Abstract
The function of Dicer’s helicase domain has been enigmatic since its discovery. Why do only some Dicers require ATP, despite a high degree of sequence conservation in their helicase domains? We discuss evolutionary considerations based on differences between vertebrate and invertebrate antiviral defense, and how the helicase domain has been co-opted in extant organisms as the binding site for accessory proteins. Many accessory proteins are double-stranded RNA binding proteins, and we propose models for how they modulate Dicer function and catalysis.
The gene we now know as Dicer was first revealed in a screen for double-stranded RNA (dsRNA) binding proteins (dsRBPs) (Bass et al. 1994) before the discovery of the surprising phenomena of RNA interference (RNAi) (Fire et al. 1998). Once RNAi was discovered, the unique features of the mysterious gene, which included an amino-terminal helicase domain and tandem ribonuclease III (RNase III) domains (Fig. 1A), led to the proposal that it was key to the process (Bass 2000). Indeed, subsequent studies in Drosophila melanogaster S2 cells (Bernstein et al. 2001) and Caenorhabditis elegans (Knight and Bass 2001) confirmed that Dicer was key to RNAi.
Figure 1.
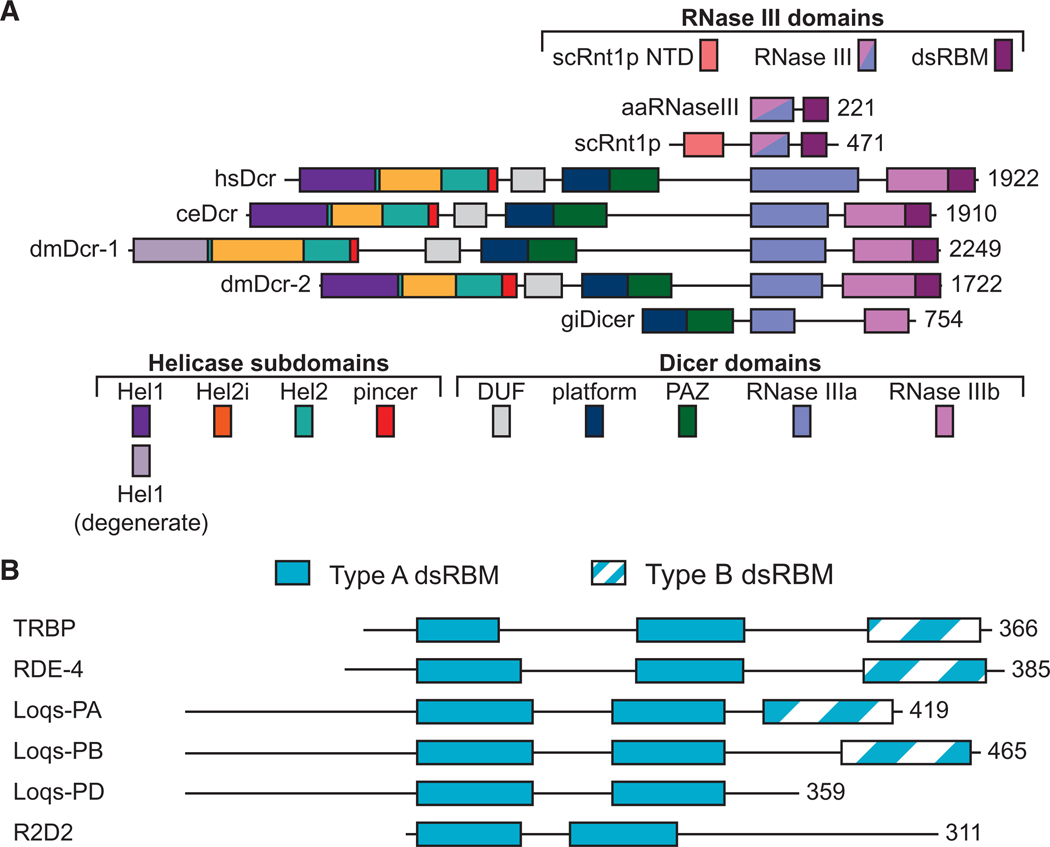
(A) Domains of Dicer and RNase III open-reading frames including Aquifex aeolicus RNase III (aaRNaseIII), Saccharomyces cerevisiae Rnt1p (scRnt1p), Homo sapiens Dicer (hsDcr), Caenorhabditis elegans Dicer (ceDcr), Drosophila melanogaster Dicers (dmDcr-1 and dmDcr-2), and Giardia intestinalis Dicer (giDicer). Domains are to scale and anchored at the amino terminus of aaRNase III, with the carboxy-terminal amino acid numbered on the right. (Top) color coding of RNase III domains, including amino-terminal domain (NTD). (Bottom) Color coding of helicase subdomains and Dicer domains. (B) Domain organization of Dicer-interacting dsRBPs noting Type A (solid) and Type B (striped) double-stranded RNA binding motifs (dsRBMs). Open-reading frames are to scale and anchored by the amino terminus of the first dsRBM. Numbers at the far right indicate the carboxy-terminal amino acid.
The observed RNase III domains placed Dicer in a well-characterized family of endoribonucleases, first discovered in Escherichia coli (Robertson et al. 1968) and now known to exist in Bacteria and Eukarya, but only infrequently in Archaea (Court et al. 2013; Nicholson 2014). Because RNAi had been correlated with the cleavage of dsRNA into smaller pieces (Zamore et al. 2000), the function of the RNase III domains was immediately predictable, and we now know that they catalyze cleavage of dsRNA into microRNAs (miRNAs) and short interfering RNAs (siRNAs) (Wilson and Doudna 2013; Ha and Kim 2014). In contrast to the RNase III domains, the function of the amino-terminal helicase domain was enigmatic when Dicer was discovered and, in many respects, remains so today. However, in recent years progress has been made in understanding this domain, and one goal of this review is to summarize recent insights.
Soon after the discovery of RNAi, it was recognized that, in plants, the mysterious phenomena of gene silencing in response to transgene expression and of cosuppression involved dsRNA and were based in an antiviral defense pathway (Waterhouse et al. 2001; Ding and Voinnet 2007). In addition, studies in invertebrates showed that RNAi targeted transposons (Ketting et al. 1999), remnants of ancient viruses. These observations paved the way for the realization that Dicer’s helicase domain is most similar to helicases that act as viral sensors in mammalian innate immunity, the RIG-I-like receptors (RLRs; e.g., RIG-I, MDA5, LGP2) (Fairman-Williams et al. 2010; Ahmad and Hur 2015), and the demonstration for invertebrate Dicers that mutations in the helicase domain increase susceptibility to viral infection (Deddouche et al. 2008; Marques et al. 2013).
Here we take the view that the ancestral function of the helicase domain was in antiviral defense, and in certain extant organisms this domain has been co-opted for other cellular functions, such as regulation of miRNA processing, and production of endogenous siRNAs (endosiRNAs). These modern-day functions often require cellular dsRNA binding proteins (dsRBPs) that directly interact with the helicase domain to modulate its functions. So far, these accessory dsRBPs all contain multiple dsRNA binding motifs (dsRBMs; Fig. 1B): an ~65-amino acid motif that folds into an αβββα topology (Masliah et al. 2013; Gleghorn and Maquat 2014). Although there are many enzymes that include dsRBMs in conjunction with catalytic domains, such as adenosine deaminases that act on RNA (ADARs), RNA-dependent protein kinase (PKR), Dicer, and most RNase III enzymes, others simply consist of multiple dsRBMs separated by linkers. For the most part, it is the latter that have been found to interact with Dicer, and these will be our main focus. Each of these dsRBPs contains two amino-terminal “Type A” dsRBMs, which are more conserved and competent to bind dsRNA (Gleghorn and Maquat 2014). As indicated (Fig. 1B), some of these proteins also contain a carboxy-terminal “Type B” dsRBM, divergent motifs that frequently lack residues important for binding dsRNA, display little or no affinity for dsRNA, and typically mediate protein–protein interactions (Gleghorn and Maquat 2014).
THE HELICASE DOMAINS OF VERTEBRATE AND INVERTEBRATE DICERS HAVE DISTINCTACTIVITIES
Like other RLRs, Dicer’s helicase domain belongs to the Superfamily 2 group of helicases and is comprised of three subdomains: Hel1, Hel2i, and Hel2 (Fig. 1A; Fairman-Williams et al. 2010; Luo et al. 2013; Ahmad and Hur 2015). Helicase domains have diverse functions, including the unwinding of nucleic acid double helices, promoting conformational changes, and translocation along nucleic acids (Singleton et al. 2007; Jarmoskaite and Russell 2014; Ahmad and Hur 2015). These functions are all fueled by ATP hydrolysis, and indeed, initial studies in D. melanogaster and C. elegans extracts showed a clear requirement for ATP (Zamore et al. 2000; Ketting et al. 2001). Thus, it came as a surprise when Homo sapiens Dicer (hsDcr) was purified to homogeneity and found to cleave dsRNA in the complete absence of ATP (Provost et al. 2002; Zhang et al. 2002).
Indeed, so far, a vertebrate Dicer has never been observed to require ATP. In contrast, there are multiple examples of invertebrate Dicers, those of plants, and that of Schizosaccharomyces pombe that show an ATP requirement in vitro (Liu et al. 2003; Colmenares et al. 2007; Welker et al. 2011; Fukudome and Fukuhara 2017). Further, a mutation in the helicase Walker A motif of D. melanogaster Dicer-2 (dmDcr-2), a motif critical for ATP binding and hydrolysis, eliminates processing of siRNAs, establishing an ATP dependence in vivo (Lee et al. 2004). Similarly, point mutations in any of three different helicase motifs in another invertebrate Dicer, C. elegans DCR-1 (ceDCR-1), produced animals that were defective for processing endo-siRNAs (Welker et al. 2010).
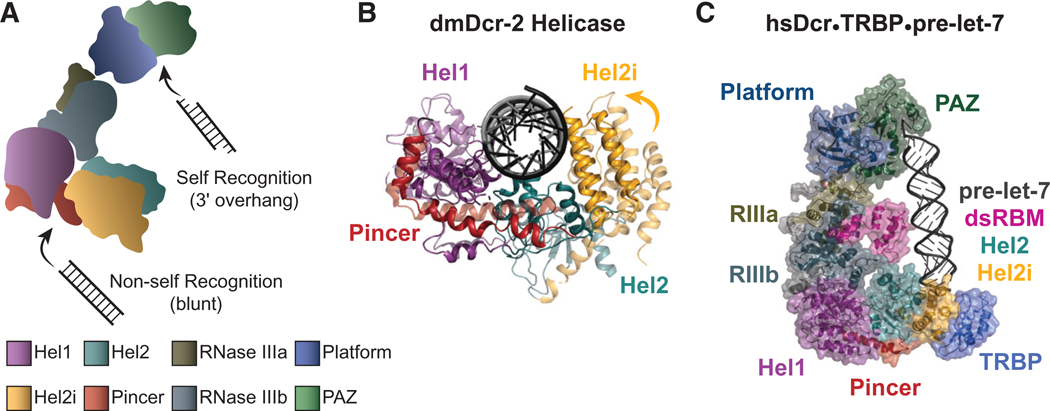
An important clue to understanding the function of Dicer’s helicase domain came with the observation that, although C. elegans with mutations in the helicase domain of ceDCR-1 were defective for processing endo-siRNAs, these animals contained normal levels of miRNAs (Pavelec et al. 2009; Welker et al. 2010). Although termini of endo-siRNA precursors are not well characterized, miRNA precursors have a distinct 3′ overhang, and, thus, the C. elegans studies led to in vitro studies with purified dmDcr-2 and C. elegans extracts that showed these invertebrate Dicers cleave dsRNA differently depending on its termini (Welker et al. 2011; Sinha et al. 2015). When dmDcr-2 encounters a dsRNA with a blunt terminus, an optimal reaction ensues, whereby the helicase domain enables ATP-dependent, processive cleavage of the dsRNA. In contrast, dsRNA with 3′-overhanging termini promote a suboptimal, distributive cleavage that does not require ATP. The different termini-dependent reaction modes were confusing until a cryo-electron microscopy (cryo-EM) study of dmDcr-2 was reported (Sinha et al. 2018). Although numerous studies had implicated the Platform•PAZ domains in mediating Dicer’s recognition of dsRNA termini (Fig. 1A), the cryo-EM study showed that dsRNA with blunt termini binds to the helicase domain (Fig. 2A). Because viral dsRNA in some cases has a blunt terminus (Schlee 2013), this led to the simple model that the helicase domain recognizes non-self dsRNA, such as viral dsRNA, threading it through the helicase domain to processively cleave the dsRNA, whereas the Platform•PAZ domains recognizes termini of cellular, or “self,” dsRNA, such as miRNAs, acting distributively to make a single dsRNA cleavage with each binding event.
Figure 2.
(A) A cartoon illustrating that dmDcr-2 uses its Platform•PAZ domains to recognize substrates with 3′ overhangs (self) and the helicase domain to recognize dsRNA with blunt termini (non-self). (Bottom) Color coding of domains, used throughout the figure. (B) Superimposition of dmDcr-2 helicase conformations shows an open, C-shaped, conformation in the apo structure (light, PDB: 6BUA) and a closed conformation in the presence of blunt dsRNA and an ATP analog (dark, PDB: 6BU9). In the closed conformation, Hel2 and Hel2i swivel toward Hel1 to clamp on the dsRNA terminus. (C) Structure of the hsDcr•TRBP•pre-let-7 complex with domains labeled (PDB: 5ZAL). Only the carboxy-terminal Type B motif of TRBP was resolved in the structure (Liu et al. 2018).
Based on phylogenetic studies (Zou et al. 2009; Mukherjee et al. 2013, 2014), it seems possible that ancient Dicers, which predated the split of invertebrate and vertebrate lineages, had helicase domains that discriminated blunt dsRNA as “non-self.” The emergence of RLRs allowed for loss of “non-self” recognition by the helicase domain of certain Dicers, and this occurred in different ways. In H. sapiens, recognition of blunt dsRNA was taken over by RIG-I, and as interferon signaling evolved, the helicase domain of hsDcr lost the ability to recognize blunt dsRNA and hydrolyze ATP. Instead, hsDcr was coopted for miRNA processing, possibly enabled by interacting with dsRBPs such as TRBP. (However, we note that the helicase domains of mammalian Dicers are highly conserved, and it remains possible that ATP-dependent functions of mammalian Dicers will be discovered.)
At the other extreme, in arthropods like D. melanogaster, gene duplication allowed dmDcr-2 to exclusively provide antiviral functions and optimize these by coupling recognition of blunt viral dsRNA to processivity (Welker et al. 2011; Sinha et al. 2015). A second Dicer in D. melanogaster, dmDcr-1, became dedicated to the miRNA pathway, and its Hel1 domain became degenerate (Fig. 1A). Like mammals, C. elegans encode only a single Dicer, but unlike mammals, C. elegans do not have an interferon pathway, and a single Dicer participates in processing miRNA and endo-siRNA precursors, and the antiviral response. ceDCR-1 has not been characterized as a pure protein, and it is unclear whether it is capable of discriminating non-self or, alternatively, is assisted by other proteins, such as the Dicer-related helicase, DRH-1, a RIG-I ortholog required for the C. elegans antiviral response (Thivierge et al. 2011; Ashe et al. 2013; Guo et al. 2013; Sowa et al. 2020).
HOW DO dsRBPs MODULATE DICER FUNCTION?
All of the Dicers that have been purified to homogeneity are able to process dsRNA in vitro, in the complete absence of dsRBP accessory factors. Our thesis is that dsRBPs evolved to lend new activities to an enzyme that was specialized for processing viral dsRNA, and indeed, Dicer-interacting dsRBPs confer additional activities to Dicer, often by using their carboxy-terminal Type B dsRBM to interact with the helicase domain (Fig. 1B). Below we review some of the most well-studied dsRBPs known to interact with Dicer and speculate on the mechanisms by which they modulate Dicer activity. We consider two general mechanisms. First, for the PD isoform of Loquacious, we discuss a mechanism whereby “tethering” of the carboxy-terminal region to the helicase domain promotes a conformational change in the helicase domain itself (Fig 2B). Second, and possibly applicable to all Dicer-interacting dsRBPs, we consider a mechanism whereby the carboxy-terminal region of the dsRBP (e.g., TRBP in Fig. 2C) acts as a tether to bring in amino-terminal dsRBMs to engage with the dsRNA substrate and directly modulate catalysis by the RNase III domains.
Mechanistic Insights from RNase III Enzymes
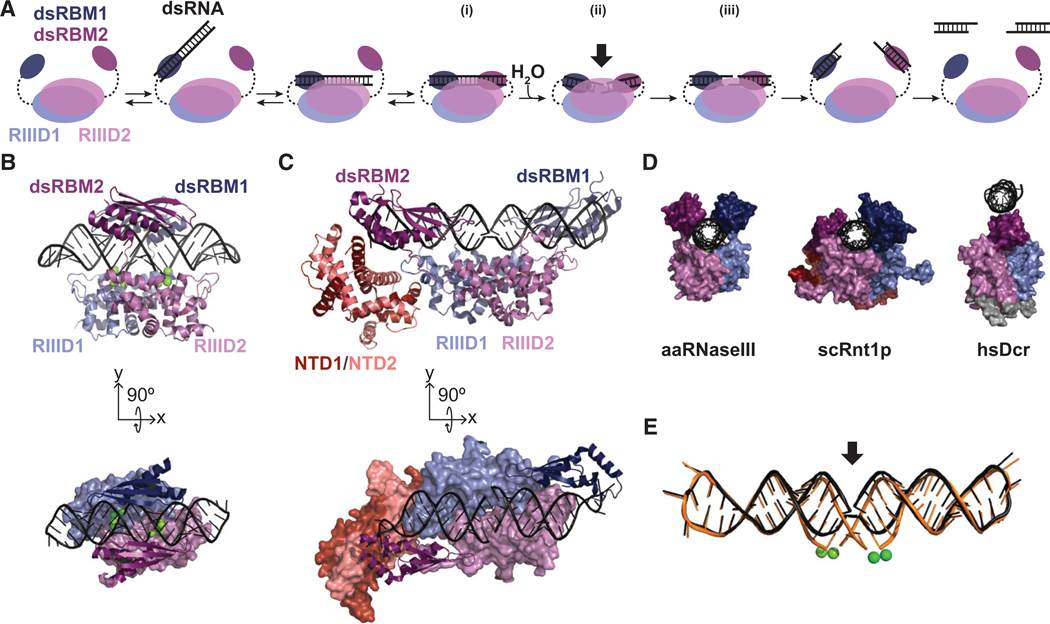
In the proposed catalytic mechanism of RNase III (Fig. 3A), the dsRNA substrate is initially recognized by a single Type A dsRBM. The dsRBM moves the dsRNA into the “catalytic valley” formed between the two RNase III monomers, and the second dsRBM associates to form the precatalytic complex, where two dsRBMs secure the dsRNA at the active site (Fig 3A, i). Substrate-loaded and postcatalytic structures of Aquifex aeolicus RNase III (aaRNaseIII) and Saccharomyces cerevisiae RNase III (scRnt1p) reveal two conformations of dsRNA in the catalytic valley: a bent and atypical (unbent) A-form RNA helix (Fig. 3B–E; Gan et al. 2005; Liang et al. 2014; Song et al. 2017). Presumably, the bent helix observed postca-talytically also represents the catalytic site arrangement immediately before cleavage by hydrolysis (Fig. 3A, ii), whereas the straightened helix represents the conformation when dsRNA is first bound and then as the first step toward product release (Fig. 3A, iii; Gan et al. 2005; Song et al. 2017). Analysis of these structures suggests that protein and RNA components undergo conformational changes during catalysis causing the scissile bond to be “pulled” into the active sites. Although the dsRBMs do not undergo a significant conformational change between these steps, there is a change in their relative positions (Gan et al. 2008), raising the possibility that the dsRBMs “slide” along the bound dsRNA to “squeeze” the scissile bonds into the active site to enable catalysis.
Figure 3.
(A) The proposed catalytic pathway for RNase III adapted for Dicer models (Nicholson 2014). Double-stranded RNA binding motifs (dsRBMs) and RNase III domains are numbered and colored with similar shades for each monomer. Dashed lines represent covalent linkage of dsRBMs in RNase III or Dicer or noncovalent linkage for those provided in trans by accessory dsRBPs. The dsRBMs engage the dsRNA (i), then “squeeze” the helix into the dimeric active sites, creating a bend in the dsRNA helix (ii) and promoting hydrolysis of the phosphodiester bonds on opposite sides of the same minor groove (iii). The arrow in ii signifies the bent helix state at the dimeric active site. For Dicer, the pathway proceeds with its carboxy-terminal dsRBM or, possibly, with dsRBMs bent byan accessory dsRBP. (B,C) Crystal structures of aaRNase III (B; PDB: 2NUF) and scRnt1p (C; PDB: 5T16) with dsRNA positioned in the catalytic valley (Gan et al. 2008; Song et al. 2017). In the side view the proteins are shown as cartoons, and in the top-down view (below) the domains beneath the dsRNA are shown as surfaces. (D) Looking down the helical axis of dsRNA for aaRNaseIII (PDB: 2NUF), scRnt1p (PDB: 5T16), and the hsDcr•TRBP•pre-let-7 cryo-EM structure (PDB: 5ZAL). For hsDcr, only let-7 pre-miRNA and the RNase IIIa, RNase IIIb, and dsRBM domains are shown (residues 1293–1913), with the structured linker between RNase IIIa and RNase IIIb shown in gray (Gan et al. 2008; Song et al. 2017; Liu et al. 2018). (E) scRnt1p crystal structures were aligned by their RNase III domains to illustrate the straight (black) and bent (orange) conformations of the dsRNA helix (PDB: 5T16 and 4OOG). Active site Mg2+ ions that facilitate cleavage are represented by green spheres (two per monomer, observed only in PDB: 4OOG).
Although recent cryo-EM structures of dmDcr-2 and hsDcr represent a significant advance in resolution (Liu et al. 2018; Sinha et al. 2018), neither structure shows dsRNA engaged at the RNase III active sites and, thus, does not offer insight into the cleavage-competent state. In fact, in the cryo-EM structure of the hsDcr•TRBP•pre-miRNA complex (Fig. 2C), Dicer’s dsRBM occupies the catalytic valley where the dsRNA substrate would be predicted to bind for cleavage, and instead, the pre-let-7 substrate interacts with the opposite face of hDcr’s carboxy-terminal dsRBM (Fig. 3D, hsDcr; Liu et al. 2018). The absence of dsRNA in the catalytic valley of hsDcr is not surprising as the complex was intentionally reconstituted in the absence of Mg2+, and based on studies of aaRNase III, magnesium is necessary for a catalytically competent state (Blaszczyk et al. 2001). In fact, the arrangement of pre-let-7 in the hsDcr•TRBP•pre-miRNA complex is similar to that in the crystal structure of dsRNA bound by aaRNase III outside of the catalytic valley in the absence of Mg2+ (Gan et al. 2008).
Based on existing RNase III structures, we propose a mechanism for how some accessory dsRBPs might contribute to Dicer activity. To form a structure analogous to aaRNaseIII and scRnt1p, the dsRNA substrate would need to be engaged by two RNase III domains and two dsRBMs. Metazoan Dicers contain two RNase III domains, one dsRBM at the carboxyl terminus, and the DUF283 domain, which adopts a dsRBM-like αβββα fold (Fig. 1; Qin et al. 2010). Although the DUF283 domain may function as one of the dsRBMs that contribute to an RNase III–type mechanism (Liu et al. 2018), it has been shown to interact with ssRNA, not dsRNA (Kurzynska-Kokorniak et al. 2016). However, certain observations are consistent with the carboxy-terminal dsRBM of hsDcr playing a role. In vitro, hsDcr lacking the carboxyterminal dsRBM shows a fourfold reduction in initial rates of cleavage of miRNA and siRNA precursors (Ma et al. 2008), but this truncation has no effect on binding affinity for the miRNA precursor and only causes a slight increase in the affinity for the siRNA precursor. This is consistent with the idea that hsDcr’s carboxy-terminal dsRBM functions similarly to one of the RNase III dsRBMs, playing a direct role in catalysis by promoting the transition state (Fig. 3A).
It seems feasible that, at least in some cases, Dicer accessory dsRBPs also contribute to catalysis by lending one or more dsRBMs to the transition state. This role is not absolutely required, as purified Dicer is catalytically active in vitro in the absence of binding partners and its carboxy-terminal dsRBM (Ma et al. 2008). Additionally, Giardia intestinalis Dicer lacks the helicase, DUF283, and dsRBM domains, yet is active in vitro in the absence of accessory proteins (Fig. 1; MacRae et al. 2006). Instead, one role of dsRBMs in Dicer and its accessory proteins would be to enhance the rate of cleavage, consistent with hsDcr truncation experiments (Ma et al. 2008). This contribution may be particularly required for certain suboptimal dsRNA substrates.
TRBP
In humans, the most well-studied cofactor of Dicer is the HIV-transactivating response RNA-binding protein (TRBP), first discovered as a binding partner for the TAR RNA structure of HIV (Gatignol et al. 1991). Later, immunoprecipitation experiments showed that TRBP interacts with hsDcr and Argonaute 2 (Ago2) as part of the RNA-induced silencing complex (RISC), which escorts the small RNA to its complementary mRNA (Chendrimada et al. 2005). TRBP is important for normal development, and ablation of the tarbp2 gene in mice causes increased mortality before weaning, and surviving mice are generally smaller in size with spermatogenesis defects (Zhong et al. 1999; Ding et al. 2015). Conditional knockout of TRBP in cardiac and skeletal muscle leads to defective development of these tissues (Ding et al. 2015, 2016). It seems likely that most of these defects derive from aberrant miRNA processing, and this is supported by studies in both mice and human cells (Kim et al. 2014; Ding et al. 2016). In vitro studies using synthetic miRNA and siRNA precursors show that TRBP modulates Dicer function by promoting optimal substrate selection, affecting cleavage rate and fidelity, and strand selection, and loading the strand into RISC (Noland et al. 2011; Lee and Doudna 2012; Noland and Doudna 2013; Kim et al. 2014; Wilson et al. 2015; Fareh et al. 2016). However, so far, there are no mechanistic explanations for how TRBP coordinates with Dicer to perform these reported functions.
As illustrated (Fig. 1B), the first two dsRBMs of TRBP are Type A motifs, and the third is a Type B motif, consistent with observations that TRBP binds dsRNA with the first two motifs (Parker et al. 2008) and interacts with Dicer’s helicase domain with its third dsRBM (Wilson et al. 2015; Liu et al. 2018). According to our model (Fig. 3A), and consistent with structural information (Liu et al. 2018), the carboxy-terminal Type B dsRBM of TRBP tethers it to the helicase domain (Fig. 2C), whereas the amino-terminal Type A dsRBMs engage the dsRNA substrate to bring it to the RNase III catalytic sites. Although only dsRBM3 was resolved in the Dicer-TRBP cryo-EM structure, a structure of dsRBM1 and dsRBM2 in complex with siRNA was solved by nuclear magnetic resonance (NMR), electron paramagnetic resonance (EPR), and single-molecule spectroscopy (Masliah et al. 2018). This structure shows that dsRBM1 and dsRBM2 bind on the same side of the dsRNA helix, leaving a large accessible surface area of the dsRNA for interaction with the RNase III active site.
One or both of TRBP’s amino-terminal dsRBMs, possibly in collaboration with Dicer’s carboxy-terminal dsRBM, could assist in docking the dsRNA substrate into the catalytic valley of the RNase III domains, stimulating cleavage activity by “squeezing” the dsRNA into the active site (Fig. 3A, ii). Indeed, in vitro studies show that TRBP•hsDcr complexes have a higher affinity for both miRNA and siRNA precursors compared to hsDcr alone, and similarly, TRBP increases the rate of dicing for both substrates in multiple-turnover experiments (Chakravarthy et al. 2010). However, single-turnover experiments in the same study show that the addition of TRBP increases the cleavage rate for an siRNA precursor, without a significant effect on cleavage of the miRNA precursor. The latter may suggest that dsRBMs of accessory proteins participate in catalysis for some substrates, but not all. Although these in vitro observations are consistent with our model (Fig. 3A), without more detailed kinetic analyses, as well as information from additional structural and mutagenesis studies, it is difficult to deconvolute effects on substrate binding from those affecting the chemical step of catalysis.
The model of Figure 3A is also consistent with the observation that TRBP enhances cleavage site fidelity in vitro and in vivo (Kim et al. 2014; Wilson et al. 2015). Bending the dsRNA helix into the catalytic valley and locking the substrate in place using dsRBMs of TRBP could reduce substrate movement that leads to cleavage at the wrong sites. Further, Dicer’s increased affinity for siRNA-sized products in the presence of TRBP (Chakravarthy et al. 2010) might not be so surprising given the stable interactions observed with the cleaved dsRNA in postcatalytic RNase III structures (Fig. 3B; Gan et al. 2005; Liang et al. 2014). This scenario might also explain the observation that TRBP participates in siRNA strand selection and loading into Ago2 after Dicer cleavage (Noland et al. 2011; Noland and Doudna 2013). At the extreme, we envision that TRBP could have a role in the precleavage, cleavage, and postcleavage phases of Dicer-mediated RNA processing in vivo (Fig. 3A).
Loquacious
The Loquacious (Loqs) proteins were discovered as interactors of dmDcr-1 and dmDcr-2 and are required for miRNA and siRNA production, respectively (Lee et al. 2004; Jiang et al. 2005; Liu et al. 2007). Because of alternative splicing, there are four Loqs isoforms: LoqsPA and Loqs-PB, which interact with dmDcr-1 and affect miRNA levels; Loqs-PC, whose function remains elusive; and Loqs-PD, which interacts with dmDcr-2 and affects endo-siRNA biogenesis (Hartig et al. 2009; Zhou et al. 2009). All four isoforms contain two amino-terminal Type A dsRBMs, whereas only Loqs-PA and Loqs-PB contain an additional Type B dsRBM3 that interacts with dmDcr1’s helicase domain (Fig. 1B; Ye et al. 2007).
Our thesis is that dsRBPs evolved to lend new activities to a Dicer enzyme that was specialized for processing viral dsRNA, and this idea is exemplified by Loqs-PD. At least for the viruses tested, Loqs-PD is not necessary for dmDcr-2 to target viral dsRNA (Marques et al. 2013), and although not definitively proven, we presume this is because viral dsRNA has termini that can be recognized by dmDcr-2 alone (Fig. 2A). However, Loqs-PD is required for processing a subset of endo-siRNA precursors (Hartig et al. 2009; Zhou et al. 2009). In D. melanogaster, endo-siRNAs map to dsRNA arising from convergent transcription, inverted repeats, and transposons (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008; Watanabe et al. 2008), and the precursors for these siRNAs are predicted to have ill-defined termini, such as long, frayed ends. Consistent with the requirement of Loqs-PD for production of endo-siRNAs, in vitro studies show that addition of Loqs-PD to dmDcr-2 allows it to function in a termini-independent manner, and this property is dependent on ATP (Sinha et al. 2015). Based on in vitro studies, the simple model is that Loqs-PD allows dmDcr-2 to bind internal regions of a dsRNA in a manner that promotes the closed, cleavage-competent conformation of the helicase domain (Fig. 2B). The latter is supported by limited proteolysis experiments (Sinha et al. 2015) in which Loqs-PD is observed to promote a protease-resistant dmDcr-2 fragment predicted to correlate with the closed conformation of the helicase domain observed in a cryo-EM structure of a complex containing dmDcr-2, blunt dsRNA, and an ATP analog (Sinha et al. 2018).
Interestingly, Loqs-PD is one of the few dsRBPs that interact with the helicase domain that does not have a Type-B domain at its carboxyl terminus (Fig. 1B). In fact, the region carboxy-terminal to the second dsRBM of Loqs-PD is too short to form the αβββα fold of other dsRBMs, although it is possible that some elements of the dsRBM secondary structure may remain. At the carboxyl terminus of Loqs-PD are 22 amino acids specific to this isoform that are necessary for efficient endo-siRNA biogenesis in vivo (Hartig and Förstemann 2011) and that have been shown to cross-link to the Hel2 domain of dmDcr-2 (Trettin et al. 2017).
The “tethering” interaction with Dicer occurs at the carboxyl terminus of all Loqs isoforms, and it is also possible that the Type A motifs, dsRBM1 and dsRBM2, facilitate binding of the dsRNA substrate and promote catalysis by analogy to RNase III enzymes (Fig. 3A). Indeed, Loqs-PD greatly increases dmDcr-2 cleavage rates in vitro (Trettin et al. 2017). Interestingly, efficient cleavage of an optimal dmDcr-2 substrate, blunt dsRNA, does not require dsRBM1, but cleavage of a suboptimal substrate, dsRNA with a 3′ overhang, requires both amino-terminal motifs (Trettin et al. 2017). Again, this raises the possibility that the requirement of dsRBMs in Dicer’s accessory dsRBPs is substrate-dependent.
R2D2
All of the dsRBPs discussed so far act in facilitating cleavage by Dicer. At least for metazoan Dicers, after cleavage, the miRNA or siRNA product must be passed to an Argonaute protein to form the RISC, which escorts the small RNA to its complementary mRNA. We know little about the mechanism of this process, although studies of D. melanogaster R2D2 offer the most insight and reiterate the key role played by dsRBPs in facilitating Dicer functions. In early RNAi studies, siRNA-generating activity was monitored in fractionated extracts from S2 cells, and dmDcr-2 and R2D2 copurified through six chromatographic steps (Liu et al. 2003); the stability of R2D2 relied on the formation of the dmDcr-2•R2D2 complex. Embryos from r2d2 deletion mutant flies were defective in silencing, but in vitro assays using recombinant proteins showed similar cleavage activity by dmDcr-2 alone and the dmDcr-2•R2D2 complex (Liu et al. 2003, 2006). These observations suggested R2D2 was important downstream from the cleavage step, and indeed, although R2D2 was not required for siRNA biogenesis in whole fly lysates, without R2D2, only minimal mRNA could be properly cleaved by RISC (Liu et al. 2006). Further, efficient transfer of a biotinylated siRNA to Argonaute 2 (dmAgo2) was observed with a dmDcr-2•R2D2 complex, but not with dmDcr-2 alone (Liu et al. 2003). Together these results indicate that R2D2 functions with dmDcr-2 downstream from siRNA production to facilitate siRNA loading into RISC.
R2D2 has an extended carboxy-terminal region (Fig. 1B) that interacts with dmDcr-2’s helicase domain (Nishida et al. 2013), but the helicase subdomain involved in the interaction is unknown. Although there is no annotated dsRBM in this region, Type B dsRBMs are sometimes difficult to detect by sequence alignment and current motif search algorithms, because of their lack of conservation (Gleghorn and Maquat 2014). Individually, dmDcr-2 and R2D2 bind poorly to siRNA, but as a complex bind strongly (Liu et al. 2006). For the guide strand to be properly loaded onto dmAgo2, the siRNA needs to unwind. However, the dmDcr-2•R2D2 complex lacks siRNA unwinding activity, suggesting the pre-RISC complex, including dmAgo2, must be assembled for unwinding and transfer to occur (Tomari et al. 2004). Although the dmDcr-2•R2D2 complex cannot unwind siRNA on its own, R2D2 appears to aid in strand selection for eventual loading, like TRBP with hsDcr (Noland et al. 2011). As both TRBP and R2D2 have been reported to be involved in guide strand selection and loading into RISC (Tomari et al. 2004), perhaps R2D2 binds dmDcr-2 similarly via the Hel2i subdomain (Noland et al. 2011; Noland and Doudna 2013).
Although R2D2 has been detected in immunoprecipitated complexes of dmDcr-2•Loqs-PD (Miyoshi et al. 2010), it is unclear if both proteins can interact with dmDcr-2 simultaneously. If R2D2 binds the Hel2i subdomain of dmDcr-2, like TRBP for hsDcr, could LoqsPD bind Hel2 at the same time or would R2D2 and Loqs-PD compete for binding? If Loqs-PD and R2D2 could bind at the same time, the dmDcr-2•R2D2•Loqs-PD complex could act as a well-oiled machine, efficiently cleaving dsRNA into siRNA products, with the help of Loqs-PD, and then allowing transfer to dmAgo2, possibly in one fluid motion. Alternatively, if these two cofactors compete for binding the helicase domain, they might act as a switch for dmDcr-2’s functions, as either an siRNA biogenesis factory or a stable siRNA transporter.
OUTSTANDING QUESTIONS
We focused on a few dsRBPs that interact with Dicer’s helicase domain to modulate its functions. We expect that other dsRBPs not covered here will emphasize paradigms we have discussed, as well as point to currently unimagined methods of regulation. Indeed, it is exciting to imagine the future biochemical and structural studies that undoubtedly will offer additional insight.
As mentioned for R2D2 and Loqs-PD, of interest will be the question of whether accessory dsRBPs interact simultaneously with Dicer or have mutually exclusive interactions. Although R2D2 and Loqs-PD may bind to different helicase subdomains, TRBP and PACT both bind to the same interface of the Hel2i subdomain of hsDcr and thus interact in a mutually exclusive manner (Wilson et al. 2015). Although reported associations of dsRBPs are sometimes indirect, mediated by their common ligand, dsRNA, in many cases interactions are direct. Further, it seems possible that even dsRNA-mediated interactions could have biological relevance. The RNA editing enzyme ADAR1 interacts directly with hsDcr to modulate processing of pre-miRNA (Ota et al. 2013) and has been reported to have direct as well as indirect interactions with other dsRBPs (Herbert 2019). As a further layer of complexity, for mammals, a subset of the dsRBPs that interact with the helicase domain of mammalian Dicer have also been shown to interact with other RLRs (Takahashi et al. 2018; van der Veen et al. 2018). Understanding this complexity is an important goal for future studies.
ACKNOWLEDGMENTS
We thank Dr. Nels Elde for helpful discussions on the evolution of Dicer. This work was supported by funding from the National Institute of General Medical Sciences (R01GM121706) to B.L.B. H.M.D was supported by a National Institutes of Health Chemical Biology training grant (T32GM122740).
REFERENCES
- Ahmad S, Hur S. 2015. Helicases in antiviral immunity: dual properties as sensors and effectors. Trends Biochem Sci 40: 576–585. doi: 10.1016/j.tibs.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Bélicard T, Le Pen J, Sarkies P, Frézal L, Lehrbach NJ, Félix M-A, Miska EA. 2013. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. eLife 2: e00994. doi: 10.7554/eLife.00994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. 2000. Double-stranded RNA as a template for gene silencing. Cell 101: 235–238. doi: 10.1016/S0092-8674(02)71133-1 [DOI] [PubMed] [Google Scholar]
- Bass BL, Hurst SR, Singer JD. 1994. Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr Biol 4: 301–314. doi: 10.1016/S0960-9822(00)00069-5 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. doi: 10.1038/35053110 [DOI] [PubMed] [Google Scholar]
- Blaszczyk J, Tropea JE, Bubunenko M, Routzahn KM, Waugh DS, Court DL, Ji X. 2001. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 9: 1225–1236. doi: 10.1016/S09692126(01)00685-2 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. 2010. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol 404: 392–402. doi: 10.1016/j.jmb.2010.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744. doi: 10.1038/nature03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W-J, Okamura K, Martin R, Lai EC. 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol 18: 795–802. doi: 10.1016/j.cub.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakić M, Moazed D. 2007. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461. doi: 10.1016/j.molcel.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Court DL, Gan J, Liang Y-H, Shaw GX, Tropea JE, Costantino N, Waugh DS, Ji X. 2013. RNase III: genetics and function; structure and mechanism. Annu Rev Genet 47: 405–431. doi: 10.1146/annurev-genet-110711-155618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802. doi: 10.1038/nature07007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deddouche S, Matt N, Budd A, Mueller S, Kemp C, GalianaArnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler J-L. 2008. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in Drosophila. Nat Immunol 9: 1425–1432. doi: 10.1038/ni.1664 [DOI] [PubMed] [Google Scholar]
- Ding S-W, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130: 413–426. doi: 10.1016/j.cell.2007.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Chen J, Wang Y, Kataoka M, Ma L, Zhou P, Hu X, Lin Z, Nie M, Deng Z-L, et al. 2015. Trbp regulates heart function through microRNA-mediated Sox6 repression. Nat Genet 47: 776–783. doi: 10.1038/ng.3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Nie M, Liu J, Hu X, Ma L, Deng Z-L, Wang D-Z. 2016. Trbp is required for differentiation of myoblasts and normal regeneration of skeletal muscle. PLoS One 11: e0155349. doi: 10.1371/journal.pone.0155349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther U-P, Jankowsky E. 2010. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 20: 313–324. doi: 10.1016/j.sbi.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fareh M, Yeom K-H, Haagsma AC, Chauhan S, Heo I, Joo C. 2016. TRBP ensures efficient Dicer processing of precursor microRNA in RNA-crowded environments. Nat Commun 7: 13694. doi: 10.1038/ncomms13694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. doi: 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Fukudome A, Fukuhara T. 2017. Plant dicer-like proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis. J Plant Res 130: 33–44. doi: 10.1007/s10265-016-0877-1 [DOI] [PubMed] [Google Scholar]
- Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. 2005. Intermediate states of ribonuclease III in complex with double-stranded RNA. Structure 13: 1435–1442. doi: 10.1016/j.str.2005.06.014 [DOI] [PubMed] [Google Scholar]
- Gan J, Shaw G, Tropea JE, Waugh DS, Court DL, Ji X. 2008. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol Microbiol 67: 143–154. doi: 10.1111/j.13652958.2007.06032.x [DOI] [PubMed] [Google Scholar]
- Gatignol A, Buckler-White A, Berkhout B, Jeang KT. 1991. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science (New York, NY) 251: 1597–1600. doi: 10.1126/science.2011739 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science (New York, NY) 320: 1077–1081. doi: 10.1126/science.1157396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn ML, Maquat LE. 2014. “Black sheep” that don’t leave the double-stranded RNA-binding domain fold. Trends Biochem Sci 39: 328–340. doi: 10.1016/j.tibs.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhang R, Wang J, Ding S-W, Lu R. 2013. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc Natl Acad Sci 110: 16085–16090. doi: 10.1073/pnas.1307453110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. doi: 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Hartig JV, Förstemann K. 2011. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res 39: 3836–3851. doi: 10.1093/nar/gkq1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Esslinger S, Böttcher R, Saito K, Förstemann K. 2009. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J 28: 2932–2944. doi: 10.1038/emboj.2009.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A 2019. ADAR and immune silencing in cancer. Trends Cancer 5: 272–282. doi: 10.1016/j.trecan.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R. 2014. RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem 83: 697–725. doi: 10.1146/annurev-biochem-060713-035546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. 2005. Dicer1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 19: 1674–1679. doi: 10.1101/gad.1334005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. 2008. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453: 793–797. doi: 10.1038/nature06938 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. 1999. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99: 133–141. doi: 10.1016/S0092-8674(00)81645-1 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659. doi: 10.1101/gad.927801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yeo J, Lee JH, Cho J, Seo D, Kim J-S, Kim VN. 2014. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep 9: 1061–1074. doi: 10.1016/j.celrep.2014.09.039 [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science (New York, NY) 293: 2269–2271. doi: 10.1126/science.1062039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzynska-Kokorniak A, Pokornowska M, Koralewska N, Hoffmann W, Bienkowska-Szewczyk K, Figlerowicz M. 2016. Revealing a new activity of the human Dicer DUF283 domain in vitro. Sci Rep 6: 23989–13. doi: 10.1038/srep23989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Doudna JA. 2012. TRBP alters human precursor microRNA processing in vitro. RNA 18: 2012–2019. doi: 10.1261/rna.035501.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. doi: 10.1016/S0092-8674(04)00261-2 [DOI] [PubMed] [Google Scholar]
- Liang Y-H, Lavoie M, Comeau M-A, Abou Elela S, Ji X. 2014. Structure of a eukaryotic RNase III postcleavage complex reveals a double-ruler mechanism for substrate selection. Mol Cell 54: 431–444. doi: 10.1016/j.molcel.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim H-E, Smith DP, Wang X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science (New York, NY) 301: 1921–1925. doi: 10.1126/science.1088710 [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q. 2006. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 12: 1514–1520. doi: 10.1261/rna.101606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Park JK, Jiang F, Liu Y, McKearin D, Liu Q. 2007. Dicer1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA 13: 2324–2329. doi: 10.1261/rna.723707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang J, Cheng H, Ke X, Sun L, Zhang QC, Wang H-W. 2018. Cryo-EM structure of human dicer and its complexes with a pre-miRNA substrate. Cell 173: 1191–1203.e12. doi: 10.1016/j.cell.2018.03.080 [DOI] [PubMed] [Google Scholar]
- Luo D, Kohlway A, Pyle AM. 2013. Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol 10: 111–120. doi: 10.4161/rna.22706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. 2008. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol 380: 237–243. doi: 10.1016/j.jmb.2008.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. 2006. Structural basis for double-stranded RNA processing by Dicer. Science (New York, NY) 311: 195–198. doi: 10.1126/science.1121638 [DOI] [PubMed] [Google Scholar]
- Marques JT, Wang J-P, Wang X, de Oliveira KPV, Gao C, Aguiar ERGR, Jafari N, Carthew RW. 2013. Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog 9: e1003579. doi: 10.1371/journal.ppat.1003579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah G, Barraud P, Allain FH-T. 2013. RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell Mol Life Sci 70: 1875–1895. doi: 10.1007/s00018-012-1119-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah G, Maris C, König SL, Yulikov M, Aeschimann F, Malinowska AL, Mabille J, Weiler J, Holla A, Hunziker J, et al. 2018. Structural basis of siRNA recognition by TRBP double-stranded RNA binding domains. EMBO J 37: e97089. doi: 10.15252/embj.201797089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC. 2010. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA 16: 506–515. doi: 10.1261/rna.1952110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Campos H, Kolaczkowski B. 2013. Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol 30: 627–641. doi: 10.1093/molbev/mss263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Korithoski B, Kolaczkowski B. 2014. Ancient origins of vertebrate-specific innate antiviral immunity. Mol Biol Evol 31: 140–153. doi: 10.1093/molbev/mst184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson AW. 2014. Ribonuclease III mechanisms of double-stranded RNA cleavage. WIREs RNA 5: 31–48. doi: 10.1002/wrna.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Miyoshi K, Ogino A, Miyoshi T, Siomi H, Siomi MC. 2013. Roles of R2D2, a cytoplasmic D2 body component, in the endogenous siRNA pathway in Drosophila. Mol Cell 49: 680–691. doi: 10.1016/j.molcel.2012.12.024 [DOI] [PubMed] [Google Scholar]
- Noland CL, Doudna JA. 2013. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA 19: 639–648. doi: 10.1261/rna.037424.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland CL, Ma E, Doudna JA. 2011. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell 43: 110–121. doi: 10.1016/j.molcel.2011.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W-J, Ruby JG, Guo H, Bartel DP, Lai EC. 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806. doi: 10.1038/nature07015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H, Sakurai M, Gupta R, Valente L, Wulff B-E, Ariyoshi K, Iizasa H, Davuluri RV, Nishikura K. 2013. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 153: 575–589. doi: 10.1016/j.cell.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GS, Maity TS, Bass BL. 2008. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J Mol Biol 384: 967–979. doi: 10.1016/j.jmb.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S. 2009. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183: 1283–1295. doi: 10.1534/genetics.109.108134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O. 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21: 5864–5874. doi: 10.1093/emboj/cdf578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Chen F, Huan X, Machida S, Song J, Yuan YA. 2010. Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein–protein interaction. RNA 16: 474–481. doi: 10.1261/rna.1965310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HD, Webster RE, Zinder ND. 1968. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem 243: 82–91. [PubMed] [Google Scholar]
- Schlee M 2013. Master sensors of pathogenic RNA—RIG-I like receptors. Immunobiology 218: 1322–1335. doi: 10.1016/j.imbio.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76: 23–50. doi: 10.1146/annurev.biochem.76.052305.115300 [DOI] [PubMed] [Google Scholar]
- Sinha NK, Trettin KD, Aruscavage PJ, Bass BL. 2015. Drosophila Dicer-2 cleavage is mediated by helicase- and dsRNA termini-dependent states that are modulated by Loquacious-PD. Mol Cell 58: 406–417. doi: 10.1016/j.molcel.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NK, Iwasa J, Shen PS, Bass BL. 2018. Dicer uses distinct modules for recognizing dsRNA termini. Science (New York, NY) 359: 329–334. doi: 10.1126/science.aaq0921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Fang X, Jin L, Shaw GX, Wang Y-X, Ji X. 2017. The functional cycle of Rnt1p: five consecutive steps of double-stranded RNA processing by a eukaryotic RNase III. Structure 25: 353–363. doi: 10.1016/j.str.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa JN, Jiang H, Somasundaram L, Tecle E, Xu G, Wang D, Troemel ER. 2020. The Caenorhabditis elegans RIG-I homolog DRH-1 mediates the intracellular pathogen response upon viral infection. J Virol 94: e01173–19. doi: 10.1128/JVI.01173-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Nakano Y, Onomoto K, Murakami F, Komori C, Suzuki Y, Yoneyama M, Ui-Tei K. 2018. LGP2 virus sensor regulates gene expression network mediated by TRBP-bound microRNAs. Nucleic Acids Res 46: 9134–9147. doi: 10.1093/nar/gky575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thivierge C, Makil N, Flamand M, Vasale JJ, Mello CC, Wohlschlegel J, Conte D, Duchaine TF. 2011. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat Struct Mol Biol 19: 90–97. doi: 10.1038/nsmb.2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. 2004. A protein sensor for siRNA asymmetry. Science (New York, NY) 306: 1377–1380. doi: 10.1126/science.1102755 [DOI] [PubMed] [Google Scholar]
- Trettin KD, Sinha NK, Eckert DM, Apple SE, Bass BL. 2017. Loquacious-PD facilitates Drosophila Dicer-2 cleavage through interactions with the helicase domain and dsRNA. Proc Natl Acad Sci 114: E7939–E7948. doi: 10.1073/pnas.1707063114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Maillard PV, Schmidt JM, Lee SA, Deddouche Grass S, Borg A, Kjær S, Snijders AP, Reis e Sousa C. 2018. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J 37: e97479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543. doi: 10.1038/nature06908 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T. 2001. Gene silencing as an adaptive defence against viruses. Nature 411: 834–842. doi: 10.1038/35081168 [DOI] [PubMed] [Google Scholar]
- Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S, Bass BL. 2010. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16: 893–903. doi: 10.1261/rna.2122010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. 2011. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell 41: 589–599. doi: 10.1016/j.molcel.2011.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. 2013. Molecular mechanisms of RNA interference. Annu Rev Biophys 42: 217–239. doi: 10.1146/annurev-biophys-083012-130404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA. 2015. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell 57: 397–407. doi: 10.1016/j.molcel.2014.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Paroo Z, Liu Q. 2007. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem 282: 28373–28378. doi: 10.1074/jbc.M705208200 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. doi: 10.1016/S0092-8674(00)80620-0 [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885. doi: 10.1093/emboj/cdf582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Peters AH, Lee K, Braun RE. 1999. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet 22: 171–174. doi: 10.1038/9684 [DOI] [PubMed] [Google Scholar]
- Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ. 2009. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 15: 1886–1895. doi: 10.1261/rna.1611309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Chang M, Nie P, Secombes CJ. 2009. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol 9: 85–14. doi: 10.1186/1471-2148-9-85 [DOI] [PMC free article] [PubMed] [Google Scholar]