Abstract
Chondrosarcoma is a malignant bone tumor characterized by the production of a modified cartilage-type extracellular matrix (ECM). In the present study, the expression levels of the small leucine-rich proteoglycans (SLRPs), decorin, biglycan and lumican, were examined in the HTB94 human chondrosarcoma cell line. HTB94 cells were found to express and secrete the 3 SLRP members. RT-qPCR and western blot analysis demonstrated that lumican was the most abundantly secreted SLRP, whereas decorin and biglycan expression levels were low. The utilization of short interfering RNA specific for the decorin, biglycan, and lumican genes resulted in the efficient downregulation of the respective mRNA levels (P≤0.001). The growth of the HTB94 cells was stimulated by lumican (P≤0.001), whereas their migration and adhesion were not affected (P=NS). By contrast, these cellular functions were not sensitive to a decrease in low endogenous levels of decorin and biglycan. Lumicandeficiency significantly inhibited both basal and insulin-like growth factor I (IGF-I)-induced HTB94 cell growth (P≤0.001 andP≤0.01, respectively). These effects were executed through the insulin-like growth factor I receptor (IGF-IR), whose activation was markedly attenuated (P≤0.01) in lumican-deficient HTB94 cells. The downregulation of lumican induced the substantial inhibition of extracellular regulated kinase (ERK1/2) activation (P≤ 0.01), indicating that ERK1/2 is a necessary component of lumican/IGF-IR-mediated HTB94 cell proliferation. Moreover, the lumican-deficient cells exhibit increased mRNA levels of p53 (P≤0.05), suggesting that lumican facilitates HTB94 cell growth through an IGF-IR/ERK1/2/p53 signaling cascade. On the whole, the findings of the present study demonstrate that endogenous lumican is a novel regulator of HTB94 cell growth.
Keywords: chondrosarcoma, lumican, small leucine-rich proteoglycans, cell growth, insulin-like growth factor receptor I, extracellular regulated kinase 1/2
Introduction
Chondrosarcoma is a malignant bone tumor characterized by the production of a modified cartilage-type extracellular matrix (ECM). It is a heterogeneous, mesenchymal origin tumor that exhibits different histopathology and clinical behavior. Chondrosarcoma is the second most common bone tumor after osteosarcoma (1). The primary treatment for localized chondrosarcomas is surgical resection (2), as these tumors of mesenchymal origin exhibit resistance to classical chemotherapy and radiotherapy. The possible mechanisms of resistance to chemotherapy are a low mitotic rate and attenuated penetration into the tumor microenvironment resulting from low vascularity and the specific structure of the tumor-derived ECM (3). However, in some rare subtypes, such as mesenchymal chondrosarcomas, chemotherapy may be useful. Thus, a retrospective study, suggests that the combination of surgery with chemotherapy results in a better outcome for undifferentiated chondrosarcoma treatment in comparison to surgery alone (4). The generation of efficient conjunctive therapy for chondrosarcoma is an unmet medical need.
As cancer progresses, significant changes occur in the structural and mechanical properties of ECM constituents (5). The ECM provides a scaffold on which cancer cells adhere and migrate. However, by regulating a myriad of signaling pathways, the ECM components likewise affect critical cellular events, such as cellular motility, adhesion, differentiation, invasion and metastasis (6,7). The tumor ECM is extensively remodeled by enzymatic digestion, releasing active mediators that facilitate tumor cell growth and spreading (8,9).
Small leucine-rich proteoglycans (SLRPs) are diverse and multifaceted matrix constituents contributing to matrix organization and crucial mediators of ECM-cell signal transduction (10,11). SLRPs are composed of a core protein undergoing post-translational modifications, including substitution with glycosaminoglycan (GAG) side chains of various types (10,12). The GAG chains bind covalently into the protein core through serine or threonine residues (10). The protein core of these proteoglycans (PGs) is in the molecular weight range between 36 and 77 kDa, characterized by a variable number of central leucine-rich repeat (LRR) domains; whereas, the total PG molecular weight depends on the level of its glycosylation (13). The SLRP family consists of 17 members distributed into 5 classes based on characteristics, such as conserved leucine-rich repeats (LRR), N-terminal cysteine-rich clusters and unique chromosomal organization (14). Upon synthesis, SLRPs are secreted into the pericellular space, where they are sequestered through binding to cell membrane receptors or diffuse and incorporate into the tissue ECM by tethering to collagen fibers (15). Thus, in the cellular milieu, the SLRPs are distributed among the pericellular matrix, bound into 'proper' ECM, and also present as a pool of free molecules (12). SLRPs are an essential constituent of mesenchymal origin tissues, including bone and cartilage, as well as intimately involved in these tissue growth processes (16-19). Importantly, it is well determined that an abnormal SLRP expression, as well as structure, conclude in abnormal function of the ECMs and disease (13). Indeed, the SLRPs are implicated in the carcinogenesis of various solid tumors (20). Furthermore, the SLRPs contribute to cartilage pathologies (19), including degenerative cartilage disease (21,22).
The putative participation of SLRPs in the processes of chondrosarcoma tumorigenesis is unknown, with only a few reports addressing the issue (23,24). In the present study, the expression of the SLRPs members, decorin, biglycan and lumican, in an in vitro model of chondrosarcoma was examined and the main focus was paid to the putative effects of these mediators on chondrosarcoma cell biological functions.
Materials and methods
Materials
Recombinant human insulin-like growth factor I (IGF-I; 291-G1; 10 ng/ml) and transforming growth factor-β2 (TGF-β2; 302-B2-010; 10 ng/ml) were obtained from R&D Systems. A selective inhibitor of ERK1/2 (U0126; 10 μM; Cell Signaling Technology, Inc.) and allosteric inhibitor of IGF-IR (AG1024; 1 µM; Sigma-Aldrich; Merck KGaA) were used in the present study for 1 h. Primary antibodies from Santa Cruz Biotechnology, Inc. were used, and these included anti-lumican (sc166871; mouse monoclonal; 1/100 dilution for western blot analysis or 1/50 for immunofluorescence), anti-β-catenin (sc7963; mouse monoclonal; 1/300 dilution), anti-ERK1/2 (sc514302; mouse monoclonal; 1/200 dilution), anti-IGF-IR (sc81464; mouse monoclonal; 1/100 dilution), anti-pERK1/2 (sc136521; mouse monoclonal; 1/100 dilution), anti-Smad2 (sc6200; goat polyclonal; 1/200 dilution) and anti-pSmad2 (sc101801; rabbit polyclonal; 1/200 dilution). In addition, anti-actin (MAB1501; mouse monoclonal; 1/5,000 dilution; EMD Millipore), anti-p-IGF-IR (PA5-37602; polyclonal rabbit; 1/500 dilution; Thermo Fisher Scientific, Inc.), keratan sulfate (KS; 270427; mouse monoclonal; 1/1,000 dilution; Seikagaku Corporation) and keratanase II (100812; 0,005 µ/ml; Seikagaku Corporation) were utilized. Secondary-HRP antibodies anti-rabbit (AP182PR) and anti-mouse (AP192PM) were used at a 1/5,000 dilution and obtained from Millipore.
Cells and cell culture
In the present study, the HTB94 (ATCC® HTB-94™) human chondrosarcoma cell line was utilized. Cells were grown in DMEM (Biosera AG; LM-D1111) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.; 10500-064; heat-inactivated), gentamycin (Invitrogen; Thermo Fisher Scientific, Inc.; 15710-049) and penicillin/streptomycin (100units/ml; Biosera LMA4118). Prior to the addition of treatments, cells were cultured in serum-free medium for 24 h at 37°C and 5% CO2. Inhibitors, when used (ERK inhibitor or IGF-IR inhibitor), were added 1h prior to growth factor treatment.
Cell adherence assay
Following the treatments, the cells were harvested, and 10,000 cells/well were seeded onto fibronectin (FN)-coated 96-well plates for 1 h. According to the manufacturer's instructions, the number of adherent cells was measured using the CyQUANT fluorometric assay (Molecular Probes; Thermo Fisher Scientific, Inc.). Fluorescence was measured on a Fluorometer (BioTek Instruments, Inc.) using the proposed excitation (485 nm) and emission filters (528 nm), as previously described (25). A separate standard curve was used to convert fluorescence units to cell numbers. All adhesion experiments were repeated at least 3 times and performed in triplicate.
Cell migration assay
HB94 cells were seeded in 24-well culture plates at a concentration of 10×104 cells per well. The optimal concentration for plating was selected so that the cells would be confluent at almost 100% after 72 h of culture at 37°C and 5% CO2. RNA interference was performed according to the protocol described in the section below entitled 'Transfection with siRNA' using siRNAs specific for lumican, or decorin, or biglycan. Serum-free medium was utilized for culture. The cell layer was scratched with a sterile 10 µl pipette tip. Detached cells were removed by washing the cell layer twice with serum-free medium. The wound closure was monitored at 6 and 12 h with a digital image processor connected to a microscope (Leica DMIL), at 5 different positions across the wound. Cell motility was quantified by ImageJ analysis (ImageJ 1.4.3.67 launcher; Symmetry Software).
Proliferation assay
Growing cells from confluent cultures were harvested and seeded in black 96-well plates (3603; Corning, Inc.) at a density of 5,000 cells per well in 200 µl of DMEM (10% FBS). The cell density number was selected from optimization experiments (data not shown). The cells were allowed to rest overnight. If necessary, transfection with short interfering RNAs (siRNAs) was performed in a serum-free medium without antibiotics for 24 h. This was then replaced with fresh medium (0% FBS) with antibiotics. All treatments (inhibitors and/or growth factors) were added for the next 24 h at 37°C and 5% CO2 in 0% FBS. The cells were then lysed, and their number was calculated using the CyQUANT fluorometric assay (C7026; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Fluorescence was measured in a Fluorometer (BioTek Instruments, Inc.) using the proposed excitation (485 nm) and emission filters (528 nm). A separate standard curve was used to convert fluorescence units to cell numbers. All experiments were performed in triplicate.
MTT assay
Growing cells from confluent cultures were harvested and seeded in 96-well plates at a density of 12,000 cells per well in 200 µl of DMEM (10% FBS). The cells were allowed to rest overnight. Transfection with siRNAs was performed in a serum-free medium without antibiotics for 24 h (siLum) or 12 h (siDec, siBig). This medium was then replaced with fresh medium (0% FBS) and cells were incubated at 37°C and 5% CO2 in 0% FBS for the following 24 h. The Vybrant MTT Cell Proliferation Assay (cat. no. 11465007001; Roche Diagnostics) was performed according to the manufacturer's instructions. In brief, the medium was replaced with 100 μl of fresh medium (0% FBS) and 10 µl of MTT stock solution (12 Mm) were added to each well. Following 4-h incubation at 37°C, 50 µl of DMSO was added to cells for the next 10 min. The absorbance was measured at 540 nm using a Synergy HTX multimode microplate reader (BioTek). A separate standard curve was used to convert absorbance to cell number. All experiments were performed in triplicate.
Transfection with siRNA
For transfection experiments, the cells were plated in serum- and antibiotic-free medium in either 96-well plates (5,000 cells/well) or T25 flasks (1:8 dilution of a 90% confluent T75 flask). siRNA specific for lumican (siLum; stealth siRNAs HSS106200; Invitrogen; Thermo Fisher Scientific, Inc.), decorin (siDec; (stealth siRNAs HSS102673; Invitrogen; Thermo Fisher Scientific, Inc.) and biglycan (siBig; stealth siRNAs HSS184531; Invitrogen; Thermo Fisher Scientific, Inc.) and RNAi negative control (siScr; medium GC content negative control; Invitrogen; Thermo Fisher Scientific, Inc.). For transfection, siRNA, and Lipofectamine 2000 (11668-027; Invitrogen; Thermo Fisher Scientific, Inc.) were diluted in Opti-MEM I Reduced Serum Medium (31985-070; Invitrogen; Thermo Fisher Scientific, Inc.). Following 5 min of incubation at room temperature, diluted Lipofectamine 2000 was mixed with diluted siRNA (100 nm) for 20 min at room temperature to allow siRNA-liposome complexes to form and added to cell layers. Transfection was allowed to take place during 24 h for siLum or 12 h for siDec and siBig, when the medium was replaced with fresh (0% FBS) containing antibiotics and the incubation period continued for 24 or 36 h, respectively. Cells were then harvested, and mRNA or protein was extracted. When necessary, treatments were performed at the 24 h point after the initial transfection period. All transfection experiments were repeated at least 3 times and performed in triplicate.
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
According to the manufacturer's instructions, total ribonucleic acid isolation was performed using TRIzol (15596026; Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA (1 µg) was added for cDNA synthesis using the Takara (RR037A) RT cDNA synthesis kit. For semi-quantification of the genes of interest, qPCR reactions were performed on a Mx300P cycler using the Universal qPCR kit (KK4602; KAPA Biosystems) in a total volume of 20 µl. The thermocycling conditions were as follows: 94°C for 15 min, 40 cycles at 94°C for 20 sec, 55°C for 30 sec, 72°C for 30 sec, 72°C for 10 min. The PCR primer sequences were as follows: GAPDH forward, 5′-GGAAGGTGAAGGTCGGAGTCA-3′ and reverse, 5′-GTCATTGATGGCAACAATATCCACT-3′; lumican forward, 5′-CTTCAATCAGATAGCCAGACTGC-3′ and reverse, 5′-AGCCAGTTCGTTGTGAGATAAAC-3′; decorin forward, 5′-TCAATGGACTGAACCAGATGA-3′ and reverse, 5′-CCTTGA GGAATGCTGGTGAT-3′; biglycan forward, 5′-TCTGAA GTCTGTGCCCAA-3′ and reverse, 5′-TCTGAGATGCGCAG GTA-3′; p53 forward, 5′-CGTCGTGGCTTCTTGCAT TC-3′ and reverse, 5′-AAGACCTGCCCTGTGCAGC-3′; p21WAF1/ CIP1 forward, TGGAGACTCTCAAGGTCGAAA-3′ and reverse, 5′-AAGATCAGCCGGCGTTTG-3′. Standard curves were run in each optimized assay, which produced a linear plot of the threshold cycle Ct (dRn) against initial quantity (copies). The amount of each target was semi-quantified based on the concentration of the standard curve and was presented as arbitrary units. GAPDH was utilized as a housekeeping gene.
Western blot analysis
The total protein secreted into the serum-free culture medium was concentrated using Amicon Ultra 15ml (UFC901024; 10 kDa cut-off) centrifugal concentrator tubes. The initial volume of 3ml serum-free medium collected from culture, to isolate secreted proteins, was concentrated to a final volume of 500 µl whereas, harvested cells were lysed with RIPA solution (50 mM Tris-HCl, 1% NP-40, 0.25% Na-Deoxycholate, 150 mM NaCl, 1 mM EDTA with protease and phosphatase inhibitors). Equal amounts of protein, either cell extracts or secreted, were subjected to SDS-PAGE using 10% polyacrylamide gels under reducing conditions. Separated protein bands were transferred to nitrocellulose membranes in 10 mM CAPS (pH 11), containing 10% methanol. Membranes were blocked overnight at 4°C with PBS containing 0.1% Tween-20 (PBS-Tween) and 5% (w/v) low-fat milk powder. The membranes were incubated for 1 h at room temperature with the primary antibodies in PBS containing 0.1% Tween-20 (PBS-Tween) and 1% (w/v) low-fat milk powder. The immune complexes were detected following incubation with the appropriate peroxidase-conjugated secondary antibody diluted (1:5,000) in PBS-Tween, 2% low-fat milk for 1 h at room temperature, using the LumiSensor Chemiluminescent HRP substrate kit (Genscript; L00221V500), according to the manufacturer's instructions. The protein expression of actin was used to correct for the amount of each sample analyzed using ImageJ Analysis Software.
Immunofluorescence
HTB94 cells were seeded on round coverslips placed in 24-well plates, at a concentration of 70,000 cells/well, and incubated in complete medium for 24 h. Subsequently, the cells were incubated for 48 h at 37°C and 5% CO2 in 0% FBS medium. The cells were fixed with 5% formaldehyde and 2% sucrose in PBS for 10 min at room temperature. Following 3 washes with PBS supplemented with 1% FBS, the permeabilizing agent Triton X-100 was added for 10 min and then washed prior to the addition of the primary antibody for 1h at room temperature. Coverslips not incubated with the primary antibody were utilized as negative controls. The coverslips were rewashed and incubated for 1 h, in the dark at room temperature, with anti-mouse Alexa Fluor 488 (A21206; 1/200 dilution; Molecular Probes). TO-PRO-3 iodide (Molecular Probes; T3605) diluted 1:300 in de-ionized H2O was applied for 40 min to stain the nuclei. The coverslips were then placed onto slides using glycerol as a mountant and visualized using confocal microscopy.
Statistical analysis
Statistical significance was evaluated using a Student's t-test, or one-way ANOVA analysis of variance with Tukey's post-test, using GraphPad Prism (version 4.0) software.
Results
Expression of decorin, biglycan and lumican in HTB94 chondrosarcoma cells
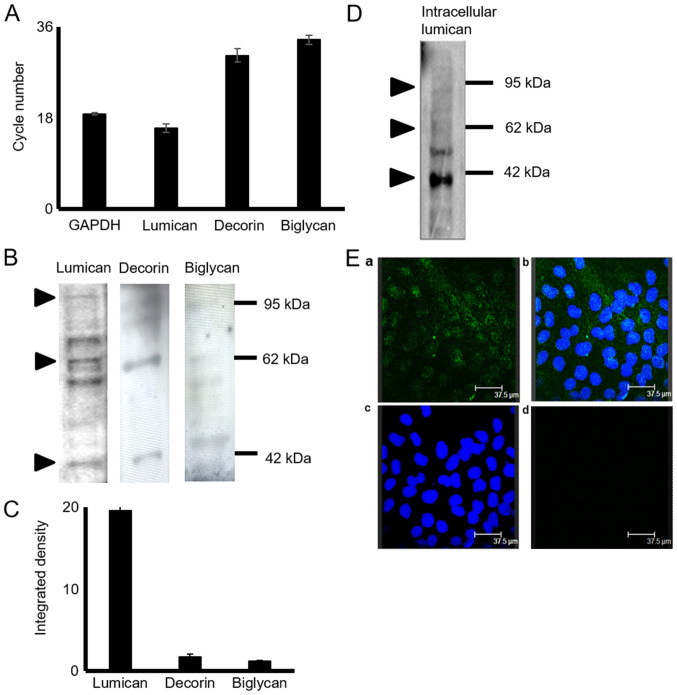
The expression of decorin, biglycan and lumican in HTB94 chondrosarcoma cells was estimated at the mRNA and protein level. The results of RT-qPCRdemonstrated that the HTB94 chondrosarcoma cells expressed lumican, decorin and biglycan at the mRNA level (Fig. 1A). The lumican transcripts were several fold higher than the low expression levels of decorin and biglycan (Fig. 1A). GAPDH was utilized as a housekeeping gene. To determine SLRP expression in HTB94 cells at the protein level, total protein was extracted from the cell culture medium, as well as from harvested cells. Western blot analysis of the proteins secreted by the HTB94 cells using specific antibodies revealed that the 3 SLRPs were secreted at discrete levels (Fig. 1B). The most abundant SLRP secreted by the HTB94 cells was lumican, with decorin and biglycan exhibiting low levels of expression, as demonstrated by densitometric analysis (Fig. 1C) and by transcript data. In all cases, in addition to the approximately 40 kDa band representative of the protein core, bands of higher molecular weight were present, indicating that the 3 SLRPs are also secreted as variously glycosylated protein products (Fig. 1B). In continuation, the present study focused on lumican, the main member of the SLRPs, secreted by the HTB94 chondrosarcoma cells. As shown in Fig. 1C, the bands representing glycosylated lumican were mostly localized in the 55 to 80 kDa molecular weight range. The monoclonal antibody (5D4) specific for KS chains was utilized to characterize the type of lumican substitution. To further evaluate the production of lumican by the HTB94 cells, respective cell extracts were probed with an anti-lumican antibody. Western blot analysis (Fig. 1D) revealed lumican specific bands of various molecular weights indicative of different stages of protein glycosylation. The utilization of fluorescence likewise demonstrated the deposition, of under synthesis, lumican to the cytoplasm of HTB94 cells (Fig. 1E). As shown in Fig. 2, the use of the 5D4 antibody gives the specific 55-80 kDa band pattern, identical to that obtained upon probing with the anti-lumican antibody. In continuation, secreted proteins were subjected to enzymic treatments with keratanase II, which cleaves within sulfated lactosamine residues, and subjected to SDS-PAGE and western blot analysis with anti-KS antibody to confirm the nature of the carbohydrate component. The specific KS-reaction was markedly attenuated in the samples treated with keratanase II, demonstrating that lumican secreted by HTB94 was partially substituted with KS (Fig. 2).
Figure 1.
Expression of SLRPs in HTB94 chondrosarcoma cells. (A) Specific primers for each gene were used and normalized against GAPDH as the housekeeping gene. Mean Ct values are presented from 6 replicates (SEM, standard error of the mean). (B) Representative western blots showing the expression of lumican, decorin and biglycan in the culture medium of HTB94 chondrosarcoma cells. (C) Protein bands of the 3 secreted SLRPs were densitometrically measured utilizing the ImageJ program and expressed as arbitrary units. (D) Following protein extraction from HTB94 harvested cells, the protein sample was loaded in an acrylamide gel. An anti-lumican antibody was used to determine the expression of lumican in the cell extracts. The position of the nearest respective protein marker band is depicted to the right. (Ea) Lumican (green; anti-mouse Alexa Fluor 488) protein staining and (Eb) nuclear staining (using TO-PRO-3) were evaluated in culture; (Ec) superimposed image; (Ed) in the negative control, the primary antibody was omitted, but the anti-mouse secondary antibody was used. Slides were analyzed by confocal microscopy, and images were obtained at ×40 magnification. Results represent the average of 3 separate experiments. Representative plots are presented. Data are the means ± SEM. SLRPs, small leucine-rich proteoglycans.
Figure 2.
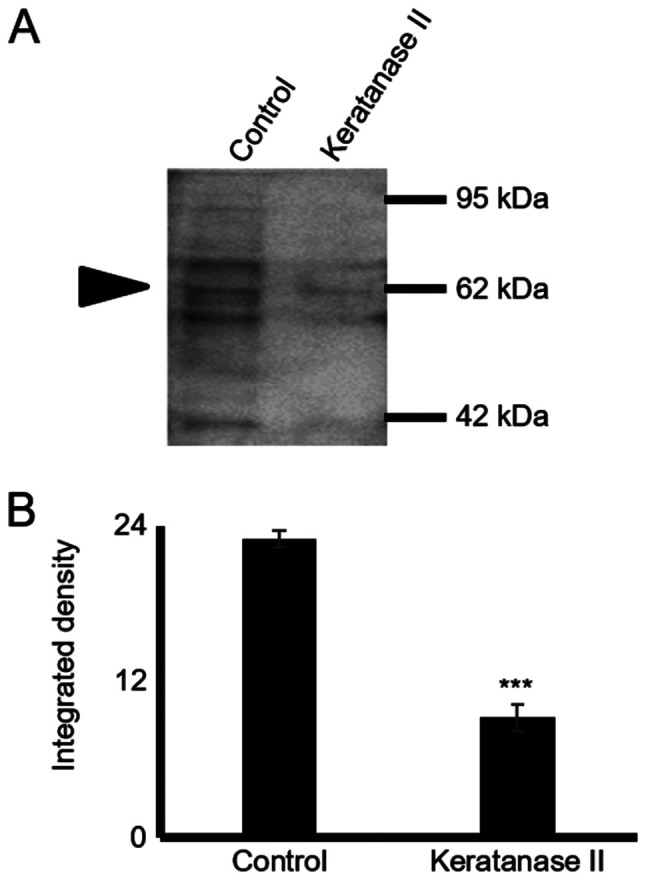
Glycosylation pattern of lumican secreted by HTB94 cells. (A) Following protein extraction from the culture medium, equal amounts of protein were treated with keratanase II for 16 h at 37°C and blotted with the antibody against keratan sulfate (cloneBD4). Staining with an anti-KS antibody is specific for the KS chains. (B) Protein bands were densitometrically measured utilizing the ImageJ program and expressed as arbitrary units. The position of the nearest respective protein marker band is depicted to the right. Results represent the average of 3 separate experiments. Representative plots are presented. Data are the means ± SEM; ***P≤0.001, statistically significant difference compared with the respective control samples. KS, keratan sulfate.
SLRP expression levels are efficiently downregulated following transfection with specific siRNAs
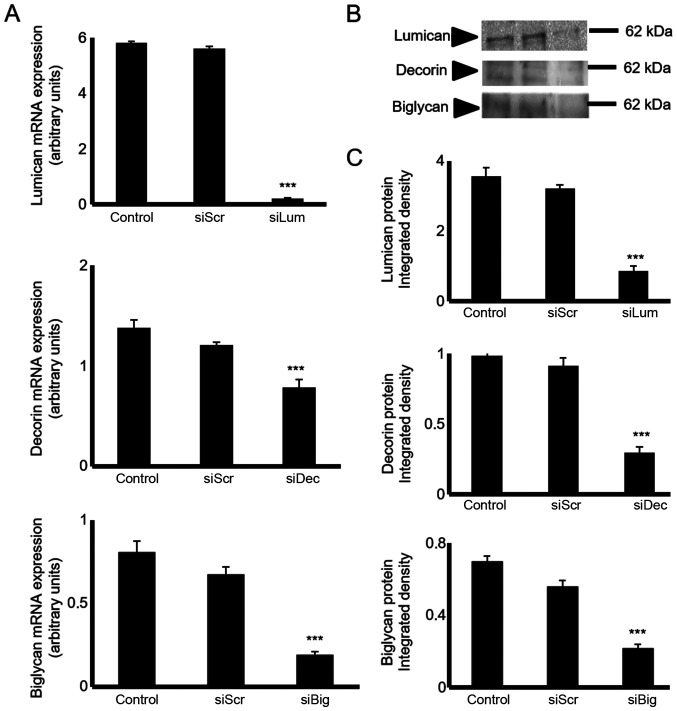
The HTB94 cells were transfected with lumican-, decorin- and biglycan-specific siRNAs to estimate their putative biological roles. The downregulation of SLRP mRNA expression was verified by RT-qPCR. Transfection of the HTB94 cells with siLum, siDec, and siBig for 24h resulted in the potent inhibition of mRNA expression (P≤0.001), respectively (Fig. 3A). The downregulation of lumican, decorin and biglycan transcripts was followed by a significant decrease in lumican protein secretion, as demonstrated by western blot analysis (Fig. 3B and C).
Figure 3.
Transfection with siRNA. HTB94 cells were transfected with siRNAs specific for 3 SLRPs, lumican, decorin and biglycan for 48 h, using scramble siRNA (siScr) as a control. (A) Lumican, decorin and biglycan expression levels were evaluated by RT-qPCR. The inhibition of lumican, decorin and biglycan mRNA expression was verified by RT-qPCR relative to the siScr control and expressed as arbitrary units. The results represent the average of 3 separate experiments in triplicate, and are plotted as the mean percentage ± standard error of the mean; statistical significance: ***P≤0.001. (B) Equal amounts of secreted protein were extracted from siScr and siLum, siDec and siBgn culture media and blotted against the anti-lumican antibody. Representative 60 kDa bands are presented. The position of the nearest respective protein marker band is depicted to the right. Results represent the average of three separate experiments. (C) Densitometric analysis of specific lumican protein bands. Means ± SEM were plotted; statistical significance: ***P≤0.001, statistically significant difference compared with the respective control samples. SLRPs, small leucine-rich proteoglycans.
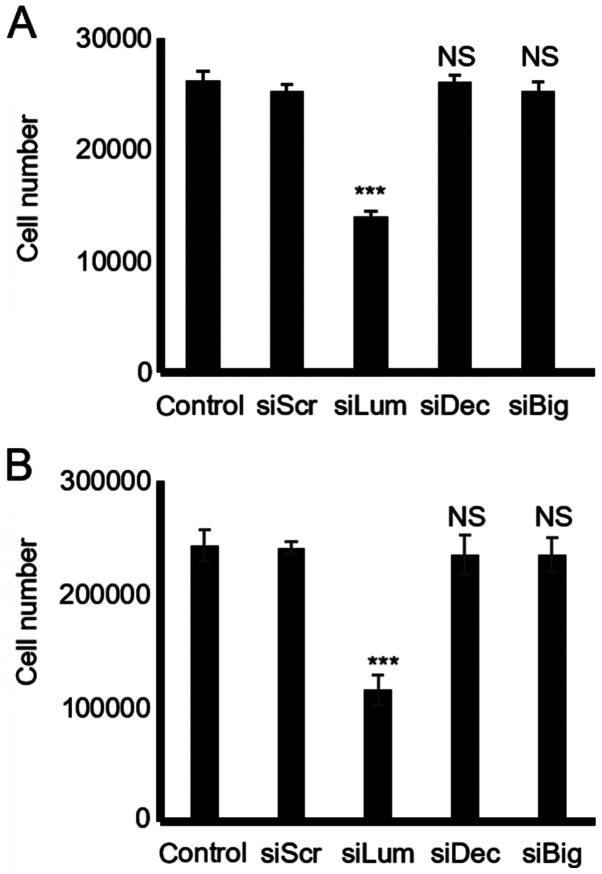
Effect of endogenous lumican, decorin and biglycan on HTB94 chondrosarcoma cell growth
The downregulation of lumican secretion following transfection of the HTB94 cells with lumican siRNA resulted in the potent inhibition of growth as compared to the cells transfected with scramble siRNA (P≤0.001; Fig. 4). However, the downregulation of the low endogenous expression of decorin and biglycan did not affect the growth of the HTB94 cells, as shown by the CyQUANT fluorometric and MTT assays (P=NS) (Fig. 4).
Figure 4.
Effect of SLRP downregulation on chondrosarcoma cell growth. (A) Cells were harvested and seeded (5,000 cells/well) in 96-well plates. HTB94 cells were transfected with siRNAs specific for lumican, decorin and biglycan, for 48 h. A non-specific RNA sequence was used as a control (siScr). Cell number was determined using a fluorometric CyQUANT Assay kit. (B) Cells were harvested and seeded (12,000 cells/well) in 96-well plates. Following incubation with siRNAs specific for lumican, decorin and biglycan for 48 h, cell number was determined using a Vybrant MTT Assay kit. Results represent the average of 3 separate experiments performed in triplicate. Data are the means ± SEM; ***P≤0.001, statistically significant difference compared with the respective control samples. NS, not significant; SLRPs, small leucine-rich proteoglycans.
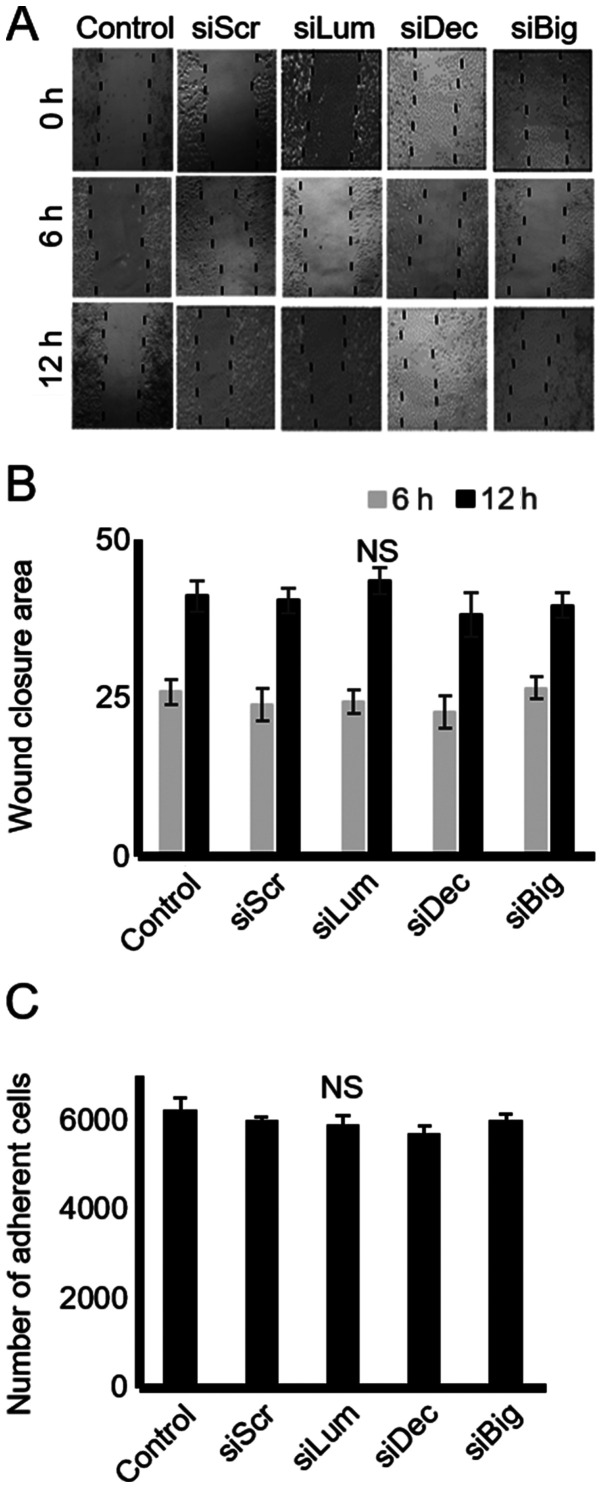
Effect of endogenous lumican, decorin and biglycan on HTB94 chondrosarcoma cell migration and adhesion
SLRPs have previously been shown to affect cell motility and adhesion by modulating the cell-matrix interaction of cells (26). The present study thus examined the effect of endogenous decorin, biglycan and lumican production on the motility of HTB94 cells using a 'wound healing' assay and transfection with siRNA, as previously demonstrated (26). In the present study, the HTB94 cells, however, migrated with equal efficiency in the presence or absence of decorin, biglycan and lumican siRNA, indicating that these SLRP members do not affect their migratory capabilities (Fig. 5A and B). In continuation, the adhesion ability of these cells was examined utilizing a 96-well plate adhesion assay, as previously described (27). The results demonstrated that transfection with decorin, biglycan and lumican siRNA did not affect the ability of the cells to attach to the fibronectin substrate (Fig. 5C).
Figure 5.
Effect of SLRP downregulation on chondrosarcoma cell migration and adhesion. (A) Cells were harvested and seeded (100,000 cells/well) in 24-well plates, to be treated with the siRNAs specific for 3 SLRPs, lumican, decorin and biglycan for 48 h (at which point they were confluent). Following treatment, the cell layer was 'scratched' with a 10 µl sterile pipette tip, and the wound surface area was measured at the 0, 6 and 12 h points. (B) The cell substratum surface area was measured utilizing the ImageJ program and expressed as arbitrary units. (C) HTB94 cells were treated with siRNAs specific for 3 SLRPs, lumican, decorin and biglycan for 48 h. A non-specific RNA sequence was used as a control (siScr). Following treatment, cells were harvested and seeded (10,000 cells/well) for 1 h at 37°C in 96-well plates coated with fibronectin. The number of attached cells was determined using fluorometric CyQUANT Assay kit Results represent the average of 3 separate experiments. Data are the means ± SEM. NS, not significant; SLRPs, small leucine-rich proteoglycans.
Mechanisms of action of lumican
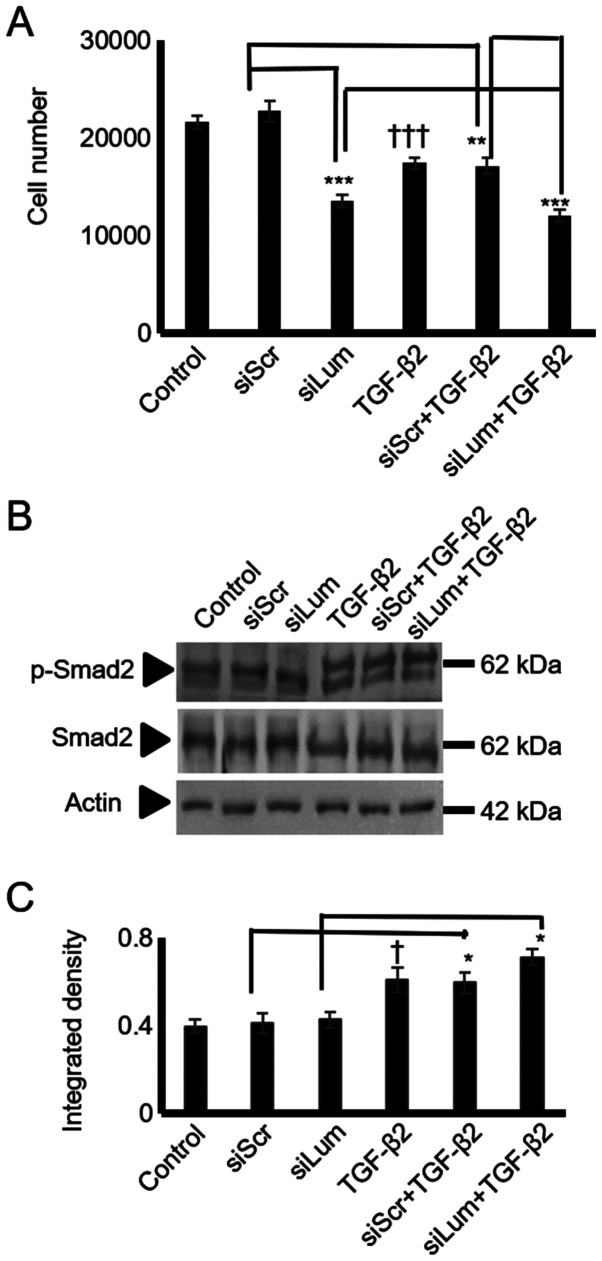
TGF-β2 has previously been shown to negatively mediate the growth of physiological chondrocytes (28,29) and human chondrosarcoma cells (30). In the present study, treatment of the HTB94 cells with TGF-β2 resulted in a potent decrease in growth (P≤0.001; Fig. 6A). As SLRP members may affect cell proliferation by modulating TGF-β2 signaling, both lumican and scramble siRNA-transfected cells were treated with TGF-β2, and cell proliferation was measured. The lumican siRNA-transfected HTB94 cells treated with TGF-β2 exhibited lower growth rates than the siScr-transfected cells treated with TGF-β2, suggesting an additive effect of lumican on the TGF-β2-dependent decrease in cell growth. Therefore, the difference in growth between the lumican- and scramble siRNA-transfected cells treated with TGF-β was attributed to the effect of lumican. These results suggest that the regulation of HTB94 cell proliferation by lumican is TGF-β2-independent.
Figure 6.
Effect of lumican downregulation and exogenous TGF-β2 on chon-drosarcoma cell growth and Smad2 activation. HTB94 cells were treated for 48 h with specific siRNA against lumican. A non-specific RNA sequence was used as a control (siScr). Following 24 h of transfection with siLum, cells were treated with TGF-β2 (10 ng/ml) in 0% FBS DMEM. (A) Cell number was determined using fluorometric CyQUANT Assay kit (B) Representative blots of Smad2 (60 kDa), p-Smad2 (60 kDa), and actin (42 kDa) are presented. (C) Smad2 and p-Smad2 protein bands were densitometrically analyzed and adjusted against actin, and the ratio of p-Smad2/Smad2 was measured and presented. The position of the nearest respective protein marker band is depicted to the right. Results represent the average of 3 separate experiments. Data are the means ± SEM; *P≤0.05, **P≤0.01 and ***P≤0.001, statistically significant difference between siScr and siLum treatments. †P≤0.05 and †††P≤0.001, statistically significant difference between control and TGF-β2 treatment groups.
To further examine the putative interaction between lumican and TGF-β2 in HTB94 cells growth, the TGF-β2 activation of Smad2, an established downstream mediator of TGF-β2 action (31), was assessed. To characterize Smad2 expression and phosphorylation levels, specific anti-Smad2 and anti-phospho-Smad2antibodies were used. As was expected, treatment ofthe HTB94 cells with TGF-β2 enhanced the Smad2 phosphorylation levels (Fig. 6B and C). In continuation, we examined the possible effects of lumican downregulation on Smad2 activation. Western blot analysis showed that neither Smad2 protein expression (specific 65 kDa band) nor the expression of phospho-Smad2 (specific 60 kDa band) was affected by lumican downregulation (Fig. 6B and C). Exogenously added TGF-β2 increased Smad 2 phosphorylation to a similar extent in both scramble and lumican-deficient cells (Fig. 6B and C). Densitometric analysis of the respective bands is presented in Fig. 6C. Protein amounts were normalized against actin. These results revealed that lumican affected neither the TGF-β2 receptor-restricted, Smad2 signaling, or consecutively the TGF-β2-dependent growth of HTB94 cells.
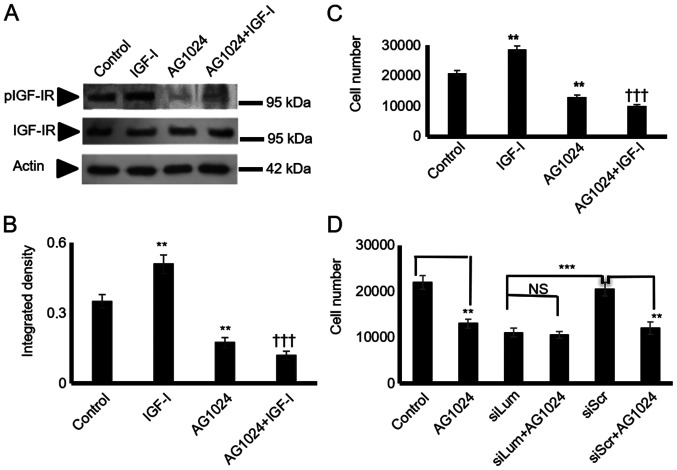
IGF-I is a well-established stimulator of chondroblast growth (32) and a moderate stimulator of chondrosarcoma proliferation (33). In initial experiments, it was verified, utilizing western blot analysis, that the HTB94 cells expressed a functional IGF-IR receptor with the ability to respond to IGF-I stimulation (Fig. 7A and B). Subsequently, the HTB94 cells were treated with IGF-I and IGF-IR inhibitor or their combination. IGF-I treatment significantly enhanced cell growth (P≤0.01), whereas the blockage of IGF-IR induced a marked attenuation of HTB94 cell basal and IGF-I-dependent growth (P≤0.01; Fig. 7C). When the lumican siRNA-transfected HTB94 cells were exposed to an IGF-IR inhibitor, no further attenuation of cell growth was detected (Fig. 7D). As SLRP members have earlier been shown to affect IGF-IR-dependent growth processes (12), both lumican and scramble siRNA-transfected cells were treated with IGF-I, and cell proliferation was measured. The lumican siRNA-transfected HTB94 cells treated with IGF-I exhibited complete abolishment of IGF-I-dependent growth stimulation (Fig. 8A), suggesting that the effects of lumican are partially mediated through IGF-IR-dependent signaling. The lumican-deficient cells were probed with IGF-IR and pIGF-IR antibodies to verify the contribution of lumican to IGF-IR signaling in the regulation of HTB94 cell growth. This approach revealed that IGF-IR activation was significantly downregulated in the lumican-deficient cells (P≤0.01; Fig. 8B and C).
Figure 7.
HTB94 chondrosarcoma cell IGF-IR expression and effect on cell growth. Following 24 h of starvation, the cells in each well were incubated with serum-free medium (control), IGF-I (10 ng/ml) in 0% FBS DMEM, 1 µM AG1024 (IGF-IR inhibitor) and 10 ng/ml IGF-I + 1 µM AG1024 for 24 h. (A) Representative western blots showing the expression of IGF-IR total protein (IGF-IR) (95 kDa) and phosphorylated IGF-IR protein (pIGF-IR) (95 kDa). (B) Protein bands were densitometrically analyzed and adjusted against actin (42 kDa), and the ratio of pIGF-IR/IGF-IR was measured and presented. The position of the nearest respective protein marker band is depicted to the right. Results represent the average of 3 separate experiments. Data are the means ± SEM; **P≤0.01, statistically significant difference between control and IGF-I and AG1024 treatments. †††P≤0.01, statistically significant difference between IGF-I and AG1024+IGF-I treatment groups. (C) HTB94 cells were harvested and seeded (5,000 cells/well) on 96-well plates. Following 24 h of starvation, cells in each well were incubated with serum-free medium (control), IGF-I (10 ng/ml) in 0% FBS DMEM, 1 µM AG1024 (IGF-IR inhibitor) and 10 ng/ml IGF-I + 1 µM AG1024 for 24 h. Cell number was determined using a fluorometric CyQUANT Assay kit. Results represent the average of 3 separate experiments. Data are the means ± SEM; **P≤0.01, statistically significant difference between control and IGF-I, AG1024 treatments. †††P≤0.01, statistically significant difference between IGF-I and AG1024+IGF-I treatment groups. (D) HTB94 cells were treated for 48 h with siRNA specific for lumican. A non-specific RNA sequence was used as a control (siScr). Following 24 h of transfection with siLum, cells were treated with 1 µM AG1024 for 24 h in 0% FBS DMEM. Cell number was determined using a fluorometric CyQUANT Assay kit. Results represent the average of 3 separate experiments. Data are the means ± SEM; **P≤0.01, statistically significant difference between siScr and AG1024 treatments. ***P≤0.001, statistically significant difference between siLum and siScr; NS, no significance between the siLum and siLum + AG1024 + treatment groups.
Figure 8.
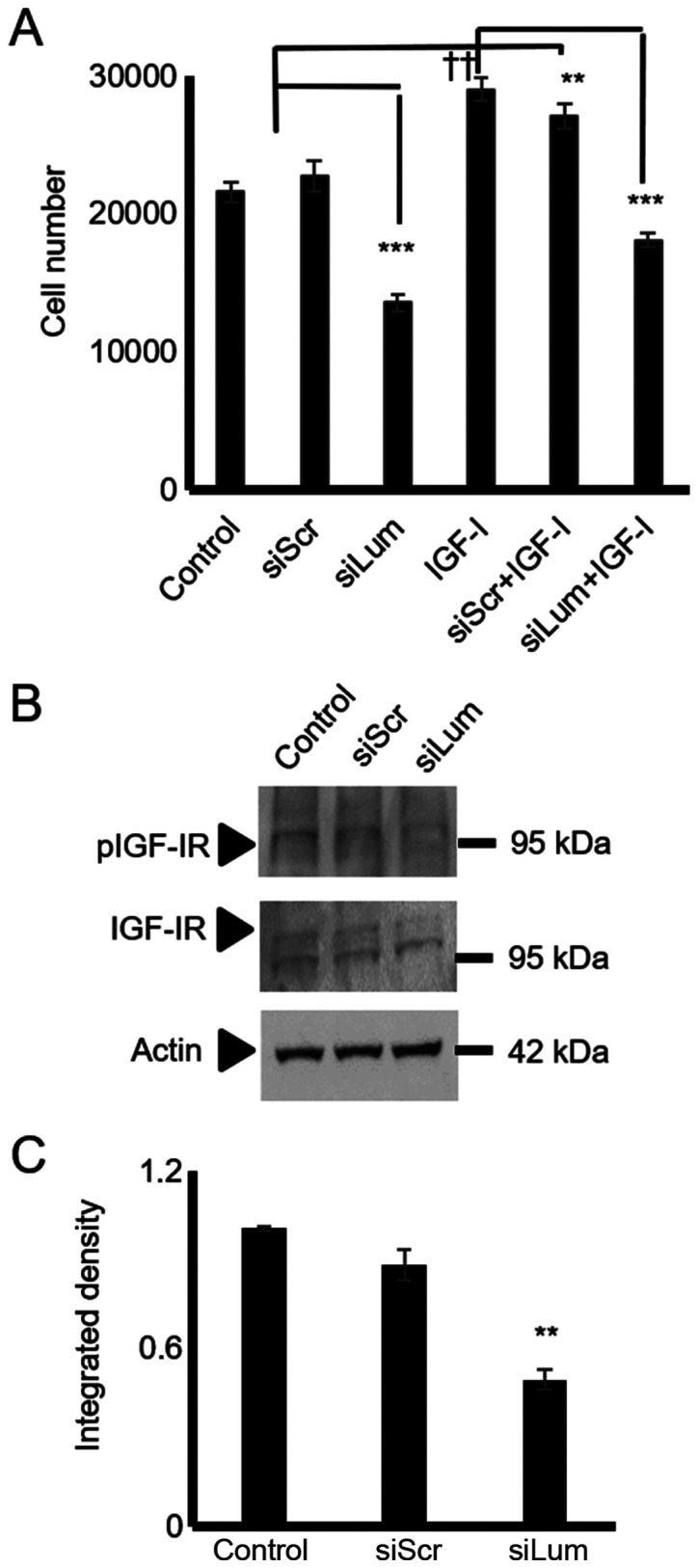
Effect of lumican downregulation and exogenous IGF-I on chon-drosarcoma cells growth and IGF-IR activation. HTB94 cells were treated for 48 h with specific siRNA against lumican. A non-specific RNA sequence was used as a control (siScr). Following 24h of incubation with siRNAs, cells were treated with IGF-I (10 ng/ml) in 0% FBS DMEM. (A) Cell number was determined using a fluorometric CyQUANT Assay kit. (B) Representative blots of IGF-IR (95 kDa), pIGF-IR (95 kDa), and actin (42 kDa) are presented. (C) IGF-IR and pIGF-IR protein bands were densitometrically analyzed and adjusted against actin, and the ratio of pIGF-IR/IGF-IR was measured and presented. The position of the nearest respective protein marker band is depicted to the right. Results represent the average of 3 separate experiments. Data are the means ± SEM; **P≤0.01 and ***P≤0.001, statistically significant difference between siScr and siLum treatments. ††P≤0.01, statistically signifi-cant difference between control and IGF-I treatments.
Erk1/2 is a downstream mediator of lumican/IGF-IR growth regulation
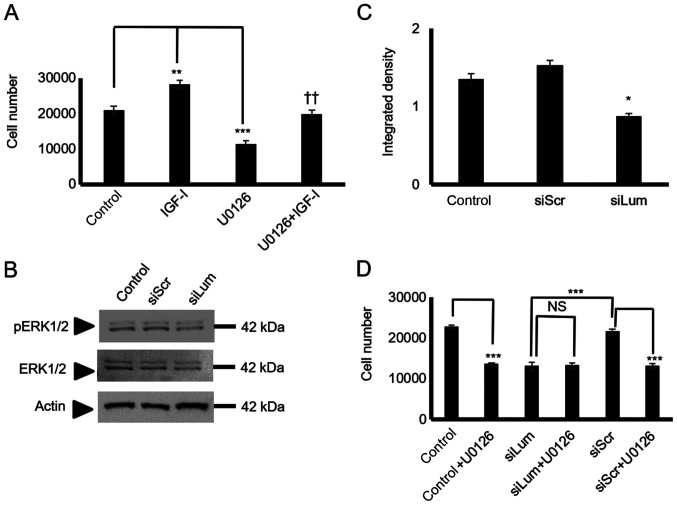
Subsequently, the present study examined critical IGF-I downstream mediators focusing on ERK1/2, a well-established IGF-I downstream conduit in the regulation of tumor cell growth, migration, and adhesion (27,34). The utilization of a specific ERK1/2 inhibitor led to the significant suppression of the HTB94 basal level of cell proliferation (P<0.001), and the complete abolishment of IGF-I-dependent growth (P<0.01; Fig. 9A). To determine the effects of lumican on IGF-I-dependent ERK1/2 activation in HTB94 cells, the ERK1/2 phosphorylation levels in both the control and IGFI-treated lumican-deficient cellswere examined. As shown in Fig. 9B and C, lumican participation was found to be crucial for ERK1/2 phosphorylation, as the lumican-deficient cells exhibited an attenuated ERK1/2 activation (P<0.05). Furthermore, when the lumican-deficient cells were treated with the ERK1/2 inhibitor, no further downregulation of cell growth was evident, demonstrating that ERK1/2 participation is necessary for the lumican effect (Fig. 9D). Upon activation of the upstream effectors the degradation of β-catenin was inhibited, resulting in its increased expression in the cytoplasm and enhanced translocation to the nucleus. No changes in the total β-catenin expression in the lumican-deficient cells were detected by western blot analysis and immunofluorescence (data not shown). On the other hand, β-catenin signaling did not participate in the lumican/IGF-IR-mediated growth effects (Fig. S1).
Figure 9.
Effect of lumican on ERK1/2 activation. (A) HTB94 cells were harvested and seeded (5,000 cells/well) in 96-well plates. Cells in each well were incubated with serum-free medium (control), IGF-I (10 ng/ml) in 0% FBS DMEM, 10 µM U0126 (ERK1/2 inhibitor) and 10 µM U0126 + 10 ng/ml IGF-I for 24 h. Cell number was determined using a fluorometric CyQUANT Assay kit. (B) Expression of ERK 1/2 total protein (ERK1/2) (42-44 kDa) and phos-phorylated ERK1/2 protein (pERK1/2) (42-44 kDa) of cells transfected for 48 h with specific siRNA against lumican (siLum) were determined by western blot analysis. A non-specific RNA sequence was used as a control (siScr). (C) ERK1/2 and pERK1/2 protein bands were densitometrically analyzed and adjusted against actin, and the ratio of pERK1/2/ERK1/1 was measured and presented. The position of the nearest respective protein marker band is depicted to the right. Representative plots are presented. Results represent the average of 3 separate experiments. Data are the means ± SEM; *P≤0.05, **P≤0.01 and ***P≤0.01, statistically significant difference between control and IGF-I and U0126 treatments. ††P≤0.01, statistically significant difference between IGF-I and U0126 + IGF-I treatment groups. (D) HTB94 cells were treated for 48 h with siRNA specific for lumican. A non-specific RNA sequence was used as a control (siScr). Following 24 h of transfection with siLum, cells were treated with 10 µM UO126 for 24 h in 0% FBS DMEM. Cell number was determined using a fluorometric CyQUANT Assay kit. (D) Results represent the average of 3 separate experiments. Data are the means ± SEM; ***P≤0.01, statistically significant difference between siScr and UO126 treatments or between siLum and siScr; NS, no significance between the siLum and siLum + U0126 treatment groups.
Lumican affects p53 cell cycle regulator expression
The expression of 3 cell cycle-related genes was then analyzed to characterize the intracellular molecular mechanisms involving the Erk1/2 pathway on lumican-induced chondrosarcoma cell growth. The mRNA levels of p21WAF1/CIP1 and p53 were estimated in lumican-deficient cells to investigate the possible effects of lumican. RT-qPCR demonstrated that the levels of p53 tumor suppressor were significantly upregulated in the siLum-treated cells as compared to the siScr cells (P≤0.05). On the other hand, no change was demonstrated in the expression of p21WAF1/CIP1. These results collectively suggest that lumican is involved in transcriptional control of cell cycle-related genes (Fig. 10).
Figure 10.
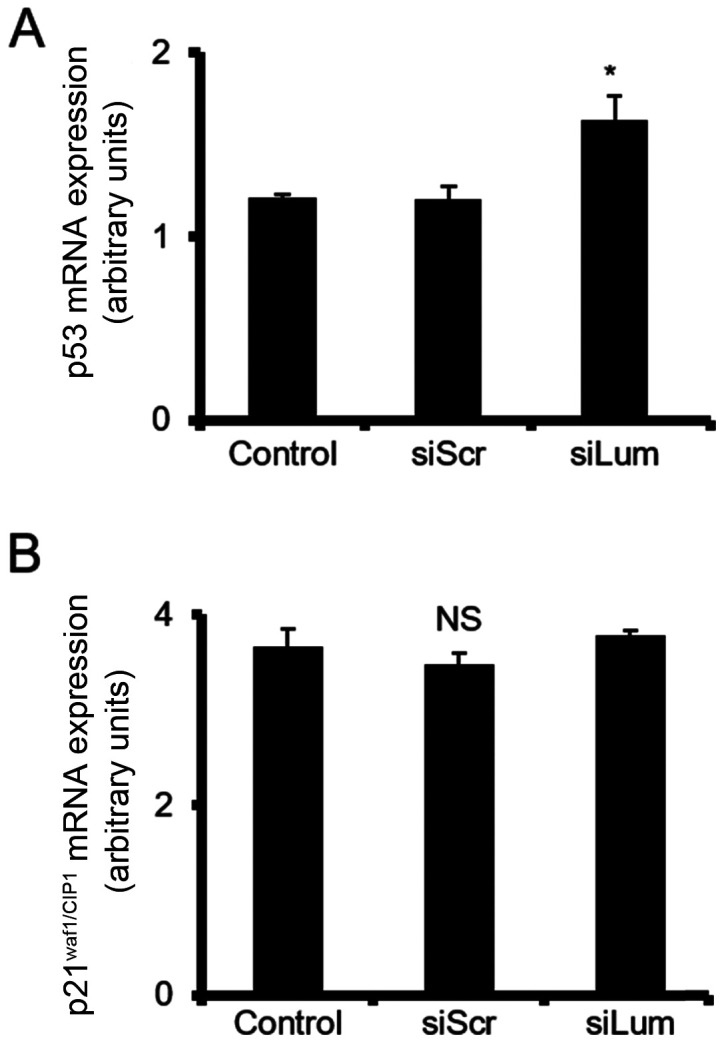
Effect of lumican downregulation on the expression of cellcycle regulators at mRNA levels. HTB94 cells were transfected for 48 h with specific siRNA against lumican. A non-specific RNA sequence was used as a control (siScr). (A) p53 and (B) p21WAF1/CIP1mRNA levels in HTB94 cells were determined by RT-qPCR using primers specific for each gene and normalized against GAPDH. Results represent the average of 3 separate experiments. Data are the means ± SEM; *P≤0.05, statistically significant difference compared with the respective controls; NS, not significant.
TGF-β2 and IGF-I mediate lumican expression in HTB94 chondrosarcoma cells
In separate experiments, the HTB94 cells were treated with TGF-β2 (10 ng/ml) and IGF-I (10 ng/ml), and the expression of lumican in HTB94 cells was estimated. The data obtained by western blot analysis revealed that TGF-β2 decreased (P≤0.001) and IGF-I enhanced (P≤0.01) lumican protein expression (Fig. 11).
Figure 11.
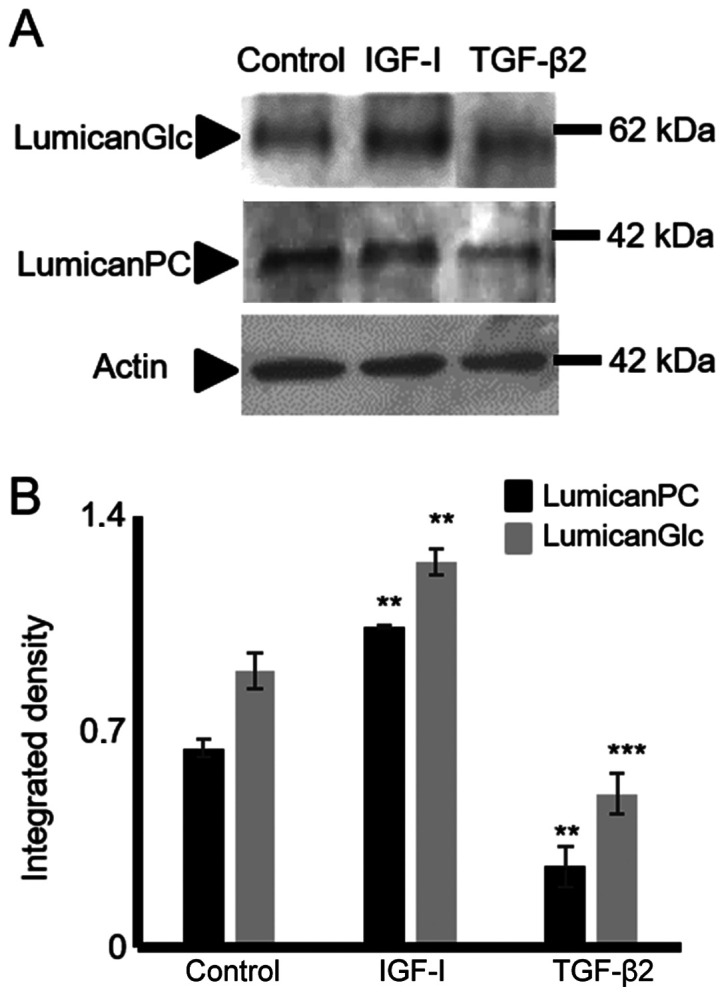
Effect of IGF-I and TGF-β2 on lumican expression in HTB94 cells. Following starvation for 24 h with 0% FBS DMEM, HTB94 cells were treated with IGF-I (10 ng/ml) and TGF-β2 (10 ng/ml) in 0% FBS DMEM for 24 h before harvesting. (A) Representative blots of glycosylated lumican (LumicanGlc) (60 kDa), lumican protein core (lumicanPC) (38 kDa) and actin (42 kDa) are presented. (B) Lumican protein bands were densito-metrically analyzed and adjusted against actin. The position of the nearest respective protein marker band is depicted to the right. Representative plots are presented. Data are the means ± SEM; statistical significance: **P≤0.01 and ***P≤0.001 compared with the control.
Discussion
Alterations in the expression of ECM molecules affect tumor cell functions, but also modify the behavior of stromal cells, exerting tumor-dependent pro-angiogenesis and pro-inflammation effects thus, facilitating the creation of a tumor-derived microenvironment (8). Notably, chondrosarcoma cells exhibit an extensive remodeling of ECM constituents, including hyalectan PGs, collagen fibers, fibronectin and laminin (16). In the present study, the HTB94 human chondrosarcoma cells with a high metastatic capacity were analyzed for decorin, biglycan and lumican expression and possible regulation of cellular function(s) by the respective ECM constituents. The present study demonstrated, as a novel finding, that the chon-drosarcoma cells synthesize and efficiently secrete the class II SLRP, lumican. This SLRP was found to be the most abundantly expressed SLRP, as compared to the low expression levels of decorin and biglycan, in the HTB94 chondrosarcoma model. Both the lumican protein core and its glycosylated forms were found to be secreted. Furthermore, the utilization of a specific anti-KS antibody and digestion with keratanase demonstrated that the secretion oflumican by chondrosarcoma cells was partially glycosylated with KS chains. Indeed, this finding is similar to the glycosylation pattern of lumican secreted by osteosarcoma cells (35).
The putative contribution of lumican has been examined in the growth and metastasis of several types of cancer (6,20,36-38), and tumor-enhancing, as well as tumor-inhibitory functions have been indicated and are dependent on tumor type, and abundance and the stage of the disease (20,39).
Lumican-contingent effects have been found to be partly dependent on the type and extent of lumican glycosylation (6). In the present study, siRNA was utilized to examine the putative role of lumican in chondrosarcoma growth, migration and adhesion. This approach resulted in the efficient downregulation of lumican secretion. The attenuated lumican secretion was associated with the decreased growth of HTB94 cells, whereas their migratory and adhesion ability were not affected. Previous studies have demonstrated that lumican enhances the growth potential of lung cancer (40), bladder cancer (37) and gastric cancer (41), whereas it was found to have inhibitive effects on breast cancer (42) and melanoma growth (43). In continuation, the present study examined the possible mechanisms of lumican action in the regulation of chondrosarcoma growth. The regulation of the TGF-β2 signaling pathway is one previously established mechanism of action of lumican (6,25). In the present study, and in agreement with previous findings, exogenous TGF-β2 was found to decrease chondrosarcoma cell growth (44). Treatment of lumican-deficient cells with TGF-β2 increased HTB94 cell growth inhibition. The pathway restricted Smad2 activation (45) was assessed in lumican and scramble siRNA-transfected cells to examine the potential contribution of lumican to the TGF-B2 signaling pathway. Neither the basal nor the TGF-β2-dependent levels of Smad2 phosphorylation were affected, as exogenously added TGF-β2 enhanced Smad2 phosphorylation to a similar extent in both scramble control-transfected and lumican-deficient cells. These results demonstrate that the effect of lumican on HTB94 cell growth was independent of TGF signaling.
Decorin and biglycan have previously been implicated in the regulation of IGF/IGF-IR signaling (46), which is a crucial player in the regulation of both physiological growth (47,48) and malignant transformation (46). Recently, a 'Phase I trial of the IGF-1R antibody ganitumab (AMG 479) in combination with everolimus (RAD001) and panitumumab in patients with advanced cancer' exhibited benefits in the case of chondrosar-coma patients (49). On the other hand, in a separate study, 'no preclinical rationale for IGF1R directed therapy in chondro-sarcoma of bone' was indicated (50).
Importantly, decorin has been shown to bind IGF-IR and attenuate its signaling (51), whereas we have recently demonstrated that biglycan modulates the growth of MG63 osteosarcoma cells through an LPR6/β-catenin/IGFR-IR signaling axis (52). Upon activation of the upstream effectors, the degradation of β-catenin is inhibited, resulting in the increased expression in the cytoplasm and the enhanced trans-location to the nucleus (52). No changes in the total β-catenin expression in lumican-deficient HTB94 cells were detected in the present study, suggesting that the effects of lumican did not involve β-catenin signaling. The present study demonstrated that the IGF-I/IGF-IR axis positively modulated HTB94 cell growth, similar to previous findings (53). Notably, lumican deficiency induced an abolishment of IGF-I-dependent HTB94 cell growth. It should be noted that the lumican-deficient cells exhibited an attenuation of IGF-IR basal level activation, whereas no effect on the expression levels of the receptor was evident. Addressing the putative contribution of β-catenin signaling on lumican/IGF-IR-mediated growth effects revealed that β-catenin was not a downstream regulator. ERK1/2 is an established downstream effector of IGF-IR signaling (54). Indeed, the IGF-IR/ERK1/2 signaling axis has been implicated in the propagation of fibrosarcoma migration (34) and breast cancer adhesion (27), as well as adrenocortical cancer cell proliferation (55). In the present study, ERK1/2 was verified as an IGF-IR downstream mediator, as well as a necessary component in IGF-IR-dependent facilitation of HTB94 cell growth. The downregulation of lumican induced a substantial attenuation of ERK1/2 activation, indicating that ERK1/2 is a necessary component of lumican/IGF-IR-mediated HTB94 cell proliferation. Lumican-deficient cells did not exhibit the further downregulation of cell growth when treated with the ERK1/2 inhibitor, indicating that ERK1/2 participation was necessary for the lumican effect. Previously it has been suggested that ERK1/2 exerts pro-oncogenic effects in chon-drosarcoma cells (56,57).
Moreover, ERK1/2, along with PI3K and p38 signaling pathways, has been shown to promote chondrosarcoma cell motility, invasion and lung colonization (58). The ERK1/2 pathway affects cell cycle progression and apoptosis through the modulation of specific cell cycle regulators expression, including p53 and p21 in OVCAR-3 human ovarian and MCF-7 breast cancer cells (59,60). In the present study, p53 expression was elevated in the HTB94 cells, in a manner dependent on active ERK1/2 signaling. The fold change in p53 expression was not marked, yet it was significant, and it is suggested that p53 is one of the downstream targets of the determined signaling pathway. Importantly, p53 has been implicated in the progression of chondrosarcoma to a higher grade (61) and was suggested, among others, as a novel therapeutic target (62).
At this point, the mechanisms through which lumican enhances IGF-IR activation remain obscure. Taking into account that lumican is a secreted protein, it is likely to engage extracellularly either with IGF-IR or other components of the IGF system (63). Previously, another SLRP member, decorin, was shown to bind and to either enhance (64) or attenuate (46) IGF-IR activation in different models. The results of the present study demonstrate for the first time, to the best of our knowledge, that lumican can affect IGF-IR activation. Of note, IGF-I was shown to enhance lumican secretion, suggesting a feedback loop supporting chondrosarcoma growth. Therefore, further studies on the mechanisms through which lumican enhances IGF-IR activation are required. Furthermore, the utilization of a single cell line is a limitation of the present study, justified by the scarcity of data on chondrosarcomas. Thus, an assessment of the proposed mechanism in other model systems, including extrapolation to in vivo experimentation, will be the focus of a future study.
In the present study, the class I SLRPs decorin and biglycan were expressed at low levels at both the mRNA and protein level. In two previously available studies on decorin and biglycan expression in chondrosarcoma, decorin mRNA was isolated in some chondrosarcoma samples (23), whereas no decorin protein and low biglycan protein was detected in a human chondrosarcoma cell line (24). In the present study, the low levels of HTB94 cell decorin and biglycan expression were inadequate to affect cell growth, migration and adhesion. To the archetypal SLRP member decorin, a tumor-suppressive effect has been designated, and this SLRP is not expressed by the majority of tumors, which is in accordance with the findings of the present study (65,66).
The role of biglycan in carcinogenesis is not clear, with the majority of studies suggesting that biglycan overexpression in the tumor microenvironment facilitates cancer cell growth, migration and angiogenesis by regulating downstream intracellular signaling and biglycan-dependent modifications of the ECM milieu (50,67-70).
In conclusion, in the present study, human chondrosarcoma cells were shown to express the SLRPs lumican, decorin and biglycan. Lumican was identified as the major secreted SLRP, whereas decorin and biglycan exhibited low levels of expression. The secreted lumican was shown to be partly substituted with KS glycosaminoglycans. Importantly, HTB94 cell growth was enhanced, whereas migration and adhesion were not affected by lumican. Basal IGF-IR and IGF-IR-dependent cell growth and phosphorylation levels were positively associated with lumican expression, suggesting that this SLRP may affect the activation of IGF-IR. The downregulation of lumican induced a substantial attenuation of the IGF-IR downstream mediator, ERK1/2, activation, indicating that ERK1/2 is a necessary component of lumican/IGF-IR-mediated HTB94 cell growth. Moreover, lumican-deficient cells specifically exhibited increased mRNA levels of p53, suggesting that lumican facilitated HTB94 cell growth through an IGF-IR/ERK1/2/p53 signaling cascade. Further studies on the mechanistic aspects of lumican/ IGF-IR interactions in chondrosarcoma and the association between lumican expression and chondrosarcoma progression are essential.
Supplementary Data
Acknowledgments
Not applicable.
Abbreviations
- SLRPs
small leucine-rich proteoglycans
- ECM
extracellular matrix
- IGF-I
insulin-like growth factor I
- IGF-IR
insulin-like growth factor I receptor
- ERK1/2
extracellular regulated kinase 1/2
- TGF-β2
transforming growth factor-β2
- siRNA
short interfering RNA
- PG
proteoglycan
- KS
keratan sulfate
Funding
The present study was supported by a Special Fund for Research Grants (ELKE) with grant no. KA: 10028 of the University of Crete to DN.
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
AP and DN designed the outline of the study. AP, EMG, AB and IS conducted the experiments, and contributed to data interpretation and manuscript preparation. AP wrote the manuscript. DAS, AT and GNT contributed to data interpretation, and manuscript preparation and revision. DN supervised the study, contributed to data interpretation and manuscript preparation/revision. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article. The other authors declare that they have no competing interests.
References
- 1.International Agency for Research on Cancer (IARC) In: WHO Classification of Tumours of Soft Tissue and Bone. 4th edition. Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, editors. Vol. 5. IARC; Lyon: 2013. [Google Scholar]
- 2.Heck RK, Jr, Peabody TD, Simon MA. Staging of primary malignancies of bone. CA Cancer J Clin. 2006;56:366–375. doi: 10.3322/canjclin.56.6.366. [DOI] [PubMed] [Google Scholar]
- 3.Chow WA. Update on chondrosarcomas. Curr Opin Oncol. 2007;19:371–376. doi: 10.1097/CCO.0b013e32812143d9. [DOI] [PubMed] [Google Scholar]
- 4.Tsuda Y, Ogura K, Hakozaki M, Kikuta K, Ae K, Tsuchiya H, Iwata S, Ueda T, Kawano H, Kawai A. Mesenchymal chon-drosarcoma: A Japanese Musculoskeletal Oncology Group (JMOG) study on 57 patients. J Surg Oncol. 2017;115:760–767. doi: 10.1002/jso.24567. [DOI] [PubMed] [Google Scholar]
- 5.Theocharis AD, Manou D, Karamanos NK. The extracellular matrix as a multitasking player in disease. FEBS J. 2019;286:2830–2869. doi: 10.1111/febs.14818. [DOI] [PubMed] [Google Scholar]
- 6.Nikitovic D, Papoutsidakis A, Karamanos NK, Tzanakakis GN. Lumican affects tumor cell functions, tumor-ECM interactions, angiogenesis and inflammatory response. Matrix Biol. 2014;35:206–214. doi: 10.1016/j.matbio.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Varol C, Sagi I. Phagocyte-extracellular matrix crosstalk empowers tumor development and dissemination. FEBS J. 2018;285:734–751. doi: 10.1111/febs.14317. [DOI] [PubMed] [Google Scholar]
- 8.Yuzhalin AE, Lim SY, Kutikhin AG, Gordon-Weeks AN. Dynamic matrisome: ECM remodeling factors licensing cancer progression and metastasis. Biochim Biophys Acta Rev Cancer. 2018;1870:207–228. doi: 10.1016/j.bbcan.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neill T, Schaefer L, Iozzo RV. Decoding the Matrix: Instructive Roles of Proteoglycan Receptors. Biochemistry. 2015;54:4583–4598. doi: 10.1021/acs.biochem.5b00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karamanou K, Perrot G, Maquart FX, Brézillon S. Lumican as a multivalent effector in wound healing. Adv Drug Deliv Rev. 2018;129:344–351. doi: 10.1016/j.addr.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Nikitovic D, Aggelidakis J, Young MF, Iozzo RV, Karamanos NK, Tzanakakis GN. The biology of small leucine-rich proteoglycans in bone pathophysiology. J Biol Chem. 2012;287:33926–33933. doi: 10.1074/jbc.R112.379602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Birk DE. The regulatory roles of small leucine-rich proteoglycans in extracellular matrix assembly. FEBS J. 2013;280:2120–2137. doi: 10.1111/febs.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaefer L, Iozzo RV. Biological functions of the small leucine-rich proteoglycans: From genetics to signal transduction. J Biol Chem. 2008;283:21305–21309. doi: 10.1074/jbc.R800020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merline R, Schaefer RM, Schaefer L. The matricellular functions of small leucine-rich proteoglycans (SLRPs) J Cell Commun Signal. 2009;3:323–335. doi: 10.1007/s12079-009-0066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grogan SP, Chen X, Sovani S, Taniguchi N, Colwell CW, Jr, Lotz MK, D'Lima DD. Influence of cartilage extracellular matrix molecules on cell phenotype and neocartilage formation. Tissue Eng Part A. 2014;20:264–274. doi: 10.1089/ten.tea.2012.0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis JL, Krawczak DA, Oegema TR, Jr, Westendorf JJ. Effect of decorin and dermatan sulfate on the mechanical properties of a neocartilage. Connect Tissue Res. 2010;51:159–170. doi: 10.3109/03008200903174342. [DOI] [PubMed] [Google Scholar]
- 18.Burdan F, Szumiło J, Korobowicz A, Farooquee R, Patel S, Patel A, Dave A, Szumiło M, Solecki M, Klepacz R, et al. Morphology and physiology of the epiphyseal growth plate. Folia Histochem Cytobiol. 2009;47:5–16. doi: 10.2478/v10042-009-0007-1. [DOI] [PubMed] [Google Scholar]
- 19.Melrose J, Shu C, Whitelock JM, Lord MS. The cartilage extracellular matrix as a transient developmental scaffold for growth plate maturation. Matrix Biol. 2016;52-54:363–383. doi: 10.1016/j.matbio.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Appunni S, Anand V, Khandelwal M, Gupta N, Rubens M, Sharma A. Small Leucine Rich Proteoglycans (decorin, biglycan and lumican) in cancer. Clin Chim Acta. 2019;491:1–7. doi: 10.1016/j.cca.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Xu L, Li Z, Liu SY, Xu SY, Ni GX. Asporin and osteoarthritis. Osteoarthritis Cartilage. 2015;23:933–939. doi: 10.1016/j.joca.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Ni GX, Li Z, Zhou YZ. The role of small leucine-rich proteoglycans in osteoarthritis pathogenesis. Osteoarthritis Cartilage. 2014;22:896–903. doi: 10.1016/j.joca.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Söderström M, Böhling T, Ekfors T, Nelimarkka L, Aro HT, Vuorio E. Molecular profiling of human chondrosarcomas for matrix production and cancer markers. Int J Cancer. 2002;100:144–151. doi: 10.1002/ijc.10457. [DOI] [PubMed] [Google Scholar]
- 24.Chansky H, Robbins JR, Cha S, Raskind WH, Conrad EU, Sandell LJ. Expression of cartilage extracellular matrix and potential regulatory genes in a new human chondrosarcoma cell line. J Orthop Res. 1998;16:521–530. doi: 10.1002/jor.1100160502. [DOI] [PubMed] [Google Scholar]
- 25.Nikitovic D, Chalkiadaki G, Berdiaki A, Aggelidakis J, Katonis P, Karamanos NK, Tzanakakis GN. Lumican regulates osteosarcoma cell adhesion by modulating TGFβ2 activity. Int J Biochem Cell Biol. 2011;43:928–935. doi: 10.1016/j.biocel.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Berdiaki A, Datsis G, Nikitovic D, Tsatsakis A, Katonis P, Karamanos N, Tzanakakis G. Parathyroid hormone (PTH) peptides through the regulation of hyaluronan metabolism affect osteosarcoma cell migration. IUBMB Life. 2010;62:377–386. doi: 10.1002/iub.320. [DOI] [PubMed] [Google Scholar]
- 27.Voudouri K, Nikitovic D, Berdiaki A, Kletsas D, Karamanos NK, Tzanakakis GN. IGF-I/EGF and E2 signaling crosstalk through IGF-IR conduit point affects breast cancer cell adhesion. Matrix Biol. 2016;56:95–113. doi: 10.1016/j.matbio.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Im GI, Jung NH, Tae SK. Chondrogenic differentiation of mesenchymal stem cells isolated from patients in late adulthood: The optimal conditions of growth factors. Tissue Eng. 2006;12:527–536. doi: 10.1089/ten.2006.12.527. [DOI] [PubMed] [Google Scholar]
- 29.Khaghani SAB, Akbarova G, Soon CF, Dilbazi G. Effect of transforming growth factor-β2 on biological regulation of multilayer primary chondrocyte culture. Cell Tissue Bank. 2018;19:763–775. doi: 10.1007/s10561-018-9732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boumédiene K, Takigawa M, Pujol JP. Cell density-dependent proliferative effects of transforming growth factor (TGF)-β 1, β 2, and β 3 in human chondrosarcoma cells HCS-2/8 are associated with changes in the expression of TGF-β receptor type I. Cancer Invest. 2001;19:475–486. doi: 10.1081/CNV-100103846. [DOI] [PubMed] [Google Scholar]
- 31.Boeuf S, Bovée JV, Lehner B, van den Akker B, van Ruler M, Cleton-Jansen AM, Richter W. BMP and TGFbeta pathways in human central chondrosarcoma: Enhanced endoglin and Smad 1 signaling in high grade tumors. BMC Cancer. 2012;12:488. doi: 10.1186/1471-2407-12-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Luca F. Regulatory Role for Growth Hormone in Statural Growth: IGF-Dependent and IGF-Independent Effects on Growth Plate Chondrogenesis and Longitudinal Bone Growth. Pediatr Endocrinol Rev. 2018;16(Suppl 1):33–38. doi: 10.17458/per.vol16.2018.l.igfeffectschondrogenesis. [DOI] [PubMed] [Google Scholar]
- 33.Hiraoka K, Zenmyo M, Komiya S, Kawabata R, Yokouchi M, Suzuki R, Hamada T, Kato S, Nagata K. Relationship of p21 (waf1/cip1) and differentiation in chondrosarcoma cells. Virchows Arch. 2002;440:285–290. doi: 10.1007/s004280100495. [DOI] [PubMed] [Google Scholar]
- 34.Mytilinaiou M, Nikitovic D, Berdiaki A, Papoutsidakis A, Papachristou DJ, Tsatsakis A, Tzanakakis GN. IGF-I regulates HT1080 fibrosarcoma cell migration through a syndecan-2/Erk/ezrin signaling axis. Exp Cell Res. 2017;361:9–18. doi: 10.1016/j.yexcr.2017.09.035. [DOI] [PubMed] [Google Scholar]
- 35.Nikitovic D, Berdiaki A, Zafiropoulos A, Katonis P, Tsatsakis A, Karamanos NK, Tzanakakis GN. Lumican expression is positively correlated with the differentiation and negatively with the growth of human osteosarcoma cells. FEBS J. 2008;275:350–361. doi: 10.1111/j.1742-4658.2007.06205.x. [DOI] [PubMed] [Google Scholar]
- 36.Coulson-Thomas VJ, Coulson-Thomas YM, Gesteira TF, Andrade de Paula CA, Carneiro CR, Ortiz V, Toma L, Kao WW, Nader HB. Lumican expression, localization and antitumor activity in prostate cancer. Exp Cell Res. 2013;319:967–981. doi: 10.1016/j.yexcr.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao W, Luo M, Huang X, Wang Q, Fan J, Gao L, Zhang Y, Geng J. Knockdown of lumican inhibits proliferation and migration of bladder cancer. Transl Oncol. 2019;12:1072–1078. doi: 10.1016/j.tranon.2019.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L, Zhang Y, Zuo Y, Ma F, Song H. Lumican expression in gastric cancer and its association with biological behavior and prognosis. Oncol Lett. 2017;14:5235–5240. doi: 10.3892/ol.2017.6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsiao KC, Chu PY, Chang GC, Liu KJ. Elevated Expression of Lumican in Lung Cancer Cells Promotes Bone Metastasis through an Autocrine Regulatory Mechanism. Cancers (Basel) 2020;12:233. doi: 10.3390/cancers12010233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang CT, Hsu PC, Chow SE. Downregulation of lumican enhanced mitotic defects and aneuploidy in lung cancer cells. Cell Cycle. 2020;19:97–108. doi: 10.1080/15384101.2019.1693189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Li X, Hu X, Jiang F, Shen Y, Xu R, Wu L, Wei P, Shen X. LUM Expression and Its Prognostic Significance in Gastric Cancer. Front Oncol. 2020;10:605. doi: 10.3389/fonc.2020.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karamanou K, Franchi M, Piperigkou Z, Perreau C, Maquart FX, Vynios DH, Brézillon S. Lumican effectively regulates the estrogen receptors-associated functional properties of breast cancer cells, expression of matrix effectors and epithelial-to-mesenchymal transition. Sci Rep. 2017;7:45138. doi: 10.1038/srep45138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeanne A, Untereiner V, Perreau C, Proult I, Gobinet C, Boulagnon-Rombi C, Terryn C, Martiny L, Brézillon S, Dedieu S. Lumican delays melanoma growth in mice and drives tumor molecular assembly as well as response to matrix-targeted TAX2 therapeutic peptide. Sci Rep. 2017;7:7700. doi: 10.1038/s41598-017-07043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahimi RA, Leof EB. TGF-β signaling: A tale of two responses. J Cell Biochem. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- 45.Herpin A, Cunningham C. Cross-talk between the bone morphogenetic protein pathway and other major signaling pathways results in tightly regulated cell-specific outcomes. FEBS J. 2007;274:2977–2985. doi: 10.1111/j.1742-4658.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- 46.Iozzo RV, Buraschi S, Genua M, Xu SQ, Solomides CC, Peiper SC, Gomella LG, Owens RC, Morrione A. Decorin antagonizes IGF receptor I (IGF-IR) function by interfering with IGF-IR activity and attenuating downstream signaling. J Biol Chem. 2011;286:34712–34721. doi: 10.1074/jbc.M111.262766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. doi: 10.1016/S0092-8674(05)80085-6. [DOI] [PubMed] [Google Scholar]
- 48.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 49.Vlahovic G, Meadows KL, Hatch AJ, Jia J, Nixon AB, Uronis HE, Morse MA, Selim MA, Crawford J, Riedel RF, et al. A Phase I Trial of the IGF-1R Antibody Ganitumab (AMG 479) in Combination with Everolimus (RAD001) and Panitumumab in Patients with Advanced Cancer. Oncologist. 2018;23:782–790. doi: 10.1634/theoncologist.2016-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterse EF, Cleven AH, De Jong Y, Briaire-de Bruijn I, Fletcher JA, Danen EH, Cleton-Jansen AM, Bovée JV. No preclinical rationale for IGF1R directed therapy in chondro-sarcoma of bone. BMC Cancer. 2016;16:475. doi: 10.1186/s12885-016-2522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schönherr E, Sunderkötter C, Iozzo RV, Schaefer L. Decorin, a novel player in the insulin-like growth factor system. J Biol Chem. 2005;280:15767–15772. doi: 10.1074/jbc.M500451200. [DOI] [PubMed] [Google Scholar]
- 52.Aggelidakis J, Berdiaki A, Nikitovic D, Papoutsidakis A, Papachristou DJ, Tsatsakis AM, Tzanakakis GN. Biglycan regulates MG63 osteosarcoma cell growth through a LPR6/ β-catenin/IGFR-IR signaling axis. Front Oncol. 2018;8:470. doi: 10.3389/fonc.2018.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsumura T, Whelan MC, Li XQ, Trippel SB. Regulation by IGF-I and TGF-beta1 of Swarm-rat chondrosarcoma chon-drocytes. J Orthop Res. 2000;18:351–355. doi: 10.1002/jor.1100180305. [DOI] [PubMed] [Google Scholar]
- 54.Metalli D, Lovat F, Tripodi F, Genua M, Xu SQ, Spinelli M, Alberghina L, Vanoni M, Baffa R, Gomella LG, et al. The insulin-like growth factor receptor I promotes motility and invasion of bladder cancer cells through Akt- and mitogen-activated protein kinase-dependent activation of paxillin. Am J Pathol. 2010;176:2997–3006. doi: 10.2353/ajpath.2010.090904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cantini G, Lombardi A, Piscitelli E, Poli G, Ceni E, Marchiani S, Ercolino T, Galli A, Serio M, Mannelli M, et al. Rosiglitazone inhibits adrenocortical cancer cell proliferation by interfering with the IGF-IR intracellular signaling. PPAR Res. 2008;2008:904041. doi: 10.1155/2008/904041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin J, Shaukat I, Mainard D, Netter P, Barré L, Ouzzine M. Constitutive activation of EGFR is associated with tumor progression and plays a prominent role in malignant phenotype of chondrosarcoma. Oncotarget. 2019;10:3166–3182. doi: 10.18632/oncotarget.26899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamemura N, Murakami S, Komatsu H, Sawanoi M, Miyamoto K, Ishidoh K, Kishimoto K, Tsuji A, Yuasa K. Type II cGMP-dependent protein kinase negatively regulates fibroblast growth factor signaling by phosphorylating Raf-1 at serine 43 in rat chondrosarcoma cells. Biochem Biophys Res Commun. 2017;483:82–87. doi: 10.1016/j.bbrc.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 58.Guan PP, Yu X, Guo JJ, Wang Y, Wang T, Li JY, Konstantopoulos K, Wang ZY, Wang P. By activating matrix metalloproteinase-7, shear stress promotes chondrosarcoma cell motility, invasion and lung colonization. Oncotarget. 2015;6:9140–9159. doi: 10.18632/oncotarget.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee KS, Kim SW, Lee HS. Orostachys japonicus induce p53-dependent cell cycle arrest through the MAPK signaling pathway in OVCAR-3 human ovarian cancer cells. Food Sci Nutr. 2018;6:2395–2401. doi: 10.1002/fsn3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boroumand Moghaddam A, Moniri M, Azizi S, Abdul Rahim R, Bin Ariff A, Navaderi M, Mohamad R. Eco-Friendly Formulated Zinc Oxide Nanoparticles: Induction of Cell Cycle Arrest and Apoptosis in the MCF-7 Cancer Cell Line. Genes (Basel) 2017;8:281. doi: 10.3390/genes8100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dai X, Ma W, He X, Jha RK. Review of therapeutic strategies for osteosarcoma, chondrosarcoma, and Ewing's sarcoma. Med Sci Monit. 2011;17:RA177–RA190. doi: 10.12659/MSM.881893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mery B, Espenel S, Guy JB, Rancoule C, Vallard A, Aloy MT, Rodriguez-Lafrasse C, Magné N. Biological aspects of chon-drosarcoma: Leaps and hurdles. Crit Rev Oncol Hematol. 2018;126:32–36. doi: 10.1016/j.critrevonc.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 63.Philippou A, Christopoulos PF, Koutsilieris DM. Clinical studies in humans targeting the various components of the IGF system show lack of efficacy in the treatment of cancer. Mutat Res Rev Mutat Res. 2017;772:105–122. doi: 10.1016/j.mrrev.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 64.Fiedler LR, Schönherr E, Waddington R, Niland S, Seidler DG, Aeschlimann D, Eble JA. Decorin regulates endothelial cell motility on collagen I through activation of insulin-like growth factor I receptor and modulation of α2β1 integrin activity. J Biol Chem. 2008;283:17406–17415. doi: 10.1074/jbc.M710025200. [DOI] [PubMed] [Google Scholar]
- 65.Santra M, Eichstetter I, Iozzo RV. An anti-oncogenic role for decorin. Down-regulation of ErbB2 leads to growth suppression and cytodifferentiation of mammary carcinoma cells. J Biol Chem. 2000;275:35153–35161. doi: 10.1074/jbc.M006821200. [DOI] [PubMed] [Google Scholar]
- 66.Zhang W, Ge Y, Cheng Q, Zhang Q, Fang L, Zheng J. Decorin is a pivotal effector in the extracellular matrix and tumour micro-environment. Oncotarget. 2018;9:5480–5491. doi: 10.18632/oncotarget.23869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xing X, Gu X, Ma T. Knockdown of biglycan expression by RNA interference inhibits the proliferation and invasion of, and induces apoptosis in, the HCT116 colon cancer cell line. Mol Med Rep. 2015;12:7538–7544. doi: 10.3892/mmr.2015.4383. [DOI] [PubMed] [Google Scholar]
- 68.Schaefer L, Tredup C, Gubbiotti MA, Iozzo RV. Proteoglycan neofunctions: Regulation of inflammation and autophagy in cancer biology. FEBS J. 2017;284:10–26. doi: 10.1111/febs.13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu B, Xu T, Xu X, Cui Y, Xing X. Biglycan promotes the chemotherapy resistance of colon cancer by activating NF-κB signal transduction. Mol Cell Biochem. 2018;449:285–294. doi: 10.1007/s11010-018-3365-1. [DOI] [PubMed] [Google Scholar]
- 70.Schulz GB, Grimm T, Sers C, Riemer P, Elmasry M, Kirchner T, Stief CG, Karl A, Horst D. Prognostic value and association with epithelial-mesenchymal transition and molecular subtypes of the proteoglycan biglycan in advanced bladder cancer. Urol Oncol. 2019;37:530.e9–530.e18. doi: 10.1016/j.urolonc.2019.05.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.