Abstract
SET and MYND domain-containing protein 3 (SMYD3) is a lysine methyltransferase, and its aberrant expression has been implicated in several malignancies. However, its clinical and biological roles in non-small cell lung cancer (NSCLC) remain unclear. In the present study, it was revealed that SMYD3 was significantly upregulated in NSCLC tissues, as compared with paired adjacent normal tissues. A high SMYD3 expression was associated with aggressive clinicopathological characteristics, as well as poor disease-free survival and overall survival (OS) in NSCLC patients. Multivariate analysis revealed that SMYD3 overexpression was an independent predictor of poor OS in NSCLC patients. In addition, SMYD3 knockdown inhibited cell proliferation, triggered apoptosis, and blocked migration and invasion in NSCLC cells in vitro, whereas stable SMYD3 overexpression promoted NSCLC cell proliferation. Furthermore, the SMYD3-silenced NSCLC cells became more sensitive, whereas the SMYD3-overexpressed NSCLC cells became more resistant to the apoptosis induced by cisplatin. Mechanistic analysis revealed that SMYD3 knockdown led to the upregulation of Bim, Bak and Bax, and the downregulation of Bcl-2, Bcl-xl, MMP-2 and MMP-9 in NSCLC cells. In combination, the present findings indicated that SMYD3 has oncogenic potential in the context of NSCLC, providing evidence that may be exploited for both prognostic and therapeutic purposes in the future.
Keywords: SET and MYND domain-containing protein 3, non-small cell lung cancer, prognostic marker, proliferation, cisplatin resistance, migration, invasion
Introduction
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer-related mortality worldwide, with approximately 2.1 million new cases and 1.8 million deaths in 2018 (1). Non-small cell lung cancer (NSCLC) is the major histological subtype of lung cancer, which accounts for 80-85% of all cases (2). Despite recent advances in the treatment of NSCLC, such as targeted therapy and immunotherapy, the long-term prognosis for NSCLC patients remains poor, and the 5-year survival rate is less than 15% (3). Therefore, it is urgently required to elucidate the detailed molecular pathogenesis of NSCLC, so as to identify more accurate prognostic markers and therapeutic targets.
SET and MYND domain-containing protein 3 (SMYD3) is a member of the SET and MYND domain family, a special class of histone and non-histone lysine methyltransferase (4). SMYD3 functions by catalyzing lysine methylation, mainly through the bi/trimethylation of histone H3 at lysine 4 (H3K4), to render chromatin more accessible and activate downstream gene transcription (5,6). In addition, SMYD3 can transactivate downstream targets by binding to the 5′-CCCTCC-3′ or 5′-GCAGGG-3′ motif in the promoter region of target genes, and interacting with RNA polymerase II and RNA helicase (5). In the past decade, emerging evidence has suggested an oncogenic role of SMYD3 in human cancer. The overexpression of SMYD3 has been observed in breast, hepatocellular, colorectal, gastric, bladder, prostate, cervical, glioma and esophageal squamous cell carcinomas, where it was revealed to be involved in cell proliferation, migration, invasion, epithelial-mesenchymal transition and autophagic activation (7-14). Furthermore, a variable number of tandem repeat (VNTR) polymorphism in the SMYD3 promoter region, namely homozygosity for an allele with three tandem repeats of a CCGCC (3/3), was found to be associated with an increased risk of breast, hepatocellular and colorectal carcinomas (15). As for NSCLC, Barlesi et al found that the SMYD3 VNTR genotype was not correlated with either the tested clinicopathological characteristics or the risk of NSCLC (16). However, the expression of SMYD3 on protein levels in clinical NSCLC samples, as well as the functional role of SMYD3 in NSCLC progression, remain unknown.
In the present study, the expression of SMYD3 in NSCLC and matched adjacent normal tissues was evaluated by immunohistochemistry (IHC), and its clinical and prognostic significance was analyzed. Moreover, the biological roles of SMYD3 in NSCLC cell proliferation, apoptosis, migration, invasion and drug resistance, as well as the molecular basis for such effects, were also characterized. The present findings demonstrated that SMYD3 may serve as an oncogene in NSCLC development and progression, recommending SMYD3 as a promising prognostic marker and therapeutic target for NSCLC.
Materials and methods
Patients and tissue microarray (TMA)
Primary NSCLC and corresponding pericarcinous lung tissues were consecutively collected from patients (89 men, 66 women; mean ± SD age 62.35 ± 10 years, range 38 to 81 years) who underwent surgical resection at Fudan University Shanghai Cancer Center (Shanghai, China) between 2010 and 2012. TMAs were constructed as previously described (17). Two cores with a 1.6-mm diameter were obtained from the original paraffin block of each sample. Clinicopathological information was retrospectively reviewed, and survival data was recorded on the basis of follow-up clinic visits or telephone calls. The pathological stages were determined based on the 7th edition of the lung cancer staging system proposed by the International Association for the Study of Lung Cancer (IASLC) (18). Patients who had received neoadjuvant chemotherapy or radiotherapy, or had a history of other malignancies were excluded. In total, 155 NSCLC cases were enrolled. The present study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center. All experiments were in accordance with the declaration of Helsinki and the approved guidelines of our institution. Written informed consent was obtained from all participants.
IHC
The TMAs were subjected to IHC using anti-SMYD3 antibody (1:800; cat. no. ab187149, Abcam) as previously described (9). Concentration-matched nonspecific rabbit IgG was used as an isotype control. The IHC results were reviewed again by two trained pathologists. Staining intensity was scored as follows: Negative (0), weak (1), moderate (2) and intense (3). The percentage of positive cells was scored as follows: 0% (0), 1-25% (1), 26-50% (2), 51-75% (3) and 76-100% (4). The final SMYD3 IHC score was the multiplication of these two scores, which were defined as low (scores of 0-3) or high (scores of 4-12).
Cell culture and cell proliferation assay
Human NSCLC H727, A549, NCI-H1299, H1650, NCI-H1975, HCC4006, PC-9, HCC827, H2170 and H226 cell lines, and human bronchial epithelial BEAS-2B cell line were obtained from the American Type Culture Collection (ATCC), and cultured in RPMI-1640 or DMEM (Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS). All cells were authenticated by short tandem repeat profiling (Thermo Fisher Scientific, Inc.) and confirmed to be mycoplasma-free. The in vitro cell proliferation was examined by sulforhodamine B (SRB) assay. Briefly, the cells in 96-well plates were fixed by gently adding cold trichloroacetic acid (100 µl; 10%) and incubated for 30 min at 4°C. The supernatant was discarded, and the plates were washed three times with tap water and air dried. SRB solution (100 µl; 0.4% in 1% acetic acid) was added, and the plates were incubated for 20 min at room temperature. The unconjugated SRB was washed by 1% acetic acid, and the conjugated SRB was dissolved in Tris solution (100 µl; 10 mM). The absorbance was measured at 560 nm using a microplate reader.
siRNA Transfection
Cells (1×105 cells/well in 6-well plates) were transfected with siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Inc.), following the manufacturer's instructions. The target sequences of double-stranded nucleotides used in siRNA transfection were 5′-GCAGAGTTGTCTTCAAACT-3′(SMYD3siRNA-1) and 5′-GGATGCTGATATGCTAACT-3′(SMYD3siRNA-2) for SMYD3 knockdown, and 5′-TTCTCCGAACGTGTCACGT-3′ as a negative control (NC) (Shanghai GenePharma Co., Ltd.).
Lentiviral vectors and cell infection
Lentiviral vectors were constructed with OBiO Biotechnology, using pRLe nti-CMV-SMYD3-3FLAG-PGK-Puro plasmids, which carry the full-length human SMYD3 (GenBank accession NM_001167740.2) coding sequence. Cell infection was performed as in our previous study (19).
Western blotting
Western blot analysis was performed as previously described (9). The primary antibodies, including anti-SMYD3 (cat. no. ab187149, Abcam), anti-Bcl-2-like protein 11 (Bim; cat. no. 2933S), anti-Bcl-2 homologous antagonist/killer (Bak; cat. no. 6947S), anti-Bax (cat. no. 14796S), anti-B-cell lymphoma 2 (Bcl-2; cat. no. 15071S), anti-Bcl-xl (cat. no. 2764S), anti-matrix metalloproteinase-2 (MMP-2; cat. no. 13132S), anti-MMP-9 (cat. no. 13667S) and anti-β-actin (cat. no. 3700S; all 1:1,000; all from Cell Signaling Technology, Inc.) were used at the manufacturer's recommended dilution. The secondary antibodies were peroxidase AffiniPure goat anti-rabbit IgG (cat. no. 111-035-003) or anti-mouse IgG (cat. no. 115-035-003; both 1:3,000; both from Jackson ImmunoResearch, Inc.). Densitometric analysis was performed using ImageJ software (v1.47, Rawak Software, Inc.). The measured intensities for all samples were first normalized to their respective internal controls and then were compared with BEAS-2B cells, which was set to 100%.
Cell apoptosis analysis
NSCLC cells (1×105 cells per tube) were harvested with EDTA-free trypsin 72 h after transfection, stained using a FITC Annexin V Apoptosis Detection kit (BD Biosciences) following the manufacturer's protocol, and then examined using a flow cytometer (BD Biosciences). For the evaluation of cisplatin resistance, SMYD3-silenced or -over-expressed NSCLC cells were treated with cisplatin (Majorbio Bio-Pharm) at various doses (1, 2 and 4 µM for A549; 0.7, 1.5 and 3 µM for PC-9; 1, 2 and 5 µM for H1299) for 72 h, and then harvested for cell apoptosis analysis.
Cell migration and invasion assays
Cell migration and invasion assays were conducted using the Transwell method, as previously described (12). The number of migrated or invaded cells were counted using a bright-field imaging system on a microscope (Olympus BX51; Olympus Corporation) with five randomly selected fields (magnification, ×200).
Statistical analysis
Statistical analysis was conducted with SPSS v19.0 (IBM Corp.). Comparisons between two groups were performed using Student's t-test. Associations between two categorical variables were examined using Chi-square test or Fisher's exact test. The survival curves were plotted using the Kaplan Meier method, and compared using the log-rank test. Univariate and multivariate Cox proportional hazard models were employed to identify independent predictors. All tests were two sided and P<0.05 was considered to indicate a statistically significant difference.
Results
SMYD3 is upregulated in NSCLC and associated with aggressive clinicopathological characteristics
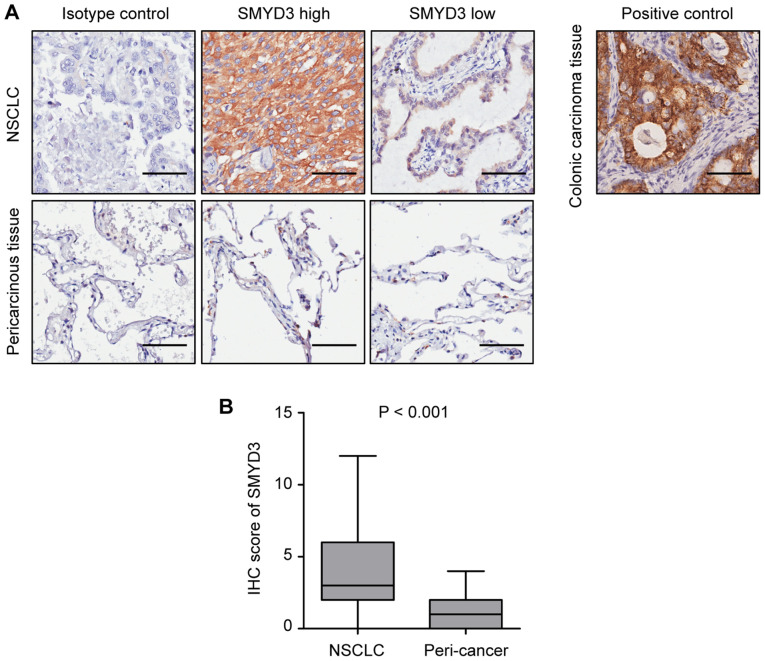
The expression of SMYD3 was assessed in 155 pairs of NSCLC and corresponding pericarcinous lung tissues by IHC. Representative images revealed that SMYD3 expression was mainly localized in the cytoplasm of NSCLC cells (Fig. 1A), which was consistent with previous studies (20,21). It was determined that SMYD3 was significantly upregulated in NSCLC tissues compared with that in pericarcinous lung tissues (Fig. 1B).
Figure 1.
SMYD3 is upregulated in NSCLC tissues. (A) Representative IHC images of high or low SMYD3 expression in 155 NSCLC tissues (upper panel; scale bar, 50 µm). Images of SMYD3 expression in corresponding pericarcinous lung tissues are also presented (lower panel). Isotype control: The primary antibody was replaced with a non-specific rabbit IgG antibody. Positive control: SMYD3 expression in human colonic carcinoma tissue. (B) The IHC score of SMYD3 was higher in NSCLC tissues than in pericarcinous lung tissues (P<0.001). SMYD3, SET and MYND domain-containing protein 3; NSCLC, non-small cell lung cancer; IHC, immunohistochemistry.
To analyze the association between SMYD3 and clinicopathological characteristics, 155 NSCLC specimens were divided into 'SMYD3 high' (scores of 4-12) and 'SMYD3 low' (scores of 0-3) groups, according to the median expression value. Consistent with the aforementioned findings, the proportion of 'SMYD3 high' samples was significantly higher in NSCLC than in paired normal lung tissues (48.4 vs. 3.2%, P<0.001) (Table I). The present results revealed that a high SMYD3 expression was significantly associated with never-smoked history, more advanced pathological stage, larger tumor size, and the presence of lymphovascular invasion (LVI), pleural invasion (PI) and distant metastasis (Table II).
Table I.
Protein expression levels of SMYD3 in NSCLC and corresponding pericarcinous lung tissues.
| Tissue sample | No. of patients | SMYD3
|
P-value | |
|---|---|---|---|---|
| Low (%) | High (%) | |||
| NSCLC | 155 | 80 (51.6) | 75 (48.4) | <0.001a |
| Peri-cancer | 155 | 150 (96.8) | 5 (3.2) | |
Data were analyzed using Chi-square test. SMYD3, SET and MYND domain-containing protein 3; NSCLC, non-small cell lung cancer.
Table II.
Clinicopathological characteristics according to SMYD3 protein expression in NSCLC.
| Variables | No. of patients | SMYD3
|
P-value | |
|---|---|---|---|---|
| High (%) | Low (%) | |||
| Sex | ||||
| Male | 89 | 39 (43.8) | 50 (56.2) | 0.186a |
| Female | 66 | 36 (54.5) | 30 (45.5) | |
| Age (years) | ||||
| >60 | 94 | 41 (43.6) | 53 (56.4) | 0.14a |
| ≤60 | 61 | 34 (55.7) | 27 (44.3) | |
| Smoking history | ||||
| Ever | 68 | 26 (38.2) | 42 (61.8) | 0.025a |
| Never | 87 | 49 (56.3) | 38 (43.7) | |
| Stage | ||||
| I | 66 | 25 (37.9) | 41 (62.1) | 0.024a |
| II-IV | 89 | 50 (56.2) | 39 (43.8) | |
| Tumor size (cm) | ||||
| > 3.0 | 76 | 43 (56.6) | 33 (43.4) | 0.045a |
| ≤ 3.0 | 79 | 32 (40.5) | 47 (59.5) | |
| LVI | ||||
| Present | 41 | 34 (82.9) | 7 (17.1) | <0.001a |
| Absent | 114 | 41 (36.0) | 73 (64.0) | |
| PI | ||||
| Present | 65 | 38 (58.5) | 27 (41.5) | 0.033a |
| Absent | 90 | 37 (41.1) | 53 (58.9) | |
| N status | ||||
| N0 | 79 | 33 (41.8) | 46 (58.2) | 0.093a |
| N1/N2N3 | 76 | 42 (55.3) | 34 (44.7) | |
| M status | ||||
| M0 | 150 | 70 (46.7) | 80 (53.3) | 0.025b |
| M1 | 5 | 5 (100) | 0 (0) | |
| Differentiation | ||||
| Well/moderate | 109 | 55 (50.5) | 54 (49.5) | 0.427a |
| Poor | 46 | 20 (43.5) | 26 (56.5) | |
Data were analyzed using Chi-square test.
Data were analyzed using Fisher's exact test. SMYD3, SET and MYND domain-containing protein 3; NSCLC, non-small cell lung cancer; LVI, lymphovascular invasion; PI, pleural invasion.
High SMYD3 expression predicts poor outcomes in NSCLC patients
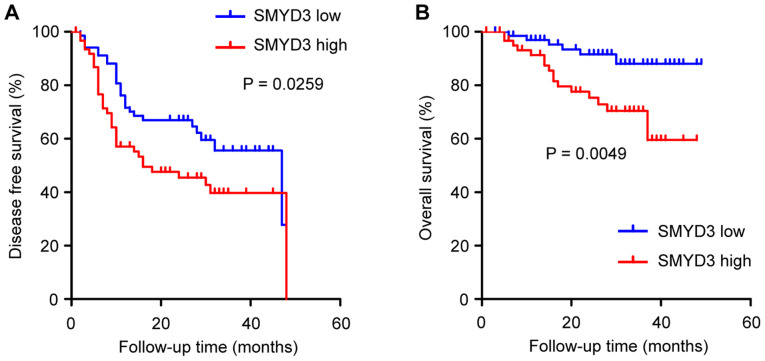
The prognostic significance of SMYD3 in NSCLC was analyzed. One hundred and thirty-one NSCLC patients qualified for survival analysis, due to sufficient follow-up. It was revealed that disease-free survival (DFS) and overall survival (OS) were significantly worse in patients with a high than in patients with a low SMYD3 expression (Fig. 2A and B).
Figure 2.
Kaplan-Meier analysis of DFS and OS in NSCLC patients according to SMYD3 expression. (A) DFS and (B) OS were significantly worse in high-SMYD3 expression patients than in low-SMYD3 expression patients. DFS, disease-free survival; OS, overall survival; NSCLC, non-small cell lung cancer.
It was also determined whether SMYD3 has predictive value for clinical outcomes in NSCLC patients. Univariate Cox regression analysis revealed that age at diagnosis (>60 years), advanced pathological stage and PI were correlated with poor DFS in NSCLC patients, and that age at diagnosis (>60 years), advanced pathological stage and high SMYD3 expression were correlated with poor OS (Tables III and IV). Furthermore, multivariate Cox regression analysis demonstrated that advanced pathological stage and PI were independent predictors for poor DFS, and that age at diagnosis (>60 year), advanced pathological stage and high SMYD3 expression were independent predictors for poor OS (Table III and IV).
Table III.
Univariate and multivariate Cox regression analysis for DFS in NSCLC.
| Variables | Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Sex | ||||||
| Male | 1 | |||||
| Female | 1.586 | 0.665-3.78 | 0.298 | |||
| Age (years) | ||||||
| ≤60 | 1 | 1 | ||||
| >60 | 1.819 | 1.022-3.24 | 0.042 | 1.712 | 0.998-2.938 | 0.051 |
| Smoking history | ||||||
| Never | ||||||
| Ever | 1.689 | 0.72-3.96 | 0.228 | |||
| Stage | ||||||
| I | 1 | 1 | ||||
| II-IV | 4.912 | 2.612-9.237 | <0.001 | 5.342 | 2.924-9.759 | <0.001 |
| Tumor size (cm) | ||||||
| ≤3.0 | 1 | |||||
| >3.0 | 1.202 | 0.695-2.076 | 0.511 | |||
| LVI | ||||||
| Absent | 1 | |||||
| Present | 1.221 | 0.639-2.331 | 0.545 | |||
| PI | ||||||
| Absent | 1 | |||||
| Present | 1.812 | 1.054-3.114 | 0.031 | 1.927 | 1.142-3.251 | 0.014 |
| Differentiation | ||||||
| Well/moderate | 1 | |||||
| Poor | 0.919 | 0.492-1.717 | 0.792 | |||
| SMYD3 | ||||||
| Low | 1 | |||||
| High | 1.462 | 0.788-2.714 | 0.228 | |||
DFS, disease-free survival; NSCLC, non-small cell lung cancer; HR, hazard ratio; CI, confidence interval; LVI, lymphovascular invasion; PI, pleural invasion; SMYD3, SET and MYND domain-containing protein 3.
Table IV.
Univariate and multivariate Cox regression analysis for OS in NSCLC.
| Variables | Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Sex | ||||||
| Male | 1 | |||||
| Female | 1.559 | 0.374-6.498 | 0.542 | |||
| Age (years) | ||||||
| ≤60 | 1 | 1 | ||||
| >60 | 5.321 | 1.651-17.142 | 0.005 | 4.474 | 1.485-13.479 | 0.008 |
| Smoking history | ||||||
| Never | 1 | |||||
| Ever | 1.719 | 0.4-7.391 | 0.467 | |||
| Stage | ||||||
| I | 1 | 1 | ||||
| II-IV | 3.246 | 1.19-8.852 | 0.021 | 3.129 | 1.244-7.871 | 0.015 |
| Tumor size (cm) | ||||||
| ≤3.0 | 1 | |||||
| >3.0 | 0.646 | 0.26-1.603 | 0.346 | |||
| LVI | ||||||
| Absent | 1 | |||||
| Present | 0.864 | 0.302-2.47 | 0.785 | |||
| PI | ||||||
| Absent | 1 | |||||
| Present | 1.369 | 0.548-3.421 | 0.501 | |||
| Differentiation | ||||||
| Well/moderate | 1 | |||||
| Poor | 1.581 | 0.561-4.457 | 0.387 | |||
| SMYD3 | ||||||
| Low | 1 | 1 | ||||
| High | 4.092 | 1.302-12.864 | 0.016 | 3.338 | 1.283-8.684 | 0.013 |
OS, overall survival; NSCLC, non-small cell lung cancer; HR, hazard ratio; CI, confidence interval; LVI, lymphovascular invasion; PI, pleural invasion; SMYD3, SET and MYND domain-containing protein 3.
In combination, the present findings indicated that SMYD3 was upregulated in NSCLC and associated with aggressive clinicopathological characteristics. SMYD3 can serve as an adverse prognostic marker for NSCLC, and may be involved in NSCLC development and progression.
SMYD3 promotes the proliferation of NSCLC cells in vitro
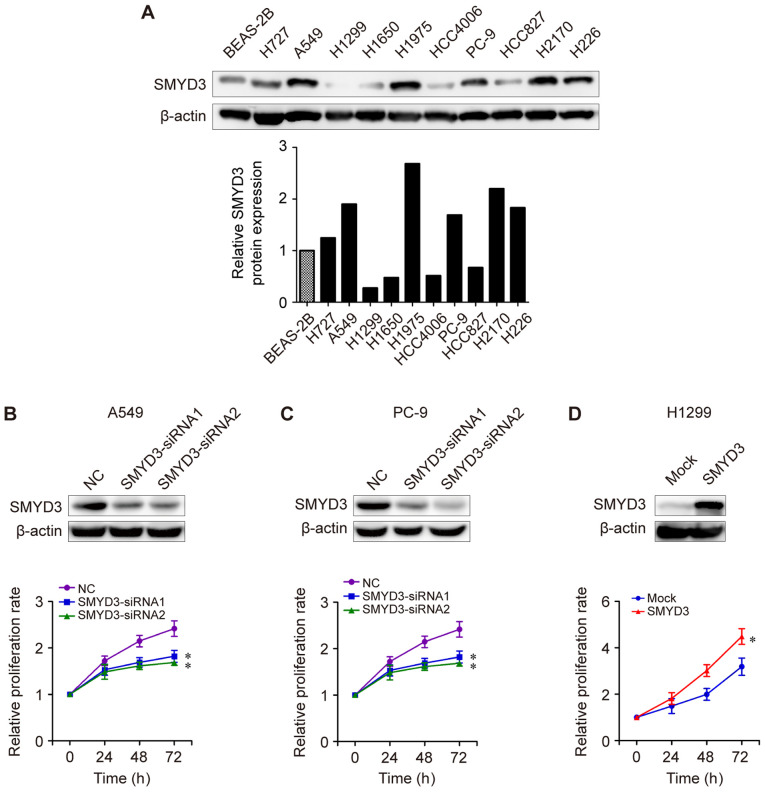
Next, the potential biological role of SMYD3 in NSCLC was investigated. The expression levels of SMYD3 protein in NSCLC cell lines were examined by western blot analysis. As presented in Fig. 3A, SMYD3 protein expression was higher in most NSCLC cell lines (H727, A549, H1975, PC-9, H2170 and H226) compared with that in human bronchial epithelial BEAS-2B cells. H727, A549, H1975, PC-9, H2170 and H226 cells exhibited a higher SMYD3 expression than that of H1299, H1650, HCC4006 or HCC827 cells (Fig. 3A). Next, SMYD3-siRNA transfections were performed in endogenous high-SMYD3-expressing cells. It was determined that SMYD3 knockdown led to a significant reduction in the proliferation rate of A549 and PC-9 cells in vitro (Fig. 3B and C). By contrast, enforced SMYD3 in endogenous low-SMYD3-expressing cells H1299 significantly promoted cell proliferation compared to a mock group (Fig. 3D), indicating an important role of SMYD3 in NSCLC cell proliferation.
Figure 3.
SMYD3 promotes NSCLC cell proliferation in vitro. (A) Western blot analysis of SMYD3 protein expression in a human bronchial epithelial BEAS-2B cells and the listed NSCLC cell lines (upper panel). Relative SMYD3 protein expression in each cell line was examined using densitometric analysis (lower panel). β-actin was used as an internal control. (B) A549 and (C) PC-9 cells were transfected with NC- or SMYD3-siRNAs. (D) H1299 cells were infected with lentiviral vectors to establish mock or SMYD3-overexpressed stable cell lines. Expression of SMYD3 protein was confirmed by western blotting (upper images). Cell proliferation was examined using SRB assay at different time-points (lower curves). All experiments were repeated at least three times. Data are presented as the mean ± SD. *P<0.05, compared with the NC or mock group. SMYD3, SET and MYND domain-containing protein 3; NSCLC, non-small cell lung cancer; SRB, sulforhodamine B.
SMYD3 knockdown triggers apoptosis by affecting pro-apoptotic and anti-apoptotic proteins in NSCLC cells
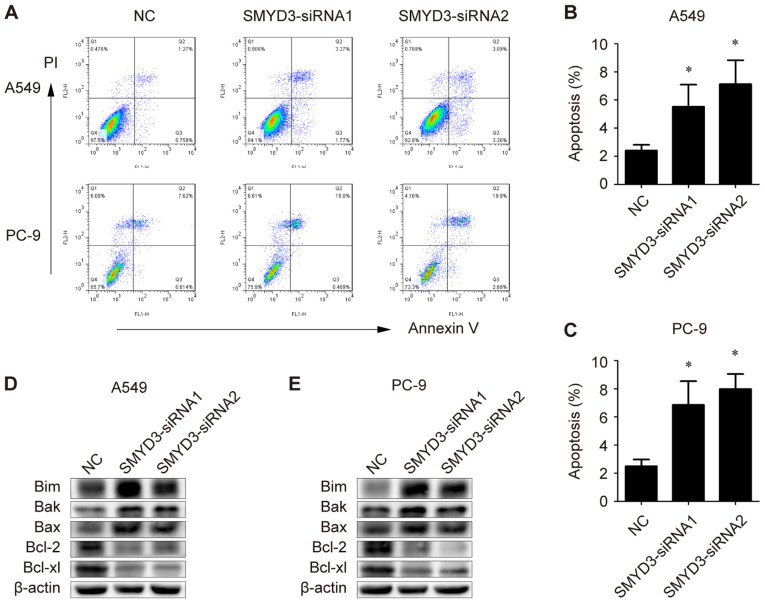
Apoptosis is considered an important approach in the inhibition of cell proliferation. In order to elucidate the mechanism underlying SMYD3 knockdown-induced proliferation reduction in NSCLC cells, the apoptotic rate of SMYD3-siRNA-transfected A549 and PC-9 cells was examined. Flow cytometric data revealed that SMYD3 silencing significantly induced apoptosis in both A549 and PC-9 cells (Fig. 4A-C).
Figure 4.
Effects of SMYD3 knockdown on apoptosis and apoptosis-associated genes in NSCLC cells. A549 and PC-9 cells were transfected with NC- or SMYD3-siRNAs. (A) The apoptotic rate was measured using Annexin V-FITC/PI double staining method in a flow cytometer 72 h after transfection. (B and C) Statistical analysis of the apoptotic population in the transfected (B) A549 and (C) PC-9 cells. (D and E) Expression of Bim, Bak, Bax, Bcl-2 and Bcl-xl in the transfected (D) A549 and (E) PC-9 cells were examined by western blotting. Data are presented as the mean ± SD of at least 3 independent experiments. *P<0.05, compared with the NC group. SMYD3, SET and MYND domain-containing protein 3; NSCLC, non-small cell lung cancer; NC, negative control; Bim, Bcl-2-like protein 11; Bak, Bcl-2 homologous antagonist/killer; Bcl-2, B-cell lymphoma 2.
To further explore the molecular basis for the induced apoptosis, the expression of pro-apoptotic and anti-apoptotic genes was evaluated. As revealed, the levels of Bim, Bak and Bax were markedly increased in SMYD3-silenced A549 and PC-9 cells, while Bcl-2 and Bcl-xl proteins were decreased upon SMYD3-siRNA transfection in A549 and PC-9 cells (Fig. 4D and E). Collectively, these results indicated that SMYD3 knockdown triggered apoptosis in NSCLC cells, probably by affecting key pro- and anti-apoptotic proteins.
Effect of SMYD3 on cisplatin resistance in NSCLC cells
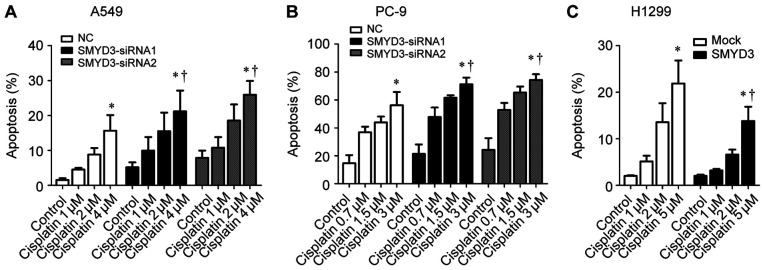
Since the dysregulated apoptotic signals can result in resistance to anticancer drugs in cancer cells (22), it was speculated that SMYD3 may be involved in cisplatin resistance, a chemotherapy drug for advanced NSCLC (23). As revealed, cisplatin induced apoptosis in A549, PC-9 and H1299 cells in a concentration-dependent manner (Fig. 5A-C). Specifically, SMYD3 silencing sensitized A549 and PC-9 cells to the apoptosis induced by cisplatin (Fig. 5A and B), whereas stable SMYD3 overexpression rendered H1299 cells more resistant to cisplatin-induced apoptosis (Fig. 5C). These data indicated that SMYD3 was associated with resistance to cisplatin in NSCLC cells.
Figure 5.
Effect of SMYD3 on cisplatin resistance in NSCLC cells. (A) A549 and (B) PC-9 cells were transfected with NC- or SMYD3-siRNAs for 24 h, and then treated with increasing concentrations of cisplatin (1, 2, and 4 µM for A549; 0.7, 1.5, and 3 µM for PC-9) for 72 h. (C) H1299 stable cell lines were treated with increasing concentrations of cisplatin (1, 2, and 5 µM) for 72 h. The apoptotic rate was measured using Annexin V-FITC/PI double staining method. Data are presented as the mean ± SD of at least 3 independent experiments. *P<0.05, compared with the control group. †P<0.05, compared with the NC or mock group. SMYD3, SET and MYND domain-containing protein 3; NSCLC, non-small cell lung cancer; NC, negative control.
SMYD3 knockdown blocks NSCLC cell migration and invasion in vitro
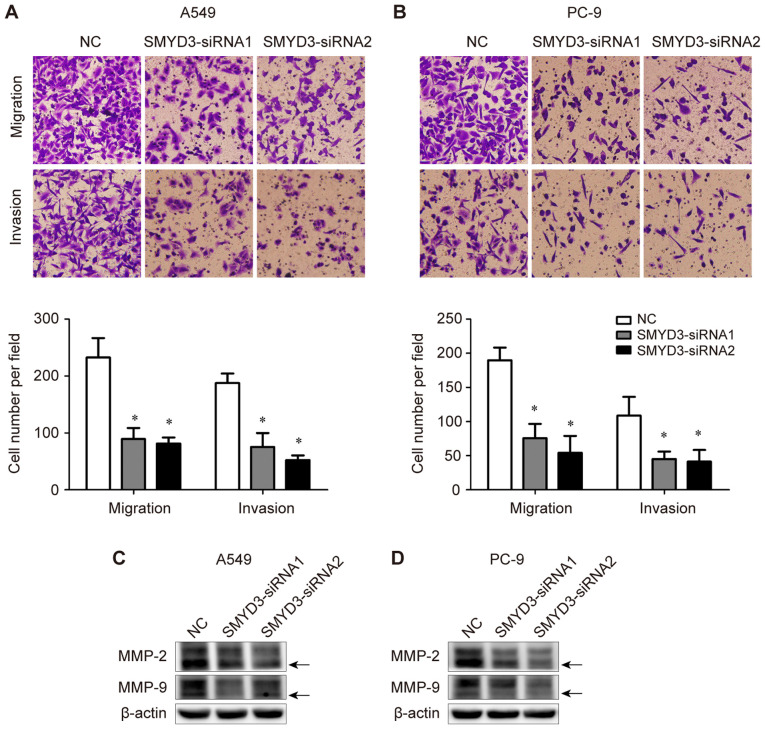
The effects of SMYD3 on NSCLC cell migration and invasion were determined using Transwell assays. It was determined that SMYD3 knockdown significantly suppressed the migratory and invasive abilities of both A549 and PC-9 cells (Fig. 6A and B). These data indicated the critical role of SMYD3 in the migration and invasion of NSCLC cells.
Figure 6.
SMYD3 knockdown suppresses NSCLC cell migration, invasion and expression of MMP-2 and MMP-9 in vitro. A549 and PC-9 cells were trans-fected with NC- or SMYD3-siRNAs. (A and B) The migratory and invasive abilities of transfected (A) A549 and (B) PC-9 cells were measured using Transwell assays. (C and D) Expression of MMP-2 and MMP-9 in the transfected (C) A549 and (D) PC-9 cells were examined by western blotting. The active form of MMP-2 or MMP-9 is indicated by an arrow. Data are presented as the mean ± SD of at least 3 independent experiments. *P<0.05, compared with the NC group. SMYD3, SET and MYND domain-containing protein 3; NSCLC, non-small cell lung cancer; MMP, matrix metalloproteinase.
In addition, the expression of invasion-associated MMP-2 and MMP-9 was assessed in the same transfection experiments. It was observed that the expression levels of both pro- and active-form MMP-2 and MMP-9 were down-regulated upon SMYD3 silencing in A549 and PC-9 cells (Fig. 6C and D).
Discussion
SMYD3 is a lysine methyltransferase that directs the fine regulation of chromatin structure and gene expression (4,24). Recently, it has been revealed that SMYD3 plays significant roles in human carcinogenesis and tumor progression, including in breast, hepatocellular, colorectal and gastric carcinomas (7,8,10). In the present study, it was demonstrated that SMYD3 was upregulated in NSCLC tissues, and associated with aggressive clinicopathological characteristics and poor survival. Multivariate analysis revealed that SMYD3 served as an independent predictor of poor OS in NSCLC patients. It was also revealed that SMYD3 played a critical role in NSCLC cell proliferation, migration, invasion and cisplatin resistance, as well as the regulation of associated genes. The present study is the first, to the best of our knowledge, to comprehensively analyze the expression of SMYD3 in clinical NSCLC samples, and offer insights into the oncogenic potential of SMYD3 in NSCLC carcinogenesis.
Barlesi et al have reported that the SMYD3 VNTR genotype was not correlated with either clinicopathological features or the risk of NSCLC in a Caucasian population (16). However, in the same cohort of NSCLC, there was no correlation between the proposed VNTR genotype and the level of H3K4 dimethylation either, implying that the SMYD3 VNTR genotype may not be equal to the expression or activity of SMYD3 protein in NSCLC. Herein, it was determined that SMYD3 protein was significantly upregulated in NSCLC tissues when compared with paired adjacent normal tissues. Notably, a high SMYD3 expression was associated with aggressive clinicopathological features, such as more advanced pathological stage, larger tumor size and distant metastasis, as well as poor DFS and OS in NSCLC patients. Multivariate Cox regression analysis revealed that SMYD3 overexpression was an independent predictor for poor OS, indicating that SMYD3 may be involved in NSCLC development and progression. In line with our findings, Mazur et al have revealed that abrogating SMYD3 catalytic activity inhibited the formation of Ras-driven lung adenocarcinomas in mouse models (25).
It was previously reported that SMYD3 is essential for the proliferation of cervical, breast, esophageal, hepatocellular and colorectal cancer cells (7,8,14,20). Consistently, the present data revealed that SMYD3 knockdown by siRNA led to a decreased proliferation rate in NSCLC cells in vitro, whereas stable SMYD3 overexpression significantly promoted NSCLC cell proliferation. These data confirmed our hypothesis that SMYD3 behaves as an oncogene in NSCLC carcinogenesis. Given the fact that apoptosis induction is a pivotal mechanism underlying the inhibition of cancer cell proliferation, the apoptosis of SMYD3-silenced NSCLC cells was examined, and it was revealed that SMYD3 silencing significantly induced apoptosis in NSCLC cells. The Bcl-2 protein family members, which can be divided into two classes (pro-apoptotic and anti-apoptotic), have been acknowledged as key regulators of caspase activation and apoptosis (26). Further experiments were then performed to explore the molecular basis for the induced apoptosis in SMYD3-silenced NSCLC cells. It was determined that the levels of pro-apoptotic Bim, Bak and Bax were markedly increased in SMYD3-silenced NSCLC cells, while anti-apoptotic Bcl-2 and Bcl-xl were decreased by SMYD3-siRNA. Our study extends the knowledge derived from Ren et al, who reported that the downregulation of SMYD3 decreased the ratio of Bcl-2/Bax in MDA-MB-231 cells (27), indicating that SMYD3 has a broad effect on apoptosis-associated genes.
Defective or inefficient apoptosis has been considered as a critical mechanism of resistance to anticancer drugs in cancer cells (22). It was then determined whether SMYD3 is involved in resistance to cisplatin, which remains a standard chemotherapy drug for advanced NSCLC (23). The present data revealed that SMYD3 silencing sensitized NSCLC cells to the apoptosis induced by cisplatin, whereas stable SMYD3 overexpression rendered NSCLC cells more resistant to cisplatin-induced apoptosis. This is probably attributed to the change of apoptotic signals in SMYD3-silenced or -over-expressed NSCLC cells. Consistently, previous studies have indicated that cisplatin resistance can be mediated by an inhibition of the apoptotic machinery derived from intracellular events (HER-2/neu amplification, or p53 mutation) in human malignancies (28,29).
In the present study, it was observed that SMYD3 knockdown significantly suppressed NSCLC cell migration and invasion in vitro, highlighting the oncogenic role of SMYD3 in NSCLC progression. Similarly, recent studies revealed that SMYD3 promoted the migration and invasion of multiple cancer types, including hepatocellular, breast and gastric carcinomas (30-32). MMPs are zinc-dependent endopeptidases that participate in tumor progression and metastasis by degrading the extracellular matrix, as well as regulating adherin, cytoskeletal proteins, growth factors, chemokines and cytokines (33). To date, 24 MMPs have been recognized in human beings. In the present study, it was determined that both pro- and active-form MMP-2 and MMP-9 proteins were decreased upon SMYD3 silencing in NSCLC cells, which probably contributed to the impaired invasive activities in those cells. The present results are supported by a previous study, which demonstrated that SMYD3 promoted the trimethylation of H3K4 in the MMP-9 promoter in a reversible model of cancer, thereby inducing MMP-9 expression and increasing cell migration and invasion (6). Based on the present data, it was reasoned that other MMPs, except for MMP-2 and MMP-9, may also be regulated by SMYD3 and involved in SMYD3-mediated cancer migration and invasion, although further studies are required to establish this hypothesis.
The present study has some limitations. First, we were unable to reveal the detailed mechanisms of SMYD3-modulated apoptosis- or invasion-associated genes in NSCLC cells. In addition, another limitation is that although SMYD3 has been recognized to regulate multiple downstream target genes, which are implicated in cell adhesion, proliferation, invasion, apoptosis, cell cycle regulation and signal transduction (5,34,35), only the effects of SMYD3 on NSCLC cell proliferation, apoptosis, migration and invasion were evaluated in the present study. More studies are required to elucidate the effects of other functions of SMYD3 in NSCLC development and progression.
In conclusion, it was demonstrated that SMYD3 was upregulated in NSCLC tissues, and associated with aggressive clinicopathological characteristics and poor survival. SMYD3 overexpression independently predicted poor OS in NSCLC patients. Functionally, SMYD3 played a critical role in NSCLC cell proliferation, migration, invasion and cisplatin resistance, as well as the regulation of apoptosis- and invasion-associated genes. The present study suggested SMYD3 as a novel oncogene in NSCLC carcinogenesis, which is anticipated to be a potential prognostic marker and therapeutic target for NSCLC patients.
Acknowledgments
Not applicable.
Abbreviations
- NSCLC
non-small cell lung cancer
- SMYD3
SET and MYND domain-containing protein 3
- H3K4
histone H3 at lysine 4
- VNTR
variable number of tandem repeats
- IHC
immunohistochemistry
- TMA
tissue microarray
- IASLC
International Association for the Study of Lung Cancer
- ATCC
American Type Culture Collection
- FBS
fetal bovine serum
- SRB
sulforhodamine B
- NC
negative control
- MMP
matrix metalloproteinase
- LVI
lymphovascular invasion
- PI
pleural invasion
- DFS
disease-free survival
- OS
overall survival
Funding
The present study was supported in part by grant nos. 81902325 and 81903633 from the National Natural Science Foundation of China, grant no. 2017WS026 from the Health and Family Planning Commission of Shandong Province (China), and grant nos. 2019GSF107042 and 2019GSF107051 from the Key Research and Development Plan of Shandong Province (P.R. China).
Availability of data and materials
All data generated or analyzed in this study are available from the corresponding author on reasonable request.
Authors' contributions
WY devised the conceptual idea. JL, YP and XM acquired and analyzed the clinical data. JL, LZ and LL performed the in vitro experiments. JL, LZ and WW analyzed the data and prepared the figures. JL, LZ and WY wrote, reviewed, and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center. All experiments complied with the Helsinki declaration and the approved guidelines of our institution. Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Zheng M. Classification and Pathology of Lung Cancer. Surg Oncol Clin N Am. 2016;25:447–468. doi: 10.1016/j.soc.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Karachaliou N. Lung cancer: Maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol. 2013;10:549–550. doi: 10.1038/nrclinonc.2013.152. [DOI] [PubMed] [Google Scholar]
- 4.Spellmon N, Holcomb J, Trescott L, Sirinupong N, Yang Z. Structure and function of SET and MYND domain-containing proteins. Int J Mol Sci. 2015;16:1406–1428. doi: 10.3390/ijms16011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyl-transferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 6.Cock-Rada AM, Medjkane S, Janski N, Yousfi N, Perichon M, Chaussepied M, Chluba J, Langsley G, Weitzman JB. SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Cancer Res. 2012;72:810–820. doi: 10.1158/0008-5472.CAN-11-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarris ME, Moulos P, Haroniti A, Giakountis A, Talianidis I. Smyd3 Is a transcriptional potentiator of multiple cancer-promoting genes and required for liver and colon cancer development. Cancer Cell. 2016;29:354–366. doi: 10.1016/j.ccell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Zhu MX, Zhang XD, Xu XE, Wu ZY, Liao LD, Li LY, Xie YM, Wu JY, Zou HY, et al. SMYD3 stimulates EZR and LOXL2 transcription to enhance proliferation, migration, and invasion in esophageal squamous cell carcinoma. Hum Pathol. 2016;52:153–163. doi: 10.1016/j.humpath.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Deng J, Luo X, Pan Y, Zhang L, Zhang R, Liang H. Overexpression of SMYD3 was associated with increased STAT3 activation in gastric cancer. Med Oncol. 2015;32:404. doi: 10.1007/s12032-014-0404-y. [DOI] [PubMed] [Google Scholar]
- 11.Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y, Fan Y. Upregulated SMYD3 promotes bladder cancer progression by targeting BCLAF1 and activating autophagy. Tumour Biol. 2016;37:7371–7381. doi: 10.1007/s13277-015-4410-2. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Wang C, Wang K, Liu L, Shen Q, Yan K, Sun X, Chen J, Liu J, Ren H, et al. SMYD3 as an oncogenic driver in prostate cancer by stimulation of androgen receptor transcription. J Natl Cancer Inst. 2013;105:1719–1728. doi: 10.1093/jnci/djt304. [DOI] [PubMed] [Google Scholar]
- 13.Dai B, Wan W, Zhang P, Zhang Y, Pan C, Meng G, Xiao X, Wu Z, Jia W, Zhang J, et al. SET and MYND domain-containing protein 3 is overexpressed in human glioma and contributes to tumorigenicity. Oncol Rep. 2015;34:2722–2730. doi: 10.3892/or.2015.4239. [DOI] [PubMed] [Google Scholar]
- 14.Wang SZ, Luo XG, Shen J, Zou JN, Lu YH, Xi T. Knockdown of SMYD3 by RNA interference inhibits cervical carcinoma cell growth and invasion in vitro. BMB Rep. 2008;41:294–299. doi: 10.5483/BMBRep.2008.41.4.294. [DOI] [PubMed] [Google Scholar]
- 15.Tsuge M, Hamamoto R, Silva FP, Ohnishi Y, Chayama K, Kamatani N, Furukawa Y, Nakamura Y. A variable number of tandem repeats polymorphism in an E2F-1 binding element in the 5′ flanking region of SMYD3 is a risk factor for human cancers. Nat Genet. 2005;37:1104–1107. doi: 10.1038/ng1638. [DOI] [PubMed] [Google Scholar]
- 16.Barlési F, Giaccone G, Gallegos-Ruiz MI, Span SW, Lefesvre P, Kruyt FA, Rodriguez JA. Genotype analysis of the VNTR polymorphism in the SMYD3 histone methyltransferase gene: Lack of correlation with the level of histone H3 methylation in NSCLC tissues or with the risk of NSCLC. Int J Cancer. 2008;122:1441–1442. doi: 10.1002/ijc.23227. [DOI] [PubMed] [Google Scholar]
- 17.Fedor HL, De Marzo AM. Practical methods for tissue microarray construction. Methods Mol Med. 2005;103:89–101. doi: 10.1385/1-59259-780-7:089. [DOI] [PubMed] [Google Scholar]
- 18.Detterbeck FC, Boffa DJ, Tanoue LT. The new lung cancer staging system. Chest. 2009;136:260–271. doi: 10.1378/chest.08-0978. [DOI] [PubMed] [Google Scholar]
- 19.You W, Chen B, Liu X, Xue S, Qin H, Jiang H. Farnesoid X receptor, a novel proto-oncogene in non-small cell lung cancer, promotes tumor growth via directly transactivating CCND1. Sci Rep. 2017;7:591. doi: 10.1038/s41598-017-00698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong SW, Zhang H, Wang BL, Sun P, Wang YG, Zhang P. Effect of the downregulation of SMYD3 expression by RNAi on RIZ1 expression and proliferation of esophageal squamous cell carcinoma. Oncol Rep. 2014;32:1064–1070. doi: 10.3892/or.2014.3307. [DOI] [PubMed] [Google Scholar]
- 21.Kunizaki M, Hamamoto R, Silva FP, Yamaguchi K, Nagayasu T, Shibuya M, Nakamura Y, Furukawa Y. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 2007;67:10759–10765. doi: 10.1158/0008-5472.CAN-07-1132. [DOI] [PubMed] [Google Scholar]
- 22.Indran IR, Tufo G, Pervaiz S, Brenner C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim Biophys Acta. 2011;1807:735–745. doi: 10.1016/j.bbabio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Rossi A, Di Maio M. Platinum-based chemotherapy in advanced non-small-cell lung cancer: Optimal number of treatment cycles. Expert Rev Anticancer Ther. 2016;16:653–660. doi: 10.1586/14737140.2016.1170596. [DOI] [PubMed] [Google Scholar]
- 24.Giakountis A, Moulos P, Sarris ME, Hatzis P, Talianidis I. Smyd3-associated regulatory pathways in cancer. Semin Cancer Biol. 2017;42:70–80. doi: 10.1016/j.semcancer.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Mazur PK, Reynoird N, Khatri P, Jansen PW, Wilkinson AW, Liu S, Barbash O, Van Aller GS, Huddleston M, Dhanak D, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283–287. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cory S, Adams JM. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 27.Ren TN, Wang JS, He YM, Xu CL, Wang SZ, Xi T. Effects of SMYD3 over-expression on cell cycle acceleration and cell proliferation in MDA-MB-231 human breast cancer cells. Med Oncol. 2011;28(Suppl 1):S91–S98. doi: 10.1007/s12032-010-9718-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 29.Perego P, Giarola M, Righetti SC, Supino R, Caserini C, Delia D, Pierotti MA, Miyashita T, Reed JC, Zunino F. Association between cisplatin resistance and mutation of p53 gene and reduced bax expression in ovarian carcinoma cell systems. Cancer Res. 1996;56:556–562. [PubMed] [Google Scholar]
- 30.Zhou Z, Jiang H, Tu K, Yu W, Zhang J, Hu Z, Zhang H, Hao D, Huang P, Wang J, et al. ANKHD1 is required for SMYD3 to promote tumor metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:18. doi: 10.1186/s13046-018-1011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo XG, Zhang CL, Zhao WW, Liu ZP, Liu L, Mu A, Guo S, Wang N, Zhou H, Zhang TC. Histone methyltransferase SMYD3 promotes MRTF-A-mediated transactivation of MYL9 and migration of MCF-7 breast cancer cells. Cancer Lett. 2014;344:129–137. doi: 10.1016/j.canlet.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 32.Zou JN, Wang SZ, Yang JS, Luo XG, Xie JH, Xi T. Knockdown of SMYD3 by RNA interference down-regulates c-Met expression and inhibits cells migration and invasion induced by HGF. Cancer Lett. 2009;280:78–85. doi: 10.1016/j.canlet.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Avila G, Sommer B, Mendoza-Posada DA, Ramos C, Garcia-Hernandez AA, Falfan-Valencia R. Matrix metal-loproteinases participation in the metastatic process and their diagnostic and therapeutic applications in cancer. Crit Rev Oncol Hematol. 2019;137:57–83. doi: 10.1016/j.critrevonc.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Sims RJ, III, Reinberg D. From chromatin to cancer: A new histone lysine methyltransferase enters the mix. Nat Cell Biol. 2004;6:685–687. doi: 10.1038/ncb0804-685. [DOI] [PubMed] [Google Scholar]
- 35.Huang L, Xu AM. SET and MYND domain containing protein 3 in cancer. Am J Transl Res. 2017;9:1–14. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed in this study are available from the corresponding author on reasonable request.