Abstract
Glycerophospholipids (GPLs), one of the main components of bacterial cell membranes, exhibit high levels of structural complexity that are directly correlated with biophysical membrane properties such as permeability and fluidity. This structural complexity arises from the substantial variability in the individual GPL structural components such as the acyl chain length and headgroup type and is further amplified by the presence of modifications such as double bonds and cyclopropane rings. Here we use liquid chromatography coupled to high-resolution and high-mass-accuracy ultraviolet photodissociation mass spectrometry for the most in-depth study of bacterial GPL modifications to date. In doing so, we unravel a diverse array of unexplored GPL modifications, ranging from acyl chain hydroxyl groups to novel headgroup structures. Along with characterizing these modifications, we elucidate general trends in bacterial GPL unsaturation elements and thus aim to decipher some of the biochemical pathways of unsaturation incorporation in bacterial GPLs. Finally, we discover aminoacyl-PGs not only in Gram-positive bacteria but also in Gram-negative C. jejuni, advancing our knowledge of the methods of surface charge modulation that Gram-negative organisms may adopt for antibiotic resistance.
Graphical Abstract
Lipids form the bilayer structure of the cell membrane and, in conjunction with proteins, are responsible for most of the membrane’s function including molecular import and export, responses to environmental variations, and cellular recognition.1 Additionally, phospholipids are recognized for their role as molecular chaperones, as they can directly impact protein folding.2,3 Owing to these key roles and the high prevalence in membranes, lipids are often also implicated in malfunctions in cellular processes and pathways including the upregulated biosynthesis of lipids in cancer and the remodeling of lipids in antibiotic resistance.4,5 The growing field of lipidomics has emerged to investigate the diversity of lipids, their modifications, and the resulting biological implications.6,7 One powerful and widely used tool in ex vivo lipidomics is mass spectrometry (MS) that, when performed using high-resolution tandem MS coupled to liquid chromatography (LC), reveals structural differences and modifications of lipids in unprecedented detail.8–10
One subfield of lipidomics that has benefitted from advances in mass spectrometry is the broad-scale analysis of lipids in bacteria. Bacterial cell membranes are primarily composed of amphiphilic glycerophospholipids (GPLs), which play major roles in both cellular structure and function.11 The complex structure of GPLs comprises fatty acid acyl chain(s), a glycerol group, a phosphate group, and a headgroup. Among GPL subclasses, lyso lipids can contain one acyl chain, whereas cardiolipins can contain up to four acyl chains.12 Here we focus on the characterization of some of the most common bacterial GPLs including phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylserine (PS), and phosphatidylcholine (PC), all containing two fatty acid acyl chains. The identities of many of these structural features are variable in their size, stereochemistry, or type, all features that further amplify the degree of complexity. The headgroup defines the lipid class, which for bacterial membranes primarily includes PEs and PGs.13 The level of structural classification is commonly limited by the level of MS data acquired; MS1 spectra allow accurate elemental composition, but owing to the possibility of variation in both acyl chains, MS/MS methods are required to provide higher levels of structural refinement.9,14
A further facet of GPL structural complexity arises from the presence of acyl chain modifications such as double bonds and cyclopropane rings. These motifs vary in the type, stereochemistry, and location of unsaturation within the GPL acyl chain. Because of the high prevalence of unsaturation elements, their features are integrated into the shorthand GPL nomenclature conventions. For example, GPL nomenclature often contains headgroup abbreviations in conjunction with the number of carbon atoms in the acyl chains and the number of unsaturations to define lipid descriptors (e.g., PG 18:1_18:1 revealed by MS/MS or PG 36:2 for MS1 limited data).15 If known, the type of unsaturation can be indicated by c for cyclopropane ring unsaturation or delta (Δ) for carbon double bonds, with the further distinction of cis or trans, which can be differentiated only with advanced MS and related techniques.16,17 The position of each acyl chain in relation to the headgroup (sn-position) can also be designated (if confidently determined) by using a forward slash in place of the underscore (e.g., PG 18:0_18:1 vs PG 18:0/18:1), but like cis/trans identifications, methods for this distinction are complicated and are often not yet sufficiently robust to implement in the analysis of complex bacterial phospholipid extracts.18 Lipidomics nomenclature has not yet been expanded to include GPL chirality (i.e., sn-3 vs sn-1 phosphorylated), largely owing to the lack of MS-based methods for the differentiation of this structural feature.19
While elaborate, this nomenclature system is required in classifying bacterial GPLs because a plethora of possible GPL modifications confound the precise profiling of variations in lipid composition that may provide critical insight into the pathophysiology of diseases. In addition to the possibility of unsaturation elements previously described, acyl chain branching and the addition of functional groups, such as hydroxyl groups, may also occur in bacterial GPLs.20–22 Although these modifications might seem to be inconsequential, numerous studies have confirmed that many GPL acyl chain modifications influence the response of bacteria to environmental stressors and the presence of antimicrobial agents by changing the membrane fluidity and charge.23–28
Beyond the acyl chain, headgroup modifications such as aminoacylation have also been found to play key roles in cellular functions.29 This type of modification is a prime example of the crucial importance of advanced analytical methods in the elucidation of subtle structural alterations. For example, aminoacyl-GPLs increase the overall charge of the lipid membrane, thus resulting in increased resistance to antibiotics, cationic antimicrobial peptides (CAMPs), and other cationic compounds, which rely on the highly negative membrane charge to penetrate bacterial cells.30 Antibiotic resistance via the aminoacylation of GPLs primarily occurs in Gram-positive bacteria, as Gram-negative bacteria have alternative methods of charge modulation via lipopolysaccharide (LPS) modification.11,31,32 Although rare, there have been reports indicating that Gram-negative species may also be able to modulate their membrane charge using this GPL aminoacylation pathway.33,34
Aside from cataloging the presence of modifications, lipidomics also focuses on deciphering the biochemical pathways and genetic information that produce such structural variations, which, in turn, may afford a better understanding and treatment of lipid-related diseases.35–38 Enzymes responsible for the incorporation of common modifications such as carbon–carbon double bonds (desaturases) have been found in bacterial species such Bacillus subtilis (B. subtilis),39 whereas the less common cyclopropane synthases such as cyclopropane fatty acyl phospholipid synthase and cyclopropane mycolic acid synthase, responsible for the incorporation of cyclopropane unsaturations, have been found in species such as Escherichia coli (E. coli) and Mycobacterium tuberculosis (M. tuberculosis).40 The acyl chain site specificity of these enzymes41 as well as the effect of environmental stressors on the enzyme activity have also been investigated.39,42,43 Despite these findings, GPL pathways and, to a large extent, even the presence of GPL modifications in the vast majority of bacterial species remain uncharted.
To address the gaps in lipidomics previously described, a variety of MS approaches have been developed to tease out nuances in lipid structures, such as multistage MSn and ion/ion chemistry methods.44–47 Other common MS-based techniques in this field have utilized derivatization reactions that add new functional groups across the carbon–carbon double bonds to provide diagnostic fragment ions.48–54 Structure-specific fragment ions can also be produced using ozonolysis, which requires the infusion of ozone to break double bonds prior to MS analysis and has also shown wide success in distinguishing unsaturations, even in GPLs.55–57 However, despite the powerful characterization capabilities of these techniques, the quest for more robust and more widely versatile methods remains a top priority, especially in the case of highly complex mixtures.9
Our group has developed a powerful MS technique for GPL characterization by using ultraviolet photodissociation (UVPD).10,58 UVPD has been shown to reveal minor structural differences in even the most complex lipid classes, ranging from fatty acids59 and simple GPLs17,18,60 to the more complex GPLs such as cardiolipins61 and even LPSs.62 The strategy has characterized a number of structural features of GPLs, such as the presence and locations of cyclopropane17 and double bond18,60,63,64 unsaturations in acyl chains as well as sn-positions.18,65,66 Here we employ high-resolution 193 nm UVPD-MS coupled to LC to uncover novel GPL modifications including hydroxyl-modified acyl chains and methylated PS headgroups. Additionally, we use this approach to elucidate specific trends in the unsaturation patterns of bacterial GPLs using an array of 12 bacterial extracts and provide the first direct evidence of aminoacyl-PGs (aa-PGs) in Gram-negative Campylobacter jejuni. In doing so, we gain information on the biochemical pathways and enzymes that are responsible for some GPL modifications, thus enhancing the comprehensive characterization of the complex bacterial lipidome and expanding our knowledge of the roles that this plethora of structural modifications play within these systems.
EXPERIMENTAL SECTION
Materials.
All bacteria except two were grown in 250 mL of LB Miller broth at 37 °C to an OD600 of 2.5 (stationary growth phase). Lactobacillus plantarum was cultured in Lactobacilli MRS broth at 37 °C to an OD600 of 2.5 (stationary growth phase). C. jejuni was grown in Mueller Hinton broth at 37 °C under microaerophilic conditions (5% oxygen, 10% CO2, and 85% nitrogen) and harvested at OD600 of 0.6 (log phase). See Table S1 for a list of bacteria used in experiments, including 10 Gram-negative and 2 Gram-positive bacteria. Bacteria pellets were washed once with 1× phosphate-buffered saline (PBS). Lipids were extracted using a chloroform/methanol-based Bligh and Dyer procedure, as previously described.67,68 In brief, cell pellets were resuspended in a 95 mL single-phase extraction mixture of chloroform/methanol/water (1:2:0.8 v/v/v) and incubated at room temperature for 20 min. The single-phase mixtures were centrifuged at 2400g for 15 min, and the supernatant was removed to a clean tube and converted to a 145 mL two-phase Bligh–Dyer system (chloroform/methanol/water 2:2:1.8 v/v/v). Mixtures were vortexed and centrifuged at 2400g for 15 min. The lower organic phase was transferred to a new tube, and a second extraction was performed on the remaining upper phase, as previously described. The pooled lower phases were “washed” by adding 190 mL of pre-equilibrated upper phase, mixed by vortexing, and centrifuged at 2400g for 15 min. The final extracted lower phases were dried by rotary evaporation, resuspended in chloroform/methanol (4:1 v/v), and transferred to 0.3 mL microvials. The samples were then dried under nitrogen and stored at −20 °C until further analysis.
Liquid Chromatography and Mass Spectrometry.
Dried lipid extracts were diluted to ~3 ug/uL in chloroform to generate stock solutions. Stock solutions were further diluted to ~30 ng/uL under mobile-phase starting conditions prior to LC-MS experiments. Chromatographic separations were performed using a reversed-phase Acquity UPLC CSH C18 column (pore size 130 Å, 1.7 μm particle size, 2.1 mm × 100 mm, Waters, Milford, MA) on a Dionex Ultimate 3000 UHPLC system (Thermo Fisher Scientific, Sunnyvale, CA). All data were collected with a heated electrospray ionization (H-ESI) source (Thermo Fisher Scientific, San Jose, CA) with a sheath gas of 5 and an auxiliary gas of 10 operated in negative mode at a spray voltage of −3.8 kV. The LC system was coupled to a Thermo Fisher Orbitrap Fusion Lumos mass spectrometer (San Jose, CA) modified with a 193 nm Coherent Excistar XS excimer laser (Santa Clara, CA) for UVPD experiments, as previously described.62 The ion transfer tube temperature was set to 300 °C, whereas the vaporizer temperature was set to 40 °C. LC experiments were performed as previously reported.17,69 In brief, mobile phase compositions consisted of (A) 60:40 ACN/water and (B) 90:10 IPA/ACN, both with 0.1% formic acid and 10 mM ammonium formate. GPLs were eluted at a flow rate of 260 μL/min using the following 62 min gradient: 10% B (0–2 min), up to 45% B (2–6 min), up to 60% B (6–46 min), up to 95% B (46–47 min), held at 95% B (47–53 min), down to 10% B (53–54 min), and held at 10% B (54–62 min) for a total of 8 min of column re-equilibration. For each run, ~300 ng (10 uL) of GPL extract was injected, and the column compartment was heated to 50 °C during the entirety of all LC-MS runs. MS1 spectra (m/z 300–1200) were collected using a resolving power of 30 000 (m/z 200), an AGC target of 1e6, a maximum injection time (MIT) of 200 ms, and 2 μscans/scan. Prior to MS/MS events, a monoisotopic peak selection (MIPS) filter was applied in the small-molecule mode in addition to an intensity filter with a threshold of 1e6. Alternating high-energy collisional dissociation (HCD) and UVPD scans were collected in a data-dependent manner with a total cycle length of eight MS2 scans (four HCD and four UVPD scans) between MS1 scans. HCD was performed at 2 μscans/scan with a normalized collision energy (NCE) of 25, an AGC target of 2e5, and a MIT of 200 ms. 193 nm UVPD was performed by the application of eight laser pulses with 2.5 mJ per pulse, 7 μscans/scan, an AGC target of 1e6, and a MIT of 500 ms. All MS2 spectra were collected using a resolving power of 15 000 (m/z 200) and an isolation width of 1 m/z. UVPD experiments were performed in the high-pressure trap (HPT) of the dual linear ion trap.
RESULTS AND DISCUSSION
Although shotgun approaches (direct infusion without chromatographic separation) have been widely used to profile lipids in complex mixtures, an LC workflow was adopted in the present study to allow a broader characterization of low-abundance lipids. These low-abundance lipids might otherwise be overlooked owing to suppressed ionization during the direct infusion of mixtures containing multiple constituents. In addition, chromatographic separation helps alleviate the inadvertent production of chimeric MS/MS spectra generated when multiple precursor ions of the same m/z are coisolated and coactivated, a more common occurrence in shotgun methods. We interrogated a panel of phospholipid extracts from 12 types of bacteria, including 10 Gram-negative and 2 Gram-positive bacteria (Table S1) by using LC-HCD-MS and LC-UVPD-MS methods. We aimed to probe the distribution of unsaturation elements of the lipids, including both the type and location of unsaturation, to map the general patterns of unsaturation across bacterial GPLs via analysis of the abundant [M – H]− precursor ions in negative ion mode. Shown in Figure 1 is an example of a typical LC trace from a Salmonella enterica serovar Typhimurium (S. typhimurium) phospholipid extract, along with the HCD and UVPD mass spectra acquired for one representative GPL of m/z 714.51. This unsaturated lipid is quite abundant, as seen in the base peak LC trace (tr = 23.9 min) and extracted ion chromatogram (XIC) in Figure 1a, and, in fact, many of the most abundant species in the 12 bacterial lipid extracts surveyed in this study are unsaturated species. The HCD mass spectrum (Figure 1b) confidently identifies the lipid as PE 16:1_18:1, as illustrated in the companion fragmentation map (Figure 1d), but does not provide any insight into the locations or types of unsaturation elements contained within the two acyl chains. However, 193 nm UVPD provides a pair of diagnostic ions spaced 24.0 Da apart (Figure 1c) of m/z 604 and 628 that reveal the types (16:1 double bond, 18:1 double bond) and positions (16:1(9Δ), 18:1(11Δ)) of unsaturation. Previous work from our group has demonstrated that the detection of double-bond positions via LC/UVPD-MS is possible for GPLs in the low picomolar concentration range (limit of detection (LOD) 25 pmol).18 This complementary information from HCD and UVPD is integrated to identify this GPL as PE 16:1(9Δ)_18:1(11Δ) (Figure 1d). This LC-MS/MS workflow does not decode sn-positions nor does it identify unsaturation element stereochemistry (cis vs trans), although the former feature can be interrogated by using a more elaborate MS3 strategy18 at the expense of throughput. Unsaturation element stereochemistry (cis vs trans) is indicated in ChemDraw structures within figures based on existing LipidMAPS entries. A lack of unsaturation element stereochemistry assignment is designated in the nomenclature assigned to each lipid identification (“_” indicates unknown sn-stereochemistry, whereas “Δ” indicates unknown unsaturation element stereochemistry). This GPL identification process was performed for every collected MS/MS scan throughout the chromatographic separation, and a complete table of identified unsaturated lipids for the S. typhimurium phospholipid extract is summarized in Figure 1e. Complete tabulations of all identified unsaturated lipids for all 12 bacterial extracts along with mass error values (within ±10 ppm error) for all fragment ions and base-peak LC-MS chromatograms are shown in Figure S1 and Tables S2–S13.
Figure 1.
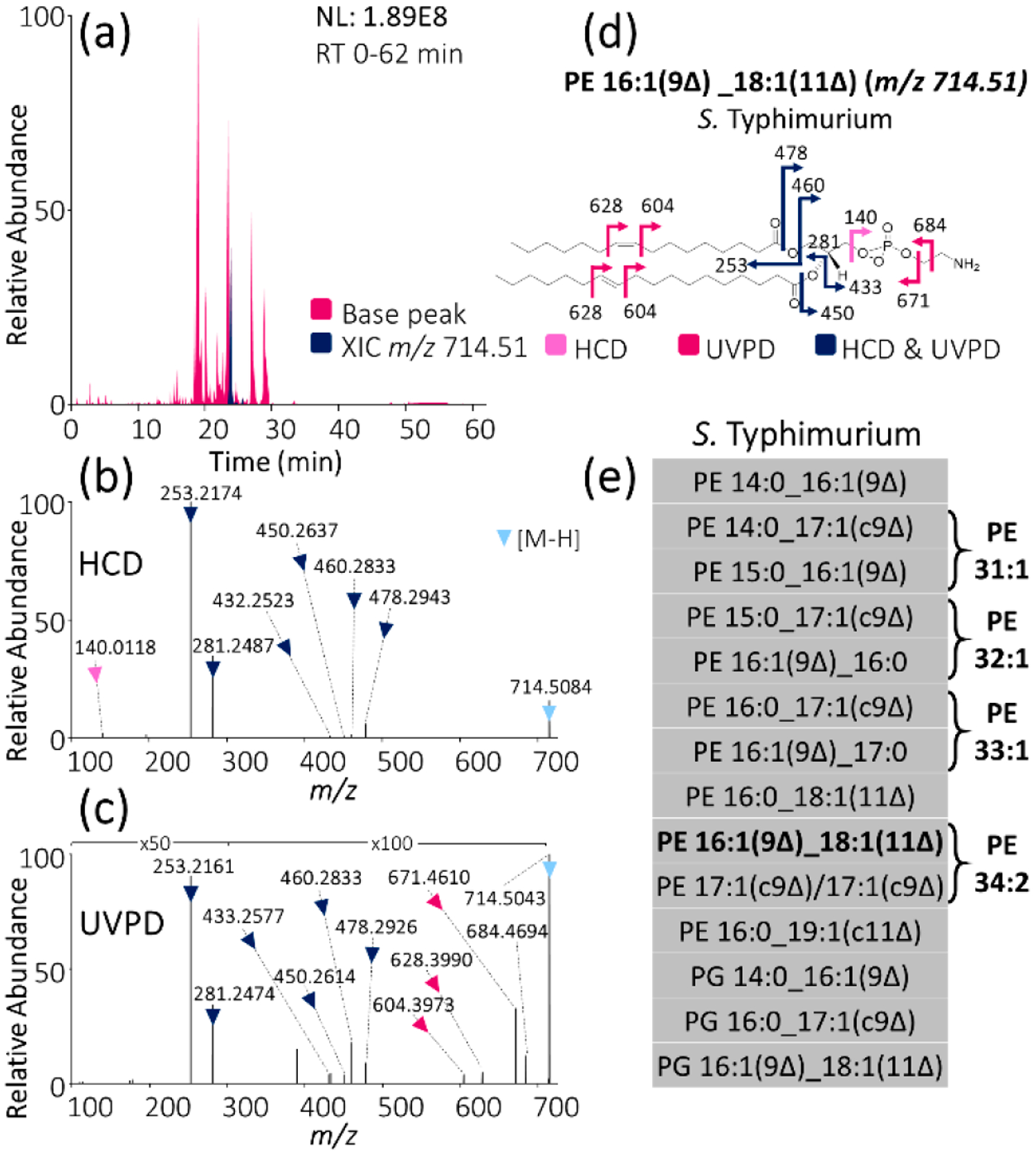
MS analysis of unsaturated GPLs from bacterial extracts: (a) Base peak LC-MS trace of Salmonella enterica serovar Typhimurium (S. Typhimurium) lipid extract with XIC of m/z 714.51 highlighted. (b) HCD mass spectrum of m/z 714.51. (c) 193 nm UVPD mass spectrum (eight pulses, 2.5 mJ per pulse) of m/z 714.51. (d) Fragment ion map of m/z 714.51 identified as PE 16:1(9Δ)_18:1(11Δ). (Fragmentation sites are color-coded to correspond to the ions identified in panels b and c.) (e) List of all identified unsaturated GPLs in S. typhimurium lipid extract using LC-MS/MS with alternating HCD and UVPD.
Using this systematic approach, we aimed to elucidate qualitative information as well as relative differences in GPL patterns between bacteria. First, we classified the identified unsaturated GPLs by their location of unsaturation. To enable this classification, we used the omega unsaturation element nomenclature, which counts the position of an unsaturation element from the methyl end instead of the more conventional carboxylic acid end (delta nomenclature). This omega nomenclature is particularly useful because bacterial desaturation mechanisms often incorporate unsaturation elements at a fixed position from the methyl terminus,70–72 and thus the omega numeration allows easier recognition of unsaturation trends in bacterial GPLs (Figure S2).
Distributions of lipids, categorized based on the locations of unsaturations, the type of unsaturation elements, and the headgroup identities, are summarized in bar graph format in Figure 2 for each of the 12 bacteria. As seen in Figure 2a (charting the position of unsaturation), almost all investigated bacterial lipid extracts primarily contained GPLs with unsaturation elements (both double bonds and cyclopropane rings) at the ω-7 position. This alignment between double bonds and cyclopropane rings agrees with previous reports that indicate that cyclopropane rings occur via methylation of the double bond in vivo, thus retaining the stereochemistry and position of the original double-bond unsaturation.73 Additionally, a few unsaturation elements were identified at other locations, including the ω-8, ω-6, and ω-9 positions, which, interestingly, are all close in proximity to the major ω-7 unsaturation position. Whereas the GPL unsaturation pathway is well-studied in E. coli and unsaturation positions have been found to occur exclusively at the ω-7 position,17,18,72 this phenomenon is largely uninvestigated in many other bacteria. A previous study from our group revealed the identification of exclusively ω−7 unsaturation elements in the GPLs of E. coli using UVPD-MS.17 The comprehensive data set in the present work shows that this trend of ω-7 unsaturation positions generally holds true across all investigated bacteria and that other unsaturation positions (ω-8, ω-6, and ω-9) are generally the minority. Additionally, the bar graph shown in Figure 2a provides insight into the relative differences in the number of unsaturated lipids identified per extract. For example, B. subtilis had a low number of identified unsaturated lipids (only 3) compared with K. pneumonia (26). This finding aligns with previous reports indicating that only traces of unsaturated fatty acids (UFAs) are found in B. subtilis, although cold shock can induce UFA synthesis.74–76
Figure 2.
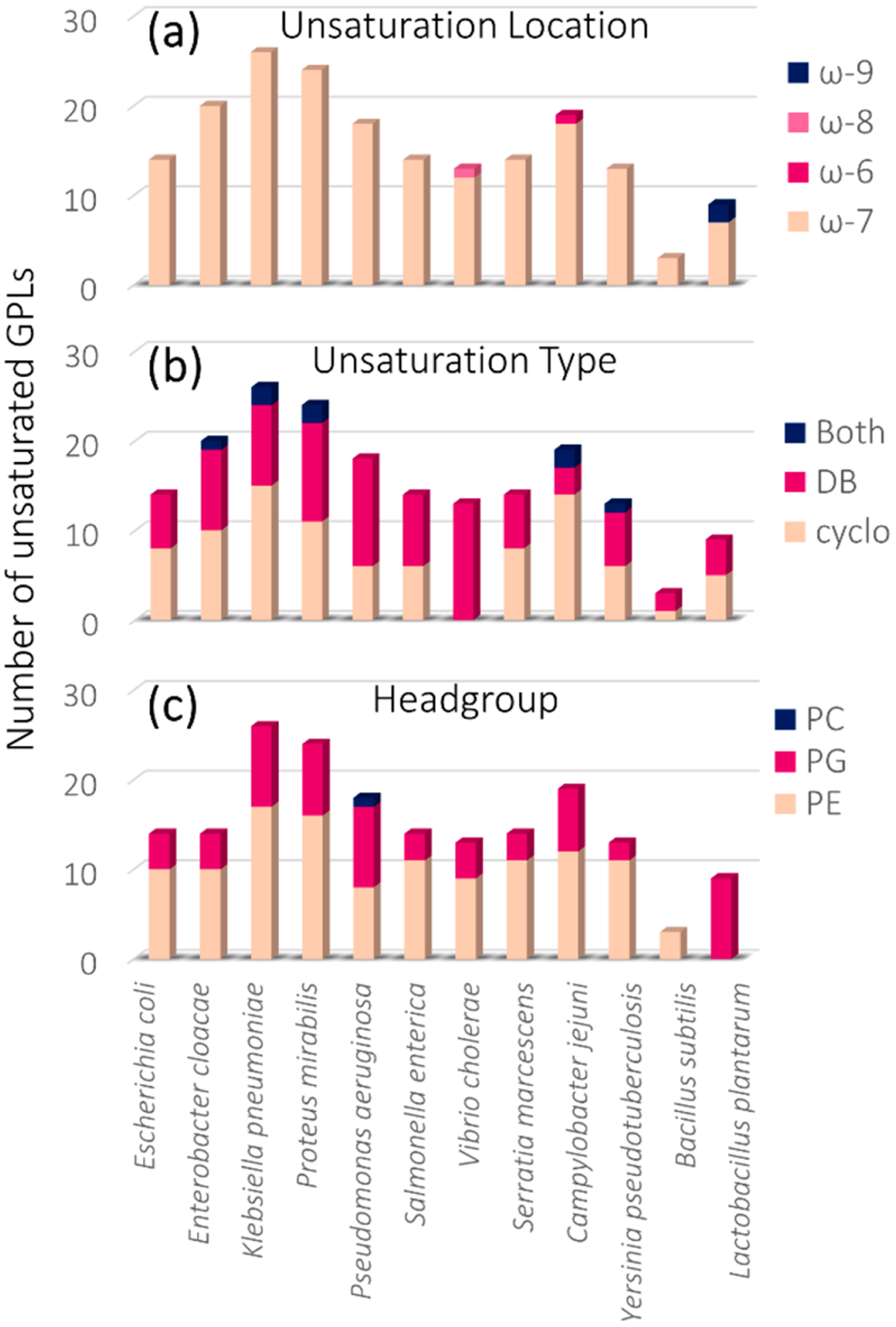
Bar graphs showing the total number of identified unsaturated GPLs for each of the 12 bacterial extracts by (a) unsaturation location, (b) unsaturation type, and (c) GPL headgroup.
Relative trends in unsaturation content were investigated by grouping the identified unsaturated GPLs by unsaturation type, that is, double bond versus cyclopropane ring. Both cyclopropane rings and double bonds can be identified and localized using UVPD-MS, as each unsaturation motif provides a diagnostic pair of fragment ions spaced either 14 or 24 Da apart, respectively.17,60 As seen in Figure 2b, most of the 12 bacteria showed a mix of cyclopropane and double-bond content, with some samples containing GPLs that had both a double-bond acyl chain and a cyclopropane acyl chain. Most notably, only unsaturated lipids containing double bonds were identified in the V. cholerae lipid extract, indicating that among the 12 bacteria, V. cholerae is unique in that it contains no cyclopropane lipids. This was a surprising finding, as the cyclopropane fatty acid (CFA) synthase gene has been previously identified in V. cholerae.77 However, only the genomic analysis of V. cholerae, and not the phenotypic outcome, was reported.77 Finally, the unsaturated lipids were grouped by GPL headgroup to reveal the headgroup-specific unsaturation trends (Figure 2c). Most samples showed a mix of unsaturated PEs and PGs, whereas P. aeruginosa was the only bacterium that additionally contained PCs, which were analyzed as [M + formate]− adducts in the negative mode. This aligns with previous reports indicating that P. aeruginosa is one of the few known prokaryotes that produces PCs.78 For the two Gram-positive bacterial extracts, we identified only unsaturated PEs in B. subtilis and only unsaturated PGs in L. plantarum. However, we saw evidence of unsaturated PGs in B. subtilis and unsaturated PEs in L. plantarum, but the key structure-specific diagnostic fragment ions generated by UVPD were not confidently identified because mass errors fell slightly above 10 ppm error due to low precursor abundances. Overall, fewer unsaturated lipids were identified in the 2 Gram-positive bacterial lipid extracts compared with the 10 Gram-negative ones, an outcome that aligns with previous studies reporting that species such as B. subtilis almost exclusively produce saturated fatty acids.79
Another striking trend evident from this comprehensive data set is the high frequency of odd-carbon saturated acyl chains (15:0, 17:0, etc.) as shown in Tables S2–S13. Like most other biomolecules, fatty acids (which comprise the two acyl chains in GPLs) are synthesized from the two-carbon-atom-containing acetyl-CoA,5,80 so each fatty acid must initially contain an even number of carbon atoms. An odd number of carbon atoms in a saturated acyl chain (15:0, 17:0, etc.) is typically indicative of branching of the acyl chain, whereas an odd number of carbon atoms in an unsaturated acyl chain (15:1, 17:1, etc.) is typically indicative of a cyclopropane element. Branched GPL acyl chains are known to affect the membrane fluidity owing to their disruptive effect on the packing of the phospholipid bilayer, and the position and geometry of branching have distinct effects on the membrane fluidity; iso-chains decrease the membrane fluidity, in contrast with anteiso-chains, which are known to increase the membrane permeability.5 CFAs have similar effects on the membrane fluidity as UFAs, although they provide more resistance to environmental stressors such as acid stress.5 Whereas UVPD has previously been shown to localize cyclopropane rings as well as double bonds within the acyl chains of GPLs,17,60 locating branching positions in the acyl chain remains an elusive challenge.
Although all unsaturated GPLs with identifiable unsaturation positions contained only monounsaturated acyl chains, we noted the additional presence of multiply unsaturated GPLs (known as polyunsaturated fatty acids (PUFAs)) within some of the bacterial extracts. As seen in Figure 3, 8 of the 12 bacterial extracts showed evidence of phospholipids with 18:2 acyl chains, whereas a less commonly observed PUFA of composition 20:4 was observed in only 1 of the 12 samples. Of the 12 types of identified PUFAs, 4 (19:2, 17:2, 18:3, and 20:4) were identified in only one bacterial extract and could thus be used as potential markers for molecular phenotyping in future studies. Some of these PUFAs were also determined to contain a hydroxyl (OH) group, shown in more detail in Figure 4. PUFAs were identified via the matching of abundant acyl chain product ions to elemental formulas and the total number of unsaturation elements, and these species were down-shifted in mass by 2 Da per additional unsaturation element relative to their monounsaturated counterparts. A full tabulation of the identified PUFAs is shown in Table S14. Whereas monounsaturated GPLs are common in bacteria,11,81 PUFAs are generally unheard of, although they have been previously detected in bacteria that inhabit psychrophilic environments (low temperature and high pressure) such as the deep ocean82,83 as well as in Gram-negative V. cholerae in the presence of bile.68 For the samples in the present study, bacterial GPL PUFAs, while detected, were of too low abundance to allow confident localization of the unsaturation elements using UVPD (i.e., no characteristic double bond or cyclopropane ions separated by 24 or 14 Da, respectively, were identified). Despite the lack of site localization of unsaturation elements, the detection of endogenous multiply unsaturated acyl chains in bacterial GPLs in the current study is an intriguing finding about the bacterial lipidome, as PUFA acyl chains have been shown to influence the lateral organization and thus the biophysical properties of cell membranes.84
Figure 3.
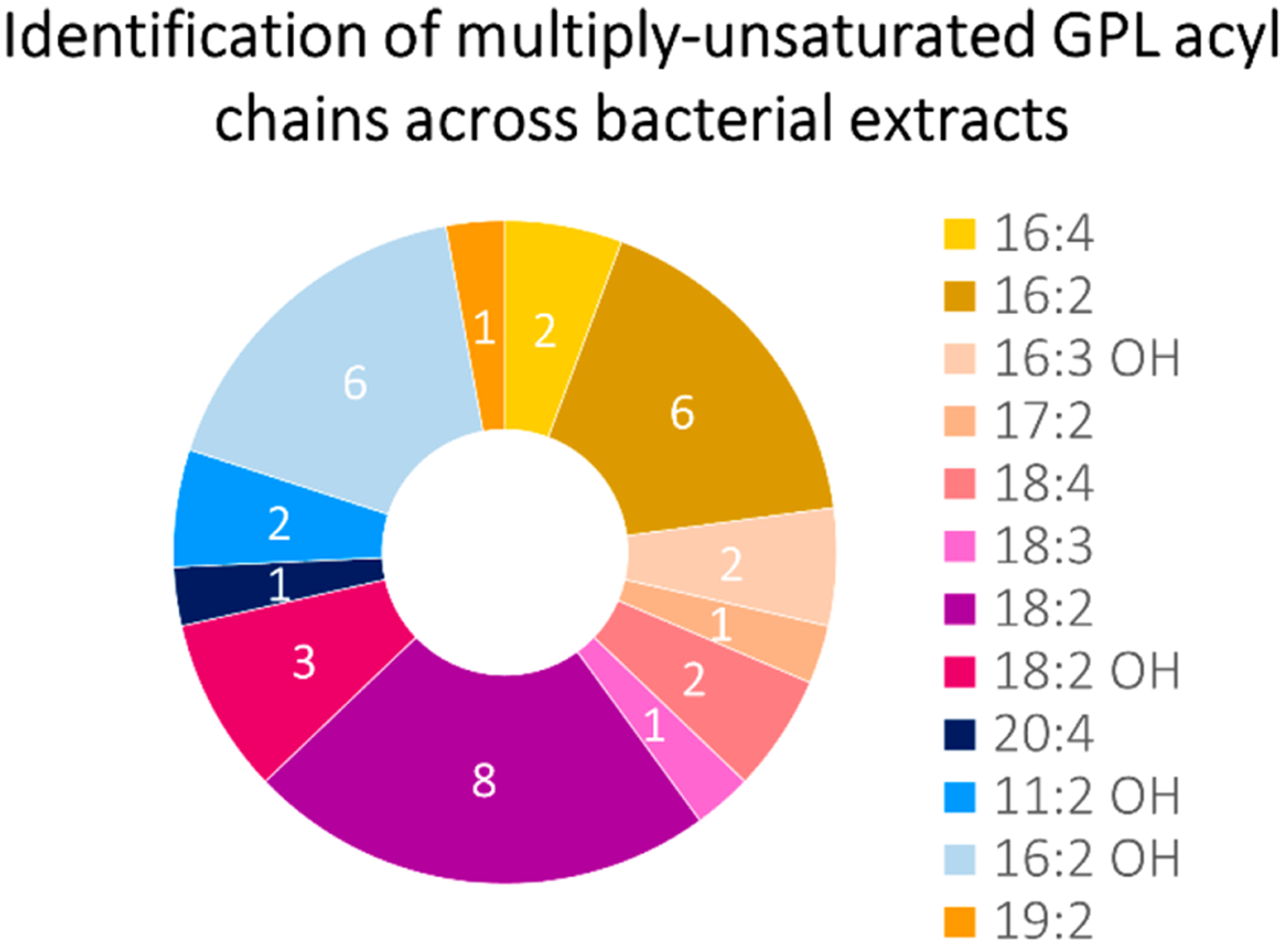
Doughnut plot displaying the frequency of the occurrence of multiply unsaturated GPL acyl chains by sample. For example, acyl chain 18:2 was the most common unsaturated acyl chain because it was identified in 8 of the 12 investigated bacterial extract samples, whereas acyl chain 20:4 was identified in only 1 of the 12 bacterial extracts.
Figure 4.
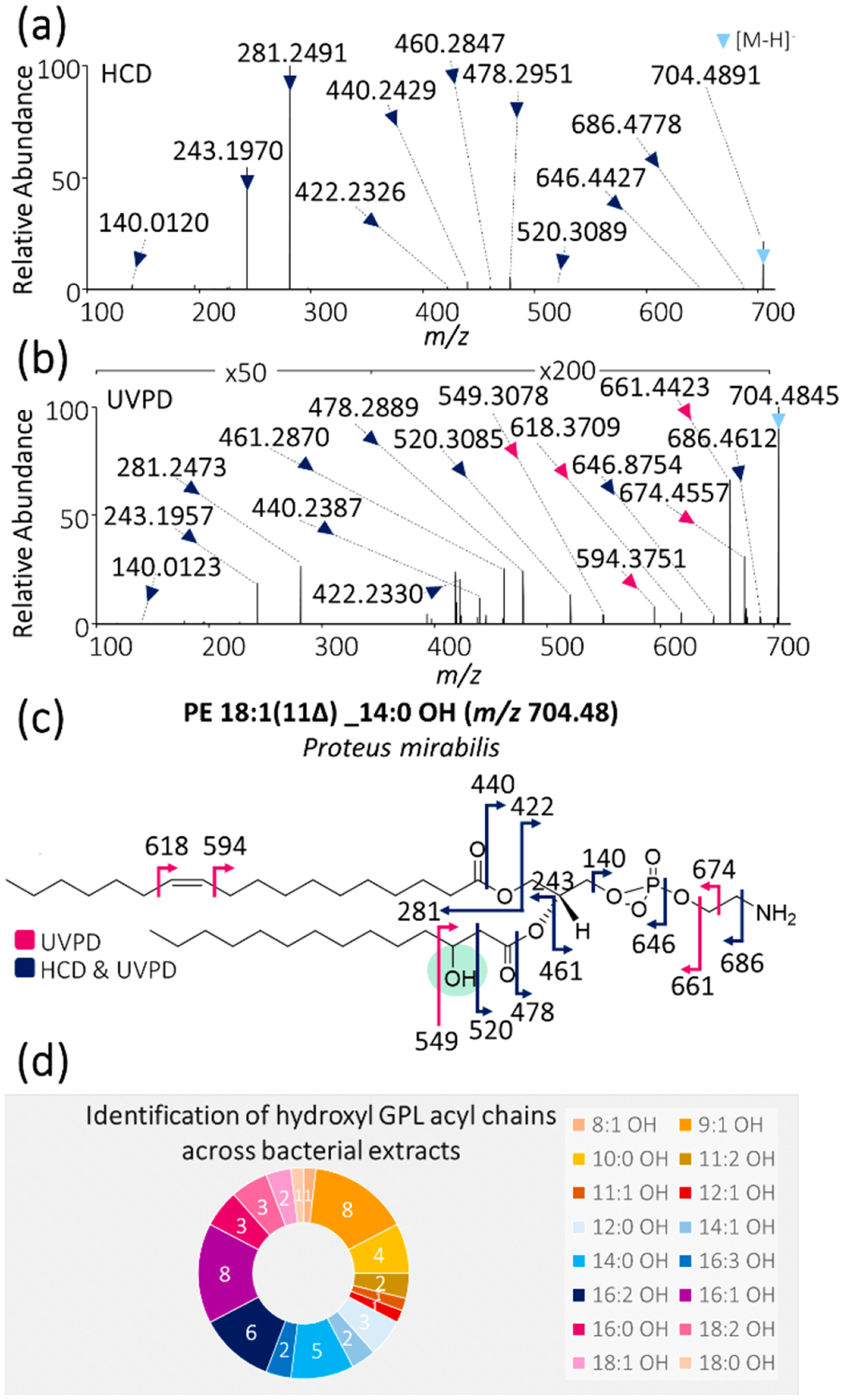
(a) HCD and (b) 193 nm UVPD (8 pulses, 2.5 mJ per pulse) mass spectra of m/z 704.49 from Proteus mirabilis lipid extract. (c) Fragment ion map of m/z 704.49 identified as PE 18:1(11Δ) _14:0 OH with the hydroxyl moiety highlighted in green. (Fragmentation sites are color-coded to correspond to the ions identified in panels a and b.) (d) Doughnut plot displaying the frequency of the occurrence of hydroxyl GPL acyl chains by sample. For example, acyl chain 16:1 OH was the most common unsaturated acyl chain because it was identified in 8 of the 12 investigated bacterial extract samples, whereas acyl chain 18:0 OH was identified in only 1 of the 12 bacterial extracts.
In addition to acyl chain unsaturation motifs and branching, we have identified a number of GPLs containing hydroxyl groups in their acyl chains. For example, HCD and UVPD mass spectra of PE 18:1(11Δ)_14:0 OH are shown in Figure 4a,b. Once again, HCD provides abundant acyl chain product ions, whereas UVPD exclusively pinpoints the position of the double bond (Δ24 Da fragment ion pair). In addition, UVPD also produces two diagnostic fragment ions of m/z 520 and 549 that bracket the OH modification, thus localizing it to the three-carbon position, as shown in the fragment ion map in Figure 4c. In contrast, HCD produces only one of these two fragments, namely, the fragment closer to the carbonyl group, thus making this lone fragment of m/z 520 insufficient to localize the hydroxyl modification with confidence. Hydroxy fatty acids have also recently been characterized via chemical derivatization coupled to 266 nm photodissociation85 as well as via Paternò–Büchi functionalization followed by collisional activation;86 however, both methods rely on chemical derivatization prior to analysis, whereas the latter additionally merely restricts but does not unambiguously identify the hydroxylation site. In contrast, 193 nm UVPD of GPLs provides a robust method for the direct and unambiguous identification and localization of hydroxylation sites within acyl chains, thus enabling the differentiation of OH-position isomers and alleviating the need for chemical derivatization strategies. A detailed tabulation of the identified hydroxyl acyl chains (identified via abundant hydroxyl acyl chain product ions) along with ppm mass error values are summarized in Table S15.
These results build upon and expand the unique capability of UVPD to localize multiple types of modifications in individual GPLs (in this case, a double bond and a hydroxyl group). 3-Hydroxy fatty acids (3-OH FAs) are known to comprise part of the immunogenic LPS in Gram-negative bacteria20,23 and have previously been employed as biomarkers for the quantification and characterization of endotoxins in Gram-negative bacteria.87 These prior findings are consistent with the lack of 3-hydroxy acyl chains detected in the two Gram-positive samples in the present study, as Gram-positive bacteria do not contain LPS as part of their cellular envelope. As seen in Figure 4d, UVPD enabled the identification of 16 different hydroxyl-containing acyl chains among the 12 bacterial extracts, of which 16:1 OH and 9:1 OH were the most common. In the context of these key results, it is crucial to emphasize the importance of the high-resolution and high-accuracy capabilities of the mass spectrometry instrumentation because without these features, it is impossible to differentiate hydroxyl-type and saturated/unsaturated nonhydroxyl acyl chains. As displayed in Table S16, for any hydroxyl-containing acyl chain with at least one unsaturation, there is a corresponding “standard” (nonhydroxyl) acyl chain counterpart that yields acyl chain product ions very close in m/z. These isobaric acyl chains differ in mass by only 0.04 Da, and thus the confirmation of hydroxyl acyl chains is not feasible using low-resolution mass analyzers alone. However, GPLs possessing hydroxylated acyl chains elute earlier than their isobaric nonhydroxyl counterparts in chromatographic separations and thus conceivably could be discerned (but not characterized) based on the retention time, as demonstrated in Figure S3. In this example, a resolving power of at least 16 900 is needed to distinguish the two species based solely on the m/z values of the intact lipids, thus necessitating high-resolution mass analyzers such as Orbitrap and Fourier transform ion cyclotron resonance (FT-ICR) MS systems. To our knowledge, this is the first direct localization of 3-hydroxyl acyl chains in bacterial GPLs using mass spectrometry with UVPD playing a pivotal role.
In addition to the elucidation of unsaturation features and hydroxyl modifications, UVPD combined with high-resolution high-mass-accuracy measurements enabled the identification of a novel GPL headgroup modification consisting of a PS with an additional methyl group at the carboxylic acid terminus, termed “Me-PS” (methyl-PS). Representative HCD and UVPD mass spectra of one such Me-PS from P. aeruginosa lipid extract are shown in Figure 5a,b, respectively. Both spectra show a characteristic fragment ion of m/z 740.49, which represents the cleavage of the methoxy group (−OCH3) from the methylated carboxylic terminus of the Me-PS corresponding to a 32.02 Da loss. UVPD generates an additional unique fragment ion of m/z 713.49 corresponding to loss of the entire methylated carboxylic acid moiety, whereas HCD generates no unique fragment ions. The UVPD fragmentation patterns confidently identify the GPL as Me-PS 16:1(9Δ)_18:1(11Δ). On the basis of the fragmentation pattern, a second slightly different structural possibility for this lipid exists and is shown in Figure S4. This Me-PS was quite abundant, as seen from the base peak LC trace and highlighted XIC in Figure S5. Both saturated and unsaturated Me-PS lipids were identified across all 12 bacterial lipid extracts (data not shown), indicating that these lipids are not unique to any one bacterium. A 2011 study performed by Garrett and coworkers provided evidence of the presence of a novel nonenzymatically derived PE-ethylcarbamate derivative termed PE-EC,88 which is structurally extremely similar to the proposed Me-PS structure. These lipids were concluded to have been produced during the lipid extraction procedure via the reaction with phosgene (a decomposition product of chloroform), as the use of methylene chloride in place of chloroform showed no evidence of PE-ECs.88 For the present study, chloroform was used in the lipid extraction process for all 12 bacterial samples. However, our analysis of a commercially available bacterial E. coli extract (Avanti Polar Lipids) showed no evidence of Me-PS lipids (data not shown), despite the use of a similar lipid extraction procedure involving chloroform. Interestingly, the commercial extract is obtained at the 3/4 log growth phase, whereas our extracts are acquired at the stationary growth phase. Thus these observed Me-PS lipids may be a signature of the growth phase, indicating that they may, in fact, be endogenous and enzymatically derived. Because anionic PS is an intermediate in the synthesis of zwitterionic PE,11,89 the production of endogenous methylated PS lipids is plausible. A more detailed and systematic investigation is needed to determine whether these Me-PS lipids are, in fact, endogenous and, if so, whether they are a signature of growth phase.
Figure 5.

(a) HCD and (b) 193 nm UVPD (eight pulses, 2.5 mJ per pulse) mass spectra of m/z 772.51 from Pseudomonas aeruginosa lipid extract. (c) Fragment ion map of m/z 772.51 identified as a methylated PS (Me-PS) 16:1(9Δ)_18:1(11Δ) with the Me-PS moiety highlighted in green. (Fragmentation sites are color-coded to correspond to the ions identified in panels a and b.)
Beyond the characterization of methylation, UVPD has the ability to pinpoint other types of GPL headgroup modifications as well. One such modification of high biological importance is the aminoacylation of GPLs. Bacteria employ the aminoacylation of negatively charged GPLs (in general, PG and cardiolipin) to increase the net charge of their membrane bilayer, which, in turn, decreases their susceptibility to antibiotics, CAMPs, and other molecules that rely on the negative charge of the bacterial membrane to adhere to and penetrate the membrane bilayer.30,90 Thus surface charge modulation via the aminoacylation of bacterial GPLs is an important mechanism of antibiotic and antimicrobial resistance and is found almost exclusively in Gram-positive bacteria.91–93 The multiple peptide resistance factor (MprF) has been found to be responsible for the addition of either lysine or alanine to PG in Gram-positive bacteria.90,94 Gram-negative bacteria have an analogous mechanism of surface charge modulation in which lipid A, which comprises a substructure of the bacterial immunogenic LPS in Gram-negative bacteria, is modified via the addition of phosphoethanolamine, glycine, or amino sugars.31,32,94 Both pathways of surface charge modulation ultimately result in an increased net charge on the bacterial membrane. Here we provide the first direct evidence of aa-PGs from a Gram-negative bacterium, namely, C. jejuni. Lysyl-PG 15:0_13:0 of m/z 793.53 was identified from C. jejuni, as illustrated in Figure 6. Figure S6 shows the abundance of the lipid of m/z 793.53 relative to the base peak chromatogram, showing that it is a rather minor component. Figure 6a,b shows HCD and UVPD mass spectra of m/z 793.53, which provide highly diagnostic fragmentation patterns, thus pinpointing the identity of the lipid as lysyl-PG 15:0_13:0, as seen in the fragment ion map in Figure 6c. In the case of this lysyl-PG, HCD and UVPD afford complementary information for complete structural characterization. HCD yields the abundant diagnostic ion of m/z 145.10 originating from C–O bond cleavage, which clearly indicates the addition of a lysine moiety to the PG lipid (fragmentation at this bond is not observed for conventional PG headgroups), whereas UVPD confirms this lysyl modification via an array of fragments that offer a detailed fingerprint of the entire headgroup. A large array of lysyl-PGs were identified in both Gram-negative C. jejuni as well as Gram-positive L. plantarum and B. subtilis. A complete listing of all identified lysyl-PGs is given in Table S17. To our knowledge, this represents the first direct evidence of aa-PGs in both Gram-positive L. plantarum and Gram-negative C. jejuni. The latter finding has vast implications in that it suggests that Gram-negative bacteria may be adapting not only by modulating their membrane surface charge via lipid A/LPS modifications but also via the aminoacylation of GPLs. This finding is corroborated by previous reports that have indicated that some Gram-negative bacteria may possess MprF homologues that are capable of synthesizing aa-PGs such as P. aeruginosa, Agrobacterium tumefaciens, and Rhizobium tropici.29,33,95–97 Studies are currently underway to identify lysyl-PG formation in C. jejuni.
Figure 6.
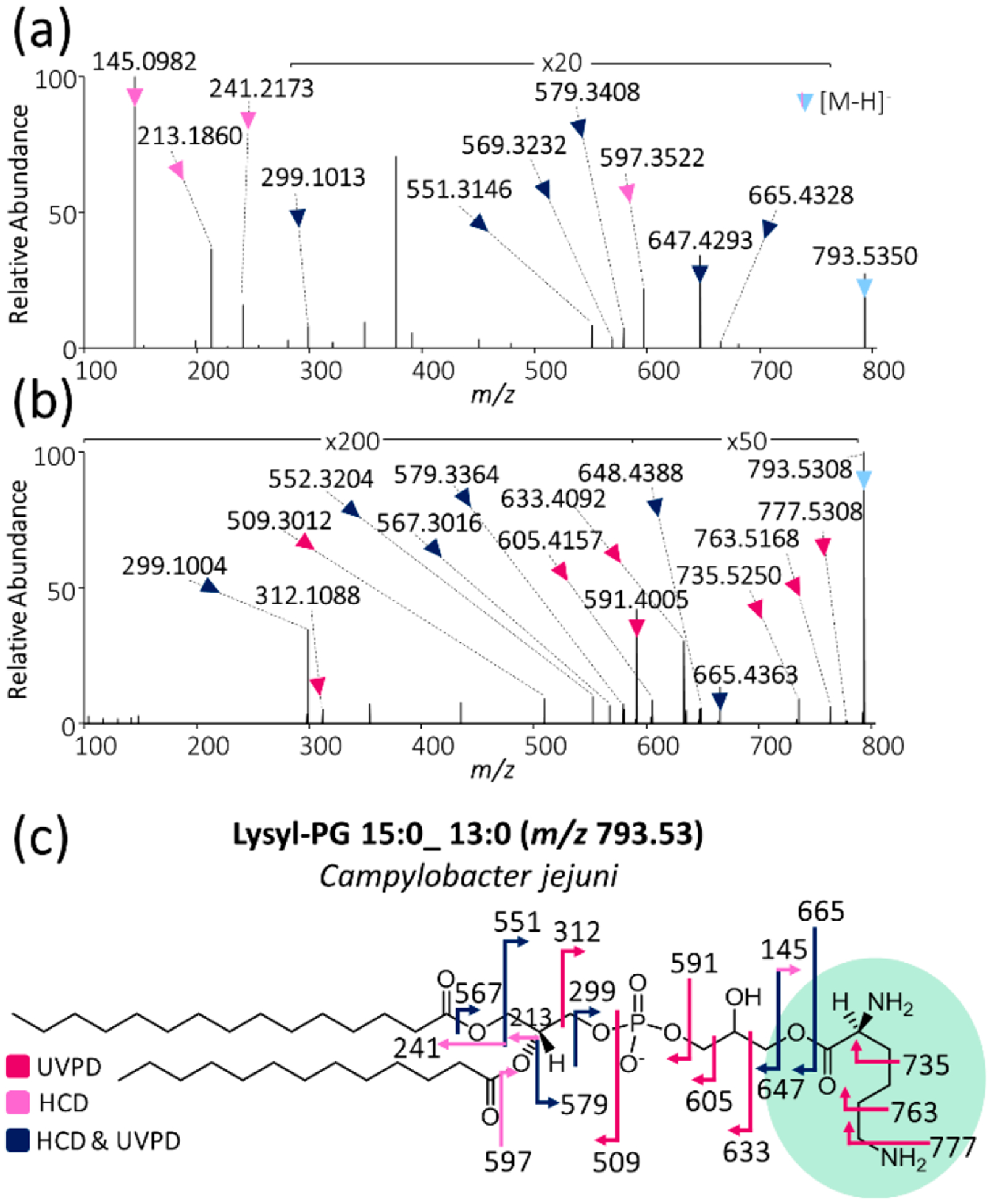
(a) HCD and (b) 193 nm UVPD (eight pulses, 2.5 mJ per pulse) mass spectra of m/z 793.53 from Campylobacter jejuni lipid extract. (c) Fragment ion map of m/z 793.53 identified as lysyl-PG 13:0_15:0 with the lysyl moiety highlighted in green. (Fragmentation sites are color-coded to correspond to the ions identified in panels a and b.)
CONCLUSIONS
Here we demonstrate the utility of high-resolution high-mass-accuracy LC-MS coupled to 193 nm UVPD for the elucidation of bacterial GPL modifications. We present UVPD as a frontier method to investigate the structure of bacterial GPLs in detail and to reveal the presence of subtle but highly consequential modifications that are frequently uncharted. This approach allowed the elucidation of patterns of bacterial GPL unsaturation motifs across both Gram-positive and Gram-negative bacteria and enabled the direct characterization and identification of novel GPL modifications, including both acyl chain and headgroup modifications. Whereas HCD data can confirm the fatty acid chain composition and suggest the presence of acyl chain and headgroup modifications, UVPD is needed to facilitate the unambiguous identification and localization of these modifications via diagnostic fragment ions, as has been demonstrated for the characterization of hydroxyl acyl chains, Me-PS type headgroups, branched odd-carbon saturated acyl chains, and aminoacyl-GPLs. Of notable significance was the finding of lysyl-PGs in Gram-negative C. jejuni, expanding our knowledge of the methods harnessed by Gram-negative bacteria to enhance antibiotic resistance via surface charge modulation. Because lipid modifications are often growth-state-dependent, investigating the impact of the growth phase on bacterial GPL modifications is also an important avenue. Many of the acyl chain and headgroup modifications identified in this work are absent in the common phospholipid MS/MS databases, tools, and literature, and thus there is a vital need for recognition of the vast and highly abundant repertoire of modifications that exist in bacterial GPLs, as they appear to be the norm rather than the exception. We are currently extending the limits of lipid LC separation (in particular, for isomeric lipids within complex mixtures) to implement this strategy in a quantitative manner as well as exploring the development of tools to automate the identification of GPLs using HCD and UVPD data from high-throughput LC experiments.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The Welch Foundation (F-1155 to J.S.B.) and the National Institutes of Health (NIGMS: R01 GM103655 to J.S.B.; NIAID: R01 AI150098, R01 AI129940, and R01 AI138576 to M.S.T.). A.B.P. was supported by the National Science Foundation under Grant No. DGE-1545433.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c01449.
Figure S1: Base peak LC-MS traces of bacterial lipid extracts. Figure S2: Omega versus delta nomenclature for classification of GPL unsaturation elements. Figure S3: Chromatographic separation of hydroxyl lipids and their nonhydroxyl isobaric counterparts. Figure S4: Possible headgroup structures for Me-PS headgroup. Figure S5: P. aeruginosa lipid extract base peak LC-MS trace with XIC of Me-PS lipid. Figure S6: C. jejuni lipid extract base peak LC-MS trace with XIC of lysyl-PG lipid. Table S1: List of investigated bacterial lipid extracts. Tables S2–S13: Lists of identified unsaturated lipids in all 12 bacterial lipid extracts with m/z and ppm error values. Table S14: List of identified PUFAs within GPLs with m/z and ppm error values. Table S15: List of identified hydroxyl acyl chains within GPLs m/z and ppm error values. Table S16: List of identified OH acyl chains with acyl chain product ion m/z values and m/z values of isobaric unmodified (nonhydroxyl) acyl chains. Table S17: List of lysyl-PGs in Gram-negative C. jejuni and Gram-positive B. subtilis and L. plantarum (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.analchem.0c01449
The authors declare no competing financial interest.
Contributor Information
Molly S. Blevins, Department of Chemistry, University of Texas at Austin, Austin, Texas 78712, United States
Virginia K. James, Department of Chemistry, University of Texas at Austin, Austin, Texas 78712, United States
Carmen M. Herrera, Department of Infectious Diseases, College of Veterinary Medicine, University of Georgia, Athens, Georgia 30602, United States
Alexandria B. Purcell, Department of Infectious Diseases, College of Veterinary Medicine, University of Georgia, Athens, Georgia 30602, United States
M. Stephen Trent, Department of Infectious Diseases, College of Veterinary Medicine, Department of Microbiology, College of Arts and Sciences, and Center for Vaccines and Immunology, University of Georgia, Athens, Georgia 30602, United States.
Jennifer S. Brodbelt, Department of Chemistry, University of Texas at Austin, Austin, Texas 78712, United States.
REFERENCES
- (1).Phillips R; Ursell T; Wiggins P; Sens P Nature 2009, 459, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Bogdanov M; Sun J; Kaback HR; Dowhan WJ Biol. Chem 1996, 271, 11615–11618. [DOI] [PubMed] [Google Scholar]
- (3).Dowhan W; Vitrac H; Bogdanov M Protein J 2019, 38, 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Park NH; Cheng W; Lai F; Yang C; Florez de Sessions P; Periaswamy B; Wenhan Chu C; Bianco S; Liu S; Venkataraman S; Chen Q; Yang YY; Hedrick JL J. Am. Chem. Soc 2018, 140, 4244–4252. [DOI] [PubMed] [Google Scholar]
- (5).Zhang Y-M; Rock CO Nat. Rev. Microbiol 2008, 6, 222–233. [DOI] [PubMed] [Google Scholar]
- (6).Shevchenko A; Simons K Nat. Rev. Mol. Cell Biol 2010, 11, 593–598. [DOI] [PubMed] [Google Scholar]
- (7).Saliba A-E; Vonkova I; Gavin A-C Nat. Rev. Mol. Cell Biol 2015, 16, 753–761. [DOI] [PubMed] [Google Scholar]
- (8).Ibáñez C; Mouhid L; Reglero G; Ramírez de Molina AJ Agric. Food Chem 2017, 65, 7827–7842. [DOI] [PubMed] [Google Scholar]
- (9).Rustam YH; Reid GE Anal. Chem 2018, 90, 374–397. [DOI] [PubMed] [Google Scholar]
- (10).Brodbelt JS; Morrison LJ; Santos I Chem. Rev 2020, 120, 3328–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sohlenkamp C; Geiger O FEMS Microbiol. Rev 2016, 40, 133–159. [DOI] [PubMed] [Google Scholar]
- (12).Lopalco P; Stahl J; Annese C; Averhoff B; Corcelli A Sci. Rep 2017, 7, 2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Fahy E; Subramaniam S; Brown HA; Glass CK; Merrill AH; Murphy RC; Raetz CRH; Russell DW; Seyama Y; Shaw W; Shimizu T; Spener F; van Meer G; VanNieuwenhze MS; White SH; Witztum JL; Dennis EA J. Lipid Res 2005, 46, 839–861. [DOI] [PubMed] [Google Scholar]
- (14).Hu T; Zhang J-LJ Sep Sci 2018, 41, 351–372. [DOI] [PubMed] [Google Scholar]
- (15).Pauling JK; Hermansson M; Hartler J; Christiansen K; Gallego SF; Peng B; Ahrends R; Ejsing CS PLoS One 2017, 12, No. e0188394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wojcik R; Webb IK; Deng L; Garimella SVB; Prost SA; Ibrahim YM; Baker ES; Smith RD Int. J. Mol. Sci 2017, 18, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Blevins MS; Klein DR; Brodbelt JS Anal. Chem 2019, 91, 6820–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Williams PE; Klein DR; Greer SM; Brodbelt JS J. Am. Chem. Soc 2017, 139, 15681–15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Taniguchi T; Manai D; Shibata M; Itabashi Y; Monde KJ Am. Chem. Soc 2015, 137, 12191–12194. [DOI] [PubMed] [Google Scholar]
- (20).Szponar B; Krásnik L; Hryniewiecki T; Gamian A; Larsson L Clin. Chem 2003, 49, 1149–1153. [DOI] [PubMed] [Google Scholar]
- (21).Minnikin DE; Kremer L; Dover LG; Besra GS Chem. Biol 2002, 9, 545–553. [DOI] [PubMed] [Google Scholar]
- (22).Sjögren J; Magnusson J; Broberg A; Schnürer J; Kenne L Appl. Environ. Microbiol 2003, 69, 7554–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kutschera A; Dawid C; Gisch N; Schmid C; Raasch L; Gerster T; Schäffer M; Smakowska-Luzan E; Belkhadir Y; Vlot AC; Chandler CE; Schellenberger R; Schwudke D; Ernst RK; Dorey S; Hückelhoven R; Hofmann T; Ranf S Science 2019, 364, 178–181. [DOI] [PubMed] [Google Scholar]
- (24).Barkan D; Liu Z; Sacchettini JC; Glickman MS Chem. Biol 2009, 16, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zheng CJ; Yoo J-S; Lee T-G; Cho H-Y; Kim Y-H; Kim W-G FEBS Lett 2005, 579, 5157–5162. [DOI] [PubMed] [Google Scholar]
- (26).Jackson M; Stadthagen G; Gicquel B Tuberculosis (Oxford, U. K.) 2007, 87, 78–86. [DOI] [PubMed] [Google Scholar]
- (27).Poger D; Mark AE J. Phys. Chem. B 2015, 119, 5487–5495. [DOI] [PubMed] [Google Scholar]
- (28).Liu Y; Hazzard C; Eustáquio AS; Reynolds KA; Moore BS J. Am. Chem. Soc 2009, 131, 10376–10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Arendt W; Groenewold MK; Hebecker S; Dickschat JS; Moser JJ Biol. Chem 2013, 288, 24717–24730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Roy H IUBMB Life 2009, 61, 940–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Simpson BW; Trent MS Nat. Rev. Microbiol 2019, 17, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hankins JV; Madsen JA; Giles DK; Brodbelt JS; Trent MS Proc. Natl. Acad. Sci. U. S. A 2012, 109, 8722–8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Sohlenkamp C; Galindo-Lagunas KA; Guan Z; Vinuesa P; Robinson S; Thomas-Oates J; Raetz CRH; Geiger O Mol. Plant-Microbe Interact 2007, 20, 1421–1430. [DOI] [PubMed] [Google Scholar]
- (34).Roy H; Dare K; Ibba M Mol. Microbiol 2009, 71, 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yang L; Li M; Shan Y; Shen S; Bai Y; Liu HJ Sep. Sci 2016, 39, 38–50. [DOI] [PubMed] [Google Scholar]
- (36).Dennis EA FASEB J 2016, 30, 114.3. [Google Scholar]
- (37).Lydic TA; Goo Y-H Clin. Transl. Med 2018, 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lv J; Zhang L; Yan F; Wang X Clin. Transl. Med 2018, 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Mansilla MC; de Mendoza D Arch. Microbiol 2005, 183, 229–235. [DOI] [PubMed] [Google Scholar]
- (40).Cronan JE Curr. Opin. Microbiol 2002, 5, 202–205. [DOI] [PubMed] [Google Scholar]
- (41).Altabe SG; Aguilar P; Caballero GM; de Mendoza DJ Bacteriol 2003, 185, 3228–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Neidleman SL Biotechnol. Genet. Eng. Rev 1987, 5, 245–268. [DOI] [PubMed] [Google Scholar]
- (43).Suutari M; Laakso S Biochim. Biophys. Acta, Lipids Lipid Metab 1992, 1126, 119–124. [DOI] [PubMed] [Google Scholar]
- (44).Randolph CE; Blanksby SJ; McLuckey SA Anal. Chem 2020, 92, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Tatituri RVV; Wolf BJ; Brenner MB; Turk J; Hsu F-F Anal. Bioanal. Chem 2015, 407, 2519–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Hsu F-F; Kuhlmann FM; Turk J; Beverley SM J. Mass Spectrom 2014, 49, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Becher S; Esch P; Heiles S Anal. Chem 2018, 90, 11486–11494. [DOI] [PubMed] [Google Scholar]
- (48).Ma X; Xia Y Angew. Chem., Int. Ed 2014, 53, 2592–2596. [DOI] [PubMed] [Google Scholar]
- (49).Ryan E; Reid GE Acc. Chem. Res 2016, 49, 1596–1604. [DOI] [PubMed] [Google Scholar]
- (50).Xu T; Pi Z; Song F; Liu S; Liu Z Anal. Chim. Acta 2018, 1028, 32–44. [DOI] [PubMed] [Google Scholar]
- (51).Jeck V; Korf A; Vosse C; Hayen H Rapid Commun. Mass Spectrom 2019, 33, 86–94. [DOI] [PubMed] [Google Scholar]
- (52).Zhang W; Zhang D; Chen Q; Wu J; Ouyang Z; Xia Y Nat. Commun 2019, 10, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Cao W; Cheng S; Yang J; Feng J; Zhang W; Li Z; Chen Q; Xia Y; Ouyang Z; Ma X Nat. Commun 2020, 11, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Franklin E; Shields S; Manthorpe J; Smith JC; Xia Y; McLuckey SA J. Am. Soc. Mass Spectrom 2020, 31, 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Harris RA; May JC; Stinson CA; Xia Y; McLean JA Anal. Chem 2018, 90, 1915–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Harrison KA; Murphy RC Anal. Chem 1996, 68, 3224–3230. [DOI] [PubMed] [Google Scholar]
- (57).Batarseh AM; Abbott SK; Duchoslav E; Alqarni A; Blanksby SJ; Mitchell TW Int. J. Mass Spectrom 2018, 431, 27–36. [Google Scholar]
- (58).Macias LA; Santos IC; Brodbelt JS Anal. Chem 2020, 92, 227–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Fang M; Rustam Y; Palmieri M; Sieber OM; Reid GE Anal. Bioanal. Chem 2020, 412, 2339–2351. [DOI] [PubMed] [Google Scholar]
- (60).Klein DR; Brodbelt JS Anal. Chem 2017, 89, 1516–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Macias LA; Feider CL; Eberlin LS; Brodbelt JS Anal. Chem 2019, 91, 12509–12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Klein DR; Holden DD; Brodbelt JS Anal. Chem 2016, 88, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Klein DR; Blevins MS; Macias LA; Douglass MV; Trent MS; Brodbelt JS Anal. Chem 2020, 92, 5986–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Ryan E; Nguyen CQN; Shiea C; Reid GE J. Am. Soc. Mass Spectrom 2017, 28, 1406–1419. [DOI] [PubMed] [Google Scholar]
- (65).Mishra VK; Buter J; Blevins MS; Witte MD; Van Rhijn I; Moody DB; Brodbelt JS; Minnaard AJ Org. Lett 2019, 21, 5126–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Becher S; Esch P; Heiles S Anal. Chem 2018, 90, 11486–11494. [DOI] [PubMed] [Google Scholar]
- (67).Bligh EG; Dyer WJ Can. J. Biochem. Physiol 1959, 37, 911–917. [DOI] [PubMed] [Google Scholar]
- (68).Giles DK; Hankins JV; Guan Z; Trent MS Mol. Microbiol 2011, 79, 716–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Damen CWN; Isaac G; Langridge J; Hankemeier T; Vreeken RJ J. Lipid Res 2014, 55, 1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Nakamura MT; Nara TY Annu. Rev. Nutr 2004, 24, 345–376. [DOI] [PubMed] [Google Scholar]
- (71).Lee JM; Lee H; Kang S; Park WJ Nutrients 2016, 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Kassab E; Fuchs M; Haack M; Mehlmer N; Brueck TB Microb. Cell Fact 2019, 18, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Wang AY; Grogan DW; Cronan JE Biochemistry 1992, 31, 11020–11028. [DOI] [PubMed] [Google Scholar]
- (74).Aguilar PS; Cronan JE; de Mendoza DJ Bacteriol 1998, 180, 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Aguilar PS; Lopez P; de Mendoza DJ Bacteriol 1999, 181, 7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Cybulski LE; Albanesi D; Mansilla MC; Altabe S; Aguilar PS; de Mendoza D Mol. Microbiol 2002, 45, 1379–1388. [DOI] [PubMed] [Google Scholar]
- (77).Valentine RC; Valentine DL Omega-3 Fatty Acids and the DHA Principle; CRC Press, 2009. [Google Scholar]
- (78).Wilderman PJ; Vasil AI; Martin WE; Murphy RC; Vasil ML J. Bacteriol 2002, 184, 4792–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Grau R; de Mendoza D Mol. Microbiol 1993, 8, 535–542. [DOI] [PubMed] [Google Scholar]
- (80).Ohlrogge J; Browse J Plant Cell 1995, 7, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Fulco AJ Prog. Lipid Res 1983, 22, 133–160. [DOI] [PubMed] [Google Scholar]
- (82).Moi IM; Leow ATC; Ali MSM; Rahman RNZRA; Salleh AB; Sabri S Appl. Microbiol. Biotechnol 2018, 102, 5811–5826. [DOI] [PubMed] [Google Scholar]
- (83).Yoshida K; Hashimoto M; Hori R; Adachi T; Okuyama H; Orikasa Y; Nagamine T; Shimizu S; Ueno A; Morita N Mar. Drugs 2016, 14, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Shaikh SR; Edidin M Am. J. Clin. Nutr 2006, 84, 1277–1289. [DOI] [PubMed] [Google Scholar]
- (85).Narreddula VR; Sadowski P; Boase NRB; Marshall DL; Poad BLJ; Trevitt AJ; Mitchell TW; Blanksby SJ Rapid Commun. Mass Spectrom 2020, 34, No. e8741. [DOI] [PubMed] [Google Scholar]
- (86).Esch P; Heiles S Analyst 2020, 145, 2256–2266. [DOI] [PubMed] [Google Scholar]
- (87).Lee AKY; Chan CK; Fang M; Lau AP S. Atmos. Environ 2004, 38, 6307–6317. [Google Scholar]
- (88).Garrett TA; Raetz CRH; Son JD; Richardson TD; Bartling C; Guan Z Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2011, 1811, 827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Cassilly CD; Reynolds TB J. Fungi (Basel) 2018, 4, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Slavetinsky C; Kuhn S; Peschel A Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2017, 1862, 1310–1318. [DOI] [PubMed] [Google Scholar]
- (91).Atila M; Katselis G; Chumala P; Luo YJ Am. Soc. Mass Spectrom 2016, 27, 1606–1613. [DOI] [PubMed] [Google Scholar]
- (92).Kilelee E; Pokorny A; Yeaman MR; Bayer AS Antimicrob. Agents Chemother 2010, 54, 4476–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Andrä J; Goldmann T; Ernst CM; Peschel A; Gutsmann T J. Biol. Chem 2011, 286, 18692–18700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Nuri R; Shprung T; Shai Y Biochim. Biophys. Acta, Biomembr 2015, 1848, 3089–3100. [DOI] [PubMed] [Google Scholar]
- (95).Slavetinsky CJ; Peschel A; Ernst CM Antimicrob. Agents Chemother 2012, 56, 3492–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Groenewold MK; Hebecker S; Fritz C; Czolkoss S; Wiesselmann M; Heinz DW; Jahn D; Narberhaus F; Aktas M; Moser J Mol. Microbiol 2019, 111, 269–286. [DOI] [PubMed] [Google Scholar]
- (97).Klein S; Lorenzo C; Hoffmann S; Walther JM; Storbeck S; Piekarski T; Tindall BJ; Wray V; Nimtz M; Moser J Mol. Microbiol 2009, 71, 551–565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


