Abstract
ARID3A and ARID3B are paralogs from the AT-Rich interactive Domain (ARID) family. ARID3A and ARID3B associate to regulate genes in B-cells and cancer. We were the first to demonstrate that ARID3B regulates stem cell genes and promotes the cancer stem cell phenotype. Importantly, different knockout phenotypes in mice and distinct patterns of expression in adult animals suggests that ARID3A and ARID3B may have unique functions. In addition, high levels of ARID3B but not ARID3A induce cell death. Our goal was to express ARID3A, ARID3B, or both genes at a moderate level (as can be observed in cancer) and then identify ARID3 regulated genes. We transduced ovarian cancer cells with ARID3A-GFP, ARID3B-RFP, or both. RNA-sequencing was conducted. ARID3A and ARID3B regulated nearly identical sets of genes. Few genes (<5%) were uniquely regulated by ARID3A or ARID3B. ARID3A/B induced genes involved in cancer and stem cell processes including: Twist, MYCN, MMP2, GLI2, TIMP3, and WNT5B. We found that ARID3A and ARID3B also induced expression of each other, providing evidence of the cooperativity. While ARID3A and ARID3B likely have unique functions in distinct contexts, they are largely capable of regulating the same stem cell genes in cancer cells. This study provides a comprehensive list of genes and pathways regulated by ARID3A and ARID3B in ovarian cancer cells.
Keywords: ARID3A, ARID3B, transcription factors, stemness, cancer
1. Introduction
The AT-Rich Interactive Domain (ARID) family of DNA binding proteins and involved in chromatin remodeling and the regulation of gene expression. These proteins are characterized by the ARID DNA-binding domain, a highly conserved sequence of ~100 amino acids (Gregory et al., 1996). ARID3B has an ARID domain that shares 89.9% amino acid identity with its paralogue ARID3A (also known as Bright) (Herrscher et al., 1995; Raney et al., 2011; Rhee et al., 2014). The ARID proteins include histone deacetylases (ARID4A/B), members of the Switch/Snf complex (ARID1A and ARID1B), and other transcriptional regulators(Wilsker et al., 2005). In particular the ARID3 subfamily acts as transcriptional regulators by binding to specific DNA consensus sites to regulate gene expression. There are 3 ARID3 proteins: ARID3A, ARID3B, and ARID3C (Kim et al., 2007b; Tidwell et al., 2011; Samyesudhas et al., 2014). ARID3A is the most widely studied of the family member and plays a role in B cell immunity (Ratliff et al., 2014). Moreover, we published that ARID3A cooperates with ARID3B in the regulation of genes in B cells (Kurkewich et al., 2016). Arid3a and Arid3b are imperative for development. Arid3b−/− null embryos die mid-gestation with defects the heart, neural tissue, craniofacial structures, limb buds, and the apical endodermal ridge (Takebe et al., 2006; Casanova et al., 2011; Webb et al., 2011). Nearly 100% of Arid3a−/− embryos die mid-gestation with hematopoietic defects (Webb et al., 2011). During embryonic development ARID3A and ARID3B are not functionally redundant (Webb et al., 2011).
ARID3A is required for placental development as Arid3a−/− embryos have intrauterine growth restriction and aberrant placental structures (Rhee et al., 2014; Rhee et al., 2017). ARID3A is mostly restricted to hematopoietic tissue in the adult, embryonic stem cells (ESCs), and trophectoderm (Ratliff et al., 2014; Rhee et al., 2014; Rhee et al., 2017). ARID3B is widely expressed in adult tissues including differentiated epithelium (Samyesudhas et al., 2014). Additionally, ARID3B is expressed in stem cell populations. ARID3A and ARID3B are jointly expressed stem cell populations including ESCs, induced pluripotent stem cells (iPSCs) and cancer stem cells (CSCs) (Wang et al., 2006; Kobayashi et al., 2012; Samyesudhas et al., 2014; Liao et al., 2016). In fact, both proteins are associated with pluripotency factors in ESCs specifically in Nac1 containing complexes (Wang et al., 2006). We were the first to demonstrate that ARID3B induces stemness in cancer cells by inducing expression of stem cell genes including Prom1 (Roy et al., 2014; Bobbs et al., 2015; Roy et al., 2018). Liao et al confirmed our results and showed that ARID3A and ARID3B co-immunoprecipitate to regulatory region of stem cell genes to promote cancer stemness (Liao et al., 2016).
However, how the ARID3 proteins cooperate in regulating stemness has not been evaluated. We wanted to ascertain if co-expression of ARID3A and ARID3B is required to regulate stem cell genes or if ARID3A and ARID3B have unique functions. In this study we perform the first genome wide screen for ARID3A, ARID3B, and ARID3A/B regulated genes and pathways. We found that over 96% of the ARID3 regulated genes can be regulated by either ARID3A or ARID3B.
2. Materials and Methods
Cell Culture
Cell lines were grown at 37°C with 5% CO2. OVCA429 and OVCA433 cells (provided by Dr. Bast, MD Anderson Cancer Center, Houston, TX) (Bast et al., 1981) were grown in Minimal Essential Medium (MEM);. Media was supplemented with 10% fetal bovine serum (FBS) (Atlas, Ft. Collins, CO), 0.1 mM L-glutamine, 1mM sodium pyruvate, 50 U/mL penicillin, and 50 μg/mL streptomycin. OVCA433, OVCA429, and Kuramochi cells were lentivirally transduced with ARID3A-GFP (Genecopia, Rockville, MD), ARID3B (Gentarget), both ARID3A and ARID3B, GFP (Gentarget, San Diego, CA), Red Fluorescent Protein (RFP) (Gentarget), or both RFP and GFP as previously described (Joseph et al., 2012). Kuramochi cells (provided by Anirban Mitra, Indiana University) (Hamilton et al., 1983) were grown in RPMI media with 20% FBS and 10mg/mL insulin. Cell lines were authenticated on October 1, 2018, at ATCC by STR profiling. For Figure 4, OVCA429 cells were transduced with ARID3B-GFP (ARID3B fused to GFP to monitor localization and expression).
Figure 4:
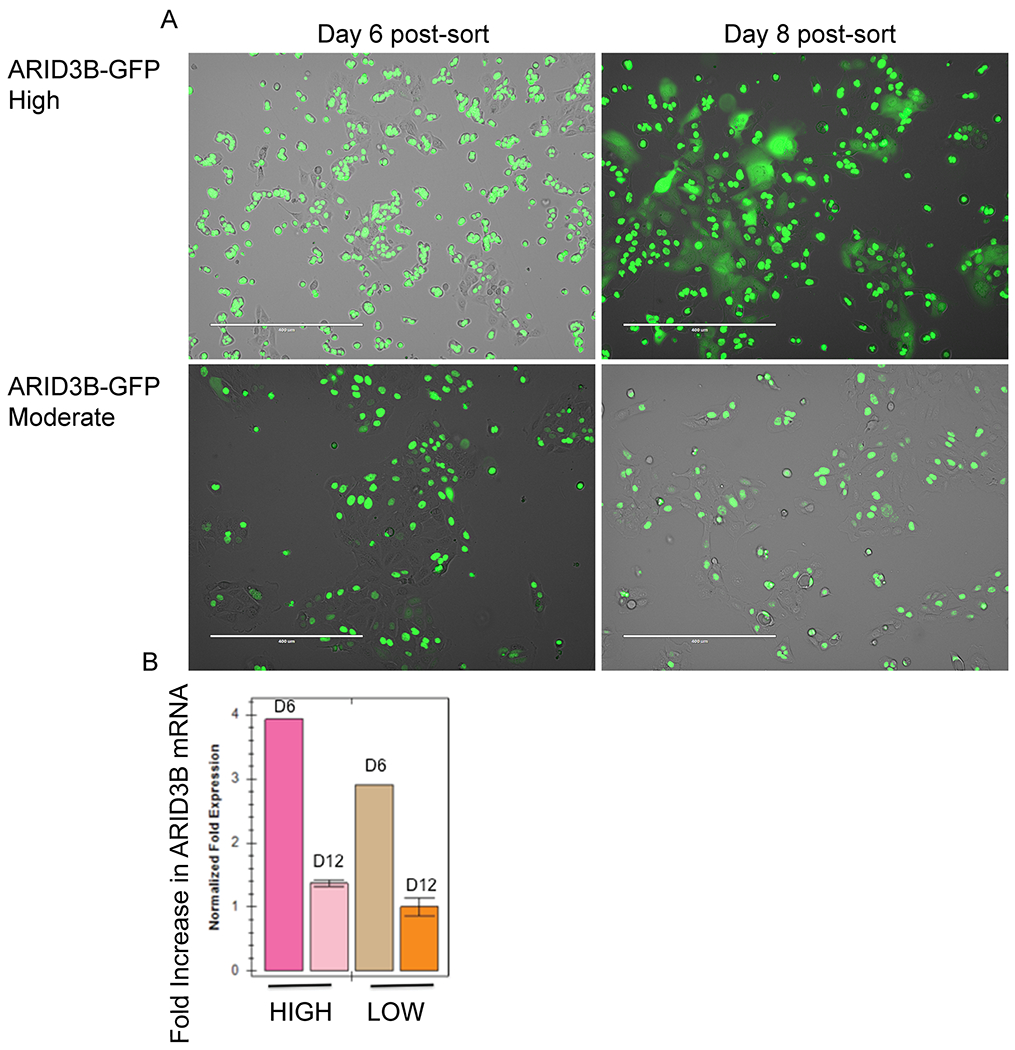
Ovarian cancer cells sorted for high and low expression of exogenous ARID3B. (A) OVCA429 cells were sorted for high or low expression of ARID3B-GFP. Fluorescence microscopy demonstrates localization of ARID3B 6- and 8-days post-sort. (B) RT-qPCR was conducted for ARID3B in OVCA429 cells sorted for high and low GFP after 6 and 12 days.
Fluorescence activated cell sorting (FACS)
FACS analysis was conducted at the Indiana University School of Medicine-South Bend at the Imaging and Flow cytometry Core Facility by Dr. Charles Tessier on the BD Biosciences FACSAria III cell sorter. Sorted cells were cultured and RNA was extracted with the Qiagen RNAeasy kit. For the cells used in RNA-seq, OVCA433 cells were transduced with RFP, GFP, RFP and GFP, ARID3A-GFP, ARID3B-RFP, or ARID3A and ARID3B. Cells were sorted and collected for GFP, RFP, or dual GFP and RFP fluorescence. In Figure 4, OVCA429 cells were transduced with ARID3B fused with GFP (ARID3B-GFP) in order to monitor expression and localization of ARID3B. RNA was isolated from cells using Trizol (Invitrogen, Carlsbad, CA ) 6, 8, and 12 days after sorting. RT-qPCR was conducted for TNF, TNFRSF1B, TNFSF10, and RIPK at day 6.
RNA-seq
RNA-seq was conducted on OVCA433 cells (expressing ARID3A, ARID3B, or GFP/RFP) in triplicate. RNA-seq libraries were prepared at the University of Notre Dame Genomics and Bioinformatics Core Facility. Total RNA samples were diluted 5 times prior to analysis. Sample concentration was measured using Qubit RNA HS Assay Kit (PN: Q32855; Invitrogen, Carlsbad, CA, USA). Total RNA evaluate with Agilent Bioanlayzer 2100 System and Agilent RNA 6000 Nano Kit (PN: 5067-1511; Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA Integrity Number (RIN) of 7 or higher were qualified for library preparation. Sample 2 and Sample 4 with RIN of 6.9 were included in the study. Total RNA input was normalized to 150 ng. Polyadendylated RNA molecules were selected for using NEBNext Poly(A) mRNA Magnetic Isolation Module (PN: E7490S/L; New England BioLabs, Ipswich, MA, USA). Enriched polyadendylated RNA was converted into an Illumina library using NEBNext Ultra II RNA Library Prep with Sample Purification Beads (PN: E7775S/L; New England BioLabs, Ipswich, MA, USA) and barcoded with NEBNext Multiplex Oligos for Illumina (Index Primers Set 1) (PN: E7335S/L; New England BioLabs, Ipswich, MA, USA) or NEBNext Multiplex Oligos for Illumina (Index Primers Set 2) (PN: E7500S/L; New England BioLabs, Ipswich, MA, USA). Indexed libraries were quantitated with Qubit dsDNA HS Assay Kit (PN: Q32854; Invitrogen, Carlsbad, CA, USA). Library quality assessment with Agilent DNA 7500 Kit (PN: 5067-1506; Agilent Technologies, Santa Clara, CA, USA). The individual libraries were normalized, and equal molar amounts were multiplexed into a single pool. Molar concentration of the multiplex pool was determined with KAPA Library Quantification Kits for Illumina (PN: KK4824; KAPA Biosystems, Boston, MA, USA). Materials used:
NEBNext Ultra II RNA Library Prep with Sample Purification Beads (PN: E7775S/L; New England BioLabs, Ipswich, MA, USA), NEBNext Poly(A) mRNA Magnetic Isolation Module (PN: E7490S/L; New England BioLabs, Ipswich, MA, USA), NEBNext Multiplex Oligos for Illumina (Index Primers Set 1) (PN: E7335S/L; New England BioLabs, Ipswich, MA, USA), NEBNext Multiplex Oligos for Illumina (Index Primers Set 2) (PN: E7500S/L; New England BioLabs, Ipswich, MA, USA), Agilent RNA 6000 Nano Kit (PN: 5067-1511; Agilent Technologies, Santa Clara, CA, USA). Agilent DNA 7500 Kit (PN: 5067-1506; Agilent Technologies, Santa Clara, CA, USA), Qubit RNA HS Assay Kit (PN: Q32855; Invitrogen, Carlsbad, CA, USA), Qubit dsDNA HS Assay Kit (PN: Q32854; Invitrogen, Carlsbad, CA, USA), and KAPA Library Quantification Kits for Illumina (PN: KK4824; KAPA Biosystems, Boston, MA, USA). Libraries were sequenced on an Illumina NextSeq 500, paired-end, 75 total cycles to obtain greater than 20 million reads per sample. Reads were aligned to the human genome (hg19) using STAR (PMC3530905). Gene level expression was calculated (Fragments per Kilobase per Million-FPKM) using Cufflinks (PMC3146043) and differentially expressed genes identified using DE-seq (PMC3218662). For all algorithms, default parameters were utilized unless otherwise noted.
Gene Expression
RNA from OVCA429, OVCA433, or Kuramochi cells was isolated using RNAeasy (Qiagen, Germantown, MD), according to the manufacturer’s instructions. Complementary DNA (cDNA) for RT-qPCR was prepared from 500 ng of RNA using High Capacity cDNA Reverse Transcription Kit (Life Technologies, Waltham, MA) as directed. Reactions were run either using iTaq Universal Probes Supermix (Bio-Rad) or Sso Fast EvaGreen Supermix (Bio-Rad). All gene expression primer sets were obtained from Integrated DNA Technologies (Coralville, IA) (Table 6) quantitative reverse transcribed polymerase chain reaction (RT-qPCR) reactions were run in triplicate and normalized to expression of GAPDH. Trizol (Thermo Fisher, Waltham, MA) was used to isolate RNA from ARID3B-GFP expressing cells (Fig. 4). RT-qPCR was conducted as described for TNF, TNFRSF1B, TNFSF10, and RIPK and normalized to GAPDH.
Table 6:
QRT-PCR Primer Information
| NCBI Gene Symbol | IDT Assay ID | Transcript | Location |
|---|---|---|---|
| ARID3A | Hs.PT.58.21479602 | NM_005224 | exon 4-5 |
| ARID3B | Hs.PT.58.40614873 | NM_006465 | exon 7-9 |
| GAPDH | Hs.PT.39a.22214836 | NM_002046(1) | exon 2-3 |
| GLI2 | Hs.PT.58.3039554 | NM_005270(1) | exon 10-11 |
| MMP2 | Hs.PT.58.39114006 | NM_001127891(2) | exon 6-7 |
| MYC | Hs.PT.58.26770695 | NM_002467 | exon 2-3 |
| MYCN | Hs.PT.58.23025106 | NM_005378(1) | exon 2-3 |
| NES | Hs.PT.58.40894423 | NM_006617 | exon 1-2 |
| RIPK1 | Hs.PT.58.15545621 | NM_003804 | exon 7-8 |
| SNAI2 | Hs.PT.58.1772559 | NM_003068 | exon 2-3 |
| TIMP3 | Hs.PT.58.1756331 | NM_000362 | exon 1-3 |
| TNF | Hs.PT.58.45380900 | NM_000594(1) | exon 1b - 4a |
| TNFRSF1B | Hs.PT.58.40638488 | NM_001066 | exon 2-3 |
| TNFSF10 | Hs.PT.58.20372853 | NM_001190943(1) | exon 1b - 2 |
| TP53 | Hs.PT.58.39676686 | NM_001126113 | exon 1-2 |
| TWIST1 | Hs.PT.58.18940950 | NM_000474 | exon 1-2 |
| WNT5B | Hs.PT.58.40348451 | NM_030775 | exon 4-5 |
Western Blots
Whole-cell lysates were obtained by lysing OVCA429, OVCA433, or Kuramochi cells (parental and expressing GFP, ARID3A, or ARID3B) in RIPA (50mM Tris pH7.5, 150mM NaCl, 1% NP-40, 0.5% EDTA, and 1X Halt Protease Inhibitor Cocktail (Pierce, Rockford, IL)). Protein concentration was determined using BCA assay according to standard protocol (Pierce, Rockford, IL). Proteins were detected using the following antibodies: ARID3B (Rabbit Polyclonal, Bethyl Laboratories, Montgomery, TX), Histone H3 (Rabbit polycolonal, Cell Signaling Technology, Danvers, MA), and ARID3A (Rabbit Polyclonal, Active Motif, Carlsbad, CA) followed by a secondary anti-rabbit HRP-conjugated antibody (Cell Signaling Technology, Danvers, MA). Imaging and quantitation were conducted using a Bio-Rad ChemiDoc Touch Imaging System, running Imager Lab Software (Hercules, CA).
Chromatin immunoprecipitation (ChIP)
ChIP was conducted on OVCA429 cells expressing GFP, ARID3A, or ARID3B using the Epiquik Chromatin Immunoprecipitation Kit (Epigentek, Farmingdale, NY). Briefly, cells were collected and chromatin-protein complexes were fixed via formaldehyde. Nuclear lysates were prepared and DNA was sheared via sonication. IPs were conducted using mouse IgG (supplied with kit), anti-ARID3B from Bethyl Laboratories (Montgomery, TX), or anti-ARID3A (Active Motif, Carlsbad, CA). After immunoprecipitation cross-links were reversed, DNA was purified, and qPCR was conducted on genes of interest including GAPDH (negative control-supplied with kit), GLI2, NES, TIMP3, and MYCN. For MYCN we used two different sets of primers that surround regions that contain either a canonical ARID3A binding motif or an ARID3B binding motif (Bobbs et al., 2015). qPCR is represented as a percentage of the total amplification of the input.
Primers for ChIP:
TIMP3 Forward 5’-TACAGACGGGGTTTCACCAT-3’
TIMP3 Reverse 5’-AAGCTAGGTGGGGTGAACCT-3’
NES Forward 5’-AGCACCTTGGAGGCTGATTA-3’
NES Reverse 5’-ATGAGACGGAGGGGATCTTT-3’
GLI2 Forward 5’-CACCATGCCCAGCTAATTTT-3’
GLI2 Reverse 5’-TCCCTTCTGGCTTCCAAATA-3’
MYCN ARID3A site Forward 5’- CCCCCTTTGGTGTGAGTC-3’
MYCN ARID3A site Reverse 5’ -ACCTAGACCCCAGCCAGTGT-3’
MYCN ARID3B site Forward 5’-CATCTGCCCTCCTCAGACTC-3’
MYCN ARID3B site Reverse 5’-CTGGCTAGGAGAGCAACAGC-3’
Statistics
Statistics were conducted on qPCR (RT-qPCR and ChIP-qPCR) data using Student t-tests on Prism GraphPad . Statistical significance was assigned to comparisons with a p-value of 0.05 or lower. Over-representation of Gene Ontology (GO) terms was calculated using a Chi-Squared Test.
3. Results
Previously we found that ARID3A and its paralog ARID3B cooperate in gene regulation in B cells and that ARID3A and ARID3B are co-expressed in ovarian cancer cells (Bobbs et al., 2015; Kurkewich et al., 2016). Additionally, we demonstrated that ARID3A and ARID3B are co-expressed with the stem cell marker CD133 in the same regions of tumors in human ovarian cancer (Roy and Cowden Dahl, 2018). We found that ARID3A and ARID3B dimerize in order to induce expression of target genes (Kurkewich et al., 2016). Others also concluded that ARID3B is important for nuclear retention of ARID3A (Kim et al., 2007a; Liao et al., 2016). Since we showed that ARID3B regulates stem cell genes and promotes a cancer stem cell phenotype and ARID3B is also expressed in ovarian cancer cells, we tested the hypothesis that ARID3A and ARID3B both regulate stem cell genes in ovarian cancer cell lines. Importantly, we previously demonstrated that high levels of ARID3B expression induces cell death through the TNF and TRAIL pathways(Joseph et al., 2012). Therefore, we wanted to identify ARID3A and ARID3B regulated genes induced by moderate levels that do not lead to cell death in ovarian cancer cell lines. We lentivirally transduced OVCA433 cells with ARID3A fused with GFP (ARID3A-GFP) first. Then we transduced cells with ARID3B co-expressing RFP (ARID3B-RFP). The order of transductions was critical as we have reported that 70% of the cell die via the TNF/TRAIL pathways when cells are transduced with ARID3B(Joseph et al., 2012). The surviving cells have lower levels of ARID3B expression; therefore it is very high expression of ARID3B that induces cell death (Joseph et al., 2012). Cell death was not seen when cells were transduced with ARID3A (not shown), and is consistent with published data(Pratama et al., 2015). ARID3A/ARID3B expressing cells were fluorescence activated cell sorted (FACSed) for RFP, GFP, or both RFP and GFP.. Cells were expanded and RNA was collected in triplicate. We generated cDNA libraries and performed RNA-sequencing. We based our cut-off for significance genes that were significantly changed in triplicates at least 3-fold (Table 1). Surprisingly we found that ARID3A, ARID3B, and the combination of ARID3A and ARID3B resulted in activation of the same genes (391-491 genes were regulated by ARID3A or ARID3B). There were very few genes (13-23) that were induced by one protein alone (or the combination of ARID3A/B) (Table 2). Therefore, over 96% of the ARID3 regulated genes were regulated by both proteins (ARID3A and ARID3B). We are still investigating the mechanism. One possibility is that ARID3A and ARID3B induce the same genes in ovarian cancer cells and are almost completely functionally redundant. Another possibility is that ARID3A and B dimerize to regulate genes and that there is a feedforward mechanism. We found that expression of ARID3A induced ARID3B by 2.2-fold. ARID3B induced ARID3A by 8.8-fold. This implies that heterodimerization may be one mechanism for ARID3A/B target gene regulation.
Table 1:
Genes induced by more than 3 fold by ARID3 proteins
| ARID3A induced genes | log2 FC | p value | ARID3B induced genes | log2 FC2 | p value3 | ARID3A/B induced genes | log2 FC3 | p value2 |
|---|---|---|---|---|---|---|---|---|
| CCND2 | 8.320 | 8.33E-161 | CCND2 | 8.962 | 5.818E-115 | TIMP3 | 8.999 | 1.80E-51 |
| TIMP3 | 8.092 | 1.35E-126 | TIMP3 | 8.825 | 9.578E-88 | GLI2 | 8.570 | 3.95E-37 |
| GLI2 | 6.848 | 1.68E-74 | GRB10 | 7.841 | 3.608E-61 | CCND2 | 8.390 | 1.99E-46 |
| GRB10 | 6.847 | 3.40E-76 | GLI2 | 7.539 | 1.455E-49 | GRB10 | 8.278 | 9.21E-36 |
| ARID3A | 6.658 | 0.00E+00 | FAM155B | 7.365 | 3.556E-65 | BGN | 7.956 | 1.24E-38 |
| TNS1 | 6.351 | 2.98E-130 | TNS1 | 7.138 | 9.825E-116 | FAM155B | 7.349 | 4.82E-28 |
| FAM155B | 6.350 | 2.12E-73 | XYLT1 | 7.073 | 1.269E-41 | XYLT1 | 7.230 | 1.49E-20 |
| MMP2 | 6.126 | 2.19E-149 | BGN | 6.243 | 3.260E-37 | TNS1 | 7.141 | 6.24E-52 |
| VCAN | 5.972 | 1.87E-95 | UNC13A | 6.216 | 1.189E-45 | MMP2 | 7.050 | 1.10E-61 |
| XYLT1 | 5.545 | 2.36E-41 | MMP2 | 6.209 | 4.896E-118 | UNC13A | 6.856 | 1.26E-32 |
| ALPPL2 | 5.540 | 1.71E-158 | MT1E | 6.116 | 8.096E-38 | ARID3A | 6.714 | 2.63E-97 |
| MCF2L | 5.472 | 1.44E-69 | ARID3A | 5.938 | 0.000E+00 | TRPV3 | 6.512 | 1.92E-25 |
| UNC13A | 5.453 | 9.66E-52 | NES | 5.872 | 6.665E-140 | MT1E | 6.443 | 2.86E-19 |
| BGN | 5.369 | 3.61E-40 | SLIT3 | 5.636 | 3.595E-48 | SLIT3 | 6.357 | 4.86E-35 |
| SPOCK1 | 5.303 | 4.32E-138 | TRPV3 | 5.572 | 1.065E-29 | CGA | 6.111 | 3.39E-19 |
| FGF13 | 5.275 | 2.91E-164 | RUNDC3A | 5.321 | 2.238E-35 | RUNDC3A | 6.028 | 1.58E-29 |
| MT1E | 5.241 | 3.37E-42 | MCF2L | 5.279 | 1.228E-39 | MIR4697HG | 5.982 | 4.88E-13 |
| RUNDC3A | 5.206 | 5.19E-60 | CLSTN2 | 5.209 | 3.165E-45 | MCF2L | 5.897 | 1.40E-37 |
| ALPP | 5.088 | 0.00E+00 | AOC1 | 5.130 | 3.784E-44 | CELSR3 | 5.846 | 8.45E-50 |
| SLIT3 | 5.069 | 4.79E-52 | WNT5B | 5.126 | 2.834E-61 | IGSF9B | 5.753 | 1.06E-11 |
| TRPV3 | 5.021 | 9.84E-38 | FABP5 | 5.069 | 1.226E-17 | SAG | 5.635 | 1.45E-11 |
| NES | 4.979 | 1.76E-205 | CGA | 5.047 | 5.258E-23 | RORC | 5.613 | 6.61E-19 |
| SCARA3 | 4.844 | 5.35E-248 | IGSF9B | 5.032 | 2.354E-17 | NES | 5.404 | 1.06E-44 |
| MAP1LC3C | 4.842 | 5.29E-105 | FGF13 | 5.001 | 6.051E-52 | SARDH | 5.403 | 2.06E-20 |
| CELSR3 | 4.814 | 4.11E-78 | SARDH | 4.984 | 1.392E-28 | ZNF469 | 5.402 | 1.12E-13 |
| FLI1 | 4.803 | 9.10E-73 | CELSR3 | 4.963 | 1.093E-56 | CKMT1B | 5.393 | 4.75E-15 |
| SEMA5A | 4.801 | 4.87E-62 | SPOCK1 | 4.943 | 7.364E-34 | ACKR3 | 5.355 | 7.22E-12 |
| KRT17 | 4.772 | 2.17E-75 | SCN2B | 4.918 | 2.698E-16 | NOS3 | 5.300 | 1.77E-34 |
| CGA | 4.698 | 2.31E-29 | ACKR3 | 4.918 | 5.201E-18 | KRT17 | 5.288 | 4.86E-46 |
| FAT3 | 4.640 | 2.03E-51 | SH3RF3 | 4.893 | 2.621E-53 | ALOX15 | 5.275 | 6.71E-33 |
| SFMBT2 | 4.566 | 7.29E-29 | SFMBT2 | 4.859 | 1.492E-20 | IGDCC3 | 5.262 | 8.57E-21 |
| CLSTN2 | 4.538 | 6.34E-60 | SYT17 | 4.843 | 4.002E-19 | SYT17 | 5.200 | 1.75E-12 |
| LOC101929331 | 4.478 | 6.60E-57 | ALPPL2 | 4.842 | 7.708E-68 | PHF21B | 5.191 | 4.89E-12 |
| ALOX15 | 4.434 | 1.70E-48 | FLI1 | 4.833 | 2.432E-53 | LOC101929331 | 5.180 | 1.10E-40 |
| WNT5B | 4.412 | 1.26E-55 | SCARA3 | 4.829 | 1.886E-75 | SFMBT2 | 5.162 | 6.77E-13 |
| ACOXL | 4.387 | 1.23E-39 | ANK1 | 4.822 | 5.627E-38 | PDCD1 | 5.152 | 1.14E-12 |
| KCNH7 | 4.338 | 4.42E-51 | NGFR | 4.791 | 2.882E-28 | ASIC2 | 5.119 | 1.91E-18 |
| HEPH | 4.317 | 9.84E-22 | KRT17 | 4.782 | 2.012E-40 | SPOCK1 | 5.081 | 7.97E-38 |
| SDPR | 4.315 | 1.12E-122 | C15orf48 | 4.747 | 3.059E-23 | TMIGD2 | 4.995 | 2.11E-08 |
| NRXN3 | 4.305 | 2.14E-27 | MIR4697HG | 4.726 | 1.310E-14 | WNT5B | 4.992 | 2.75E-23 |
| PAPPA | 4.301 | 5.15E-79 | NOS3 | 4.722 | 1.199E-50 | MAP1LC3C | 4.978 | 1.45E-34 |
| SCG2 | 4.291 | 2.28E-58 | SCNN1G | 4.708 | 1.368E-20 | SCARA3 | 4.969 | 6.91E-57 |
| SH3RF3 | 4.256 | 1.18E-51 | ZNF469 | 4.668 | 4.526E-16 | SH3RF3 | 4.964 | 1.17E-21 |
| ANK1 | 4.233 | 1.12E-91 | ALPP | 4.663 | 1.804E-169 | MEIS3 | 4.949 | 1.06E-16 |
| MGP | 4.211 | 1.12E-54 | ALOX15 | 4.640 | 1.308E-37 | ALPPL2 | 4.937 | 7.71E-24 |
| AOC1 | 4.200 | 1.45E-40 | LOC101929331 | 4.620 | 2.218E-44 | ANK1 | 4.928 | 1.40E-20 |
| RNF175 | 4.194 | 4.13E-42 | MAP1LC3C | 4.590 | 3.499E-59 | CKMT1A | 4.914 | 1.04E-15 |
| MEIS3 | 4.126 | 3.60E-25 | RNF175 | 4.574 | 3.731E-36 | ZCCHC18 | 4.905 | 3.64E-08 |
| SARDH | 4.122 | 3.06E-27 | CKMT1B | 4.544 | 4.147E-17 | FGF13 | 4.855 | 7.07E-24 |
| RORC | 4.068 | 1.46E-21 | ASIC2 | 4.471 | 3.180E-26 | GSTO2 | 4.811 | 2.07E-20 |
| LRRC17 | 4.062 | 1.28E-35 | MEIS3 | 4.437 | 1.714E-19 | FABP5 | 4.809 | 8.44E-08 |
| MIR4697HG | 4.036 | 1.83E-18 | CKMT1A | 4.426 | 2.900E-21 | CLSTN2 | 4.796 | 2.33E-33 |
| SPOCK2 | 4.024 | 7.87E-291 | LHX6 | 4.390 | 1.059E-106 | AP3B2 | 4.751 | 4.07E-09 |
| C15orf48 | 4.015 | 3.59E-24 | HEPH | 4.389 | 2.496E-12 | CHGA | 4.654 | 2.19E-09 |
| ZNF469 | 4.007 | 5.34E-19 | ACOXL | 4.387 | 2.131E-27 | FOXS1 | 4.649 | 1.28E-08 |
| CKMT1B | 4.007 | 2.90E-21 | RORC | 4.354 | 4.530E-18 | AR | 4.607 | 1.89E-07 |
| IGDCC3 | 4.001 | 3.79E-24 | APCDD1L-AS1 | 4.335 | 2.147E-33 | ACOXL | 4.605 | 3.63E-22 |
| GSTO2 | 3.978 | 6.35E-35 | PRSS3P2 | 4.332 | 9.934E-12 | HIF3A | 4.565 | 1.17E-35 |
| OSR2 | 3.968 | 0.00E+00 | VCAN | 4.306 | 2.188E-18 | ALPP | 4.552 | 9.38E-50 |
| PAPPA-AS1 | 3.963 | 3.03E-24 | SCG2 | 4.260 | 9.455E-39 | AOC1 | 4.469 | 9.56E-19 |
| HIF3A | 3.957 | 6.56E-35 | GSTO2 | 4.240 | 3.845E-24 | SYBU | 4.458 | 7.96E-14 |
| CKMT1A | 3.863 | 2.98E-24 | ZCCHC18 | 4.211 | 3.506E-11 | RNF175 | 4.441 | 3.23E-24 |
| RGMA | 3.851 | 7.01E-39 | CDH16 | 4.199 | 2.223E-13 | ONECUT3 | 4.412 | 8.14E-20 |
| MFSD7 | 3.786 | 2.70E-45 | PHF21B | 4.161 | 4.024E-13 | CALY | 4.341 | 2.49E-08 |
| ELAVL2 | 3.745 | 5.81E-122 | NOG | 4.119 | 5.898E-63 | LINC00885 | 4.327 | 3.90E-07 |
| EYA1 | 3.745 | 4.73E-67 | SULF2 | 4.117 | 7.327E-14 | MFSD7 | 4.310 | 4.14E-24 |
| DEPDC7 | 3.736 | 3.50E-41 | MFSD7 | 4.080 | 1.306E-40 | DGCR5 | 4.304 | 1.62E-06 |
| IGSF9B | 3.727 | 2.43E-15 | NRXN3 | 4.071 | 2.845E-14 | C15orf48 | 4.281 | 3.31E-11 |
| APCDD1L-AS1 | 3.716 | 1.18E-32 | PRSS2 | 4.049 | 4.464E-10 | TGM4 | 4.281 | 5.09E-17 |
| SYT17 | 3.698 | 1.63E-16 | MYCN | 4.043 | 9.632E-58 | SCNN1G | 4.273 | 1.33E-07 |
| CACNG4 | 3.645 | 2.43E-51 | LRRC17 | 4.043 | 6.739E-25 | FAM178B | 4.273 | 3.57E-18 |
| NOS3 | 3.633 | 4.83E-25 | SH3RF3-AS1 | 4.008 | 1.731E-26 | BCL11A | 4.268 | 3.27E-10 |
| PEG10 | 3.626 | 9.57E-59 | TGM4 | 4.004 | 9.888E-21 | FLI1 | 4.252 | 6.19E-19 |
| FLRT2 | 3.621 | 3.05E-70 | CSF2RB | 3.996 | 1.314E-11 | OSR2 | 4.201 | 9.93E-56 |
| NDN | 3.606 | 1.31E-105 | RGMA | 3.948 | 8.112E-34 | TMEM145 | 4.189 | 2.45E-13 |
| WNT6 | 3.583 | 3.39E-37 | MN1 | 3.912 | 6.927E-21 | MGP | 4.189 | 2.75E-12 |
| PRSS23 | 3.581 | 3.57E-156 | TMC5 | 3.909 | 4.657E-14 | HRK | 4.178 | 7.33E-08 |
| LHX6 | 3.551 | 1.14E-82 | BCL11A | 3.903 | 2.867E-16 | EPDR1 | 4.158 | 8.10E-08 |
| FIBCD1 | 3.547 | 9.67E-87 | PAPPA | 3.868 | 1.050E-19 | APCDD1L-AS1 | 4.153 | 1.07E-16 |
| ASIC2 | 3.544 | 9.59E-19 | OSR2 | 3.852 | 2.266E-122 | LOC283070 | 4.140 | 5.26E-13 |
| PHF21B | 3.520 | 2.39E-14 | GDNF-AS1 | 3.846 | 6.117E-09 | CACNG4 | 4.104 | 8.28E-38 |
| CTGF | 3.518 | 6.91E-120 | VGF | 3.825 | 1.817E-112 | RGMA | 4.102 | 3.28E-19 |
| FABP5 | 3.503 | 4.14E-13 | DTNA | 3.801 | 1.581E-10 | SH3RF3-AS1 | 4.052 | 1.06E-12 |
| LSAMP-AS1 | 3.502 | 7.13E-15 | MGP | 3.790 | 1.430E-26 | OASL | 4.004 | 5.39E-27 |
| ONECUT3 | 3.477 | 2.55E-29 | AR | 3.785 | 1.983E-09 | PRODH | 3.936 | 1.64E-06 |
| RYR2 | 3.474 | 1.24E-32 | CACNG4 | 3.784 | 1.100E-52 | VGF | 3.897 | 1.72E-16 |
| LOC283070 | 3.474 | 1.74E-18 | HIF3A | 3.772 | 2.970E-34 | C5AR1 | 3.885 | 1.10E-06 |
| ACKR3 | 3.459 | 1.88E-13 | NDN | 3.769 | 2.845E-42 | FIBCD1 | 3.880 | 1.94E-35 |
| RTN4RL1 | 3.451 | 4.92E-24 | CSF1R | 3.756 | 5.981E-80 | NDN | 3.862 | 5.04E-26 |
| AR | 3.429 | 3.46E-13 | LSAMP-AS1 | 3.749 | 8.379E-11 | NRXN3 | 3.839 | 1.68E-05 |
| LINC00889 | 3.418 | 1.82E-16 | SDPR | 3.745 | 2.704E-44 | LHX6 | 3.838 | 2.26E-24 |
| OASL | 3.394 | 9.09E-102 | SPOCK2 | 3.716 | 1.564E-91 | TMEM240 | 3.821 | 1.68E-08 |
| SH3RF3-AS1 | 3.379 | 5.93E-23 | RPP25 | 3.697 | 4.118E-57 | NEURL1 | 3.786 | 2.71E-30 |
| FBN1 | 3.364 | 2.92E-75 | NEURL1 | 3.693 | 1.914E-165 | RPP25 | 3.779 | 1.67E-17 |
| NEURL1 | 3.358 | 1.94E-206 | CTGF | 3.682 | 1.180E-61 | DTNA | 3.741 | 2.42E-05 |
| NEBL | 3.355 | 1.57E-44 | SYBU | 3.681 | 8.361E-14 | RTN4RL1 | 3.733 | 1.09E-15 |
| PRSS3P2 | 3.350 | 4.87E-12 | ADRA1B | 3.678 | 4.239E-50 | SYT5 | 3.689 | 3.16E-10 |
| SCNN1G | 3.343 | 5.54E-14 | LOC283070 | 3.661 | 2.038E-14 | CSF1R | 3.686 | 2.43E-18 |
| GFRA1 | 3.338 | 1.08E-159 | PDCD1 | 3.656 | 1.702E-09 | LSAMP-AS1 | 3.681 | 5.66E-05 |
| BCL11A | 3.322 | 6.15E-19 | IGDCC3 | 3.655 | 2.351E-13 | INHBE | 3.668 | 2.87E-07 |
| TGM4 | 3.309 | 4.25E-18 | FAM178B | 3.621 | 3.939E-17 | CTGF | 3.655 | 3.19E-40 |
| TP53I11 | 3.308 | 5.60E-65 | TNS4 | 3.607 | 9.411E-26 | SPOCK2 | 3.652 | 1.83E-63 |
| TMEM145 | 3.300 | 6.54E-19 | LARGE | 3.573 | 2.130E-52 | PLIN5 | 3.639 | 2.75E-06 |
| DTNA | 3.299 | 8.82E-14 | TMEM145 | 3.528 | 1.460E-16 | LARGE | 3.620 | 1.23E-18 |
| MYCN | 3.270 | 1.07E-48 | GPR17 | 3.473 | 2.191E-11 | ADRA1B | 3.598 | 2.02E-14 |
| LARGE | 3.269 | 1.46E-61 | KRTAP2-3 | 3.464 | 5.371E-26 | TMC5 | 3.595 | 2.57E-07 |
| SHC3 | 3.234 | 4.05E-167 | ADAMTS17 | 3.456 | 3.043E-17 | TMEM158 | 3.566 | 2.11E-43 |
| SCN8A | 3.229 | 2.44E-54 | PRSS23 | 3.455 | 4.180E-104 | HES7 | 3.561 | 1.68E-07 |
| TMEM158 | 3.224 | 2.26E-92 | FBLL1 | 3.444 | 3.908E-08 | S100A4 | 3.560 | 6.67E-70 |
| ADRA1B | 3.221 | 6.93E-67 | ONECUT3 | 3.443 | 2.465E-19 | SCAMP5 | 3.536 | 1.69E-21 |
| VGF | 3.192 | 9.89E-107 | PAPPA-AS1 | 3.428 | 9.642E-09 | NDRG4 | 3.523 | 1.03E-40 |
| ZCCHC18 | 3.191 | 1.27E-12 | MUC16 | 3.419 | 8.153E-12 | CAPN8 | 3.509 | 1.89E-17 |
| NOG | 3.176 | 2.07E-136 | TP53I11 | 3.419 | 7.542E-76 | TMEM105 | 3.501 | 3.23E-20 |
| RARB | 3.138 | 1.33E-34 | CLU | 3.404 | 1.047E-64 | DHH | 3.492 | 6.43E-05 |
| PRSS2 | 3.122 | 2.45E-10 | DENND2A | 3.401 | 6.525E-63 | TP53I11 | 3.483 | 2.37E-34 |
| NTRK2 | 3.111 | 7.41E-38 | SEMA5A | 3.392 | 4.693E-14 | DENND2A | 3.481 | 5.68E-27 |
| LOC100506178 | 3.110 | 2.01E-80 | VIM-AS1 | 3.373 | 9.451E-48 | FXYD1 | 3.479 | 1.01E-09 |
| CLU | 3.106 | 4.04E-47 | CACNA2D4 | 3.369 | 2.961E-14 | OGDHL | 3.472 | 2.05E-18 |
| PRODH | 3.078 | 1.23E-10 | FIBCD1 | 3.357 | 3.927E-43 | CHST4 | 3.467 | 1.07E-06 |
| ACE | 3.060 | 2.06E-21 | HSD11B1 | 3.355 | 1.988E-17 | SLC47A2 | 3.411 | 1.19E-14 |
| RPP25 | 3.054 | 5.36E-49 | SHC3 | 3.346 | 6.048E-128 | HCAR1 | 3.371 | 2.29E-20 |
| DGCR5 | 3.033 | 4.70E-10 | TMEM158 | 3.344 | 3.098E-71 | MYCN | 3.371 | 5.37E-17 |
| ACHE | 3.030 | 3.01E-46 | EPDR1 | 3.338 | 3.065E-08 | AGAP2 | 3.370 | 3.76E-20 |
| AP3B2 | 3.024 | 5.12E-10 | HCAR1 | 3.336 | 1.903E-54 | PAPPA | 3.364 | 2.30E-08 |
| TMC5 | 3.024 | 1.08E-11 | WNT6 | 3.333 | 2.409E-27 | ADSSL1 | 3.361 | 3.78E-17 |
| S100A4 | 3.012 | 0.00E+00 | KCNH7 | 3.319 | 5.377E-14 | FAM71D | 3.354 | 5.73E-07 |
| ADSSL1 | 3.010 | 9.60E-32 | VIM | 3.310 | 5.432E-53 | LHB | 3.349 | 2.08E-05 |
| ABAT | 3.001 | 1.26E-42 | OGDHL | 3.308 | 1.790E-24 | P2RX5 | 3.339 | 1.09E-12 |
| CSF1R | 2.987 | 1.30E-64 | P2RX5 | 3.303 | 1.434E-58 | PPFIBP2 | 3.335 | 6.70E-12 |
| SYBU | 2.985 | 2.49E-12 | S100A4 | 3.287 | 2.551E-37 | FAM115C | 3.329 | 6.43E-12 |
| IL7R | 2.985 | 5.10E-27 | SBK2 | 3.259 | 4.190E-07 | NOG | 3.325 | 4.70E-18 |
| NDRG4 | 2.977 | 2.03E-108 | DHH | 3.257 | 6.933E-08 | CECR6 | 3.301 | 2.16E-16 |
| PDCD1 | 2.972 | 5.13E-10 | GDNF | 3.249 | 1.231E-29 | CACNA2D4 | 3.298 | 2.17E-06 |
| HCAR1 | 2.958 | 1.19E-51 | EYA1 | 3.232 | 1.115E-21 | GPR17 | 3.298 | 2.27E-05 |
| LINC01468 | 2.947 | 1.83E-77 | TRIM29 | 3.224 | 6.595E-11 | ELFN1 | 3.289 | 2.02E-05 |
| TENM1 | 2.943 | 4.90E-09 | SCAMP5 | 3.209 | 9.157E-49 | DYSF | 3.285 | 8.82E-33 |
| EPDR1 | 2.931 | 3.73E-10 | OASL | 3.197 | 5.015E-28 | PRSS23 | 3.282 | 1.88E-14 |
| DENND2A | 2.917 | 6.07E-128 | ADSSL1 | 3.190 | 3.455E-22 | GDF15 | 3.256 | 1.36E-54 |
| TMEM105 | 2.899 | 7.00E-28 | NTRK2 | 3.159 | 4.005E-16 | ACE | 3.248 | 9.27E-19 |
| SCAMP5 | 2.896 | 9.71E-86 | GFRA1 | 3.144 | 6.656E-45 | IGFBP3 | 3.242 | 1.11E-47 |
| IGFBP3 | 2.895 | 0.00E+00 | APLN | 3.135 | 6.880E-09 | PAPPA-AS1 | 3.230 | 0.000158502 |
| ENOX1 | 2.889 | 6.08E-23 | ARID3B | 3.134 | 1.105E-107 | LGALS7B | 3.226 | 5.87E-14 |
| OGDHL | 2.874 | 2.45E-27 | CRB2 | 3.127 | 2.663E-19 | CPLX1 | 3.225 | 2.66E-11 |
| CCL5 | 2.869 | 2.55E-44 | TMEM105 | 3.116 | 3.330E-24 | NOX5 | 3.219 | 1.93E-08 |
| SULF2 | 2.864 | 1.46E-09 | APCDD1L | 3.086 | 4.203E-58 | KRTAP2-3 | 3.215 | 1.07E-08 |
| GUCY1A3 | 2.853 | 1.12E-65 | DYSF | 3.085 | 1.026E-48 | VIM-AS1 | 3.210 | 1.22E-15 |
| PLCE1 | 2.850 | 3.35E-76 | LURAP1 | 3.081 | 1.266E-09 | NUPR1 | 3.202 | 2.85E-13 |
| KIAA1549L | 2.815 | 9.82E-66 | GGT5 | 3.056 | 2.921E-10 | SHC3 | 3.168 | 3.21E-52 |
| FBLL1 | 2.811 | 7.40E-09 | SYT5 | 3.042 | 1.505E-09 | ACOX2 | 3.165 | 1.03E-05 |
| ASS1 | 2.808 | 1.18E-134 | ACHE | 3.032 | 1.988E-36 | VIM | 3.163 | 3.03E-17 |
| FAM178B | 2.796 | 8.35E-12 | FSCN1 | 3.031 | 2.770E-69 | ACHE | 3.154 | 1.85E-23 |
| PRR16 | 2.779 | 2.59E-43 | NOX5 | 3.014 | 2.038E-13 | TNIK | 3.152 | 9.05E-09 |
| TNIK | 2.777 | 1.33E-70 | CAPN8 | 3.013 | 3.281E-16 | WNT6 | 3.133 | 3.91E-12 |
| ACSS1 | 2.750 | 4.75E-15 | RTN4RL1 | 3.002 | 1.897E-12 | NAT16 | 3.128 | 5.26E-05 |
| TMEM45A | 2.749 | 2.43E-163 | LOC100506178 | 2.997 | 2.679E-48 | CD22 | 3.127 | 3.10E-11 |
| DYSF | 2.746 | 8.57E-100 | HECW1 | 2.994 | 1.001E-19 | GDNF | 3.126 | 5.25E-13 |
| UST | 2.738 | 2.32E-43 | UNC5A | 2.988 | 6.403E-07 | MUC16 | 3.124 | 3.97E-17 |
| ABI3BP | 2.733 | 1.29E-24 | TRIB1 | 2.976 | 1.763E-65 | SCG2 | 3.120 | 8.78E-05 |
| VIM-AS1 | 2.723 | 1.37E-107 | AGAP2 | 2.975 | 2.206E-51 | CLU | 3.099 | 1.47E-25 |
| P2RX5 | 2.721 | 1.25E-55 | KIAA1549L | 2.973 | 3.950E-21 | CAMK1D | 3.095 | 3.66E-08 |
| FSCN1 | 2.716 | 1.29E-128 | UST | 2.970 | 1.595E-23 | HECW1 | 3.088 | 2.95E-13 |
| SNAP25 | 2.704 | 4.07E-08 | CPLX1 | 2.968 | 8.179E-23 | RENBP | 3.077 | 4.50E-07 |
| FAM115C | 2.701 | 7.77E-16 | NDRG4 | 2.968 | 5.101E-50 | WIPF3 | 3.075 | 6.06E-14 |
| FGF11 | 2.698 | 5.25E-25 | RARB | 2.955 | 8.203E-18 | ACSS1 | 3.057 | 3.84E-10 |
| CECR6 | 2.685 | 2.28E-15 | FAT3 | 2.953 | 3.162E-07 | CCL5 | 3.042 | 1.98E-18 |
| HOTAIR | 2.684 | 3.10E-11 | TNIK | 2.946 | 1.846E-11 | LOC440028 | 3.034 | 6.81E-08 |
| VIM | 2.675 | 1.79E-185 | FAM115C | 2.917 | 7.447E-15 | MN1 | 3.028 | 1.30E-09 |
| AGAP2 | 2.666 | 3.29E-123 | TMEM240 | 2.880 | 6.420E-07 | KCNAB3 | 3.025 | 3.91E-07 |
| MUC16 | 2.658 | 4.33E-09 | TNNC1 | 2.873 | 9.544E-23 | ANKRD24 | 3.024 | 1.03E-15 |
| ITGB6 | 2.655 | 1.21E-61 | LOC440028 | 2.871 | 5.958E-09 | GPC4 | 3.019 | 2.95E-38 |
| TMEM130 | 2.652 | 2.16E-82 | LINC00889 | 2.870 | 4.576E-06 | BSN | 3.010 | 2.43E-15 |
| HRK | 2.648 | 4.39E-08 | GDF15 | 2.869 | 2.026E-38 | TRIB1 | 2.979 | 7.68E-22 |
| HECW1 | 2.633 | 9.13E-18 | GYPC | 2.865 | 7.169E-27 | FGF11 | 2.976 | 2.75E-14 |
| LOC101929680 | 2.633 | 7.27E-30 | HRK | 2.853 | 1.828E-05 | ATOH8 | 2.967 | 2.20E-09 |
| LOC440028 | 2.631 | 1.37E-11 | IFI27 | 2.833 | 1.748E-09 | FSCN1 | 2.965 | 7.94E-20 |
| CPNE4 | 2.630 | 1.97E-52 | CHST4 | 2.826 | 1.444E-06 | MAPK8IP2 | 2.958 | 5.01E-13 |
| SLC47A2 | 2.628 | 1.34E-15 | KCNMA1 | 2.817 | 5.679E-47 | LURAP1 | 2.946 | 5.27E-06 |
| C5AR1 | 2.610 | 8.31E-08 | DEPDC7 | 2.816 | 4.467E-12 | UST | 2.936 | 3.53E-23 |
| TRIB1 | 2.605 | 2.61E-93 | CECR6 | 2.811 | 5.911E-16 | CHRM4 | 2.933 | 1.43E-06 |
| HMCN1 | 2.605 | 2.39E-36 | CPNE4 | 2.810 | 2.015E-41 | ANKRD13B | 2.922 | 7.73E-17 |
| CD180 | 2.605 | 4.74E-08 | NHSL2 | 2.807 | 1.807E-10 | NTN1 | 2.892 | 9.01E-18 |
| PLIN5 | 2.595 | 6.40E-08 | MFNG | 2.803 | 4.281E-15 | GFRA1 | 2.888 | 5.42E-28 |
| CACNA2D4 | 2.595 | 8.69E-11 | CHRM4 | 2.794 | 1.248E-11 | ASS1 | 2.887 | 5.16E-47 |
| PTGER4 | 2.592 | 1.70E-70 | FGF11 | 2.792 | 1.014E-22 | LOC100506178 | 2.883 | 1.01E-19 |
| SBF2-AS1 | 2.584 | 2.30E-55 | CDSN | 2.781 | 8.496E-14 | SPTBN4 | 2.881 | 5.97E-08 |
| GYPC | 2.581 | 8.34E-93 | SCN8A | 2.781 | 1.998E-11 | ZFYVE28 | 2.880 | 1.37E-08 |
| GPC4 | 2.580 | 5.11E-62 | ACE | 2.778 | 2.317E-17 | INPP5D | 2.876 | 1.08E-07 |
| TMEM2 | 2.577 | 4.02E-219 | FAM83A | 2.768 | 1.446E-40 | STAC3 | 2.871 | 2.08E-15 |
| COL4A5 | 2.573 | 4.16E-176 | ELAVL2 | 2.762 | 2.465E-19 | STMN3 | 2.862 | 5.00E-10 |
| ARMCX4 | 2.570 | 3.18E-15 | WIPF3 | 2.754 | 3.531E-30 | SDPR | 2.852 | 8.42E-05 |
| TNNC1 | 2.566 | 2.57E-64 | PEG10 | 2.744 | 1.656E-12 | COL4A2-AS1 | 2.838 | 4.82E-10 |
| APCDD1L | 2.563 | 2.70E-49 | LGALS7B | 2.732 | 2.112E-09 | LYPD3 | 2.830 | 8.21E-32 |
| GDF15 | 2.554 | 2.12E-13 | KPNA7 | 2.728 | 7.736E-07 | HSPA2 | 2.822 | 4.37E-06 |
| CHRM4 | 2.549 | 8.73E-12 | ANKRD24 | 2.728 | 4.668E-30 | LMCD1 | 2.816 | 1.16E-11 |
| LURAP1 | 2.543 | 7.22E-09 | ABAT | 2.723 | 3.789E-27 | CPNE4 | 2.815 | 7.86E-26 |
| GDNF | 2.542 | 1.27E-20 | ACSS1 | 2.713 | 1.840E-10 | KIAA1549L | 2.810 | 1.50E-34 |
| MN1 | 2.539 | 2.92E-10 | TRPV4 | 2.704 | 4.792E-44 | CRIP1 | 2.808 | 2.71E-07 |
| CAMK1D | 2.527 | 6.05E-12 | ST3GAL1 | 2.696 | 4.022E-51 | TH | 2.807 | 3.12E-07 |
| ELFN1 | 2.506 | 2.82E-08 | LMCD1 | 2.689 | 1.032E-26 | TMEM63C | 2.802 | 9.71E-17 |
| CAPN8 | 2.505 | 3.63E-13 | SLC47A2 | 2.678 | 3.305E-13 | KPNA7 | 2.800 | 0.000453299 |
| FZD4 | 2.499 | 6.80E-79 | SBSN | 2.674 | 7.606E-09 | COL17A1 | 2.787 | 1.22E-13 |
| NUPR1 | 2.498 | 2.16E-11 | NTN1 | 2.674 | 5.398E-102 | EYA1 | 2.786 | 5.74E-05 |
| LGALS7B | 2.473 | 7.72E-13 | PRSS1 | 2.672 | 7.233E-08 | ZG16B | 2.781 | 2.19E-07 |
| MAPK8IP2 | 2.470 | 7.25E-19 | CCL5 | 2.672 | 7.989E-17 | EHD2 | 2.780 | 1.00E-20 |
| TMEM240 | 2.456 | 3.52E-07 | INHBE | 2.670 | 1.080E-06 | MAPT | 2.778 | 8.35E-06 |
| LINC01279 | 2.445 | 5.96E-61 | MAPK8IP2 | 2.659 | 2.571E-17 | TNNC1 | 2.752 | 7.62E-30 |
| RSAD2 | 2.444 | 4.89E-77 | PPARGC1B | 2.657 | 3.947E-24 | TRPV4 | 2.751 | 2.41E-11 |
| EPHB6 | 2.437 | 2.88E-51 | CAMK1D | 2.649 | 3.417E-13 | CTB-113P19.1 | 2.751 | 0.000287867 |
| TRPV4 | 2.436 | 4.23E-56 | RYR2 | 2.647 | 6.508E-07 | KCND1 | 2.742 | 2.43E-14 |
| NHSL2 | 2.431 | 1.44E-13 | IGFBP3 | 2.620 | 1.547E-35 | RCOR2 | 2.735 | 4.37E-07 |
| CPLX1 | 2.422 | 1.08E-15 | FLRT2 | 2.609 | 2.888E-10 | GYPC | 2.722 | 1.16E-16 |
| LYPD3 | 2.422 | 1.67E-183 | ANKRD13B | 2.608 | 2.663E-57 | ASAP3 | 2.711 | 1.73E-34 |
| FXYD1 | 2.418 | 1.16E-08 | GPC4 | 2.606 | 8.631E-49 | APCDD1L | 2.693 | 1.56E-10 |
| COL5A1 | 2.416 | 7.04E-129 | STMN3 | 2.602 | 6.829E-27 | ADAMTS17 | 2.676 | 3.02E-06 |
| LINC00482 | 2.416 | 3.84E-07 | ATG9B | 2.590 | 1.297E-21 | SLC22A20 | 2.659 | 3.28E-07 |
| SYT5 | 2.387 | 1.06E-07 | FXYD1 | 2.588 | 2.129E-07 | CDC42EP5 | 2.657 | 1.40E-22 |
| ASAP3 | 2.381 | 5.82E-75 | GOLGA7B | 2.575 | 4.507E-12 | CALHM3 | 2.656 | 8.14E-14 |
| PNMA2 | 2.375 | 8.72E-66 | BSN | 2.575 | 1.968E-11 | THBS3 | 2.656 | 1.22E-22 |
| CNKSR3 | 2.368 | 3.76E-35 | ZG16B | 2.573 | 2.474E-09 | LIMS2 | 2.643 | 6.75E-06 |
| STAC3 | 2.362 | 3.58E-14 | EPHB6 | 2.545 | 1.587E-34 | COL5A1 | 2.622 | 7.28E-32 |
| CRIP1 | 2.360 | 4.39E-09 | ELFN1 | 2.541 | 1.595E-05 | TMEM130 | 2.621 | 3.06E-34 |
| NTN1 | 2.356 | 1.37E-137 | ENOX1 | 2.541 | 2.343E-10 | BCAN | 2.615 | 3.34E-11 |
| ANKRD24 | 2.350 | 7.82E-27 | DPF3 | 2.540 | 2.121E-09 | HOTAIR | 2.613 | 3.94E-05 |
| NOX5 | 2.334 | 1.62E-10 | LINC01315 | 2.539 | 5.085E-07 | MFNG | 2.610 | 5.34E-09 |
| SLC9A7P1 | 2.332 | 1.29E-08 | CALHM3 | 2.537 | 2.559E-21 | PRR16 | 2.602 | 7.53E-10 |
| FAM71D | 2.331 | 1.11E-07 | KCNAB3 | 2.518 | 1.847E-06 | PTGER4 | 2.598 | 1.60E-14 |
| KRTAP2-3 | 2.320 | 2.01E-12 | LOC284344 | 2.518 | 6.099E-06 | RHBDL1 | 2.596 | 1.57E-08 |
| TRPC4 | 2.320 | 6.47E-20 | NCF2 | 2.514 | 1.086E-21 | VSIG10L | 2.595 | 1.33E-14 |
| STEAP4 | 2.319 | 6.23E-21 | HMOX1 | 2.512 | 4.407E-24 | C2CD4C | 2.586 | 1.49E-13 |
| INHBA | 2.307 | 4.32E-107 | LIMS2 | 2.508 | 2.901E-09 | PDZD7 | 2.584 | 1.69E-07 |
| STXBP6 | 2.301 | 1.44E-37 | SPHK1 | 2.499 | 8.315E-36 | AQP3 | 2.578 | 8.76E-33 |
| WIPF3 | 2.300 | 5.89E-28 | ASS1 | 2.482 | 7.836E-21 | APOE | 2.544 | 1.16E-23 |
| CASC10 | 2.300 | 1.74E-71 | COL17A1 | 2.480 | 2.972E-49 | P2RX2 | 2.542 | 0.000243848 |
| INPP5D | 2.296 | 6.85E-08 | NCR3LG1 | 2.476 | 3.426E-20 | FLRT2 | 2.541 | 0.002928445 |
| LOC284344 | 2.293 | 6.78E-07 | PNMA2 | 2.453 | 2.923E-44 | CNKSR3 | 2.527 | 4.47E-24 |
| TNFSF15 | 2.290 | 2.74E-30 | TMEM130 | 2.444 | 6.396E-43 | SEZ6L2 | 2.526 | 5.01E-18 |
| IFITM1 | 2.288 | 2.58E-30 | FBN1 | 2.438 | 3.071E-13 | KCNH7 | 2.522 | 0.010675877 |
| BSN | 2.276 | 7.56E-15 | LOC102724094 | 2.437 | 3.138E-06 | WTIP | 2.517 | 4.11E-14 |
| EPSTI1 | 2.262 | 1.05E-88 | BCAN | 2.426 | 4.121E-42 | SLC23A3 | 2.515 | 0.000239171 |
| LOC101927151 | 2.261 | 1.68E-33 | TBC1D30 | 2.424 | 2.893E-28 | FAT3 | 2.507 | 0.015356083 |
| ANKRD13B | 2.254 | 5.96E-58 | STAC3 | 2.421 | 7.111E-12 | PPARGC1B | 2.501 | 1.28E-12 |
| AQP3 | 2.253 | 4.89E-44 | G0S2 | 2.415 | 1.199E-21 | MIR4709 | 2.496 | 0.001704675 |
| UNC5A | 2.251 | 5.07E-06 | INPP5D | 2.403 | 5.552E-07 | FZD8 | 2.493 | 2.93E-06 |
| HES7 | 2.226 | 2.24E-06 | WNT4 | 2.401 | 7.704E-10 | AIFM3 | 2.484 | 1.98E-05 |
| MFNG | 2.220 | 2.11E-11 | SLC25A53 | 2.398 | 1.325E-09 | KCNC3 | 2.484 | 8.43E-15 |
| INHBE | 2.215 | 7.46E-06 | TMEM63C | 2.397 | 5.329E-28 | SPHK1 | 2.472 | 1.85E-06 |
| SNTB1 | 2.203 | 1.68E-09 | CORO6 | 2.393 | 5.623E-23 | ATF3 | 2.458 | 8.48E-15 |
| PRSS1 | 2.203 | 4.76E-08 | CTB-113P19.1 | 2.388 | 1.196E-05 | NCF2 | 2.448 | 2.33E-08 |
| COL17A1 | 2.202 | 1.99E-53 | FZD8 | 2.383 | 1.295E-14 | PRRT4 | 2.441 | 3.53E-30 |
| SLC23A3 | 2.200 | 1.07E-06 | MAPT | 2.379 | 7.237E-07 | ABAT | 2.437 | 2.34E-09 |
| PSG8 | 2.191 | 3.13E-06 | TMEM45A | 2.373 | 5.632E-68 | NDRG1 | 2.434 | 5.37E-10 |
| LMCD1 | 2.187 | 2.50E-37 | COL4A5 | 2.362 | 1.695E-76 | ARTN | 2.428 | 9.50E-10 |
| KCND1 | 2.181 | 2.62E-17 | IL7R | 2.360 | 4.644E-08 | CDK5R2 | 2.425 | 3.81E-12 |
| PPFIBP2 | 2.171 | 9.04E-09 | LINC01468 | 2.349 | 1.378E-22 | EPHB6 | 2.422 | 2.26E-21 |
| APLN | 2.167 | 5.57E-06 | TH | 2.337 | 4.316E-06 | CASC10 | 2.421 | 5.68E-27 |
| CDC42EP5 | 2.160 | 1.07E-50 | SPARC | 2.335 | 1.778E-32 | CCR10 | 2.418 | 8.24E-05 |
| ITGBL1 | 2.160 | 1.22E-72 | PRR16 | 2.327 | 3.527E-21 | DMPK | 2.403 | 4.38E-31 |
| THBS3 | 2.148 | 2.61E-51 | C2orf54 | 2.324 | 3.754E-29 | COL4A1 | 2.399 | 7.67E-12 |
| ADAMTS17 | 2.148 | 1.93E-07 | SPTBN5 | 2.323 | 2.546E-11 | ST3GAL1 | 2.398 | 1.88E-07 |
| IL1RAP | 2.147 | 5.56E-58 | EHD2 | 2.322 | 1.153E-71 | COL7A1 | 2.395 | 2.58E-05 |
| NCR3LG1 | 2.146 | 6.16E-22 | GUCY1A3 | 2.311 | 2.297E-15 | EDA | 2.394 | 1.99E-06 |
| FGFR3 | 2.141 | 1.30E-38 | NDRG1 | 2.305 | 1.261E-24 | C19orf83 | 2.392 | 9.58E-06 |
| PPARGC1B | 2.124 | 7.85E-30 | IFITM1 | 2.303 | 3.333E-18 | BTBD19 | 2.391 | 1.04E-16 |
| FILIP1L | 2.124 | 2.13E-08 | FZD4 | 2.299 | 1.812E-40 | SAMD14 | 2.377 | 1.24E-10 |
| TMEM63C | 2.123 | 1.13E-24 | COL5A1 | 2.297 | 2.294E-34 | KALRN | 2.375 | 3.49E-09 |
| SPTBN5 | 2.118 | 1.79E-17 | PVRL1 | 2.292 | 7.128E-16 | ASPHD1 | 2.369 | 1.58E-10 |
| CALHM3 | 2.116 | 3.68E-29 | PPFIBP2 | 2.290 | 7.189E-07 | FGFR3 | 2.361 | 5.89E-21 |
| LINC00662 | 2.115 | 3.12E-26 | ASAP3 | 2.285 | 4.523E-29 | CYP26B1 | 2.358 | 3.20E-34 |
| APOE | 2.105 | 3.86E-139 | TNFSF15 | 2.282 | 3.172E-12 | LOC101927911 | 2.355 | 4.60E-05 |
| KALRN | 2.099 | 5.69E-13 | STAC | 2.273 | 4.712E-26 | TUBA4A | 2.354 | 1.97E-08 |
| LINC00346 | 2.096 | 2.67E-64 | APOE | 2.261 | 1.630E-21 | ANGPT4 | 2.350 | 0.00040507 |
| PRRT4 | 2.096 | 3.61E-71 | HOTAIR | 2.261 | 1.229E-05 | SERPINF2 | 2.347 | 3.65E-06 |
| C9orf84 | 2.095 | 1.10E-24 | OAS2 | 2.261 | 3.281E-23 | KLHDC7B | 2.343 | 7.13E-07 |
| TRHDE | 2.086 | 8.90E-32 | LYPD3 | 2.244 | 1.223E-35 | DOK3 | 2.341 | 1.11E-06 |
| SH3BGRL | 2.082 | 3.83E-47 | LINC00346 | 2.238 | 1.593E-29 | SCN8A | 2.326 | 0.000310342 |
| LBH | 2.080 | 1.52E-11 | CD22 | 2.233 | 2.649E-07 | FAM83A | 2.325 | 2.80E-06 |
| ZFYVE28 | 2.078 | 6.98E-10 | COL4A1 | 2.230 | 2.955E-96 | PARVG | 2.325 | 0.000517755 |
| EHD2 | 2.069 | 4.77E-33 | ACTL8 | 2.229 | 9.231E-05 | PRSS1 | 2.322 | 0.00025244 |
| ZNF711 | 2.066 | 1.68E-16 | CNKSR3 | 2.226 | 9.289E-25 | CNTNAP1 | 2.308 | 1.17E-10 |
| WTIP | 2.065 | 1.75E-60 | GSN-AS1 | 2.221 | 5.165E-14 | EFNA2 | 2.308 | 8.73E-10 |
| SLC16A12 | 2.061 | 4.87E-07 | RAMP1 | 2.221 | 9.535E-17 | DNAJC22 | 2.307 | 1.24E-21 |
| TNS4 | 2.060 | 3.85E-09 | DLL1 | 2.220 | 1.047E-27 | LOC284344 | 2.305 | 0.020386634 |
| RASSF2 | 2.058 | 5.31E-81 | TUBA4A | 2.211 | 8.574E-22 | CPAMD8 | 2.296 | 4.90E-14 |
| TIAM2 | 2.057 | 4.16E-49 | NUPR1 | 2.194 | 1.166E-06 | CORO6 | 2.293 | 1.17E-09 |
| SKIDA1 | 2.056 | 4.69E-15 | SLC23A3 | 2.187 | 2.715E-04 | GSN | 2.288 | 7.21E-30 |
| ZNF365 | 2.051 | 1.46E-42 | LOC101929680 | 2.187 | 1.780E-16 | BTG2 | 2.285 | 1.01E-12 |
| CD22 | 2.042 | 1.16E-07 | PRCD | 2.185 | 8.559E-05 | COL4A5 | 2.279 | 1.59E-28 |
| CPAMD8 | 2.036 | 1.79E-14 | CPAMD8 | 2.179 | 4.209E-22 | NNMT | 2.270 | 6.40E-07 |
| FLRT3 | 2.034 | 4.97E-05 | THBS3 | 2.173 | 4.449E-41 | KCNMA1 | 2.267 | 1.14E-05 |
| GPR17 | 2.033 | 1.59E-05 | GSN | 2.171 | 6.781E-18 | COL4A2 | 2.266 | 3.62E-08 |
| GSN | 2.032 | 1.10E-95 | KCNK3 | 2.159 | 2.403E-07 | HCN2 | 2.265 | 2.67E-11 |
| DMPK | 2.032 | 1.12E-52 | AQP1 | 2.159 | 9.584E-09 | TNFRSF21 | 2.251 | 1.11E-53 |
| DPP4 | 2.031 | 3.96E-12 | PARVG | 2.158 | 5.175E-05 | FBXO27 | 2.250 | 7.23E-10 |
| LEPREL1 | 2.031 | 7.42E-175 | DOK3 | 2.156 | 2.830E-07 | LEPREL1 | 2.249 | 3.36E-47 |
| GUCY1B2 | 2.030 | 4.56E-09 | KCND1 | 2.150 | 7.458E-14 | IFITM2 | 2.248 | 2.15E-09 |
| VSIG10L | 2.028 | 3.07E-21 | SAMD14 | 2.134 | 1.409E-25 | GSN-AS1 | 2.244 | 4.62E-11 |
| LINC00052 | 2.027 | 1.01E-07 | HSPA2 | 2.129 | 8.340E-05 | DLL1 | 2.243 | 6.87E-08 |
| PTCH1 | 2.025 | 4.79E-46 | CD79A | 2.128 | 1.280E-05 | TGFBI | 2.236 | 4.51E-12 |
| FAM198B | 2.025 | 3.99E-48 | RASSF2 | 2.127 | 4.097E-23 | DNM1 | 2.226 | 1.97E-12 |
| ZG16B | 2.024 | 2.83E-07 | COL4A2-AS1 | 2.124 | 5.839E-18 | HDAC5 | 2.224 | 4.91E-11 |
| ST3GAL1 | 2.020 | 1.23E-32 | COL7A1 | 2.121 | 3.312E-05 | SH2B2 | 2.216 | 5.74E-18 |
| NCF2 | 2.006 | 1.00E-24 | ADPRHL1 | 2.115 | 1.296E-11 | ETHE1 | 2.212 | 2.51E-13 |
| IFIT2 | 2.005 | 4.59E-45 | CCR1 | 2.105 | 2.739E-04 | STAC | 2.212 | 4.95E-11 |
| DNAJC22 | 2.000 | 1.84E-52 | PDZD7 | 2.105 | 1.265E-10 | SPARC | 2.209 | 2.27E-14 |
| DLL1 | 2.000 | 2.85E-29 | SBF2-AS1 | 2.097 | 3.055E-26 | C9orf3 | 2.199 | 2.92E-25 |
| GS1-259H13.2 | 1.997 | 4.27E-09 | CASC10 | 2.096 | 2.205E-21 | CEL | 2.196 | 3.17E-05 |
| AIFM3 | 1.990 | 3.42E-08 | LOC101927911 | 2.094 | 7.370E-08 | PNMA2 | 2.193 | 4.75E-14 |
| SPARC | 1.990 | 9.38E-103 | HDAC5 | 2.089 | 3.935E-20 | LINC00346 | 2.189 | 2.51E-27 |
| C9orf3 | 1.987 | 7.21E-68 | ZFYVE28 | 2.087 | 9.868E-09 | ATG9B | 2.187 | 1.29E-12 |
| SLC25A53 | 1.984 | 1.86E-08 | PPP2R2C | 2.082 | 8.097E-18 | ATP1A3 | 2.184 | 4.88E-07 |
| RCOR2 | 1.984 | 1.35E-06 | EDA | 2.081 | 7.417E-07 | FLJ42969 | 2.180 | 1.34E-05 |
| TH | 1.982 | 2.41E-06 | SLC22A20 | 2.079 | 3.761E-06 | RASSF2 | 2.177 | 6.30E-32 |
| BCAS1 | 1.975 | 1.27E-09 | PRPS1 | 2.079 | 1.472E-19 | ARHGEF19 | 2.175 | 3.16E-13 |
| ATOH8 | 1.973 | 3.99E-08 | ARHGAP44 | 2.079 | 6.824E-08 | TIMP4 | 2.171 | 2.66E-06 |
| ADAM22 | 1.973 | 1.65E-38 | SERPINF2 | 2.078 | 4.186E-07 | BEGAIN | 2.168 | 8.80E-06 |
| KCNMA1 | 1.970 | 1.44E-25 | TRHDE-AS1 | 2.078 | 6.720E-11 | LTBP4 | 2.167 | 6.81E-20 |
| CORO6 | 1.968 | 2.03E-16 | GSDMA | 2.073 | 1.086E-04 | UCN | 2.164 | 1.85E-05 |
| TRHDE-AS1 | 1.966 | 1.60E-19 | CDC42EP5 | 2.071 | 1.516E-17 | ARID3B | 2.162 | 8.10E-09 |
| HSD11B1 | 1.965 | 2.01E-06 | RENBP | 2.046 | 2.318E-04 | ADM | 2.161 | 1.99E-16 |
| C2CD4C | 1.964 | 2.54E-22 | SFTA1P | 2.044 | 3.568E-13 | HSD11B1 | 2.159 | 0.000191586 |
| STMN3 | 1.963 | 7.63E-32 | FBXO27 | 2.044 | 1.974E-20 | ENOX1 | 2.147 | 0.004249116 |
| BCAN | 1.962 | 5.18E-20 | PTGER4 | 2.044 | 1.026E-27 | PVRL1 | 2.145 | 7.21E-05 |
| ARID5B | 1.962 | 6.98E-39 | KALRN | 2.043 | 5.820E-09 | PPP2R2C | 2.137 | 2.79E-06 |
| TBC1D30 | 1.961 | 1.20E-21 | MYO7B | 2.041 | 1.652E-04 | HMOX1 | 2.131 | 1.66E-05 |
| OAS2 | 1.958 | 4.14E-23 | COL4A2 | 2.029 | 2.533E-96 | PTCH1 | 2.128 | 1.74E-26 |
| PDE1C | 1.957 | 1.12E-64 | NEBL | 2.027 | 1.149E-06 | PITPNM3 | 2.126 | 1.32E-13 |
| PIK3AP1 | 1.956 | 1.73E-69 | TIMP4 | 2.024 | 3.546E-07 | LINC01315 | 2.123 | 0.002616831 |
| FZD8 | 1.949 | 1.42E-10 | EDN2 | 2.012 | 4.343E-20 | LOC101927151 | 2.121 | 2.56E-22 |
| CYP26B1 | 1.945 | 5.90E-68 | BTBD19 | 2.007 | 9.534E-10 | SPTBN5 | 2.120 | 2.72E-07 |
| MST4 | 1.943 | 3.17E-89 | SLC9A7P1 | 2.006 | 3.429E-04 | ADAMTS15 | 2.116 | 2.02E-06 |
| PSG5 | 1.942 | 1.73E-17 | ARMCX4 | 2.004 | 8.240E-05 | ST6GALNAC1 | 2.111 | 5.71E-05 |
| ADPRHL1 | 1.936 | 8.74E-13 | SYT7 | 2.003 | 3.863E-17 | TMEM255B | 2.110 | 4.61E-08 |
| EFHD1 | 1.935 | 2.38E-07 | FAM71D | 2.001 | 6.576E-04 | STXBP6 | 2.110 | 3.33E-13 |
| EML6 | 1.931 | 2.24E-16 | HTR1D | 1.997 | 1.756E-08 | RAMP1 | 2.109 | 1.26E-05 |
| CDK5R2 | 1.929 | 3.60E-18 | TRPC4 | 1.996 | 1.697E-08 | ADPRHL1 | 2.106 | 2.24E-09 |
| SERPINF2 | 1.923 | 1.53E-09 | FGFR1 | 1.993 | 7.616E-35 | GRIN1 | 2.105 | 4.00E-05 |
| ARMCX3 | 1.914 | 9.50E-63 | SEZ6L2 | 1.992 | 6.372E-54 | NXPH4 | 2.104 | 1.68E-12 |
| GSN-AS1 | 1.908 | 5.72E-15 | ETHE1 | 1.989 | 1.120E-11 | APLP1 | 2.102 | 5.72E-16 |
| COL4A6 | 1.906 | 1.71E-35 | ITGBL1 | 1.984 | 2.224E-33 | BEX5 | 2.089 | 0.001269072 |
| TNFRSF21 | 1.905 | 3.34E-27 | LOC100507346 | 1.984 | 7.261E-10 | BCAS1 | 2.086 | 0.000201639 |
| INHBA-AS1 | 1.902 | 3.59E-05 | STXBP6 | 1.982 | 1.831E-21 | RAP1GAP | 2.084 | 2.51E-06 |
| PLLP | 1.901 | 7.91E-25 | CACNA1H | 1.981 | 1.672E-07 | LOC100507346 | 2.082 | 3.91E-08 |
| ST6GALNAC5 | 1.894 | 8.69E-57 | TPM1 | 1.981 | 4.329E-62 | PRPS1 | 2.075 | 7.79E-09 |
| KCNAB3 | 1.893 | 4.83E-05 | C19orf83 | 1.981 | 1.513E-09 | MEF2B | 2.075 | 1.74E-09 |
| ANKDD1A | 1.892 | 3.01E-18 | LBH | 1.981 | 7.375E-09 | LOC102724094 | 2.073 | 0.038921813 |
| SEZ6L2 | 1.890 | 3.82E-41 | C2CD4C | 1.977 | 1.908E-16 | IGF1R | 2.073 | 6.61E-32 |
| TPM1 | 1.887 | 3.90E-111 | TAGLN3 | 1.977 | 7.680E-07 | GOLGA7B | 2.070 | 0.000836578 |
| LIPA | 1.885 | 7.95E-99 | RRAD | 1.977 | 4.595E-08 | LRG1 | 2.067 | 1.39E-11 |
| HDAC5 | 1.885 | 2.95E-52 | DNAJC22 | 1.975 | 2.213E-25 | MSI1 | 2.065 | 5.44E-09 |
| LIMS2 | 1.883 | 1.29E-06 | LY6D | 1.972 | 4.469E-07 | DPF3 | 2.063 | 0.008258627 |
| POF1B | 1.883 | 1.05E-13 | VSIG10L | 1.968 | 1.797E-13 | CCDC64 | 2.062 | 6.08E-10 |
| SCN9A | 1.876 | 4.76E-29 | CNTNAP1 | 1.965 | 1.218E-15 | ECSIT | 2.061 | 1.48E-06 |
| MAMDC2 | 1.868 | 9.73E-44 | PLXNA2 | 1.965 | 1.184E-18 | CCDC159 | 2.055 | 4.34E-05 |
| ATF3 | 1.866 | 1.06E-35 | CDK5R2 | 1.964 | 2.639E-20 | FBXL16 | 2.052 | 6.99E-09 |
| PDE1A | 1.863 | 3.08E-05 | RCOR2 | 1.956 | 4.797E-05 | G0S2 | 2.051 | 4.67E-05 |
| SH2B2 | 1.855 | 3.74E-22 | DMPK | 1.954 | 3.723E-49 | CHADL | 2.050 | 0.009004621 |
| FBXO27 | 1.853 | 9.57E-59 | ADAMTS2 | 1.948 | 2.870E-05 | GS1-259H13.2 | 2.049 | 7.51E-06 |
| MAPT | 1.852 | 3.66E-05 | SPTBN4 | 1.937 | 7.331E-05 | PDLIM1 | 2.046 | 2.44E-15 |
| NDUFA4L2 | 1.850 | 3.59E-06 | AQP3 | 1.936 | 2.149E-18 | CAMK2N2 | 2.043 | 8.64E-09 |
| STAC | 1.850 | 9.18E-43 | DNM1 | 1.932 | 2.082E-58 | HSD11B2 | 2.041 | 0.000123533 |
| ETHE1 | 1.849 | 1.65E-52 | ENDOD1 | 1.929 | 5.924E-37 | FBN1 | 2.040 | 0.003981023 |
| MIR24-1 | 1.844 | 1.09E-04 | SNTB1 | 1.927 | 4.355E-05 | RBAKDN | 2.039 | 2.35E-08 |
| SLC22A20 | 1.838 | 5.97E-07 | AVIL | 1.926 | 9.816E-09 | AVIL | 2.038 | 4.35E-07 |
| TUBA4A | 1.835 | 7.42E-35 | ECSIT | 1.923 | 1.581E-13 | SLC25A53 | 2.033 | 0.000157179 |
| RHBDL1 | 1.835 | 5.14E-06 | TMEM255B | 1.922 | 1.926E-08 | C1QTNF6 | 2.030 | 1.67E-12 |
| COL4A1 | 1.834 | 1.40E-97 | OAS1 | 1.921 | 8.163E-11 | SLC7A7 | 2.029 | 1.24E-06 |
| IFIT1 | 1.828 | 7.04E-43 | ESPN | 1.914 | 2.296E-07 | ARMCX6 | 2.023 | 2.40E-10 |
| C8orf46 | 1.819 | 4.05E-17 | BEGAIN | 1.914 | 1.263E-15 | CYP27B1 | 2.021 | 7.90E-06 |
| KCNC3 | 1.817 | 8.41E-07 | TGFBI | 1.907 | 2.598E-24 | CSDC2 | 2.001 | 3.49E-11 |
| PTHLH | 1.803 | 1.18E-08 | ATP1A3 | 1.901 | 1.846E-20 | IL6R | 1.991 | 3.35E-08 |
| IGF1R | 1.801 | 1.04E-76 | ZDHHC9 | 1.893 | 1.001E-29 | TWIST1 | 1.988 | 4.09E-10 |
| TNFSF18 | 1.796 | 3.13E-04 | TNC | 1.892 | 1.711E-48 | SH3PXD2B | 1.985 | 1.02E-08 |
| COL4A2-AS1 | 1.790 | 1.43E-14 | CDHR5 | 1.890 | 2.355E-03 | TAGLN3 | 1.985 | 0.000103474 |
| EFEMP1 | 1.788 | 4.31E-191 | ATF3 | 1.889 | 6.527E-24 | LMF1 | 1.978 | 3.59E-12 |
| FMNL2 | 1.788 | 8.57E-26 | ARTN | 1.885 | 4.997E-25 | SERPINE1 | 1.974 | 1.49E-08 |
| BTBD19 | 1.781 | 3.49E-12 | TMEM2 | 1.883 | 2.146E-12 | LBH | 1.971 | 1.03E-06 |
| SNAI2 | 1.779 | 2.75E-68 | MTRNR2L9 | 1.882 | 1.005E-03 | B3GNT3 | 1.964 | 3.64E-09 |
| SPTBN4 | 1.778 | 2.47E-05 | PDE1A | 1.880 | 5.923E-04 | ARMCX4 | 1.964 | 0.003075744 |
| LOC100507346 | 1.774 | 9.71E-10 | PLCE1 | 1.876 | 5.659E-06 | ATP2A1 | 1.959 | 9.60E-05 |
| CCDC85A | 1.772 | 4.16E-06 | EML6 | 1.867 | 8.562E-07 | MAG | 1.958 | 0.006136636 |
| SAMD14 | 1.770 | 1.14E-21 | CALHM1 | 1.859 | 2.843E-04 | ACTA2-AS1 | 1.955 | 0.000386814 |
| BTG2 | 1.766 | 3.86E-15 | LINC00052 | 1.857 | 5.046E-05 | DDIT4 | 1.954 | 1.16E-26 |
| SYT1 | 1.760 | 7.28E-19 | PTCH1 | 1.856 | 1.307E-20 | SLC22A18 | 1.952 | 2.53E-12 |
| LINC01315 | 1.756 | 5.89E-05 | EPSTI1 | 1.855 | 4.789E-38 | TMEM159 | 1.950 | 8.51E-09 |
| TMEM64 | 1.756 | 4.93E-76 | ST6GALNAC5 | 1.851 | 9.169E-32 | DGCR6 | 1.946 | 3.79E-13 |
| TGFBI | 1.745 | 6.17E-162 | PYGL | 1.849 | 8.612E-35 | C1orf226 | 1.946 | 4.57E-07 |
| METTL7A | 1.744 | 8.45E-38 | LINC01272 | 1.849 | 5.539E-05 | SLC9A7P1 | 1.944 | 0.018168032 |
| PVRL1 | 1.743 | 2.09E-10 | TRAF1 | 1.846 | 9.404E-13 | NHSL2 | 1.943 | 7.32E-05 |
| PPP2R2C | 1.737 | 8.71E-17 | IGF1R | 1.845 | 2.357E-20 | FARP1 | 1.941 | 7.45E-08 |
| LHX9 | 1.734 | 2.65E-04 | C8orf46 | 1.843 | 1.010E-14 | PTPRH | 1.940 | 6.40E-07 |
| AVIL | 1.731 | 3.55E-08 | COL4A6 | 1.842 | 3.265E-32 | BNIPL | 1.940 | 0.009815729 |
| OAS1 | 1.725 | 3.22E-43 | ARHGEF19 | 1.841 | 2.035E-24 | IFITM1 | 1.939 | 6.24E-08 |
| MYPN | 1.718 | 5.71E-19 | DPH6-AS1 | 1.841 | 4.116E-04 | HSF4 | 1.937 | 1.27E-11 |
| ARHGEF19 | 1.708 | 2.78E-68 | LOC100128531 | 1.839 | 1.088E-04 | ZDHHC9 | 1.936 | 3.48E-08 |
| BNIPL | 1.707 | 9.28E-05 | PTPRB | 1.838 | 2.739E-14 | STAB1 | 1.933 | 0.001081894 |
| TBC1D4 | 1.706 | 1.28E-35 | TNFRSF21 | 1.838 | 4.518E-51 | SLC29A3 | 1.925 | 3.80E-19 |
| ANTXR1 | 1.705 | 1.27E-112 | SYNPO2L | 1.838 | 6.003E-04 | VAV2 | 1.918 | 1.42E-08 |
| DOK3 | 1.704 | 4.04E-06 | PIK3AP1 | 1.838 | 4.525E-38 | ABI3BP | 1.917 | 0.005936399 |
| ST6GALNAC1 | 1.700 | 1.33E-05 | PRRT4 | 1.835 | 1.812E-31 | ANKRD2 | 1.911 | 4.69E-07 |
| PLLP | 1.824 | 1.058E-07 | ADAMTS10 | 1.903 | 2.88E-10 | |||
| SH3PXD2B | 1.824 | 5.193E-51 | CYP2S1 | 1.903 | 4.00E-08 | |||
| LIPA | 1.822 | 1.217E-37 | RARB | 1.898 | 0.012910091 | |||
| LINC00341 | 1.821 | 1.550E-04 | TNC | 1.896 | 3.18E-17 | |||
| ELF4 | 1.820 | 3.753E-65 | ENO3 | 1.887 | 6.74E-05 | |||
| TIAM2 | 1.818 | 2.586E-22 | EBI3 | 1.887 | 2.22E-06 | |||
| SPNS2 | 1.818 | 1.382E-29 | TRPC4 | 1.885 | 0.002404155 | |||
| SFN | 1.817 | 3.061E-16 | PNPLA7 | 1.883 | 0.000335928 | |||
| AIFM3 | 1.816 | 9.545E-05 | ADM5 | 1.882 | 2.02E-05 | |||
| CYP27B1 | 1.815 | 1.279E-06 | SFTA1P | 1.876 | 2.45E-05 | |||
| WTIP | 1.815 | 2.382E-27 | RNF113A | 1.876 | 1.23E-08 | |||
| ASPHD1 | 1.813 | 5.493E-21 | SKIDA1 | 1.874 | 8.80E-06 | |||
| C1QTNF6 | 1.812 | 1.164E-30 | GCHFR | 1.860 | 7.79E-07 | |||
| PTHLH | 1.811 | 1.501E-06 | RNF139-AS1 | 1.860 | 0.010801165 | |||
| JAG1 | 1.810 | 2.609E-37 | SOX12 | 1.857 | 5.66E-09 | |||
| LMF1 | 1.810 | 4.159E-16 | ELF4 | 1.853 | 6.84E-06 | |||
| GS1-259H13.2 | 1.809 | 2.053E-04 | NRARP | 1.849 | 1.20E-05 | |||
| CYFIP1 | 1.805 | 2.113E-18 | CYFIP1 | 1.844 | 2.47E-08 | |||
| PITPNM3 | 1.802 | 1.026E-22 | PPP1R26-AS1 | 1.843 | 4.34E-08 | |||
| DUSP5 | 1.799 | 5.678E-11 | NGF | 1.843 | 0.039137171 | |||
| RASSF4 | 1.799 | 2.197E-17 | GSDMA | 1.840 | 0.029606583 | |||
| LTBP2 | 1.795 | 2.285E-14 | SFN | 1.839 | 3.02E-05 | |||
| ITPRIPL1 | 1.792 | 2.775E-13 | LRP1 | 1.836 | 2.34E-14 | |||
| ACTA2-AS1 | 1.790 | 9.696E-05 | TMEM253 | 1.830 | 0.028104575 | |||
| C9orf3 | 1.789 | 1.733E-17 | SNORD86 | 1.829 | 9.59E-05 | |||
| ABI3BP | 1.784 | 3.155E-04 | MIR3074 | 1.820 | 0.017375964 | |||
| CCR10 | 1.775 | 3.213E-04 | METTL7B | 1.820 | 0.00016621 | |||
| HSD11B2 | 1.765 | 6.776E-06 | TPM1 | 1.819 | 2.11E-19 | |||
| KCNC3 | 1.757 | 1.385E-04 | COL4A6 | 1.817 | 4.29E-10 | |||
| MYD88 | 1.753 | 1.148E-13 | METTL7A | 1.815 | 1.83E-10 | |||
| MIR4709 | 1.752 | 2.117E-03 | LRRC26 | 1.814 | 0.000483156 | |||
| EBI3 | 1.751 | 5.364E-20 | SEMA7A | 1.813 | 4.01E-06 | |||
| ZNF488 | 1.751 | 3.568E-13 | SLC12A8 | 1.805 | 5.92E-07 | |||
| DIRAS1 | 1.749 | 2.942E-10 | AHNAK2 | 1.803 | 2.66E-14 | |||
| TRHDE | 1.749 | 1.417E-06 | LOXL1 | 1.803 | 5.02E-06 | |||
| BEX5 | 1.748 | 1.922E-03 | ITGB5 | 1.802 | 3.80E-06 | |||
| SNAI2 | 1.747 | 3.800E-36 | IQCH-AS1 | 1.802 | 0.002140359 | |||
| TAS1R3 | 1.742 | 1.034E-06 | LYPD1 | 1.802 | 3.92E-08 | |||
| SLC12A8 | 1.738 | 4.678E-20 | EFHD1 | 1.799 | 0.005441587 | |||
| ACVR2B-AS1 | 1.734 | 3.285E-04 | SSC5D | 1.797 | 0.000578189 | |||
| CCDC64 | 1.733 | 2.855E-35 | FAM110D | 1.795 | 6.79E-06 | |||
| SLAIN1 | 1.728 | 1.867E-04 | TM4SF19 | 1.791 | 0.012297142 | |||
| SERPINE1 | 1.728 | 5.438E-36 | PKD1L2 | 1.789 | 0.000723345 | |||
| ASIC3 | 1.727 | 5.340E-06 | LOC102723703 | 1.786 | 0.001389492 | |||
| GRAMD2 | 1.725 | 3.863E-06 | IGSF1 | 1.786 | 3.67E-06 | |||
| APLP1 | 1.721 | 1.397E-27 | RYR2 | 1.783 | 0.081712464 | |||
| MAMDC2 | 1.721 | 7.870E-16 | TRHDE-AS1 | 1.783 | 1.25E-05 | |||
| CYP2S1 | 1.720 | 7.774E-24 | LINC01123 | 1.781 | 6.93E-07 | |||
| BNIPL | 1.717 | 4.095E-04 | TMEM198 | 1.778 | 1.84E-08 | |||
| VAV2 | 1.716 | 1.420E-46 | NPM2 | 1.776 | 1.32E-05 | |||
| CEL | 1.715 | 7.378E-05 | C10orf11 | 1.776 | 9.20E-08 | |||
| DOCK2 | 1.711 | 3.371E-06 | ACVR2B-AS1 | 1.774 | 0.003545902 | |||
| IL6R | 1.704 | 7.098E-09 | MIR3615 | 1.774 | 0.006410784 | |||
| F12 | 1.703 | 3.151E-08 | ACTA2 | 1.773 | 5.26E-06 | |||
| SHISA9 | 1.703 | 7.226E-38 | METRNL | 1.773 | 1.34E-09 | |||
| TMEM190 | 1.770 | 0.00868946 | ||||||
| CYP1A1 | 1.767 | 0.000333413 | ||||||
| TMEM45A | 1.761 | 0.001012484 | ||||||
| LOC284837 | 1.761 | 0.01050783 | ||||||
| LINC00662 | 1.759 | 8.47E-06 | ||||||
| TUBB3 | 1.758 | 1.99E-05 | ||||||
| MANEAL | 1.756 | 1.55E-05 | ||||||
| TBC1D30 | 1.755 | 9.03E-09 | ||||||
| LOC100128531 | 1.752 | 0.004519439 | ||||||
| QTRT1 | 1.752 | 2.38E-05 | ||||||
| SPR | 1.749 | 7.14E-06 | ||||||
| SDSL | 1.748 | 6.79E-07 | ||||||
| SNORA65 | 1.748 | 0.007575303 | ||||||
| LTBP3 | 1.744 | 1.31E-06 | ||||||
| ITGBL1 | 1.737 | 4.58E-07 | ||||||
| CPT1B | 1.737 | 3.95E-07 | ||||||
| PCBP3 | 1.736 | 1.11E-05 | ||||||
| F12 | 1.736 | 0.00038732 | ||||||
| KRT13 | 1.732 | 0.00217359 | ||||||
| GFAP | 1.731 | 0.000186248 | ||||||
| FZD4 | 1.729 | 0.004625213 | ||||||
| DIRAS1 | 1.729 | 3.03E-06 | ||||||
| GLA | 1.726 | 1.27E-05 | ||||||
| MYD88 | 1.725 | 6.11E-07 | ||||||
| NDUFA4L2 | 1.721 | 0.010881735 | ||||||
| SPNS2 | 1.717 | 2.19E-05 | ||||||
| KREMEN2 | 1.716 | 5.26E-06 | ||||||
| SH2D3C | 1.715 | 0.008907916 | ||||||
| NXPH3 | 1.710 | 0.000609435 | ||||||
| LOC101928525 | 1.709 | 0.000208399 | ||||||
| DOCK2 | 1.706 | 9.41E-06 | ||||||
| RTN2 | 1.704 | 2.87E-08 | ||||||
| ANO2 | 1.703 | 2.34E-05 | ||||||
| PLXNA2 | 1.702 | 0.000659444 | ||||||
| KIRREL3 | 1.701 | 7.84E-05 | ||||||
| PLLP | 1.701 | 1.08E-07 | ||||||
| SLC25A5-AS1 | 1.700 | 6.78E-06 |
Table 2:
Uniquely induced genes
| ARID3A only | ARID3B only | ARID3A/B only |
|---|---|---|
| TENM1 | SCN2B | SAG |
| SNAP25 | NGFR | TMIGD2 |
| NMCN1 | CDH16 | CHGA |
| CD180 | CSF2RB | FOXS1 |
| LINC00482 | GDNF-AS1 | CALY |
| PSG8 | SBK2 | LINC00885 |
| SLC16A12 | TRIM29 | LHB |
| FLRT3 | GGT5 | ACOX2 |
| ARID5B | CDSN | NAT16 |
| INHBA-AS1 | SBSN | P2RX2 |
| MIR24-1 | ACTL8 | CHADL |
| TNFSF18 | PRCD | IQCH-AS1 |
| CCDC85A | KCNK3 | TM4SF19 |
| AQP1 | MIR3615 | |
| CD79A | CYP1A1 | |
| CCR1 | LOC284837 | |
| MYO7B | SNORA65 | |
| RRAD | ||
| ADAMTS2 | ||
| ESPN | ||
| CDHR5 | ||
| LINC01272 | ||
| SYNPO2L | ||
| RASSF4 |
We selected several genes to validate for regulation by ARID3A and ARID3B. For validation we chose to use three ovarian cancer cell lines: OVCA429, OVCA433, and Kuramochi cells. We chose cell lines that represent a couple of different types of ovarian cancer. OVCA429 and OVCA433 are likely not high-grade serous (HGSOC) as they do not have mutations in TP53. Kuramochi cells have genetic alterations consistent with HGSOC, which is the type of ovarian cancer that is most common (Mitra et al., 2015). For validation purposes we identified ten genes induced by ARID3 that ranged from highly induced to modestly induced. In particular we chose to validate the modestly induced TP53 and MYC as we have already validated those as direct ARID3B targets(Bobbs et al., 2015). Interestingly, on the RNA-seq, there was an additive effect of expressing both ARID3A and ARID3B for about half of the genes (values in red on Table 3). OVCA429, OVCA433, and Kuramochi cells were transduced with ARID3A and ARID3B. Expression was confirmed by RT-qPCR and western blot (Fig. 1 and Supplemental Fig. 1). RT-qPCR was conducted on cells for the ARID3A/B regulated genes in Table 3. For all RT-qPCR gene expression was normalized GAPDH and expressed as a fold change over the endogenous gene of interest in the vector control cells (using the ΔΔCT method). We were able to confirm that in OVCA429 cells ARID3A and ARID3B induced Twist, MYCN, MMP2, GLI2, TIMP3, and WNT5B. ARID3B, but not ARID3A, induced NES (Fig. 1). ARID3A, but not ARID3B, induced MYC (Fig. 1). In OVCA433 cells, ARID3A and ARID3B induced NES, MYCN, MMP2, GLI2, WNT5B, and SNAI2. In contrast to OVCA429 cells, TWIST was only induced by ARID3B. Like the OVCA429 cells, MYC was induced by ARID3A. We did not see an additive effect of ARID3A and ARID3B expression. One reason for this may be the way the cells were selected. For the RNA-seq experiment it took 2 rounds of FACSing to obtain enough cells that were both RFP+ and GFP+. We found that most cells preferred to express either ARID3A-GFP or ARID3B-RFP and they rarely would express both. Therefore, the cells used for validation may have had more of a mixed population than those used for RNA-seq. ARID3A/B induction of genes in Kuramochi cells varied somewhat from the OVCA429 and OVCA433 cells, suggesting that cellular context is important for the regulation of ARID3A/B target genes. MYC and NES were induced by ARID3A, ARID3B, and both proteins in Kuramochi cells (Fig. 1). Intriguingly, TIMP3 and MYCN were only induced by ARID3B. SNAI2 was only induce by ARID3A. Cellular context impacted ARID3A/B regulation of genes.
Table 3:
Genes selected for validation:
| Gene | Fold induction by ARID3A | Fold induction by ARID3B | Fold induction by ARID3A/B | Identified by ChIP-CHIP |
|---|---|---|---|---|
| TIMP3 | 272 | 453 | 511 | Yes |
| GLI2 | 115 | 186 | 380 | No |
| MMP2 | 70 | 74 | 132 | No |
| NES | 32 | 59 | 42 | Yes |
| WNT5B | 21 | 35 | 32 | Yes |
| MYCN | 9.6 | 16.5 | 10.3 | No |
| SNAI2 | 3.4 | 3.4 | 2.44 | No |
| TWIST | 2.6 | 2.5 | 4 | No |
| TP53 | 1.7 | 1.8 | 2.4 | No |
| MYC | 1.39 | 1.6 | 2.2 | Yes |
| ARID3A | 101 | 61 | 105 | No |
| ARID3B | 2.2 | 8.8 | 4.5 | No |
Figure 1:
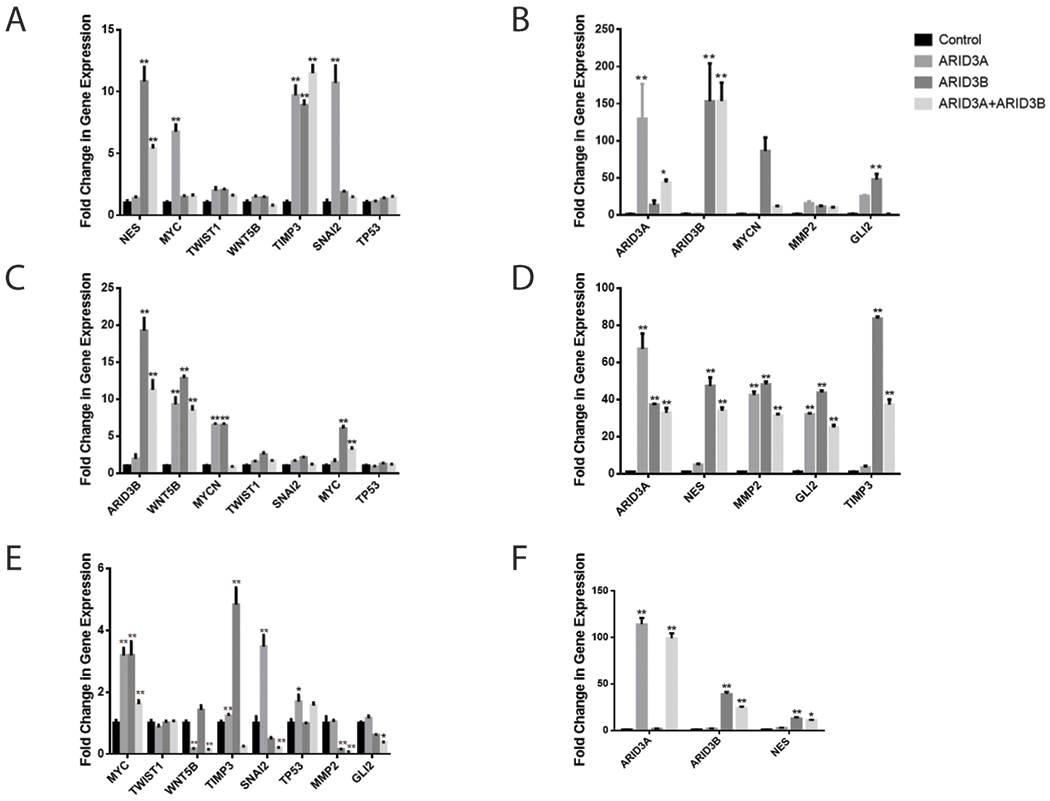
Validation of RNA-seq data via RT-qPCR in three ovarian cancer cell lines. (A and B) Ovarian cancer cell line OVCA429 was lentivirally transduced with GFP, ARID3A, ARID3B or ARID3A + ARID3B. RT-qPCR was performed to verify fold change in ARID3A, ARID3B and ARID3A+ARID3B regulated genes. (C and D) Ovarian cancer cell line OVCA433 was lentivirally transduced with GFP, ARID3A, ARID3B, or ARID3A+ARID3B. RT-qPCR was performed for ARID3 regulated genes. (E and F) The ovarian cancer cell line Kuramochi was lentivirally transduced with GFP, ARID3A, ARID3B or ARID3A+ARID3B. RT-qPCR was performed for ARID3 regulated genes. ΔΔct was used to calculate relative expression over the endogenous expression of each gene of interest (control samples). Expression was normalized to GAPDH. Statistical (Student t test) comparisons are made between control and ARID3A or ARID3B expressing cells. *P < 0.05, **P < 0.001
Next we determined if ARID3A and ARID3B could bind to promoter/enhancer regions of MYCN, TIMP3, NES, and GLI2. First, we identified ARID3A and ARID3B binding sites in the promoters (Fig. 2). Then we performed chromatin immunoprecipitation (ChIP) using protein-chromatin complexes isolated from OVCA429-GFP, OVCA429 ARID3A-GFP, and OVCA429 ARID3B-RFP cells (Fig. 3). We used antibodies for IgG (control IP), ARID3A, and ARID3B to immunoprecipitated protein-DNA complexes. After immunoprecipitating protein complexes, reversing crosslinks, and purifying DNA, qPCR was conducted for GAPDH (control), TIMP3, NES, GLI2, and MYCN promoter regions containing ARID3 consensus binding sites (Bobbs et al., 2015). Statistical (Student t test) comparisons are made between GFP control and ARID3A or ARID3B expressing cells. For MYCN we amplified two different promoter regions. One had a traditional ARID3A binding site and one had a more distinct ARID3B binding site as described (Bobbs et al., 2015). Of note, we previously identified that ARID3B binds to TIMP3, NES, WNT5B, and MYC regulatory regions by ChIP followed by microarray (ChIP-CHIP) (Bobbs et al., 2015) in SKOV3IP cells. We now shown that ARID3A and ARID3B are able to bind ARID3 sites in MYCN, NES, and TIMP3 in OVCA429 cells. We did not detect binding of ARID3A or ARID3B to a binding site in GLI2 by ChIP or ChIP-CHIP(Bobbs et al., 2015). Therefore, GLI2 may be and indirect target of ARID3 regulation or more extensive ChIP studies need to be conducted to identify which binding sites are most relevant to GLI2 gene regulation.
Figure 2:
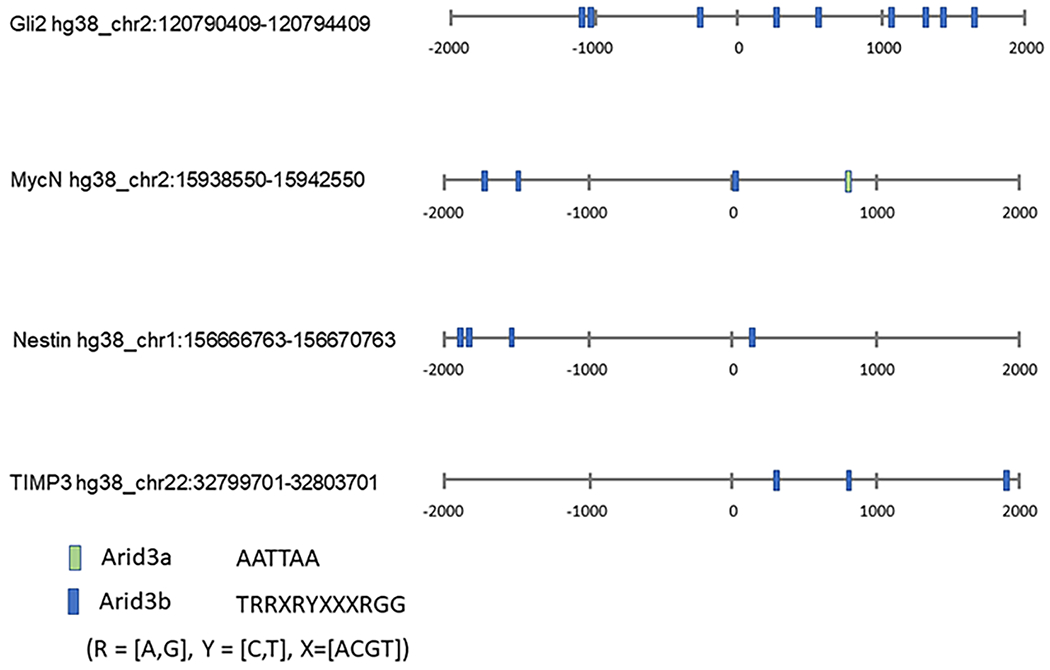
Identification of ARID3A and ARID3B binding sites in ARID3 regulated genes of interest. 2000 base pairs upstream and downstream of the transcription starts sites of the indicated genes were scanned for binding sites for Arid3a and Arid3b using DNA Pattern Find (https://www.bioinformatics.org/sms2/dna_pattern.html). Genomic sequences were downloaded from UCSC Genome Browser.
Figure 3:
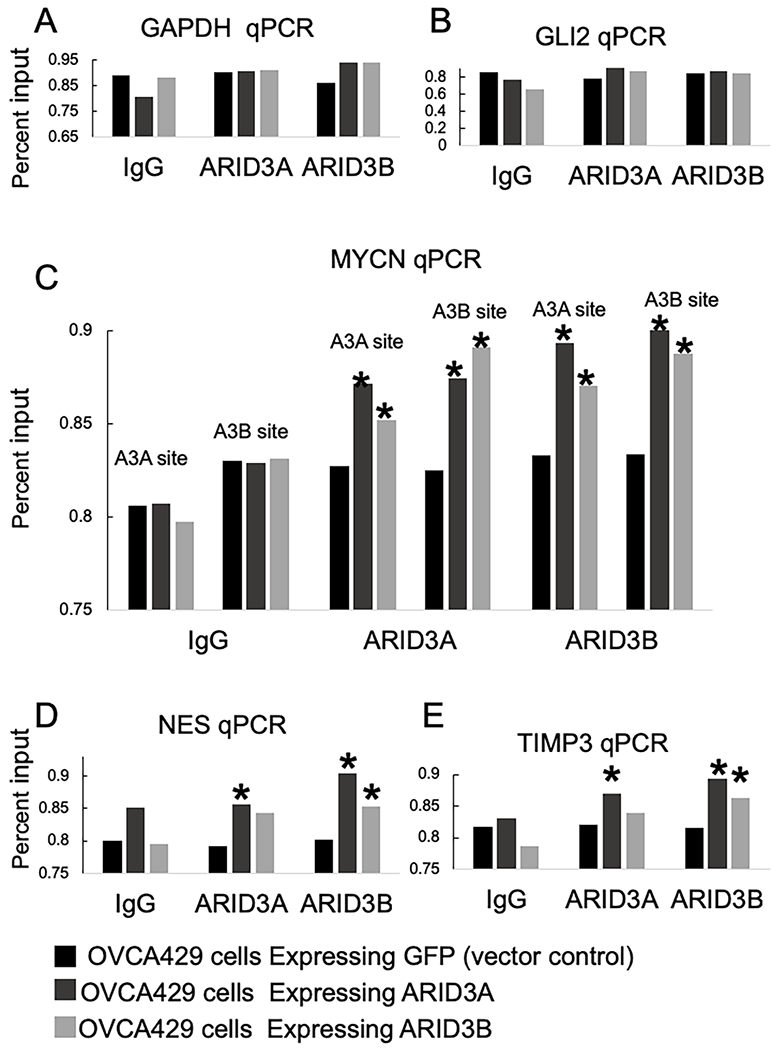
ARID3A and ARID3B bind gene regulatory regions. Chromatin immunoprecipitation (ChIP) followed by qPCR was performed on OVCA429 cells expressing GFP, ARID3A, or ARID3B. CHIP was conducted using antibodies for IgG (negative control), ARID3A, or ARID3B. All values are reported as a percent input of the amplification of input DNA. qPCR is conducted for (A) GAPDH (negative control), (B) GLI2, (C) MYCN (two sets of primers-one for an ARID3A specific site and one for an ARID3B specific site), (D) NES, and (E) TIMP3. * p<0.05 Statistical (Student t test) comparisons are made between GFP control and ARID3A or ARID3B expressing cells.
We performed IPA analysis on the ARID3A/B regulated genes. The top 10 induced pathways are presented in Table 4. The top pathways identified were Axonal Guidance Signaling and Molecular Mechanisms of Cancer. Wnt/β-catenin Signaling and Human Embryonic Stem Cell Pluripotency Pathways were also found to be induced by the ARID3 family.
Table 4:
IPA Pathways
| Top ARID3A/B IPA iduced pathways |
|---|
| Axonal Guidance Signaling |
| Molecular Mechanisms of Cancer |
| EIF2 Signaling |
| Wnt/β-catenin Signaling |
| Sirtuin Signaling Pathway |
| Basal Cell Carcinoma Signaling |
| Cell Cycle: G1/S Checkpoint Regulation |
| Human Embryonic Stem Cell Pluripotency |
| Ephrin B Signaling |
| Integrin Signaling |
Some of the discrepancies between cell lines or between the RNA-seq and the validation studies may be due to the levels of ARID3A and ARID3B expression in cells. As we previously published, it is hard to maintain ARID3B levels in cells as the cells will induce TNF/TRAIL apoptosis. Interestingly, the cells that do survive long enough for us to make RNA and sequence, have reduced TNF/TRAIL pathways genes. TNFSF10, TNFRSF1B, TNFSF11B, TNFAIP2, and TNFAIP3 are all reduced in the cells used for RNA-seq (Table 5).
Table 5:
ARID3A/B represses TNF/TRAIL pathways
| Gene | Fold decrease by ARID3A | Fold decrease by ARID3B | Fold decrease by ARID3A/B |
|---|---|---|---|
| TNFSF10 (TRAIL) | 2.193 | 4.739 | 4.167 |
| TNFRSF1b (TNFR2) | 5.236 | 3.030 | 5.556 |
| TNFRSF11b | 8.065 | 12.987 | 27.778 |
| TNFAIP2 | 5.556 | 5.000 | 4.545 |
| TNFAIP3 | 1.471 | 1.266 | 1.832 |
4. Discussion
Our intent was to discover shared and unique target genes of ARID3A and ARID3B. Surprisingly we found that in ovarian cancer cells very few (13-23) genes were regulated uniquely by either ARID3A or ARID3B. In part this may be due to ARID3A and ARID3B inducing expression of each other. We did find that some genes were more induced when both ARID3A and ARID3B were co-expressed (Table 2). Additionally, in some cell lines ARID3A or ARID3B were better at regulating genes (Fig. 1). Therefore, there may be some selectivity in gene regulation by ARID3A and ARID3B that our study design did not identify.
We thought that ARID3A and ARID3B might have unique target genes because ARID3B induces cell death and ARID3A does not (Joseph et al., 2012; Pratama et al., 2015). Around seventy percent of cells with high ARID3B mRNA undergo TNFα induced cell death within 3 days of transduction with ARID3B lentivirus(Joseph et al., 2012). Moreover, we found that for cells overexpressing ARID3B to survive, the cells decrease expression of apoptosis promoting pathways (Table 5). Additionally, as cells adapt to high levels of ARID3B, more of the ARID3B becomes sequestered in the cytoplasm (Fig. 4A). This suggests that the net ARID3B levels available to regulate gene expression, may not be accurately reflected in RT-qPCR (Fig. 1). Importantly we found that in human ovarian tumors, ARID3B was nuclear when co-expressed with ARID3A (Roy and Cowden Dahl, 2018). Yet ARID3B was more diffuse when ARID3A was not present (Roy and Cowden Dahl, 2018). This suggests that subcellular localization of ARID3B in addition to localization of ARID3A may be important for target gene regulation in vivo.
In this study our goal was to analyze cells with moderately high expression of ARID3A and ARID3B that may reflect levels of ARID3B seen in ovarian cancer. Moderate, but not high levels of ARID3B correlate with increased relapse after chemotherapy in ovarian cancer (Roy et al., 2014). Additionally, we previously published that ARID3B induces cell death when overexpressed at high levels (Joseph et al., 2012). Importantly, ARID3A does promote apoptosis (Pratama et al., 2015). To further demonstrate that the levels of ARID3B are critical to target gene regulation, we transduced OVCA429 cells with ARID3B-GFP and then sorted for high and moderate expression of GFP. After 6 days, the cells selected for high ARID3B expression had a 25% higher expression of ARID3B that the cells selected for moderate expression (Fig. 4A). By day 12 there was no difference in expression of ARID3B (Fig. 4B). Additionally, cells expressing moderate ARID3B had a 6-fold increase in TNF and a 3-fold increase pro-survival TNF receptor TNFRSF1B compared to parental cell lines. While cells with high ARID3B exhibited a 3-fold increase in proapoptotic TNFSF10 and RIPK. This data is relevant because it demonstrates that in order to see differences in gene regulation between ARID3A and ARID3B we will have to examine target gene expression with different doses of ARID3A and ARID3B and likely in different cell types. Since ARID3A does not induce cell death, apoptotic genes could be unique targets of ARID3B, that were not captured in our experimental design (due to having moderate and not high ARID3B expression). Additionally, ARID3A may regulate genes differently in different cell types such as trophoblast cells or B-cells. More studies will need to be conducted to identify differential target gene regulation by context. What this study was able to show is that moderate expression of ARID3A and ARID3B results in regulation of the same cohort of stem cell genes in ovarian cancer cell lines.
5. Conclusion
ARID3A and ARID3B proteins regulate cancer stem cell genes and may contribute to a stem cell phenotype.
Supplementary Material
Supplemental Figure 1: Ovarian Cancer cells engineered to express ARID3A and ARID3B. OVCA433 and OVCA429 cells were lentivirally transduced with GFP, ARID3A, or ARID3B. Western blot demonstrates ARID3A, ARID3B, and Histone H3 protein expression.
Acknowledgements:
The authors would like Dr. Lynn Roy for reviewing statistical analysis. This publication was made possible, in part, with support from the Notre Dame Genomics and Bioinformatics Core Facility through Genomics Services utilizing Bioanalyzer analysis and Illumina Library Preparation. We specifically acknowledge the assistance of Jackie Lopez-Erickson. Fluorescence activated cell sorting (FACS) was conducted at the Indiana University School of Medicine-South Bend at the Imaging and Flow cytometry Core Facility by Dr. Charles Tessier.
Funding: This project was supported by the funds from the Enhanced Mentoring Program with Opportunities for Ways to Excel in Research (EMPOWER) at the Indiana University-Purdue University Indianapolis.
Abbreviations:
- ARID
AT-Rich Interactive Domain
- GFP
green fluorescent protein
- RFP
red fluorescent protein
- RNA-seq
ribonucleic acid sequencing
- RT-qPCR
reverse transcribed quantitative polymerase chain reaction
- FACS
fluorescence activated cell sorting
- ChIP
Chromatin Immunoprecipitation
References
- Bast RC Jr., Feeney M, Lazarus H, Nadler LM, Colvin RB and Knapp RC, 1981. Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest 68, 1331–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobbs A, Gellerman K, Hallas WM, Joseph S, Yang C, Kurkewich J and Cowden Dahl KD, 2015. ARID3B Directly Regulates Ovarian Cancer Promoting Genes. PLoS One 10, e0131961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JC, Uribe V, Badia-Careaga C, Giovinazzo G, Torres M and Sanz-Ezquerro JJ, 2011. Apical ectodermal ridge morphogenesis in limb development is controlled by Arid3b-mediated regulation of cell movements. Development 138, 1195–205. [DOI] [PubMed] [Google Scholar]
- Gregory SL, Kortschak RD, Kalionis B and Saint R, 1996. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol Cell Biol 16, 792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TC, Young RC, McKoy WM, Grotzinger KR, Green JA, Chu EW, Whang-Peng J, Rogan AM, Green WR and Ozols RF, 1983. Characterization of a human ovarian carcinoma cell line (NIH:OVCAR-3) with androgen and estrogen receptors. Cancer Res 43, 5379–89. [PubMed] [Google Scholar]
- Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R and Tucker PW, 1995. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev 9, 3067–82. [DOI] [PubMed] [Google Scholar]
- Joseph S, Deneke VE and Cowden Dahl KD, 2012. ARID3B Induces Tumor Necrosis Factor Alpha Mediated Apoptosis While a Novel ARID3B Splice Form Does Not Induce Cell Death. PLoS One 7, e42159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Probst L, Das C and Tucker PW, 2007a. REKLES is an ARID3-restricted multifunctional domain. J Biol Chem 282, 15768–77. [DOI] [PubMed] [Google Scholar]
- Kim D, Probst L, Das C and Tucker PW, 2007b. REKLES is an ARID3-restricted multifunctional domain. The Journal of biological chemistry 282, 15768–77. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Jakt LM and Nishikawa SI, 2012. Epigenetic regulation of the neuroblastoma genes, Arid3b and Mycn. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkewich JL, Klopfenstein N, Hallas WM, Wood C, Sattler RA, Das C, Tucker H, Dahl R and Cowden Dahl KD, 2016. Arid3b Is Critical for B Lymphocyte Development. PLoS One 11, e0161468. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liao TT, Hsu WH, Ho CH, Hwang WL, Lan HY, Lo T, Chang CC, Tai SK and Yang MH, 2016. let-7 Modulates Chromatin Configuration and Target Gene Repression through Regulation of the ARID3B Complex. Cell Rep 14, 520–33. [DOI] [PubMed] [Google Scholar]
- Mitra AK, Davis DA, Tomar S, Roy L, Gurler H, Xie J, Lantvit DD, Cardenas H, Fang F, Liu Y, Loughran E, Yang J, Sharon Stack M, Emerson RE, Cowden Dahl KD, V.B M., Nephew KP., Matei D. and Burdette JE., 2015. In vivo tumor growth of high-grade serous ovarian cancer cell lines. Gynecol Oncol 138, 372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratama E, Tian X, Lestari W, Iseki S, Ichwan SJ and Ikeda MA, 2015. Critical role of ARID3B in the expression of pro-apoptotic p53-target genes and apoptosis. Biochem Biophys Res Commun 468, 248–54. [DOI] [PubMed] [Google Scholar]
- Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, Suh BB, Hinrichs AS, Clawson H, Zweig AS, Kirkup V, Fujita PA, Rhead B, Smith KE, Pohl A, Kuhn RM, Karolchik D, Haussler D and Kent WJ, 2011. ENCODE whole-genome data in the UCSC genome browser (2011 update). Nucleic Acids Res 39, D871–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratliff ML, Templeton TD, Ward JM and Webb CF, 2014. The Bright Side of Hematopoiesis: Regulatory Roles of ARID3a/Bright in Human and Mouse Hematopoiesis. Front Immunol 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee C, Edwards M, Dang C, Harris J, Brown M, Kim J and Tucker HO, 2017. ARID3A is required for mammalian placenta development. Dev Biol 422, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee C, Lee BK, Beck S, Anjum A, Cook KR, Popowski M, Tucker HO and Kim J, 2014. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genes Dev 28, 2219–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy L, Bobbs A, Sattler R, Kurkewich JL, Dausinas PB, Nallathamby P and Cowden Dahl KD, 2018. CD133 Promotes Adhesion to the Ovarian Cancer Metastatic Niche. Cancer Growth Metastasis 11, 1179064418767882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy L and Cowden Dahl KD, 2018. Can Stemness and Chemoresistance Be Therapeutically Targeted via Signaling Pathways in Ovarian Cancer? Cancers (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy L, Samyesudhas SJ, Carrasco M, Li J, Joseph S, Dahl R and Cowden Dahl KD, 2014. ARID3B increases ovarian tumor burden and is associated with a cancer stem cell gene signature. Oncotarget 5, 8355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samyesudhas SJ, Roy L and Cowden Dahl KD, 2014. Differential expression of ARID3B in normal adult tissue and carcinomas. Gene 543, 174–80. [DOI] [PubMed] [Google Scholar]
- Takebe A, Era T, Okada M, Martin Jakt L, Kuroda Y and Nishikawa S, 2006. Microarray analysis of PDGFR alpha+ populations in ES cell differentiation culture identifies genes involved in differentiation of mesoderm and mesenchyme including ARID3b that is essential for development of embryonic mesenchymal cells. Dev Biol 293, 25–37. [DOI] [PubMed] [Google Scholar]
- Tidwell JA, Schmidt C, Heaton P, Wilson V and Tucker PW, 2011. Characterization of a new ARID family transcription factor (Brightlike/ARID3C) that co-activates Bright/ARID3A-mediated immunoglobulin gene transcription. Mol Immunol 49, 260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW and Orkin SH, 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–8. [DOI] [PubMed] [Google Scholar]
- Webb CF, Bryant J, Popowski M, Allred L, Kim D, Harriss J, Schmidt C, Miner CA, Rose K, Cheng HL, Griffin C and Tucker PW, 2011. The ARID family transcription factor bright is required for both hematopoietic stem cell and B lineage development. Mol Cell Biol 31, 1041–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsker D, Probst L, Wain HM, Maltais L, Tucker PW and Moran E, 2005. Nomenclature of the ARID family of DNA-binding proteins. Genomics 86, 242–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Ovarian Cancer cells engineered to express ARID3A and ARID3B. OVCA433 and OVCA429 cells were lentivirally transduced with GFP, ARID3A, or ARID3B. Western blot demonstrates ARID3A, ARID3B, and Histone H3 protein expression.


