Abstract
ENL is a transcriptional co-activator that recruits elongation machinery to active cis-regulatory elements upon binding of its YEATS domain – a chromatin reader module – to acylated lysine side chains. Discovery chemistry for the ENL YEATS domain is highly motivated by its significance in acute leukemia pathophysiology, but cell-based assays able to support large-scale screening or hit validation efforts do not presently exist. Here, we report on the discovery of a target engagement assay that allows for high-throughput ligand discovery in living cells. This assay is based on the cellular thermal shift assay (CETSA) but does not require exposing cells to elevated temperatures, as small-molecule ligands are able to stabilize the ENL YEATS domain at 37 °C. By eliminating temperature shifts, we developed a simplified target engagement assay that requires just two steps: drug treatment and luminescence detection. To demonstrate its value for higher throughput applications, we miniaturized the assay to a 1,536-well format and screened 37,120 small molecules, ultimately identifying an acyl-lysine-competitive ENL/AF9 YEATS domain inhibitor.
The recognition that transcription factors and transcriptional co-regulators are among some of the most common proto-oncogenes and cancer dependencies has motivated ongoing efforts to discover drug-like small molecules that target transcriptional processes.1 While DNA-binding transcription factors are challenging targets for ligand discovery, many transcriptional co-regulators feature enzymatic activity and/or protein-protein interaction surfaces that can be modulated by potent and selective small-molecules.2 Notably, the interactions between chromatin reader domains and modified histone side chains are now habitually targeted by successful discovery chemistry campaigns.
In pursuit of novel targets for pharmacological development, we and others recently identified ENL (eleven nineteen leukemia), encoded by MLLT1 (multiple lineage leukemia translocated to), as a cancer-specific acute leukemia dependency.3, 4 ENL is a transcriptional co-activator and chromatin reader protein that interacts with the elongation-associated histone methyltransferase, DOT1L, and also forms part of a higher-order transcriptional elongation complex that contains P-TEFb (positive transcription elongation factor b, a heterodimer of CDK9 and cyclin T1).5-8 Toward a mechanistic understanding of the anti-leukemic effects elicited by ENL loss, we previously developed the dTAG system to enable rapid pharmacological degradation of ENL in AML cells.3 The dTAG system revealed a direct role for ENL in recruiting P-TEFb to the promoters of leukemic proto-oncogenes with disproportionately high enrichment of chromatin-bound ENL.3 Consequently, degradation of ENL results in selective downregulation of these leukemic drivers, rationalizing its role in acute leukemia pathogenesis and establishing it as a compelling target for drug discovery.3
ENL features an amino-terminal YEATS domain that binds specifically to acylated lysine side-chains found within the unstructured tail of histone H3.3,4,9 This domain has emerged as an attractive target for pharmacological development due to its well-formed binding pocket and established role in mediating chromatin contacts by ENL that are essential to sustain ENL-dependent leukemic growth.3,4 Moreover, recent disclosures of acetyl-lysine-competitive small-molecules and peptide-based tools confirm that this protein fold is pharmacologically tractable.10-14 In the pursuit of optimized chemical tools for the ENL YEATS domain, high-throughput screening (HTS)-compatible biochemical assays are easily accessed, but cell-based assays capable of supporting large-scale chemical screening in living systems have not been reported.
The cellular thermal shift assay (CETSA) is a well-established and facile technique for assaying small molecule target engagement via ligand-induced thermal stabilization.15,16 While the use of immunoblot detection as the end-point measurement for CETSA inherently limits the scale at which it can be implemented, multiple groups have worked to adapt the CETSA protocol to homogenous formats able to support higher throughput applications.17,18 In our efforts to prosecute the ENL YEATS domain with coordinated discovery chemistry, we sought to develop a homogenous cell-based assay for intracellular target engagement using split NanoLuc technology. Originally intending for this work to support medium-throughput hit validation studies downstream of biochemical-based HTS, we serendipitously discovered a simplified CETSA protocol that allows for highly reliable, unbiased, and high-throughput ligand discovery in cells.
RESULTS AND DISCUSSION
CETSA is widely used to assess pharmacological target engagement in complex living systems. Procedurally, intact cells or cell lysates are treated with drug and temporarily exposed to elevated temperatures, which induces protein unfolding and irreversible aggregation. Subsequent isolation of the soluble fraction after heating enables quantitation of ligand-induced thermal stabilization by immunoblot or mass spectrometry.15,16,19-22 While these methods of detection are inherently limited in scale, recent adaptation of the CETSA protocol to use a split NanoLuc system for protein quantification (similar to the HiBiT system from Promega23) has enabled high-throughput screening in 1,536-well plates.18 The key advantage afforded by this system is the ability to detect soluble protein levels remaining in cell lysates without need of a separation step to remove aggregated isoforms.
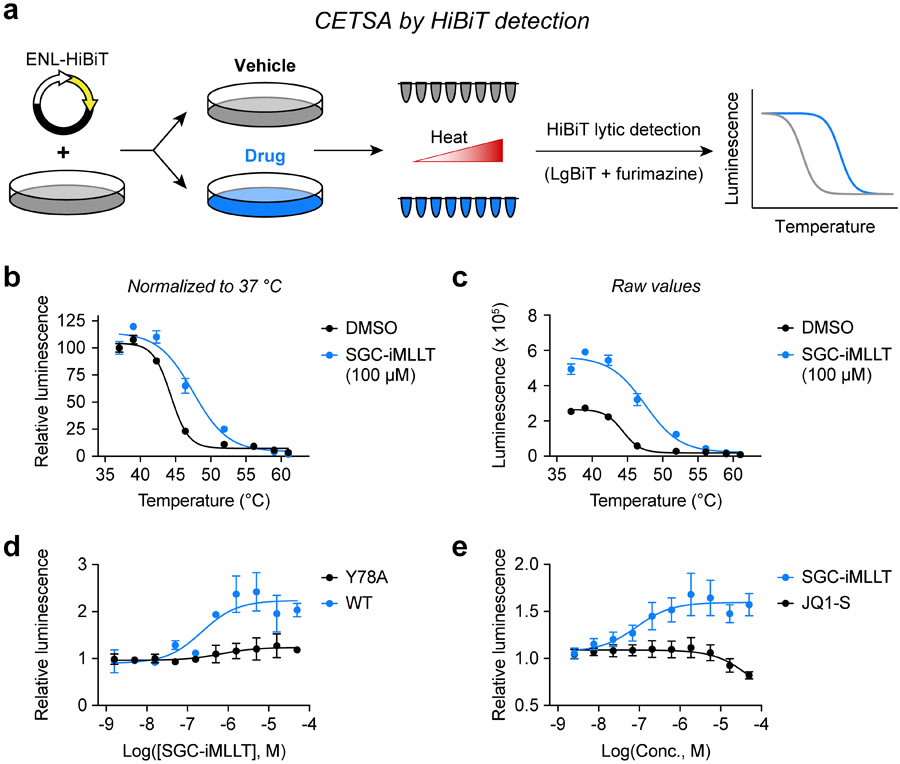
We encountered this study in the course of our own efforts to adapt a CETSA protocol for the ENL YEATS domain with the commercially available HiBiT tag (Figure 1a), an 11 amino-acid tag that self-associates with its NanoLuc complement, LgBiT, producing a luminescent signal in the presence of furimazine. We began by expressing full-length ENL or ENL(YEATS) with carboxy-terminal HiBiT fusions in HEK293T cells and then performed CETSA melt curves with the small-molecule ENL/AF9 YEATS domain inhibitor, SGC-iMLLT.11 While SGC-iMLLT did not affect the luminescence-based melt curve of full-length ENL-HiBiT (Figure S1a,b), we observed a rightward shift for ENL(YEATS) (Figure 1b), confirming HiBiT tagged ENL(YEATS) can be applied toward measurements of intracellular target engagement. Interestingly, CETSA melt curves are typically normalized to 37 °C, but we noticed that drug treatment elicited a consistent and dramatic increase of ENL(YEATS)-HiBiT luminescence at that temperature (Figure 1c). Compelled by this finding, we considered that to perform CETSA-based experiments on ENL YEATS, it might not be necessary to subject cells to elevated temperatures. Using CETSA for high throughput screening or isothermal dose response (ITDR) assays requires treating cells or lysates with drugs and then uniformly heating samples to a temperature that induces aggregation in the absence of ligand binding. Previous implementation of SplitLuc CETSAs in 1,536-well plates required a custom-machined aluminum plate block to supply conductive heat transfer, due to inconsistent results using convection heating.18 We hypothesized that in some instances, it would be possible to bypass this requirement, performing target engagement assays at physiologic temperature with just two steps: drug treatment followed by luminescence detection. We confirmed this by performing ITDR assays with SGC-iMLLT at 37 °C, observing dose-responsive increases in luminescence for wild-type ENL YEATS, but not ENL YEATS-Y78A (Figure 1d), a mutant isoform that prevents compound binding.11
Figure 1.
Discovery of a simplified CETSA protocol for YEATS domains. (a) Schematic depiction of CETSA melt curves using the HiBiT tag to enable detection of soluble protein from homogenous cell lysates. (b) CETSA melt curve of ENL(YEATS)-HiBiT in HEK293T cells after 1-hour treatments with drug or vehicle (N = 4, mean ± standard error of the mean, s.e.m.). Luminescence is normalized to 37 °C. (c) Raw luminescence values from the experiment depicted in (b). (d) ITDR at 37 °C for HEK293T cells expressing wild-type or mutant ENL(YEATS)-HiBiT and treated for 1 hour with SGC-iMLLT (N = 2, mean ± s.e.m.). Luminescence is normalized to DMSO control. (e) ITDR at 37 °C for OCI/AML-2 cells stably expressing AF9(YEATS)-HiBiT and treated for 1 hour (N = 4, mean ± s.e.m.). Luminescence is normalized to DMSO control.
Uncertain of the mechanism by which SGC-iMLLT might be able to increase HiBiT luminescence, we first tested whether the assay could be applied to its other known target, AF9 YEATS. Indeed, we observed dose-dependent increases in luminescence when treating cells that express AF9(YEATS)-HiBiT with SGC-iMLLT (Figure 1e). In contrast, the compound had no such effect on the bromodomain (BD) of SMARCA4 (Figure S1c), which it does not bind.11 Together, these data confirm that the increased luminescence is specifically related to the biomolecular interaction between ENL/AF9 YEATS domains and SGC-iMLLT. However, it was unclear whether thermal stabilization or some other biophysical phenomena was responsible for these observations.
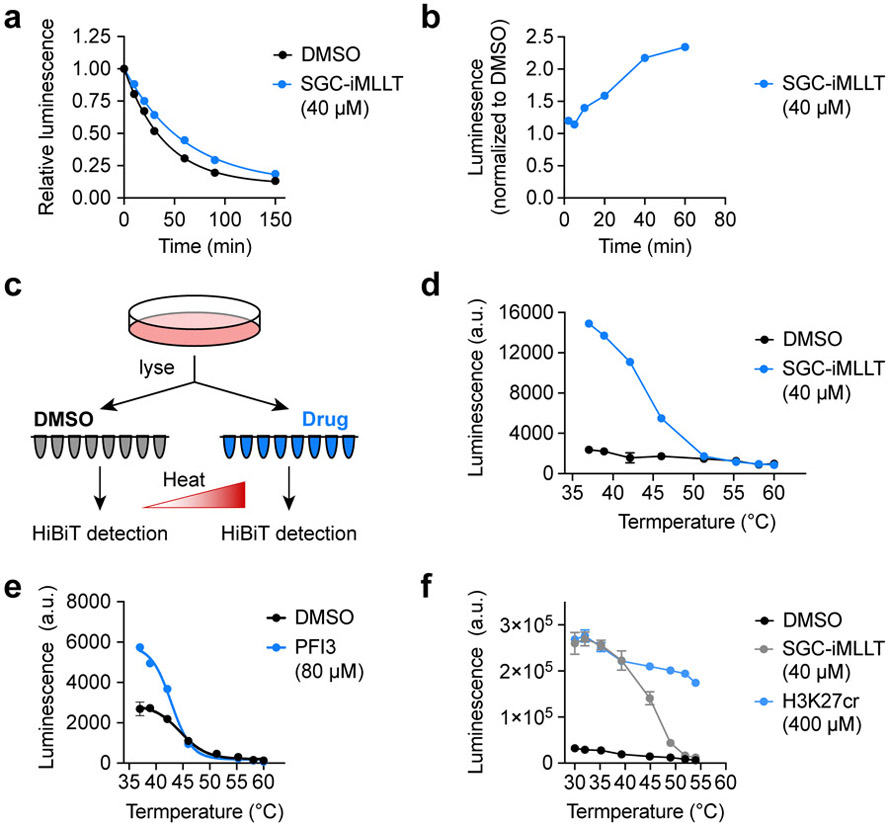
We considered several possibilities other than thermal stabilization that might explain ligand induced HiBiT luminescence. First, we reasoned that ENL and AF9, expressed as their YEATS domain alone, might be susceptible to proteolytic degradation in a manner that is rescued by ligand binding. However, while SGC-iMLLT did increase the half-life of ENL(YEATS)-HiBiT from 28 minutes (R2 = 0.997) to 41 minutes (R2 = 0.998, Figure 1a), we concluded that this would not account for the rapid stabilization observed within 60 minutes of drug exposure (Figure 1b). We also wondered whether a change in sub-nuclear localization was responsible for the increase in luminescence, whereby the translocation of ENL(YEATS)-HiBiT from chromatin to the nucleoplasm might make the protein more soluble upon cell lysis. To test this, we first lysed cells and then treated with SGC-iMLLT so that the amount of protein in the insoluble chromatin-bound fraction would remain fixed between vehicle- and drug-treated samples (Figure 1c). Still, luminescence was increased by SGC-iMLLT at 37° C (Figure 1d), so we next considered the possibility that the YEATS domain and HiBiT tag interact in a manner that obscures LgBiT recognition and is disrupted by drug treatment. For example, if the HiBiT tag binds intramolecularly into the acyl-lysine channel on the YEATS domain, then SGC-iMLLT might displace the HiBiT peptide and make it more easily detected by LgBiT. If such interactions do occur between HiBiT and YEATS domains, we would not necessarily expect them to be preserved in the structurally unrelated bromodomain of SMARCA4. Therefore, we stably expressed SMARCA4(BD)-HiBiT in OCI/AML-2 cells and tested the effects of PFI-3, a potent and selective SMARCA2/4 ligand.24 Inspecting the raw values, PFI-3 was able to elevate luminescence at 37 °C in cells expressing SMARCA4(BD)-HiBiT but not ENL(YEATS)-HiBiT (Figure 1e, S1e). Together, these data suggest that a direct-acting mechanism of thermal stabilization is the most likely explanation for the observed increase in luminescence at physiologic temperature.
It is important to note that it would have been difficult to appreciate that ENL YEATS can be stabilized at 37 °C if a positive control compound had not been available, complicating whether or not this approach can be explored for as-yet unliganded targets at the earliest stages of drug discovery. However, prior art has already established that natural small-molecule metabolites (e.g. 2′3′-cGAMP21 and ATP22) can be used to assay for ligand-induced thermal stabilization, so proteins or domains known to bind small-molecule cofactors should be amenable to testing even if chemical tools are not available. Similarly, since the assay works in cell lysates, we reasoned that peptides, proteins, or other biomolecules known to bind with high affinity could initially be used in cell lysates to test whether a given target can be stabilized at physiological temperature. We tested this with ENL(YEATS)-HiBiT by first lysing cells and then treating with a crotonylated histone peptide and as expected, luminescence was increased over a wide range of temperatures, including 37 °C (Figure 1f).
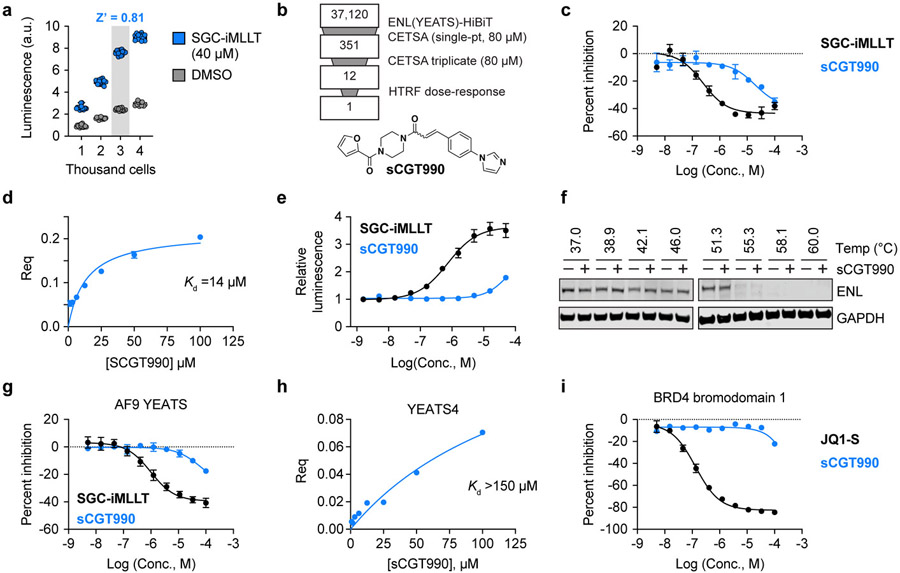
While we originally sought to develop an ENL YEATS CETSA as a secondary assay to remove false-positive hits identified in high-throughput screening, the simplicity of the resulting assay motivated us to consider its use as a primary, cell-based screening platform. Toward that end, we pursued miniaturization and optimization of the assay in 1,536-well plates. We began by engineering OCI/AML-2 cells to stably express ENL(YEATS)-HiBiT, which eliminated the need for multi-day transfection protocols and, being a suspension cell line, afforded maximum flexibility and ease of use. Comparing the assay performance after transient expression in HEK293T cells (Figure S2a) to stable expression in OCI/AML-2 cells (Figure 2a), we also noticed that stable expression produced much less well-to-well variability. Performing the assay on 3,000 cells in a 4 μL assay volume afforded the highest signal to background ratio (greater than 3) and Z-prime factor (greater than 0.8) in 1,536-well plates (Figure 2a, Figure S2b,c), reflecting a well-behaved assay that is suitable for HTS.25
Figure 2.
Mechanism of ligand-dependent luminescence at physiological temperature. (a) Cycloheximide-enabled determination of ENL(YEATS)-HiBiT half-life when treated with vehicle or drug. The depicted HiBiT luminescence measures of protein abundance are normalized to pre-treatment levels (N = 3, mean ± s.e.m.). (b) Kinetics of ENL(YEATS)-HiBiT luminescence after SGC-iMLLT treatment with values normalized to 24-hour DMSO treatment (N = 10, mean ± s.e.m.). (c) Schematic depiction of HiBiT CETSA performed on pre-lysed cells. (d) Raw luminescence values for HiBiT CETSA performed on cell lysates treated with vehicle or SGC-iMLLT (N = 4, mean ± s.e.m.). (e) CETSA melt curve of SMARCA4(bromodomain)-HiBiT in OCI/AML-2 cells after 1-hour treatments with drug or vehicle (N = 3, mean ± s.e.m.). (f) CETSA melt curve for ENL(YEATS)-HiBiT performed by treating cell lysates with a peptide substrate, H3K27cr (N = 3, mean ± s.e.m.).
We next considered the appropriate concentration at which to screen compounds in this assay. As was elaborated in the index manuscript,15 a general phenomenon of CETSA is that half-maximal effective concentrations (EC50) of compounds in ITDR assays are typically much higher than would be expected based on their affinity. For example, PFI-3 requires a concentration of 80 μM to elevate the luminescence from SMARCA4(BD)-HiBiT (Figure S2d), despite its reported affinity of 97 nM.24 With this in mind, we reasoned that significantly higher concentrations should be used for CETSA-based screening (as high as 100 μM) than are typically desired in cell-based screening assays. However, DMSO sensitivity tests revealed a gain of HiBiT signal at concentrations equal to or above 1% (Figure S2e), so all assays were limited to 0.8%, permitting a top screening concentration of 80 μM from libraries at a typical format of 10 mM in 100% DMSO.
Once miniaturized, we proceeded to benchmark the performance of this assay by executing a screen of 37,120 compounds for stabilization of the ENL YEATS domain in intact cells (Figure 2b). Only 10 compounds fell beyond a hit threshold of 3 z-scores from the mean, supporting previous observations by Henderson and colleagues that the HiBiT CETSA assay is durable against false-positives elicited by non-specific effects.18 Wanting to capture as many true-positives as possible, we lowered the threshold used to call hits from ≥ 3 z-scores to the top 1% of all compounds screened (equal to 24% activation compared to 40 μM SGC-iMLLT positive control). Of the top 1%, 331 compounds were available for further studies and were retested in triplicate, again at 80 μM, with 12 eliciting ≥ 24% activation in 2 or more replicates. One of these compounds, sCGT990 (Figure 2b), inhibited acyl-lysine recognition by the ENL YEATS domain and was chosen for follow-up studies (Figure 2c).
To conclude that sCGT990 was not a false-positive screening hit, we sought to obtain biophysical evidence of target engagement using biolayer interferometry (BLI). In agreement with its potency by HTRF (IC50 = 19 μM, Figure 2c, S3a), we confirmed that sCGT990 binds to ENL YEATS with 14 μM affinity (Figure 2d). Returning to the primary screening assay, we observed dose-responsive target engagement in living cells and note that concentrations as high as 80 μM were required to reliably elevate luminescence (Figure 2e). Together, these data provide three orthogonal measures validating sCGT990 as a true-positive screening hit and support our decision to screen at such an unusually high concentration. Interestingly, even at 80 μM, immunoblot-based CETSAs were only able to detect weak, if any, stabilization by sCGT990 (Figure 2f). Taken together, we were encouraged by these data, as they suggest that the HiBiT-based assay is sufficiently sensitive to detect weakly active compounds that are difficult to validate in cells by other methods.
Finally, to determine the potential for sCGT990 to serve as a novel scaffold for the elaboration of potent and selective ENL YEATS domain inhibitors, we tested its activity against other acyl-lysine chromatin readers. As expected, sCGT990 also inhibited crotonyl-lysine recognition by the highly homologous AF9 YEATS domain (IC50 = 62 μM), albeit at high micromolar concentrations (Figure 2g). While sCGT990 appears to inhibit ENL YEATS more potently than AF9 YEATS, we also note that our assays report similar behavior for SGC-iMLLT (Figure 2c,g), which is known to possess higher affinity for AF9 than ENL.11 Thus, this is likely a reflection of assay performance and should not be interpreted to suggest that sCGT990 is selective for ENL over AF9. Further testing revealed that sCGT990 does not efficiently bind to YEATS4/GAS41 (Figure 2h, S3b) and does not inhibit BRD4 bromodomain 1 (BD1) association with tetra-acetyl H4 (Figure 2i). Therefore, sCGT990 is positioned as a promising new scaffold for the development of potent and selective ENL/AF9 YEATS domain inhibitors by medicinal chemistry optimization. Such efforts would benefit to know whether sCGT990 is a covalent inhibitor of ENL YEATS, as its acrylamide might react with cysteine side chains. Using high-resolution matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry, we were unable to identify covalent adducts between sCGT990 and ENL YEATS (Figure S3c), suggesting the acrylamide can likely be replaced with isosteres for which electrophilicity is less of a concern.
Here, we have reported a novel cell-based assay for facile interrogation of target engagement in living cells, which can be implemented in any laboratory capable of measuring luminescence by simply expressing a HiBiT fusion protein. Previous cellular assays reported for ENL YEATS have included measures of ENL chromatin localization by fluorescence recovery after photobleaching (FRAP) and NanoLuc bioluminescence resonance energy transfer (nanoBRET).11 While these provide effective methods to detect cellular activity, the HiBiT assay described here offers significant advantages in scale, providing a complementary assay that reports on target engagement in high throughput. Notably, it also features the advantage of being a gain-of-signal assay, which is particularly attractive for limiting false-positives when screening diverse chemical structures in large-scale screens. Indeed, we have demonstrated the fitness of this assay for high-throughput implementation in 1,536-well plates, identifying a novel scaffold that inhibits ENL/AF9 YEATS domains from a screen of over 37,000 small molecules.
The available evidence suggests that elevated luminescence at physiological temperature reflects ligand-dependent thermal stabilization. If this is true, it represents a novel assay that should, in principle, be capable of identifying ligands that bind outside the acyl-lysine-binding channel, perhaps supporting the elaboration of ENL-specific allosteric inhibitors or proteolysis targeting chimeras (PROTACs). PROTACs are heterobifunctional small molecules that elicit target-specific protein degradation by enforcing proximity between a target protein and an E3 ubiquitin ligase complex.26 While chromatin-competitive ENL YEATS inhibitors are unlikely to achieve selectivity against AF9 YEATS due to their highly similar structures,4,9,11,27 multiple reports of isoform-selective PROTACs elaborated from non-selective parent ligands motivate pursuing this chemistry for ENL YEATS.28-35 We posit that this approach is likely to benefit from diverse parent ligands that bind a variety of YEATS domain sites, which might be possible to identify by the unbiased screening platform reported herein. However, the outlook for unbiased ligand discovery is complicated by the recognition that we cannot conclusively rule out mechanisms other than thermal stabilization. For instance, if displacement of the HiBiT tag from the acyl-lysine pocket of YEATS domains is responsible for the increased luminescence, one would not expect to identify neutral binders without bias. We attempted to study this mechanism by performing the assay with the SMARCA4 bromodomain, which shares no structurally homology to YEATS domains. Nevertheless, bromodomains also bind acyl-lysine residues and may present similar behavior toward the HiBiT tag. Over time, attempting this assay for a wider scope of target proteins will help to conclusively address this question.
The hypothesis we favor for the observed behavior at 37 °C is that ENL/AF9 YEATS domains and the SMARCA4 bromodomain may feature compromised thermal stability outside the context of their full protein structures. If this is true, then it may provide an inroad to extend our findings to other proteins, as other domains may be similarly unstable when isolated from their full protein structures. Additionally, it might be possible to mimic such behavior by engineering mutant isoforms with compromised stability at 37 °C. While it remains unclear what fraction of targets might behave similarly to the acyl-lysine readers we studied here, we ultimately hope that the demonstrated ease of implementation for this assay and its convenience for HTS applications will motivate its consideration for diverse drug discovery programs.
METHODS
Cell lines.
HEK293T cells were purchased from American Type Culture Collection (ATCC) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotic/antimycotic solution (Corning, Cat. No. 30-004-CI). OCI/AML-2 cells were a provided by Prof. James E. Bradner and cultured in alpha-MEM supplemented with 20% FBS and antibiotic/antimycotic solution. Both cell lines were periodically (2-4 times annually) tested for mycoplasma infections and never tested positive.
Small molecules.
SGC-iMLLT was a kind gift from O. Fedorov (Structural Genomics Consortium & Target Discovery Institute, University of Oxford) and later purchased from Sigma-Aldrich (Cat. No. SML2440). sCGT990 was purchased from Enamine (Product ID: Z61016540), PFI-3 was purchased from Sigma-Aldrich (Cat. No. SML0939), and JQ1 was purchased from Selleck Chemicals (Cat. No. S7110).
Plasmids.
For mammalian expression, MLLT1 and MLLT1-YEATS (amino acids 1-138) sequences in pENTR223 were supplied by Prof. James E. Bradner. pLEX_306 was a gift from David Root (Addgene plasmid #41391; http://n2t.net/addgene:41391; RRID: Addgene_41391) and PGK-gateway-HiBiT was constructed by exchange of the V5 tag on pLEX_306 with HiBiT using Gibson assembly (New England Biolabs, NEBuilder HiFi DNA Assembly Master Mix, Cat. No. E2621). MLLT1 sequences were cloned into PGK-gateway-HiBiT by Gateway Recombination Cloning Technology (Invitrogen, LR Clonase II Enzyme Mix, Cat. No. 11791020). SMARCA4(1426-1531) was cloned out of pCMV5 BRGI-Flag (a gift from Joan Massague,36 Addgene plasmid #19143; http://n2t.net/addgene:19143; RRID: Addgene_19143) into pDONR223 and then pLEX306-HiBiT using Gateway Recombinant Cloning Technology.
Lentivirus.
Lentivirus was produced in HEK293T cells by polyethylenimine-mediated (Polysciences Inc., PEI MAX 40K, Cat. No. 24765-1) co-transfection of transfer plasmids with pMD2.G (gift from Didier Trono; Addgene plasmid #2259; http://n2t.net/addgene:12259; RRID: Addgene_12259) and psPAX2 (gift from Didier Trono; Addgene plasmid #12260; http://n2t.net/addgene:12260; RRID:Addgene_12260). Viral supernatants were collected 48 and 72 h after transfection, combined and filtered through a 0.45 μm PVDF membrane (EMD Millipore, Steriflip-HV Sterile Centrifuge Tube Top Filter Unit, Cat. No. SE1M003M00), and concentrated 20-fold with Lenti-X Concentrator (Takara, Cat. No. 631232). Infection of OCI/AML-2 cells with lentivirus was accomplished by centrifugation at 2,000 rpm for 1 hour at room temperature with 8 μg/mL polybrene (EMD Millipore, TR-1003-G).
CETSA.
ENL YEATS HiBiT CETSA assays were performed on HEK293T cells transiently expressing ENL-HiBiT fusions or OCI/AML-2 cells stably expressing ENL(YEATS)-HiBiT. For HEK29T cells, 80-90% confluent 10 cm plates were transfected with 5 μg of the expression plasmid and 15 μg polyethylenimine. Media was refreshed after 12 hours and cells were collected 24 hours later. For melt curves, cells were treated with vehicle or drug for 1 hour, transferred to PCR strip tubes, and heated at the specified temperature for 3 min in a pre-heated thermocycler (Biorad C1000 Touch). Samples were then cooled to room temperature in the thermocycler, transferred to white 384-well tissue culture plates (20,000 cells in 20 μL per well), and combined with 20 μL Nano-Glo HiBiT Lytic Reagent (Promega, HiBiT Lytic Detection System, Cat. No. N3040) per well. Luminescence was measured after 30 min incubation using an EnVision multilabel plate reader (Perkin Elmer, Model No. 2104). ITDR assays were performed similarly with the following differences. Transfected cells were transferred immediately to white tissue culture plates (20,000 cells in 20 μL per well) and then 100 nL of drug or vehicle was transferred to each well using a pin tool transfer instrument (Biomek FX Laboratory Automation Workstation). After a 1-hour incubation with drug, HiBiT was detected as described above. For OCI/AML-2 cells stably expressing ENL(YEATS)-HiBiT, cells were transferred to 384-well (20,000 cells per well in 20 μL) or 1,536-well (3,000 cells per well in 4 μL) plates and drug was transferred using a 100 nL pin tool for 384-well plates or acoustic transfer (Labcyte, Echo Liquid Handler) for 1,536-well plates. HiBiT detection was performed as described above with 20 μL Lytic Reagent for 384-well plates and 2 μL for 1,536-well plates. CETSA on endogenous ENL protein by immunoblot detection was performed identically to the protocol described previously,11 using an anti-ENL rabbit monoclonal antibody (Cell Signaling; D9M4B; Cat. No. 14893) and an anti-GAPDH mouse monoclonal antibody (Santa Cruz Biotechnology; 6C5; Cat. No. SC-32233).
Protein Production.
Double stranded DNA fragments encoding the protein sequence of ENL YEATS(1-148) were synthesized (IDT DNA) for assembly-based cloning into a modified expression vector backbone derived from pET21a (Novagen). The protein was fused to an N-terminal StrepII-SUMO or SUMO-AviTag and a C-terminal 6xHis tag. Transformed BL21(DE3) cells (New England Biolabs) were grown at 37°C in 2xYT growth medium to an OD600 of 0.6. Protein expression was induced at 25°C with the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.4mM. The culture was harvested by centrifugation following a 20-hour incubation and subsequently purified by immobilized metal affinity chromatography. Cells were resuspended 50mM Tris-HCl pH 8.0, 300mM NaCl, 10% glycerol, 10mM imidazole, 1mM DTT containing Complete EDTA-free protease inhibitor (Roche) and lysed by two passes through a MicroFluidizer at 10,000 psi. The lysate was clarified by centrifugation at 16,000 rpm for 30 minutes at 4°C. The supernatant was passed over NiNTA agarose (QIAGEN) and the protein was eluted in 50mM Tris-HCl pH 8.0, 300mM NaCl, 10% glycerol, 250mM imidazole, 1mM DTT. Fractions containing the ENL YEATS(1-148) fusion protein were buffer exchanged into 100mM Tris-HCl pH 8.0, 150mM NaCl, 1mM EDTA, 1mM DTT using PD10 desalting columns (GE Life Sciences). Ulp1 protease was added to cleave the StrepII-SUMO fusion and the reaction mixture was incubated overnight at 4°C. The cleavage reaction mixture was loaded onto a StrepTrap column (GE Life Sciences) and ENL YEATS(1-148) was recovered in the flow through. AF9 YEATS was purchased from ActiveMotif (Cat. No. 81124) and BRD4 BD1 were purchased from Cayman (Cat. No. 11720). Double stranded DNA fragments encoding the protein sequence of YEATS4(13-158) were synthesized (IDT DNA) for assembly-based cloning into a pET-28a expression vector. The protein was fused to an amino-terminal 6xHis tag and a carboxy-terminal AviTag. To produce biotinylated protein for biolayer interferometry, BL21(DE3) cells were co-transformed with the AviTag fusion constructs and a biotin ligase expression vector, pBirAcm (Avidity). Cells were grown at 37°C in LB to an OD600 of 0.6. Protein expression was induced at 16°C with the addition of IPTG to a final concentration of 0.25 mM. The culture was harvested by centrifugation following a 16-hour incubation and subsequently purified by immobilized metal affinity chromatography. Cells were resuspended with 50mM sodium phosphate (pH 7.4), 500 mM NaCl, 10 mM imidazole, 1x Halt Protease Inhibitor Cocktail (Thermo Scientific) and lysed by probe sonication. The lysate was clarified by centrifugation at 16,000 xg for 20 minutes at 4°C. The supernatant was passed over a TALON Cobalt Resin (Takara) and the protein was eluted in 50mM sodium phosphate (pH 7.4), 500 mM NaCl, 200 mM imidazole. Fractions containing the YEATS4(13-158) fusion protein were buffer exchanged into 50 mM Sodium Phosphate (pH 7.4) ,150 mM NaCl using Amnicon Ultracel 10K desalting columns (Millipore). The protein was further purified by size exclusion chromatography through a HiLoad Superdex column (GE) on an AKTA pure (GE).
Biolayer interferometry (BLI).
BLI experiments were performed using a BLI OctetRED. Superstreptavidin biosensors (ForteBio biologics by molecular devices) were first incubated in assay buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 0.05% TWEEN-20, 1% DMSO) for 15 minutes. Sensors were coated with 40-60 μg/mL of biotinylated protein for 30 minutes at 25 °C. Compounds were serially diluted across a 96-well plate in a volume of 180 uL. Association of samples to either ENL- or YEATS4-coated and uncoated reference sensors were measured over 180 seconds and dissociation over 180 seconds with baseline measurements (buffer only) for 60 seconds between each sample concentration. All assays were run with continuous 1,000 rpm shaking. Data was analyzed using the OctetRED data analysis software with a reference subtraction in which reference uncoated sensors and reference samples were subtracted. Kd values were fitted using a 1:1 binding model in GraphPad Prism 8. As a technical control, we also tested ENL and YEATS4 against H3K27cr, custom synthesized by Genscript (amino-terminal acetyl, residues 13-32, Ac-GKAPRKQLATKAARKcrSAPAT).
Homogenous time-resolved FRET (HTRF) histone-binding assays.
All HTRF assays were performed by combining recombinant protein and synthetic histone peptide in assay buffer (25 mM HEPES pH 7, 20 mM NaCl, 0.2% Pluronic F-127, and 0.05% BSA) with 1 nM LanthaScreen Eu-anti-His Tag antibody (ThermoFisher, Cat. No, PV5597) and 8.9 nM SureLight allophycocyanin-streptavidin (Perkin Elmer, APC-SA, Cat. No. CR130-100). ENL YEATS, AF9 YEATS, and YEATS4 were used at 5 nM and BRD4 BD1 was used at 10 nM. ENL and AF9 assays were performed with H3(13-32)K27cr (13.3 and 100 nM, respectively), custom synthesized at ABclonal (N-terminal biotin, C-terminal amide); YEATS4 assay was performed with 25 nM H3(21-43)K27ac from Anaspec (Cat. No. AS-64846-1); and BRD4 BD1 assay was performed with 13.3 nM tetra-acetylated H4 (BioVision, Cat. No. 7144-01). Once all reagents were combined (with or without peptide), 10 μL was dispensed per well into black 384-well low-volume plates (Corning, Cat. No. 3821) and drug was added by pin tool transfer (Biomek FX). Assays were incubated for 2 or more hours before measurement of HTRF signal on a PHERAstar plate reader (BMG Labtech; simultaneous dual emission; excitation = 337 nm, emission 1 = 665 nm, emission 2 = 620 nm). HTRF signal (ratio of emission 1 to emission 2) from vehicle-treated wells (maximum signal) and no-peptide-control wells (minimum signal) were used to calculate percent inhibition for drug-treated wells.
Supplementary Material
Figure 3.
High-throughput ligand discovery for ENL YEATS in living cells. (a) Optimization of cell density for 1536-well formatted assay. Luminescence values are shown for vehicle and drug-treated OCI/AML-2 cells stably expressing ENL(YEATS)-HiBiT at increasing cell concentrations (N = 15). (b) Summary of high-throughput screen to identify ENL YEATS domain inhibitors. Chemical structure of sCGT990 is depicted at bottom. (c) Dose-dependent inhibition of homogenous time-resolved FRET (HTRF) signal generated by ENL YEATS association with H3K27cr. SGC-iMLLT is included as a positive control (N = 2, mean ± s.e.m.). (d) Biolayer interferometry validation of sCGT990 binding to immobilized biotin-ENL(YEATS). (e) Dose-dependent stabilization of ENL(YEATS)-HiBiT by sCGT990 and positive control compound, SGC-iMLLT. Luminescence values are depicted relative to DMSO (N = 3, mean ± s.e.m.). (f) Immunoblot analysis of CETSA melt curve for endogenous ENL protein in OCI/AML-2 cells treated with sCGT990 (80 μM) or vehicle (DMSO) for 1 hr. (g) Dose-dependent inhibition of HTRF signal generated by AF9 YEATS association with H3K27cr. SGC-iMLLT is included as a positive control (N = 2, mean ± s.e.m.). (h) Biolayer interferometry measures of sCGT990 binding to immobilized biotin-YEATS4(YEATS). (i) Same as in (g), but for BRD4 bromodomain 1 and tetra-acetyl H4 (JQ1-S included as a positive control, (N = 2, mean ± s.e.m.).
ACKNOWLEDGMENT
The authors would like to gratefully acknowledge D. Remillard, G. Winter, and R. Zeid for critical reading of the manuscript while it was in preparation; L.L. Lairson, P.G. Schultz, and I.A. Wilson for access to instrumentation; and The Scripps Research Institute Center for Metabolomics and Mass Spectrometry for performing mass spectrometry experiments. We would also like to thank P.G. Schultz and members of the Schultz Laboratory, especially L. Supekova, for technical support and helpful discussions. We thank O. Fedorov for providing SGC-iMLLT, used throughout the manuscript. This research was supported by the National Institutes of Health (NIH) through a NIH Director’s Early Independence Award (DP5-OD26380) to M.A.E.
Footnotes
Supporting Information
Supplemental figures for assay development, assay miniaturization, and biophysical validation of screening hits
REFERENCES
- 1.Bradner JE, Hnisz D & Young RA Transcriptional Addiction in Cancer. Cell 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop TR, Zhang Y & Erb MA Pharmacological Modulation of Transcriptional Coregulators in Cancer. Trends Pharmacol Sci 40, 388–402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erb MA, Scott TG, Li BE, Xie H, Paulk J, Seo H-S, Souza A, Roberts JM, Dastjerdi S, Buckley DL, Sanjana NE, Shalem O, Nabet B, Zeid R, Offei-Addo NK, Dhe-Paganon S, Zhang F, Orkin SH, Winter GE & Bradner JE Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan L, Wen H, Li Y, Lyu J, Xi Y, Hoshii T, Joseph JK, Wang X, Loh Y-HE, Erb MA, Souza AL, Bradner JE, Shen L, Li W, Li H, Allis CD, Armstrong SA & Shi X ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 543, 265–269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller D, Bach C, Zeisig D, García-Cuéllar M-P, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL & Slany RK A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 110, 4445–4454 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama A, Lin M, Naresh A, Kitabayashi I & Cleary ML A Higher-Order Complex Containing AF4 and ENL Family Proteins with P-TEFb Facilitates Oncogenic and Physiologic MLL-Dependent Transcription. Cancer Cell 17, 198–212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC & Shilatifard A AFF4, a Component of the ELL/P-TEFb Elongation Complex and a Shared Subunit of MLL Chimeras, Can Link Transcription Elongation to Leukemia. Mol Cell 37, 429–437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He N, Chan CK, Sobhian B, Chou S, Xue Y, Liu M, Alber T, Benkirane M & Zhou Q Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc National Acad Sci 108, E636–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, Wan L, Huang H, Tang Z, Zhao Y, Roeder RG, Shi X, Allis CD & Li H Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol Cell 62, 181–193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heidenreich D, Moustakim M, Schmidt J, Merk D, Brennan PE, Fedorov O, Chaikuad A & Knapp S Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL). J Med Chem 61, 10929–10934 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Moustakim M, Christott T, Monteiro OP, Bennett J, Giroud C, Ward J, Rogers CM, Smith P, Panagakou I, Díaz-Sáez L, Felce SL, Gamble V, Gileadi C, Halidi N, Heidenreich D, Chaikuad A, Knapp S, Huber KVM, Farnie G, Heer J, Manevski N, Poda G, Al-awar R, Dixon DJ, Brennan PE & Fedorov O Discovery of an MLLT1/3 YEATS Domain Chemical Probe. Angewandte Chemie Int Ed 57, 16302–16307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christott T, Bennett J, Coxon C, Monteiro O, Giroud C, Beke V, Felce SL, Gamble V, Gileadi C, Poda G, Al-awar R, Farnie G & Fedorov O Discovery of a Selective Inhibitor for the YEATS Domains of ENL/AF9. Slas Discov 110, 247255521880990–9 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Li X, Li X-M, Jiang Y, Liu Z, Cui Y, Fung KY, van der Beelen SHE, Tian G, Wan L, Shi X, Allis CD, Li H, Li Y & Li XD Structure-guided development of YEATS domain inhibitors by targeting π-π-π stacking. Nat Chem Biol 14, 1140–1149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni X, Heidenreich D, Christott T, Bennett J, Moustakim M, Brennan PE, Fedorov O, Knapp S & Chaikuad A Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1. Acs Med Chem Lett 10, 1661–1666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molina DM, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y & Nordlund P Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundbäck T, Nordlund P & Molina DM The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc 9, 2100–22 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Almqvist H, Axelsson H, Jafari R, Dan C, Mateus A, Haraldsson M, Larsson A, Molina DM, Artursson P, Lundbäck T & Nordlund P CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil. Nat Commun 7, 11040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez NJ, Asawa RR, Cyr MG, Zakharov A, Urban DJ, Roth JS, Wallgren E, Klumpp-Thomas C, Coussens NP, Rai G, Yang S-M, Hall MD, Marugan JJ, Simeonov A & Henderson MJ A widely-applicable high-throughput cellular thermal shift assay (CETSA) using split Nano Luciferase. Sci Rep-uk 8, 9472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savitski MM, Reinhard FBM, Franken H, Werner T, Savitski MF, Eberhard D, Molina DM, Jafari R, Dovega RB, Klaeger S, Kuster B, Nordlund P, Bantscheff M & Drewes G Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Franken H, Mathieson T, Childs D, Sweetman GMA, Werner T, Tögel I, Doce C, Gade S, Bantscheff M, Drewes G, Reinhard FBM, Huber W & Savitski MM Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat Protoc 10, 1567–1593 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Huber KVM, Olek KM, Müller AC, Tan CSH, Bennett KL, Colinge J & Superti-Furga G Proteome-wide drug and metabolite interaction mapping by thermal-stability profiling. Nat Methods 12, 1055–1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinhard FBM, Eberhard D, Werner T, Franken H, Childs D, Doce C, Savitski MF, Huber W, Bantscheff M, Savitski MM & Drewes G Thermal proteome profiling monitors ligand interactions with cellular membrane proteins. Nat Methods 12, 1129–1131 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Schwinn MK, Machleidt T, Zimmerman K, Eggers CT, Dixon AS, Hurst R, Hall MP, Encell LP, Binkowski BF & Wood KV CRISPR-Mediated Tagging of Endogenous Proteins with a Luminescent Peptide. Acs Chem Biol 13, acschembio.7b00549–8 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Fedorov O, Castex J, Tallant C, Owen DR, Martin S, Aldeghi M, Monteiro O, Filippakopoulos P, Picaud S, Trzupek JD, Gerstenberger BS, Bountra C, Willmann D, Wells C, Philpott M, Rogers C, Biggin PC, Brennan PE, Bunnage ME, Schüle R, Günther T, Knapp S & Müller S Selective targeting of the BRG/PB1 bromodomains impairs embryonic and trophoblast stem cell maintenance. Sci Adv 1, e1500723 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J-H, Chung TDY & Oldenburg KR A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Cromm PM & Crews CM Targeted Protein Degradation: from Chemical Biology to Drug Discovery. Cell Chem Biol 24, 1181–1190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SYR, Li W, Li H & Shi X AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell 159, 558–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zengerle M, Chan K-H & Ciulli A Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. Acs Chem Biol 10, 1770–1777 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gadd MS, Testa A, Lucas X, Chan K-H, Chen W, Lamont DJ, Zengerle M & Ciulli A Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol 13, 514–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remillard D, Buckley DL, Paulk J, Brien GL, Sonnett M, Seo H-S, Dastjerdi S, Wühr M, Dhe-Paganon S, Armstrong SA & Bradner JE Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands. Angewandte Chemie Int Ed 56, 5738–5743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, Zhang T, Kwiatkowski N, Boukhali M, Green JL, Haas W, Nomanbhoy T, Fischer ES, Young RA, Bradner JE, Winter GE & Gray NS Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol 14, 163–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, Wang J, Hamman BD, Ishchenko A & Crews CM Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem Biol 25, 78–87.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H-T, Dobrovolsky D, Paulk J, Yang G, Weisberg EL, Doctor ZM, Buckley DL, Cho J-H, Ko E, Jang J, Shi K, Choi HG, Griffin JD, Li Y, Treon SP, Fischer ES, Bradner JE, Tan L & Gray NS A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell Chem Biol 25, 88–99.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brand M, Jiang B, Bauer S, Donovan KA, Liang Y, Wang ES, Nowak RP, Yuan JC, Zhang T, Kwiatkowski N, Müller AC, Fischer ES, Gray NS & Winter GE Homolog-Selective Degradation as a Strategy to Probe the Function of CDK6 in AML. Cell Chem Biol 26, 300–306.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowak RP, DeAngelo SL, Buckley D, He Z, Donovan KA, An J, Safaee N, Jedrychowski MP, Ponthier CM, Ishoey M, Zhang T, Mancias JD, Gray NS, Bradner JE & Fischer ES Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat Chem Biol 14, 706–714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xi Q, He W, Zhang XHF, Le H-V & Massagué J Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor beta transcriptional program. J Biol Chem 283, 1146–1155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.