Graphical abstract
Keywords: COVID-19, SARS-CoV-2, Protocol, Quality Control (QC), Wastewater-Based Epidemiology (WBE)
Highlights
-
•
COVID-19 pandemic has been an impetus to sewage-based epidemiology development.
-
•
There is urgent need for optimised SARS-CoV-2 detection/quantification protocols.
-
•
Appropriate quality controls must accompany all steps of the analytical process.
-
•
Global efforts of method optimization should include inter-laboratory result comparison.
Abstract
COVID-19 is an ongoing global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). As of July 29th 2020, more than 16,6 million cases have been reported in more than 188 countries/territories, leading to more than 659000 deaths. One of the main challenges facing health authorities has been testing for the virus on a sufficiently comprehensive scale. The pandemic has been an impetus for the wastewater community as it has inspired scientists to look to wastewater to help fill in the gap of measuring the prevalence of SARS-CoV-2 within a given community. Testing the wastewater may serve as an early warning system allowing timely interventions. Although viral shedding varies among individuals and over the course of their infection, the sewage system can blend these variations into an average that represents the wider studied community. The urgent need has led to a lack of coherent reporting of data regarding the analysis, as these huge and remarkable efforts by the wastewater scientific community were made in a very short time. Important information on the analytical approach is often lacking, while there is still no optimisation of the methodology, including sampling, sample storage and concentration, RNA extraction and detection/quantification. This review aims at identifying the main issues for consideration, relating to the development of validated methodological protocols for the virus quantitative analysis in wastewater. Their inclusion will enable the methodological optimisation of SARS-CoV-2 wastewater analyses, transforming the wastewater infrastructure into a source of useful information for the health sector.
1. Wastewater-Based Epidemiology: a litre of wastewater, an ocean of information
In the framework of the COVID-19 pandemic, the development of an early-warning and surveillance system through the utilisation of a Wastewater-Based Epidemiology (WBE) approach aims at tackling important community public health questions that arise due to the COVID-19 or any other future pandemic, which can be answered through the investigation and surveillance of selected indicators of community health and behaviour, reflected in the composition of urban wastewater. As wastewater of an area is collected at an urban wastewater treatment plant (UWTP), sampling and analysing the wastewater composition can reveal the presence of the SARS-CoV-2 genetic fingerprint in wastewater. Given sufficient information is provided on the population served by the treatment plant, the fingerprint can be attributed to it, providing a cross-section of the public health status of the society.
Currently, public health interventions are initiated in a broad manner; potentially not considering communities that would benefit from them and burdening areas where the virus may currently not pose a risk, thereby making hardship-inducing containment measures economically and socially disruptive. The clinical diagnostic tests currently used for COVID-19 are already proven insufficient for rapid and cost-effective monitoring of the incidence of the virus at a community-wide level [1]. Another problem concerns the worldwide high demand of consumables (e.g. swabs, reagents) needed for the collection and screening of samples for COVID-19. From the social point of view, there were cases that revealed that people of a community were sceptical to take the COVID-19 clinical test due to the fear of social stigma attached to the pandemic.
With limitations on COVID-19 testing making it hard to know how many people actually have the disease, turning to the sewer systems for a fast snapshot seems to be promising and a useful method to provide complementary and additional information to the clinical testing. The SARS-CoV-2’s faecal signature could actually turn out to be very useful, helping track how and where disease is spreading among the population. WBE has been used for decades to detect polio in countries like Brazil and Israel, where the disease remains endemic. Israel’s sewage surveillance system, set up in 1989 by the Israeli health authorities to detect poliovirus in wastewater, enabled to track polio in sewage trunk lines and UWTPs during polio re-emergence in 2013, and the response of the public authorities to the epidemic was immediate [2]. More recently, efforts have been made to set up a surveillance system for other viruses via wastewater, such as the Zika virus, whose large-scale outbreak was reported in the Americas followed by 87 countries worldwide in 2015, when Brazil first confirmed a novel febrile illness outbreak to WHO and the virus emerged as a cause of serious birth defect microcephaly and of the Guillain-Barre syndrome neurological disorder [3].
Sewage is a source of information on human health and habits and can be transformed into a public health observatory and used as an instrument for refining public health response to a pandemic caused by a pathogen. Public health authorities could use this information to refine their response and to help them evaluate when and how to start scaling up or back quarantine-style policies and recommendations. Among the various methods of public health and infectious disease assessment and surveillance, WBE provides significant advantages to face obstacles faced by other commonly applied techniques, such as reliable provision of spatio-temporal trends in human behaviour and infection, near-real-time and whole population data, including asymptomatic people and those with mild symptoms resembling other common viral infections and relatively low cost.
One critical point in relation to the application of WBE monitoring program is the conduction of sampling from the sewer system in specific neighbourhoods. Sewer systems offer near-real-time outbreak data, because they continually receive human excreta that contain viral particles shed by infected humans, regardless of their symptomatology status (symptomatic; asymptomatic [no symptoms]; paucisymptomatic or subclinical [mild symptoms]; and presymptomatic [no symptoms for the first few days before exhibiting COVID-19 symptoms]). Furthermore, interestingly, during air travel and cruises, wastewater monitoring may provide public health officials with an additional tool of assessing the prevalence of SARS-CoV-2 infections among passengers using on-board facilities, which can be spread in this way internationally [4]. This is important also in relation to the fact that at least one COVID-19 patient was found to be positive after faecal specimen examination, despite being negative after pharyngeal and sputum analysis [5]. In this way, it is expected that the collected samples from both the sewer lines and the wastewater treatment facilities will enable the tracing of viral outbreaks to a more accurate location making possible the identification of urban areas of concern. In addition, it is envisaged that the virus spread and fate may vary among wastewater treatment facilities in urban settings, which utilise enclosed underground sewer pipes and rural areas, which use septic tanks and catchments. WBE can also enable tracking the silent circulation of the virus due to the detection of low levels of the viral RNA before cases appear. [6] ([preprint]), who evaluated the impact of lockdown on the dynamics of SARS-CoV-2 via the quantification of the viral RNA in wastewater, revealed that genome units were concomitantly decreased along with the amount of the recorded COVID-19 cases as a result of the lockdown measures. Moreover, [7] ([preprint]) have shown the incidence of SARS-CoV-2 in wastewater since November 2019, before the recording of the first COVID-19 symptomatic cases in Santa Catalina, Brazil. This finding suggests the shedding of the virus from paucisymptomatic and asymptomatic persons in the community, months before the reporting of the first cases by the national authorities ([7] [preprint]).
One important feature of WBE relies on the fact that it can detect variations in the viral strains via phylogenetic analysis, providing a substantial advantage for recognising virus trees that have evolved over time and among various regions. In the case of SARS-CoV-2, there are only two studies available on the phylogenetic analysis of wastewater samples. According to Nemudryi et al. [8] ([preprint]), the most prevalent strains of SARS-CoV-2 detected in wastewater of Bozeman, Montana were associated with those previously observed in Europe (France and Iceland). Genome sequencing and phylogenetic analysis carried out by [9] ([preprint]), revealed similarities among the isolated viral strains with those found in Europe and in the Lombardy region in northern Italy.
WBE can also track seasonal fluctuations in viral concentrations in wastewater, reflecting the epidemiological patterns in a community. To date, there is no information about the effect of the season on the SARS-CoV-2 concentration in wastewater, while such information may be available if we consider previous relevant analyses performed for other viruses. For example, Nordgren et al. [10] reported seasonal variations of noroviruses (NoV GGI and NoV GGII), with the highest concentration of these viruses being recorded during winter and summer, respectively. Also, NoV GGII exhibited higher seasonal peaks compared to NoV GGI. The increase in NoV GGI during summer, which is a ‘low season’ for clinically-reported norovirus-caused gastroenteritis, gave way to milder or asymptomatic infections as compared to the NoV GGII strains in circulation during winter. Another study conducted by Li et al. [11] in China, has demonstrated that the concentration levels of rotavirus were higher during November to March, corresponding to the clinical data of the virus reported in the country [12]. The norovirus concentration in wastewater was reportedly higher during November to April [13], whereas the concentrations of adenovirus and enterovirus were largely consistent throughout the year. Since a correlation between the COVID-19 spread and temperature exists [14], it is envisaged that seasonal variations of the viral genetic material will also occur in the sewage, with the detection levels being higher at the locations falling inside of the climatologically favoured zones (4-12 °C).
Given the fact that SARS-CoV-2 is a novel coronavirus strain that was not formerly identified in human excreta (and thus in sewage), research in the WBE field is in its infancy and the position of the scientific community is still unclear, especially when considering the establishment and validation of a methodology for the isolation, detection and quantification of the virus genetic material in wastewater.
2. Main considerations related to the development of a methodological protocol for detecting and quantifying SARS-CoV-2 RNA in wastewater
The potential transmission via the fecal-oral pathway was recently underlined by Foladori et al. [15], due to the detection of SARS-CoV-2 in the human gastrointestinal tract. Besides, potential transmission through bioaerosols from stool through toilet flushing was demonstrated in Hong Kong for the SARS-CoV epidemic cluster in Amoy Gardens [16,17] and was recently proposed for SARS-CoV-2 [18]. During this outbreak, SARS-CoV shed in the feces of an infected building visitor has been suggested to have spread the virus to other building inhabitants via droplets and aerosols of virus-contaminated commode water, which was transmitted to multiple flats through faulty toilet plumbing and floor drains [19]. This outbreak scenario proposes that wastewater of infected persons may be a means of transmission of the infectious virus in stools. As a result, it may be deduced that the fecal droplet–respiratory route is potentially possible. However, different aspects require further and deeper investigation, such as the viability and infectivity of the virus in stools and urban wastewater. If SARS-CoV-2 is capable of surviving for long periods of time in stools and wastewater, exposure and transmission via faecally-contaminated water droplets and aerosols may be more probable. In order to assess the risks posed by this exposure pathway more effectively, more data are needed on the survival and persistence of the virus in sewage, in combination with evidence on the survival of the virus inside the gastrointestinal system and human excreta. Rimoldi et al. ([9] [preprint]) have reported the examination of the infectious virus in UWTP influents and effluents, as well as in two rivers, without any positive samples for infectious SARS-CoV-2. However, this study used a small volume of wastewater (2 mL) and no positive assay controls. As a result, negative infectivity results do not necessarily mean absence of the infectious virus, as the lack of adequate quality control measures and other methodological considerations may complicate the determination of the infectious virus in wastewater [20]. Moreover, because working with this type of infectious virus requires specifically trained personnel working in Biological Safety Level 3 (BSL-3) laboratory containment, there are substantial analytical challenges involved in studying the virus survival, while there is still currently limited available information [15].
To this end, even though more information is needed on the exact aspects of transmission of the alive virus through faeces via the oral-faecal pathway and of its infectivity in sewage, the presence of SARS-CoV-2 RNA in wastewater was described worldwide; in the Netherlands ([21] [preprint]; [38]), Italy ([[22], [23]]; [24] [preprint]; [9] [preprint]), France ([25] [preprint]; [6] [preprint]), Spain [26], Israel ([27] [preprint]), Turkey ([28] [preprint]), USA ([29] [preprint]; [8] [preprint]; [30]; [31] [preprint]), India ([32] [preprint]), Japan ([33] [preprint]), Brazil ([7] [preprint]) and Australia [4,34,35].
The studies reviewed herein, dedicated to the detection of SARS-CoV-2 in urban wastewater, were published between the 30th of March 2020 to the 15th of July 2020, with the majority of them not being certified by formal peer review. More specifically, at the time of preparing this review, thirteen (13) out of the twenty (20) studies were published in the literature as preprints ([27] [preprint]; [7] [preprint]; [29] [preprint]; [33] [preprint]; [28] [preprint]; [32] [preprint]; [21] [preprint]; [8] [preprint]; [9] [preprint]; [25] [preprint]; [31] [preprint]; [6] [preprint]; [24] [preprint]). It is clear that there is an increased scientific and public engagement with the COVID-19 preprints. Even though the possibility exists that some will not be eventually published, the present review solely discusses methodological aspects of the sewage analysis reported in them, that most probably will not be impacted by the peer review process. Hence, even though some may not eventually appear in the literature as peer-reviewed manuscripts, the aspects for methodological consideration reported by this review paper will still be relevant.
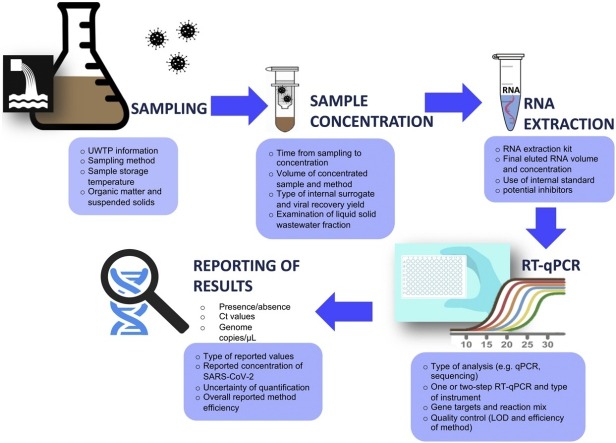
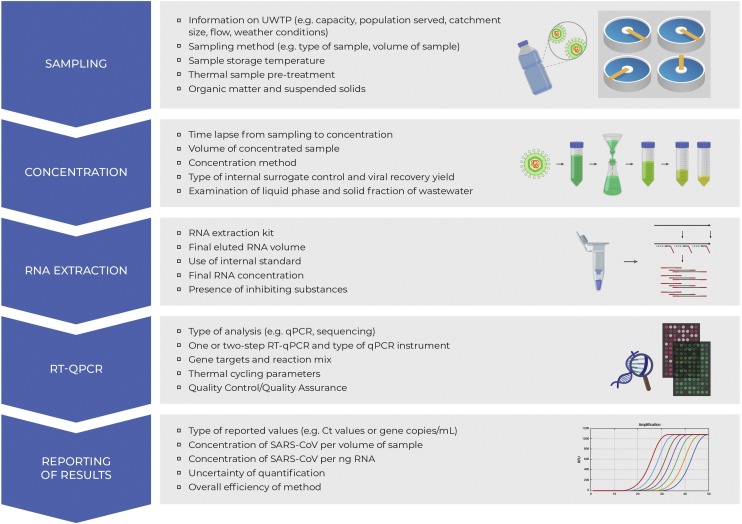
Complementary information with respect to the location, UWTPs and corresponding inhabitant/population equivalents, type of sample, sample storage and transfer to the laboratory, sample pre-treatment and concentration, RNA extraction, detection and quantification are presented in Table 1, Table 2, Table 3,5 and 6 . Also, since there is limited literature surrounding the detection of the RNA of SARS-CoV-2 in wastewater, and considering that the SARS-CoV-2 virus may behave similarly to other coronaviruses, an effort was made to provide information about previously identified coronaviruses that may be relevant to the findings as defined by each study. The studies concerning the prevalence of SARS-CoV-2 in sewage sludge ([36,37] [preprint]) were not included in the tables but their major findings are discussed in the manuscript. Special emphasis was given to the main parameters, processes, and challenges that are associated with the efficiency and credibility of each methodological protocol applied for the isolation, detection and quantification (where available) of the virus RNA in wastewater.
Table 1.
Information on the sampling, storage/transport conditions and sample pre-treatment used for the detection of SARS-CoV-2 RNA in wastewater.
| Reference The papers marked with an asterisk (*) were not certified by peer review (preprints) | Location given in alphabetical order | Information on UWTPs (Inhabitant/Population Equivalents, treatment, etc.) | Dates of sampling | Type of sample | Type of sampling container | Freezing and transfer of samples | Storage conditions at the laboratory | Days to analysis | Thermal sample treatment |
|---|---|---|---|---|---|---|---|---|---|
| [34] | Australia | Suburban pumping station and 2 UWTPs of urban catchments in Southeast Queensland | March-April 2020 | collection of untreated wastewater (grab samples) using conventional refrigerated autosampler or a submersible high frequency autosampler | not reported | transfer on ice | storage at 4 °C | not reported | × |
| [35] | Australia | UWTP in Brisbane, Australia (PE = 325000) urban and industrial wastewater; primary treatment, CAS, and disinfection with chlorine and UV |
not reported | 2 L of untreated wastewater | not reported | transfer on ice | storage at 4 °C | 24 h | × |
| [4] | Australia | Cruise ship and 3 aircrafts | Cruise ship: 23/4/2020 Aircrafts:
|
Cruise ship: two grab samples from influent and effluent of MBR of the ship Aircrafts: three grab wastewater samples from each aircraft |
not reported | Maintenance at 4 °C for transport | Maintenance at 4 °C for transport | 6-24 hours | × |
| [7]* | Brazil | UWTP in Florianopolis, Santa Catalina, Brazil (serving 5000 inhabitants) Raw sewage |
30th October 2019 to 4th of March 2020 | 200 mL of urban sewage | not reported | not reported | storage at 80 °C | not reported | × |
| [25]* | France | Influent wastewater of UWTP in Montpellier metropolitan area in Lattes | May 7th, 18th, 26th and June 4th, 15th and 25th 2020 | Composite samples | not reported | Maintenance at 4 °C for transport | Centrifugation at 4500xg for 30 min at 4 °C andsieving of supernatant through 40 μm Corning strainer followed by storage at -20 °C | Samples were processed immediately upon arrival in the laboratory | × |
| [6]* | France | 3 UWTPs (>100000 IE) in Paris | 5th of March 2020 (start of the epidemic) - 23th of April, 2020. | not available | not reported | not reported | storage at 4 °C | Samples were processed in less than 24 h after sampling | × |
| [32]* | India | UWTP in Ahmedabad, Gujarat (106 million L/day) Upflow Anaerobic Sludge Blanket (UASB); receives the wastewater from a governmental hospital treating COVID-19 patients |
8th and 27th of May, 2020 | influent and treated effluents (after UASB and aeration pond) | sterilized bottles | transfer on ice | refrigeration of the samples taken on the 8th of May at 4 °C till the 27th of May when next batch of samples were transported to the laboratory and analysed immediately | not reported | × |
| [27]* | Israel | 11 UWTPs in Haifa, Shafdan, Rahat, Arara, Beer Sheva, Ayalon, Zfat, El Hamra, Kidron, Sorek, Og | 10th March, 25th March, 30th March, 3rd April, 5th April, 13th April, 15th April, 16th April, 21th April, 2020 | collection of 200 mL of sample every 30 min for 24 hours | not reported | transfer of 6-10 L of sample to the laboratory, with transfer of 2 L samples to clean plastic bottles | storage at -20 °C or−80 °C. | not reported | × |
| [22] | Italy | 3 UWTPs in Milan (Plant A and B; PE = 1.050.000) and Rome (Plant C1 and C2; PE = 900.000) | 3rd of February - 2nd of April, 2020 | 24-h composite raw wastewater samples | not reported | not reported | storage at -20 °C | not reported | √ 56 °C, 30 min |
| [24]* | Italy | 5 UWTPs in Milan (Plants A and B; PE = 1116928 and 750863, respectively), Turin (Plants C and D; PE = 2297000 and 180000, respectively) and Bologna (Plant E; PE = 653809) | 9th of October 2019 – 28th of February 2020 | 24-h composite raw wastewater samples | not reported | storage at -20 °C | not reported | √ 56 °C, 30 min | |
| [9]* | Italy | 3 UWTPs (UWTP-A in Monza/Brianza; UWTP-B and C in Milano); flow = 11 m3/s, IE = 2 million secondary treatment; tertiary disinfection by peracetic acid or high intensity UV disinfection. UWTP-A and UWTP-B disposal into the Lambro River, and UWTP-C into the Lambro Meridionale River |
14th and 22th April, 2020 | grab wastewater and river water samples (at 1.00 p.m.) | polypropylene bottles for wastewater, dark glass bottles for river water | transfer of samples under refrigeration | not reported | not reported | × |
| [33]* | Japan | UWTP and river in Yamanashi Prefecture CAS |
17th of March-7th of May, 2020 | collection of samples from influent and secondary-treated wastewater before chlorination and river water samples | 1-L sterilized plastic bottles | transfer on ice | not reported | 6 h after sampling | × |
| [26] | Spain | 6 UWTPs in Murcia (PE = 530499; CAS, disinfection [NaClO]), Cartagena (PE = 163969; CAS, disinfection), Molina de Segura (PE = 150545; CAS, coagulation, flocculation, sand filtration, disinfection [UV, NaClO]), Lorca (PE = 101161; CAS, coagulation, flocculation, sand filtration, disinfection [UV, NaOCl]), Cieza (PE = 69502; CAS, coagulation, flocculation, sand filtration, disinfection [UV]), Totana (PE = 28289; CAS, disinfection [UV]) | 12th March - 14th April, 2020 | grab samples (at 7-12 am) 42 influent, 18 secondary and 12 tertiary treated effluent samples | HDPE plastic containers | transfer of samples on ice to the laboratory. | storage at 4 °C | 24 h | × |
| [21]* [38] |
The Netherlands | UWTPs in Amsterdam (IE = 1014000), Hague, Utrecht (IE = 530000), Apeldoorn (IE = 350000), Amersfoort (IE = 335000), Tilburg (IE = 375000), Schiphol airport (IE = 54000) | 5th, 6th and 7th of February, 2020 (3 weeks before the first reported COVID-19 case in the Netherlands), 4th and 5th March, 2020 (38 and 82 reported COVID-19 cases, resp.), 15th, 16th and 25th of March, 2020 (1135, 1413, and 6412 reported COVID-19 cases, resp.) | 24 h flow-dependent composite wastewater sample (250 mL) | not reported | freezing of samples at 4 °C andtransfer on melting ice | not reported | not reported | × |
| [28]* | Turkey | 7 UWTPs in Istanbul, Turkey and 2 manholes nearby Istanbul hospitals Preliminary treatment; CAS; advanced nutrient removal |
21st and 25th of April, 2020 | grab samples | 250-mL sterilized bottles | not reported | not reported | not reported | × |
| [29]* | USA | 11 points of sampling (i.e., 6 wastewater treatment plants, and 5 inlet pumping stations, or interceptor lines) in Syracuse, NY and other locations in Onondaga County, NY. PE = 12800-223900 (UWTPs) PE = 3841-112747 (interceptor lines) |
6th and 13th May, 2020 | 24 hour-composite wastewater samples | not reported | storage of samples at 4 °C andtransportation on ice to Upstate Medical University (Syracuse, NY) | not reported | Next morning | × |
| [8]* | USA | UWTP in Bozeman, Montana | 18th-25th of May, 2020 | Collection of 0.5-L samples of pre-treated wastewater on 7 days within a 17-day period, using two collection methods (grab and 24 h-composite samples) | not reported | not reported | not reported | not reported | × |
| [30] | USA | 9 composite and 6 grab wastewater samples from two UWTPs in Southern Louisiana, respectively (UWTP A = 244627 PE; UWTP B = 45694 PE) 7 raw wastewater samples 4 secondary treated samples 4 final effluent samples after chlorination |
January 13th 2020 February 3rd 2020 March 2nd 2020, April 8th and April 29th 2020 |
not reported | Sterile 1 L Nalgene bottles | Transport on ice to the laboratory | not reported | not reported | × |
| [31]* | USA | UWTP in Massachusetts | 18th March-25th of March, 2020 | 24 hour-composite samples of raw sewage | not reported | not reported | storage at 4 °C | not reported |
√ 60 °C, 90 min |
Abbreviations (given in alphabetical order): CAS: Conventional Activated Sludge; IE: Inhabitant Equivalent; PE: Population Equivalent; UASB: Upflow Anaerobic Sludge Blanket
Table 2.
Information on the concentration method used for the detection of SARS-CoV-2 RNA in wastewater.
| Reference The papers marked with an asterisk (*) were not certified by peer review (preprints) | Location given in alphabetical order | Main sample concentration steps | Surrogate control | Recovery |
|---|---|---|---|---|
| [34] | Australia |
(i) electronegative membranes:
|
- | - |
| [35] | Australia |
|
MHV | 26.7-65.7%, |
| [4] | Australia |
|
- | - |
| [7]* | Brazil |
|
Murine Norovirus | 1.6 – 2.6 % |
| [25]* | France |
|
- | - |
| [6]* | France |
|
- | - |
| [32]* | India |
|
- | - |
| [27]* | Israel |
|
- | - |
| [22] | Italy |
|
- | - |
| [24]* | Italy |
|
Alphacoronavirus HCoV 229E | 2.04 ± 0.70% |
| [9]* | Italy |
|
- | - |
| [33]* | Japan |
(i) electronegative membrane vortex method:
|
- | - |
| [26] | Spain |
|
PEDV strain CV777 MgV |
influent: PEDV = 11 ± 2.1% MgV = 11 ± 3.5% effluent: PEDV = 3.3 ± 1.6% MgV = 6.2 ± 1.0% |
| [21]* [38] |
The Netherlands |
|
F-specific RNA | 73 ± 50% |
| [28]* | Turkey |
|
- | - |
| [29]* | USA |
|
- | - |
| [30] | USA |
Method A:
|
- | - |
| [31]* | USA |
|
- | - |
| [8]* | USA |
|
- | - |
Abbreviations (given in alphabetical order): MgV: Mengovirus; MHV: Murine hepatitis virus; PBS: Phosphate Buffered Saline; PEDV: Porcine Epidemic Diarrhea Virus; PEG: Polyethelene glycol
Table 3.
RNA extraction conditions of the available studies.
| Reference The papers marked with an asterisk (*) were not certified by peer review (preprints) | Location given in alphabetical order | RNA extraction kit | Use of surrogate control | Final elution volume | Storage of extracted samples |
|---|---|---|---|---|---|
| [34] | Australia |
Method A
|
Not reported | 100 μL | −80 °C |
| [35] | Australia |
|
Not reported | 100 μL | Not reported |
| [4] | Australia |
|
Not reported | 100 μL | Not reported |
| [7]* | Brazil |
|
Not reported | 200 μL | Not reported |
| [25]* | France |
|
VP40-encoding RNA of Ebola virus | Not reported | Not reported |
| [6]* | France |
|
Not reported | Not reported | Not reported |
| [32]* | India |
|
MS2 phage | Not reported | Not reported |
| [27]* | Israel | RNeasy mini kit and easyMAG kit | Not reported | Not reported | −80 °C |
| [22] | Italy |
|
Not reported | Not reported | Not reported |
| [24]* | Italy |
|
Not reported | Not reported | Not reported |
| [9]* | Italy | QIamp viral RNA mini kit | Not reported | Not reported | Not reported |
| [33]* | Japan |
|
Not reported |
|
Not reported |
| [26] | Spain |
|
Porcine epidemic diarrhea virus (PEDV) and mengovirus vMC0 (MgV) | 100 μL | Not reported |
| [21]* [38] |
The Netherlands |
|
Not reported | 100 μL | Not reported |
| [28]* | Turkey |
|
Infectious Bronchitis Virus | Not reported | Not reported |
| [29]* | USA | AllPrep PowerViral DNA/RNA Kit | Not reported | 50 μL | Not reported |
| [30] | USA | ZR Viral RNA kit | Pseudomonas bacteriophage Φ6 | 100 μL | Not reported |
| [31]* | USA | Trizol | Not reported | Not reported | Not reported |
| [8]* | USA | RNeasy Mini Kit | Not reported | 40 μL | Not reported |
Table 5.
RT-PCR reaction conditions of the available studies.
| Reference The papers marked with an asterisk (*) were not certified by peer review (preprints) | Location given in alphabetical order | Template volume used | Reaction mixture volume | Selected primers | PCR reaction Mix | Instrumentation | Cycling parameters | Quality control measures | Other analyses |
|---|---|---|---|---|---|---|---|---|---|
| [34] | Australia | 6 μL | 40 μL |
|
iTaq Universal Probes One-Step Reaction Mix | BioRad CFX 96 thermal cycler |
N_Sarbeco 10 min at 50 °C 3 min at 95 °C X 45 cycles of: 15 sec at 95 °C 30 sec at 58 °C NIID_2019-nCOV_N 10 min at 50 °C 15 min at 95 °C X 45 cycles of: 15 sec at 95 °C 60 sec at 60 °C |
|
Sequencing using Sanger and Illumina sequencing platforms |
| [35] | Australia | 5 μL | 25 μL | MHV virus | iTaq Universal Probes One-Step Reaction Mix | BioRad CFX 96 thermal cycler | 10 min at 50 °C 5 min at 95 °C X 40 cycles of: 15 sec at 95 °C 60 sec at 60 °C |
|
Not reported |
| [4] | Australia | 3 μL | 20 μL | CDC N1, N2 and E NIID_2019-nCoV N assay |
iTaq Universal Probes One-Step Reaction Mix | BioRad CFX 96 thermal cycler | Not reported |
|
|
| [7]* | Brazil | Not reported | Not reported | N1, RdRp, S | OneStep qPCR Quantinova kit | Not reported | Not reported | Murine Norovirus | Not reported |
| [25]* | France | Not reported | Not reported | N1 and N3 | Not reported | Not reported | Not reported | VP40-encoding RNA of Ebola virus as normalisation standard | Not reported |
| [6]* | France | 5 μL | 25 μL | E and RdRp | Fast virus 1-step Master mix | Viaa7 Real Time PCR System | 10 min at 55 °C 3 min at 95 °C X 45 cycles of: 15 sec at 95 °C 58 sec at 30 °C |
Internal positive control | Not reported |
| [32]* | India | Not reported | Not reported | ORF1ab, N, S | TaqPath Covid-19 RT-PCR Kit | Not reported | Not reported | Positive controls Negative controls |
Not reported |
| [27]* | Israel | Not reported | Not reported | E gene | Fast Start Universal Probe Master | Step One Plus real -time PCR system | Not reported |
|
Not reported |
| [22] | Italy | 2.5 μL 5 μL of first PCR product |
25 μL 50 μL |
ORF1ab (broad range and SARS-CoV-2 specific) and NIID_WH-1 |
|
Not reported | 30 sec at 98 °C X 35 cycles of: 10 sec at 95 °C 10 sec at 50 °C 10 sec at 54 °C 30 sec at 72 °C 5 min at 72 °C 30 sec at 98 °C X 45 cycles of: 10 sec at 95 °C 10 sec at 50 °C 10 sec at 54 °C 30 sec at 72 °C 5 min at 72 °C |
Negative controls | Sequencing |
| [24]* | Italy | 2 μL and 5 μL | 25 μL | ORF1ab, E and RdRp |
|
Not reported | 30 sec at 98 °C X 35 cycles of: 10 sec at 98 °C 10 sec at 54 °C 30 sec at 72 °C 5 min at 72 °C 30 min at 50 °C X 45 cycles of: 15 sec at 95 °C 60 or 95 sec at 58 °C 30 sec at 60 °C |
|
Sequencing |
| [9]* | Italy | 2.5 μL | Not reported | N1, N2, N3, ORF1ab, E | Not reported | Not reported | Not reported | Not reported |
|
| [33]* | Japan | 2.5 μL | 25 μL | N_Sarbeco and NIID_2019-nCOV_N, N1, N2 | Probe qPCR Mix with UNG | Dice Real Time System TP800 | 10 min at 25 °C 30 sec at 95 °C X 45 cycles of: 5 sec at 95 °C 60 sec at 60 °C |
|
Not reported |
| [26] | Spain | Not reported | 25 μL | N1, N2, N3 | OneStep PrimeScript RT-PCR kit | LightCycler 480 system | 10 min at 50 °C 3 min at 95 °C X 45 cycles of: 3 sec at 95 °C 30 sec at 55 °C |
|
Not reported |
| [21]* [38] |
The Netherlands | 5 μL | 20 μL | N1, N2, N3 and E | TaqMan Fast Virus 1-step Master Mix | BioRad CFX 96 thermal cycler | 5 min at 50 °C X 45 cycles of: 10 sec at 95 °C 30 sec at 60 °C |
|
Not reported |
| [28]* | Turkey | 5 μL | 20 μL | RdRp | QuantiNova Pathogen + IC Kit | BioRad CFX 96 thermal cycler | Not reported |
|
Not reported |
| [29]* | USA | 2.5 μL | 25 μL | IP2 and IP4 of RdRp | Reliance One-Step Multiplex RT-qPCR Supermix | QuantStudio3 or QuantStudio5 | 10 min at 50 °C 10 min at 95 °C X 45 cycles of: 10 sec at 95 °C 30 sec at 59 °C |
|
Not reported |
| [30] | USA | 2.5 μL | 25 μL | N1, N2 | PerfecTa qPCR ToughMix | BioRad CFX 96 thermal cycler | 10 min at 95 °C X 45 cycles of: 10 sec at 95 °C 30 sec at 55 °C |
|
Not reported |
| [31]* | USA | Not reported | Not reported | N1, N2, N3 | Not reported | Not reported | Not reported |
|
Sanger sequencing |
| [8]* | USA | 5 μL | 40 μL | 2019 nCoV CDC EUA Kit (N1 and N2) | TaqPath 1-step RT-qPCR Master Mix | ABI 7500 Fast Real-Time PCR system | 2 min at 25 °C 15 min at 50 °C 2 min at 2 °C X 45 cycles of: 3 sec at 95 °C 30 sec at 55 °C |
|
Sanger Sequencing |
Table 6.
Quantification of the RT-qPCR results in the available studies.
| Reference The papers marked with an asterisk (*) were not certified by peer review (preprints) |
Location | Quantification of targets (yes/no) | Units of reporting | Positive sample Ct values and quantification results |
|---|---|---|---|---|
| [34] | Australia | Yes | Ct values | 37.5 Ct = 12 copies/100 mL of WW 39 Ct = 1.9 copies/100 mL of WW (1 sample positive for N_Sarbeco and 1 positive for NIID_2019-nCOV_N) |
| [35] | Australia | Yes | Copies/MHV recovered | Recovery efficiency: Method C > Method B > Method D > Method F > Method G > Method E > Method A |
| [4] | Australia | Yes | Ct values Copies/100 mL | 4/5 aircraft samples positive for N or E target Both concentration methods recovered SARS-CoV-2 RNA from aircraft wastewater (N_Sarbeco and E_Sarbeco) Cq values of positive samples: 36-39 CDC N1, N2 and NIID_2019-nCoV N assays did not provide any positive results 7/21 cruise ship samples were positive for all assays 14/21 samples were positive for at least one assay |
| [7]* | Brazil | No | Ct values Genome copies/L |
1 log increase observed from November 2020 to March 2020 5.49 log10 genome copies/L (November 2019) to 6.68 log10 genome copies/L (March 2020) |
| [25]* | France | Yes | Ct values RNA copies/100 mL |
No direct temporal relationship between SARS-CoV-2 detection and epidemiological features of COVID-19 |
| [6]* | France | Yes | Genome units/L | 5.4 × 104 – 3 × 106 genome units/L |
| [32]* | India | Yes | Ct values Copies/L |
27.92-29.52 Ct 2.42 × 108 copies/L |
| [27]* | Israel | No | Ct values | 32.76-38.5 Ct |
| [22] | Italy | No | Ct values | 4/8 days positive signals in plant A 4/8 days positive signals in plant B 2/8 days positive signals in plant C1 2/8 days positive signals in plant C2 |
| [24]* | Italy | Yes | Genomic copies/μL | 15/40 samples positive signals LOD to 5.9 × 103 genomic copies/L to 5.6 × 104 genomic copies/L |
| [9]* | Italy | No | Ct values | ORF1ab, N and E positive signal in raw influent No positive signal in treated wastewater |
| [33]* | Japan | Yes | Copies/L | 1/5 secondary treated WW were positive 0/5 influent samples were positive for SARS-CoV-2 |
| [26] | Spain | Yes | Ct values Genomic copies/L |
35/42 influent positive samples for at least one gene target 2/18 secondary treated positive samples for at least one gene target Concentration: 5.1-5.5 log10 genomic copies/L |
| [21]*, [38] | The Netherlands | Yes (N1-N3) No (E) |
Ct values Gene copies/mL |
Concentration of N1, N2 and N3: 1.2 × 101 genome copies/mL E: 18/29 UWTPs positive signals |
| [28]* | Turkey | Yes | Ct values Copies/L |
34.67-39.54 Ct 3.11 × 102 – 7.78 × 103 copies/L |
| [29]* | USA | Yes | Copies/mL | 18/22 positive samples 42.7 ± 32.9-112.35 ± 8.01 genomes/mL |
| [30] | USA | Yes | Ct values Copies/L |
Positive samples during April (Method A) 2.5-3.2 log10 copies/L |
| [31]* | USA | No | Ct values | 33.87-38.39 Ct (southern-filtrate and northern-unfiltered samples positive for N1, N2 and N3, and N1 and N3 respectively) |
| [8]* | USA | Yes | Viral genomes/L | Not provided |
The major focus of the studies conducted in the field of WBE relating to SARS-CoV-2 was the isolation and detection of the viral RNA at the inlet of UWTPs (raw wastewater), with very few of them providing information on the prevalence of the virus during the various stages of treatment applied at the UWTP. In general, there is lack of data on the effect of various wastewater treatment technologies applied at the UWTPs on SARS-CoV-2 as the first efforts were to firstly identify the virus in the influent of UWTPs. The removal efficiency of SARS-CoV-2 with traditional biological treatment processes such as the Conventional Activated Sludge (CAS) and the Membrane BioReactor (MBR) process, remains unclear due to the absence of experimental data (40[preprint]). More specifically, the majority of the available data focuses on a large range of surrogate viruses of bacteriophages and laboratory-cultured viruses such as Enterovirus, Adenovirus and human polyomavirus JC, with a large variety of removal exhibited, from 0.9 to 5.8 logs [41]. At the same time however, there is no confirmation that SARS-CoV-2 behaves in a different manner than other coronaviruses [42]. The impact of the presence of SARS-CoV-2 on the microbial community in the sewage sludge during CAS or MBR treatment is another issue that remains to be clarified, as any effects may lead to alterations of the microbial community that performs the biological degradation of contaminants in incoming wastewater. Previous work has shown that introduced or ‘foreign’ viruses may significantly affect bacterial populations, via bacterial cell lysis and horizontal gene transfer (HGT) [43]. Viruses are suggested to selectively lyse bacteria whose populations in their habitat is large, influencing in this way the richness of their population, while their concentrations in activated sludge have been shown to increase in the course of CAS treatment of wastewater, suggesting the promotion of viral reproduction in the presence of bacterial hosts [44]. However, the exact degree to which specific viruses have such an impact on host bacterial communities still remains unclear, as more trials and experimental work is needed to prove the action of specific viral species such as the SARS-CoV-2 on sludge microbial communities.
Gundy et al. [45] compared the viability (percentage of live viruses in a whole viral population) of the human coronavirus 229E (HCoV) and the Feline Infectious Peritonitis Virus (FIPV) in tap water and wastewater, and their findings indicated that coronaviruses survived longer when present in primary-treated wastewater compared to secondary-treated wastewater, a fact that may be attributed to the higher solids content that may offer viral protection from inactivation. Another study by [39] ([preprint]) which was available on the 17th of April 2020 has indicated the reduction of SARS-CoV-2 RNA load by 100 times in treated effluents of three Parisian UWTPs compared to the raw sewage wastewater. However, this finding was not discussed in the 2nd version (May 6th, 2020) of the same manuscript ([6] [preprint]).
The disposal of SARS-CoV-2 in untreated sewage wastewater is of concern in countries where untreated sewage is disposed in rivers, due to the high risk of infection of the population and animals (livestock and wildlife) in contact with downstream wastewater. Swimming in sewage-contaminated water has previously been linked to respiratory disease, with earlier studies of respiratory infection in the Great Lakes being associated to adenoviruses [46]. More recently, researchers in Ecuador, a country that commonly practices this type of disposal, have shown the presence of SARS-CoV-2 RNA in three locations along Quito’s river, at concentrations of 2.84 × 105 to 3.19 × 106 gene copies/L for N1 target and 2.07 × 105 to 2.23 × 106 gene copies/L for N2 target [47]. The authors suggest that the measured concentrations reflect a large undiagnosed fraction of COVID-19 patients, as well as asymptomatic or pre-symptomatic cases. Another study by Haramoto et al. [33] ([preprint]) has shown the absence of SARS-CoV-2 from river water in the Yamanashi Prefecture Japan on three different sampling occasions between 17th and 7th of May, 2020, using four quantitative (N_Sarbeco, NIID_2019-nCoV_N, CDC N1 and N2) and two nested PCR assays (ORF1a and S protein). Rimoldi et al. [9] ([preprint]) also examined river water for the presence of SARS-CoV-2. Samples from the Lambro river and the Lambro Meridionale River were taken on April 14th and April 22nd 2020. Positive signals were obtained on the 14th of April 2020 in samples from both rivers, while only samples from Labro river were positive for the virus on the 22nd of April. The presence of the virus RNA in river samples has been attributed, according to the authors, to discharge of untreated sewage into the rivers, a situation that was exacerbated during an anomalous and long drought observed within the sampling period and to the lack of separation of the urban runoff waters from domestic effluents, leading to combined sewer overflows.
According to the World Health Organisation (WHO), SARS-CoV-2 is likely to possess a poor stability in wastewater and to be more susceptible to disinfectants (e.g. chlorine) compared to non-enveloped human enteric viruses (e.g. adenoviruses, rotavirus, norovirus, and hepatitis A) ([48,49]). The physicochemical conditions prevailing in wastewater such as pH, solids and the presence of micropollutants [15] may have a significant impact on the stability and survival of viruses such as noroviruses, astroviruses, rotaviruses and hepatitis viruses in wastewater, with alkalinity showing a strong detrimental effect on virus persistence in the solid and liquid wastewater component, due to inactivation of a large fraction of the viral population at high alkalinity levels [50].
In a study by Wang et al. [51], the virus most closely linked to SARS-CoV-2, being SARS-CoV-1, was shown to be very vulnerable to chlorine and chlorine dioxide disinfection, compared to E. coli and coliphage [51]. Chlorine dioxide (40 mg/L, 5 min) was found to be less efficient for the inactivation of SARS-CoV-1 than chlorine (20 mg/L, 1 min). The effect of wastewater disinfection on SARS-CoV-2 has not been elucidated yet, as there have been no reports to date on whether this virus is susceptible or persistent during the application of such processes. In the study of Randazzo et al. [26], 11% of samples collected after secondary treatment were found to be positive to SARS-CoV-2, while none of the tertiary effluent samples (sand filtration, flocculation/coagulation, NaClO disinfection coupled, is some cases, with UV) tested positive for the virus. Although these findings do not decipher the effect of the dose of the disinfectant and contact time on virus survival, they show that tertiary treatment and disinfection process may be adequate. In addition, the inactivation kinetics of SARS-CoV-2 (log inactivation vs Ct values) during various disinfection processes such as chlorination, ozonation, peracetic acid treatment, and UV irradiation in combination with oxidants (e.g., hydrogen peroxide) should be examined so that solid knowledge on the specific virus is obtained. Silverman and Boehm [52] provided a comprehensive review on the decay rates of human coronaviruses and of their viral surrogates (animal coronaviruses and the enveloped Pseudomonas phage Φ6) during disinfection with chlorine and UV irradiation in water/wastewater, suggesting that even though there is limited available data on the inactivation of coronaviruses upon their exposure to disinfectants, it is expected that the inactivation of coronaviruses may be efficient when doses of disinfectant recommended for non-enveloped viruses are applied. It is also critical that wastewater treatment facilities implement effective disinfection to ensure the virus does not spread via wastewater discharge or reuse schemes. Furthermore, a reduction in the used amounts of disinfectants can be achieved through the use of membrane technologies such as the Membrane Bioreactor (MBR) utilising ultrafiltration to separate virions of a size of 60-140 nm [15]. Currently, the potential route of the transmission of SARS-CoV-2 to humans in the wastewater-receiving environments via the reuse practice for agricultural irrigation has not been elucidated. Oliver et al. [40] ([preprint]) reported that the selection of irrigation technique is critical for minimising the spread of the virus to the environment and suggested that alternative irrigation techniques, e.g. micro-irrigation, should be considered. Taking into account that enveloped viruses are more likely to partition to solids and more susceptible to wastewater treatment than their non-enveloped enteric counterparts, the authors of this review paper are of the opinion that multi-barrier wastewater treatment processes will be effective in removing SARS-CoV-2, so that the associate environmental- and public health-associated risks for wastewater reuse are likely to be negligible.
2.1. Effect of wastewater sampling, storage conditions and sample pre-treatment on the vitality of SARS-CoV-2 RNA
2.1.1. Effect of sampling method
Both grab ([8][preprint];[34]; [9][preprint]; [26]) and composite([27][preprint];[29][preprint];[21][preprint];[23,38]; [8][preprint];[31][preprint]) sampling methods have been reported for the collection of wastewater samples for the detection of SARS-CoV-2. In the case of composite samples, both time- (i.e. fixed aliquot volume at defined time interval, e.g. 24 h) and flow-proportional (i.e. fixed aliquot volume at defined flow volume interval) samples were used. Among the studies conducted, ([8] [preprint]) have demonstrated that the most reliable average concentration of the virus RNA in wastewater was provided by the composite samples compared to grab samples. The viral titers in composite samples were found to be lower (∼1500 viral genomes/L) than those in the grab samples (∼2 × 104 viral genomes/L), while the variation between replicates was considerably lower. In general, there is limited literature regarding the impact of the type of sampling on the detection of viruses in wastewater. Gerba et al. [53] suggested that 24-h composite samples can enable catching the peak flows, and in the case where untreated wastewater is used for determining the level of viral inactivation requirements, peak concentrations of viruses should be considered rather than the average ones. The selection of the sampling time is also of crucial significance to the methodology applied for the detection of the virus. For example, in the case where grab samples are used, the viral concentration will be a mere snapshot of the particular sample. In most cities, the flow rate of sewage is highest in the morning and evening hours. It is noted that only few studies reported the time of grab sampling for the detection of SARS-CoV-2 in wastewater; 7-12 pm [26] and 1.00 pm ([9] [preprint]). Moreover, recording grab sampling time enables the accurate portrayal of the peak daily faecal load in wastewater via the measurement of indicators which are abundant in the wastewater specifically because of human shedding, are highly abundant in urban wastewater and are of specific geographic variability [54] (i.e. through the enumeration of faecal load indicators such as: i) faecal coliforms, ii) cross-assembly phages (CrAssphages), iii) norovirus, iv) Pepper Mild Mottle Virus and v) enterovirus concentrations), which may potentially be linked to the highest SARS-CoV-2 shedding within a day, from the infected persons within a community. On the other hand, the collection of composite samples will represent the average concentration of the virus RNA during the collection period, without being able to discriminate any peak values recorded within the sampling duration.
2.1.2. Effect of temperature of sample storage
Samples collected for viral determinations in environmental matrices are usually analysed within a short time span, but, in most cases, there is a need for storage at the laboratory prior to further processing and analysis. According to the virology and microbiology guidelines [55,56], samples intended for viral analysis within less than 48 h are usually kept at 4 °C in the dark, whereas storage at lower temperatures (20 °C or−80 °C) is deemed necessary for longer periods in order to maintain sample integrity. Generally, there is a paucity of information available on the effect of freezing process on the virus vitality (physiological capability of the live examined viruses [57]. The study of Olson et al. [58] on the effect of storage temperature on the viability of the MS2 bacteriophage in wastewater, revealed that viral degradation does not seem to occur when samples are stored at 4 °C for one week before degradation of the virus equalled the initial virus loss due to freezing at −80 °C. It was also observed that the virus titers were substantially lower after sample storage for an approximately 40-day period at 4 °C compared to those observed upon sample storage at −80 °C. Interestingly, viral degradation was shown to increase at -20 °C compared to 4 °C and−80 °C, due to the formation of large ice crystal, which provokes viral damage. Cryoprotectants (e.g. glycerol) have also been employed to retain phage infectivity over time [59]. Nevertheless, these agents have not been used in environmental samples (e.g. wastewater) and it remains unclear whether such preservatives will provide protective shield to the viruses from the detrimental effects of freezing and storage without affecting sample’s integrity. In the case of SARS-CoV-2, the storage of wastewater samples was performed at 4 °C ([34,35]; [21] [preprint]; [29] [preprint]; [26]; [6] [preprint]), -20 °C [23] and −80 °C ([27] [preprint]) with no information provided on the effect of temperature on the viral viability and vitality. Gundy et al. [45] have shown that coronaviruses (human coronavirus 229E, and feline infectious peritonitis virus) were more sensitive to temperature than poliovirus 1 (PV-1) and their survivability in the water matrix was shown to be affected by the level of the organic content, and the presence of antagonistic bacterial microorganisms. The inactivation of coronaviruses (decrease by 99.9%; T99.9) was found to be more rapid in tap water at 23 °C (10 days) than in the same medium at lower temperature (4 °C; >100 days), while they were completely inactivated in wastewater, with T99.9 values being 2 to 4 days.
2.1.3. Effect of thermal sample pre-treatment
In some studies, samples were subjected to thermal treatment (56 °C for 30 min or 60 °C for 90 min), prior to viral concentration, to increase the safety of the laboratory personnel during sample handling ([22]; [31] [preprint]). The thermal treatment of the sample was shown to reduce the infectivity of SARS-CoV-2 with over 5 logs without affecting its RNA structure ([60] [preprint]). Experiments using a surrogate virus (Mengovirus), confirmed that no loss of the viral RNA occurred when samples were treated at 56 °C for 30 min [23]. Similarly, raw sewage samples were pasteurized at 60 °C for 90 min to inactivate SARS-CoV-2 ([31] [preprint]). The thermal treatment of samples is consistent with previous studies dealing with enveloped virus survival in pasteurized wastewater [61]. Ye et al. [62] reported discrepancies in the inactivation of the non-enveloped MS2 virus compared to the enveloped viruses (MHV and ϕ6) in pasteurised and non-pasteurised wastewater, suggesting that this may be attributed to the bacterial extracellular enzyme activity as well as protozoan or metazoan predation. The time needed for 90% viral inactivation (T90) ranged between 7-13 h for the enveloped viruses ϕ6 and MHV in unpasteurised wastewater at 25 °C, whereas an increase in the T90 values to 28-36 h was observed at 10 °C. This suggests that enveloped viruses excreted in faeces may therefore reach UWTPs in an infective state, especially in regions with cool climate zones.
2.1.4. Effect of organic matter and suspended solids
Coronavirus (FIPV and HCoV) inactivation was shown to be higher in tap water which was subjected to filtration compared to unfiltered water and the survival of the coronaviruses was found to be affected by the level of suspended solids and organic matter as the viruses survived longer in primary-treated wastewater than secondary-treated wastewater [45]. In addition, it was observed that the titer of the coronaviruses significantly decreased by 99.9% in wastewater, compared to PV-1 (10% decrease) possibly due to the presence of organic compounds that may interact with the viral envelope and provoke inactivation. This observation also indicates that coronaviruses adsorb more readily than PV-1 to solids originally present in the wastewater, due to the hydrophobic character of the viral envelope, which renders coronaviruses less soluble in water and could therefore increase the propensity of these viruses to adsorb to the solids. Ye et al. [62] reported that the MHV virus and the Pseudomonas phage Φ6, which possess an enveloped structure, exhibited higher partitioning to solids present in wastewater compared to non-enveloped viruses (26% of MHV and Φ6 was adsorbed to solids compared to the 6% of the two non-enveloped viruses). Based on these findings, it can be inferred that a significant portion of SARS-CoV-2, may be adsorbed to solids and sewage sludge. Also, the adsorption kinetic experiments performed by Ye et al. [62] in both solids-containing and solids-free samples revealed that once equilibrium is reached, enveloped viruses seem to have greater affinity to solids than the non-enveloped viruses, and thus it may be assumed that the latter would be removed to a lesser extent than the former during primary treatment. The heterogeneity of collected wastewater samples in aircrafts due to a large fraction of particulate matter such as toilet paper, was also reported by Ahmed et al. [4], who consider this presence to act as a limiting factor for obtaining representative samples. Sewage sludge has been also shown to act as carrier of SARS-CoV-2 viral particles. Peccia et al. [37] ([preprint]) have shown that the concentration of SARS-CoV-2 RNA ranged from two to three orders of magnitude higher in primary sludge compared to raw wastewater due to the higher content of solids. The presence of the virus RNA in both primary and secondary sludge was also reported in [36] ([preprint]), who found that the copy numbers of SARS-CoV-2 in both types of sludge were similar (Ct ranging from 33.5 to 35.8 corresponding to titers of SARS-CoV-2 ranging from 1.17 × 104 to 4.02 × 104 per liter). It should be noted at this point, that the presence of the genetic sequence of the virus in the sludge solids, does not warrant the virulence of the virus itself [42]. Moreover, on the basis of the data currently available, it is currently not feasible to precisely define the level of the virus contamination for untreated sludge, or to specify a storage period beyond which the virus is inactivated. To date, there is also lack of information in relation to the sampling, storage and processing of sewage sludge for the detection of SARS-CoV-2.
2.2. Virus RNA concentration in wastewater as a key step in the detection methodology
2.2.1. Concentration in environmental matrices
Various sample concentration methods have been used in relation to virus detection and quantification in complex environmental matrices such as wastewater. The main challenge of the application of such methods concerns the low abundance of viral particles and the establishment of low enough Limits of Detection (LOD), requiring thus the utilisation of reliable concentration methods prior to viral RNA extraction [63]. The wastewater, due to its high chemical and biological complexity, may result in low viral recovery yields, or poorly reproducible yields or both [64], which may hinder the association of waterborne viruses to specific disease outbreaks. In addition, the complex composition of wastewater hinders the easy detection of viruses in such matrices, as both particulate and dissolved constituents inherently present in wastewater get concentrated along with the target virus and can influence the virus recovery yield of the concentration method. It is thus crucial to methods that yield low enough LOD that reflects the lowest possible concentrations of the virus that may be present in wastewater, which will accurately estimate very low prevalence of COVID-19 cases within the community. Various methods, either individual or combined (i.e. primary and secondary), were reported in the scientific literature for the concentration of viruses from aquatic matrices [65,66], including among others polyethylene glycol (PEG) precipitation [67,68], ferric chloride (FeCl3) precipitation [69], skimmed milk flocculation (SMF) [70], glass wool (GW) filtration [71] or monolithic adsorption filtration (MAF) [72], ultrafiltration (UF) [73], and ultracentrifugation [74].
The concentration method to be considered effective and applicable should be technically simple and fast, be able of processing large volume of water, provide a high viral recovery yield, be applicable for a variety of viruses, be repeatable (within a laboratory) and be reproducible (between laboratories), and be cost- and time-effective. No single concentration method was shown to fulfil all these requirements so far. One important observation made is that the effect on viral diversity, specificity, detection and viral community composition was found to be strongly affected by the type of concentration and the extraction method, and vice versa (Hjelmsø et al., 2016). Also, it was clearly demonstrated that the recovery yields during concentration differ significantly between the non-enveloped and enveloped viruses, with the studies focusing on the enveloped viruses, such as SARS-CoV-2, being limited. Since non-enveloped and enveloped viruses possess distinct structural characteristics, it is logical to be assumed that both viral types will not behave similarly. Thus, it is expected that the recovery of SARS-CoV-2 will be different from that of non-enveloped viruses, a fact that may result in high discrepancies (e.g. an order of magnitude) in the virus concentration in untreated wastewater. Given that the recovery of non-enveloped viruses was reported to be varied among the type of virus and the matrix under investigation [75], it is apparent that viral concentration controls are necessary to be used for assessing recovery efficiencies. The recovery of various viruses and of their surrogate controls in various matrices, including wastewater and sewage sludge, is provided in the comprehensive review of Haramoto et al. [75]. Nevertheless, relevant information on enveloped viruses is currently lacking.
PEG precipitation was found to be effective for concentrating human viruses in environmental samples, with recovery yield being 86% for hepatitis A virus, 87% for simian rotavirus, and 68% for poliovirus [68]. The PEG concentration method was also shown to have a remarkably higher proportion of viral reads compared to the SMF and GW methods, and the highest recovery of murine norovirus (MNV) and adenovirus 35 (HAdV) was obtained with PEG followed by MAF, GW, and SMF [63]. Instead, Falman et al. [69] reported that SMF resulted in a higher recovery of poliovirus type 1 (106 ± 24.8%) when compared to PEG/NaCl precipitation (59.5 ± 19.4%) in wastewater.
UF, employing tangential flow (i.e. cross-flow), has been also successfully utilised for the concentration of viruses in wastewater, but challenges of engineering such as membrane fouling and non-reversible adsorption of viruses to filtration components unit may affect the duration of the sample concentration and result in low recovery yields [64]. The use of pre-filtration prior to UF to minimise the fouling phenomena may result in the loss of viruses, especially when the latter are present in low concentrations in the effluent organic matter that may be retained by the membranes [46]. Fumian et al. [76] employed ultracentrifugation (100000×g for 1 h), as well as an electronegative membrane followed by secondary concentration with a centrifugal ultrafilter. The former concentration method resulted in a mean recovery of 47% (range of 34-60%) of rotavirus A from wastewater, while a lower mean recovery of the virus was observed (3.5%, range of 1.5-5.5%) using the combination of the membrane and the ultrafilter. In the study of Prata et al. [74], an average viral (Adenoviruses, Rotaviruses) recovery of 69% and 76% was observed for wastewater and recreational water samples, respectively, whereas the SMF flocculation method led to a much lower recovery of both viruses (38 and 22%, respectively).
Results also showed that GW filtration resulted in higher recoveries of the non-enveloped virus Poliovirus 3 (57.9%) compared to the other non-enveloped viruses (Bovine Coronavirus = 18.1%, Bovine Rotavirus group A = 22.1%, Bovine Viral Diarrhea Virus [type 1 and 2] = 15.6-19.7%) [77]. Blanco et al. [78] employed adsorption/elution onto GW and PEG precipitation for the concentration of the porcine coronavirus Transmissible Gastroenteritis Virus (TGEV) and the non-enveloped Hepatitis A virus (HAV) in environmental samples. The results have shown that the recovery of TGEV was improved by increasing the GW and eluent contact time and the elution pH, increasing PEG concentration, or performing the elution either by recirculation or under agitation. Also, it was reported that the addition of a detergent (Tween 80) hindered the TGEV recovery, by degrading the lipid-containing envelope of the virus.
Ye et al. [62] assessed PEG precipitation, ultracentrifugation, and ultrafiltration for concentrating the enveloped MHV virus and the non-enveloped phage MS2 in wastewater. Their findings indicated that the ultracentrifugation method resulted in negligible recovery yields (∼5%) for both studied viruses possibly due to the effect of the high g-force applied during ultracentrifugation on the viral survival. Also, it was found that 26% of the murine coronavirus was adsorbed to solids compared to 6% for MS2, suggesting that a proportion of the viruses particles may have been removed by the centrifugation step. The PEG precipitation method also yielded low recovery (∼5%) for the enveloped MHV virus, whereas in the case of MS2, the recovery was significantly higher (43.1%). However, the ultrafiltration method resulted in 25.1% recovery of MHV and 55.6% of MS2, indicating that higher recoveries may be achieved for the non-enveloped viruses using this concentration method. Following the SARS-CoV-1 outbreak of 2003, Wang et al. [51] assessed the recovery of SARS-CoV-1 and of a surrogate virus, bacteriophage f2, in both urban and hospital wastewater, using an electropositive filter media particle (Al(OH)3) packed in a glass column. Interestingly, the virus recovery ranged from 0% (sewage from a housing estate) to 21.4% (sewage from the hospital), while the recovery of phage f2 under the same conditions was found to be significantly higher (33.6% - >100%).
2.2.2. Concentration of SARS-CoV-2 in wastewater
In the case of SARS-CoV-2, a variety of concentration methods has been used, with ultracentrifugation being the most studied method ([29] [preprint]; [21] [preprint]; [38]; [6] [preprint]). In general, the different wastewater matrices, the different concentration methods, and the fact that few studies exist that provide the recovery yield of the virus with the concentration methods used, do not allow for a systematic comparison among the various studies performed.
The volume of wastewater to be concentrated can influence the viral recovery yield and it seems that there is a discrepancy in the scientific literature on the appropriate volume that each concentration method requires. According to Haramoto et al. [79], concentrating a volume of <100 mL of untreated wastewater seems to be considered adequate for detecting enteric viruses, whereas higher volumes (1 L) of sample were suggested as able to obtain high concentration of enteric viruses in both untreated and treated wastewater [80], depending of course on the concentration method. In the case of the SARS-CoV-2, up to 500 mL (minimum volume used: 100 mL) of raw wastewater were concentrated ([21] [preprint]; [38]; [8] [preprint]; [26]; [31] [preprint]; [6] [preprint]), whereas in only one study, 2 L of untreated wastewater were collected [35]. It is highlighted that a higher volume of wastewater sample should be used for sample concentration in the regions where the number of COVID-19 recorded cases is low and thus the prevalence of SARS-CoV-2 in wastewater is expected to be low as well.
A surrogate virus possessing similar structural/molecular characteristics (e.g. shape, functional groups, surface charge, etc.) as SARS-CoV-2 was used as an indicator for assessing the recovery yield of the concentration methods. The enveloped murine hepatitis virus (MHV), which belongs to the Coronaviridae family as SARS-CoV-2, has been used as surrogate virus for assessing the recovery yield of SARS-CoV-2 in wastewater [35]. Murine norovirus (Caliciviridae family) has been also used as model virus for both enveloped and non-enveloped viruses [62,81]. In addition, the porcine epidemic diarrhea virus (PEDV), an enveloped virus belonging to the Coronaviridae family, as well as the mengovirus (MgV) vMC0 (CECT 100000), a non-enveloped member of the Picornaviridae family, have been also utilised to evaluate the recovery of SARS-CoV-2 in wastewater [26]. There are many points that need to be taken into consideration when using surrogate viruses, such as the level of the surrogate especially when high volumes of wastewater are processed.
The recovery values reported so far in relation to the surrogates of SARS-CoV-2 vary greatly (3.3-73%), and currently, there is no consensus on the threshold recovery yield. This great variability of recoveries by the different applied methods in the available studies, dictates for optimization of the detection and quantification method for more reliable measurements that are comparable among them, as a method with a 5% of spiked surrogate virus recovery cannot be directly comparable with a method that has shown a recovery of 73% of the same surrogate. Randazzo et al. [26] used the porcine epidemic diarrhea virus (PEDV) strain CV777 and the mengovirus (MgV) vMC0 (CECT 100000) to evaluate the aluminum hydroxide adsorption-precipitation method followed by ultracentrifugation (1700×g for 20 min and 1900×g for 30 min). Both MgV and PEDV in wastewater influent yielded similar recovery values of 11 ± 2.1% and 11 ± 3.5% for PEDV and MgV, respectively, indicating that more trials are needed for the improvement of the recovery value of the enveloped viruses and their surrogates in complex matrices such as wastewater. On the other hand, there was a significant difference between the recovery of PEDV (3.3 ± 1.6%) and MgV (6.2 ± 1.0%) in wastewater effluents. The recovery of F-specific RNA phages by ultracentrifugation (4654×g for 30 min followed by 1500×g for 15 min) was found to be 73 ± 50% [38]. According to Medema et al. [38], no specific trends were observed for the sample volume processed and the phage recovery, and it was suggested that the non-enveloped F-specific RNA phages may lead to overestimation of the recovery efficiency of enveloped SARS-CoV-2. In another study conducted by Ahmed et al. [35], various concentration methods (i.e. adsorption-extraction, PEG precipitation, centrifugal filter device method, and ultracentrifugation), were assessed in relation to their efficiency to recover MHV from untreated wastewater. The MHV recovery was calculated based on the quantified copies by RT-qPCR by dividing the total viral RNA gene copies recovered with those seeded. The findings have shown that the recovery of MHV was in the range of 26.7-65.7%, with the most effective methods being the adsorption-extraction method (in both the presence and absence of MgCl2 pre-treatment) followed by the Amicon® Ultra-15 centrifugal filter device. Adsorption-extraction method with acidification and PEG resulted in the lowest MHV recovery. An interesting observation made was that the MHV recovery obtained using PEG precipitation (44%) was much higher than the value reported in Ye et al. [62] (∼5%), and this may be attributed to the fact that MHV was concentrated from both liquid and solid wastewater fraction, whereas in Ye et al. [62], the concentration of the virus was performed only from the liquid portion. The results of this study highlighted that virus concentration should be carried out not only in the liquid phase of wastewater but also the solid fraction should not be overlooked.
[27]([preprint]) successfully applied centrifugation to remove large particles in wastewater samples, and secondary concentration using alum or PEG (20 mg L-1), followed by additional centrifugation, resulting in positive Ct values of SARS-CoV-2 of 33.6 and 33 for alum and PEG, respectively. In the study of La Rosa et al. [22], the concentration of samples was performed using the PEG-dextran method, according to the WHO guidelines for poliovirus (the latter was adapted to enveloped viruses, and the chloroform treatment included in the protocol was neglected to retain the integrity of the viruses). The PEG concentration method in conjunction with centrifugation (40 mL, 12000×g for 120 min) was also used by [31] ([preprint]). Sample concentration with electronegative membranes and ultrafiltration produced inconsistent results according to the study of Ahmed et al. [34], as different values (positive/negative) were observed for SARS-CoV-2. According to the authors, the rationale behind the use of the electronegative membrane was that enveloped viruses (e.g. MHV, phage Φ6) exhibit higher adsorption to the solids compared to non-enveloped viruses [62]. This is also in agreement with the findings of Haramoto et al. [79], who observed high adsorption of the enveloped koi herpesvirus virus (KHV) to the electronegative membranes [79].
It is obvious that a tailored to the SARS-CoV-2 method is required for its concentration in the wastewater, and optimisation should take place in terms of sample characteristics and effective volume, whilst considering both organic and inorganic inhibitors that could affect viral recovery efficiency and subsequently virus detection.
2.3. Concerns in relation to RNA extraction of SARS-CoV-2 in wastewater samples
Following the concentration of wastewater samples, the viral RNA extraction process aims at obtaining the RNA from the sample matrix, without damaging it. The choice of an appropriate protocol can be a challenge, as the breakage of the viral particles must be considered without damaging the nucleic acids, whilst maximising nucleic acid recovery. The three main utilized techniques used for RNA extraction currently include organic extraction with the use of solutions such as phenol-guanidine isothiocyanate, silica-membrane based spin column techniques and the use of paramagnetic particles. The first one being the most popular, has the disadvantage of sample contamination with proteins and other substances such as organic solvents like phenol-chloroform, salts and ethanol [82]. Silica columns and paramagnetic particle-based RNA extraction techniques do not make use of organic solvents and are simpler to use, efficient and low cost while they have lower levels of contamination from proteinic and other compounds. Nevertheless, despite their advantages, they bear the disadvantage of potentially high levels of genomic DNA contamination [82].
The main steps of RNA extraction in any type of sample, are the following (other methods may utilize some of the steps or similar ones) (Table 3):
-
1
Cell lysis: The step of cell lysis or cellular disruption leads to the breaking down of the cell membrane outer boundary for the release of RNA from the cell. Ceramic, steel or silica beads (magnetic or not) with sharp edges are particularly useful for physical damage of the viral membrane, for the release of the nucleic acids contained inside viral particles. Currently, a lysis step is incorporated into the extraction process, either after a cell extraction step from the matrix or directly within the matrix, followed by released nucleic acid recovery. Viral cell lysis can be otherwise achieved with the use of buffers or reagents such as guanidinium isothiocyanate, guanidinium chloride, sodium dodecyl sulphate (SDS) and others. To this end, solutions such as TRIzol can be used for the maintenance of RNA integrity. Various steps may be added by each manufacturer, aiming at improving purity, yield and analyte detection [83].
-
2
Denaturation of DNA and proteins: DNase may be used for DNA degradation, while commonly, proteinase K is used for protein digestion. Otherwise, organic extraction using phenol and chloroform or dissolving the sample in buffers which contain guanidium salts can be used for protein removal.
-
3
Denaturation and inactivation of RNases: the use of any organic chaotropic agent such as phenol and chloroform can be efficient for RNase inactivation and denaturation.
-
4
Separation or removal of cellular components: In order to separate RNA from the rest of the cellular components present in a solution, chloroform may be added followed by centrifugation, in order to separate the organic from the aqueous phases (RNA component).
-
5
RNA recovery: RNA recovery from the aqueous phase is done using isopropyl alcohol or with ammonium acetate or lithium chloride for selective precipitation of RNA from DNA.
-
6
RNA elution: The final treatment of RNA is done in the elution step, where the total RNA obtained during the RNA extraction is eluted into a 40-100 μL of eluent buffer.
Regarding SARS-CoV-2, various extraction systems have been qualified and validated by the USA Center for Disease Control (CDC) for use with the 2019-nCoV real time RT-PCR diagnostic panel. However, the rapid increase in testing during the COVID-19 pandemic, has led to a global shortage in commercially available extraction kits. Thus, other, non-CDC-validated RNA extraction kits have been studied by various authors in respect to SARS-CoV-2 detection and enumeration by RT-PCR. However, no studies so far, have compared different RNA extraction methods in order to establish their RNA extraction efficiency in influent wastewater. To this end, no standardization of RNA extraction protocols exists, to allow for comparable extraction of the SARS-CoV-2 from influent wastewater, a matrix that is highly complex that also contains a high variety of compounds, organic and inorganic, that may be inhibitory towards RT-qPCR analysis.
The main difficulties that may be faced during RNA extraction and subsequently the detection of SARS-CoV-2 in influent wastewater using a variety of RNA extraction protocols and commercially available kits, include obtaining sufficient nucleic acid amounts which may arise from incomplete cell lysis and surface binding of nucleic acids, low yields of nucleic acids and inter- and intra-process variability [83]. Besides, a potentially high degree of secondary RNA folding leading to low yield and difficult downstream analysis, inaccurate copying during replication leading to high mutation rates may lead to under-estimation of RNA content and to inaccurate RT-PCR analyses.
Besides matrix-introduced interfering substances, care must be taken during RNA extraction of complex matrices such as influent wastewater, which contains various enzymatic molecules including RNases that degrade RNA, due to the fact that RNA, a single-stranded molecule is more prone to damage and disintegration than DNA, a double stranded molecule. Besides the fact that RNases are abundant in environmental matrices and also on hands and surfaces, they are difficult to remove or destroy completely. Other introduced inhibitors include glove powder, salts such as sodium chloride and potassium chloride, detergent molecules such as EDTA, ethanol, phenol and isopropyl alcohol [84]. Therefore, careful handling of samples and utilisation of aseptic techniques is crucial, along with the use of RNase-free reagents and solutions as well as RNase-free glassware and pipette tips.
All substances that may cause problems of the RT-PCR process have been collectively called PCR inhibitors, and the main impact of partial or total reaction inhibition is decreased sensitivity or false-negative results, respectively [85]. PCR inhibitor compounds include calcium ions, bile salts, urea, phenol, ethanol, polysaccharides, sodium dodecyl sulphate (SDS) and other proteins including collagen, myoglobin, haemoglobin and proteinases [85,86]. Different process steps may be affected by the presence of inhibitors. Nucleases may degrade template RNA produced after RNA extraction while phenols may cross-link to RNA under oxidising conditions, leading to hindering of the RNA extraction process. Polysaccharides present in wastewater may limit resuspension of precipitated RNA capacity, while melanin may inhibit reverse transcription. Competitive binding of inhibitors to the template RNA instead of primer annealing leads to the need for careful primer design which aims at higher melting temperatures [87]. The detection of low viral concentrations in treated wastewater or fresh waters may require the concentration of large volumes of samples. However, increasing the sample volume also means that the concentration of the sample leads to the concentration of different inhibitors in the sample, which interfere with RT-PCR reactions, such as humic and fluvic acids.
Suggested methods to deal with the presence of inhibitors in samples include solvent extraction, column chromatography and silica columns, cation exchange resins and magnetic silica beads which purify complex samples such as wastewater. The rationale behind treating samples before RNA extraction is to use the viral capsid to protect the viral nucleic acids. Another widely accepted method of interference reduction, is the dilution of samples of extracted nucleic acids, resulting in an immediate dilution of inhibitory substances. Despite the advantage of the reduction of inhibition, there is a decrease in sensitivity due to the dilution of the nucleic acid concentration in extracts. For this reason, the addition of compounds in the extracted RNA that may counteract inhibition such as betaine, bovine serum albumin (BSA), glycerol, non-ionic detergents, polyethylene glycol (PEG) and proteinase inhibitors has been suggested [85,88].
2.4. RT-qPCR aspects during SARS-CoV-2 analysis
The main detection and quantification method of SARS-CoV-2 in water and wastewater is the RT-qPCR, which is based on the presence of TaqMan detection probe assay. This type of assay is specifically designed for the increase of specificity of qPCR reactions and relies on: a) the 5’-3’exonuclease activity of Taq polymerase for cleaving a dual-labelled probe during the hybridization step of qPCR and b) fluorophore-based detection. During the exponential phase of the PCR process, quantitation of the product is made possible through the fluorescent signal emitted. Furthermore, this assay involves the labelling of the probes with two different fluorescent dyes that emit at different wavelengths. More specifically, the probe is a sequence (DNA or RNA oligo) that has the purpose of hybridising in the DNA target region between two PCR primers. Normally, the annealing temperature of the probe is higher compared to the one of the PCR primers (forward, F and reverse, R primers). In this way, the probe hybridizes along with the start of extension of the primers. The two different fluorescent dyes are the ‘reporter’ (R) dye that is attached to the 5’-end of the sequence of the probe, and on the other end of the sequence, a ‘quencher’ (Q) dye is synthesized. Common dyes for the TaqMan detection probe assay include among others 6-carboxyfluorescein (FAM), carboxyrhodamine (ROX) and tetrachlorofluorescein (TET) reporter dyes [89]. Quenchers, whose role is the quenching of the emitted fluorescence by the reporter when excited by a light source when the fluorophore is in close proximity to the quencher, include the Black Hole Quencher (BHQ), TAMRA and Deep Dark Quenchers I and II [90].
TaqMan assays that have been utilised for the detection and quantification of the SARS-CoV-2 nucleic acids in wastewater so far, target the virus nucleocapsid (N1, N2 and N3) protein genes, the RNA-dependent RNA polymerase (RdRp) and the envelope (E) of the virus [91]. The discrimination of SARS-CoV-2 genetic material from other types of SARS-CoV including bat-related SARS-CoV has been made possible through the availability of two different fluorescent probes for the RdRp gene. However, it was shown that the RT-qPCR assay for the E gene can react with SARS-CoV-2 and with SARS-CoV [46]. There are reports that only the three N protein gene detection assays have worked well for the detection of SARS-CoV-2 [46,92]. Ahmed et al. [4] report that among five different RT-qPCR assays (CDCN1, CDC N2, N_Sarbeco, NIID_2019-nCoV_N and E_Sarbeco), the CDC N1 assay has been shown to be the most sensitive one while the N_Sarbeco showed the least sensitivity, while positive samples among wastewater samples of a cruise ship and three aircrafts were near the assay LOD (37 to 40 cycles), providing inconsistent results among the tested assays. To this end, various protocols and target gene regions have been recommended for research use by national and international organizations, including the Charité, Berlin (WHO) protocol primer and probe panel which includes the RdRp and the E_Sarbeco (E gene) primers and probes and the CDC has published a Real-Time RT-PCR Diagnostic Panel including all materials, reagents and guidelines needed for qualitative detection of SARS-CoV-2 [48,49]. The primers and probes included in the CDC guidelines include the primers and probes for specific detection of the N1, N2 and RdRp genes, whose quality control and authorization was completed (US CDC, 2020). However, it must be highlighted that these assays have been developed for clinical samples and not for environmental ones, making their application one that requires additional attention in wastewater-based analyses.
It is important to note that a positive result for one out of the whole assay of gene targets, does not guarantee the presence of SARS-CoV-2 in the analyzed samples. False positive results may influence the analytical process in the absence of sound and coherent quality control procedures, due to potential cross-contamination of the qPCR assays. Moreover, the lack of confirmation of the positive result of one gene target with a second gene target, may cause doubt about the actual presence of the virus. To avoid the misinterpretation of laboratory contamination of samples as false positive results, the use of positive controls (synthetic or natural) species-specific sequences is recommended, which contain a readily detectable sequence with the applied RT-qPCR methodology [93]. On the other hand, [24] ([preprint]) suggest the confirmation of the specificity of the reaction and thus eliminating any concerns about cross-reactivity of the SARS-CoV-2 assays with untested organisms or uncharacterized viruses, through the absence of amplification in negative control or ‘blank’ samples.
A complete list of the available primer and probe assays available by acknowledged institutions and organizations and adopted by the World Health Organisation (WHO), is provided in Table 4 . Despite the availability of only few primer and probe assay sets available for the detection and quantification of the SARS-CoV-2, there is a lack of coherence in the combinations of assay sets used, in each available study, so far. In more detail, each available study uses a different combination of primer and probe assays (Table 5 ). This analytical heterogeneity also bears the inherent disadvantage of the differentiated sensitivity of each assay, especially to low numbers of SARS-CoV-2 copies/volume of sample, which is determined by the LOD of the RT-qPCR reaction, the efficiency of sample processing (sample concentration and RNA extraction) and the cycling conditions present. A difference in performance of SARS-CoV-2 assays has been observed due to differences in priming efficiency, the protocols used and the viral RNA secondary structure and/or stability. As stated by Bustin and Nolan [94], primers are the single most critical component of a reliable RT-qPCR assay, due to the fact that their properties influence the specificity and sensitivity of the method. As a result, poor experimental design in combination to poorly optimized reactions may lead to false negative results [95].
Table 4.
Adopted from the World Health Organisation (2020).
| Institution | Gene targets |
|---|---|
| US CDC, USA | N1, N2, N3 |
| Charité, Berlin, Germany | RdRp, E, N |
| China CDC, China | ORF1ab and N |
| Institute Pasteur, France | Two targets in RdRp (IP2 and IP4) |
| National Institute of Infectious Diseases, Japan | Pancorona and multiple targets, Spike protein |
| HKU, Hong Kong | ORF1b-nsp14, N |
| National Institute of Health, Thailand | N |
As already stated in previous work, a low enough LOD is required for testing samples with low virus concentration, mainly due to virus concentration dilution effects and low prevalence of the COVID-19. This may be achieved through further improvement of the analytical sensitivity of the existing SARS-CoV-2 assays in wastewater, by employing concentration and extraction methods able to recover more than 50% of the SARS-CoV-2 RNA from wastewater [4]. However, many of the studies ineffectively document the quality control measures of their study, including the LOD of their method. Rapid developments during the COVID-19 pandemic and the need to apply rapid testing and screening of samples, has led to a lack of coherent gathering of data in produced studies and thus to a lack of LOD and quality control information in published works.
The use of process controls in order to keep track of the levels of target recovery and measurement efficiency is crucial. The application of such quality control measures prevents the appearance of false negative results and assures the analyst that even very low concentrations of SARS-CoV-2 in complex matrices such as influent wastewater can be detected with the utilized methods, to accurately detect low incidences of COVID-19 within examined communities. Potential inhibition of measurement arising from each step of the sample treatment/analysis process is also more obvious after the use of process controls, which include according to Haramoto et al. [75] three types: i) whole process surrogate controls to be inoculated in a water sample before virus concentration, ii) molecular process controls inoculated into the viral concentrate and iii) RT-qPCR controls which must be inoculated before RT-qPCR. Especially inoculation before RT-qPCR of a positive surrogate control virus, along with the appropriate negative controls and non-template controls, provides adequate evidence of potential sample and reagent contamination, low reaction efficiency as well as need for further optimisation of the process.
To increase the robustness of RT-qPCR results and assure the absence of false positive results of such a highly diverse selection of assays applied in the different research work so far, sequencing of positive samples has been highly recommended. Besides confirmation of the presence of the specific virus, the phylogenetic origin and mutations of the SARS-CoV-2 virus strains present in positive samples can be identified by amplifying phylogenetically informative regions ([8] [preprint]).
The check of the process performance and efficiency is also critical in the quality assurance procedure, as comparison of two or more reactions where a condition is changed (e.g. different reaction mixes, different instruments etc.) is only enabled once it is assured that all process steps and their associated parameters have been checked. To achieve the successful intra- and inter-laboratory comparison of processes and results, it is thus important to evaluate the following process efficiency-associated parameters [96]:
-
i)
Linear dynamic range is the mathematical variation of the slope or efficiency when testing serial dilutions of the same sample, in replicates. Repeated analysis of the same dilution provides a standard deviation that provides information on the ability to repeat a single measurement.
-
ii)
R2value is a statistical term which indicates how good a value is at predicting another, and ranges from 0 to 1. An R2 value above 0.99 gives good statistical confidence in the correlation between two values.
-
iii)
Precision is estimated through the standard deviation among measurements. The closer that the majority of measurements are to the mean value, the smaller the standard deviation. For example, in a 100% efficient PCR analysis, the difference among successive serial dilution measurements is close to 1.
-
iv)
Sensitivity is the ability of a system to successfully amplify and detect one copy of the starting template. To achieve high sensitivity of measurement, a high number of replicates would be required for high statistical significance.
Regarding the RT-qPCR analytical aspect of the SARS-CoV-2 in wastewater, there was shown over the past few months, an enormous variety in selected reagents used for the detection and enumeration of the virus nucleic acids. After RNA extraction, the quality of the RNA (either concentration or 260/280 and 230/260 ratios). As shown in Table 5, it is evident that each study uses different PCR reaction mixes, which may have a different reaction efficiency with the specific primers. The composition of each PCR reaction mix (salt concentration, pH level) may have an impact on the threshold of the Ct values and thus of related viral concentrations, in cases of quantification. The variety observed in reagents and methodology, highlights the need for method and reagent optimisation among testing laboratories, to assure that analyses are being done in a coherent and correct way, with all the necessary controls and checks of quality all along the process.
2.5. Reporting of results
One key aspect among conducted studies on the presence and quantity of SARS-CoV-2 in wastewater is the type of reporting, and the ability to perform inter-laboratory comparisons among studies. So far, the results of the available studies have been reported in two distinct ways, based on the type of result sought (Table 6 ): i) absence or presence of the virus in the form of Ct values reported directly by the used qPCR instrument and ii) gene copies/volume of sample, with the use of a quantitative calibration curve of Ct values against known concentrations of the virus for the calculation of the gene copies present in a certain sample volume (relative quantification).
The Ct value comprises an intersection between the qPCR amplification curve and a threshold line and is a relative measure of gene target concentration, through fluorescence emission based on the concentration of the target gene fragment in the analysed samples. The fluorescence signal is recorded during every cycle and represent the amount of product amplified during the exponential phase of the qPCR reaction up to that point, while a higher amount of template leads to fewer cycles (Ct values) needed to record a fluorescent signal above the background. However, important aspects of the Ct value results are not always presented in the available studies, so far. More specifically, according to Bustin and Nolan [94], Ct values are subjected to inter-run variation and should not be conveyed without proper calibration standards. Further, artefacts in the prepared reaction mix may change the fluorescence associated with Ct and the subsequent quantification calculations, resulting in template-independent Ct shifts. Besides, quantification made in low-efficiency conditions may lead to a different calibration curve with a different slope to one under high-efficiency conditions, for the same target concentration. Low concentrations of the target sequence may lead to inadequate pairing of primers with the template during the first qPCR cycles. As a result, different fractions of the template may get amplified, leading to large variability giving a high margin of uncertainty.
The calculation of gene copies/volume of sample must be carefully prepared, initially with the calculation of mean Ct from associated technical replicates, followed by the calculation of the relative quantity of gene copies after the estimation of the efficiency of the process and finally by the enumeration of gene copies per volume of samples alongside the performance of statistical analysis.
Beyond applying the quality control measures necessary for the assurance of the quality of the results, it is important when publishing qPCR experimental data, to make sure that the data complies to the Minimum Information for Publication of Quantitative real-time PCR experiments (MIQE) guidelines [97]. These guidelines provide a clear framework of qPCR experiment conduction and guidelines on the use of qPCR results in scientific manuscripts, with the purpose of achieving consistent, comparable and homogenised high-quality reported data. According to the MIQE guidelines, it is important to provide detailed information on the data analysis methods, the estimation of confidence and software used. The reporting of assay precision with the use of statistical methods for analysis of variances is also key, along with the report of the concentration of gene copies/volume of sample, for quantitative and qualitative reported results.
Qualitative analysis (reporting of presence/absence of the SARS-CoV-2 gene targets) has been shown to be a highly utilised reporting method. However, a qualitative assessment of the presence of the virus target genes still necessitates the assessment of the sensitivity of the qPCR assay to very low target concentrations to prove if indeed the applied method can detect even a few gene copies of the virus. Thus, the reporting in the scientific literature of assay performance characteristics, including the linear dynamic range, the R2, the precision, the sensitivity and the LOD of the applied method is essential. Moreover, differences in abundance and recovery efficiency of SARS-CoV-2 among different studies, may not be due to the actual concentration of the virus found in wastewater, but because of methodological discrepancies that currently exist, including the concentration methods, RNA extraction strategies and used RT-qPCR assays [30]. Unfortunately, the COVID-19 pandemic restrictions to laboratory access, to purchase of standards and appropriate reagents to effectively validate the applied assays, have made the comparison of different studies difficult, while at the same time improvement in the quality controls used within each laboratory need to be improved according to validated standards such as the MIQE guidelines.
The currently available studies provide a good basis for the preliminary assessment the concentration of SARS-CoV-2 in wastewater, in relation to the number of COVID-19 patients in the catchment area. In order to be able to utilise the obtained information to the SARS-CoV-2 WBE framework, it is necessary to be able to precisely associate the gene copies/volume of sample results, to the prevalence of COVID-19 cases within the WWTP-served community. This estimation requires background knowledge which remains to be answered to date, on the number of diseased individuals, the rate of shedding of the virus in human faeces and the range of time within this shedding takes place, to reach the sewage network. An effort was made by Medema et al. ([21] [preprint]; [38]) to estimate the prevalence of infected persons among a UWTP population which may yield positive signals during wastewater analysis for the virus. The results showed a prevalence of 0.1 case per 100000 when using N1 and N2 targets [38], while N3 and E targets provided a positive signal at 3.5 cases per 100000 people [21] [preprint]. Another study by Jorgensen et al. [99][preprint] has estimated that a positive signal of the viral RNA in wastewater may be produced if there are around 3 cases per 10000 people. However, it is highlighted by the authors that this estimation is based on assumptions that may likely convey lower than normally accepted precision. Moreover, the amount of travel time once in the sewage network, to the UWTP is needed. However, an important aspect of the COVID-19 to consider is the residence time of the virus in each diseased individual, keeping in mind that asymptomatic or paucisymptomatic (few symptoms) people that have not been included in COVID-19 estimations also contribute to the SARS-CoV-2 in wastewater. At a community level, to drive the development of the analytical methodology of SARS-CoV-2 in wastewater forward, the determination of the minimum number of COVID-19 cases present within a community that allows the detection and exact quantification of SARS-CoV-2 in wastewater remains to be explored.
In the currently available studies, a change in SARS-CoV-2 RNA concentration has been observed as the number of COVID-19 cases changed ([27] [preprint]; [38]). Interestingly, in the study by Medema et al. [38], the slopes of the quantified gene targets N1 and N2 were shown to change according to the prevalence of COVID-19, by 0.1 case per 100000 person equivalent, something that was not noticed in the case of the N3 gene target, suggesting a reduced sensitivity of the concentration of this gene to the prevalence of cases in the served UWTP area. Moreover, in a study by [27] ([preprint]), the Ct values of SARS-CoV-2 RNA (E gene) has been correlated to the general number of COVID-19 positive individuals in Tel Aviv, with a high R2 correlation value (R2 = 0.998). [6] ([preprint]) also proved an increase in SARS-CoV-2 concentration along with COVID-19 cases, providing also indirect evidence of significant reduction of virus transmission (via SARS-CoV-2 concentrations in wastewater) as a result of lockdown measures in Paris. On the other hand, Randazzo et al. [26] showed an average concentration of 5.4 ± 0.2 log10 gene copies/L (N1, N2, N3) in influent wastewater in Spain without an association to the COVID-19 cases in the UWTP catchment area, highlighting the need for an improved quantitative model that includes further information on the variables that affect wastewater data, for a better interpretation of the available information. However, the limitations that span the detection methodology in general as discussed herein, impose limitations to the causation that may prevail by the COVID-19 cases, to the SARS-CoV-2 concentration. Despite the current findings, all available studies acknowledge the fact that an absolute comparison between COVID-19 cases with SARS-CoV-2 RNA is still difficult, due to the variability in COVID-19 case testing and reporting in each country, as well as the regulatory framework in place regarding testing of COVID-19.
3. Next steps in methodology development
The discussion provided herein has presented the main aspects of the methodology of SARS-CoV-2 detection and quantification in wastewater to be considered. The review of the available information has thus led to the conception and proposal of key points that, if considered collectively, may lead to significant improvements of the produced research work, worldwide. The suggested key points, according to the process step to which they belong, are provided in Fig. 1 .
Fig. 1.
Suggested key points according to each process step of the SARS-CoV-2 methodology for consideration.
Systematic evaluation of each step of the applied methodology must take place to assure the quality of results as well as their accuracy. This can be achieved through the application of recovery efficiency controls of the examined virus and of its related surrogates, and of quality controls at each step of the way along the methodology (LOD, LOQ, positive controls, negative controls). The evaluation of process efficiency-associated parameters is also of crucial importance for process and result credibility. These parameters (i.e. linear dynamic range, value of R2, precision, sensitivity) provide the base for intra- and inter-laboratory comparisons of different processes and results, as crude results are evaluated. Moreover, homogeneity of reporting of results is another key aspect that needs to be optimized among research groups and studies. Homogeneous reporting enables global comparison and assessment of results despite the use of different methods among research groups, providing also a basis for further collaborations and creation of databases that will target the gathering of all relevant information in one place for all interested parties. Increased methodological reliability and reliable estimation of infected cases in a given population that are needed for a positive analytical signal in wastewater, will furthermore provide an additional tool to the health sector for the assessment of the status of an infectious disease such as COVID-19 before its spread among the community, especially in areas including vulnerable infectious disease zones such as refugee camps, elderly residences and medical facilities [98]. In this way, zones of infection ‘peaks’ which may not have symptomatic patients may be detected in a timely manner, allowing for early measures and precautions to prevent the further spread of the disease, making WBE a truly powerful tool for the protection of public and environmental health.
4. Concluding remarks and outlook
Taking into account the current situation and the various conditions created due to the COVID-19 pandemic, as well as the need for a swift response to deal with its health- and social-side implications, WBE may transform the wastewater infrastructure into a public health observatory. Currently, as confirmed in the available literature, there is an absence of an optimised and univocal methodological framework concerning the detection and quantification of SARS-CoV-2 in wastewater. Testing and comparing various processes for achieving the same methodological step by single laboratories, with the use of appropriate process controls and quality assurance, and also inter-laboratory comparisons should take place. The wastewater community is to be commented on the huge efforts it made during the current pandemic. As history has shown, necessity is the mother of invention. And in this case, the need is pushing science and research towards important advances in relation to the current state of knowledge as to what wastewater monitoring can achieve, and at the same time is opens new directions toward transforming the wastewater infrastructure into a source of obtaining credible information for the benefit of the health sector and our societies.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. The Science of the total environment. 2020;726 doi: 10.1016/j.scitotenv.2020.138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brouwer A.F., Eisenberg J.N., Pomeroy C.D., Shulman L.M., Hindiyeh M., Manor Y., Grotto I., Koopman J.S., Eisenberg M.C. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proceedings of the National Academy of Sciences. 2018;115(45):E10625–E10633. doi: 10.1073/pnas.1808798115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muirhead A., Zhu K., Brown J., Basu M., brinton M.A., Costa F., Hayat M.J., Stauber C.E. Zika Virus RNA persistence in sewage. Environmental Science and Technology Letters. 2020 doi: 10.1021/acs.estlett.0c00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travelers. Journal of Travel Medicine, taaa116. 2020 doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L., Lou J., Bai Y., Wang M. COVID-19 disease with positive fecal and negative pharyngeal and sputum viral tests. American Journal of Gastroenterology. 2020:115. doi: 10.14309/ajg.0000000000000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wurtzer S., Marechal V., Mouchel J., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fongaro G., Stoco P.H., Sobral Marques Souza D., Grisars E.C., Magri M.E., Rogovski P., Schörner A.M., Barazzetti F.H., Christoff A.P., de Oliveira L.F.V., Bazzo M.L., Wagner G., Hernández M., Rodriguez-Lázaro D. SARS-CoV-2 in human sewage in Santa Catalina, Brazil, November 2019. medRxiv. 2020 doi: 10.1101/2020.06.26.20140731. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. medRxiv. 2020 doi: 10.1101/2020.04.15.20066746. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. medRxiv. 2020 doi: 10.1101/2020.05.01.20086009. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordgren J., Matussek A., Mattsson A., Svensson L., Lindgren P. Prevalence of norovirus and factors influencing virus concentrations during one year in a full-scale wastewater treatment plant. Water Research. 2009;43(4):1117–1125. doi: 10.1016/j.watres.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 11.Li D., Gu A., Zeng S., Yang W., He M., Shi H. Monitoring and evaluation of infectious rotaviruses in various wastewater effluents and receiving waters revealed correlation and seasonal pattern of occurrences. Journal of applied microbiology. 2011;110(5):1129–1137. doi: 10.1111/j.1365-2672.2011.04954.x. [DOI] [PubMed] [Google Scholar]
- 12.Orenstein E.W., Fang Z.Y., Xu J., Liu C., Shen K., Qian Y., Jiang B., Kilgore P.E., Glass R.I. The epidemiology and burden of rotavirus in China: a review of the literature from 1983 to 2005. Vaccine. 2007;25(3):406–413. doi: 10.1016/j.vaccine.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 13.Katayama H., Haramoto E., Oguma K., Yamashita H., Tajima A., Nakajima H., Ohgaki S. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Research. 2008;42(6-7):1441–1448. doi: 10.1016/j.watres.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 14.Poole L. Seasonal Influences on the Spread of SARS-CoV-2 (COVID19), Causality, and Forecastabililty (3-15-2020) Causality and Forecastabililty. 2020 (3-15-2020) (March 15, 2020) [Google Scholar]
- 15.Foladori P., Cutrupi F., Segat N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: What do we know? A review. Science of the Total Environment. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watts J. Report details lessons from SARS outbreak. Lancet. 2003;362(9391):1207. doi: 10.1016/S0140-6736(03)14561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu I., Qiu H., Tse L., Wong T. Severe Acute Respiratory Syndrome Beyond Amoy Gardens: Completing the Incomplete Legacy. Clinical Infectious Diseases. 2013;58(5):683–686. doi: 10.1093/cid/cit797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong S., Tan Y., Chia P., Lee T., Ng O., Wong M., Marimuthu K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA Research Letter. 2020 doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinney K.R., Gong Y.Y., Lewis T.G. Environmental transmission of SARS at Amoy Gardens. Journal of Environmental Health. 2006;68:26–30. [PubMed] [Google Scholar]
- 20.Singer A., Wray R. Detection and Survival of SARS in Human Stool, Urine, Wastewater and Sludge. Europe BMC. 2020 doi: 10.20944/preprints202006.0216.v1. [DOI] [Google Scholar]
- 21.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. MedRxiv. 2020 doi: 10.1101/2020.03.29.20045880. [preprint] [DOI] [PubMed] [Google Scholar]
- 22.La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Science of The Total Environment. 2020 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: Occurrence, persistence and concentration methods-A scoping review. Water Research. 2020 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since December 2019: evidence from environmental monitoring. MedRxiv. 2020 doi: 10.1101/2020.06.25.20140061. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trottier J., Darques R., Mouheb N.A., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. medRxiv. 2020 doi: 10.1101/2020.07.08.20148882. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water research. 2020 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar-Or I.B., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. 2020 doi: 10.1101/2020.04.26.20073569. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kocamemi B.A., Kurt H., Hacioglu S., Yarali C., Saatci A.M., Pakdemirli B. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. medRxiv. 2020 doi: 10.1101/2020.05.03.20089417. [preprint] [DOI] [Google Scholar]
- 29.Green H., Wilder M., Middleton F.A., Collins M., Fenty A., Gentile K., Kmush B., Zeng T., Larsen D.A. Quantification of SARS-CoV-2 and cross-assembly phage (crAssphage) from wastewater to monitor coronavirus transmission within communities. medRxiv. 2020 doi: 10.1101/2020.05.21.20109181. [preprint] [DOI] [Google Scholar]
- 30.Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Science of the Total Environment. 2020 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.05.20051540. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. The first proof of the capability of wastewater surveillance for COVID-19 in India through the detection of the genetic material of SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.06.16.20133215. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. medRxiv. 2020 doi: 10.1101/2020.06.04.20122747. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Science of The Total Environment. 2020 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed W., Bertsch P., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Science of The Total Environment. 2020 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kocamemi B.A., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. medRxiv. 2020 doi: 10.1101/2020.05.12.20099358. [preprint] [DOI] [Google Scholar]
- 37.Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Weinberger D.M. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv. 2020 doi: 10.1101/2020.05.19.20105999. [preprint] [DOI] [Google Scholar]
- 38.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environmental Science & Technology Letters. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 39.Wurtzer S., Marechal V., Mouchel J.M., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters with COVID-19 confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. [preprint] [DOI] [Google Scholar]
- 40.Oliver M.M.H., Hewa G.A., Pezzaniti D., Haque M.A., Haque S., Haque M.M., Moniruzzaman M., Rahman M.M., Saha K.K., Kadir M.N. COVID-19 and Recycled Wastewater Irrigation: A Review of Implications. Preprints. 2020 doi: 10.20944/preprints202006.0105.v1. [preprint] [DOI] [Google Scholar]
- 41.Qiu Y., Lee B.E., Neumann N., Ashbolt N., Craik S., Maal-Bared R., Pang X.L. Assessment of human virus removal during municipal wastewater treatment in Edmonton, Canada. Journal of Applied Microbiology. 2015 doi: 10.1111/jam.12971. [DOI] [PubMed] [Google Scholar]
- 42.Colivignarelli M.C., Colivignarelli C., Miino M.C., Abba A., Pedrazzani R., Bertanza G. SARS-CoV-2 in sewer systems and collected facilities. Process Safety and Environmental Protection. 2020 doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wommack K.E., Colwell R.R. Virioplankton: viruses in aquatic ecosystems. Microbiology and Molcular Biology Reviews. 2000;64(1):69–114. doi: 10.1128/MMBR.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otawa K., Lee S.H., Yamazoe A., Onuki M., Satoh H., Mino T. Abundance, Diversity, and Dynamics of Viruses on Microorganisms in Activated Sludge Process. Microbial Ecology. 2006;53:143–152. doi: 10.1007/s00248-006-9150-9. [DOI] [PubMed] [Google Scholar]
- 45.Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food and Environmental Virology. 2009;1(1):10. [Google Scholar]
- 46.Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Science of The Total Environment. 2020 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: Implications in low sanitation countries. Science of the Total environment. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.WHO . 2020. Water, sanitation, hygiene and waste management for COVID-19. [Google Scholar]
- 49.WHO . 2020. WHO lists two COVID-19 tests for emergency use.https://www.who.int/news-room/detail/07-04-2020-who-lists-two-covid-19-tests-for-emergency-use Available at: [Google Scholar]
- 50.Auffret M.D., Brassard J., Jones T.H., Gagnon N., Gagné M., Muehlhauser V., Masser L., Topp E., Talbot G. Impact of seasonal temperature transition, alkalinity and other abiotic factors on the persistence of viruses in swine and dairy manures. Science of the total Environment. 2019;659:640–648. doi: 10.1016/j.scitotenv.2018.12.306. [DOI] [PubMed] [Google Scholar]
- 51.Wang X., Li J., Jin M., Zhen B., Kong Q., Song N., Xiao W., Yin J., Wei W., Wang G. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. Journal of Virological Methods. 2005;126(1-2):171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silverman A.I., Boehm A.B. Systematic review and meta-analysis of the persistence and disinfection of human coronaviruses and their viral surrogates in water and wastewater. Environmental Science & Technology Letters. 2020 doi: 10.1021/acs.estlett.0c00313. [DOI] [PubMed] [Google Scholar]
- 53.Gerba C.P., Betancourt W.Q., Kitajima M. How much reduction of virus is needed for recycled water: A continuous changing need for assessment? Water research. 2017;108:25–31. doi: 10.1016/j.watres.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crank C., Xiang L., North D., Bonanno Ferraro G., Iaconelli M., Mancini P., La Rosa G., Bibby K. CrAssphage abundance and correlation with molecular viral markers in Italian wastewater. Water Research. 2020 doi: 10.1016/j.watres.2020.116161. [DOI] [PubMed] [Google Scholar]
- 55.USEPA . EPA 821-R-01-029; Washington, D.C: 2001. Method 1602: Male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure; p. 30. [Google Scholar]
- 56.APHA . 19th ed. American Public Health Association; Washington, DC: 2002. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- 57.Kwolek-Mirek M., Zadrag-Tecza R. Comparison of methods used for assessing the viability and vitality of yeast cells. FEMS Yeast Research. 2014;14(7):1068–1079. doi: 10.1111/1567-1364.12202. [DOI] [PubMed] [Google Scholar]
- 58.Olson M.R., Axler R.P., Hicks R.E. Effects of freezing and storage temperature on MS2 viability. Journal of Virological Methods. 2004;122(2):147–152. doi: 10.1016/j.jviromet.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 59.Mendez J., Jofre J., Lucena F., Contreras K., Mooijman K., Araujo R. Conservation of phage reference materials and water samples containing bacteriophages of enteric bacteria. Journal of Virological Methods. 2002;106:215–224. doi: 10.1016/s0166-0934(02)00163-5. [DOI] [PubMed] [Google Scholar]
- 60.Pastorino B., Touret F., Gilles M., de Lamballerie X., Charrel R.N. Evaluation of Heating and Chemical Protocols for Inactivating SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.11.036855. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casanova L.M., Weaver S.R. Inactivation of an enveloped surrogate virus in human sewage. Environmental Science & Technology Letters. 2015;2(3):76–78. [Google Scholar]
- 62.Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environmental Science & Technology. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- 63.Hjelmsø M.H., Hellmér M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., Elsässer D., Aarestrup F.M., Löfström C., Bofill-Mas S. Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PloS one. 2017;12(1) doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi H., Pasco E.V., Tarabara V.V. Membrane-based methods of virus concentration from water: a review of process parameters and their effects on virus recovery. Environmental Science: Water Research & Technology. 2017;3(5):778–792. [Google Scholar]
- 65.Wyn-Jones A., Sellwood J. Enteric viruses in the aquatic environment. Journal of Applied Microbiology. 2001;91(6):945–962. doi: 10.1046/j.1365-2672.2001.01470.x. [DOI] [PubMed] [Google Scholar]
- 66.Ikner L.A., Gerba C.P., Bright K.R. Concentration and recovery of viruses from water: a comprehensive review. Food and Environmental Virology. 2012;4(2):41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- 67.Schlindwein A., Rigotto C., Simões C., Barardi C. Detection of enteric viruses in sewage sludge and treated wastewater effluent. Water Science and Technology. 2010;61(2):537–544. doi: 10.2166/wst.2010.845. [DOI] [PubMed] [Google Scholar]
- 68.Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environmental Science & Technology. 2013;47(4):1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falman J.C., Fagnant-Sperati C.S., Kossik A.L., Boyle D.S., Meschke J.S. Evaluation of secondary concentration methods for poliovirus detection in wastewater. Food and environmental virology. 2019;11(1):20–31. doi: 10.1007/s12560-018-09364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calgua B., Rodriguez-Manzano J., Hundesa A., Suñen E., Calvo M., Bofill-Mas S., Girones R. New methods for the concentration of viruses from urban sewage using quantitative PCR. Journal of virological methods. 2013;187(2):215–221. doi: 10.1016/j.jviromet.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 71.Cashdollar J.L., Wymer L. Methods for primary concentration of viruses from water samples: a review and meta‐analysis of recent studies. Journal of applied microbiology. 2013;115(1):1–11. doi: 10.1111/jam.12143. [DOI] [PubMed] [Google Scholar]
- 72.Pei L., Rieger M., Lengger S., Ott S., Zawadsky C., Hartman N.M., Selinka H.C., Tiehm A., Niessner R., Seidel M. Combination of Crossflow Ultrafiltration, Monolithic Affinity Filtration, and Quantitative Reverse Transcriptase PCR for Rapid Concentration and Quantification of Model Viruses in Water. Environmental Science & Technology. 2012;46(18):10073–10080. doi: 10.1021/es302304t. [DOI] [PubMed] [Google Scholar]
- 73.Hill V.R., Kahler A.M., Jothikumar N., Johnson T.B., Hahn D., Cromeans T.L. Multistate Evaluation of an Ultrafiltration-Based Procedure for Simultaneous Recovery of Enteric Microbes in 100-Liter Tap Water Samples. Applied and Environmental Microbiology. 2007;73(13):4218–4225. doi: 10.1128/AEM.02713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prata C., Ribeiro A., Cunha Â., Gomes N.C., Almeida A. Ultracentrifugation as a direct method to concentrate viruses in environmental waters: virus-like particle enumeration as a new approach to determine the efficiency of recovery. Journal of Environmental Monitoring. 2012;14(1):64–70. doi: 10.1039/c1em10603a. [DOI] [PubMed] [Google Scholar]
- 75.Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Research. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Fumian T.M., Leite J.P.G., Castello A.A., Gaggero A., Caillou M.S.L., Miagostovich M.P. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods. 2010;170:42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 77.Abd-Elmaksoud S., Spencer S.K., Gerba C.P., Tamimi A.H., Jokela W.E., Borchardt M.A. Simultaneous concentration of bovine viruses and agricultural zoonotic bacteria from water using sodocalcic glass wool filters. Food and Environmental Virology. 2014;6(4):253–259. doi: 10.1007/s12560-014-9159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blanco A., Abid I., Al-Otaibi N., Pérez-Rodríguez F.J., Fuentes C., Guix S., Pintó R.M., Bosch A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food and environmental virology. 2019;11(2):184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haramoto E., Kitajima M., Katayama H., Ito T., Ohgaki S. Development of virus concentration methods for detection of koi herpesvirus in water. Journal of Fish Diseases. 2009;32(3):297–300. doi: 10.1111/j.1365-2761.2008.00977.x. [DOI] [PubMed] [Google Scholar]
- 80.Farkas K., McDonald J.E., Malham S.K., Jones D.L. Two-step concentration of complex water samples for the detection of viruses. Methods and protocols. 2018;1(3):35. doi: 10.3390/mps1030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patel S.J., Zhao G., Penna V.R., Park E., Lauron E.J., Harvey I.B., Beatty W.L., Plougastel-Douglas B., Poursine-Laurent J., Fremont D.H., Wang D., Yokoyama W.M. A Murine Herpesvirus Closely Related to Ubiquitous Human Herpesviruses Causes T-Cell Depletion. Journal of Virology. 2017:91. doi: 10.1128/JVI.02463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tavares L., Alves P.M., Ferreira R.B., Santos C.N. Comparison of different methods for DNA-free RNA isolation from SK-N-MC neuroblastoma. BMC Research Notes. 2011;4(3) doi: 10.1186/1756-0500-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mumy K.L., Findlay R.H. Convenient determination of DNA extraction efficiency using an external DNA recovery standard and quantitative-competitive PCR. Journal of Microbiological Methods. 2004;57(2):259–268. doi: 10.1016/j.mimet.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 84.Demeke T., Jenkins G.R. Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Analytical and bioanalytical chemistry. 2010;396:1977–1990. doi: 10.1007/s00216-009-3150-9. [DOI] [PubMed] [Google Scholar]
- 85.Schrader C., Schielke A., Ellerbroek L., Johne R. PCR Inhibitors – occurrence, properties and removal. Journal of Applied Microbiology. 2012;113(5) doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 86.Rådström P., Knutsson R., Wolffs P., Lövenklev M., Löfström C. Pre-PCR processing. Molecular Biotechnology. 2004;26:133–146. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 87.Opel K.L., Chung D., McCord B.R. A study of PCR inhibition mechanisms using real time PCR. Journal of Forensic Sciences. 2010;55(1) doi: 10.1111/j.1556-4029.2009.01245.x. [DOI] [PubMed] [Google Scholar]
- 88.Eckhart L., Bach J., Ban J., Tschachler E. Melanin Binds Reversibly to Thermostable DNA Polymerase and Inhibits Its Activity. Biochemical and Biophysical Research Communications. 2000;271(3):726–730. doi: 10.1006/bbrc.2000.2716. [DOI] [PubMed] [Google Scholar]
- 89.Tuomi J.M., Voorbraak F., Jones L.J., Ruijter J.M. Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods. 2010;50(4):313–322. doi: 10.1016/j.ymeth.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 90.Dymond J.S. Explanatory Chapter: Quantitative PCR. Methods in Enzymology. 2013;529:279–289. doi: 10.1016/B978-0-12-418687-3.00023-9. [DOI] [PubMed] [Google Scholar]
- 91.Corman V.M., Landt O., Kalser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van Brink S., Wijsman L., Goderski G., roomette J., ellis J., Zambon M., Peiris M., Gooss H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of Genetic Diagnostic Methods for Novel Coronavirus 2019 (nCoV-2019) in Japan. Japanese Journal of Infectious Diseases. 2020 doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 93.Wilson C.C., Wozney K.M., Smith C.M. Recognizing false positives: synthetic oligonucleotide controls for environmental DNA surveillance. Methods in Ecology and Evolution. 2016;7:23–29. [Google Scholar]
- 94.Bustin S.A., Nolan T. RT-qPCR testing of SARS-CoV-2: A primer. MDPI International Journal of Molecular Science. 2020;21(8) doi: 10.3390/ijms21083004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Svec D., Tichopad A., Novosadova V., Pfaffl M.W., Kubista M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomolecular Detection and Quantification. 2015;3:9–16. doi: 10.1016/j.bdq.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chemistry. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 98.Amirian E.S. Potential fecal transmission of SARS-CoV-2: Current evidence andimplications for public health. International Journal of Infectious Diseases. 2020 doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jorgensen A.U., Gamst J., Hansen L.V., Hogh Knudsen I.I., Skovgaard Jensen S.K. Eurofins COVID-19 Sentinel TM Wastewater Test provide early warning of a potential COVID-19 outbreak. medRxiv. 2020 doi: 10.1101/2020.07.10.20150573. [preprint] [DOI] [Google Scholar]