Abstract
Objectives
Emerging evidence suggests the microbiome plays an important role in the pathogenesis of osteoarthritis (OA). We aimed to test the two-hit model of OA pathogenesis and potentiation in which one ‘hit’ is provided by an adverse gut microbiome that activates innate immunity; the other ‘hit’ is underlying joint damage.
Methods
Medical history, fecal and blood samples were collected from human healthy controls (OA-METS-, n=4), knee OA without metabolic syndrome (OA+METS-, n=7) and knee OA with metabolic syndrome (OA+METS+, n=9). Each group of human fecal samples, whose microbial composition was identified by 16S rRNA sequencing, was pooled and transplanted into germ-free mice 2 weeks prior to meniscal ligamentous injury (MLI) (n≥6 per group). Eight weeks after MLI, mice were evaluated for histological OA severity and synovitis, systemic inflammation and gut permeability.
Results
Histological OA severity following MLI was minimal in germ-free mice. Compared with the other groups, transplantation with the OA+METS+ microbiome was associated with higher mean systemic concentrations of inflammatory biomarkers (interleukin-1β, interleukin-6 and macrophage inflammatory protein-1α), higher gut permeability and worse OA severity. A greater abundance of Fusobacterium and Faecalibaterium and lesser abundance of Ruminococcaceae in transplanted mice were consistently correlated with OA severity and systemic biomarkers concentrations.
Conclusion
The study clearly establishes a direct gut microbiome-OA connection that sets the stage for a new means of exploring OA pathogenesis and potentially new OA therapeutics. Alterations of Fusobacterium, Faecalibaterium and Ruminococcaceae suggest a role of these particular microbes in exacerbating OA.
Keywords: Osteoarthritis, germ-free, Gut microbiome, Inflammation, lipopolysaccharide
Introduction
The past decade has witnessed a gradual but fundamental shift in our understanding of the mechanisms underlying OA toward the concept that it is a complex multi-tissue pathology in which low-grade, chronic inflammation has a central role[1]. A number of risk factors playing important roles in OA have been identified, including age[2], obesity[2], metabolic syndrome (MetS)[3], joint trauma[2] and genetic predisposition[2]. Among these factors, MetS has attracted great research interest[4–6]. Schott et al.[7] reported that oligofructose, a nondigestible prebiotic fiber, could restore the gut microbiota of obese OA mice to that of a lean gut microbial profile, suggesting that systemic inflammation in OA is influenced by the gut microbiome. A role of the gut microbiome has been shown for several diseases with inflammatory components including type 2 diabetes[8], non-alcoholic steatohepatitis[9] and cardiovascular diseases[10]. A subtype of metabolic OA has been proposed[11], with some studies proposing that metabolic syndrome in OA is largely driven by BMI[12]. However, the mechanisms underlying metabolic syndrome are still unclear. Although the gut microbiome contributes to activation of low-grade inflammation in MetS conditions[13–15], the link is currently speculative in OA[16].
Our group has proposed a two-hit model of OA pathogenesis in which, as one hit, factors associated with an adverse gut microbiome activate the innate immune response. The other hit results from joint damage that releases damage associated with molecular pattern (DAMP) molecules, such as hyaluronan and fibronectin fragments that result in a synergistic activation of inflammatory pathways and the inflammasome leading to OA[16]. The goal of the current study was to evaluate our two-hit hypothesis model of OA pathogenesis. We hypothesized that an adverse microbiome would exacerbate the histopathological severity of OA induced by joint injury in a murine model. To generate a ‘first-hit’, we transplanted these mice with human fecal samples from healthy controls, or individuals with knee OA without MetS, or knee OA with MetS. To generate a ‘second-hit’, we created a meniscal/ligamentous injury (MLI) in murine knee joints.
Methods
Study cohort, patient characteristics and sample collection
Human blood sample collection and all experimental procedures were approved by the Institutional Review Board of SiChuan University (WCHSIRB-D-2017–111), in accordance with the guidelines provided by the Health Sciences Authority of China. Informed written consent was obtained from participants for this study in accordance with Declaration of Helsinki. MetS was defined according to criteria developed by the International Diabetes Federation (IDF)[17] (see online supplementary Methods). Primary symptomatic knee OA for this study was defined by the American College of Rheumatology criteria[18], together with radiographic severity grade III or IV of the Kellgren/Lawrence scale[19]. Based on these criteria, participants (all female) were classified as: (1) Healthy controls without evidence of OA or MetS (OA-METS-, n=4); (2) OA without MetS (OA+METS-, n=7); (3) OA with MetS (OA+METS+, n=9).
Stool samples were freshly collected from each participant within 15 minutes after excretion, were immediately frozen and stored in liquid nitrogen. Fasting plasma and serum were collected on the day of admission and stored at −80°C for further analysis.
Experimental animals and study design
Germ-free C57BL/6J mice were bred at the Department of Laboratory Animal Science of the Third Military Medical University in Chongqing, China and housed under a 12-h light-dark cycle in the gnotobiotic facilities. All mice were fed with sterile food and water ad libitum, and bacterial contamination was monitored by periodic examination of stools. Age-matched C57BL/J mice were born, raised and maintained in a specific-pathogen-free (SPF) animal facility at the same institution with the same light-dark cycle, food and water. All animal experiments were approved by the SiChuan University Animal Care and Use Committee and followed the recommendations from the Guide for the Care and Use of Laboratory Animals of Chinese Association for Laboratory Animal Science.
Forty-two germ-free mice (8 weeks old) underwent fecal transplantation over 2 weeks with or without MLI surgery 2 weeks later; they were divided into five groups as follows: (1) Germ-free with surgery (SALINE/MLI+): Germ-free mice transplanted with sterile normal saline with meniscal/ligamentous injury (MLI) surgery (3 male and 3 female); (2) Healthy with no surgery (OA-METS-/MLI-): Germ-free mice transplanted with stool samples from healthy controls without MLI surgery (3 male and 3 female); (3) Healthy with surgery (OA-METS-/MLI+): Germ-free mice transplanted with stool samples from healthy controls with MLI surgery (3 male and 3 female); (4) Non-MetS with surgery (OA+METS-/MLI+): Germ-free mice transplanted with stool samples from patients with OA without MetS with MLI surgery (6 male and 6 female); (5) MetS with surgery (OA+METS+/MLI+): Germ-free mice transplanted with stool samples from patients with both OA and MetS with MLI surgery (6 male and 6 female) (Figure 1A). Twelve SPF mice (6 male and 6 female) underwent MLI at the age of 10 weeks to serve as a SPF/MLI+ control group (Figure 1A). As germ-free mice are very sensitive to anesthesia, 1 male mouse in the OA+METS+/MLI+ group died postoperatively. We were unable to retrieve fecal samples from the gut of one male mouse from the OA+METS-/MLI+ group, thus were unable to obtain the 16S rRNA gene sequencing data for this animal. In order to provide a reference, we also evaluated the level of histological knee OA of three age-matched naïve germ-free mice (1 male and 2 females).
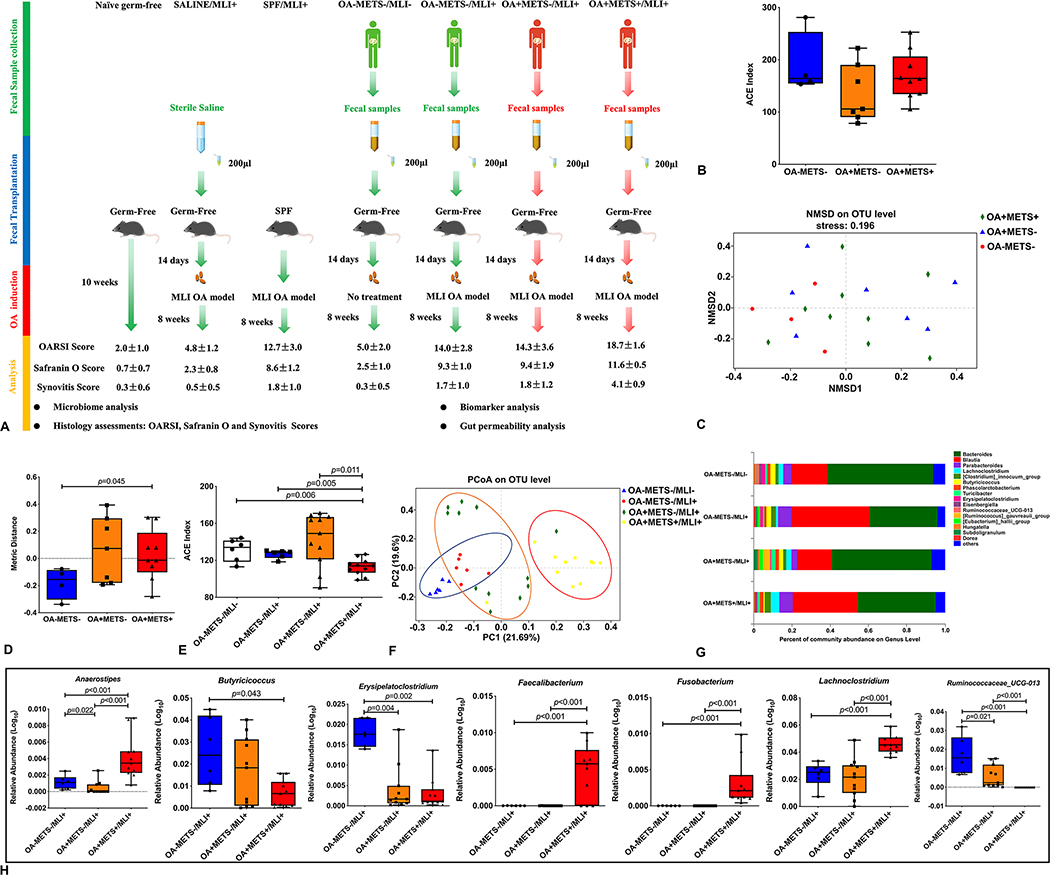
Figure 1. Gut microbiome profiles of the cohort and recipient mice.
(A) Schematic representation of fecal microbiota transplantation. Germ-free mice are orally inoculated with prepared fecal contents from three different groups of people (OA-METS-, OA+METS- and OA+METS+). After the success of gut microbiome colonization, meniscal/ligamentous injury (MLI) was induced in mice from the SALINE/MLI+, SPF/MLI+, OA-METS-/MLI+, OA+METS-/MLI+ and OA+METS+/MLI+ groups. After 8 weeks, the mice were sacrificed for further analysis. (B) Alpha diversity analysis showed no significant differences between the people from three different groups. (C) Beta diversity of fecal microbiome of people from three different groups. (D) None metric multidimensional scaling (NMDS) analysis showed significant differences between Health and OA with MetS groups. (E) Decreased Alpha diversity observed in mice from the OA+METS+/MLI+ group, compared with mice from the SALINE/MLI+, OA-METS-/MLI-, OA-METS-/MLI+ and OA+METS-/MLI+ groups. (F) Beta diversity analysis showed enterotype of mice from the OA-METS-/MLI- and OA-METS-/MLI+ groups was clustered together, enterotype of mice from the OA+METS-/MLI+ group was clustered together while enterotype of mice from the OA+METS+/MLI+ group was clustered together. (G) Community bar-plot analysis showed relative abundance of fecal microbiota in each group at the genera level. (H) Seven kinds of bacteria showed significant differences in the genera level between mice from the OA-METS/MLI+, OA+METS-/MLI+ and OA+METS+/MLI+ groups. Data were compared by the Kruskal-Wallis test (p values were adjusted by the Bonferroni correction). Each bar represents a group (x-axis) with mean and 95% confidence interval (y-axis). (OA=Osteoarthritis; METS=Metabolic Syndrome; MLI=Meniscal/Ligamentous Injury)
Microbiome transplantation
For microbiota transplantation, frozen human fecal samples were pooled from donors in the same group, were pulverized with a mortar and pestle, then resuspended with sterile saline and centrifuged to obtain the supernatant. At 8 weeks of age, germ-free mice were orally inoculated (200 μL for each mouse) twice daily for 2 weeks with the prepared fecal contents from the different groups of donors. Recipient mice transplanted with microbiota were kept in different Trexler-type film isolators, fed with sterile food and water, with bacterial contamination strictly controlled.
MLI surgery
MLI was performed at age 10 weeks after successful fecal transplantation of mice. MLI was induced by transecting the medial collateral ligament (MCL) and detaching the anterior horn of the meniscus in the right knee, as described previously[20]. The animals were sacrificed 8 weeks after MLI with collection of knee joints, blood, small intestine, colon, peritoneal fat tissue and stool samples.
Histologic assessment of OA severity
All right knees were evaluated histologically for cartilage lesions, cartilage proteoglycan content and synovitis (as described in online supplementary Methods) by two evaluators (XJW and CG) blinded to the group assignment (all mice were given a numerical code).
Multivariable regression analysis of predictors of histological outcomes
In order to assess the effects of both sex and body weight on the histological outcomes, we performed multivariable regression analysis with covariates of group, sex and weight (weight at start, weight at 18 weeks or Δweight defined as weight at 18 weeks minus weight at start).
Plasma analysis for inflammatory biomarkers
EDTA plasma samples were collected from all mice. Details on the ELISA assay for inflammatory biomarkers (G-CSF, IL-1β, IL-6, IL-10, IL-17, IP-10, MCP-1 and MIP-1α) and the LPS detection assay are provided in online supplementary Methods.
16S rRNA gene sequencing, bioinformatics and statistical analysis
Total DNA was isolated, amplified and sequenced according to standard procedures[21, 22]. The determination of bacterial species is described in online supplementary Methods.
FISH analysis for bacterial translocation in the gut
Evaluation for bacterial translocation via the gut endothelium was evaluated by fluorescence in situ hybridization (FISH) by modification of a previous protocol[23] (see online supplementary Methods).
Peritoneal fat gene expression
Peritoneal fat from each mouse was harvested and processed for RNAseq analyses as described in online supplementary Methods.
Gene expression of tight junction molecules
Colonic tissue was harvested from mice for mRNA expression of tight junction proteins (ZO-1 and occludin) by RT-PCR (details provided in online supplementary Methods).
Statistical analyses
All statistical analyses were performed using SPSS 22.5 (SPSS, Chicago, IL, USA) unless otherwise specified. Descriptive statistics are presented as means±standard deviation (SD) for continuous variables. Normal distribution was tested using the Kolmogorov-Smirnov test. Normally distributed data from multiple groups were compared by one-way ANOVA and all other data were compared by the Kruskal-Wallis test. When multiple comparisons were fewer than 10, p values were adjusted using the Bonferroni correction. Otherwise p values were adjusted by FDR correction according to the Benjamini-Hochberg procedure. The Shannon index at the genera level was calculated with QIIME (Version 1.7.0). Principle Component Analysis (PCA) was performed using the FactoMineR packages, and clusterSim package in R software (Versuib 2.15.3). PLS-DA was performed using SIMCA-P software to cluster the sample plots across groups. For correlation of inflammatory biomarker levels with OA severity scores, Spearman correlations were used. To test the null hypothesis, we defined the significance level α as 0.05. A P-value < α was considered statistically significant.
Results
Cohort Demographics
For murine transplantation, a total of 20 human female study participants provided fecal samples including 4 healthy controls without knee OA (OA-METS-), 7 with knee OA without MetS (OA+METS-) and 9 with knee OA with MetS (OA+METS+). Baseline characteristics of age and total plasma cholesterol of the 3 groups were similar (Table 1). However, the OA+METS+ group had significantly higher mean values for body mass index (BMI), low-density lipoprotein cholesterol (LDL), and TG level (Table 1) and significantly lower mean HDL level (Table 1). Metabolic syndrome criteria for each donor are provided in online supplementary Table S1.
Table 1.
Human donor demographics
| Demographics | Healthy (n=4) | OA without MetS (n=7) | OA with MetS (n=9) | p value |
|---|---|---|---|---|
| Gender (F/M) | 4/0 | 7/0 | 9/0 | 1 |
| Age (y) | 64.3±9.9 | 61.7±6.2 | 68.8±8.1 | 0.244 |
| BMI (kg/m2) | 22.19±1.37 | 21.61±2.50 | 29.58±2.82 | <0.001 |
| Waist Circumference (cm) | 72.2±1.7 | 73.1±3.2 | 83.9±3.0 | <0.001 |
| LDL (mmol/L) | 2.86±0.79 | 2.89±0.56 | 3.83±0.67 | 0.017 |
| HDL (mmol/L) | 1.96±0.41 | 1.86±0.64 | 1.28±0.42 | 0.034 |
| TG (mmol/L) | 1.12±0.32 | 0.81±0.44 | 1.85±0.85 | 0.015 |
| CHOL (mmol/L) | 4.57±0.94 | 4.89±0.81 | 4.12±1.94 | 0.588 |
F=Female; M=Male; BMI=Body Mass Index; LDL=Low-density Lipoprotein; HDL=High-density Lipoprotein; TG=Triglyceride; CHOL=Cholesterol; OA=Osteoarthritis; MetS=Metabolic Syndrome
Gut microbiome profiles of the cohort and recipient mice
By 16S rRNA gene sequencing, there were no significant differences of alpha diversity among the three groups of donor microbiomes (Figure 1B). By non-metric multidimensional scaling (NMDS) analysis, there was a significantly different beta diversity of the fecal microbiomes of the three groups (p=0.045) (Figure 1C&D). The alpha diversity at a genus level was much lower in the germ-free mice transplanted with fecal matter from the OA+METS+/MLI+ group (Figure 1E).) PCA using Jensen-Shannon distance was performed to cluster the 34 samples into three distinct enterotypes (Figure 1F). At the genus level, a total of 140 genera were identified among all samples; the five dominant genera were Bacteroides, Blautia, Parabacteroides, Lachnoclostridium and Turicibacter (Figure 1G). At the genus level, the relative abundance of ten bacteria was significantly different in the fecal samples collected from mice of the healthy with surgery (OA-METS-/MLI+), Non-MetS with surgery (OA+METS-/MLI-) and MetS with surgery (OA+METS+/MLI+) groups. Seven of the bacterial genera, including Anaerostipes, Butyricicoccus, Erysipelatoclostridium, Faecalibacterium, Fusobacterium, Lachnoclostridium and Ruminococcaceae_UCG-013, were significantly different between mice from the OA-METS-/MLI+ and OA+METS+/MLI+ groups (Figure 1H), others are shown in online supplementary Figure S1.
Highest histological joint damage observed in the OA+METS+/MLI+ group
For ease of comparison, histological scores (mean, SD) for each group are presented in Figure 1A; they are also depicted in bar graph form in Figure 2B,C&E, with p values presenting the differences across all main groups (data for naïve germ-free mice provided for reference only).
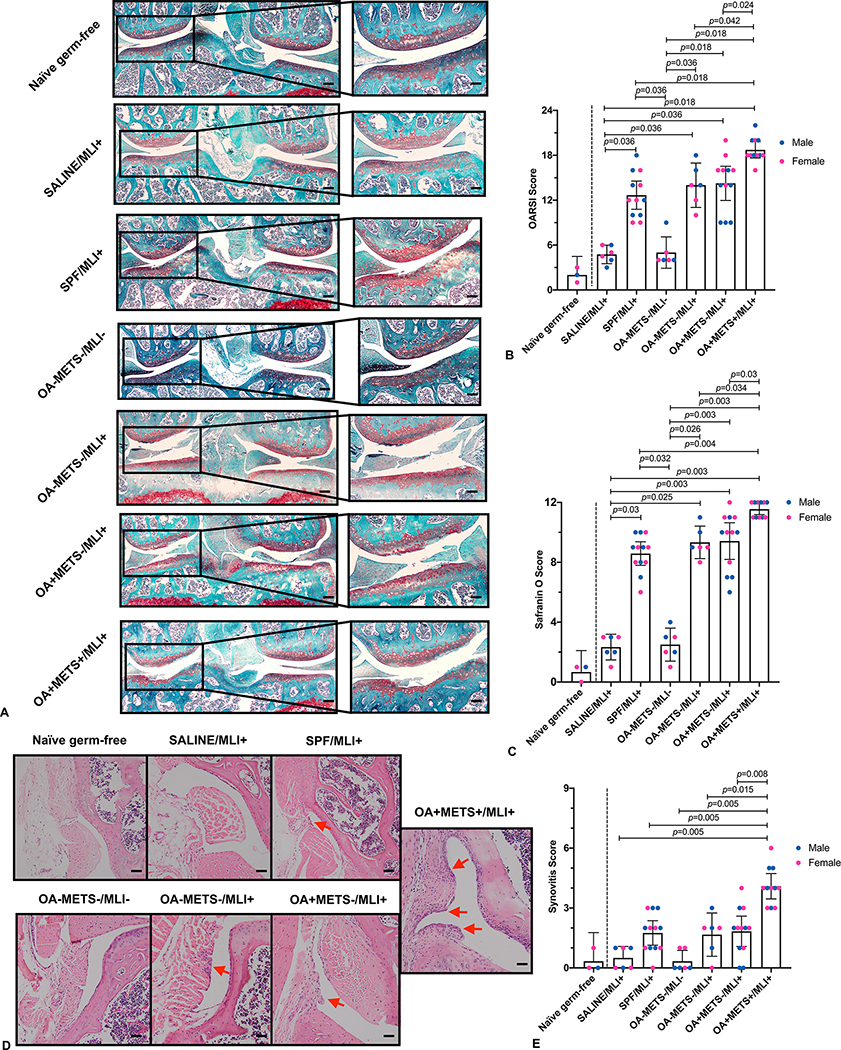
Figure 2. Histology assessments of osteoarthritis severity and synovitis 8 weeks after the surgery.
(A) Representative images of safranin O/fast green section of the mice knee joint from different groups (Scale bar in the left panels, 200μm; Scale bar in the right panels, 50μm). (B) Comparison of OARSI scores between the groups. (C) Comparison of Safranin O scores between the groups. (D) Representative images of H&E section showing synovitis in mice from different groups (arrows show synovitis) (Scale bar, 50μm). (E) Comparison of synovitis scores between the groups. With the exception of the naïve germ-free mice (provided for reference only as indicated by a vertical dashed line to the right of this group in panels B, C & E), data for all other groups were compared by the Kruskal-Wallis test, multiple comparison p values were adjusted by the Benjamini-Hochberg procedure. Each bar represents a group (x-axis) with mean and 95% confidence interval (y-axis). Green dotted lines are references for mean respective histological scores of naïve germ-free mice. (OARSI=Osteoarthritis Research Society International)
Minimal signs of OA and synovitis were found in the age-matched naïve germ-free mice (OARSI score: 2.0±1.0; Safranin O score: 0.7±0.7; Synovitis score: 0.3±0.6) (Figure 2B,C&E). Mean OARSI cartilage damage scores were significantly higher in the OA+METS+/MLI+ group (18.7±1.6) in comparison with the other groups. Mice from the OA+METS-/MLI+ (14.3±3.6), OA-METS-/MLI+ (14.0±2.8) and SPF/MLI+ (12.7±3.0) groups had significantly higher mean OARSI scores than mice from the SALINE/MLI+ (4.8±1.2) and OA-METS-/MLI- (5.0±2.0) groups. There were no significant differences between SALINE/MLI+ and OA-METS-/MLI- groups. Also, no significant differences were found among SPF/MLI+, OA-METS-/MLI+ and OA+METS-/MLI+ groups (Figure 2A&B). Similar results were observed for cartilage safranin O scores indicative of proteoglycan content; the mice from the OA+METS+/MLI+ group had a significantly higher mean score (11.6±0.5) compared with those from SALINE/MLI+ (2.3±0.8, p=0.003), SPF/MLI+ (8.6±1.2, p=0.004), OA-METS-/MLI- (2.5±1.0, p=0.003), OA-METS-/MLI+ (9.3±1.0, p=0.034) and OA+METS-/MLI+ (9.4±1.9, p=0.03) groups (Figure 2A&C).
The mean synovitis score of the OA+METS+/MLI+ group (4.1±0.9) was significantly higher than the mean scores of the SALINE/MLI+ (0.5±0.5, p=0.005), SPF/MLI+ (1.8±1.0, p=0.005), OA-METS-/MLI- (0.3±0.5, p=0.005), OA-METS-/MLI+ (1.7±1.0, p=0.015) and OA+METS-/MLI+ (1.8±1.2, p=0.008) groups (Figure 2D&E). The mean synovitis scores of these latter groups, (SALINE/MLI+, OA-METS-/MLI-, OA-METS-/MLI+ and OA+METS-/MLI+), did not differ significantly.
Multivariable regression analysis of histological outcomes
There were no significant differences in weight at 8 weeks (study start), weight at 18 weeks or Δweight among the six groups (online supplementary Table S2 & Figure S2). By multivariable regression analyses, group was strongly associated with OARSI, Safranin O and synovitis scores. However, sex, weight at start, weight at 18 weeks and Δweight were not associated with the histological outcomes (online supplementary Table S3).
Inflammatory biomarkers analysis
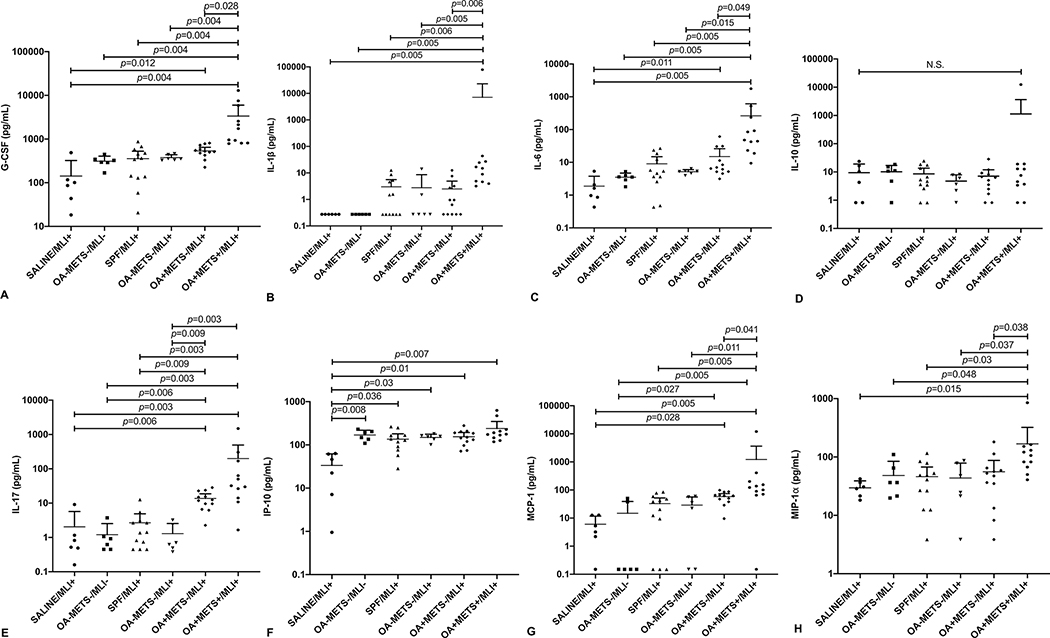
OA+METS+/MLI+ had the highest mean plasma G-CSF, IL-1β, IL-6, IL-10, IL-17, IP-10, MCP-1, MIP-1α. After FDR correction, compared with the other five groups, the OA+METS+/MLI+ group had statistically significant higher mean plasma IL-1β, IL-6 and MIP-1α (Figure 3) (online supplementary Table S2). Mean IP-10 was significantly lower in the SALINE/MLI+ group compared with the other five groups (Figure 3) (online supplementary Table S2). There were no significant differences of mean plasma IL-10 or lipopolysaccharide binding protein (LBP) among the six groups.
Figure 3. Inflammatory biomarkers analysis.
(A) G-CSF. (B) IL-1β. (C) IL-6. (D) IL-10. (E) IL-17. (F) IP-10. (G) MCP-1. (H) MIP-1α. Data were compared by the Kruskal-Wallis test, multiple comparison p values were adjusted by the Benjamini-Hochberg procedure. Each bar represents a group (x-axis) with mean and 95% confidence interval (y-axis). (G-CSF=Granulocyte-colony stimulating factor; IL-1β=Interleukin 1 beta; IL-6=Interleukin 6; IL-10=Interleukin 10; IL-17=Interleukin 17; IP-10=Interferon Gamma-Induced Protein 10; MCP-1=Monocyte Chemoattractant Protein 1; MIP-1α=Macrophage Inflammatory Protein 1α)
Gut permeability
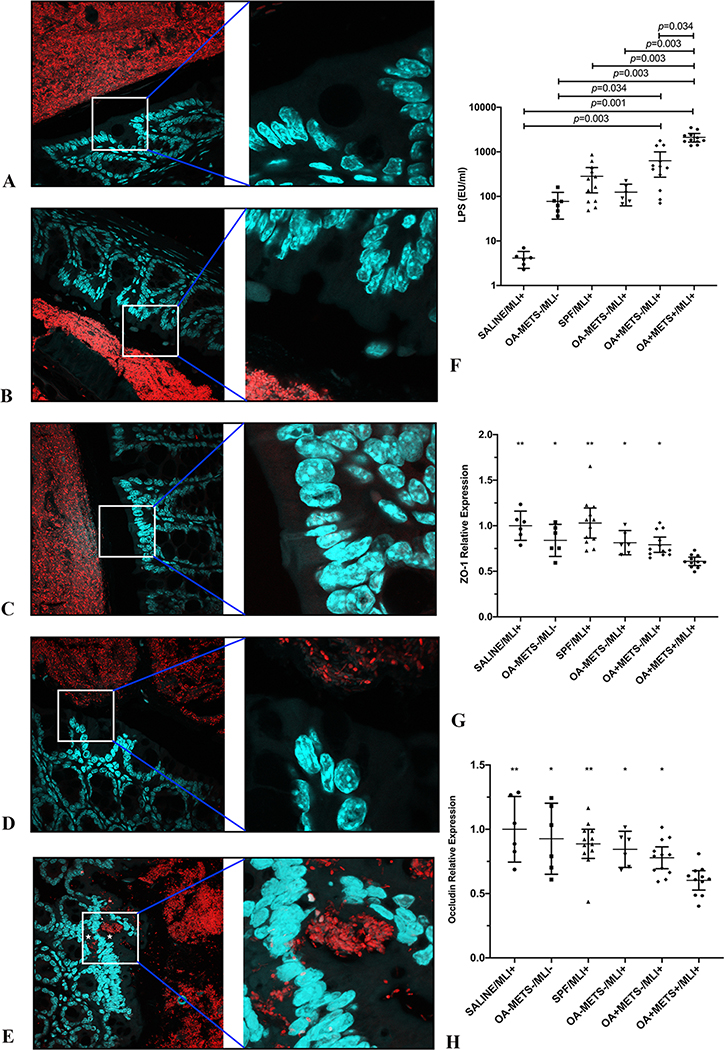
According to the two-hit hypothesis, MetS can increase gut permeability, causing systematic low-grade inflammation. In the current study, we examined gut permeability by mRNA expression of intestinal tight junction proteins (ZO-1 and occludin) in colon tissue, plasma LPS level and bacterial translocation by fluorescence FISH analyses of the gut endothelium. As expected, significantly lower mRNA levels of ZO-1 and occludin were detected in the OA+METS+/MLI+ group compared with the other five groups (Figure 4G&H). Also, OA+METS+/MLI+ group had significantly higher mean plasma LPS level compared to the other five groups (Figure 4F) with evidence of bacterial translocation via the gut endothelium confirmed by FISH (Figure 4A to E).
Figure 4. Germ-free mice transplanted with fecal samples from patients of OA with metabolic syndrome showed increased gut permeability.
(A-E) Representative images of gut barrier visualized by FISH (16S rRNA genes of all bacteria [red] and nuclei [green]) (left panels original magnification: ×60; right panels original magnification: ×240). Bacteria invasion into colonic mucosa was captured in mice from the OA+METS+/MLI+ group (E) (marked by asterisk). No signs of bacteria invasion were observed in mice from the SPF/MLI+ (A), OA-METS-/MLI- (B), OA-METS-/MLI+ (C), OA+METS-/MLI+ (D) groups. (F) Significantly increased systemic plasma LPS levels were observed in mice from the OA+METS+/MLI+ group. (G) Significantly decreased mRNA level of tight junction protein ZO-1 was observed in mice from the OA+METS+/MLI+ group, compared with mice from the SALINE/MLI+ (p<0.01), SPF/MLI+ (p<0.01), OA-METS-/MLI- (p=0.036), OA-METS-/MLI+ (p=0.036) and OA+METS-/MLI+ (p=0.036) groups. (H) Significantly increased mRNA level of tight junction protein Occludin was preserved in mice from the SALINE/MLI+ (p<0.01), SPF/MLI+ (p<0.01), OA-METS-/MLI- (p=0.02), OA-METS-/MLI+ (p=0.034) and OA+METS-/MLI+ (p=0.048) groups, compared with those from the OA+METS+/MLI+ group. ** stands for p<0.01, * stands for 0.01≤p<0.05. Data were compared by the Kruskal-Wallis test, multiple comparison p values were adjusted by the Benjamini-Hochberg procedure. Each bar represents a group (x-axis) with mean and 95% confidence interval (y-axis). (OA=Osteoarthritis; METS=Metabolic Syndrome; MLI=Meniscal/Ligamentous Injury)
Differential expression and gene set enrichment analyses of peritoneal fat tissue
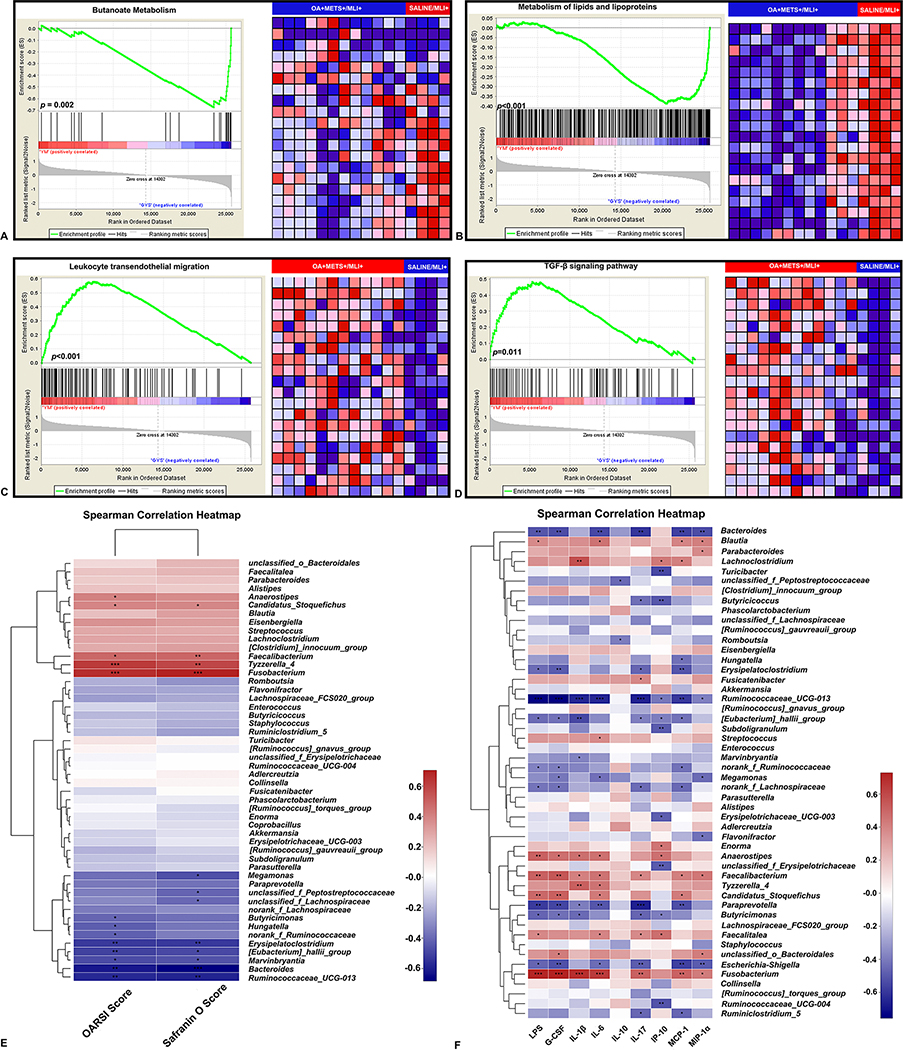
We identified 171 differentially expressed genes (DEGs) in peritoneal fat tissue between the OA+METS+/MLI+ and SALINE/MLI+ groups (see online supplementary Table S4); 101 DEGs were upregulated and 70 DEGs downregulated in the OA+METS+/MLI+ group. To obtain a deeper insight into the function of the DEGs, gene set enrichment analysis (GSEA) was used. DEGs between the two groups were highly enriched in pathways related to butanoate metabolism (highly activated in the SALINE/MLI+ group, p=0.002, Normalized Enrichment Score (NES)=−2.12, FDR q=0.010), lipids and lipoproteins metabolism (highly activated in the SALINE/MLI+ group, p<0.001, NES=−1.94, FDR q=0.029), leukocyte transendothelial migration (highly activated in the OA+METS+/MLI+ group, p<0.001, NES=1.88, FDR q=0.231) and the TGF-β signaling pathway (highly activated in the OA+METS+/MLI+ group, p=0.011, NES=1.76, FDR q=0.165) (Figure 5A to D).
Figure 5.
Gene set enrichment analysis (GSEA) of the RNA sequencing data of peritoneal fat tissue between OA+METS+/MLI+ VS. SALINE/MLI+ mice (A-D). (A) GSEA for butanoate metabolism. (B) GSEA for metabolism of lipids and lipoproteins. (C) GSEA for leukocyte transendothelial migration. (D) GSEA for TGF-β signaling pathway. The enrichment plot on the left of each panel shows the distribution of genes in the set that is correlated with the SALINE/MLI+ or OA+METS+/MLI+ groups. The heatmap on the right of each panel shows where gene expression is relatively high (red) or low (blue) for each gene in the indicated sample. Correlation between gut bacterial genera, cartilage histology scores and inflammatory biomarkers (E-F). (E) Correlations between abundance of bacterial genera and cartilage histology scores. (F) Correlations between the abundance of bacterial genera and inflammatory biomarkers. ** stands for p<0.01, * stands for 0.01≤p<0.05. Positive correlation-red, negative correlation-green. Spearman correlations were performed. (OA=Osteoarthritis; METS=Metabolic Syndrome; MLI=Meniscal/Ligamentous Injury)
Correlations between gut bacterial genera, cartilage histology scores and inflammatory biomarkers
OARSI scores reflecting overall severity of knee OA were significantly positively correlated with the abundance of 5 different gut bacterial genera and significantly negatively correlated with 8 other gut bacterial genera (Figure 5E) (online supplementary Table S5). Safranin O scores reflecting cartilage proteoglycan loss were significantly positively correlated with 4 different gut bacterial genera and significantly negatively correlated with 8 other gut bacterial genera (Figure 5E). Specifically, Candidatus_Stoquefichus, Faecalibacterium, Tyzzerlla_4 and Fusobacterium were consistently significantly positively correlated with cartilage histology scores (higher scores worse) while Erysipelatoclostridium, Eubacterium, Marvinbryantia, Bacteroides and Ruminococcaceae_UCG-013 were consistently significantly negatively correlated with cartilage histology scores.
Plasma concentrations of IL-1β, IL-6 and MIP-1α were consistently significantly positively correlated with the abundance of Fusobacterium and Faecalibacterium and negatively correlated with the abundance of Ruminococcaceae_UCG-013 (Figure 5F) (online supplementary Table S6). Thus, these three gut bacterial genera, Fusobacterium, Faecalibacterium and Ruminococcaceae_UCG-013 showed congruent correlations with both inflammatory biomarkers and cartilage histology scores.
Discussion
We previously hypothesized that the gut microbiome plays a critical role in the two-hit model of MetS related OA pathogenesis[16]. Our study results support this hypothesis showing a noticeable impact of the human gut microbiome on injury induced OA severity in a mouse model system. The MetS+ mice developed increased gut permeability (by colonic epithelial FISH analyses) with endotoxemia (elevated LPS) in association with upregulation of circulating pro-inflammatory molecules consistent with an activated innate immune system. Cani et al.[24] first showed that endotoxemia triggers the expression of inflammatory factors and functions as a stimulus of metabolic syndrome. Several of the plasma cytokines elevated in the MetS+ mice in our study, IL-1β, IL-6 and MIP-1α, are considered key mediators of OA in patients[25–27]. Emerging evidence suggests that IL-1β[28, 29], IL-6[30, 31] and MIP-1α[32] play important roles in lipid metabolism and persistence of low-grade inflammation. In the context of joint injury, the activated innate immune system would be expected to exacerbate the pathological process of OA. Despite joint injury, the germ-free mice (SALINE/MLI+) in our study had signs of mild OA that were reduced compared to the SPF/MLI+ group (62% reduction in OARSI score and 73% reduction in Safranin O score) and the OA-MetS-/MLI+ group (66% reduction in OARSI score and 75% reduction in Safranin O score). This is consistent with Ulici et al.[33] who demonstrated that germ-free DMM+ mice had reduced OA compared to SPF-DMM mice (47% reduction in sum articular cartilage structure score and 42% reduction in sum Safranin O score). Taken together, we show for the first time, to our knowledge, a direct gut microbiome-OA connection whereby the microbiome from different groups of people could change the pathological process of OA induced by surgery in mice.
In order to evaluate the metabolic status of our mice, we also performed RNA sequencing of the peritoneal fat tissue. The GSEA analysis revealed highly activated butanoate (or butyrate), lipid and lipoprotein metabolism pathways in the germ-free SALINE/MLI+ group. Butanoate or butyrate is a group of short chain fatty acids produced through fermentation of dietary fibers in the lower intestinal tract[34]. Colonocytes utilize bacterially-produced butyrate as their primary energy source; butyrate prevents deficits in mitochondrial respiration and autophagy[35]. The expression of GPR109, a butyrate receptor, and SLC5A8, a butyrate transporter, are markedly reduced in the ileum and colon of germ-free mice[36]. Thus, a germ-free mouse devoid of a gut microbiome producing butyrate could increase the capacity of the gut endothelial cells to oxidize n-butyrate, as observed in this study as a form of rescue or countermeasure to butyrate scarcity (supported by Cherbuy et al. [37]). The concurrent elevation of lipid and lipoprotein metabolism in germ-free mice[38, 39], provide one explanation for the resistance of germ-free C57BL/6J mice to high-fat-diet induced obesity[38].
Moreover, the GSEA analysis revealed highly activate leukocyte transendothelial migration and TGF-β signaling pathways in the OA+/METS+/MLI+ group. This finding might reflect elevated low-grade inflammation, which is consistent with our biomarker findings and endotoxemia. TGF-β can regulate multiple types of immune cells including T cells, B cells, macrophage, dendritic cells and natural killer cells[40]. Additionally, TGF-β signaling is highly activated during tissue repair processes such as OA[41, 42]. Thus, the highly activated TGF-β signaling found in the OA+METS+/MLI+ group might represent not only elevated low-grade inflammation but also the highly activated tissue repair processes.
In the current study, although we did not find significant differences of alpha diversity among the three human donor groups, the mice gut microbiome profile differed significantly in alpha diversity following transplantation. As shown by Turnbaugh et al.[43], only 77% of the human gut microbiome can colonize germ-free mice, likely related to differences in diet, environment and small-intestine: colon length ratio. Though humans and mice share more than 60% of their gut microbiome genera, certain genera, such as Mitsuokella, Megasohera and Dialister, are exclusive to humans[44] and therefore unlikely to be able to colonize germ-free mice.
To identify the bacteria responsible for aggravating OA induced by MLI, we evaluated the correlation of gut bacterial genera, OA histologic severity and inflammatory biomarkers. The abundance of Fusobacterium and Faecalibacterium (both increased in abundance in OA+METS+/MLI+) was consistently positively correlated while Ruminococcaceae (decreased in abundance in OA+METS+/MLI+) was consistently negatively correlated with OARSI scores, Safranin O scores, IL-1β, IL-6 and MIP-1α. Interestingly, Fusobacterium has been associated with abnormal metabolic status, with enrichment in obese Japanese[45] and Chinese[46] populations, in patients with MetS[47] and sows (pigs) with MetS[48]. One of the most famous and notorious members from Fusobacterium genus, Fusobacterium nucleatum, has been associated with the occurrence of multiple conditions including inflammatory bowel disease[49], colorectal cancer[50] and rheumatoid arthritis[51]. In contrast, Faecalibacterium, especially Faecalibacterium prausnitzii, is decreased in many chronic pathological conditions and generally considered a good organism with versatile metabolic abilities[52]. However, we observed a positive correlation of abundance of Faecalibacterium with OA histological severity and inflammatory biomarkers. Our results are supported by van Dijkhuizen et al. who demonstrated significantly greater abundance of Faecalibacterium in patients with juvenile idiopathic arthritis compared with controls[53]. There are several potential reasons underlying these differences in results. We only used 16S rRNA sequencing to identify the genus of different bacteria; this might not specifically identify the species. Faecalibacterium prausnitzii is the only species isolated from this genus till now[54]; we still know nothing about the biological behavior of the other members of the Faecalibacterium family. Even among the Faecalibacterium prausnitzii species, there are different subspecies; differing proportions of these subspecies could result in differences in metabolite production such as short-chain fatty acid production. Second, the positive correlation between Faecalibacterium abundance, OA histological severity and inflammatory biomarkers may be an effect rather than a cause of inflammation in the donors.
Ruminococcaceae was protective in our model system and reduced in abundance in the OA+METS+/MLI+ group. Decreased abundance of Ruminococcaceae has been found in patients with MetS[55] and non-alcoholic steatohepatitis[56]. Ruminococcaceae can mediate the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by a high-fat diet[57]. Its protective effect against low-grade inflammation was attributed to its production of butyrate, serving as a primary energy source for colonic intestinal bacteria and a beneficial factor for gut health by causing decreased intestinal permeability[58].
Recently, Boer et al.[59] found in the Rotterdam Study that Streptococcus species are associated with increased knee pain and inflammation. In the current study, compared with the other groups, the relative abundance of Streptococcus was increased in both the OA+MetS+ humans and OA+MetS+/MLI+ mice (online supplementary Figure S3). However, the differences in our donors were not statistically significantly different (p=0.42) and of borderline significance in mice (p=0.053). The gut microbiome is affected by many factors including diet, ethnicity and lifestyle[60]. The population of the Rotterdam Study was primarily Caucasian of European ancestry; in contrast, our human microbiome donors were all Han Chinese. This might in part explain why we did not find similar results. Study power may also have contributed to this difference since the Rotterdam study included n=1427 (with replication in n=867) participants; given the trends in our OA+/MetS+ humans and OA+MetS+/ML+ mice, with larger sample sizes we may have been able to detect an association of Streptococcal species with OA outcomes.
There are several limitations to this study. First, the sample size of the human study was relatively small and all donors were Chinese. Therefore, our results may only reflect the specific population tested. Second, we did not include an OA-MetS+ group thus, we could not evaluate the single effect of MetS on OA. Third, some studies have shown that sex has effects in surgical mouse models[61]. Although we used both female and male germ-free mice in the current study, we used only female human donors; thus, we cannot rule out the effect of sex on our study. However, the multivariable regression showed that sex effects on the histological outcomes in the current study were not statistically significant. Fourth, because of the constrained spaces of the germ-free containers during sacrifice, we could not collect other tissues, including synovial fluid or intrapatellar fat pad to explore their differences between groups. However, according to Barboza et al.[5], the intrapatellar fat pad is not the initiating factor of metabolic induced knee OA nor does it well represent the metabolic status locally in the joint. Fifth, due to the limited resolution of the 16S rRNA sequencing methodology, we were unable to identify specific species or strains of Fusobacterium, Faecalibacterium or Ruminococcaceae that could be driving the correlations between OA histologic severity and inflammatory biomarkers. Sixth, our results support the Koch’s second postulate of infectious disease in this ‘non-communicable’ disease[62] and cannot unequivocally establish the causality between these bacteria and the occurrence of OA. To prove Koch’s first and third postulates, the OA-gut microbiome connections need to be evaluated in more people with OA and the suspect microbes need to be transplanted into germ-free mice to understand how they are associated with disease in humans. Seventh, though the metabolic and inflammatory status changed in the mice after the MetS fecal sample transplantation, we did not observe any differences in body weight (weight at 8 weeks (study start), weight at 18 weeks or Δweight) or their association with histological outcomes. Previously, Ussar et al.[63] studying the interactions between the gut microbiome, host genetics, diet and obesity, found that all three elements were necessary for weight change. In the current study, we used only a normal diet in all groups, and sacrificed mice 8 weeks after confirmed colonization. Parseus et al.[64] showed that weight differences caused by different gut microbiome colonization, even fed with a high fat diet, would take as long as 10 weeks. Finally, the goal of our study was to evaluate the effects of human microbiota on trauma induced OA in the mouse. We therefore cannot speculate on the effects of the mouse microbiome on OA in this study. We also believe these results merit further analysis of the role of key microbiota in human OA.
In summary, we demonstrated that (1) OA severity induced by MLI surgery was reduced in germ-free mice; (2) Fecal transplantation from MetS patients to germ-free mice could increase gut permeability causing endotoxemia, systemic low-grade inflammation and aggravate severity of OA induced by MLI surgery; (3) The abundances of the Fusobacterium, Faecalibacterium and Ruminococcacea bacterial genera were consistently associated with both histological OA severity and inflammatory biomarkers, suggesting they might be the bacterial agents linked to the MetS that are responsible for aggravating OA severity in germ-free mice. These results support our two-hit hypothesis of OA involving the gut microbiome. Because the gut microbiome is capable of change through interventions such as diet and exercise, it could be a promising therapeutic target in OA. Future studies are needed to explore the causal mechanism and potential translation of the present findings into clinical practice.
Supplementary Material
Supplementary Figure S1. The other three kinds of bacteria showed significant differences in the genera level between mice from OA-METS-/MLI+, OA+METS-/MLI+ and OA+METS+/MLI+ groups. (A) Eisenbergiella; (B) Parabacteroides; (C) [Ruminococcus]_gauvreauii_group. (OA=Osteoarthritis; METS=Metabolic Syndrome; MLI=Meniscal/Ligamentous Injury)
Supplementary Figure S2. Weight of the mice in different groups. (A) Weight at 8 weeks; (B) Weight at 18 weeks; (C) ΔWeight (defined as weight at 18 weeks – weight at 8 weeks).
Supplementary Figure S3. Relative abundance of Streptococcus in the gut microbiome of the human donors and mouse recipients. (A) Relative abundance of Streptococcus in the gut microbiome of human donors n≥4/group; (B) Relative abundance of Streptococcus in the gut microbiome of the mouse microbiome recipients n≥6/group.
Key messages.
What is already known about this subject?
The gut-microbiome plays an important role in the pathogenesis of many different conditions, including osteoarthritis (OA). Germ-free mice have reduced susceptibility to OA following destabilization of the medial meniscus (DMM), although the mechanism is not clear.
What does this study add?
We demonstrated (1) transplanting fecal samples from metabolic syndrome (MetS) patients to germ-free mice increased gut permeability, endotoxemia and systemic low-grade inflammation and lead to aggravated severity of OA induced by MLI surgery. (2) The abundances of the Fusobacterium, Faecalibacterium and Ruminococcacea bacterial genera were consistently associated with both histological OA severity and inflammatory biomarkers, suggesting they might be some of the bacterial agents linked to the MetS capable of aggravating OA severity in germ-free mice and potentially their human donors.
How might this impact on clinical practice or future developments?
These results support treatments aimed at beneficially modifying the intestinal microbiome such as a high-fiber diet, weight loss, exercise, unabsorbable antibiotic, and as a last resort, microbiota transplantation.
Acknowledgements
Dr. ZYH would like to thank Li Li, Fei Chen and Chunjuan Bao from Laboratory of Pathology, West China Hospital, SiChuan University for helping with processing the histological sections and staining. Dr. ZYH would like to thank Dr. Ke Xiao from Department of Orthopaedic Surgery, West China Hospital for helping with drawing the schematic diagram in Figure 1A. Dr. ZYH would like to thank Dr. John Martin from Department of Orthopaedic Surgery, Duke University for his previous suggestions on the manuscript revision.
Funding
This work was supported by grants from the NSFC to ZYH (NSFC: 81702185; NSFC: 81972097), LC (NSFC: 81870759) and XDZ (NSFC: 81430011). This research was also supported by SiChuan Science and Technology Program to ZYH (No. 2018HH0071) and LC (No. 2017JQ0028) and a Claude D. Pepper Older Americans Independence Centers grant (5P30 AG028716 from NIA to VBK).
Footnotes
Competing interests
None declared.
Patient and public involvement statement
This research was done without direct patient involvement beyond sample collection. Patients did not participate in designing the study, analyzing the data, or drafting the manuscript.
Patient consent for publication
Not required.
Data availability statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the online supplementary Materials. Additional data related to this paper may be requested from the authors.
References
- 1.Robinson WH, Lepus CM, Wang Q, Raghu H, Mao R, Lindstrom TM, et al. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2016. October; 12(10):580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson VL, Hunter DJ. The epidemiology of osteoarthritis. Best Pract Res Clin Rheumatol. 2014. February; 28(1):5–15. [DOI] [PubMed] [Google Scholar]
- 3.Yoshimura N, Muraki S, Oka H, Tanaka S, Kawaguchi H, Nakamura K, et al. Accumulation of metabolic risk factors such as overweight, hypertension, dyslipidaemia, and impaired glucose tolerance raises the risk of occurrence and progression of knee osteoarthritis: a 3-year follow-up of the ROAD study. Osteoarthritis Cartilage. 2012. November; 20(11):1217–1226. [DOI] [PubMed] [Google Scholar]
- 4.Collins KH, Herzog W, MacDonald GZ, Reimer RA, Rios JL, Smith IC, et al. Obesity, Metabolic Syndrome, and Musculoskeletal Disease: Common Inflammatory Pathways Suggest a Central Role for Loss of Muscle Integrity. Front Physiol. 2018; 9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barboza E, Hudson J, Chang WP, Kovats S, Towner RA, Silasi-Mansat R, et al. Profibrotic Infrapatellar Fat Pad Remodeling Without M1 Macrophage Polarization Precedes Knee Osteoarthritis in Mice With Diet-Induced Obesity. Arthritis Rheumatol. 2017. June; 69(6):1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin TM, Huebner JL, Kraus VB, Yan Z, Guilak F. Induction of osteoarthritis and metabolic inflammation by a very high-fat diet in mice: effects of short-term exercise. Arthritis Rheum. 2012. February; 64(2):443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schott EM, Farnsworth CW, Grier A, Lillis JA, Soniwala S, Dadourian GH, et al. Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI Insight. 2018. April 19; 3(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creely SJ, McTernan PG, Kusminski CM, Fisher f M, Da Silva NF, Khanolkar M, et al. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007. March; 292(3):E740–747. [DOI] [PubMed] [Google Scholar]
- 9.Brun P, Castagliuolo I, Di Leo V, Buda A, Pinzani M, Palu G, et al. Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2007. February; 292(2):G518–525. [DOI] [PubMed] [Google Scholar]
- 10.Ahmadmehrabi S, Tang WHW. Gut microbiome and its role in cardiovascular diseases. Curr Opin Cardiol. 2017. November; 32(6):761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuo Q, Yang W, Chen J, Wang Y. Metabolic syndrome meets osteoarthritis. Nat Rev Rheumatol. 2012. December; 8(12):729–737. [DOI] [PubMed] [Google Scholar]
- 12.Niu J, Clancy M, Aliabadi P, Vasan R, Felson DT. Metabolic Syndrome, Its Components, and Knee Osteoarthritis: The Framingham Osteoarthritis Study. Arthritis Rheumatol. 2017. June; 69(6):1194–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pascale A, Marchesi N, Marelli C, Coppola A, Luzi L, Govoni S, et al. Microbiota and metabolic diseases. Endocrine. 2018. May 2. [DOI] [PubMed] [Google Scholar]
- 14.Kovatcheva-Datchary P, Arora T. Nutrition, the gut microbiome and the metabolic syndrome. Best Pract Res Clin Gastroenterol. 2013. February; 27(1):59–72. [DOI] [PubMed] [Google Scholar]
- 15.Yoo JY, Kim SS. Probiotics and Prebiotics: Present Status and Future Perspectives on Metabolic Disorders. Nutrients. 2016. March 18; 8(3):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Z, Kraus VB. Does lipopolysaccharide-mediated inflammation have a role in OA? Nat Rev Rheumatol. 2016. February; 12(2):123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alberti KG, Zimmet P, Shaw J, Group IDFETFC. The metabolic syndrome--a new worldwide definition. Lancet. 2005. September 24-30; 366(9491):1059–1062. [DOI] [PubMed] [Google Scholar]
- 18.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, et al. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum. 1986. August; 29(8):1039–1049. [DOI] [PubMed] [Google Scholar]
- 19.Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957. December; 16(4):494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamada D, Sampson ER, Maynard RD, Zuscik MJ. Surgical induction of posttraumatic osteoarthritis in the mouse. Methods Mol Biol. 2014; 1130:61–72. [DOI] [PubMed] [Google Scholar]
- 21.Sekiguchi H, Tomioka N, Nakahara T, Uchiyama HJBl. A single band does not always represent single bacterial strains in denaturing gradient gel electrophoresis analysis. 2001; 23(15):1205–1208. [Google Scholar]
- 22.Wang AH, Li M, Li CQ, Kou GJ, Zuo XL, Li YQ. Human colorectal mucosal microbiota correlates with its host niche physiology revealed by endomicroscopy. Sci Rep. 2016. February 26; 6:21952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011. October 14; 334(6053):255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007. July; 56(7):1761–1772. [DOI] [PubMed] [Google Scholar]
- 25.Mabey T, Honsawek S, Tanavalee A, Yuktanandana P, Wilairatana V, Poovorawan Y. Plasma and synovial fluid inflammatory cytokine profiles in primary knee osteoarthritis. Biomarkers. 2016. November; 21(7):639–644. [DOI] [PubMed] [Google Scholar]
- 26.Sohn DH, Sokolove J, Sharpe O, Erhart JC, Chandra PE, Lahey LJ, et al. Plasma proteins present in osteoarthritic synovial fluid can stimulate cytokine production via Toll-like receptor 4. Arthritis Res Ther. 2012. January 8; 14(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao XY, Yang ZB, Zhang ZJ, Zhang ZQ, Kang Y, Huang GX, et al. CCL3 serves as a potential plasma biomarker in knee degeneration (osteoarthritis). Osteoarthritis Cartilage. 2015. August; 23(8):1405–1411. [DOI] [PubMed] [Google Scholar]
- 28.Matsuki T, Horai R, Sudo K, Iwakura Y. IL-1 plays an important role in lipid metabolism by regulating insulin levels under physiological conditions. J Exp Med. 2003. September 15; 198(6):877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luheshi GN, Gardner JD, Rushforth DA, Loudon AS, Rothwell NJ. Leptin actions on food intake and body temperature are mediated by IL-1. Proc Natl Acad Sci U S A. 1999. June 8; 96(12):7047–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallenius V, Wallenius K, Ahren B, Rudling M, Carlsten H, Dickson SL, et al. Interleukin-6-deficient mice develop mature-onset obesity. Nat Med. 2002. January; 8(1):75–79. [DOI] [PubMed] [Google Scholar]
- 31.Peters M, Schirmacher P, Goldschmitt J, Odenthal M, Peschel C, Fattori E, et al. Extramedullary expansion of hematopoietic progenitor cells in interleukin (IL)-6-sIL-6R double transgenic mice. J Exp Med. 1997. February 17; 185(4):755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenson RS. Effect of fenofibrate on adiponectin and inflammatory biomarkers in metabolic syndrome patients. Obesity (Silver Spring). 2009. March; 17(3):504–509. [DOI] [PubMed] [Google Scholar]
- 33.Ulici V, Kelley KL, Azcarate-Peril MA, Cleveland RJ, Sartor RB, Schwartz TA, et al. Osteoarthritis induced by destabilization of the medial meniscus is reduced in germ-free mice. Osteoarthritis Cartilage. 2018. August; 26(8):1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Wang J, He T, Becker S, Zhang G, Li D, et al. Butyrate: A Double-Edged Sword for Health? Adv Nutr. 2018. January 1; 9(1):21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011. May 4; 13(5):517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cresci GA, Thangaraju M, Mellinger JD, Liu K, Ganapathy V. Colonic gene expression in conventional and germ-free mice with a focus on the butyrate receptor GPR109A and the butyrate transporter SLC5A8. J Gastrointest Surg. 2010. March; 14(3):449–461. [DOI] [PubMed] [Google Scholar]
- 37.Cherbuy C, Darcy-Vrillon B, Morel MT, Pegorier JP, Duee PH. Effect of germfree state on the capacities of isolated rat colonocytes to metabolize n-butyrate, glucose, and glutamine. Gastroenterology. 1995. December; 109(6):1890–1899. [DOI] [PubMed] [Google Scholar]
- 38.Rabot S, Membrez M, Bruneau A, Gerard P, Harach T, Moser M, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010. December; 24(12):4948–4959. [DOI] [PubMed] [Google Scholar]
- 39.Mestdagh R, Dumas ME, Rezzi S, Kochhar S, Holmes E, Claus SP, et al. Gut microbiota modulate the metabolism of brown adipose tissue in mice. J Proteome Res. 2012. February 3; 11(2):620–630. [DOI] [PubMed] [Google Scholar]
- 40.Yoshimura A, Wakabayashi Y, Mori T. Cellular and molecular basis for the regulation of inflammation by TGF-beta. J Biochem. 2010. June; 147(6):781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varela-Eirin M, Loureiro J, Fonseca E, Corrochano S, Caeiro JR, Collado M, et al. Cartilage regeneration and ageing: Targeting cellular plasticity in osteoarthritis. Ageing Res Rev. 2018. March; 42:56–71. [DOI] [PubMed] [Google Scholar]
- 42.Grol MW, Lee BH. Gene therapy for repair and regeneration of bone and cartilage. Curr Opin Pharmacol. 2018. June; 40:59–66. [DOI] [PubMed] [Google Scholar]
- 43.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009. November 11; 1(6):6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015. January; 8(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andoh A, Nishida A, Takahashi K, Inatomi O, Imaeda H, Bamba S, et al. Comparison of the gut microbial community between obese and lean peoples using 16S gene sequencing in a Japanese population. J Clin Biochem Nutr. 2016. July; 59(1):65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017. July; 23(7):859–868. [DOI] [PubMed] [Google Scholar]
- 47.Haro C, Garcia-Carpintero S, Alcala-Diaz JF, Gomez-Delgado F, Delgado-Lista J, Perez-Martinez P, et al. The gut microbial community in metabolic syndrome patients is modified by diet. J Nutr Biochem. 2016. January; 27:27–31. [DOI] [PubMed] [Google Scholar]
- 48.Cheng C, Wei H, Yu H, Xu C, Jiang S, Peng J. Metabolic Syndrome During Perinatal Period in Sows and the Link With Gut Microbiota and Metabolites. Front Microbiol. 2018; 9:1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011. September; 17(9):1971–1978. [DOI] [PubMed] [Google Scholar]
- 50.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016. December; 65(12):1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziebolz D, Rupprecht A, Schmickler J, Bothmann L, Kramer J, Patschan D, et al. Association of different immunosuppressive medications with periodontal condition in patients with rheumatoid arthritis: Results from a cross-sectional study. J Periodontol. 2018. November; 89(11):1310–1317. [DOI] [PubMed] [Google Scholar]
- 52.Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017. April; 11(4):841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dijkhuizen EHP, Del Chierico F, Malattia C, Russo A, Pires Marafon D, Ter Haar NM, et al. Microbiome Analytics of the Gut Microbiota in Patients With Juvenile Idiopathic Arthritis: A Longitudinal Observational Cohort Study. Arthritis Rheumatol. 2019. June; 71(6):1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002. November; 52(Pt 6):2141–2146. [DOI] [PubMed] [Google Scholar]
- 55.He Y, Wu W, Wu S, Zheng HM, Li P, Sheng HF, et al. Linking gut microbiota, metabolic syndrome and economic status based on a population-level analysis. Microbiome. 2018. September 24; 6(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013. July; 11(7):868–875 e861–863. [DOI] [PubMed] [Google Scholar]
- 57.Kang C, Wang B, Kaliannan K, Wang X, Lang H, Hui S, et al. Gut Microbiota Mediates the Protective Effects of Dietary Capsaicin against Chronic Low-Grade Inflammation and Associated Obesity Induced by High-Fat Diet. MBio. 2017. May 23; 8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev. 2017. December; 81(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boer CG, Radjabzadeh D, Medina-Gomez C, Garmaeva S, Schiphof D, Arp P, et al. Intestinal microbiome composition and its relation to joint pain and inflammation. Nat Commun. 2019. October 25; 10(1):4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta VK, Paul S, Dutta C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front Microbiol. 2017; 8:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma HL, Blanchet TJ, Peluso D, Hopkins B, Morris EA, Glasson SS. Osteoarthritis severity is sex dependent in a surgical mouse model. Osteoarthritis Cartilage. 2007. June; 15(6):695–700. [DOI] [PubMed] [Google Scholar]
- 62.Fedak KM, Bernal A, Capshaw ZA, Gross S. Applying the Bradford Hill criteria in the 21st century: how data integration has changed causal inference in molecular epidemiology. Emerg Themes Epidemiol. 2015; 12:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ussar S, Griffin NW, Bezy O, Fujisaka S, Vienberg S, Softic S, et al. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab. 2015. September 1; 22(3):516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parseus A, Sommer N, Sommer F, Caesar R, Molinaro A, Stahlman M, et al. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017. March; 66(3):429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. The other three kinds of bacteria showed significant differences in the genera level between mice from OA-METS-/MLI+, OA+METS-/MLI+ and OA+METS+/MLI+ groups. (A) Eisenbergiella; (B) Parabacteroides; (C) [Ruminococcus]_gauvreauii_group. (OA=Osteoarthritis; METS=Metabolic Syndrome; MLI=Meniscal/Ligamentous Injury)
Supplementary Figure S2. Weight of the mice in different groups. (A) Weight at 8 weeks; (B) Weight at 18 weeks; (C) ΔWeight (defined as weight at 18 weeks – weight at 8 weeks).
Supplementary Figure S3. Relative abundance of Streptococcus in the gut microbiome of the human donors and mouse recipients. (A) Relative abundance of Streptococcus in the gut microbiome of human donors n≥4/group; (B) Relative abundance of Streptococcus in the gut microbiome of the mouse microbiome recipients n≥6/group.