μg ml−1 free cholesterol were used as indicated. Cell culture images were taken using a Primovert microscope (Zeiss).
Isotope tracing experiments and lipid metabolite profiling
Jurkat cells were washed three times with PBS and plated as triplicates (1 × 106 cells per replicate) in 6-well plates using RPMI medium supplemented with 10% LPDS in the presence or absence of sterols (10 μg ml−1 cholesterol, 1 μg ml−1 25-hydroxycholesterol). After 24 h, media was replaced with fresh medium containing sodium acetate (10mM) or 13C1 sodium acetate (10 mM). Following an incubation of 48 hours, cell pellets were washed twice with 1 ml of 0.9% NaCl (800g for 2 minutes) and resuspended in 600 μl of cold LC-MS grade methanol. Non-polar metabolites were extracted by consecutive addition of 300 μl of LC-MS grade water followed by 400 μl of LC-MS grade chloroform. The samples were vortexed (10 min) and centrifuged for 10 min at 20,000g and 4°C. The lipid-containing chloroform layer was carefully removed and dried under liquid nitrogen. Dry lipid extracts were stored at −80°C till further analysis.
The lipid extracts were saponified in 200 ul of 2M methanolic KOH (95% methanol) for 2 hours at 60°C in a thermoblock (Eppendorf ThermoMixer). Upon cooling to room temperature, 200ul of 5% NaCl was added to the saponified extracts and the mixture was vortexed and acidified with 6N HCl (pH <2). HPLC grade hexanes was added and the mixture was vortexed vigorously for 10 seconds (3X). After a centrifugation for 10 min at 20,000g and 4°C, the hexane layer was transferred to a glass vial. The lipids were extracted with hexanes twice more, adding 300ul hexanes each time. The combined hexane layers were dried under liquid nitrogen and stored at −80 °C until LC-MS analysis.
Lipids were separated on an Ascentis Express C18 2.1 mm × 150 mm × 2.7 μm particle size column (Supelco) connected to a Vanquish UPLC system and a Q Exactive benchtop orbitrap mass spectrometer (Thermo Fisher Scientific), equipped with a heated electrospray ionization (HESI) probe. Dried lipid extracts were reconstituted in 50 μl of 65:30:5 acetonitrile: isopropanol: water (v/v/v), vortexed for 10 sec, centrifuged for 10 min (20,000 g, 4°C) and 5 μl of the supernatant was injected into the LC-MS in a randomized order, with separate injections for positive and negative ionization modes. Mobile phase A consisted of 10mM ammonium formate in 60:40 water: acetonitrile (v/v) with 0.1% formic acid, and mobile phase B consisted of 10mM ammonium formate in 90:10 isopropanol:acetonitrile (v/v) with 0.1% formic acid. Chromatographic separation was achieved using the previously described gradient51. The column oven and autosampler were held at 55 °C and 4 °C, respectively.
The mass spectrometer was operated with the following parameters; positive or negative ion polarity; spray voltage, 3500 V; heated capillary temperature, 285 °C; source temperature, 250 °C; sheath gas, 60 (arbitrary units); auxiliary gas, 20 (arbitrary units). External mass calibration was performed every five days using the standard calibration mixture.
Mass spectra were acquired in positive ionization mode, using a Top3 data-dependent MS/MS method. The full MS scan was acquired as such; 70,000 resolution, 1 × 106 AGC target, 250 ms max injection time, scan range 350 – 450 m/z. The data-dependent MS/MS scans were acquired at a resolution of 17,500, AGC target of 1 × 105, 75 ms max injection time, 1.0 Da isolation width, stepwise normalized collision energy (NCE) of 20, 30, 40 units and 8 sec dynamic exclusion.
Relative quantification of unlabeled and labeled cholesterol was performed using Skyline Daily (MacCoss Lab)52 with the maximum mass and retention time tolerance set to 2 ppm and 20 sec, respectively. The measured isotopologues of cholesterol in the unlabeled acetate experiments were used to correct for natural isotope abundance in the [13C1] acetate-treated samples. Data are presented as percentage of the labeled cholesterol in the total pool.
Real-time PCR assays
Jurkat cells were washed three times with PBS and plated as triplicates (1 × 106 cells per replicate) in 6-well plates using RPMI medium supplemented with 10% LPDS supplemented with 50uM compactin and 50uM sodium mevalonate in the presence or absence of sterols (10 μg ml−1 cholesterol, 1 μg ml−1 25-hydroxycholesterol). After an 8-hour incubation, RNA was isolated from cell pellets by a RNeasy Kit (Qiagen) according to the manufacturer’s protocol. RNA was spectrophotometrically quantified and equal amounts were used for cDNA synthesis with the Superscript II RT Kit (Invitrogen). qPCR analysis was performed on an ABI Real Time PCR System (Applied Biosystems) with the SYBR green Mastermix (Applied Biosystems). Primers for each target are provided in the supplementary files. Results were normalized to β-actin.
Immunofluorescence
For lipoprotein depletion experiments, HEK293T cells were washed three times with PBS, resuspended in DMEM supplemented with 10% LPDS and seeded (2× 105) on coverslips in 6-well plates previously coated with poly-D-lysine (Sigma). 12h later, cells were transfected with 100ug of pMXS-mCherry-SREBP1 with the XtremeGENE 9 DNA transfection reagent, according to the manufacturer’s manual. After 12 hours, cells were switched to fresh media with 10% LPDS supplemented with 50uM compactin and 50uM sodium mevalonate in the presence or absence of sterols (10 μg ml−1 cholesterol, 1 μg ml−1 25-hydroxycholesterol). Following 16-hour incubation, cells were fixed for 15 min with 4% paraformaldehyde diluted in PBS at room temperature. After three washes with PBS, cells on the coverslips were permeabilized by incubation with 0.05% Triton X-100 in PBS for 10 min at room temperature prior to another three PBS washes. Coverslips were blocked with normal donkey serum (20X diluted in PBS) at room temperature for 20 min and washed thrice with PBS. Coverslips were then blocked with 5% normal donkey serum (NDS) for 1 hour at room temperature, before an overnight incubation with the indicated primary antibodies diluted in 5% NDS at 4C. On the next day, following three washes with PBS, coverslips were then incubated with secondary antibodies (Alexa Fluor 488 and Alexa Fluor 568) in the dark for 1 hour at room temperature. Three washes with PBS were followed by an incubation with a 300 nM solution of DAPI in PBS for 5 min in dark. Coverslips were washed three times with PBS and finally mounted onto slides with Prolong Gold antifade mounting media (Invitrogen). Images were taken on a confocal microscope. For other localization experiments, HEK293T cells were cultured and transfected in DMEM with 10% FBS.
Brefeldin A treatment
Jurkat cells were in grown in RPMI supplemented with 10% serum. One day before stimulation, 1×106 cells were plated in 6-well plates. On the day of the experiment, cells were washed three times with PBS and resuspended in fresh media with 10% LPDS supplemented with 50uM compactin and 50uM sodium mevalonate in the presence or absence of sterols (10 μg ml−1 cholesterol, 1 μg ml−1 25-hydroxycholesterol) and Brefeldin 1ug/ml was added to the indicated cells. 6 hours post-induction, cell pellets were subjected to nuclear extraction as described above.
Immunoprecipitation
Before the day of immunoprecipitation, HEK293T cells overexpressing the indicated plasmids were plated (1× 107) in a 15-cm culture dish. After 15 hours, cells were washed with ice cold PBS twice and lysed in immunoprecipitation lysis buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and cOmplete EDTA-free protease inhibitor). The mixture was placed on an end-over-end rotator for 10 minutes at 4C and spun down at 1000g for 4 minutes to separate the supernatant. For anti-FLAG immunoprecipitations, the FLAG-M2 affinity gel was washed with 1 mL TBS (150 mM NaCl) twice and 40 uL of the affinity gel was then added to the lysate supernatant and incubated rotating at 4C for 3 hours. Affinity gel was placed on spin columns (Chromotek) and washed thrice with TBS. Proteins were eluted by incubating with 100 ng/uL of 3X FLAG peptide in lysis buffer for 15 min at room temperature. For the proteomics experiment, proteins were chemically crosslinked in live cells prior to lysis by adding dithiobis(succinimidyl propionate) to a working concentration of 2.5 mM and incubating for 7 min at room temperature. Crosslinking reaction was quenched by adding 1/10 volume of 1M Tris pH 8.5 to the media and incubating for 2 min at room temperature.
Proteomics
Competitively eluted (3X FLAG peptide) samples, in 1% Triton, were diluted 2-fold followed by precipitation overnight in 6 volumes ice cold acetone. Precipitates were dissolved and chemically reduced in 35uL 8M Urea/70mM ammonium bicarbionate/20mM Dithiothreitol followed by alkylation (50mM iodoacetamide). Samples were diluted and digested using Endopeptidase LysC (Wako Chemicals) followed by additional dilution and trypsinization (Promega). Acidified tryptic peptides were desalted53 and analyzed using nano-LC-MS/MS (EasyLC1200 and Fusion Lumos operated in High-High mode, ThermoFisher). Data were queried against UniProt human database (March 2016) concatenated with common contaminants and quantitated using MaxQuant v. 1.6.0.13 54. False discovery rates of 2% and 1% was applied to peptide and protein identification. The iBAQ55 values obtained from MaxQuant, were filtered, using Perseus software56, and the following filters; 80% of replicates must contain a valid value in either the ‘experiment’ (n=6) and/or ‘control’ (n=2) groups, protein must be matched to a minimum of 3 razor/unique peptides. Missing values in the ‘control’ samples were imputed (Perseus) from a normal distribution. For visualization only, a t-test was performed (Fig. 3g).
Phylogenetic analysis
Protein sequences of C12orf49 in different species (UniProtKB) were aligned using the Clustal W and MegAlign Software (DNASTAR). Phylogenetic tree was constructed automatically by applying BioNJ algorithm with uncorrected pairwise distance metrics and global gap removal.
CRISPR/Cas9 genome editing in zebrafish
CRISPR/Cas9 target sites within zebrafish c12orf49 gene (GRCz11 assembly, gene name: zgc:110063) were identified using CHOPCHOP57 web tool. Two independent genomic sites within c12orf49 locus were targeted by alternative guide RNAs (gRNAs), namely g1 and g2 with the following sequences; g1: 5’-GGTCTGAGTCCCTCGCCTCCAGG-3’ and g2: 5’-GGATGAACTTAACCTTCCACTGG-3’. Genomic locations targeted by gRNA g1 and g2 are as follows: chr5:11947798 and chr5:11947828, respectively. A cloning-free method to generate gRNA template was performed as previously described 58. Guide RNAs were synthesized with MEGAshortscript T7 transcription kit (ThermoFisher Scientific). To generate mutations with CRISPR/Cas9 system, a mixture of 500 pg purified Cas9 protein (PNA Bio Inc, # CP01) and 300 pg of either gRNA was injected into one-cell stage embryos of wild-type (AB) crosses. Efficient generation of mutations was confirmed by DNA heteroduplex formation assay59 using following primers: forward 5’-ATGTACAGGAGGAGCGAACG-3’ and reverse 5’-TGAGAAGGCTCTTTCCCTGA-3’. RNA was isolated from zebrafish embryos using TRIzol method following manufacturer’s intructions; cDNA was synthesized using oligo dT primers. Following exonic primer (reverse) was used in combination with the forward primer listed above to amplify c12orf49-g2 targeted site: Exonic Reverse:5’-CTCGAGCTGGGAGCATTAAC-3’
Sequence-confirmed mutant embryos were grown to adulthood to generate two independent germline mutant lines, c12orf49 g1 and c12org49 g2, thus establishing F0 founders. These allelic F0 lines were then crossed to each other to produce trans-heterozygous mutant F1 embryos that carry a c12orf49 g1 mutation in their maternal copy and a c12org49 g2 mutation in their paternal copy. The advantage of this cross is the ability to eliminate off-target effects that potentially might have been induced in either animal, and drive to homozygosity only the targeted site.
Dietary Lipid Clearance Assay
Injected embryos were grown to 5 dpf stage and fed with 10% organic chicken egg yolk for 4 hours, followed by 16 hours of fasting. Next, zebrafish larvae were fixed in 4% paraformaldehyde and processed for oil red O staining to assay dietary lipid clearance in the digestive system, as described previously60. Stained larvae were imaged with Zeiss Axioimager Z1 scope equipped with Axiocam HRc camera.
PrediXcan Discovery Analyses
We investigated to the association of c12orf49 with hyperlipidemia. We performed PrediXcan43 analysis, leveraging a SNP-based prediction model in colon (transverse). We estimated the genetically regulated gene expression (GReX) in the approximately twenty five thousand BioVU subjects 41,61,62 using the GTEx resource (v6p)63,64 as a reference transcriptome panel, and tested for association with hyperlipidemia41. From the weights derived from the gene expression imputation model for c12orf49 (driven by the single-nucleotide polymorphism rs10507274 with effect allele “C” with false discovery rate65 (q-value) of 0.03) and the number of effect alleles for individual i at the variant j, we estimated GReX as follows:
in the BioVU subjects. We performed logistic regression to determine the association between GReX and the disease trait. To maximize the quality of the phenome information, we required at least two ICD9 or ICD10 codes on different clinical visits to instantiate a phecode for diagnosis of the phenotype.
Analytical Validation of Method and Comparison with Alternatives
Pearson correlation is the most commonly used method for building co-essentiality networks among genes. Pan et al. (2019) has used genome-scale Pearson correlation networks to identify functional modules and protein complexes2. However, gene networks based on statistically significant Pearson correlation tend to have many edges, including many false positives, which makes it difficult to identify suitable targets for novel gene interaction discovery and wet-lab validation. Thus there is a need for computational methods with higher specificity (lower false positives) that identifies fewer but high-confidence putative genetic interactions from data. In a recent work, Wainberg et al. (2019) proposed an alternative co-essentiality network method based on generalized least squares (GLS), which explicitly accounts for non-independence of cell lines and reduces the number of false positives and has identified 93,575 significant co-essential gene pairs3. Although these comprehensive methods undoubtedly identified many novel gene functions, we wanted to create a conservative method that more easily allowed us to manually curate each individual network. As result, we looked towards alternative methods and filters that allowed us to short list putatively novel gene interactions.
In essence, both methods described above measure pairwise association between two genes, without accounting for indirect or spurious effects due to their interactions with a third gene. Partial correlation, a canonical method in classical statistics, allows explicitly accounting for such indirect associations and produces a smaller but high-confidence set of putative interactions for follow-up wet-lab validation. While clustering based on pairwise correlation allows us to zoom in on a specific module of genes, calculating partial correlation among genes within the module help us focus on gene pairs which are more likely to interact directly. As a result, we were better equipped with a manageable list of gene interactions that can be studied at an experimental scale. This is in sharp contrast with Pearson correlation based methods described above, which only analyses association between two genes at a time.
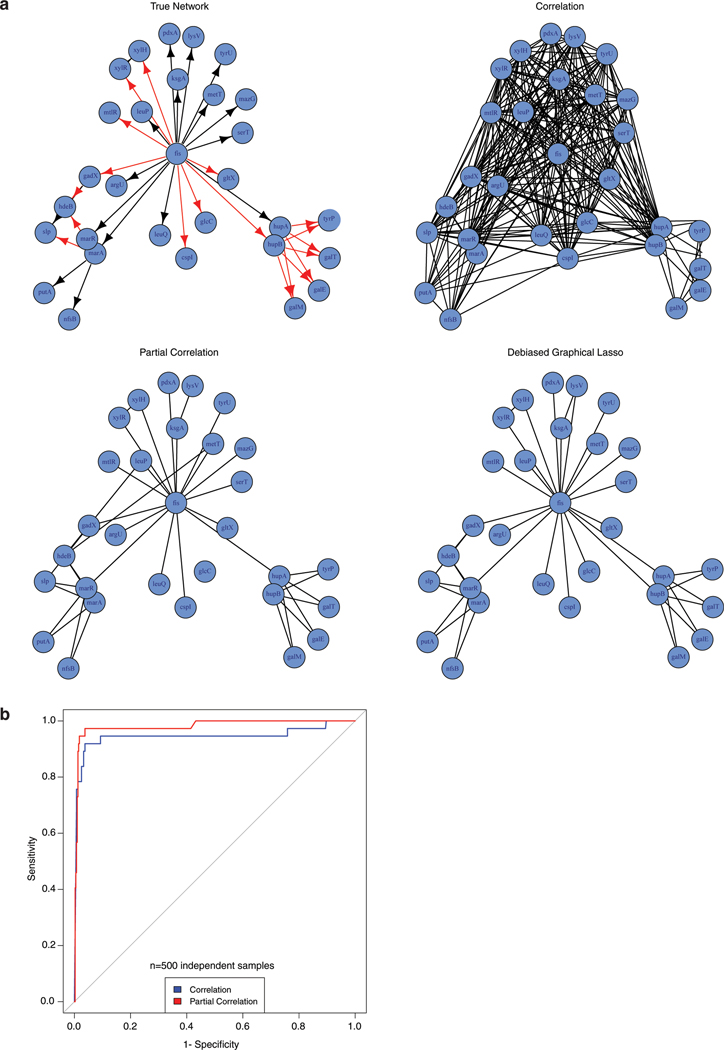
The principle of filtering out effects of other nodes in a network is at the core of graphical modeling literature in statistics and machine learning. Prior works that successfully employed this idea to build metabolic networks12,13. Here we illustrate the benefit of such a strategy using a simulation experiment based on biologically inspired network structure.
We select a subnetwork of 30 nodes from an E.Coli network using the GeneNetWeaver software66, a popular tool for benchmarking network inference methods. This network has a few hubs, with a main hub node at gene fis. We then simulated (log) co-essentiality score of every gene g (denoted by Xg) based on the following rule:
Here, pa(g) denotes the set of genes in the network which have an outgoing edge to gene g. In other words, essentiality score of gene g is influenced by the essentiality score of its parent genes pa(g), although the main hub gene fis exerts a stronger effect than other parent genes. The term e in the above equation denotes standard Gaussian noise in the structural equation system.
We simulated essentiality scores according to the above model for n=500 independent samples (cell lines), and used Pearson and partial correlation (using both ‘pcor’ and debiased graphical lasso) to reconstruct the gene networks from data (statistically significant partial correlations (FDR < 0.05) were used to construct edges in networks). Results of this experiment are displayed in Extended Data Fig. 1a. As expected, we see that gene pairs which are connected only through fis (e.g. xylR, xylH, pdxA, lysV) have high Pearson correlation, leading to false positive edges. However, such edges are rarely picked up in both partial correlation networks.
We note that building a Pearson correlation network with high cutoff (very small p-value) is not an alternative to partial correlation. In the example above, even genes having only an indirect association through fis may have higher Pearson correlation than two genes that interact directly (e.g. marA and putA) due to the strong effect of fis. So a network of large absolute correlation is likely to keep more indirect associations and miss some of the directly interacting gene pairs. This can be seen in the ROC curve of Extended Data Fig. 1b, where we calculate false positive and negatives based on a range of cut-offs on Pearson and partial correlation.
We conducted a more systematic simulation study by repeating the above experiments on N=20 replicates, varying the number of genes (p = 30, 40, 50) and number of cell lines (n = 100, 200, 300, 400, 500). Number of false positives and true positives for Pearson correlation and the two types of partial correlation methods (pcor and DGLASSO) are reported in Supplementary Table 2. Standard errors calculated over the N=20 replicates are shown in parenthesis. These results show that partial correlation networks substantially reduce the number of false positives (hence increases specificity) over Pearson correlation, while reducing the true positives to some extent. Our simulation results also show that partial correlation tends to have lower power (sensitivity) as the network size (p) increases. This is expected since calculation of partial correlation matrix requires estimation of O(p2) parameters. Therefore, we do not advocate using partial correlation at genome-scale, and only use it to filter the set of interactions in small components (modules) obtained by Pearson correlation or other pairwise association methods. Developing a one-step method that combines the strengths of both Pearson and partial correlation to make it applicable at genome-scale and possibly accounts for dependence among cell lines as in Wainberg et al (2019)3 is an interesting research question, but beyond the scope of this paper and is left for future work.
Generation of knockout and cDNA overexpression cell lines
For mix population knockout experiments in U-87 MG and MDA-MB-435, sgRNA of C12orf49 (5′-TTTCAGGCTACGTTTGCGAG-3′) was cloned into lentiCRISPR-V2-puro vector. Vector was transfected into HEK293T cells with lentiviral packaging vectors VSV-G and Delta-VPR using XtremeGene transfection reagent (Roche). Indicated cells were spin-infected in 6-well tissue culture plates using 8 μg ml−1 of polybrene at 1,200g for 1.5 h and selected by puromycin with corresponding minimum lethal dose for 3 days. For knockout experiments of TMEM41A, sgRNAs (5′-CATGCTGCTACCTGCTCTCC-3′, 5′-TCGCCTTGTACTTGCTGTCG-3′) were cloned into lentiCRISPR-v1-GFP vector. Following transduction, cells were single cell sorted using GFP. Overexpression of guide-resistant version TMEM41A and other plasmids used were cloned into pMXs retroviral expression vector and was carried on by viral transduction and selection as described.
Viral infectivity assays
The green fluorescent protein (GFP)-tagged bunyamwera virus (BUNV-GFP) 67 (generously provided by Richard M Elliott) was amplified in BHK-21 cells and titrated by median tissue culture infectious dose (TCID50). For virus replication assays, HEK293T cells (WT and C12orf49 KO) were seeded into poly-L-lysine coated 24-well plates at 2.5×104 cells/well using lipid-depleted DMEM supplemented with 10% fetal bovine serum (FBS). The following day, cells were washed with Opti-MEM (Gibco) and infected with BUNV-GFP diluted in 200 μL Opti-MEM at a multiplicity of infection (MOI) of 0.1 infectious units (IU)/mL. Cells were inoculated for 2 h at 37°C before virus inoculum was removed and washed off using Opti-MEM. For the remainder of the virus infection assay, cells were cultured in lipid-depleted DMEM. Supernatants with progeny BUNV-GFP were harvested at various timepoints (0, 24, 48, 72 hpi) and the infectious titers were determined by TCID50 assays on BHK-21 cells. At the final timepoint (72 hpi), cells were harvested into 250 μl Accumax cell dissociation medium (eBioscience) and transferred to a 96-well block containing 250 μl 4% paraformaldehyde (PFA) fixation solution. Cells were pelleted at a relative centrifugal force (RCF) of 930 for 5 min at 4°C, resuspended in cold phosphate-buffered saline (PBS) containing 3% FBS and stored at 4°C until flow cytometry analysis. Samples were analyzed using the LSRII flow cytometer (BD Biosciences) equipped with a 488 nm laser for detection of GFP, and resulting data using FlowJo software (Treestar).
Lipid metabolite profiling for TMEM41A null cells
The procedure for lipid extraction and analysis of the cellular lipidomes were adopted from previously described protocols68. Briefly, Jurkat cells were washed three times with PBS and plated as triplicates (1 × 106 cells per replicate) in 6-well plates using RPMI medium supplemented with10% FBS. After 24 hours, cell pellets were resuspended in 1 mL cold PBS. A 30 μL aliquot of the cell suspension was taken for determining protein concertation. The remaining 970 μL of cell suspension was then transferred to a homogenizer to which 2 mL of chloroform and 1 mL of methanol was added. The solution was kept on ice and homogenized 30 times. The homogenized solution was centrifuged (500 rcf, 4 °C, 10 minutes) to separate aqueous and organic layers. The organic layer was carefully transferred into a 1-dram glass vial, of which 1.5 mL was transferred into a new vial to ensure equal volume was removed from each extract. The chloroform extract was dried under vacuum. Samples were then resuspended in a calculated amount of chloroform based on total protein concentration.
Lipidomics data was acquired using an Agilent 1260 HPLC paired with an Agilent 6530 Accurate-Mass Quadrupole Time-of-Flight mass spectrometer. A Gemini C18 reversed-phase column (5 μm, 4.6×50mm, Phenomenex) with a C18 reversed-phase guard cartridge was used in negative mode. Mobile phase A was 95:5 water:methanol (v/v) and mobile phase B was 60:35:5 isopropanol:methanol:water (v/v). Mobile phases were supplemented with 0.1% (w/v) ammonium hydroxide for negative mode. The gradient used for separation began after 5 minutes, increasing from 0% B to 100% B over 60 minutes. At 65 minutes an isocratic gradient at 100% B was applied for 7 minutes, followed by equilibration of the column with 0% B for 8 minutes. The flow rate for the initial 5 minutes was 0.1 mL/min and was increased to 0.5 mL/min for the remaining gradient. A DualJSI fitted electrospray ionization source was used. Capillary voltage was set to 3500 V and fragmentor voltage set to 175 V. The drying gas temperature was set to 350 °C with a flow rate of 12 L/min. Targeted data analysis was performed using MassHunter Qualitative Analysis software (version B.06.00, Agilent). The corresponding m/z for each lipid was extracted and the peak area was manually integrated.
Lipotoxicity assays
Palmitic acid was conjugated to BSA. A 12 mM solution of the fatty acid was dissolved in 20 mL of 0.01M NaOH and stirred for 30 min at 70ºC, followed by addition into a stirring 60 mL 10% BSA solution in PBS to make a final concentration of 3 mM. Solution was stirred for 1hr at 37C to allow fatty acids to conjugate with BSA. Finally, the fatty acid-BSA solution was filtered through 0.22Um filter and stored in a glass container at 4ºC. Indicated Jurkat cells were cultured as triplicates in 96-well plates at 400 cells per well in a final volume of 0.2 ml RPMI-1640 with increasing concentrations of palmitate. A duplicate plate was setup to determine initial luminescence on the day plates were set up, without any treatment. To measure luminescence, 40 μl of Cell Titer Glo reagent (Promega) was added in each well according to the manufacturer’s instructions and data was obtained using a SpectraMax M3 plate reader (Molecular Devices). Data are presented as relative fold change in luminescence of the final measurement to the initials.
Luciferase Reporter assays
Three tandem repeats of the Sterol Regulated Element (SRE-1) in the promoter of LDRL were cloned into pGL4.20 luciferase vector. Parental, knockout and addback HEK293T cells were washed three times with PBS, resuspended in DMEM supplemented with 10% LPDS and seeded (2.5× 104) in 96-well plates previously coated with poly-D-lysine (Sigma). 12h later, cells were transfected with increasing amounts of pGL-3xSRE and pRL-SV40 (1:20 ratio of renilla: total plasmid) with the XtremeGENE 9 DNA transfection reagent, according to the manufacturer’s manual. After 12 hours, cells were switched to fresh media with 10% LPDS supplemented with 50uM compactin and 50uM sodium mevalonate in the presence or absence of sterols (10 μg ml−1 cholesterol, 1 μg ml−1 25-hydroxycholesterol). At 24h, cells were lysed and luminescence was read by using the Dual-Glo Luciferase Assay System (Promega) and SpectraMax M3 plate reader (Molecular Devices). Data is presented as Firefly/Renilla luminescence.
Cleavage assays of other site-1 protease targets
Knockout and addback HEK293T cells were plated in DMEM supplemented with 10% FBS (2× 105) in 6-well plates. 12h later, cells were transfected with 100ng of plasmids of triple tandem HA tagged GNPTAB, CREB3L2 or CREB4 with the XtremeGENE 9 DNA transfection reagent, according to the manufacturer’s manual. 24 hours post transfection, total proteins were extracted and immunoblotted as described above.
SCAP localization
HEK293T cells were plated in DMEM supplemented with 10% FBS (2× 105) on coverslips in 6-well plates previously coated with poly-D-lysine (Sigma). 12h later, cells were transfected with 100ug of GFP-SCAP with the XtremeGENE 9 DNA transfection reagent, according to the manufacturer’s manual. After 12 hours, cells were switched to fresh media with 10% LPDS supplemented with 50uM compactin and 50uM sodium mevalonate in the presence or absence of sterols (10 μg ml−1 cholesterol, 1 μg ml−1 25-hydroxycholesterol). Following 16-hour incubation, cells were fixed and processed for imaging as described above. Anti-GFP antibody (ProteinTech) was used for detection of SCAP.
Gene expression, conservation and architecture analysis
Gene expression across different tissues was obtained from GTEx. For the uncharacterized domain of unknown function (DUF2054), Hidden Markov Model (HMM) logo, different domain architectures and occurrence across different species were obtained from Pfam (EMBL-EBI).
For c12orf49, predicted motifs and post-translational modifications were obtained from UniProtKB. Prediction of transmembrane helices of human C12orf49 was performed by using the TMHMM Server v.2.0. Phylogenetic tree of C12orf49 across species is described at TreeFam (EMBL-EBI).
Statistical analysis
Sample size, mean, and significance (p-values) are indicated in the text and figure legends. Error bars in the experiments represent standard deviation (SD) from either independent experiments or independent samples. Statistical analyses were performed using GraphPad Prism 7 or reported by the relevant computational tools.
Data availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request. Source data for all figures are included with the online version of the paper.
Code availability
The code for the computational analysis that is used in this study are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Fig. 1. Comparative Simulation between partial and Pearson correlation.
A. Simulation experiment of a subnetwork from an E. coli network demonstrating the advantage of using partial correlation over Pearson correlation.
B. Receiver operating characteristic (ROC) curve based on the simulated data. (n= 500 independent samples)
Extended Data Fig. 2. Metabolic coessentiality modules.
35 Metabolic coessentiality modules. Blue line indicates a previously known interaction between the genes. Poorly characterized genes are highlighted as orange.
Extended Data Fig. 3. C12orf49 is necessary for cell growth under sterol depletion.
A. Pearson correlation values of the essentiality scores of the indicated genes across different cancer cell lines (n=558).
B. Differential sgRNA score for C12orf49 gene of Jurkat cell line in the presence or absence of sterols.
C. Fold change in cell number (log2) of U-87 MG or MDA-MB-435 c12orf49_KO cell line following a 6-day growth under lipoprotein depletion in the absence or presence of sterols. (mean ± SD, n=3 biologically independent samples). Statistical significance was determined by two-tailed unpaired t-test.
D. Immunoblots of c12orf49 in the indicated knockout cells of HEK293T. Actin was used as the loading control. The experiment was repeated independently twice with similar results.
E. (left) Immunoblots of c12orf49 knockout and addback cells in Jurkat cells. Actin was used as the loading control. The experiment was repeated independently twice with similar results. (right) Fold change in cell number (log2) of indicated knockout and rescued addback Jurkat cells following a 6-day growth under lipoprotein depletion in the absence or presence of sterols. (mean ± SD, n=3 biologically independent samples). Statistical significance was determined by two-tailed unpaired t-test.
F. Fold change in cell number (log2) of indicated knockout and rescued addback HEK293T cells following a 6-day growth under lipoprotein depletion in the absence or presence of sterols. (mean ± SD, n=3 biologically independent samples). Statistical significance was determined by two-tailed unpaired t-test.
Extended Data Fig. 4. TMEM41A is involved in lipid metabolism.
A. Pearson correlation values of the essentiality scores of the indicated genes across different cancer cell lines (n=558).
B. Localization of TMEM41A to ER. Wild type HEK293T cells expressing FLAG-TMEM41A cDNA were processed for immunofluorescence analysis using antibodies against FLAG and PDI (ER). White color indicates overlap (Scale bar, 8 μm). The experiment was repeated independently twice with similar results.
C. Heatmap showing the relative abundance of indicated lipid species in TMEM41-null Jurkat cells and those expressing sgRNA resistant TMEM41A cDNA.
D. Immunoblot of TMEM41A in Jurkat wild type cell line, TMEM41A nulls and those expressing TMEM41A cDNA. Actin was used as the loading control. The experiment was repeated independently twice with similar results.
E. Fold change in cell number (log2) of Jurkat wild type cell line, TMEM41A-null cells and those expressing TMEM41A cDNA after a 7-day growth upon treatment of indicated palmitate concentrations (0–80 uM). (mean ± SD, n=3 biologically independent samples). Statistical significance was determined by two-tailed unpaired t-test.
Extended Data Fig. 5. Role of C12orf49 in sterol synthesis and SREBP-mediated transcription.
A. (top left) Percentage of Bunyamwera virus-positive cells at 72-hours post-infection (MOI=0.1IU/Ml) in indicated knockout and addback HEK293T cells (mean ± SD, n=3 biologically independent samples). Statistical significance was determined by two-tailed unpaired t-test. (top right) Viral titer measured by TCID50 assays on BHK-21 cells with the harvested supernatant from the Bunyamwera virus infected HEK293T cells of C12orf49 knockouts and addbacks. (mean ± SD, n=3 biologically independent samples) Statistical significance was determined by two-tailed unpaired t-test. (bottom) Growth of the viral titers at different time points in the knockout and addback cells.
B. Fold change in mRNA levels (log2) of SREBP target genes in indicated Jurkat cell lines following 8h growth under lipoprotein depletion in the presence and absence of sterols (mean ± SD, n=3).
C. Relative luminescence activity (Luciferase/Renilla) in the indicated HEK293 cell lines following transfection with firefly luciferase under SRE promoter and Renilla luciferase for normalization of transfection following 24h growth under lipoprotein depletion in the presence and absence of sterols (mean ± SD, n=3 biologically independent samples). Statistical significance was determined by two-tailed unpaired t-test.
Extended Data Fig. 6. C12orf49 gene expression in various tissues.
A. Gene expression analysis across different tissues for C12orf49. Box plots are shown as median and 25th and 75th percentiles; points are displayed as outliers if they are above or below 1.5 times the interquartile range (Source: GTEx Portal).
B. DUF2054 profile hidden Markov Model (HMM) logo from Pfam shows 14 conserved cysteines, 3 of which are CC-dimers.
C. Different architectures of DUF2054 in different species. (Source: Pfam)
D. Occurrence of DUF2054 domain across different species.
E. Predicted N-glycosylation site (UniProtKB) and transmembrane domains (predicted with TMHMM v.2.0) for C12orf49.
F. Scheme for different functional domains of C12orf49.
Extended Data Fig. 7. The impact of C12orf49 loss on the cleavage of MBTPS1 targets.
A. Immunoblot analysis of OS9 in the C12orf49 immunoprecipitates of the HEK293T cell line expressing the indicated cDNAs. The experiment was repeated independently twice with similar results.
B. Immunoblot analysis of cleavage of other site-1 protease targets, GNPTAB, CREB3L2 and CREB4 at 24-hours following transfection in the C12orf49-knockout and addback HEK293T cells. Actin was used as loading control. The experiment was repeated independently twice with similar results.
C. Localization of SCAP-GFP in c12orf49 null HEK293T cells expressing control or C12orf49 cDNA under lipoprotein depletion in the presence or absence of sterols (Scale bar, 8 μm). The experiment was repeated independently twice with similar results.
Extended Data Fig. 8. Conservation of C12orf49 function in metazoa and zebrafish.
A. Phylogenetic tree of the C12orf49 genes across species (Source: TreeFam).
B. DNA gel showing the cutting efficiencies of c12orf49 sgRNAs used in the zebrafish experiments. Upper bands (smears) represent DNA heteroduplexes caused by CRISPR-Cas9 mutations; lower band is unedited DNA. This assay was repeated twice with similar results.
C. Strategy to evaluate the effect of CRISPR-Cas9-generated c12orf49 mutations at transcript level. c12orf49-g2 founder F0 fish were crossed and F1 progeny was individually analyzed. Briefly, RNA was isolated from individual larvae, then cDNA was synthesized. Using exon-specific primers g2 target site was PCR amplified and sequenced. Various mutations detected from transcripts are shown.
Extended Data Fig. 9. GReX analysis identifies C12orf49 association with mixed hyperlipidemia.
Disease traits associated with reduced c12orf49 GReX in BioVU biobank. Phecodes are indicated in parentheses. Traits are categorized into systems (y-axis), and significance is displayed on x-axis. Significance is tested by logistic regression analysis (two-sided), n = 25,000. Multiple testing adjustment is done using Bonferroni correction.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Birsoy lab for helpful suggestions. This research is supported by funds from a Merck Postdoctoral Fellowship (E.C.B.) at Rockefeller University. The project described was co-sponsored by the Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust. Research is supported by NIDDK (R01 DK123323-01 to K.B.), the Irma-Hirschl Trust (K.B.), NSF DMS-1812128 (S.B.), R01 MH113362-02 (E.W.K.), R01 GM117473-02 (E.W.K.), R35 HG010718 (E.R.G) and 1R01GM135926-01 (S.B.). K.B. is a Searle Scholar, Pew-Stewart Scholar and Basil O’Connor Scholar of the March of Dimes.
Footnotes
Competing interests
The authors declare no competing interests.
REFERENCES
- 1.Wang T et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 168, 890–903 e815, doi: 10.1016/j.cell.2017.01.013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan J et al. Interrogation of Mammalian Protein Complex Structure, Function, and Membership Using Genome-Scale Fitness Screens. Cell Syst 6, 555–568 e557, doi: 10.1016/j.cels.2018.04.011 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wainberg M et al. A genome-wide almanac of co-essential modules assigns function to uncharacterized genes. bioRxiv, 827071, doi: 10.1101/827071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanehisa M, Furumichi M, Tanabe M, Sato Y & Morishima K KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–D361, doi: 10.1093/nar/gkw1092 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rozman J et al. Identification of genetic elements in metabolism by high-throughput mouse phenotyping. Nat Commun 9, 288, doi: 10.1038/s41467-017-01995-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnoes AM, Brown SD, Dodevski I & Babbitt PC Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol 5, e1000605, doi: 10.1371/journal.pcbi.1000605 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey AK, Lu L, Wang X, Homayouni R & Williams RW Functionally enigmatic genes: a case study of the brain ignorome. PLoS One 9, e88889, doi: 10.1371/journal.pone.0088889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadadi N, MohammadiPeyhani H, Miskovic L, Seijo M & Hatzimanikatis V Enzyme annotation for orphan and novel reactions using knowledge of substrate reactive sites. Proc Natl Acad Sci U S A 116, 7298–7307, doi: 10.1073/pnas.1818877116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyers RM et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784, doi: 10.1038/ng.3984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsherniak A et al. Defining a Cancer Dependency Map. Cell 170, 564–576 e516, doi: 10.1016/j.cell.2017.06.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim E et al. A network of human functional gene interactions from knockout fitness screens in cancer cells. Life Sci Alliance 2, doi: 10.26508/lsa.201800278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krumsiek J, Suhre K, Illig T, Adamski J & Theis FJ Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst Biol 5, 21, doi: 10.1186/1752-0509-5-21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basu S et al. Sparse network modeling and metscape-based visualization methods for the analysis of large-scale metabolomics data. Bioinformatics 33, 1545–1553, doi: 10.1093/bioinformatics/btx012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher MM, Elsabrouty R, Seemann J, Jo Y & DeBose-Boyd RA The prenyltransferase UBIAD1 is the target of geranylgeraniol in degradation of HMG CoA reductase. Elife 4, doi: 10.7554/eLife.05560 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu XG et al. CHP1 Regulates Compartmentalized Glycerolipid Synthesis by Activating GPAT4. Mol Cell 74, 45–58 e47, doi: 10.1016/j.molcel.2019.01.037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallego-Garcia A et al. A bacterial light response reveals an orphan desaturase for human plasmalogen synthesis. Science 366, 128–132, doi: 10.1126/science.aay1436 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Bermudez J et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature, doi: 10.1038/s41586-019-0945-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton JD, Goldstein JL & Brown MS SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109, 1125–1131, doi: 10.1172/JCI15593 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Sato R, Brown MS, Hua X & Goldstein JL SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77, 53–62, doi: 10.1016/0092-8674(94)90234-8 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Brown MS & Goldstein JL The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340, doi: 10.1016/s0092-8674(00)80213-5 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Sakai J et al. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85, 1037–1046, doi: 10.1016/s0092-8674(00)81304-5 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Sakakura Y et al. Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis. Biochem Biophys Res Commun 286, 176–183, doi: 10.1006/bbrc.2001.5375 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Matsuda M et al. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev 15, 1206–1216, doi: 10.1101/gad.891301 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J et al. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc Natl Acad Sci U S A 98, 13607–13612, doi: 10.1073/pnas.201524598 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua X, Nohturfft A, Goldstein JL & Brown MS Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell 87, 415–426, doi: 10.1016/s0092-8674(00)81362-8 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Kleinfelter LM et al. Haploid Genetic Screen Reveals a Profound and Direct Dependence on Cholesterol for Hantavirus Membrane Fusion. mBio 6, e00801, doi: 10.1128/mBio.00801-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osuna-Ramos JF, Reyes-Ruiz JM & Del Angel RM The Role of Host Cholesterol During Flavivirus Infection. Front Cell Infect Microbiol 8, 388, doi: 10.3389/fcimb.2018.00388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pombo JP & Sanyal S Perturbation of Intracellular Cholesterol and Fatty Acid Homeostasis During Flavivirus Infections. Front Immunol 9, 1276, doi: 10.3389/fimmu.2018.01276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ericsson J, Jackson SM & Edwards PA Synergistic binding of sterol regulatory element-binding protein and NF-Y to the farnesyl diphosphate synthase promoter is critical for sterol-regulated expression of the gene. J Biol Chem 271, 24359–24364, doi: 10.1074/jbc.271.40.24359 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Vallett SM, Sanchez HB, Rosenfeld JM & Osborne TF A direct role for sterol regulatory element binding protein in activation of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene. J Biol Chem 271, 12247–12253, doi: 10.1074/jbc.271.21.12247 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Guan G, Dai PH, Osborne TF, Kim JB & Shechter I Multiple sequence elements are involved in the transcriptional regulation of the human squalene synthase gene. J Biol Chem 272, 10295–10302, doi: 10.1074/jbc.272.15.10295 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Edwards PA, Tabor D, Kast HR & Venkateswaran A Regulation of gene expression by SREBP and SCAP. Biochim Biophys Acta 1529, 103–113, doi: 10.1016/s1388-1981(00)00140-2 (2000). [DOI] [PubMed] [Google Scholar]
- 33.DeBose-Boyd RA et al. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99, 703–712, doi: 10.1016/s0092-8674(00)81668-2 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Lippincott-Schwartz J, Yuan LC, Bonifacino JS & Klausner RD Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801–813, doi: 10.1016/0092-8674(89)90685-5 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espenshade PJ, Cheng D, Goldstein JL & Brown MS Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J Biol Chem 274, 22795–22804, doi: 10.1074/jbc.274.32.22795 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Cheng D et al. Secreted site-1 protease cleaves peptides corresponding to luminal loop of sterol regulatory element-binding proteins. J Biol Chem 274, 22805–22812, doi: 10.1074/jbc.274.32.22805 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Velho RV et al. Site-1 protease and lysosomal homeostasis. Biochim Biophys Acta Mol Cell Res 1864, 2162–2168, doi: 10.1016/j.bbamcr.2017.06.023 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Asada R, Kanemoto S, Kondo S, Saito A & Imaizumi K The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J Biochem 149, 507–518, doi: 10.1093/jb/mvr041 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Shao W & Espenshade PJ Sterol regulatory element-binding protein (SREBP) cleavage regulates Golgi-to-endoplasmic reticulum recycling of SREBP cleavage-activating protein (SCAP). J Biol Chem 289, 7547–7557, doi: 10.1074/jbc.M113.545699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Passeri MJ, Cinaroglu A, Gao C & Sadler KC Hepatic Steatosis in Response to Acute Alcohol Exposure in Zebrafish requires Srebp Activation. Hepatology 49, 443–452, doi: 10.1002/hep.22667 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unlu G et al. GRIK5 Genetically Regulated Expression Associated with Eye and Vascular Phenomes: Discovery through Iteration among Biobanks, Electronic Health Records, and Zebrafish. Am J Hum Genet 104, 503–519, doi: 10.1016/j.ajhg.2019.01.017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roden DM et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 84, 362–369, doi: 10.1038/clpt.2008.89 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gamazon ER et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47, 1091–1098, doi: 10.1038/ng.3367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazumder R & Hastie T Exact Covariance Thresholding into Connected Components for Large-Scale Graphical Lasso. J Mach Learn Res 13, 781–794 (2012). [PMC free article] [PubMed] [Google Scholar]
- 45.Possemato R et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350, doi: 10.1038/nature10350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Bermudez J et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature 567, 118–122, doi: 10.1038/s41586-019-0945-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber RA et al. Maintaining Iron Homeostasis Is the Key Role of Lysosomal Acidity for Cell Proliferation. Mol Cell, doi: 10.1016/j.molcel.2020.01.003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jankova J & van de Geer S Confidence intervals for high-dimensional inverse covariance estimation. arXiv e-prints (2014). <https://ui.adsabs.harvard.edu/abs/2014arXiv1403.6752J>. [Google Scholar]
- 49.Cullot G et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun 10, 1136, doi: 10.1038/s41467-019-09006-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adikusuma F et al. Large deletions induced by Cas9 cleavage. Nature 560, E8–E9, doi: 10.1038/s41586-018-0380-z (2018). [DOI] [PubMed] [Google Scholar]
- 51.Harsha HC et al. Activated epidermal growth factor receptor as a novel target in pancreatic cancer therapy. J Proteome Res 7, 4651–4658, doi: 10.1021/pr800139r (2008). [DOI] [PubMed] [Google Scholar]
- 52.Schilling B et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics 11, 202–214, doi: 10.1074/mcp.M112.017707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rappsilber J, Mann M & Ishihama Y Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906, doi: 10.1038/nprot.2007.261 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372, doi: 10.1038/nbt.1511 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Schwanhausser B et al. Global quantification of mammalian gene expression control. Nature 473, 337–342, doi: 10.1038/nature10098 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Tyanova S et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740, doi: 10.1038/nmeth.3901 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Montague TG, Cruz JM, Gagnon JA, Church GM & Valen E CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42, W401–407, doi: 10.1093/nar/gku410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varshney GK et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res, doi: 10.1101/gr.186379.114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin L et al. Multiplex Conditional Mutagenesis Using Transgenic Expression of Cas9 and sgRNAs. Genetics 200, 431–441, doi: 10.1534/genetics.115.176917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levic DS et al. Animal model of Sar1b deficiency presents lipid absorption deficits similar to Anderson disease. J Mol Med (Berl) 93, 165–176, doi: 10.1007/s00109-014-1247-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Unlu G et al. Phenome-based approach identifies RIC1-linked Mendelian syndrome through zebrafish models, biobank associations and clinical studies. Nat Med 26, 98–109, doi: 10.1038/s41591-019-0705-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Denny JC et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nature biotechnology 31, 1102–1110, doi: 10.1038/nbt.2749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.GTEx_Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660, doi: 10.1126/science.1262110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.GTEx_Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213, doi: 10.1038/nature24277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Storey JD & Tibshirani R Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100, 9440–9445, doi: 10.1073/pnas.1530509100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaffter T, Marbach D & Floreano D GeneNetWeaver: in silico benchmark generation and performance profiling of network inference methods. Bioinformatics 27, 2263–2270, doi: 10.1093/bioinformatics/btr373 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Shi X, van Mierlo JT, French A & Elliott RM Visualizing the replication cycle of bunyamwera orthobunyavirus expressing fluorescent protein-tagged Gc glycoprotein. J Virol 84, 8460–8469, doi: 10.1128/JVI.00902-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.del Solar V et al. Differential Regulation of Specific Sphingolipids in Colon Cancer Cells during Staurosporine-Induced Apoptosis. Chem Biol 22, 1662–1670, doi: 10.1016/j.chembiol.2015.11.004 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request. Source data for all figures are included with the online version of the paper.