Abstract
Purpose:
Targeted prostate biopsy devices include a 3-dimensional digital template grid for guiding systematic biopsy locations. Following a template could better ensure uniform and well-distributed sampling of the prostate compared to the traditional free-hand biopsy approach, possibly decreasing the chance for false-negative biopsy. Thus, we determined cancer detection rates obtained by conventional free-hand systematic sampling versus template mapping sampling using an MRI/ultrasound fusion device.
Materials and Methods:
Men who underwent first-time conventional or image-guided prostate biopsy were identified retrospectively in an IRB-approved protocol. Excluded were men with prior biopsy or treatment, or fewer than 10 cores taken. Targeted cores taken from image-guided biopsy were censored from analysis to simulate systematic template biopsy. The resulting cancer detection rate was compared to that of conventional biopsies.
Results:
We identified 1582 patients who met criteria for analysis between 2006 and 2014: 1052 patients who underwent conventional biopsy and 530 who underwent template biopsy with an MRI/Ultrasound fusion device. Age, PSA, and number of systematic cores were the same in both groups. Template biopsy detected any prostate cancer in 257/530 men (48.5%) and clinically-significant cancer in 196/530 (37.0%), whereas conventional biopsy detected any cancer in 432/1052 (41.0%, p=0.005) and clinically-significant cancer in 308/1052 (29.2%, p=0.002).
Conclusions:
Template mapping systematic biopsy detects more prostate cancer than conventional sampling in biopsy-naïve men, and is a promising cost-effective alternative to MRI/Ultrasound biopsy as an upfront screening tool.
Keywords: prostatic neoplasms/diagnosis, prostate, image-guided biopsy, ultrasonography, magnetic resonance imaging
INTRODUCTION
Free-hand transrectal ultrasound-guided (TRUS-guided) prostate biopsy has been the standard of care for prostate cancer (CaP) diagnosis for a quarter-century. Regardless of the number of cores taken, biopsy techniques rely on systematically sampling different regions of the prostate, focusing on the peripheral zone, where fCaP is more likely to found. Preliminary evidence suggests that biopsies taken using a free-hand TRUS-guided approach are distributed in clusters, rather than evenly throughout the prostate, and that an optimized geometric distribution of biopsy cores can catch up to 20% more cancers.1,2 This is one possible reason why the cancer detection rate of a repeat TRUS biopsy is as high as 30%.3,4
Targeted biopsy of MRI-visible prostate lesions with fusion devices has been shown to improve the diagnosis of both indolent and clinically-significant prostate cancer (csCaP) when paired with systematic biopsy.5,6 Targeted biopsy is particularly useful for patients with prior negative biopsies or currently on active surveillance (AS). Randomized control trials have recently established its utility in biopsy-naïve patients as well.7–10 However, given the higher cost of obtaining a prostate MRI, its role as a frontline diagnostic for patients with CaP remains uncertain.11 For this reason, the American Urologic Association only recommends prostate MRI and targeted biopsy in the setting of prior negative biopsy and persistently elevated PSA.12
Targeted biopsy devices include a 3-dimensional (3D) digital template map for guiding systematic biopsy locations, even in the absence of an MRI-derived target. Furthermore, in contrast with traditional free-hand biopsy tools, fusion devices provide mechanical assistance with stabilization and targeting during biopsy. 3D template mapping biopsy (TMB) with MRI/Ultrasound (MRI/US) fusion devices may be an effective alternative for biopsy-naïve patients suspected to have CaP. Compared to the standard free-hand biopsy (FHB) approach, it has a theoretical benefit of eliminating human error that might otherwise lead to under-sampling of the prostate and also does not invoke the costs of an MRI. We hypothesize that TMB allows for more evenly-distributed prostate sampling, thus improving detection of csCaP in biopsy-naïve patients. To explore the value of 3D template mapping, we retrospectively compared the cancer detection rates (CDR) of TMB to FHB.
MATERIALS AND METHODS
Study Design
In this IRB-approved study, medical records were retrospectively analyzed for biopsy-naïve men who underwent prostate biopsy at a single institution between 2006–2014. Initial inclusion criteria were first-time biopsy and a minimum of 10 cores. Patients with previous biopsy or less than 10 cores were excluded. Biopsy cores from non-systematic locations (i.e. anterior or image-derived targets) were excluded from analysis. CDR, defined as the proportion of patients with positive biopsies, was compared between FHB and TMB groups.
Biopsy Procedure
All biopsies were taken using TRUS biopsy with an end-fire probe under local anesthesia. Biopsy of cores via the virtual template, or TMB, was carried out using an MRI/US fusion and biopsy tracking system (Artemis, Eigen, Grass Valley, CA). First, multiple TRUS images are obtained via rotation of the arm, and reconstructed into 3D. Next a physician contours the prostate capsule, producing an accurate volume representation of the prostate. Biopsy cores are sampled according to a 12-core biopsy template. Templates were derived from the fusion device software, which automatically and evenly distributes planned systematic locations throughout the 3D prostate volume based on standard sextant positions as well as the size and shape of the prostate at hand (Figure 2). During biopsy, the real-time location of the biopsy needle guide is displayed in relation to template locations, and mapped onto the US image. After each core is sampled, its 3D position within the prostate location is tracked and digitally stored. Each sample was individually bottled in formalin and read by a pathologist.
Figure 2.
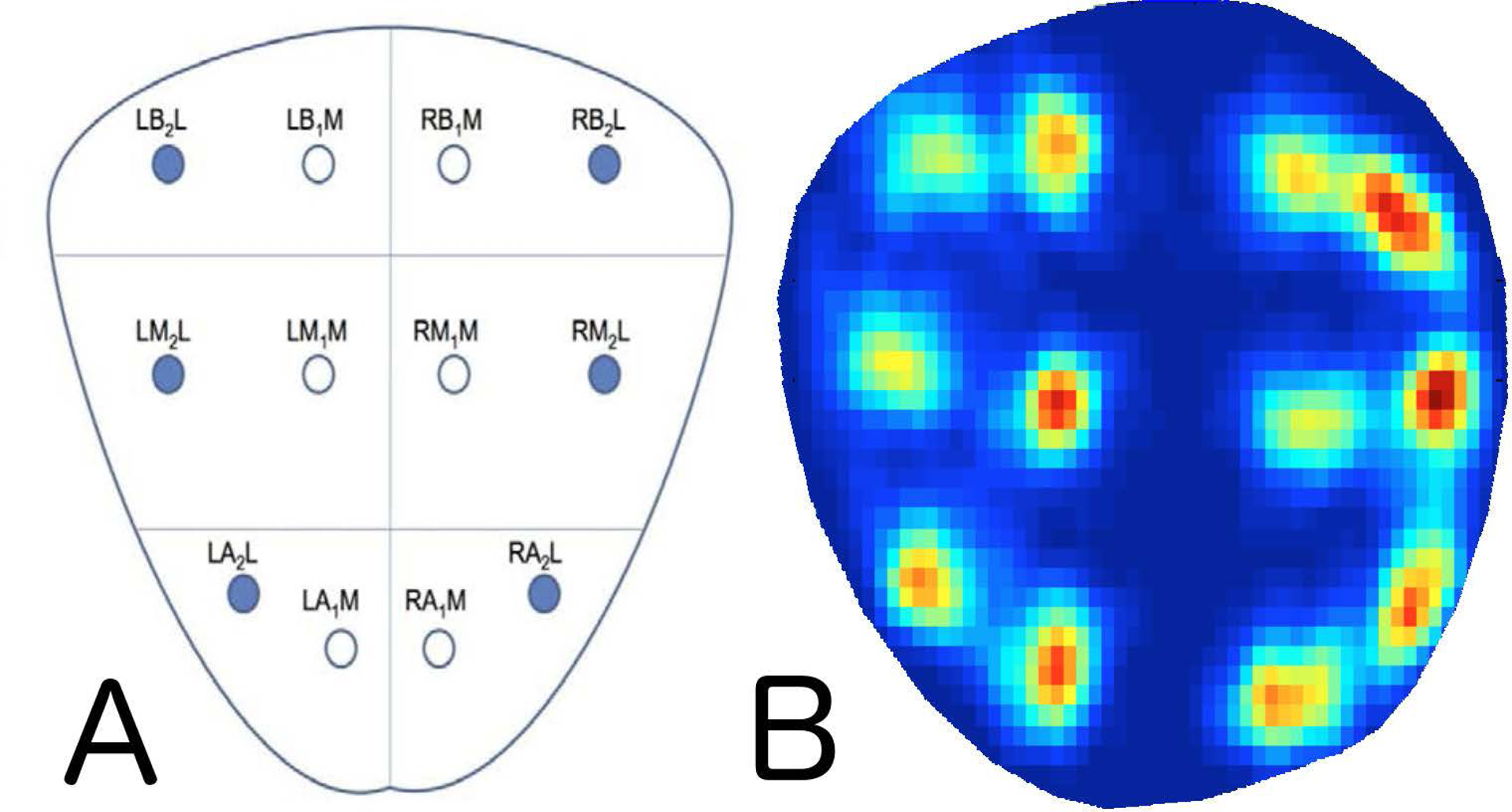
Diagram of standard systematic biopsy locations (A) and the corresponding visualization/distribution of template mapping biopsy cores from this cohort (B), scaled to a standard 25cc prostate. Dark blue represents locations that are never sampled throughout the prostate, whereas a standard heat scale from blue to red demonstrate increased frequency of sampling around template locations.
Clinical Data Extraction
Clinical information on each patient who underwent FHB was extracted from an electronic health record manually. Variables included age, race, number of systematic biopsy cores, total PSA, prostate volume (PV), previous biopsy history, and physician performing biopsy. On a per-core basis, Gleason grade (primary and secondary pattern), core positivity, and maximum cancer core length (MCCL) were extracted. For patients with multiple PSA values, the reading immediately prior to initial biopsy was used.
Information on patients who received TMB, including 3D location of biopsy cores and segmented prostate contour, was derived from a pre-existing research database. For patients who received TMB, spatial coordinates for biopsy cores stored in the fusion device were extracted, scaled, and graphed onto a prostate template heat map to visualize core distribution for the entire cohort (Figure 2).
Validation and Statistical Analyses
Data analysis was performed using MATLAB 2013a (Natick, MA) and R 3.1 (R Foundation for Statistical Computing).
CDR and csCaP detection rate were calculated for both FHB and TMB cohorts. We defined csCaP as Gleason Grade Group (GG) ≥ 2, GG1 with MCCL ≥ 4mm, GG1 with > 2 positive cores or GG1 with > 50% cancer involvement.13,14 Due to evidence suggesting that CaP can be harder to detect in larger prostates, we performed a sub-analysis stratified by prostate volume in both cohorts.15,16 Due to the recent adoption of the fusion devices, we also accounted for potential learning curve confounders by stratifying the TMB cohort sub-analysis by date of biopsy.
Descriptive statistics were used for population characteristics. Non-parametric tests were used for un-paired comparisons of binary and ordinal data (Chi-squared test, Mann-Whitney U test). Univariate and multivariate analysis was performed with the following clinical factors: age, pre-biopsy serum PSA, prostate volume, year of biopsy, number of cores taken, biopsy method (FHB versus TMB), race, and doctor performing the biopsy. Statistical significance was defined as alpha = 0.05, followed by adjustment for multiple testing with the Holm/Hochberg criterion.
RESULTS
Baseline Characteristics
In the FHB cohort, there were 12 performing physicians, who took a median (IQR) of 12 (12–13) cores per biopsy. Mean patient age was 63.8±9.0 years. Median PSA (IQR) was 5.1ng/mL (3.7–7.3). All biopsies in the TMB cohort were performed by a single physician, with a median (IQR) of 12 (12–12) cores taken per biopsy. Biopsies took between 5–10 minutes for FHB and 10 minutes for TMB due to time required for prostate scanning and segmentation by fusion software.17 Mean patient age was 63.5±8 years. Median PSA (IQR) was 5.6ng/mL (4.1–7.9). No differences were found between cohorts in baseline age and PV (Table 1). Median PSA and PSAD were 0.5 ng/mL higher (p<0.01) and 0.01 ng/mL/cc higher (p<0.01) in the TMB group, respectively. Average number of cores taken was 0.8 higher in the FHB group (p<0.01). The TMB group had a higher proportion of all recorded race categories, but this was because half of the FHB group did not have an identified race category in the patient chart, as opposed to under 10% in the TMB group (p<0.01).
Table 1.
Patient baseline characteristics.
| Characteristic | Total N = 1582 |
Free-hand N = 1052 |
Template Mapping N = 530 |
P value |
|---|---|---|---|---|
| Mean Age, years (SD) | 63.7 (8.6) | 63.8 (9.0) | 63.5 (8.0) | 0.6 |
| PSA, ng/mL, median (IQR) | 5.3 (3.8–7.5) | 5.1 (3.7–7.3) | 5.6 (4.1–7.9) | 0.002 |
| Prostate Volume, cc, median (IQR)* | 42 (32–60) | 41 (30–62) | 42 (32–58) | 0.94 |
| PSAD, ng/mL/cc, median (IQR)* | 0.12 (0.07–0.19) | 0.11 (0.07–0.17) | 0.12 (0.08–0.19) | 0.005 |
| Median Number of cores taken (IQR) | 12 (12–12) | 12 (12–13) | 12 (12–12) | <0.001 |
| Ethnicity n (%) | 1582 (100): | 1052 (100): | 530 (100): | |
| Caucasian | 804 (51) | 407 (39) | 397 (75) | <0.001 |
| Asian | 85 (5) | 51 (5) | 34 (6) | 0.19 |
| African American | 80 (5) | 44 (4) | 36 (6) | 0.03 |
| Hispanic | 26 (2) | 11 (1) | 15 (3) | 0.008 |
| Other or Unknown | 587 (37) | 539 (51) | 48 (9) | <0.001 |
Data is unavailable for 750/1052 (71%) and 12/530 (2%) of patients in the FHB and TMB groups. Baseline characteristics are reported from the sub-group with available data.
Tracked locations from the fusion device were used to generate a heat map, demonstrating a consistent sampling distribution that reflects a standard 12-core template in the TMB group (Figure 2).
Cancer Detection Rate
Overall CDR was higher with TMB (p=0.005), which detected cancer in 257 of 530 men (48.5%; 95% CI, 45.5–51.5), whereas FHB detected cancer in 432 of 1052 men (41.1%; 95% CI, 38.1–44.0). TMB also detected more csCaP (p=0.002): 196 of 530 men (37.0%; 95% CI, 34.1–39.9) with TMB versus 308 of 1052 men (29.2%; 95% CI, 26.5–32.0) with FHB. TMB did not detect more GG≥2 cancers (22.6% vs 25.7%, p=0.18). An increased detection rate of high volume or multifocal GG1 accounted for the increase in csCaP detection rate when the two groups were compared (Figure 3). CDR remained higher with TMB when only employing the number-of-cores criteria for clinically significant GG1 (31.6% vs 25.0%, p=0.005). CDR was also higher with TMB when defining clinically significant GG1 by MCCL or percent-core-involvement criteria (32.2% vs 26.0%, p=0.006).
Figure 3.
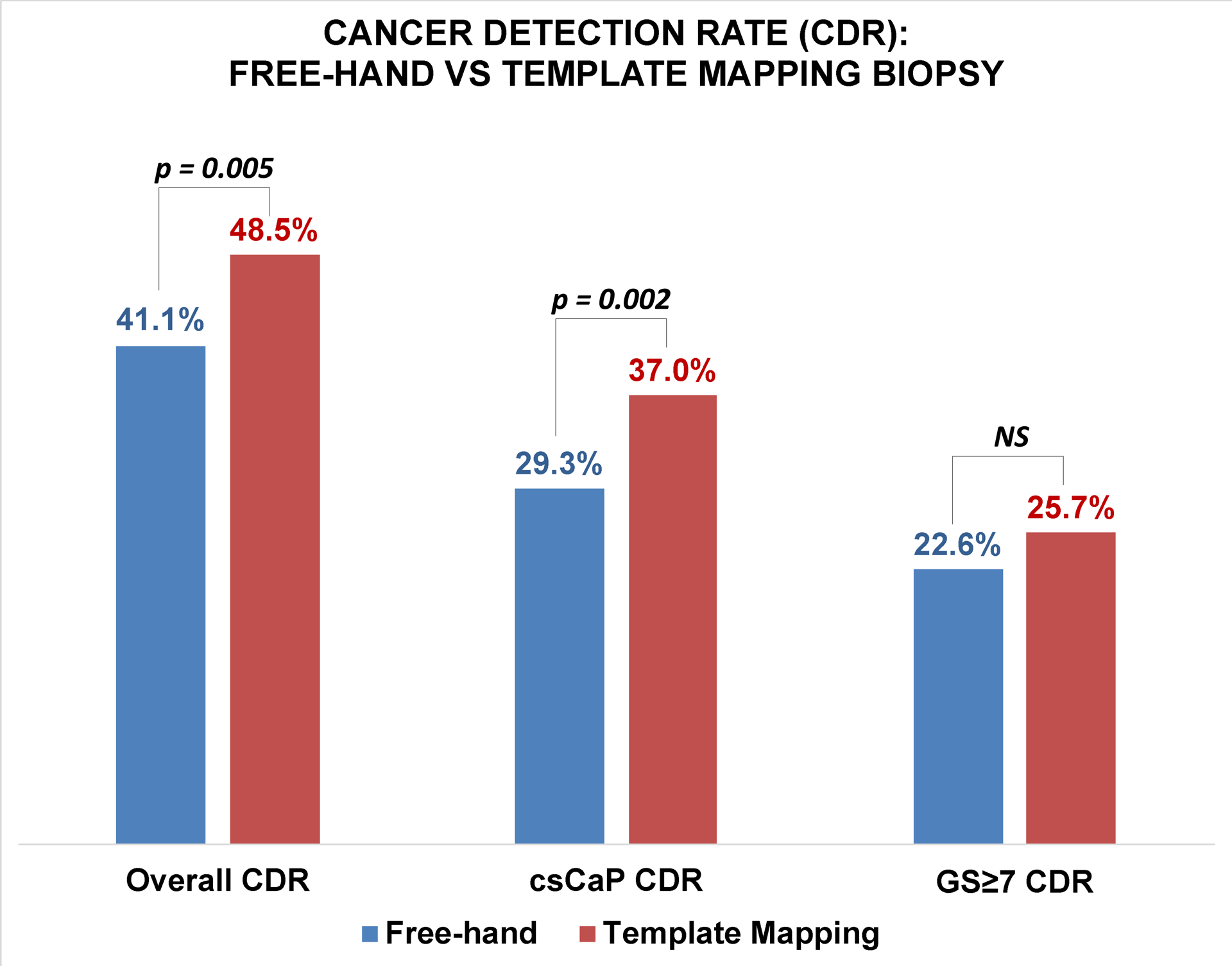
Cancer detection rate for any cancer, clinically-significant prostate cancer, and GS≥7 between free-hand versus 3D template mapping biopsy cohorts. Template mapping biopsy detected more cancers overall and more clinically-significant cancers, but did not detect more GG≥2 cancers.
Sub-analyses
Both cohorts were divided into subgroups based on median PV reported in the literature for men aged 40–75: PV≤30 cc vs PV>30 cc18. PV data were unavailable for 750/1052 (71%) and 10/530 (2%) of patients in the FHB and TMB groups, respectively. Subanalysis was performed with the 302 and 520 patients with available PV data in the FHB and TMB cohorts respectively. For this sub-population, there was no difference in baseline median PV (41cc vs 42cc, p = 0.94). PV was greater than 30 cc in 226/302 (75%) of patients in the FHB cohort, and in 410/520 (77%) of patients in the TMB cohort. TMB detected more cancer overall (45.4% vs 34.5%, p = 0.008) and more csCaP (33.4% vs 23.5%, p = 0.009) in prostates greater than 30 cc. In prostates ≤ 30 cc, there was no statistically-significant difference in CDR (TMB 62.7% vs FHB 53.9%, p=0.23) or csCaP detection rate (TMB 51.8% vs FHB 42.1%, p=0.19) (Figure 4).
Figure 4.
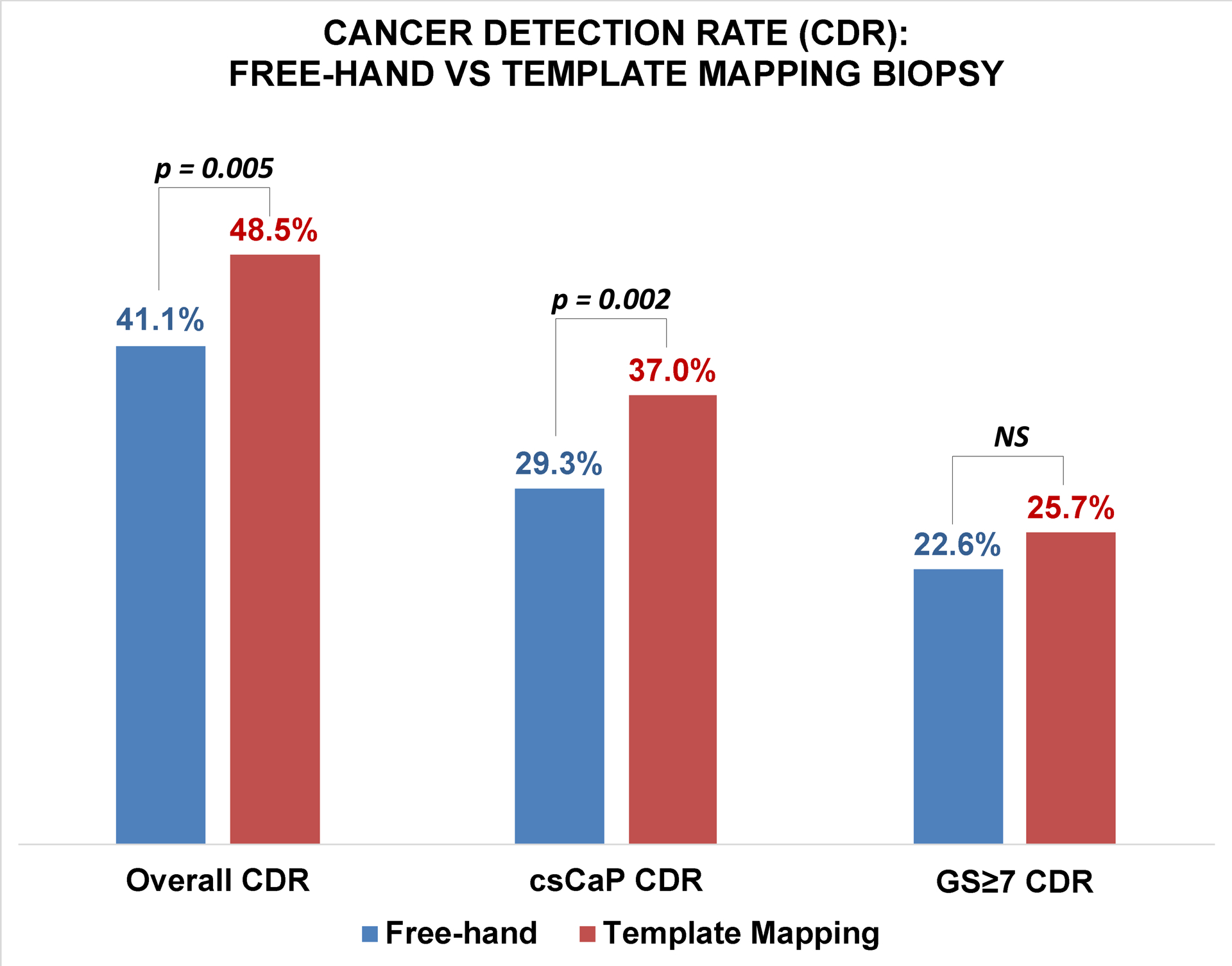
Cancer detection rate (LEFT), clinically-significant prostate cancer detection rate (MIDDLE), and GG≥2 detection rate (RIGHT) stratified by prostate volume. Template mapping biopsy detected more cancers in prostate larger than 30cc.
Because baseline PSA and PSAD was higher in the TMB cohort, we performed an additional subanalysis stratified by PSAD < 0.15 ng/mL/cc vs PSAD ≥0.15 ng/mL/cc. PSAD was 0.15 ng/mL/cc or higher in 94/302 (31%) of patients in the FHB cohort, and in 199/518 (38%) of patients in the TMB cohort (p=0.04). TMB overall detected more cancers (39.2% vs 30.3%, p=0.04) in patients with PSAD < 0.15 ng/mL/cc. TMB also detected more csCaP (26.3% vs 17.3%, p=0.02) in this group. No statistically-significant difference was found for CDR or csCaP detection in patients with PSAD ≥ 0.15 ng/mL/cc (Figure 5).
Figure 5.
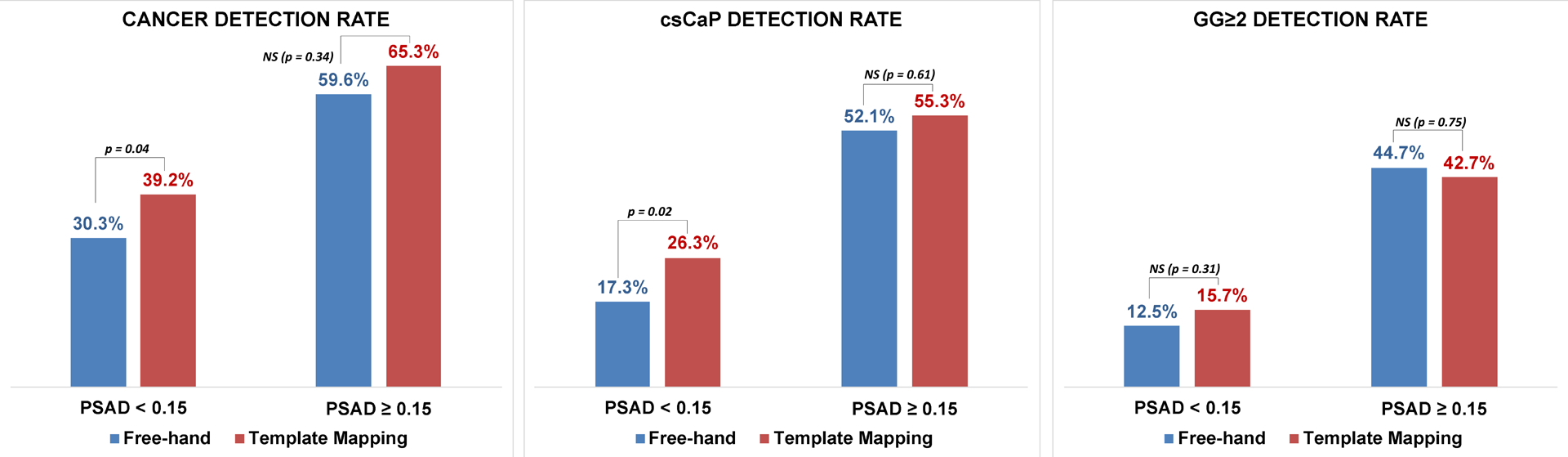
Cancer detection rate (LEFT), clinically-significant prostate cancer detection rate (MIDDLE), and GG≥2 detection rate (RIGHT) stratified by PSAD. Template mapping biopsy detected more cancers overall and more clinically-significant cancers in patients with PSAD<0.15.
The TMB cohort was also divided in half based on the chronologic order of the biopsy: either before or after May 2013. There was also no difference in CDR between the first half of the TMB cohort vs the second half (45.3% vs 51.7%, p=0.14).
Univariate and multivariate analysis was performed for all patients who had data available for the considered covariates (N=1031). The most frequent missing data was race. All covariates were scaled to values between 0 and 1. Logarithmic scale was used for PSA and number of cores, in order to obtain normal distribution. We found no significant correlation between CDR and ethnicity or diagnosing physician (Table 2). As expected, age, PSA, and biopsy cohort (TMB vs FHB) were predictive of positive biopsy result. A higher number of cores did not correlate with a higher chance for positive biopsy result.
Table 2.
Univariate and multivariate analysis for predictors of positive biopsy result. Age, PSA, and Biopsy Cohort were predictive of a positive biopsy result.
| Covariate | Univariate | P value | Holm* | Multivariate** | P value | Holm* |
|---|---|---|---|---|---|---|
| Patient age (years) | 0.98 | <0.001 | 0.002 | 0.68 | 0.001 | 0.002 |
| PSA (ng/mL) | 0.59 | <0.001 | 0.002 | 0.77 | <0.001 | 0.002 |
| Prostate Volume (cc)** | −1.22 | <0.001 | 0.002 | −2.04 | <0.001 | 0.002 |
| PSA Density (ng/mL/cc)** | 0.004 | <0.001 | 0.002 | −0.0008 | 0.67 | 0.008 |
| Year of biopsy | 0.11 | 0.014 | 0.003 | 0.02 | 0.85 | 0.013 |
| Number of cores taken | −0.33 | 0.27 | 0.007 | 0.58 | 0.26 | 0.005 |
| Biopsy cohort*** | −0.08 | 0.004 | 0.002 | −0.14 | 0.03 | 0.003 |
| Race | ||||||
| Caucasian | 0.05 | 0.04 | 0.003 | 0.19 | 0.04 | 0.003 |
| Asian | −0.24 | 0.05 | 0.004 | 0.007 | 0.94 | 0.025 |
| African American | 0.15 | 0.01 | 0.003 | 0.24 | 0.03 | 0.003 |
| Hispanic | −0.09 | 0.35 | 0.008 | 0.08 | 0.51 | 0.006 |
| Other or Unknown | −0.06 | 0.03 | 0.003 | 0.08 | 0.44 | 0.006 |
| Diagnosing physician**** | ||||||
| 1 (N=115) | 0.05 | 0.13 | 0.005 | 0.18 | 0.004 | 0.003 |
| 2 (N=18) | −0.18 | 0.01 | 0.003 | −0.12 | 0.65 | 0.007 |
| 3 (N=7) | 0.06 | .65 | 0.050 | −0.01 | 0.94 | 0.050 |
| 4 (N=69) | −0.06 | 0.18 | 0.006 | 0 | 0.14 | 0.004 |
| 5 (N=1) | −0.44 | 0.38 | 0.010 | 0 | 0.002 | 0.002 |
| 6 (N=88) | 0.06 | 0.16 | 0.005 | 0.61 | 0.07 | 0.004 |
| 7 (N=2) | 0.56 | 0.26 | 0.006 | 0 | 0.04 | 0.003 |
| 8 (N=102) | −0.15 | 0.42 | 0.013 | 0.04 | 0.84 | 0.010 |
| 9 (N=3) | −0.07 | 0.09 | 0.004 | 0 | 0.04 | 0.004 |
| 10 (N=7) | −0.22 | 0.09 | 0.004 | 0.04 | 0.91 | 0.017 |
| 11 (N=3) | −0.08 | 0.55 | 0.025 | 0 | <0.001 | 0.002 |
| 12 (N=10) | 0.23 | 0.42 | 0.017 | 0.55 | 0.23 | 0.005 |
| 13 (N=533) | −0.28 | 0.04 | 0.003 | −0.35 | 0.03 | 0.003 |
Represents statistical significance threshold of alpha = 0.05 with Holm stepdown correction.
Analysis performed with the 820 patients with prostate volume data available.
Categorical variable represented as FHB – 1, TMB – 0.
Physician 8 performed all TMB and no FHB.
DISCUSSION
In 2012, Han et al performed a study in which 5 different operators performed biopsies using a simulator either free-hand or with robotic assistance.1 There was significant variability in CDR between the different urologists in the study, with a mean targeting error of up to 9mm. Urologists were found to bunch cores together when taking a free-hand biopsy, and found that robotic assistance significantly decreased biopsy distribution errors, and increased the cancer detection rate. Chang et al performed a simulation to evaluate the effectiveness of ideal biopsy plans: a 3D model was constructed for patient prostates to optimize core distribution throughout the gland, generating a custom biopsy plan based on prostate volume.2 The study found that geometrically-optimized biopsy plans would have detected up to 20% more cancer.
The present work adds to these findings; when compared to FHB, use of a 3D template and fusion device to guide systematic biopsy detected more cancers overall and more csCaP. This sensitivity improvement was even higher in prostates greater than 30cc, suggesting that the benefit of more evenly-distributed biopsy cores is magnified in larger prostates. Notably, most patients in this cohort had prostates larger than 30cc, and prior work has demonstrated that cancers are harder to detect in larger prostates.15,16 This finding is corroborated in the present study by the higher CDR in patients with prostates smaller than 30cc. Combined with prior work, these results suggest that not all 12-core biopsies are created equal; assistance via an optimized virtual template or robotic guidance may ensure an even distribution of prostate sampling.
The benefit of template biopsies is clinically important; for every 14 additional patients screened with FHB, one patient with CaP would go undetected that would be caught by TMB. For every 13 additional patients screened with FHB, one additional patient with csCaP would go undetected that would be caught by TMB. These patients could be subject to additional biopsies, or experience delay of appropriate management. TMB specifically diagnosed more high-volume GG1 cancers. Outcomes from exemplary active surveillance programs show that up to 20% of patients with very-low-risk CaP will have biopsy reclassification by 2 years, with up to 10% reclassified to GG2 or higher.19 It is therefore important to ensure that these patients are monitored appropriately for progression.
Previous work has established the benefit of biopsy with fusion devices, both with targeted biopsies alone and combined with systematic biopsies.5–10 However, while the purported advantage is frequently attributed to use of the MRI, no studies to date have examined the contributing benefit of the 3D template and mechanical assistance additionally provided by the fusion device. Given the notable cost of MRI, widespread use of targeted biopsy as a first-line diagnostic tool remains limited.11,12 There is therefore strong incentive for continued improvement of biopsy without imaging, the subject of the present study. TMB can be done without an MRI, and the same device that is traditionally used for fusion biopsies can also be used for TMB.
There are several limitations with this study. First, as a retrospective study, these findings must be interpreted with all associated shortcomings in mind. Second, because many subjects from the FHB cohort predated the implementation of the electronic health record at our institution, certain data—especially ethnicity and PV—were much more frequently missing in the FHB cohort. However, analysis of available baseline characteristics showed very little differences compared to the TMB cohort. A key assumption is that these characteristics were largely representative of the entire FHB cohort at large. Third, the TMB cohort does have a higher PSAD. However, the benefit of TMB remained significant when analysis was stratified by PSAD. Fourth, for the TMB, only one physician performed biopsy. While using template mapping significantly improved the CDR, this trend might be limited or not hold true when compared to a number of physicians with varying skill. Nevertheless, when biopsies that were done earlier were compared to biopsies that were done later—when the physician was more experienced with the device—there was no difference in CDR or csCaP detection rate. Furthermore, no significant correlation was found between the 23 TRUS biopsy operators and CDR. Fifth, this study reports the benefit of the template software supplied by only one of many fusion devices currently on the market; unaddressed in this study is whether different fusion devices still confer a CDR advantage. The nuances between template software algorithms remains unexplored. Finally, we acknowledge that the definition for csCaP varies in practice. It is notable that the CDR difference did not reach statistical significance for GG≥2 cancers, another widely used definition of csCaP. However, we assert that the additional detection of high-volume GG1 still confers clinical benefit. Furthermore, a negative TMB provides higher confidence that cancer was not missed.
CONCLUSIONS
Systematic sampling with the guidance of virtual template mapping was found to have a higher CDR than free-hand TRUS biopsy. For every additional 13 patients screened, template mapping biopsy would detect an additional case of clinically-significant prostate cancer that would be missed by free-hand biopsy. Template mapping biopsy is a promising cost-effective alternative to MRI-guided biopsy, and provides a better characterization of tumor burden than standard free-hand biopsy.
Figure 1.
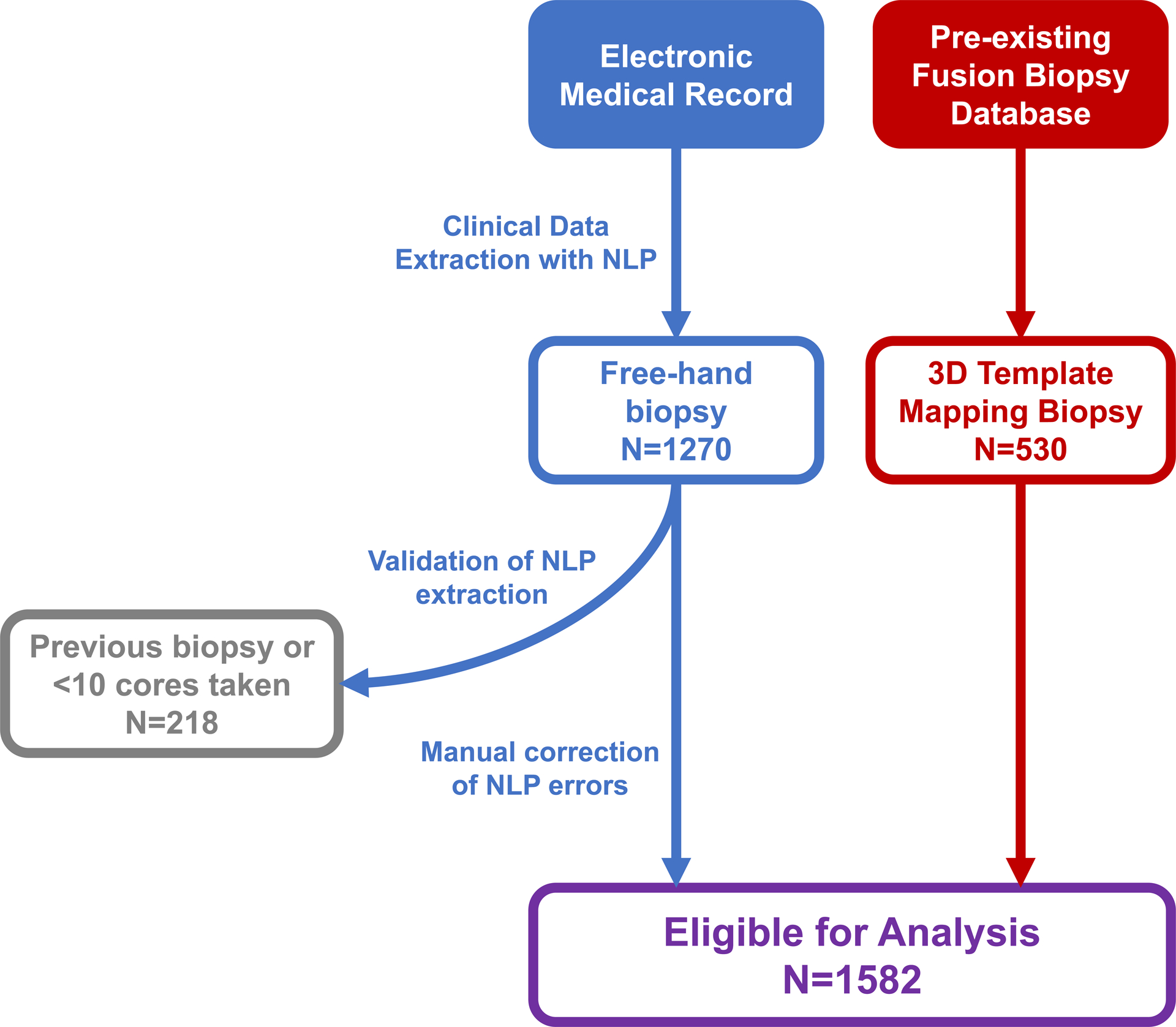
Workflow for clinical data extraction with natural language processing.
Acknowledgments
This research was supported by Award Number R01CA158627 from the National Cancer Institute. The content is solely the responsibility of the authors, and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Additional support was provided by the Beckman Coulter Foundation, the Jean Perkins Foundation, the Steven C. Gordon Family Foundation, and the UCLA Dean’s Leadership in Health and Sciences Scholarship.
Footnotes
Disclosures: Dr. Marks and Dr. Natarajan are co-founders of Avenda Health, Inc.
REFERENCES
- 1.Han M, Chang D, Kim C, et al. Geometric evaluation of systematic transrectal ultrasound guided prostate biopsy. J Urol. 2012;188(6):2404–2409. doi: 10.1016/j.juro.2012.07.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang D, Chong X, Kim C, et al. Geometric systematic prostate biopsy. Minim Invasive Ther Allied Technol. 2017;26(2):78–85. doi: 10.1080/13645706.2016.1249890 [DOI] [PubMed] [Google Scholar]
- 3.Fleshner NE, O’Sullivan M, Fair WR. Prevalence and predictors of a positive repeat transrectal ultrasound guided needle biopsy of the prostate. J Urol. 1997;158(2):505–509. doi: 10.1016/S0022-5347(01)64518-X [DOI] [PubMed] [Google Scholar]
- 4.Roehrborn CG, John Pickens G, Sanders JS. Diagnostic yield of repeated transrectal ultrasound-guided biopsies stratified by specific histopathologic diagnoses and prostate-specific antigen levels. Urology. 1996;47(3):347–352. doi: 10.1016/S0090-4295(99)80451-8 [DOI] [PubMed] [Google Scholar]
- 5.Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/Ultrasound Fusion–Guided Biopsy With Ultrasound-Guided Biopsy for the Diagnosis of Prostate Cancer. JAMA. 2015;313(4):390. doi: 10.1001/jama.2014.17942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filson CP, Natarajan S, Margolis DJA, et al. Prostate cancer detection with magnetic resonance-ultrasound fusion biopsy: The role of systematic and targeted biopsies. Cancer. 2016;122(6):884–892. doi: 10.1002/cncr.29874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonn GA, Chang E, Natarajan S, et al. Value of Targeted Prostate Biopsy Using Magnetic Resonance – Ultrasound Fusion in Men with Prior Negative Biopsy and Elevated Prostate-specific Antigen. Eur Urol. 2014;65(4):809–815. doi: 10.1016/j.eururo.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran GN, Leapman MS, Nguyen HG, et al. Magnetic Resonance Imaging–Ultrasound Fusion Biopsy During Prostate Cancer Active Surveillance. Eur Urol. 2017;72(2):275–281. doi: 10.1016/j.eururo.2016.08.023 [DOI] [PubMed] [Google Scholar]
- 9.Frye TPTP George AKAK, Kilchevsky A, et al. Magnetic Resonance Imaging-Transrectal Ultrasound Guided Fusion Biopsy to Detect Progression in Patients with Existing Lesions on Active Surveillance for Low and Intermediate Risk Prostate Cancer. J Urol. 2017;197(3 Part 1):640–646. doi: 10.1016/j.juro.2016.08.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasivisvanathan V, Rannikko AS, Borghi M, et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med. 2018;378(19):1767–1777. doi: 10.1056/NEJMoa1801993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchinson RC, Costa DN, Lotan Y. The economic effect of using magnetic resonance imaging and magnetic resonance ultrasound fusion biopsy for prostate cancer diagnosis. Urol Oncol Semin Orig Investig. 2016;34(7):296–302. doi: 10.1016/j.urolonc.2015.10.014 [DOI] [PubMed] [Google Scholar]
- 12.Rosenkrantz AB, Verma S, Choyke P, et al. Prostate Magnetic Resonance Imaging and Magnetic Resonance Imaging Targeted Biopsy in Patients with a Prior Negative Biopsy: A Consensus Statement by AUA and SAR. J Urol. 2016;196(6):1613–1618. doi: 10.1016/j.juro.2016.06.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanda Martin G. MD; Chen Ronald C. MTCSF, MD; Greene Kirsten MD; Klotz Laurence H. MD; Makarov Danil V. MJB, Nelson, MD; Reston James PhD; Rodrigues George MD; Howard M Sandler M, Mary Ellen Taplin MD; Jeffrey A Cadeddu M CLINICALLY LOCALIZED PROSTATE CANCER: AUA/ASTRO/SUO GUIDELINE; 2017. [Google Scholar]
- 14.Ahmed HU, Hu Y, Carter T, et al. Characterizing clinically significant prostate cancer using template prostate mapping biopsy. J Urol. 2011;186(2):458–464. doi: 10.1016/j.juro.2011.03.147 [DOI] [PubMed] [Google Scholar]
- 15.Ung JO, San Francisco IF, Regan MM, DeWolf WC, Olumi AF. The relationship of prostate gland volume to extended needle biopsy on prostate cancer detection. J Urol. 2003;169(1):130–135. doi: 10.1097/01.ju.0000034153.49106.b7 [DOI] [PubMed] [Google Scholar]
- 16.Uzzo RG, Wei JT, Waldbaum RS, Perlmutter AP, Byrne JC, Vaughan D. The influence of prostate size on cancer detection. Urology. 1995;46(6):831–836. doi: 10.1016/S0090-4295(99)80353-7 [DOI] [PubMed] [Google Scholar]
- 17.Jayadevan R, Zhou S, Priester AM, Delfin M, Marks LS. Use of MRI-ultrasound Fusion to Achieve Targeted Prostate Biopsy. J Vis Exp. 2019;(146):e59231. doi: 10.3791/59231 [DOI] [PubMed] [Google Scholar]
- 18.Girman CJ, Jacobsen SJ, Guess HA, et al. Natural History of Prostatism: Relationship Among Symptoms, Prostate Volume and Peak Urinary Flow Rate. J Urol. 1995;153(5):1510–1515. doi: 10.1016/S0022-5347(01)67448-2 [DOI] [PubMed] [Google Scholar]
- 19.Tosoian JJ, Trock BJ, Landis P, et al. Active surveillance program for prostate cancer: An update of the Johns Hopkins experience. Int Braz J Urol. 2011;37(2):278. doi: 10.1590/S1677-55382011000200021 [DOI] [PubMed] [Google Scholar]


