Abstract
Catecholamines stimulate the mobilization of stored triglycerides in adipocytes to provide fatty acids (FAs) for other tissues. However, a large proportion is taken back up and either oxidized or re-esterified. What controls the disposition of these FAs in adipocytes remains unknown. Here we report that catecholamines redirect FAs for oxidation through the phosphorylation of signal transducer and activator of transcription 3 (STAT3). Adipocyte STAT3 is phosphorylated upon activation of β-adrenergic receptors, and in turn suppresses FA re-esterification to promote FA oxidation. Adipocyte-specific Stat3 KO mice exhibit normal rates of lipolysis, but exhibit defective lipolysis-driven oxidative metabolism, resulting in reduced energy expenditure and increased adiposity on high fat diet. This previously unappreciated, non-genomic role of STAT3 explains how sympathetic activation can increase both lipolysis and fatty acid oxidation in adipocytes, revealing a new regulatory axis in metabolism.
Introduction
The worldwide obesity epidemic1–3 has focused much attention on the pathways that regulate energy homeostasis, particularly in adipocytes, the primary site of energy storage and mobilization. Nutrients are transported into adipocytes and stored as triglycerides (TGs) in lipid droplets. Glucose is converted into fatty acids (FAs) by de novo lipogenesis; adipocytes also take up FAs directly from the circulation. In both cases, FAs are esterified onto a glycerol backbone via the glycerol phosphate pathway. The first-committed and rate-limiting step is carried out by the glycerol-3-phosphate acyltransferase (GPAT) family of enzymes, which catalyze the conversion of glycerol-3-phosphate and long-chain fatty acyl-coenzyme A (FA-CoA) to lysophosphatidic acid4–7. There are 4 GPAT proteins; GPAT1 and GPAT2 are localized in mitochondria, while GPAT3 and GPAT4 are microsomal8–18. GPAT3 is highly induced during adipogenesis, and is the dominant isoform in white adipocytes, whereas GPAT4 is predominant in brown adipocytes11,19. Because FA-CoA can undergo either re-esterification or oxidation, GPAT activity is critical for determining the fate of FAs. In the liver, GPAT1 ensures storage of de novo synthesized FAs20, while in brown adipose tissue (BAT), GPAT4 limits basal FA oxidation, thereby reducing energy expenditure and promoting obesity21. In white adipocytes, GPAT3 is the critical rate-limiting enzyme for TG synthesis11,19.
The dynamic regulation of TG synthesis and breakdown in adipocytes is required for metabolic health. Failure to store triglycerides in adipose tissue causes lipodystrophies, associated with severe metabolic complications22–24. Parenthetically, excessive energy storage in adipocytes leads to obesity and ensuing metabolic disease25,26. Nutrient uptake and TG synthesis are increased by insulin, while TG breakdown is stimulated by epinephrine and norepinephrine-induced lipolysis, resulting in the release of FA and glycerol27–36. During lipolysis, adipocytes take back up a large fraction of the released FAs37–47. While under basal conditions, white adipocytes esterify and store the vast majority of FAs, and oxidation rates are very low48, lipolytic stimulation dramatically increases FA oxidation at the expense of re-esterification49. This lipolysis-driven oxidative metabolism represents a significant avenue for energy expenditure in white adipocytes that is not dependent on beiging or thermogenesis. Furthermore, there is evidence that lipolysis-driven oxidative metabolism, but not lipolysis, is defective in adipocytes from obese humans49.
We demonstrate here that signal transducer and activator of transcription 3 (STAT3) is a crucial effector of lipolysis-driven oxidative metabolism in adipocytes. STAT3 is phosphorylated in response to β-adrenergic activation in a lipolysis-dependent manner. STAT3 redirects FAs towards oxidation by suppressing FA re-esterification. Specific deletion of the Stat3 gene in adipocytes dramatically reduced lipolysis-driven oxidative metabolism, prevented dynamic remodeling of lipid droplets, increased lipid storage, and resulted in increased adiposity in mice on a high fat diet. Interestingly, gene expression changes produced by Stat3 deletion could not explain these effects. These data demonstrate that STAT3 is a crucial regulator of adipocyte lipid metabolism through a non-genomic mechanism that ensures proper energy balance.
Results
Stat3 is phosphorylated in response to β-adrenergic receptor activation in adipocytes
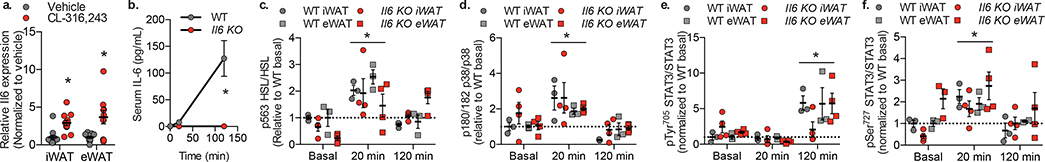
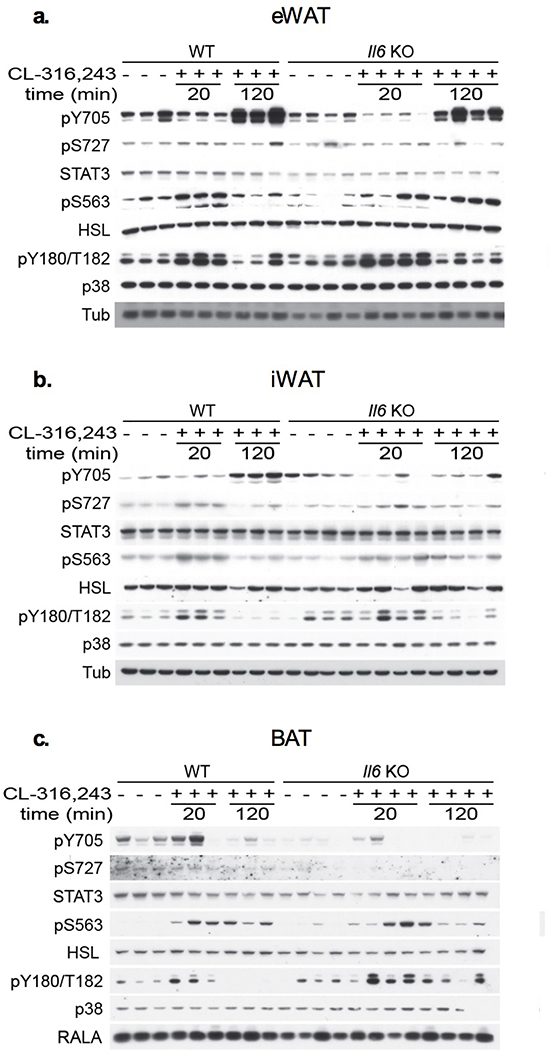
Epinephrine and norepinephrine are released from sympathetic nerve terminals in adipose tissue50–53, activating β-adrenergic receptors to stimulate the production of cAMP31–34. Subsequent activation of PKA results in the phosphorylation and activation of hormone-sensitive lipase (HSL) and perilipin 1, resulting in increased lipolysis54–59. Despite the extensive characterization of this pathway60,61, the molecular events downstream of β-adrenergic receptor activation in adipocytes remain incompletely understood. One example is the secretion of the cytokine IL-6 from adipocytes62–64, which regulates metabolic gene expression in liver65 and muscle66–68, and controls macrophage polarization to influence adipocyte metabolism69,70, but also may be associated with maladaptive responses to obesity71–74. Several studies have suggested that adipocyte-derived IL-6 may exert autocrine or paracrine effects in adipose tissue75–77. As this cytokine is known to activate STAT3 upon binding to its receptor via activation of JAKs, we sought to clarify the impact of IL-6 on adipocyte function by investigating STAT3 phosphorylation in adipocytes from Il6 KO and WT littermate mice treated with the β−3 specific adrenergic receptor agonist, CL-316,243 (Fig. 1). The β−3 adrenergic receptor is expressed at dramatically higher levels than other β-adrenergic receptors in mature mouse adipocytes, and almost exclusively expressed in these cells in mice. Thus, this drug primarily affects adipose tissue, without confounding effects on other tissues such as heart or liver78–80. CL-316,243 treatment resulted in a robust increase in Il6 mRNA levels in both inguinal and epididymal WAT within 30 minutes, as well as a dramatic increase in IL-6 secretion 2 hours after treatment in WT animals (Fig. 1a,b). As expected, serum IL-6 protein was undetectable in Il6 KO mice. Injection of the agonist increased the phosphorylation of HSL on the PKA-responsive site57, Ser563, in both inguinal and epididymal adipose tissue derived from both Il6 KO and WT mice (Fig. 1c). Phosphorylation of p38 in response to CL-316,243 was also seen in both genotypes (Fig. 1d). Thus, catecholamine signaling appeared not to be affected in adipocytes of Il6 KO animals.
Fig. 1: Stat3 is phosphorylated in response to β-adrenergic receptor activation in adipocytes.
a. Il6 expression in 12-week high fat diet (HFD) fed C57BL/6 mice treated with 1 mg/kg CL-316,243 or vehicle control for 30 minutes before sacrifice and tissue collection. Expression levels normalized to vehicle within each tissue. Individual data points plotted ± SEM (n = 12 mice per treatment group). p value = 0.0003 (iWAT) and 0.006 (eWAT) b. Serum IL-6 levels in 12-week HFD fed C57BL/6 mice treated with 1 mg/kg CL-316,243 or vehicle control before sacrifice and blood collection. Data are represented as mean ± SEM (n = 3 mice per genotype at each time point). p value < 0.0001 WT vs Il6 KO. Quantification of phosphorylation of c. HSL at serine 563 over total HSL (p value < 0.0001 20 min vs. basal) d. p38 at threonine 180 and tyrosine 182 over total p38 (p value = 0.003 20 min vs. basal) e. STAT3 at tyrosine 705 over total STAT3 (p value < 0.0001 120 min vs. basal) and f. STAT3 at serine 727 over total STAT3 (p value = 0.003 20 min vs. basal). c.-f. Western blots shown in Extended Data Fig. 1. Individual data points plotted ± SEM (n = 3 WT, 4 SAKO mice per time point). * p value < 0.05 from post hoc analysis after significant two-way ANOVA.
STAT3 activity is regulated by phosphorylation at two sites: phosphorylation of Tyr705 promotes STAT3 dimerization and transcriptional regulation81,82, while Ser727 phosphorylation is required for mitochondrial localization83–85, and also enhances transcription factor activity86,87. IL-6 is known to increase Tyr705 STAT3 phosphorylation by activation of the JAK family of tyrosine kinases81,88. Treatment of WT mice with CL-316,243 produced STAT3 Tyr705 phosphorylation after 2 hours (Fig. 1e). This phosphorylation event was blunted in the inguinal white adipose tissue (iWAT) of Il6 KO mice but occurred normally in the epididymal WAT (eWAT) of the same animals (Fig. 1e). On the other hand, Ser727 STAT3 phosphorylation was detected in both iWAT and eWAT within 20 minutes of CL-316,243 treatment, preceding the appearance of IL-6 in the serum. Moreover, this effect was observed in both WT and Il6 KO mice (Fig. 1f), indicating that this action is not dependent on the autocrine action of adipocyte-secreted IL-6. Interestingly, the regulation of STAT3 in response to catecholamine signaling appears to be white fat-specific, as neither STAT3 Tyr705 nor Ser727 phosphorylation was observed in BAT (Extended Data Fig. 1).
Changes in net STAT3 phosphorylation on either serine or tyrosine residues in whole cell lysates from 3T3-L1 adipocytes were not detectable after CL-316,243 treatment. However, subcellular fractionation (Extended Data Fig. 2) revealed Tyr705 phosphorylation in the membrane, cytosolic and nuclear fractions 30 to 60 minutes after CL-316,243 (Fig. 2a). While present in the mitochondrial fraction, STAT3 Tyr705 phosphorylation was undetectable, while Ser727 phosphorylation was detected but not changed with CL-316,243 treatment. However, increased Ser727 phosphorylation was observed specifically in the lipid droplet fraction (Fig. 2a). We also fractionated adipose tissue from mice treated with vehicle or CL-316,243 for 20 min or 2 hrs. Tyr705 phosphorylated STAT3 was localized to the nucleus and appeared later than Ser727 phosphorylated STAT3, which was found in the lipid droplet and cytosolic fractions (Fig. 2b).
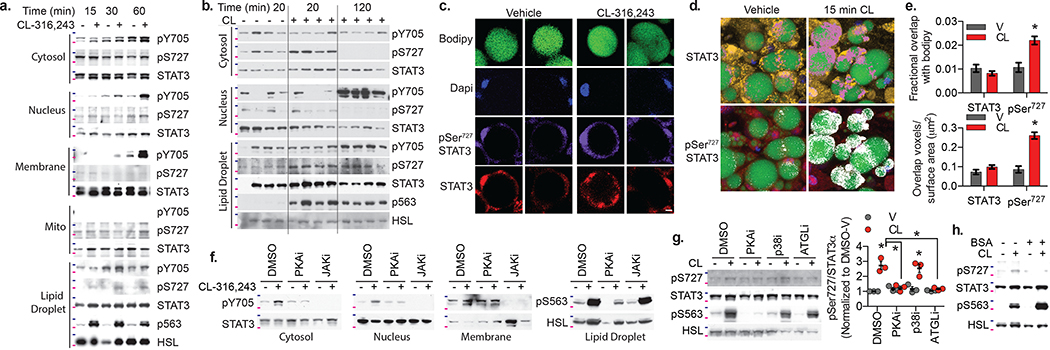
Fig. 2: Localization and phosphorylation of Stat3 in catecholamine stimulated adipocytes.
a., f.-h. Western blot analysis of STAT3 phosphorylation in fractionated 3T3-L1 adipocytes treated with 10 μM CL-316,243; HSL phosphorylation in the lipid droplet fraction also shown to demonstrate effectiveness of CL-316,243 treatment. b. Western blot analysis of STAT3 phosphorylation in fractionated adipose tissue collected after intraperitoneal injection of 1 mg/kg CL-316,243 (20 or 120 min) or vehicle control (20 min after vehicle injection). c. Confocal images of PPDIVs stained with Bodipy, DAPI, STAT3 (Alex fluor 555 conjugated secondary) and pSer727 STAT3 (Alex fluor 647 conjugated secondary). Individual channels of two representative vehicle and CL treated cells shown. Scale bar = 10 μm. d. 3-D representation of z-stack images of PPDIVs stained with bodipy (green), Dapi (blue) STAT3 (yellow) or pSer727 STAT3 (red) are shown with colocalization of Bodipy and STAT3 shown in pink, and colocalization of Bodipy pSer727 STAT3 shown in white. e. Quantification of colocalization of STAT3 and pSer727 STAT3 with bodipy, normalized to lipid droplet volume (top) or lipid droplet surface area (bottom). Data are represented as mean ± SEM (n = 61 V and 64 CL lipids droplets) p value < 0.0001 WT versus SAKO pSer727. f. Cells pretreated with H89 (PKA inhibitor) or JAK inhibitor I for 30 min, before Cl-316,243 treatment for 60 min. g. Cells pretreated with H89, SB-303,580 or atglistatin for 30 min, before Cl-316,243 treatment for 15 min. Right panel: Quantification of Ser727 STAT3 phosphorylation in three independent experiments, data are represented as mean ± SEM. p value < 0.0001 DMSO v vs CL, CL DMSO vs. PKAi/ATGLi and = 0,003 p38i V vs CL. h. Cl-316,243 treatment performed in the presence or absence of 2% BSA in the media. Blots are representative of results from three independent experiments, 75 kDa and 100 kDa protein marker locations indicated in pink and blue respectively. Results in c-e were replicated in an independent experiment. * p value < 0.05 from post hoc analysis after significant two-way ANOVA.
While the localization of Tyr705 phosphorylated STAT3 to the membrane and nucleus is consistent with classical JAK/STAT3 signaling, Ser727 phosphorylation of STAT3 at the lipid droplet has not been previously described. To further characterize the localization of Ser727 phosphorylated STAT3, we performed immunofluorescence confocal microscopy in differentiated 3T3-L1 adipocytes treated with CL-316,243 prior to fixation and staining. Consistent with our fractionation data, total STAT3 and Ser727 phosphorylated STAT3 were detected in the cytosol and nucleus, but unchanged by CL-316,243 treatment (Fig. 2c). Evaluation of the overlap of STAT3 or Ser727 phosphorylated STAT3 with bodipy, which stains the lipid droplet, revealed a significant increase in Ser727 phosphorylated STAT3 overlap with bodipy, while colocalization of total STAT3 with bodipy was unchanged (Fig. 2d,e), suggesting that the pool of STAT3 phosphorylated in response to catecholamines is adjacent to the lipid droplet in adipocytes.
We next investigated the kinases responsible for phosphorylating STAT3 in response to β-adrenergic stimulation. Both basal and stimulated STAT3 Tyr705 phosphorylation were completely blocked by pretreatment with a pan-JAK inhibitor89,90, while inhibition of PKA with H8991 blocked only the stimulation of STAT3 Tyr705 phosphorylation by CL-316,243 (Fig. 2f), consistent with canonical JAK-mediated phosphorylation of STAT3 at Tyr705 activated downstream of PKA.
Inhibition of PKA with H89 also prevented Ser727 phosphorylation of STAT3 in response to CL-316,243 (Fig. 2g). Although PKA has previously been implicated as an upstream STAT3 kinase92, the Ser727 site does not match the PKA consensus sequence93. Moreover, we were not able to produce STAT3 phosphorylation in vitro after incubation with purified active PKA, suggesting that the effects of PKA on STAT3 are indirect. The protein kinases that phosphorylate STAT3 Ser727 have been the subject of much investigation92,94–101. Although p38 has been reported to phosphorylate STAT3 at Ser727 downstream of PKA in adipocytes100–103, p38 inhibition with SB-203,58065,104,105 did not block Ser727 phosphorylation of STAT3 by CL-316,243 (Fig. 2g).
We wondered whether STAT3 Ser727 phosphorylation might result from the stimulation of lipolysis. Lipolysis in adipocytes is catalyzed by HSL and Adipocyte Triglyceride Lipase (ATGL)31,52. The ATGL inhibitor completely prevented Ser727 phosphorylation of STAT3 by CL-316,243 (Fig. 2g), while HSL phosphorylation was unaffected, strongly suggesting that lipolysis and not PKA activity per se is critical for STAT3 phosphorylation. Addition of bovine serum albumin (BSA) to the media binds to and sequesters FAs upon their release, preventing reuptake during lipolysis106. Ser727 phosphorylation of STAT3 by CL-316,243 was attenuated by addition of BSA (Fig. 2h), indicating that FA reuptake is a critical component of the lipolysis-driven signaling event, and that the serine kinase responsible for STAT3 Ser727 phosphorylation may be activated by fatty acids or their metabolites.
Adipocyte STAT3 protects against diet-induced obesity
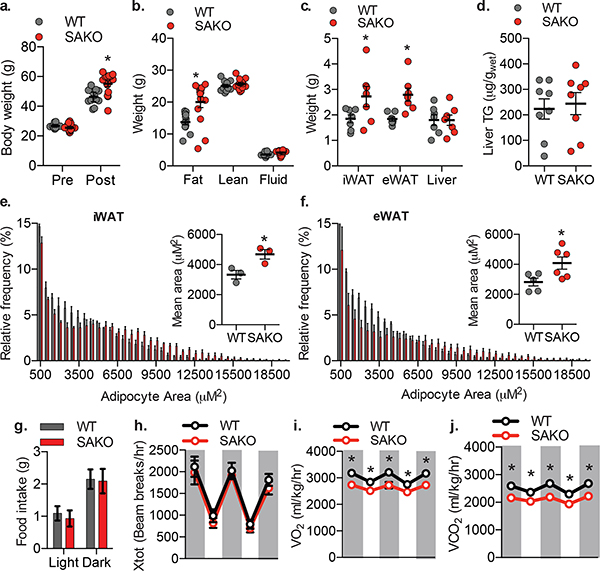
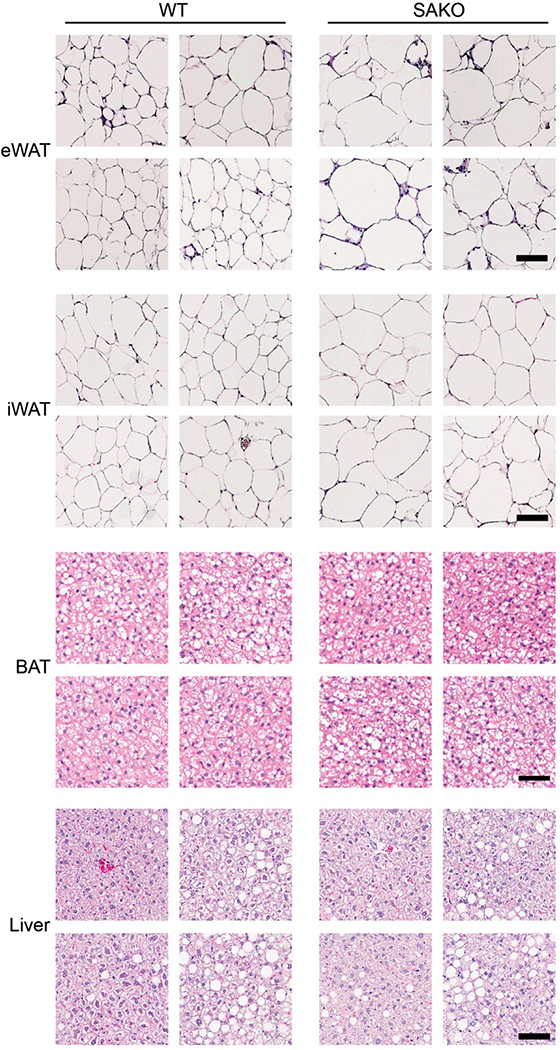
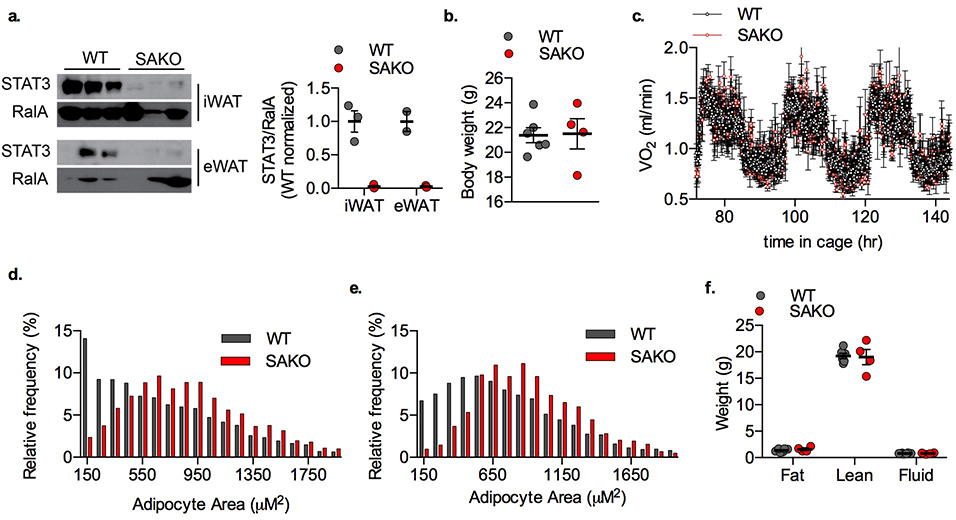
While it has been suggested that STAT3 is involved in adipogenesis88,107–109, its role in mature adipocyte metabolism is largely unknown. We created adipocyte-specific Stat3 knockout (SAKO) mice by crossing floxed Stat3 mice with Adipoq-Cre mice110,111. STAT3 protein levels in mature adipocytes were reduced by more than 95 percent in the SAKO mice (Extended Data Fig. 3a). While an Ap2-Cre-driven Stat3 KO mouse was reported to have slightly increased adiposity on a normal diet112, interpretation is confounded by Stat3 deletion in Ap2-expressing immune and other cells. Body weight and energy expenditure of the SAKO mice were equivalent to wild type floxed controls on normal diet (Extended Data Fig. 3b,c). However, both inguinal and epididymal adipocytes in SAKO mice were larger than those observed in the wildtype controls (Extended Data Fig. 3d–f). When challenged with a high fat diet (HFD), SAKO mice gained significantly more weight than the floxed littermate controls (Fig. 3a), due to increased adiposity (Fig. 3b); there was no difference in lean body mass (Fig. 3b). SAKO mice exhibited increased weight of both the iWAT and eWAT depots (Fig. 3c), while liver weight and TG content remained unchanged (Fig. 3c,d). Histological analysis of eWAT and iWAT revealed significantly larger adipocytes in the SAKO mice (Fig. 3e,f and Extended Data Fig. 4), while BAT and liver histology were identical (Extended Data Fig. 4). No differences in activity or food intake were observed between the genotypes (Fig. 3g,h). However, oxygen consumption and carbon dioxide production were reduced in SAKO relative to control mice, attributing weight gain to a reduction in energy expenditure (Fig. 3i,j).
Fig. 3: Adipocyte Stat3 protects against diet-induced obesity.
a. Body weight of WT and SAKO mice before and after 12-weeks HFD. Individual data points plotted ± SEM (n = 12 WT and 14 SAKO mice). p value < 0.0001 WT vs. SAKO post HFD. b. Body composition by NMR after 12-weeks HFD. Individual data points plotted ± SEM (n = 12 WT and 14 SAKO mice). p value < 0.0001 WT vs. SAKO body fat. c. Tissue weights after 12-weeks HFD. Individual data points plotted ± SEM (n = 7 per genotype). d. Quantification of liver triglycerides, normalized to wet liver weight. Individual data points plotted ± SEM (n = 8 per genotype). p value = 0.02 WT vs. SAKO iWAT and eWAT. e. Histogram showing adipocyte size in iWAT 12-weeks HFD. Inset: bar graph showing mean adipocyte size. Individual data points plotted ± SEM (n = 3 per genotype) p value = 0.033. f. Histogram showing adipocyte size in eWAT 12-week HFD feeding. Inset: bar graph showing mean adipocyte size. Individual data points plotted ± SEM (n = 5 per genotype) p value = 0.033. g.-j. Metabolic cage experiment in WT and SAKO mice after 12-weeks HFD, dark cycle indicated by grey shading. Data are represented as mean ± SEM (n = 6 per genotype). g. Food intake. h. Ambulatory activity (x-axis). i. Oxygen consumption, p values = 0.012, 0.048, 0.006, 0.046, 0.012. j. Carbon dioxide production p values = 0.001, 0.005, 0.0003, 0.005, 0.0008. * p value < 0.05 from post hoc analysis after significant two-way ANOVA (a-c, i, j) or two sided student’s t-test (e and f).
Loss of adipocyte STAT3 causes a defect in oxidative metabolism
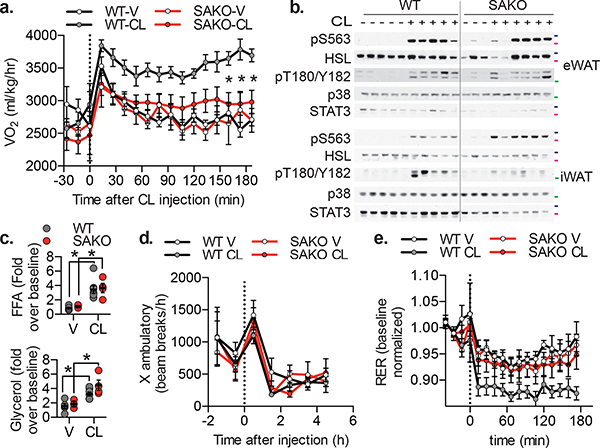
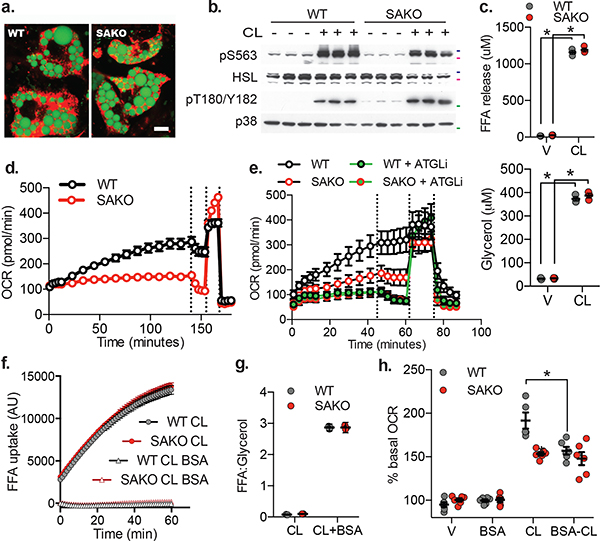
Stimulation of β−3 adrenergic receptors in mice increases energy expenditure113–115 (Extended Data Fig. 5a). While injection with CL-316,243 increased oxygen consumption in WT mice, this response was markedly blunted in SAKO mice (Fig. 4a). This defect was not the result of generalized catecholamine resistance, as there were no differences in the phosphorylation of HSL or p38 (Fig. 4b), nor in the stimulation of lipolysis between the genotypes (Fig. 4c). Additionally, neither CL-316,243 treatment nor genotype impacted physical activity after injection (Fig. 4d). The increase in oxidative metabolism following CL-316,243 treatment was associated with reduced respiratory exchange ratio (RER), presumably due to the increased utilization of FA acids released by lipolysis (Extended Data Fig. 5b). Although loss of STAT3 did not impact lipolytic rates (Fig. 4c), SAKO mice did not exhibit reduced RER after treatment with CL-316,243, as observed in WT animals (Fig. 4e). Thus, adipocyte STAT3 appears to be required for the utilization, but not generation of FAs during lipolysis.
Fig. 4: Loss of adipocyte STAT3 causes a defect in oxidative metabolism in vivo.
a. Oxygen consumption in 12 week HFD-fed mice, injected with 1 mg/kg CL-316,243 or vehicle control. p values = 0.002, 0.0003, 0.002. b. Western blot analysis of 12-week HFD fed mice injected with 1 mg/kg CL-316,243 or vehicle control 20 min before sacrifice and tissue collection. 50 kDa, 75 kDa and 100 kDa protein marker locations indicated in green, pink and blue respectively, blots are representative of three independent experiments. c. Serum free fatty acid (top panel – p value = 0.006 WT and 0.028 SAKO V vs CL) and glycerol (bottom panel – p value = 0.027 WT and 0.010 SAKO V vs CL) levels in 12-week HFD fed mice injected with 1 mg/kg CL-316,243 or vehicle control for 20 min, normalized to the value in each animal prior to injection. Individual data points plotted ± SEM (n = 8 vehicle, 10 CL-316,243). d. Ambulatory activity and e. RER in 12 week HFD-fed mice, injected with 1 mg/kg CL-316,243 or vehicle control at time zero. p value < 0.0001 WT V vs. CL, and WT vs. SAKO CL. a., d. and e. Data are represented as mean ± SEM (n = 20). * p value < 0.05 from post hoc analysis after significant two-way ANOVA.
To explore the cell autonomous effect of STAT3, we isolated primary preadipocytes from SAKO and floxed control inguinal white adipose tissue and differentiated them in vitro. Preadipocytes derived from mice with both genotypes differentiated with high efficiency (Fig. 5a). As observed in vivo, the stimulation of lipolysis and phosphorylation by CL-316,243 was equivalent in WT and SAKO primary preadipocytes differentiated in vitro (PPDIVs) (Fig. 5b,c). While WT PPDIVs exhibited a time-dependent increase in oxygen consumption rate (OCR), SAKO PPDIV OCR remained low (Fig. 5d). The increase in OCR in the WT PPDIVs was completely blocked by inhibition of the lipolytic enzyme ATGL, which also blocked the small increase in OCR observed in SAKO PPDIVs (Fig. 5e), indicating that under these conditions, oxidative metabolism in adipocytes is driven by lipolysis, but also requires STAT3.
Fig. 5: Loss of adipocyte STAT3 causes a cell autonomous defect in lipolysis driven oxidative metabolism.
a. Differentiated WT and SAKO PPDIVs, results are representative of dozens of independent experiments. Lipid droplet stained with Bodipy (shown in green). Mitochondria stained with MitoTracker (shown in red). Scale bar = 10 μm. b. Western blot analysis of WT and SAKO PPDIVs treated with 1 μM CL-316,243 for 20 min, results are representative of three independent experiments. 50 kDa, 75 kDa and 100 kDa protein marker locations indicated in green, pink and blue respectively. c. FA (top panel) and glycerol (bottom panel) secreted into the media from WT and SAKO PPDIV treated with 1 μM CL-316,243 or vehicle control for 20 min. Individual data points plotted ± SEM (n = 3 per treatment per genotype, p values < 0.0001). d., e., and h. Basal oxygen consumption rate in PPDIVs. Vertical lines indicate injection times for oligomycin, FCCP and Rotenone/Antimycin A. d. Data are represented as mean ± SEM (n = 8 wells per condition). e. Atglistatin pretreated for 15 min. e. Data are represented as mean ± SEM (n = 6 wells per condition). f. FA uptake in WT and SAKO PPDIV in the presence and absence of 2% BSA. Data are represented as mean ± SEM (n = 8 wells per condition). g. Ratio of FA to glycerol released into the media from WT and SAKO PPDIV in the presence and absence of 2% BSA. Data are represented as mean ± SEM h. Assay performed in the presence and absence of 0.2% BSA in the culture media. Oxygen consumption rates after 30 min vehicle or CL-316,243 shown as a percentage of the basal OCR prior to treatment is shown. Individual data points plotted ± SEM. (n = 6 wells per condition, p value = 0.0008). * p value < 0.05 from post hoc analysis after significant two-way ANOVA.
Once released from adipocytes by lipolysis, a significant percentage of FAs are taken back up by adipocytes37–47. FA uptake was identical in control and SAKO PPDIVs (Fig. 5f), indicating that STAT3 promotes oxidation of fatty acids after reuptake. In the presence of BSA, FA reuptake is negligible, and the ratio of released FA to glycerol during β-adrenergic-stimulated lipolysis is 3:1 (Fig. 5f,g). In the absence of BSA, this ratio approaches 0:1, as nearly all FAs are taken back up (Fig. 5g). As has been previously reported in 3T3-L1 adipocytes49, CL-316,243-stimulated oxygen consumption rate (OCR) was significantly attenuated in the presence of BSA in WT PPDIVs (Fig. 5h). CL-316,243-stimulated OCR in SAKO PPDIVs was lower than in WT cells and equal to the level observed in WT PPDIVs in the presence of BSA. Moreover, BSA did not further reduce OCR in SAKO PPDIVs (Fig. 5h). These data indicate that STAT3 is required for oxidative metabolism driven by fatty acid reuptake in lipolytic adipocytes.
The effect of STAT3 on adipocyte oxidative metabolism is non-genomic and non-mitochondrial
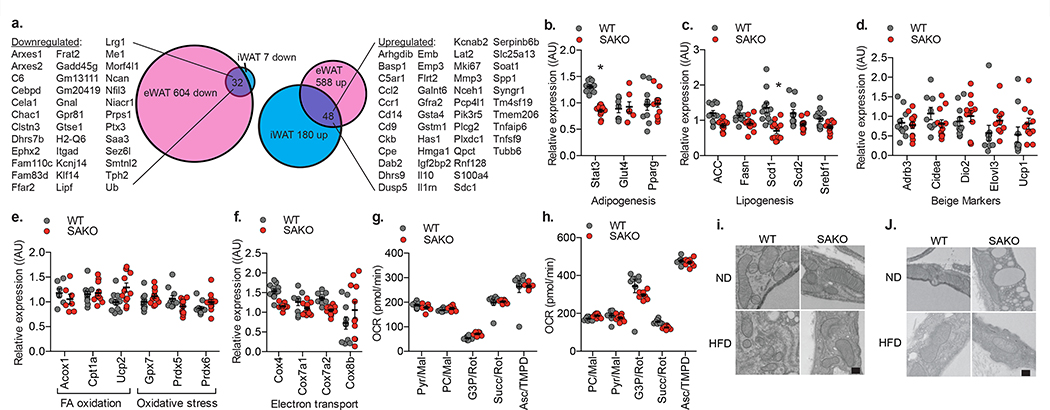
Since STAT3 is well known to function as a transcription factor, we performed RNA sequencing analysis in primary mature adipocytes isolated independently from the iWAT and eWAT of SAKO and floxed littermates. There were 143 and 228 significantly upregulated genes respectively in the epididymal and inguinal primary adipocytes from the SAKO mice compared to the floxed controls; 48 of these were upregulated in both depots. Additionally, there were 636 and 39 significantly downregulated genes respectively in the epididymal and inguinal primary adipocytes from the SAKO mice compared to the floxed controls; 32 were downregulated in both depots (Fig. 6a). There was an increase in inflammatory gene expression in the SAKO adipocytes, but this is likely secondary to the increase in adipocyte size and adiposity, and not STAT3 transcriptional regulation, as Stat3 was not deleted in immune cells. Although STAT3 has been shown to promote oxidative metabolism in other cell types116–119, we did not observe downregulation of genes involved in oxidative metabolism in SAKO adipocytes. Furthermore, directed Q-PCR analysis of numerous genes involved in oxidative phosphorylation, uncoupled respiration, β-oxidation, and lipogenesis did not reveal any changes in gene expression that would explain the profound defect in oxidative metabolism and increased adiposity observed in SAKO mice (Fig. 6b–f).
Fig. 6: The effect of STAT3 on adipocyte oxidative metabolism is non-genomic and non-mitochondrial.
a. Venn diagram showing number of differentially regulated genes in inguinal and epididymal mature adipocytes from WT and SAKO mice after 12-weeks HFD feeding (n = 3 mice per genotype). b.-f. Q-PCR analysis of gene expression in iWAT from WT and SAKO mice after 12-weeks HFD feeding. Individual data points plotted ± SEM (n = 12 mice per genotype, p value < 0.0001). g. Phosphorylating (State 3) respiration rates of oxygen consumption in mitochondria isolated from iWAT in the presence of different substrates. Individual data points plotted ± SEM (n = 8 wells per group). h. State 3 oxygen consumption rate in permeabilized PPDIV in the presence of different substrates. Individual data points plotted ± SEM (n = 8 wells per group). d.-h. No statistically significant results WT vs. SAKO. Experiments in g and h were repeated in 4 independent experiments, all with similar results; no significant difference between WT and SAKO OCR found by two-way ANOVA with post hoc analysis. Electron micrograph of iWAT (i) and eWAT (j), showing mitochondrial structure. This experiment was repeated independently with similar results. Scale bar = 200 nm. * p value < 0.05 versus WT two-sided student’s t-test.
STAT3 has also been shown to localize to mitochondria where it promotes electron transport chain (ETC) activity83–85. Because we observed STAT3 localization to mitochondria in adipocyte fractionation experiments (Fig. 2a), we isolated and evaluated mitochondria from WAT. Mitochondrial metabolism from the SAKO and WT adipocytes was identical with every substrate tested (Fig. 6g and Extended Data Fig. 6a–e). Moreover, no differences were observed in ETC activity between permeabilized WT and SAKO PPDIVs (Fig. 6h and Extended Data Fig. 6f–j). To determine whether there were mitochondrial defects in vivo, we examined WAT mitochondria by electron microscopy from in vivo fixed tissues. Mitochondrial structure was indistinguishable in the SAKO versus flox control adipocytes from both iWAT and eWAT (Fig. 6i,j). These data indicate that STAT3 promotes oxidative metabolism in adipocytes through a non-genomic and non-mitochondrial mechanism.
STAT3 suppresses FA re-esterification in adipocytes
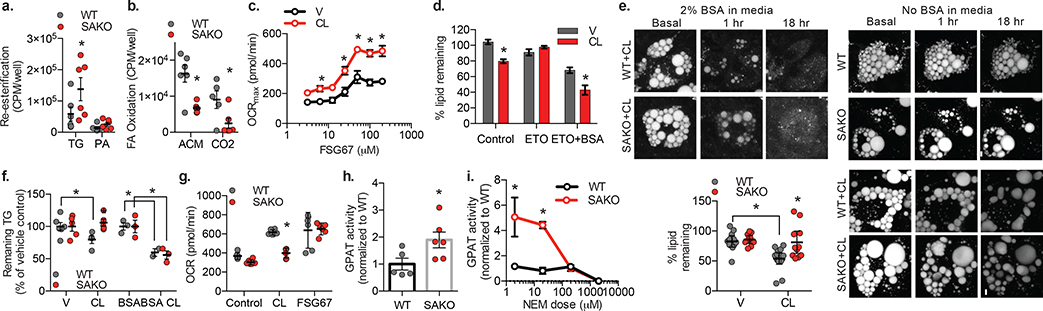
Having established that the metabolic effect of STAT3 occurs downstream of FA reuptake, we examined the fate of exogenous 14C-labeled palmitic acid in isolated adipocytes, measuring incorporation into triglycerides (TG) and phosphatidic acid (PA), or alternatively oxidization to acid soluble metabolites (comprised mainly of short chain FAs) or CO2. PPDIVs derived from SAKO mice exhibited 2-fold higher levels of 14C-palmitic acid incorporation into TG and PA than control adipocytes (Fig. 7a). This occurred at the expense of FA oxidation, as the formation of 14C-labeled CO2 and acid metabolites was substantially reduced in the SAKO cells relative to WT controls (Fig. 7b). These results indicate that STAT3 promotes FA oxidation while inhibiting fatty acid re-esterification in the adipocyte. Consistent with this idea, inhibition of GPAT activity with the pan-GPAT inhibitor, FSG67, resulted in a dose-dependent increase in oxidative metabolism in mature adipocytes (Fig 7c). Importantly, the IC50 of FSG67 (27.7 μM ± 4.4 μM) was consistent with the previously described IC50 for inhibition of GPATs (24 μM)120, strongly supporting the specific effect of the compound. Co-treatment with CL-316,243 did not change the observed IC50 (24.9 μM ± 1.8 μM), but did potentiate the effect of GPAT inhibition on oxidative rate (Fig 7c), indicating that stimulation of lipolysis is required to provide sufficient FA substrate to drive oxidative metabolism to its full potential.
Fig. 7: STAT3 suppresses FA re-esterification in adipocytes.
a. Incorporation of 14C-palmitic acid into triglycerides and phosphatidic acid (p value = 0.018), b. Oxidation of 14C-palmitic acid to form acid metabolites (ACM) and CO2 by WT and SAKO PPDIV (p values = 0.002 ACM and 0.022 CO2). Individual data points plotted ± SEM (n = 6 per genotype). Data in a. and b. are from the same experiment. c. Dose response of FSG67 impact on maximal OCR in 3T3-L1 adipocytes ± 10 μM CL-316,243 (n = 5 per treatment, p values = 0.016, 0.021 and < 0.0001 at FSG67 = 6.25, 12.5 and ≥ 25 μM respectively) d. Quantification of Bodipy stained area at 5 h relative to basal. BSA and etomoxir pretreated for 15 min (n = 20 cells per condition, p values < 0.0001). e. Representative micrographs (confocal, collapsed z-stack) of bodipy staining in WT and SAKO PPDIVs ± 2% BSA/1 μM CL-316,243. Lower-left panel: Quantification of Bodipy stained area at 18 h relative to basal. Individual data points plotted ± SEM (n = 12 vehicle and 16 CL cells per genotype, p value = 0.001 WT V vs. CL and 0.002 WT vs. SAKO CL). f. Triglyceride content in WT and SAKO PPDIVs after 24 h ± 2% BSA/1 μM CL-316,243, percent vehicle. Individual data points plotted ± SEM (n = 3 +BSA, 6 -BSA wells per genotype, p value = 0.046 V vs. CL WT, 0.035 WT vs. SAKO CL, 0.018 BSA vs. BSA-CL WT, 0.002 BSA vs. BSA-CL SAKO). g. WT and SAKO PPDIV OCR at 30 min ± 100 μM FSG67/1 μM CL-316,243. Individual data points plotted ± SEM (n = 6 per treatment, p value = 0.0002). h. and i. GPAT activity assay in homogenates from WT and SAKO PPDIV. Individual data points plotted ± SEM h. (n = 5 WT and 6 SAKO, p value = 0.042). i. NEM preincubation for 15 min. Data are represented as mean ± SEM (n = 3 per genotype, p values = 0.0002). * p value < 0.05 post hoc analysis after significant two-way ANOVA.
Since we did not observe a direct effect of STAT3 on mitochondrial capacity for fatty acid oxidation, we investigated the role of STAT3 in FA esterification. Lipid depletion by CL-316,243 in the absence of BSA is mediated by mitochondrial FA oxidation, as it was blocked in 3T3-L1 adipocytes by pretreatment with etomoxir to inhibit CPT1 activity (Fig. 7d). However, in the presence of BSA, lipolysis produces lipid droplet depletion, which is not blocked by CPT1 inhibition (Fig. 7d). We anticipated that the preference for re-esterification would preserve lipid content in the SAKO PPDIVs. Consistent with equal lipolytic rates in the WT and SAKO PPDIVs, lipid droplet disappearance was rapid in both genotypes in the presence of BSA (Fig. 7e). While CL-316,243 treatment produced lipid droplet disappearance in the WT cells in the absence of BSA, lipid droplet volume in the SAKO PPDIVs was preserved (Fig. 7e and Supplementary Video). We confirmed this result in a larger population of cells by quantifying TG levels in WT and SAKO PPDIVs after treatment with CL-316,243 (Fig. 7f). These results indicate that STAT3-mediated suppression of re-esterification is essential for lipid burning during stimulated lipolysis, which in turn has a significant impact on TG levels in adipocytes. This explains why SAKO adipocytes are significantly larger than WT adipocytes in both iWAT and eWAT under both normal diet and high fat diet feeding (Fig. 3e,f and Extended Data Fig. 3d,e).
Supplementary Video.
Time course of lipid droplet depletion (visualized by Bodipy staining) in WT and SAKO PPDIVs treated with CL-316,243.
STAT3 suppresses GPAT activity
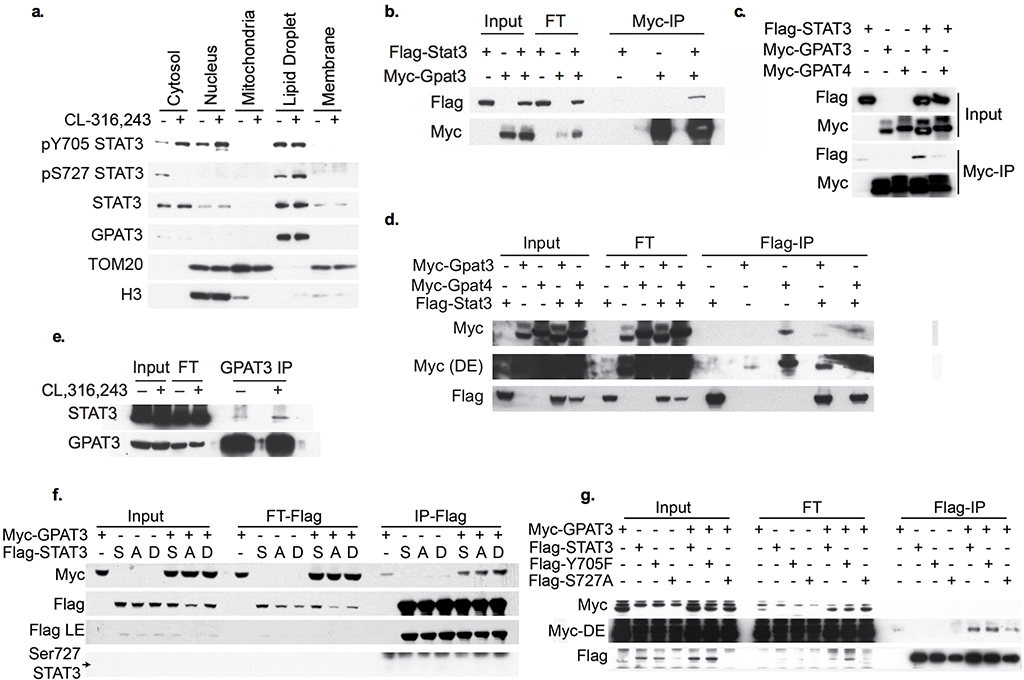
The rate-limiting step in FA esterification is catalyzed by GPATs, leading us to wonder whether STAT3 regulates the activity of these enzymes. Treatment with the GPAT inhibitor FSG67, restored oxidative metabolism in SAKO PPDIVs (Fig. 7g). GPAT activity was significantly higher in lysates from SAKO PPDIVs as compared to control lysates (Fig. 7h). The increased GPAT activity in the SAKO adipocytes was N-ethylmaleimide (NEM)-sensitive (Fig. 7i), ruling out GPAT1, which is NEM-insensitive14. STAT3 itself is NEM-insensitive121, and thus is not the cause of the NEM-sensitivity. Since GPAT2 is not expressed in adipocytes122, it seems likely that either GPAT3 or GPAT4 are affected by STAT3. Although both were originally characterized as ER-localized proteins, there is mounting evidence that GPAT3 and GPAT4 localize to the lipid droplet when present in the cell8–10. Indeed, we detected GPAT3 localization specifically in the lipid droplet fraction in adipocytes (Extended Data Fig. 7a). We thus treated cells with CL-316,243 and immunoprecipitated with a STAT3 antibody or IgG control, and performed mass spectrometry on STAT3-associated proteins corresponding to the size of GPAT1/2 (95/90 kDa) and GPAT3/4 (49/52 kDa) separated by SDS-PAGE. Among other proteins, GPAT3 was found in the anti-STAT3 immunoprecipitate, and was absent in the IgG control (Supplementary Data 1), while no other GPAT proteins were co-precipitated. To investigate a possible interaction between STAT3 and GPAT3, we overexpressed tagged versions of both proteins in HEK293T cells. STAT3 coimmunoprecipitated with GPAT3, and reciprocally, GPAT3 coimmunoprecipitated with STAT3 (Extended Data Fig. 7b–d), while specific interaction with the closely related family member GPAT4 was not observed (Extended Data Fig. 7c,d). Together these results suggest that STAT3 may interact with GPAT3, the major GPAT in white adipocytes, to suppresses GPAT activity.
Regulation of FA re-esterification by STAT3
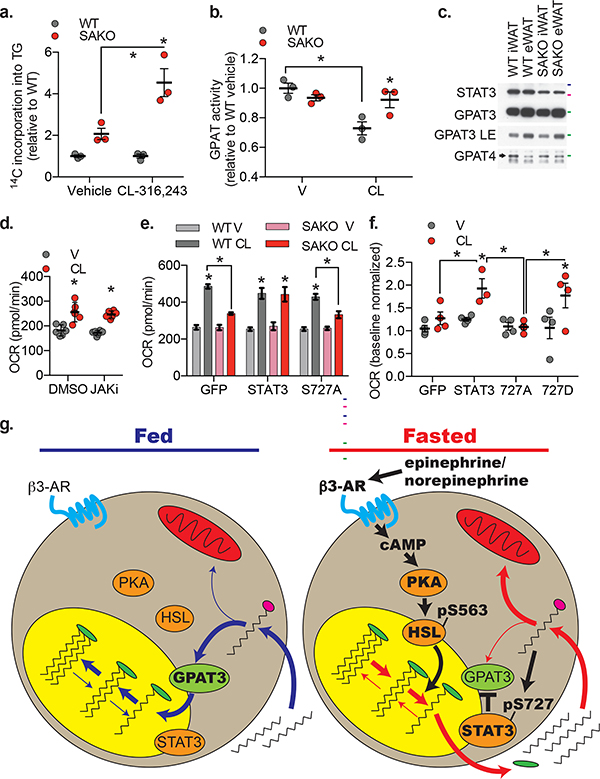
We investigated whether STAT3-mediated suppression of re-esterification is regulated by catecholamines. Basal re-esterification of 14C-palmitic acid to TG in the SAKO PPDIVs was two times the rate observed in WT cells; upon CL-316,243 stimulation re-esterification in the SAKO PPDIVs was 4.5 times the WT rate (Fig. 8a), supporting the hypothesis that STAT3-mediated suppression of re-esterification is enhanced by β-adrenergic activation. We also measured GPAT activity in WAT from animals treated with CL-316,243 or vehicle control. In WAT derived from WT mice, GPAT activity was suppressed by CL-316,243 (Fig. 8b). This GPAT suppression by CL-316,243 was STAT3-dependent, as CL-316,243 failed to suppress GPAT activity in WAT from SAKO animals (Fig. 8b). No difference in GPAT activity was detected between vehicle treated WT and SAKO WAT, nor was there a difference in GPAT3 or GPAT4 protein expression levels (Fig. 8b,c). Thus, although some suppression of GPAT activity is observed in the absence of β-adrenergic stimulation in serum-starved SAKO PPDIVs, we do not observe STAT3-mediated suppression of GPAT activity in unstimulated ad-libitum fed animals. These data suggest that catecholamines enhance STAT3-mediated suppression of GPAT activity in white adipocytes.
Fig. 8: Regulation of FA disposition by STAT3.
a. Incorporation of 14C-palmitic acid into triglycerides by SAKO PPDIV relative to WT control cells ± 1 μM CL-316,243. Individual data points plotted ± SEM (n = 3 wells per condition, p values = 0.0008). b. GPAT activity in iWAT homogenates from WT and SAKO mice treated with 1 mg/kg CL-316,243 or vehicle control for 20 minutes. Individual data points plotted ± SEM (n = 3 mice per condition, p value =0.002 WT V vs. CL, 0.019 WT vs. SAKO CL). c. Western blot analysis of GPAT3 and GPAT4 protein levels in lysates from WT and SAKO iWAT and eWAT. Results are representative of three independent experiments, 50, 75 and 100 kDa protein markers indicated in green, pink and blue respectively. d. OCR at 30 min ± 10 μM CL-316,243 in 3T3-L1 adipocytes ± JAK inhibitor I pretreatment for 30 min, normalized to baseline. Individual data points plotted ± SEM (n = 8 wells per condition, p values < 0.0001). e. and f. OCR at 30 min ± 0.5 μM CL-316,243 in WT and SAKO PPDIV with lentiviral STAT3 (WT, S727A or S727D) or GFP overexpression. e. Data are represented as mean ± SEM (n = 12 wells per over expression construct in each genotype, p value < 0.0001 V vs. CL, 0.002 WT CL vs. SAKO CL GFP, and 0.046 WT CL vs. SAKO CL S727A ). f. Individual data points plotted ± SEM (n = 4 wells per condition, p value = 0.029 V vs. CL STAT3, 0.016 V vs. CL 727D, 0.047 GFP vs. STAT3 CL, 0.011 727A vs. STAT3 CL, and 0.024 727A vs. 727D CL). g. Model of FA handling in the fed state (blue arrows) versus the fasted stated (red arrows) and the signaling regulating lipolysis-driven oxidative metabolism (black arrows) in the fasted state. * p value < 0.05 from post hoc analysis after significant two-way ANOVA.
Serine phosphorylation of STAT3 is required for lipid oxidation
These data suggest that lipolysis-driven oxidative metabolism requires STAT3-mediated suppression of re-esterification. As shown above, treatment with a pan-JAK inhibitor completely blocks STAT3 Tyr705 phosphorylation. However, inhibition of JAKs did not suppress lipolysis-driven oxidative metabolism, suggesting that this function of STAT3 is independent of its phosphorylation at Tyr705 (Fig. 8d). To test the role of STAT3 Ser727 phosphorylation, we reconstituted the SAKO PPDIVs with WT or S727A STAT3 using lentiviral overexpression. Reconstitution with WT STAT3 restored lipolysis-driven oxidative metabolism in the SAKO PPDIVs to WT levels (Fig. 8e), while reconstitution with the S727A STAT3 mutant failed to rescue lipolysis-driven oxidative metabolism (Fig. 8e). Although the S727D mutant also rescued the induction of oxidative metabolism by CL-316,243, it did not impact OCR in vehicle-treated cells, likely because lipolysis was not induced in these cells (Fig. 8f).
We hypothesize that the molecular mechanism by which STAT3 suppresses re-esterification may involve interaction with GPAT3. Consistent with this hypothesis, we found that the co-immunoprecipitation of endogenous STAT3 with GPAT3 was enhanced by CL-316,243 treatment (Extended Data Fig. 7e). Furthermore, co-immunoprecipitation of GPAT3 was enhanced by S727D phosphorylation-mimetic STAT3 mutation in HEK293T cells (Extended Data Fig. 7f), while the S727A phosphorylation-null mutant exhibited reduced co-immunoprecipitation of GPAT3 in CL-316,243-treated 3T3-L1 adipocytes (Extended Data Fig. 7g). Consistent with the dispensable role of Tyr705 phosphorylation of STAT3 in this context, GPAT3 co-immunoprecipitation was unchanged by the Y705F STAT3 mutation (Extended Data Fig. 7g). While these data suggest that STAT3 specifically blocks GPAT3 activity through a direct interaction, we cannot rule out the involvement of other proteins in this process.
These results suggest that upon sympathetic activation in the adipose tissue during conditions such as fasting, catecholamine signaling promotes oxidative metabolism in adipocytes via Ser727 phosphorylation of STAT3, which suppresses re-esterification in turn redirecting FAs from re-esterification to oxidation (Fig. 8h).
Discussion
During feeding and fasting, adipocytes are regulated by anabolic and catabolic hormones to store or mobilize energy. The dominant catabolic regulation comes after activation of adrenergic receptors due to release of epinephrine and norepinephrine from nerve terminals in adipose tissue27–30. In response, adipocytes undergo lipolysis, releasing FAs to be used as an energy substrate. A large proportion of FAs are reincorporated into the adipocyte37–47, where they can either re-esterify into triglycerides or undergo mitochondrial oxidation. Interestingly, adipocytes are metabolically reprogrammed during lipolysis to increase FA oxidation49. There is evidence that lipolysis-driven oxidative metabolism, but not lipolysis itself, is defective in adipose tissue from obese humans49, although it is unclear whether defects in this pathway lead to the development or are a consequence of obesity. However, the mechanisms by which the adipocyte is instructed to re-esterify versus oxidize FAs remain completely unknown. Data presented here indicate that catecholamines suppress re-esterification by increasing serine phosphorylation of STAT3, and suppression of re-esterification leads to increased FA oxidation due to the increased availability of FA-CoA.
We show here that β-adrenergic-stimulated fatty acid oxidation was dramatically reduced in adipocyte-specific Stat3 KO mice in vivo, and in adipocytes derived from these mice in vitro, accounting for the increased adiposity of these mice. Moreover, the defect in fatty acid oxidation observed in adipocytes from SAKO mice was rescued by WT but not S727A mutant STAT3. Thus, it appears that catecholamine-stimulated Ser727 phosphorylation of STAT3 results in the suppression of GPAT activity, and this inhibition results in the redirection of FA-CoAs to mitochondria for oxidation. In contrast, STAT3 Tyr705 phosphorylation appears to be dispensable for this process. However, this phosphorylation event could be critical for other catecholamine-stimulated pathways, or for the perpetuation of oxidative metabolism after the first few hours, as OCR remains high long after Ser727 phosphorylation returns to baseline.
The signaling pathways linking catecholamine stimulation to STAT3 Tyr705 phosphorylation are not clear, and there may be another adipokine besides IL-6, such as IL-11, that activates the JAK/STAT3 axis123. The serine/threonine kinases that phosphorylate STAT3 Ser727 have been the subject of much investigation, and include numerous MAPKs92,94–101. While we find that this phosphorylation is PKA-dependent, the Ser727 site on STAT3 does not match a PKA consensus site. Instead, PKA-induced lipolysis followed by FA reuptake appears to be required for this phosphorylation event at the lipid droplet, suggesting that the serine kinase responsible for STAT3 Ser727 phosphorylation may be activated by fatty acids or their metabolites. Future studies will be required to identify the STAT3 serine kinase, and to understand the spatial regulation of this phosphorylation event.
The first committed and rate-limiting step in the esterification of FAs with glycerol to form glycerolipids is catalyzed by the GPAT enzymes, which are crucial in the regulation of fatty acid handling in the cell. GPAT3 is the dominant isoform in WAT but is also expressed in the intestine, heart and kidney, with low levels in the brain11,124. Gpat3 global KO mice exhibit increased energy expenditure, and Gpat3 KO females are protected from diet induced obesity19. Moreover, treatment of obese mice with a GPAT inhibitor elevates energy expenditure125,126. While these data are consistent with the hypothesis presented here, it is impossible to tease out the adipose tissue-specific effects of GPATs in these experiments19. In support of an obesogenic role of adipose tissue re-esterification, adipose-specific overexpression of Pck1, which increases re-esterification via increased glyceroneogenesis, dose-dependently increased adiposity and adipocyte size127.
Suppression of FA re-esterification by STAT3 in adipocytes represents a novel non-genomic activity of STAT3. While gene expression differences were noted in adipose tissue of the SAKO mice, these were largely unrelated to fatty acid oxidation or storage. STAT3 has also been detected in mitochondria, where it is proposed to control ETC activity83,84. The mechanism of increased ETC activity by mitochondrial STAT3 remains uncertain, and the role of STAT3 as a mitochondrial transcription factor has not been eliminated116,128. However, we observed no difference in structure or function of mitochondria in white adipocytes from SAKO mice, and this potential effect of STAT3 is unlikely to contribute to the phenotype described here. Interestingly, ER-localized STAT3 has also been ascribed a non-genomic activity129, where it may interact with the calcium channel, IP3R3, and suppress its activity. This action of the signaling protein deserves further attention.
Understanding the mechanisms controlling the fate of fatty acids in white adipocytes is of great importance. Although previous studies have reported that adrenergic activation increases the oxidative step, the pathways regulating this critical event have been a complete mystery. The crucial part played by STAT3 in this process reveals yet another surprising role for this versatile signaling intermediate, and implicates a new crucial regulatory node for metabolic homeostasis.
Methods
Animals
Homozygous Il6 KO mice were obtained from Jackson Labs (Stock No: 002650). These mice were bred to C57BL/6J mice (Stock No: 000664) to generate heterozygous animals. Cohorts of WT and KO littermate controls were generated from heterozygous to heterozygous breeding. Animals were genotyped prior to weaning and WT and KO mice were cohoused after weaning.
Animals homozygous for the Stat3 floxed allele were obtained from Jackson Labs (Stock No: 016923). These mice were bred to Adipoq-promoter driven Cre mice (Stock No: 028020). The F1 generation was then intercrossed to generate mice homozygous for the Stat3 floxed allele both with and without the Adipoq-Cre. Animals with Adipoq-Cre expression lose Stat3 in mature adipocytes and are referred to in the manuscript as SAKO animals, while littermates without Adipoq-Cre are WT or floxed controls.
All strains of mice were on the C57BL/6J background. Only male mice were used for experiments. We fed mice with normal diet (ND) (7912 – Teklad) or high fat diet (HFD) consisting of 45% of calories from fat (D12451 – Research Diets Inc.) for 12–13 weeks, starting at 6–8 weeks of age. Mice were treated with CL-316,243 at a dose of 1 mg/kg by intraperitoneal injection with 1o ml/kg of a 0.1 g/l solution of CL-316,243 in PBS, or PBS vehicle control using a 28G needle. During metabolic studies, ear tag numbers were used to identify animals. Within an experiment, the genotype and or treatment groups were both littermates and cage mates. Researchers performing tests and collecting data were blinded during experiments. Animals in each cohort were produced from up to 20 breeding pairs to minimize the birthdate range. Within each genotype, mice were assigned to treatment groups, such that prior to treatment the mean body weight, as well as the standard deviation of the body weight, was equal across treatment groups, using a method similar to block randomization. Additional consideration was given to housing, such that each cage contained multiple treatment groups, to avoid confounding by cage effects. Sample size was determined based on the available cohorts and the number of treatment groups required. Mice were housed in a specific pathogen-free facility with a 12-h light, 12-h dark cycle, and given free access to food and water, except for fasting period. All animal use was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California-San Diego.
Serum IL-6 measurement
Blood was collected by submandibular bleed, and allowed to clot at room temperature for 30 min prior to serum isolation. Serum IL-6 levels were quantified using Mouse IL-6 Quantikine ELISA from R&D (SM6000B) with 50 μl of serum.
Body composition
The University of Michigan Metabolic Phenotyping Core used NMR analysis to quantify body fat, lean body mass and fluid content in HFD fed mice. Body composition of normal diet (ND) fed mice was determined at the University of California San Diego using an EchoMRI™ (3 in 1).
Metabolic cage studies
For metabolic study, mice were subject to Comprehensive Laboratory Monitoring System (Columbus Instruments) (CLAMS is an integrated open-circuit calorimeter equipped with an optical beam activity monitoring system) to measure oxygen consumption (VO2), carbon dioxide production (VCO2) by indirect calorimetry and spontaneous motor activity at ACP phenotyping core of UCSD. For metabolic study in ND fed mice, animals were subject to Promethium system metabolic cage in a temperature control cabinet. Values presented are normalized to body weight.
Adipocyte size
Adipocyte size was quantified from Hematoxylin and Eosin stained WAT histology slides, imaged with standard Texas Red excitation and emission filter. Adipocyte size was assayed using Cell Profiler 130. We developed a pipeline to exclude objects that deviated from circularity by omitting those objects with a form factor below 0.5 and an eccentricity above 0.95, which greatly improved the accurate identification of single adipocytes by the automated system. A minimum of 1,000 cells were assayed from each HFD fed animal, and a minimum of 600 cells in ND fed mice.
Lipolysis
FA concentration was measured using either 2 μl of serum or 25 μl conditioned media with the NEFA kit (WAKO), using 75 μl Reagent A and 150 μl Reagent B. Absorbance was measured at 550 nm (reference 660 nm) using the manufacturer’s protocol. Free glycerol concentration in serum was measured by reacting 10 μl of serum with 190 μl Free Glycerol Reagent (Sigma) and absorbance was measured at 540 nm using the manufacturer’s protocol. Free glycerol concentration in cell culture media was measured by reacting 50 μl of conditioned media with 150 μl Free Glycerol Reagent reconstituted to 4/3x (Sigma) and absorbance was measured at 540 nm using the manufacturer’s protocol.
Cell culture
3T3-L1: 3T3-L1 fibroblasts (American Type Culture Collection) were cultured in culture media (DMEM containing 4.5 g/l glucose, 10% FBS, 10 U/ml penicillin, 10 U/ml streptomycin and 292 mg/l glutamine). Once grown to confluence, adipocyte differentiation was initiated using a three component cocktail containing 500 μM 3-Isobutyl-1-methylxanthine, 250 nM dexamethasone and 1 μg/ml insulin for the first 3 days, followed by an additional 3 days of media containing 1 μg/ml insulin and finally differentiation was completed in the culture media. Only cultures in which >90% of cells displayed adipocyte morphology were used. Fully differentiated adipocytes, which were cultured in culture media for 5–9 days were used for experiments. CL-316,243 was treated at 10 μM in serum free DMEM, with 2% free fatty-acid free BSA when indicated. Inhibitors (50 μM H89, 10 μM JAK Inhibitor I (CAS 457081–03-7 – Calbiochem), and 10 μM SB-203,580) were pretreated in complete media for 30 min, then continued in serum free DMEM with CL or vehicle treatment. For cellular respiration studies, JAK inhibitor I pretreatment was performed in seahorse DMEM, and CL was injected via port A. CL-316,243 was dissolved in water at 1,000x, other inhibitors were dissolved in DMSO at 1,000x.
Fractionation
3T3-L1 adipocytes and adipose tissue were fractionated using differential centrifugation in fractionation buffer: HEPES (pH 7.4) 20 mM, KCI 10 mM, MgCl2 2 mM, EDTA 1 mM, EGTA 1 mM, DTT 200 μM, NaF 10 mM, NaVO4 1 mM, β-glycerophosphate 2.5 mM, phosphatase inhibitor tablet (Roche). All steps were performed at 4 °C. Cells/tissue were lysed with a glass dounce homogenizer, then spun at 1,000 g for 10 min to separate lipid droplets (floating fat cake) and nuclear pellet. Mitochondria were then pelleted from the supernatant at 10,000, and then Membrane pelleted at 21,000 g after incubation with 8 mM CaCl2. Remaining proteins in solution were cytosol. Each fraction (except cytosol) was resuspended in fractionation buffer and spun a second time to wash, before addition of lysis buffer. Each supernatant was spun a second time at the same speed, before transferring to a new tube to pellet the next fraction. Protein content in each fraction was normalized and 20 μg of protein loaded per well for western blot analysis.
PPDIV
Primary preadipocytes were isolated from inguinal fat pads as follows: Following fine mincing, the tissue was collagenase (2 mg/ml Sigma – C6885) digested in a 37 °C water bath with shaking for 15–20 min. FBS was added to 10 %, then the slurry was passed through a 100 μm filter, and spun at 500 g for 5 min. The fat cake was washed once and spun again. The two pellets were combined and washed once and plated in culture media with 2.5 mg/L amphotericin B (Sigma – A2411). The following day, the media was removed and nonadherent cells washed away with DPBS, fresh culture media was added. When the cells reached ~40 % confluence, they were split 1:3. Cells from the first and second passage were plated to confluence for experiments to ensure that all cells reached confluence at the same time. Differentiation was initiated with 500 μM 3-Isobutyl-1-methylxanthine, 250 nM dexamethasone, 1 μg/ml insulin and 1 μM Troglitazone for 4 days, followed by insulin for 3 days. Cells were used for experiment 8 or 9 day after the initiation of differentiation. Only cultures in which >90% of cells displayed adipocyte morphology were used. CL-316,243 was treated at 0.5 or 1 μM in serum free DMEM, with 0.2 or 2% Free fatty-acid free BSA as indicated. Atglistatin 50 μM pretreatment was performed in seahorse DMEM, 15 min prior to beginning OCR measurements.
Lentiviral Transduction
After differentiation cocktail, 3T3-L1 cells were exposed to concentrated virus-containing insulin media for 2 hours, then additional insulin media was added for another 14 h. After which the media was changed to fresh insulin media for an additional 72 h. 3T3-L1 cells were cultured for an additional 4–8 days in culture media before being used in an experiment. The same protocol was followed for PPDIVs, except that insulin treatment was 4 days, and cells were utilized for the experiment after 24 h in culture media.
HEK293T
HEK293T cells (American Type Culture Collection) were cultured in culture media.
Transfection
HEK293T cells were transfected with a combination of pLVX-3xFLAG-STAT3, pLVX-3xFLAG-STAT3-S727D, pLVX-3xFLAG-STAT3-S727A, pLVX-3xMYC-GPAT3, pLVX-3xMYC-GPAT4, and pLVX-GFP plasmids as designated in each experiment using Lipofectamine 3000 reagent (Invitrogen) following the manufacturer’s protocol. GFP vector was used to normalized transfected DNA amount in control samples that received only STAT3 or GPAT3/4, as well as monitor transfection efficiency. 24 to 48 hours after transfection, cells were used in a Co-IP assay.
Co-Immunoprecipitation
HEK293T and 3T3-L1 cells were lysed in a buffer containing 20 mM HEPES (pH 7.6), 100 mM NaCl, 20 mM β-glycerophosphate, 1 mM NaVO3, 10 mM MgCl2, 1 mM DTT, 1 mM PMSF, 0.5 g/l Triton X-100, 0.5 g/l NP-40, protease inhibitors [1 complete™ Protease Inhibitor Cocktail (Roche) per 50 ml of buffer] by mechanical homogenization via 10 time passage through a 28-gauge needle. Homogenate was spun at 1,000 g for 10 minutes, then at 23000 g centrifugation for 15 min, then lysates were collected and diluted 1:1 using dilution buffer containing 20 mM HEPES (pH 7.6), 20 mM β-glycerophosphate, 1 mM NaVO3, 10 mM MgCl2, 1mM DTT, 1 mM PMSF, protease inhibitors [1 cOmplete™ Protease Inhibitor Cocktail (Roche) per 50 ml of buffer]. The diluted cell lysates were incubated with either monoclonal anti-FLAG M2 magnetic beads (M8823, Sigma) or monoclonal anti-MYC magnetic beads (5698S, Cell Signaling Technology) for 2-h at 4 °C. For the endogenous Co-IP, the 3T3-L1 lysate was incubated with GPAT3 antibody (Invitrogen –PA5–38698 Lot TI2643756) overnight at 4 °C, then protein A/G beads (Santa Cruz – sc-2003) were added for 2 h, or lysates were incubated with STAT3 (Cell Signaling – 4368) or IgG (Cell Signaling − 3423) antibody coated beads overnight. Flow through was collected and the beads were washed 5 times with wash buffer containing 20 mM HEPES (pH 7.6), 50 mM NaCl, 20 mM B-glycerophosphate, 1 mM NaVO3, 10 mM MgCl2, 1mM DTT, 1 mM PMSF, 0.25% Triton X-100, 0.25% NP-40, protease inhibitors [1 cOmplete™ Protease Inhibitor Cocktail (Roche) per 50 ml of buffer]. Proteins were eluted off the beads by the addition of SDS sample buffer and 5 min incubation at 95 °C with vortexing every 30 seconds.
Lentivirus production
HEK293T cells were plated onto polylysine (0.01%) coated plate, then transfected with pLVX expression vectors along with pMDL/pLP1 (packaging plasmid), pVSVG/pLP (envelope plasmid), and pLP2/ pRev (packaging plasmid) and using Lipofectamine 3000 reagent (Invitrogen, L3000) following the manufacturer’s protocol. Media was changed 24 h later. The lentivirus containing media were collected at 48 and 72 hours post transfection and incubated overnight at 4 °C with lenti-X concentrator (TaKaRa, 631232), then spun at 1500 g at 4 °C for 45 minutes, the viral pellets were reconstituted in fresh insulin media (high glucose (4500 mg/L) DMEM with 10% FBS and 10 μg/ml insulin).
pLVX expression vectors
STAT3 constructs in the pAdCMV vector were purchased from Addgene: pAdCMV/V5-DEST-STAT3–3xFlag (99260), pAdCMV/V5-DEST-S727A-STAT3–3xFlag (99262), pAdCMV/V5-DEST-S727D-STAT3–3xFlag (99263), and pAdCMV/V5-DEST-Y705F-STAT3–3xFlag (99261). A PCR based cloning method was used to transfer the constructs into the pLVX vector. GPAT3 was cloned from C57BL/6 inguinal WAT cDNA, and GPAT4 was cloned from C57BL/6 liver cDNA and placed into the TOPO vector. From there the constructs were transferred into the pLVX vector with a C-terminal Myc Tag and N-terminal Kozak sequence.
Microscopy
Live Cell Confocal
PPDIVs were incubated in culture media containing 1 μM of Bodipy 493/503 (Life Technology) to label lipid droplets, and/or 25 nM of MitoTracker Deep Red FM (Thermo Fisher) at 37 °C for 30 min. Cells were then washed with media twice and then washed into phenol red free DMEM with and without 1 μM CL-316,243 and 2% FFA-free BSA. Live cell images were obtained using Nikon A1R confocal with 100x oil immersion objective. For live cell time-lapse microscopy, multi z-steps (15 steps, 1 μm/step) images were taken roughly every 20 minutes.
Immunofluorescence confocal microscopy
PPDIVs were differentiated on chambered cover glass (Lab-Tek). Lipid droplets were labeled by incubation with 10 μM of Bodipy 493/503 (Life Technology) at 37 °C for 15 min, followed by treatment with and without 1 μM CL-316,243, and then immediately fixed with 4% paraformaldehyde. Cells were permeabilized and blocked with 3% BSA + 0.2% Triton X-100 for 30 min at room temperature, then incubated with primary antibodies in 3% BSA overnight at 4 °C (STAT3 1:1,600 Cell Signaling 9139, p727 STAT3 1:100 Abcam ab131103). Cells were washed three times with PBS, then incubated with Secondary antibodies for 1 hour at 4 °C (Invitrogen Goat anti-mouse Alexa Fluor 555, A32732 1:1,000 and Goat anti-rabbit Alexa Fluor 647 A32733 1:2,000), then stained with Hoechst dye, and washed three more times before imaging. Multi z-step (71 steps 0.5 μm/step) images were obtained using Nikon A1R confocal with 100x oil immersion objective. Blinded analysis was performed using Volocity 6.3 software to identify lipid droplets (Bodipy stained objects in 3D reconstruction). Misidentified objects (either multiple lipid droplets identified as one object or only part of a lipid droplet selected) were eliminated from the analysis. Voxels in the object that co-localize with either STAT3 or Ser727 phosphorylated STAT3 were determined and normalized either total object voxels or surface area.
Lipid Droplet Volume
To calculate the volume of lipid droplets, cells were manually selected. The same intensity threshold adjustment was applied to every individual cell to reduce background and eliminate out of focus pixels. LD volume of each cell was then calculated using NIS-Elements (Nikon). This analysis uses the number of above threshold pixels in the selected region to calculate volume and is not affected by staining intensity. The researcher was blinded to the experimental group during analysis.
Electron Microscopy
Mice were fixed via cardiac perfusion at 37 °C with 2% paraformaldehyde (freshly made), 2.5% glutaraldehyde in pH 7.4, 0.15 M sodium cacodylate buffer with 0.03% calcium chloride. Fat pads were removed and immersion fixed overnight at 4 °C. Tissues were washed five times with ice-cold buffer consisting of 0.15 M sodium cacodylate, 0.03% calcium chloride, and postfixed with ice-cold 2% OsO4, 0.8% KFe(CN)3, 0.03% CaCl2 in 0.15 M sodium cacodylate for 1 h on ice, washed three times with ice-cold distilled water and stained overnight with 2% uranyl acetate at 4 °C. Tissues were dehydrated in an ethanol series, infiltrated and embedded in Durcupan ACM resin (Fluka). Ultrathin 80-nm-thick sections were made using a Leica EM UC6 ultramicrotome and Diatome 45° diamond knife. Sections were imaged by an FEI Spirit transmission electron microscope operated at 80 kV with a Teitz TemCam F224 2k by 2k CCD camera.
Triglyceride quantification
Lipids were extracted from PPDIV cells using a 3:2 mixture of Hexane:Isopropanol. Liver tissue was weighed and homogenized and lyophilized, then lipids were extracted with chloroform. The organic solvent was evaporated from the extracted lipids and triglycerides content was quantified using the infinity TG reagent (Thermo-Fisher) following the manufacturer’s protocol.
Cellular and Mitochondrial Respiration
Oxygen consumption rates were measured using a Seahorse XF96 or XFe96 analyzer. Data were analyzed as previously described 131.
Mitochondrial isolation
Mitochondria were isolated from inguinal primary mature adipocytes from WT and SAKO animals. Each isolation utilized tissue from at least 3 animals to reduce animal to animal variance within each genotype. Harvested tissues were minced and collagenase (1 mg/ml) digested in a 37 °C water bath with shaking for 15–25 min, followed by addition of FBS to 10% and passage through a 100 μm filter; from this point forward all steps were performed at 4 °C. The floating adipocyte fraction was collected and washed into SHE buffer pH 7.4 with 4% FA-free BSA (250 mM sucrose, 5 mM HEPES, 1 mM EGTA), then homogenized with 5 rotating strokes in a glass-Teflon homogenizer. The homogenate was spun at 800 g for 10 min, and the supernatant collected then spun again at 10,000 g at 4 °C for 10 min to pellet mitochondria, which were then washed into SHE with 0.5 % FA free BSA, 2.5 μg were plated per well in a 96 well Seahorse plate generally as previously described 132. Media was changed to MAS buffer (70 mM Sucrose, 220 mM Mannitol, 5 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1 mM EGTA and 0.2% FA-free BSA), containing various substrates (4 mM ADP + 5 mM pyruvate + 1 mM malate, 40 μM palmitoylcarnitine + 1 mM malate, 5 mM succinate + 2 μM rotenone, 5 mM glycerol 3 phosphate + 2 μM rotenone + 700 nM CaCl2, 20 mM ascorbate + 200 μM Tetramethyl-p-Phenylenediamine) before measurement of oxygen consumption. Phosphorylating (“State 3”) respiration was measured as the initial rate, followed by sequential injections of oligomycin (2 μM), and then FCCP (sequential additions of 3 μM).
PPDIV permeabilization
Cells were permeabilized by 15 min incubation in MAS with 4 mM ADP and 3 nM recombinant perfringolysin O 133 (Agilent – 102504–100). Then various substrates (4 mM ADP + 5 mM pyruvate + 1 mM malate, 40 μM palmitoylcarnitine + 1 mM malate, 5 mM succinate + 2 μM rotenone, 5 mM glycerol 3 phosphate + 2 μM rotenone 700 nM CaCl2, 20 mM ascorbate + 200 μM Tetramethyl-p-Phenylenediamine) were added before measurement of oxygen consumption. Phosphorylating (“State 3”) respiration was measured, followed by sequential injections of oligomycin (2 μM), and then FCCP (sequential additions of 2 μM) 134.
Intact cell experiments
Cells were differentiated on a 96 well seahorse plate, then washed into base media (DMEM containing 8 mM glucose, 1 mM pyruvate, 2 mM glutamine and 0.5 mM carnitine without phenol red or sodium bicarbonate), then incubated in a CO2 free incubator for 15 min prior to measurement of oxygen consumption. Inhibitor pretreatments were performed in base media. Basal rates of respiration were measured, followed by sequential injections of oligomycin (2μM), FCCP (sequential additions of 500 nM) and rotenone (1 μM)/antimycin A (1 μM), or sequential injections of CL-316,243 (0.5 μM PPDIV/10 μM 3T3-L1), oligomycin (2μM), FCCP (500 nM) and rotenone (1 μM)/antimycin A (1 μM).
Re-esterification and FA Oxidation
PPDIVs in a 12-well plate were incubated in 1 ml/well DMEM containing 28 mM (0.5 μCi well) 14C-palmitic acid with or without 1 μM CL-316,243. After a 60 min incubation, 900 μl of media was collected and 100 μl 10% BSA added. The remaining media was suctioned off and the cells washed 3x with cold PBS. Lipids were extracted with 200 μl 3:2 hexane:isopropanol three times. Lipid extracts were dried and run on a TLC plate with 80:20:1 hexane:ether:acetic acid to separate TG, FA and DG, which were cut off then the plate was run in the opposite direction with 6:35:8 chloroform:methanol:water to separate PA, FA-CoA, LPA. While all FA species could be identified based on standards and seen by I2 staining, the vast majority of the radioactivity was contained in the TG and to a lesser extent the PA bands, which were scraped off the plate for measurement of radioactivity in a liquid scintillation counter (Perkin Elmer, MicroBeta TriLux 1450).
GPAT activity assay
The GPAT activity assay was adapted from previously described assays 19,20. GPAT specific activity was assayed for 15 min at room temperature in a 200 μl reaction mixture containing 100 μg total lysate protein in GPAT assay buffer (75 mm Tris-HCl, pH 7.5, 4 mm MgCl2, 1 mg/ml BSA (FA-free), 1 mm dithiothreitol, 8 mm NaF, 77 μM (2 μCi/rxn) 14C-glycerol 3-phosphate, and 50 μM lauryl-CoA). PPDIV cell lysates were prepared by 10 passages through a 28 g needle in GPAT assay lysis buffer (250 mM sucrose, 10 mM Tris pH 7.5, 1 mM EDTA and 8 mm NaF). Inguinal WAT tissue lysates were prepared by mechanical homogenization (Roto Star) with 1 μl GPAT assay lysis buffer per mg tissue. Crude lysates were spun at 1,000 g for 10 min followed by 17,000 g for 15 min. Supernatant was transferred to a new tube and protein concentration measured before dilution to 1 mg/mL and addition of 2x GPAT assay buffer. NEM treatments were performed for 15 min prior to addition of 2x GPAT assay buffer. Reactions were terminated by addition of 2:1 chloroform:methanol to extract lipids and precipitate protein. Lipid extraction was repeated an additional two times, dried and separated by TLC with 6:35:8 chloroform:methanol:water, to isolate phosphatidic acid. The spot corresponding to phosphatidic acid was identified based on a standard 16:0PA standard (Avanti Polar Lipids, 830855), and visualized with I2 in order to be scraped off the plate and radioactivity quantified in a liquid scintillation counter (Perkin Elmer, MicroBeta TriLux 1450).
Western blot analysis
We homogenized tissues and cells in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM dithiothreitol, 1 mM Na3VO4, 5 mM NaF, 1 mM phenylmethanesulfonylfluoride, 25 mM glycerol 2-phosphate and freshly added protease inhibitor tablet) and then incubated them for 1 h at 4 °C. We centrifuged crude lysates at 17,000 g for 15 min twice and determined the protein concentration using Bio-Rad Protein Assay Dye Reagent. Mature adipocytes were isolated using the same protocol as used to isolate preadipocytes for PPDIVs, except that three spins were done at 20g to wash the floating mature adipocytes, followed by protein precipitation with 5% TCA, acetone wash and resuspension in 8M urea, 100 mM Tris pH 8.0. Samples were diluted in SDS sample buffer. Bound proteins were resolved by SDS–PAGE and transferred to nitrocellulose membranes (Bio-Rad). Individual proteins were detected with specific antibodies and visualized on film using horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) and Western Lightning Enhanced Chemiluminescence (Perkin Elmer Life Sciences). Primary antibodies were used at a 1:1,000 dilution unless otherwise specified and purchased from Cell Signaling: pS563 HSL (4139), HSL (4107), pT108.Y182 p38 (9211), p38 (9212), p727 STAT3 1:500 (9134), p705 STAT3 (9131), STAT3 1:4,000 (9139), H3 (4499), TOM20 (42406), Flag R (14793), Myc R (2278), Perilipin 1 (9349), β-tubulin (2128) or Invitrogen: GPAT3 (PA5–38698 Lot TI2643756). Goat anti-mouse (31430) and goat anti-rabbit (31460) secondary antibodies were purchased from Thermo Fisher, and Donkey anti-goat secondary antibody was purchased from R&D system (HAF 109) and used at a concentration of 1:10,000. Uncropped western blot scans with the size markers indicated are included in the Source Data files. All blots are from separate membranes unless otherwise noted, but were run, transferred and blotted in parallel using the same power source and antibody dilutions.
Real-time PCR analysis of gene expression
RNA extractions from inguinal WAT were performed using the RNeasy Lipid Tissue Kit (Qiagen). We used the Superscript First-Strand Synthesis System for reverse transcription–PCR (Invitrogen) with a 3:1 mixture of random hexamers/oligo dT primers for reverse transcription. Real-time PCR amplification was performed on samples in triplicate with Power SYBR Green PCR Master Mix (Applied Biosystems) using the Applied Biosystems QuantStudio5 real-time PCR System and quantified using an internal standard curve with Arbp as the control gene. The sequences of all primers used in this study are listed in Supplementary Table 1.
RNA sequencing analysis
RNA extractions from primary mature inguinal adipocytes were performed using the RNeasy Lipid Tissue Kit (Qiagen) according to the manufacturer’s instructions. Biological triplicates of RNA isolated from WT and SAKO primary mature adipocytes were used to prepare sequencing libraries from 100–500 ng total RNA using the TruSeq RNA Sample Preparation Kit v2 (Illumina) according to the manufacturer’s protocol.
Sample preparation
Briefly, mRNA was purified, fragmented and used for first- and second-strand complementary DNA synthesis followed by adenylation of 3′ ends. Samples were ligated to unique adapters and subjected to PCR amplification. Libraries were then validated using the 2100 BioAnalyzer (Agilent), normalized and pooled for sequencing on the Illumina HiSeq 2000 using bar-coded multiplexing and a 100 bp read length.
Data analysis
Read alignment and junction mapping was accomplished using TopHat2 v2.0.4 using a 25 bp 5′ segment seed for initial mapping followed by differential gene expression analysis using Cuffdiff v2.0.2 to map reads to the reference genome annotation, NCBI mouse build 37.2 135. Median sequencing read yield per replicate sample was 24.2 M. Data were expressed as fragments per kilobase of exon per million fragments mapped. Volcano plots were generated from Cuffdiff output using CummeRbund v2.0.0 135.
RNA-seq library construction
Single-cell complementary DNA size distribution and concentration were assessed on a capillary electrophoresis-based fragment analyzer (Advanced Analytical). Illumina libraries were constructed in 96-well plates using the Illumina Nextera XT DNA Sample Preparation kit as described previously using the protocol supplied by Fluidigm. For each C1 experiment, a bulk RNA control (about 200 cells) and a no-cell negative control were processed in parallel in PCR tubes, using the same reagent mixes as used on chip. Libraries were quantified by Bioanalyzer, using High Sensitivity DNA analysis kit, and also fluorometrically using Qubit dsDNA HS Assay kits and a Qubit 2.0 Fluorometer (Invitrogen, Life Technologies).
DNA sequencing
Single-cell Nextera XT (Illumina) libraries of one experiment were pooled and 100 bp) paired-end were sequenced on Illumina HiSeq 2000 to a depth of (2–6) × 106 reads (three replicate experiments of distal mouse lung epithelial cells at E18.5, one experiment at E14.5 and one experiment on adult AT2 cells) or 150 bp paired-end on Illumina MiSeq (one experiment at E16.5) to a depth of 100,000–550,000 reads with v3 chemistry. CASAVA 1.8.2 was used to separate out the data for each single cell by using unique barcode combinations from the Nextera XT preparation and to generate *.fastq files.
Statistical Analyses
All statistical analyses were two sided. When comparing the two groups, two-tailed unpaired Student’s t test was used to determine the significance of experimental results. For experiments with a two factorial design a two-way analysis of variance (ANOVA) was performed to establish that not all groups were equal. The Holm-Sidak post-hoc analysis was then used for specific between-group comparisons after statistical significance was established by ANOVA. In each case, significance was set at α = 0.05. Statistical analyses were performed in GraphPad Prism version 6.
Additional methodological detail on sample size, statistics, software and code used, authentication, ect can be found in the Reporting Summary included with this manuscript.
Extended Data
Extended Data Fig. 1: Catecholamine signaling in white and brown adipose tissue.
Western blot analysis of WT and SAKO mice fed a HFD for 12 weeks, then treated with 1 mg/kg CL-316,243 or vehicle control for indicated time before sacrifice and tissue collection. a. Epididymal white adipose tissue. b. Inguinal white adipose tissue. Outlier detected with Grubs outlier test removed. c. Brown adipose tissue. Blots are representative of results from three independent experiments
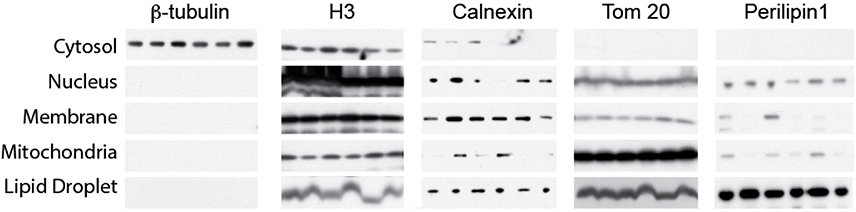
Extended Data Fig. 2: Fractionation of 3T3-L1 differentiated adipocytes.
Relative levels of β-tubulin (cytosol marker), H3 (nuclear marker), Calnexin (ER/membrane marker), TOM20 (mitochondrial marker) and Perilipin1 (lipid droplet marker) in fractionated samples from time course analysis in Fig. 2a. Cytosol, nucleus, membrane and mitochondria run on the same gel. These experiments were repeated independently four times with similar results.
Extended Data Fig. 3: Metabolic phenotype of SAKO mice on a normal diet (ND).
a. Left panel: Western blot of mature adipocytes isolated from 12-week old WT and SAKO mice. Right panel: Quantification of STAT3 protein relative to RalA loading control. Individual data points plotted ± SEM (n = 3 iWAT, 2 eWAT). b. Body weight of 12-week old ND fed WT and SAKO mice. Individual data points plotted ± SEM (n = 6 per genotype). c. Oxygen consumption rate in ND fed WT and SAKO mice at 16-weeks of age. Data are represented as mean ± SEM (n = 16). d. Adipocyte size distribution from ND-fed 12-week old WT and SAKO eWAT (n = 2 WT and 3 SAKO). e. Adipocyte size distribution from ND-fed 12-week old WT and SAKO iWAT (n = 2 WT and 3 SAKO). f. Body composition of ND fed WT and SAKO mice at 12 weeks of age. Individual data points plotted ± SEM (n =6 WT and 4 SAKO).
Extended Data Fig. 4: Histology from HFD-fed WT and SAKO mice.
From top to bottom, eWAT (scale bar = 100 μm), iWAT (scale bar = 100 μm), BAT (scale bar = 50 μm), and liver (scale bar = 100 μm). Results are representative of results from three independent experiments
Extended Data Fig. 5: Effect of CL-316,243 on metabolism.
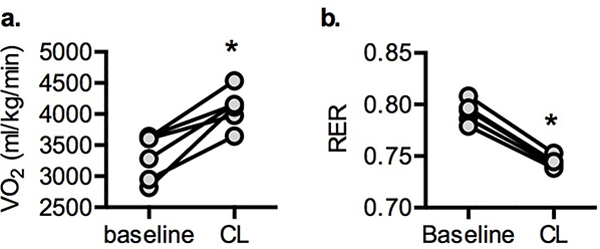
a. Oxygen consumption and b. RER before and after intraperitoneal injection with 1 mg/kg CL-316,243. Individual data points plotted ± SEM (n = 6 per treatment). * p value = 0.002 (VO2) and <0.0001 (RER) CL versus baseline (two-tailed paired t-test).
Extended Data Fig. 6: Mitochondria bioenergetics profiles.
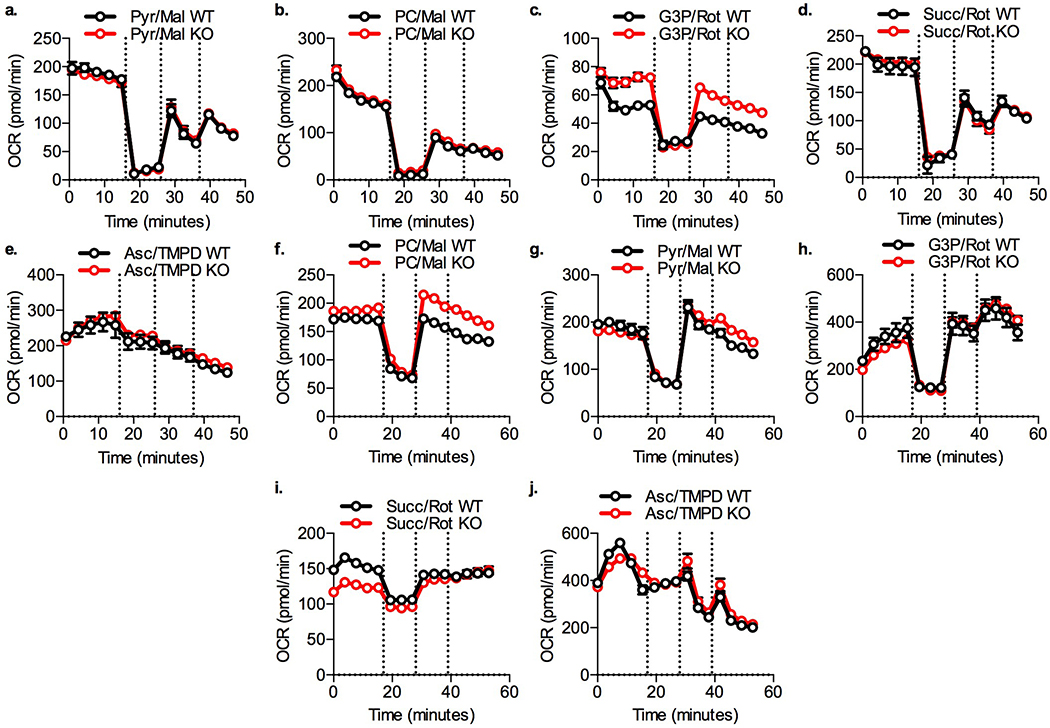
a.-e. Isolated mitochondria. Vertical lines indicate addition of oligomycin (2 μM), and FCCP (two sequential additions of 3 μM). a. 4 mM ADP + 5 mM pyruvate + 1 mM malate, b. 40 μM palmitoyl-carnitine + 1 mM malate, c. 5 mM succinate + 2 μM rotenone, d. 5 mM glycerol 3 phosphate + 2 μM rotenone 700 nM CaCl2, e. 20 mM ascorb-ate + 200 μM Tetramethyl-p-Phenylenediamine. f.-j. Permeabilized PPDIVs. Vertical lines indicate addition of oligomycin (2 μM), and FCCP (two sequential additions of 2 μM). f. 40 μM palmitoylcarnitine + 1 mM malate, g. 4 mM ADP + 5 mM pyruvate + 1 mM malate, h. 5 mM succinate + 2 μM rotenone, i. 5 mM glycerol 3 phosphate + 2 μM rotenone 700 nM CaCl2, j. 20 mM ascorbate + 200 μM Tetramethyl-p-Phenylenediamine. Data are represented as mean ± SEM (n = 8 per genotype).
Extended Data Fig. 7: STAT3/GPAT3 interaction.
a. Western blot analysis of fractionated 3T3-L1 adipocytes treated with 10 μM CL-316,243 or vehicle control for 60 min. b.-d. and f. Western blot analysis of input, flow through and immunoprecipitation using Myc-antibody coated beads (b, c) or Flag-antibody coated beads (d, f) of HEK293T cell lysates overexpressing Flag-tagged STAT3 and/or Myc-tagged GPAT3/GPAT4. Blots are representative of three independent replicates. Dark exposure (D.E.). e. Western blot analysis of input and immunoprecipitation using GPAT3 antibody in 3T3-L1 differentiated adipocytes treated with 10 μM CL-316,243 or vehicle control for 15 min. f. Western blot analysis of input, flow through and immunoprecipitation using Flag antibody coated beads of HEK293T cell lysates overexpressing Flag-tagged STAT3 (WT/S727A/S727D) and/or Myc-tagged GPAT3. Blots are representative of three independent replicates. Arrow indicates expected size of Ser727 phosphorylated STAT3; the band observed in the IP samples is a larger non-specific band. g. Western blot analysis of input, flow through, and immunoprecipitation using Flag antibody coated beads from 3T3-L1 differentiated adipocytes with lentiviral overexpression of flag-tagged STAT3 (WT/Y705F/S727A) and/or Myc-tagged GPAT3, cells treated with 10 μM CL-316,243 or vehicle control for 60 min before harvest and IP. These experiments were repeated independently twice with similar results.
Supplementary Material
Acknowledgments
We thank members of the Saltiel laboratory for helpful discussions. We thank Mason R. Mackey and Daniela Boassa at the NCMIR Core Facility for performing electron microscopy and Brechtje Vanderfeesten for blinded analysis of lipid droplet volume. This work was supported by US National Institutes of Health grants 1K01DK105075-01A1 and R03DK118195 to S.M.R., R01NS087611 to A.N.M, R01DK117551 and R01DK076906 to A.R.S., K99HL143277 to P.Z., P30DK063491 to A.R.S., S.M.R., P30 2P30CA023100-28 to the Microscopy core at UCSD Moores Cancer Center, P30NS047101 to the UCSD School of Medicine Microscopy Core, and P41 GM103412 to the NCMIR Core Facility. This work was also supported by American Diabetes Association grant 1-19-JDF-012 to S.M.R. and 1-18-PDF-094 to C.W.H. Finally, the authors dedicate this work to the memory of our inspirational friend and colleague Maryam Ahmadian.
Footnotes
Competing Interests Statement:
The authors declare no competing interests.
Data availability:
The RNA sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under accession code PRJNA557252. All additional data that support the findings of this study are available within the manuscript or supplement and from the corresponding author upon request.
References
- 1.James PT, Leach R, Kalamara E & Shayeghi M The worldwide obesity epidemic. Obes Res 9 Suppl 4, 228S–233S, doi: 10.1038/oby.2001.123 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Obesity: preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ Tech Rep Ser 894, i–xii, 1–253 (2000). [PubMed] [Google Scholar]
- 3.Swinburn BA et al. The global obesity pandemic: shaped by global drivers and local environments. Lancet 378, 804–814, doi: 10.1016/S0140-6736(11)60813-1 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Kennedy EP Biosynthesis of complex lipids. Fed Proc 20, 934–940 (1961). [PubMed] [Google Scholar]
- 5.Weiss SB, Kennedy EP & Kiyasu JY The enzymatic synthesis of triglycerides. J Biol Chem 235, 40–44 (1960). [PubMed] [Google Scholar]
- 6.Wendel AA, Lewin TM & Coleman RA Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis. Biochim Biophys Acta 1791, 501–506, doi: 10.1016/j.bbalip.2008.10.010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimeno RE & Cao J Thematic review series: glycerolipids. Mammalian glycerol-3-phosphate acyltransferases: new genes for an old activity. J Lipid Res 49, 2079–2088, doi: 10.1194/jlr.R800013-JLR200 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Wilfling F et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24, 384–399, doi: 10.1016/j.devcel.2013.01.013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H et al. Seipin is required for converting nascent to mature lipid droplets. Elife 5, doi: 10.7554/eLife.16582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagac M et al. SEIPIN Regulates Lipid Droplet Expansion and Adipocyte Development by Modulating the Activity of Glycerol-3-phosphate Acyltransferase. Cell Rep 17, 1546–1559, doi: 10.1016/j.celrep.2016.10.037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J, Li JL, Li D, Tobin JF & Gimeno RE Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc Natl Acad Sci U S A 103, 19695–19700, doi: 10.1073/pnas.0609140103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YQ et al. AGPAT6 is a novel microsomal glycerol-3-phosphate acyltransferase. J Biol Chem 283, 10048–10057, doi: 10.1074/jbc.M708151200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagle CA et al. Identification of a novel sn-glycerol-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6−/− mice. J Lipid Res 49, 823–831, doi: 10.1194/jlr.M700592-JLR200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monroy G, Kelker HC & Pullman ME Partial purification and properties of an acyl coenzyme A:sn-glycerol 3-phosphate acyltransferase from rat liver mitochondria. J Biol Chem 248, 2845–2852 (1973). [PubMed] [Google Scholar]
- 15.Yet SF, Lee S, Hahm YT & Sul HS Expression and identification of p90 as the murine mitochondrial glycerol-3-phosphate acyltransferase. Biochemistry 32, 9486–9491, doi: 10.1021/bi00087a029 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Ganesh Bhat B et al. Rat sn-glycerol-3-phosphate acyltransferase: molecular cloning and characterization of the cDNA and expressed protein. Biochim Biophys Acta 1439, 415–423, doi: 10.1016/s1388-1981(99)00103-1 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Harada N et al. Molecular cloning of a murine glycerol-3-phosphate acyltransferase-like protein 1 (xGPAT1). Mol Cell Biochem 297, 41–51, doi: 10.1007/s11010-006-9321-5 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Shan D et al. GPAT3 and GPAT4 are regulated by insulin-stimulated phosphorylation and play distinct roles in adipogenesis. J Lipid Res 51, 1971–1981, doi: 10.1194/jlr.M006304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao J et al. Mice deleted for GPAT3 have reduced GPAT activity in white adipose tissue and altered energy and cholesterol homeostasis in diet-induced obesity. Am J Physiol Endocrinol Metab 306, E1176–1187, doi: 10.1152/ajpendo.00666.2013 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Wendel AA, Cooper DE, Ilkayeva OR, Muoio DM & Coleman RA Glycerol-3-phosphate acyltransferase (GPAT)-1, but not GPAT4, incorporates newly synthesized fatty acids into triacylglycerol and diminishes fatty acid oxidation. J Biol Chem 288, 27299–27306, doi: 10.1074/jbc.M113.485219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper DE, Grevengoed TJ, Klett EL & Coleman RA Glycerol-3-phosphate Acyltransferase Isoform-4 (GPAT4) Limits Oxidation of Exogenous Fatty Acids in Brown Adipocytes. J Biol Chem 290, 15112–15120, doi: 10.1074/jbc.M115.649970 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarwal AK et al. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat Genet 31, 21–23, doi: 10.1038/ng880 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Akinci B, Meral R & Oral EA Phenotypic and Genetic Characteristics of Lipodystrophy: Pathophysiology, Metabolic Abnormalities, and Comorbidities. Curr Diab Rep 18, 143, doi: 10.1007/s11892-018-1099-9 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Garg A & Misra A Lipodystrophies: rare disorders causing metabolic syndrome. Endocrinol Metab Clin North Am 33, 305–331, doi: 10.1016/j.ecl.2004.03.003 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Alberti KG, Zimmet P, Shaw J & Group IDFETFC The metabolic syndrome--a new worldwide definition. Lancet 366, 1059–1062, doi: 10.1016/S0140-6736(05)67402-8 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Eckel RH, Grundy SM & Zimmet PZ The metabolic syndrome. Lancet 365, 1415–1428, doi: 10.1016/S0140-6736(05)66378-7 (2005). [DOI] [PubMed] [Google Scholar]
- 27.White JE & Engel FL A lipolytic action of epinephrine and norepinephrine on rat adipose tissue in vitro. Proc Soc Exp Biol Med 99, 375–378, doi: 10.3181/00379727-99-24355 (1958). [DOI] [PubMed] [Google Scholar]
- 28.Zeng W et al. Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell 163, 84–94, doi: 10.1016/j.cell.2015.08.055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartness TJ & Song CK Thematic review series: adipocyte biology. Sympathetic and sensory innervation of white adipose tissue. J Lipid Res 48, 1655–1672, doi: 10.1194/jlr.R700006-JLR200 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Youngstrom TG & Bartness TJ Catecholaminergic innervation of white adipose tissue in Siberian hamsters. Am J Physiol 268, R744–751, doi: 10.1152/ajpregu.1995.268.3.R744 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Duncan RE, Ahmadian M, Jaworski K, Sarkadi-Nagy E & Sul HS Regulation of lipolysis in adipocytes. Annu Rev Nutr 27, 79–101, doi: 10.1146/annurev.nutr.27.061406.093734 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lafontan M & Langin D Lipolysis and lipid mobilization in human adipose tissue. Prog Lipid Res 48, 275–297, doi: 10.1016/j.plipres.2009.05.001 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Jocken JW & Blaak EE Catecholamine-induced lipolysis in adipose tissue and skeletal muscle in obesity. Physiol Behav 94, 219–230, doi: 10.1016/j.physbeh.2008.01.002 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Czech MP, Tencerova M, Pedersen DJ & Aouadi M Insulin signalling mechanisms for triacylglycerol storage. Diabetologia 56, 949–964, doi: 10.1007/s00125-013-2869-1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haemmerle G et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312, 734–737, doi: 10.1126/science.1123965 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Zimmermann R et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383–1386, doi: 10.1126/science.1100747 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Vaughan M The production and release of glycerol by adipose tissue incubated in vitro. J Biol Chem 237, 3354–3358 (1962). [PubMed] [Google Scholar]
- 38.Jensen MD, Ekberg K & Landau BR Lipid metabolism during fasting. Am J Physiol Endocrinol Metab 281, E789–793, doi: 10.1152/ajpendo.2001.281.4.E789 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Ballard FJ, Hanson RW & Leveille GA Phosphoenolpyruvate carboxykinase and the synthesis of glyceride-glycerol from pyruvate in adipose tissue. J Biol Chem 242, 2746–2750 (1967). [PubMed] [Google Scholar]
- 40.Reshef L, Hanson RW & Ballard FJ A possible physiological role for glyceroneogenesis in rat adipose tissue. J Biol Chem 245, 5979–5984 (1970). [PubMed] [Google Scholar]
- 41.Gorin E, Tal-Or Z & Shafrir E Glyceroneogenesis in adipose tissue of fasted, diabetic and triamcinolone treated rats. Eur J Biochem 8, 370–375, doi: 10.1111/j.1432-1033.1969.tb00537.x (1969). [DOI] [PubMed] [Google Scholar]
- 42.Elia M, Zed C, Neale G & Livesey G The energy cost of triglyceride-fatty acid recycling in nonobese subjects after an overnight fast and four days of starvation. Metabolism 36, 251–255 (1987). [DOI] [PubMed] [Google Scholar]
- 43.Reshef L et al. Glyceroneogenesis and the triglyceride/fatty acid cycle. J Biol Chem 278, 30413–30416, doi: 10.1074/jbc.R300017200 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Edens NK, Leibel RL & Hirsch J Mechanism of free fatty acid re-esterification in human adipocytes in vitro. J Lipid Res 31, 1423–1431 (1990). [PubMed] [Google Scholar]
- 45.Vaughan M & Steinberg D Effect of Hormones on Lipolysis and Esterification of Free Fatty Acids during Incubation of Adipose Tissue in Vitro. J Lipid Res 4, 193–199 (1963). [PubMed] [Google Scholar]
- 46.Brooks B, Arch JR & Newsholme EA Effects of hormones on the rate of the triacylglycerol/fatty acid substrate cycle in adipocytes and epididymal fat pads. FEBS Lett 146, 327–330, doi: 10.1016/0014-5793(82)80945-9 (1982). [DOI] [PubMed] [Google Scholar]
- 47.Bjorntorp P, Karlsson M & Hovden A Quantitative aspects of lipolysis and reesterification in human adipose tissue in vitro. Acta Med Scand 185, 89–97 (1969). [DOI] [PubMed] [Google Scholar]
- 48.Carnicero HH Changes in the metabolism of long chain fatty acids during adipose differentiation of 3T3 L1 cells. J Biol Chem 259, 3844–3850 (1984). [PubMed] [Google Scholar]
- 49.Yehuda-Shnaidman E, Buehrer B, Pi J, Kumar N & Collins S Acute stimulation of white adipocyte respiration by PKA-induced lipolysis. Diabetes 59, 2474–2483, doi: 10.2337/db10-0245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldstein DS, Eisenhofer G & Kopin IJ Sources and significance of plasma levels of catechols and their metabolites in humans. J Pharmacol Exp Ther 305, 800–811, doi: 10.1124/jpet.103.049270 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Cannon WB & de la Paz D Emotional stimulation of adrenal secretion. American Journal of Physiology 28, 64–70 (1911). [Google Scholar]
- 52.Ahmadian M, Wang Y & Sul HS Lipolysis in adipocytes. Int J Biochem Cell Biol 42, 555–559, doi: 10.1016/j.biocel.2009.12.009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheurink AJ et al. Sympathoadrenal influence on glucose, FFA, and insulin levels in exercising rats. Am J Physiol 256, R161–168, doi: 10.1152/ajpregu.1989.256.1.R161 (1989). [DOI] [PubMed] [Google Scholar]
- 54.Egan JJ et al. Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc Natl Acad Sci U S A 89, 8537–8541, doi: 10.1073/pnas.89.18.8537 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinberg D Hormonal control of lipolysis in adipose tissue. Adv Exp Med Biol 26, 77–88, doi: 10.1007/978-1-4684-7547-0_6 (1972). [DOI] [PubMed] [Google Scholar]
- 56.Garton AJ, Campbell DG, Cohen P & Yeaman SJ Primary structure of the site on bovine hormone-sensitive lipase phosphorylated by cyclic AMP-dependent protein kinase. FEBS Lett 229, 68–72, doi: 10.1016/0014-5793(88)80799-3 (1988). [DOI] [PubMed] [Google Scholar]
- 57.Anthonsen MW, Ronnstrand L, Wernstedt C, Degerman E & Holm C Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. J Biol Chem 273, 215–221, doi: 10.1074/jbc.273.1.215 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Sztalryd C et al. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol 161, 1093–1103, doi: 10.1083/jcb.200210169 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyoshi H et al. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J Biol Chem 281, 15837–15844, doi: 10.1074/jbc.M601097200 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Brasaemle DL Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res 48, 2547–2559, doi: 10.1194/jlr.R700014-JLR200 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Carmen GY & Victor SM Signalling mechanisms regulating lipolysis. Cell Signal 18, 401–408, doi: 10.1016/j.cellsig.2005.08.009 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Mohamed-Ali V et al. beta-Adrenergic regulation of IL-6 release from adipose tissue: in vivo and in vitro studies. J Clin Endocrinol Metab 86, 5864–5869, doi: 10.1210/jcem.86.12.8104 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Tchivileva IE et al. Signaling pathways mediating beta3-adrenergic receptor-induced production of interleukin-6 in adipocytes. Mol Immunol 46, 2256–2266, doi: 10.1016/j.molimm.2009.04.008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin F et al. Noncanonical cAMP pathway and p38 MAPK mediate beta2-adrenergic receptor-induced IL-6 production in neonatal mouse cardiac fibroblasts. J Mol Cell Cardiol 40, 384–393, doi: 10.1016/j.yjmcc.2005.12.005 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Reilly SM et al. A subcutaneous adipose tissue-liver signalling axis controls hepatic gluconeogenesis. Nat Commun 6, 6047, doi: 10.1038/ncomms7047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saini A et al. Interleukin-6 in combination with the interleukin-6 receptor stimulates glucose uptake in resting human skeletal muscle independently of insulin action. Diabetes Obes Metab, doi: 10.1111/dom.12299 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Carey AL et al. Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes 55, 2688–2697, doi: 10.2337/db05-1404 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Stanford KI et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest 123, 215–223, doi: 10.1172/JCI62308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mauer J et al. Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nat Immunol 15, 423–430, doi: 10.1038/ni.2865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Braune J et al. IL-6 Regulates M2 Polarization and Local Proliferation of Adipose Tissue Macrophages in Obesity. J Immunol 198, 2927–2934, doi: 10.4049/jimmunol.1600476 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Kern PA, Ranganathan S, Li C, Wood L & Ranganathan G Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am J Physiol Endocrinol Metab 280, E745–751 (2001). [DOI] [PubMed] [Google Scholar]
- 72.Bastard JP et al. Adipose tissue IL-6 content correlates with resistance to insulin activation of glucose uptake both in vivo and in vitro. J Clin Endocrinol Metab 87, 2084–2089, doi: 10.1210/jcem.87.5.8450 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Pradhan AD, Manson JE, Rifai N, Buring JE & Ridker PM C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. Jama 286, 327–334 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Rotter V, Nagaev I & Smith U Interleukin-6 (IL-6) induces insulin resistance in 3T3-L1 adipocytes and is, like IL-8 and tumor necrosis factor-alpha, overexpressed in human fat cells from insulin-resistant subjects. J Biol Chem 278, 45777–45784, doi: 10.1074/jbc.M301977200 (2003). [DOI] [PubMed] [Google Scholar]
- 75.van Hall G et al. Interleukin-6 stimulates lipolysis and fat oxidation in humans. J Clin Endocrinol Metab 88, 3005–3010, doi: 10.1210/jc.2002-021687 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Path G et al. Human breast adipocytes express interleukin-6 (IL-6) and its receptor system: increased IL-6 production by beta-adrenergic activation and effects of IL-6 on adipocyte function. J Clin Endocrinol Metab 86, 2281–2288, doi: 10.1210/jcem.86.5.7494 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Ji C et al. IL-6 induces lipolysis and mitochondrial dysfunction, but does not affect insulin-mediated glucose transport in 3T3-L1 adipocytes. J Bioenerg Biomembr 43, 367–375, doi: 10.1007/s10863-011-9361-8 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Nahmias C et al. Molecular characterization of the mouse beta 3-adrenergic receptor: relationship with the atypical receptor of adipocytes. EMBO J 10, 3721–3727 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cannon B & Nedergaard J Brown adipose tissue: function and physiological significance. Physiol Rev 84, 277–359, doi: 10.1152/physrev.00015.2003 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Krief S et al. Tissue distribution of beta 3-adrenergic receptor mRNA in man. J Clin Invest 91, 344–349, doi: 10.1172/JCI116191 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guschin D et al. A major role for the protein tyrosine kinase JAK1 in the JAK/STAT signal transduction pathway in response to interleukin-6. EMBO J 14, 1421–1429 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schindler C, Levy DE & Decker T JAK-STAT signaling: from interferons to cytokines. J Biol Chem 282, 20059–20063, doi: 10.1074/jbc.R700016200 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Wegrzyn J et al. Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797, doi: 10.1126/science.1164551 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gough DJ et al. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324, 1713–1716, doi: 10.1126/science.1171721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Q et al. Mitochondrial localized Stat3 promotes breast cancer growth via phosphorylation of serine 727. J Biol Chem 288, 31280–31288, doi: 10.1074/jbc.M113.505057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wen Z, Zhong Z & Darnell JE Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82, 241–250 (1995). [DOI] [PubMed] [Google Scholar]
- 87.Aznar S et al. Simultaneous tyrosine and serine phosphorylation of STAT3 transcription factor is involved in Rho A GTPase oncogenic transformation. Mol Biol Cell 12, 3282–3294, doi: 10.1091/mbc.12.10.3282 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heinrich PC, Behrmann I, Muller-Newen G, Schaper F & Graeve L Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J 334 ( Pt 2), 297–314 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chrencik JE et al. Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6. J Mol Biol 400, 413–433, doi: 10.1016/j.jmb.2010.05.020 (2010). [DOI] [PubMed] [Google Scholar]
- 90.Taura M et al. TLR3 induction by anticancer drugs potentiates poly I:C-induced tumor cell apoptosis. Cancer Sci 101, 1610–1617, doi: 10.1111/j.1349-7006.2010.01567.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chijiwa T et al. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem 265, 5267–5272 (1990). [PubMed] [Google Scholar]
- 92.Liu AM et al. Activation of STAT3 by G alpha(s) distinctively requires protein kinase A, JNK, and phosphatidylinositol 3-kinase. J Biol Chem 281, 35812–35825, doi: 10.1074/jbc.M605288200 (2006). [DOI] [PubMed] [Google Scholar]
- 93.Songyang Z et al. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol 4, 973–982 (1994). [DOI] [PubMed] [Google Scholar]
- 94.Chung J, Uchida E, Grammer TC & Blenis J STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol Cell Biol 17, 6508–6516, doi: 10.1128/mcb.17.11.6508 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lo RK, Cheung H & Wong YH Constitutively active Galpha16 stimulates STAT3 via a c-Src/JAK- and ERK-dependent mechanism. J Biol Chem 278, 52154–52165, doi: 10.1074/jbc.M307299200 (2003). [DOI] [PubMed] [Google Scholar]
- 96.Lo RK & Wong YH Signal transducer and activator of transcription 3 activation by the delta-opioid receptor via Galpha14 involves multiple intermediates. Mol Pharmacol 65, 1427–1439, doi: 10.1124/mol.65.6.1427 (2004). [DOI] [PubMed] [Google Scholar]
- 97.Fung MM, Rohwer F & McGuire KL IL-2 activation of a PI3K-dependent STAT3 serine phosphorylation pathway in primary human T cells. Cell Signal 15, 625–636 (2003). [DOI] [PubMed] [Google Scholar]
- 98.Ng J & Cantrell D STAT3 is a serine kinase target in T lymphocytes. Interleukin 2 and T cell antigen receptor signals converge upon serine 727. J Biol Chem 272, 24542–24549, doi: 10.1074/jbc.272.39.24542 (1997). [DOI] [PubMed] [Google Scholar]
- 99.Su L, Rickert RC & David M Rapid STAT phosphorylation via the B cell receptor. Modulatory role of CD19. J Biol Chem 274, 31770–31774, doi: 10.1074/jbc.274.45.31770 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turkson J et al. Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol Cell Biol 19, 7519–7528, doi: 10.1128/mcb.19.11.7519 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stephens JM, Lumpkin SJ & Fishman JB Activation of signal transducers and activators of transcription 1 and 3 by leukemia inhibitory factor, oncostatin-M, and interferon-gamma in adipocytes. J Biol Chem 273, 31408–31416, doi: 10.1074/jbc.273.47.31408 (1998). [DOI] [PubMed] [Google Scholar]
- 102.Moule SK & Denton RM The activation of p38 MAPK by the beta-adrenergic agonist isoproterenol in rat epididymal fat cells. FEBS Lett 439, 287–290, doi: 10.1016/s0014-5793(98)01392-1 (1998). [DOI] [PubMed] [Google Scholar]
- 103.Cao W, Medvedev AV, Daniel KW & Collins S beta-Adrenergic activation of p38 MAP kinase in adipocytes: cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 MAP kinase. J Biol Chem 276, 27077–27082, doi: 10.1074/jbc.M101049200 (2001). [DOI] [PubMed] [Google Scholar]
- 104.Cuenda A et al. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett 364, 229–233, doi: 10.1016/0014-5793(95)00357-f (1995). [DOI] [PubMed] [Google Scholar]
- 105.Wesselborg S, Bauer MK, Vogt M, Schmitz ML & Schulze-Osthoff K Activation of transcription factor NF-kappaB and p38 mitogen-activated protein kinase is mediated by distinct and separate stress effector pathways. J Biol Chem 272, 12422–12429, doi: 10.1074/jbc.272.19.12422 (1997). [DOI] [PubMed] [Google Scholar]
- 106.Davis RJ & Martin BR The effect of beta-adrenergic agonists on the membrane potential of fat-cell mitochondria in situ. Biochem J 206, 611–618, doi: 10.1042/bj2060611 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang K, Guo W, Yang Y & Wu J JAK2/STAT3 pathway is involved in the early stage of adipogenesis through regulating C/EBPbeta transcription. J Cell Biochem 112, 488–497, doi: 10.1002/jcb.22936 (2011). [DOI] [PubMed] [Google Scholar]
- 108.Wang D et al. Signal transducer and activator of transcription 3 (STAT3) regulates adipocyte differentiation via peroxisome-proliferator-activated receptor gamma (PPARgamma). Biol Cell 102, 1–12, doi: 10.1042/BC20090070 (2010). [DOI] [PubMed] [Google Scholar]
- 109.Richard AJ & Stephens JM The role of JAK-STAT signaling in adipose tissue function. Biochim Biophys Acta 1842, 431–439, doi: 10.1016/j.bbadis.2013.05.030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eguchi J et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab 13, 249–259, doi: 10.1016/j.cmet.2011.02.005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee KY et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 62, 864–874, doi: 10.2337/db12-1089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cernkovich ER, Deng J, Bond MC, Combs TP & Harp JB Adipose-specific disruption of signal transducer and activator of transcription 3 increases body weight and adiposity. Endocrinology 149, 1581–1590, doi: 10.1210/en.2007-1148 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Atgie C, Faintrenie G, Carpene C, Bukowiecki LJ & Geloen A Effects of chronic treatment with noradrenaline or a specific beta3-adrenergic agonist, CL 316 243, on energy expenditure and epididymal adipocyte lipolytic activity in rat. Comp Biochem Physiol A Mol Integr Physiol 119, 629–636 (1998). [DOI] [PubMed] [Google Scholar]
- 114.Weyer C, Tataranni PA, Snitker S, Danforth E Jr. & Ravussin E Increase in insulin action and fat oxidation after treatment with CL 316,243, a highly selective beta3-adrenoceptor agonist in humans. Diabetes 47, 1555–1561, doi: 10.2337/diabetes.47.10.1555 (1998). [DOI] [PubMed] [Google Scholar]
- 115.Fisher MH et al. A selective human beta3 adrenergic receptor agonist increases metabolic rate in rhesus monkeys. J Clin Invest 101, 2387–2393, doi: 10.1172/JCI2496 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carbognin E, Betto RM, Soriano ME, Smith AG & Martello G Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J 35, 618–634, doi: 10.15252/embj.201592629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang W et al. Critical Roles of STAT3 in beta-Adrenergic Functions in the Heart. Circulation 133, 48–61, doi: 10.1161/CIRCULATIONAHA.115.017472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Demaria M et al. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging (Albany NY) 2, 823–842, doi: 10.18632/aging.100232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang T et al. JAK/STAT3-Regulated Fatty Acid beta-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab 27, 136–150 e135, doi: 10.1016/j.cmet.2017.11.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wydysh EA, Medghalchi SM, Vadlamudi A & Townsend CA Design and synthesis of small molecule glycerol 3-phosphate acyltransferase inhibitors. J Med Chem 52, 3317–3327, doi: 10.1021/jm900251a (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Levy DE, Kessler DS, Pine R & Darnell JE Jr. Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev 3, 1362–1371, doi: 10.1101/gad.3.9.1362 (1989). [DOI] [PubMed] [Google Scholar]
- 122.Wang S et al. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2). Arch Biochem Biophys 465, 347–358, doi: 10.1016/j.abb.2007.06.033 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Babaei R et al. Jak-TGFbeta cross-talk links transient adipose tissue inflammation to beige adipogenesis. Sci Signal 11, doi: 10.1126/scisignal.aai7838 (2018). [DOI] [PubMed] [Google Scholar]
- 124.Khatun I et al. Characterization of a Novel Intestinal Glycerol-3-phosphate Acyltransferase Pathway and Its Role in Lipid Homeostasis. J Biol Chem 291, 2602–2615, doi: 10.1074/jbc.M115.683359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kuhajda FP et al. Pharmacological glycerol-3-phosphate acyltransferase inhibition decreases food intake and adiposity and increases insulin sensitivity in diet-induced obesity. Am J Physiol Regul Integr Comp Physiol 301, R116–130, doi: 10.1152/ajpregu.00147.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McFadden JW et al. Increasing fatty acid oxidation remodels the hypothalamic neurometabolome to mitigate stress and inflammation. PLoS One 9, e115642, doi: 10.1371/journal.pone.0115642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Franckhauser S et al. Increased fatty acid re-esterification by PEPCK overexpression in adipose tissue leads to obesity without insulin resistance. Diabetes 51, 624–630 (2002). [DOI] [PubMed] [Google Scholar]
- 128.Macias E, Rao D, Carbajal S, Kiguchi K & DiGiovanni J Stat3 binds to mtDNA and regulates mitochondrial gene expression in keratinocytes. J Invest Dermatol 134, 1971–1980, doi: 10.1038/jid.2014.68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Avalle L et al. STAT3 localizes to the ER, acting as a gatekeeper for ER-mitochondrion Ca(2+) fluxes and apoptotic responses. Cell Death Differ 26, 932–942, doi: 10.1038/s41418-018-0171-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.MacDougald OA Methods in Enzymology. Methods of adipose tissue biology, part B. Preface. Methods Enzymol 538, xv, doi: 10.1016/B978-0-12-800280-3.09987-4 (2014). [DOI] [PubMed] [Google Scholar]
- 131.Divakaruni AS, Paradyse A, Ferrick DA, Murphy AN & Jastroch M Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol 547, 309–354, doi: 10.1016/B978-0-12-801415-8.00016-3 (2014). [DOI] [PubMed] [Google Scholar]
- 132.Rogers GW et al. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One 6, e21746, doi: 10.1371/journal.pone.0021746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Divakaruni AS et al. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proc Natl Acad Sci U S A 110, 5422–5427, doi: 10.1073/pnas.1303360110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Divakaruni AS, Rogers GW & Murphy AN Measuring Mitochondrial Function in Permeabilized Cells Using the Seahorse XF Analyzer or a Clark-Type Oxygen Electrode. Curr Protoc Toxicol 60, 25 22 21–16, doi: 10.1002/0471140856.tx2502s60 (2014). [DOI] [PubMed] [Google Scholar]
- 135.Trapnell C et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578, doi: 10.1038/nprot.2012.016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.