Abstract
The fate of an RNA, from its localization, translation and ultimate decay, is dictated by interactions with RNA binding proteins (RBPs). ß-actin mRNA has functioned as the classic example of RNA localization in eukaryotic cells. Studies of ß-actin mRNA over the past three decade have allowed understanding of how RBPs, such as ZBP1 (IGF2BP1), can control both RNA localization and translational status. Here, we summarize studies of ß-actin mRNA and focus on how ZBP1 serves as a model for understanding interactions between RNA and their binding protein(s). Central to the study of RNA and RBPs were technological developments that occurred along the way. We conclude with a future outlook highlighting new technologies that may be used to address still unanswered questions about RBP mediated regulation of mRNA during its life cycle, within the cell.
Introduction
Changes in protein synthesis define all aspects of a cell’s life. While most cells carry identical copy numbers of their DNA, differences in transcription and translation define both cellular fate as well as moment to moment cellular decisions. Therefore, the regulation of RNA is central to the control of protein synthesis (Vera et al. 2016). The timing and localization of protein synthesis is highly regulated at the level of RNA by RNA binding proteins (RBPs).
In eukaryotes, RBPs shuttle between nucleus and cytoplasm participating in all aspects of gene expression. RBPs can direct the processes of splicing, polyadenylation, export, localization, translation and decay (reviewed in (Glisovic et al. 2008)). RBPs allow for rapid spatiotemporal regulation of transcripts. While long term changes in steady state RNA abundance can occur during differentiation, there are several instances when more rapid modulation of transcripts is necessary. By storing transcripts in a translationally inactive state until stimulated, cellular RBPs can rapidly initiate localized translation.
RNA localization patterns appear across organisms and cell types (Buxbaum et al. 2015; Martin and Ephrussi 2009). Asymmetric distribution of poly A RNAs was first described in ascidian eggs (Jeffery et al. 1983). Shortly thereafter, specific RNAs were observed to be localized to the animal and vegetal pole in maternal xenopus eggs (Rebagliati et al. 1985). In chicken embryonic myoblasts both tritiated and biotinylated probes were used to observe cytoskeletal mRNA localization (Lawrence and Singer 1986; Singer and Ward 1982). Of the three RNAs tested (actin, vimentin and tubulin) – actin had the most distinct localization pattern. Approximately 95% of myoblasts had a nonrandom distribution of ß-actin mRNA, where RNAs are concentrated at the cell extremities.
Key to the observation of RNA localization was the development and optimization of RNA fluorescent in situ hybridization (RNA FISH) (Singer and Ward 1982; Lawrence and Singer 1985). Improvements to both the detectors and fluorophores allowed for visualization and tracking of single RNA molecules within cells (Femino et al. 2003; Bertrand et al. 1998). Building upon the advent of single molecule RNA FISH, advances in microfluidics have allowed for multiplexed re-probing of samples and transcriptome-wide studies of RNA localization in both cells and tissues (Eng et al. 2019; Lécuyer et al. 2007; Levsky et al. 2002). By increasing the throughput of approaches to study RNA localization, it has now been possible to appreciate the breadth of RNAs that undergo this process. For example, RNA localization was found to be prevalent in the developing Drosophila embryo (Lécuyer et al. 2007). RNA-FISH against 3,370 genes determined that over 70% of the RNAs tested showed different patterns of localization, not only emphasizing the prevalence of RNA localization patterns but also the different mechanisms by which localization patterns are encoded. In neurons, over half of the transcripts queried by RNA sequencing were localized in the neurite and nearly half of the localized mRNAs lead to neurite localized proteins (Zappulo et al. 2017).
Profiling of RNAs from different subcellular compartments shows that mRNA localization confers function on the sub-cellular scale. Within the cytoplasm, localization and local translation of mRNAs to the mitochondria and ER ensures correct expression of mitochondrial and secretory proteins (Gadir et al. 2011; Garcia et al. 2010; Jan et al. 2014; Williams et al. 2014). In the migrating cell, β-actin mRNA localization to the leading edge allows for directional movement in response to guidance cues (Katz et al. 2012; Lawrence and Singer 1986; Shestakova et al. 2001).
RNA transport and localization provides the means for gene expression regulation, where local sites can determine what proteins are made in response to spatiotemporal cues. In highly polarized neurons, there are clear advantages to local control of gene expression. As dendrites and axons can extend hundreds of microns away from the cell body, the process of RNA localization and local protein synthesis is particularly relevant to neuronal function (Zappulo et al. 2017). Synapses that mediate transmission of information from one neuron to another have the capacity to undergo long-term cytoskeletal remodeling, growing or shrinking the size of the dendritic spine in response to firing (Hotulainen and Hoogenraad 2010). This synaptic modulation requires newly translated proteins and underlies higher cognitive functions such as learning and memory (reviewed in (Costa-Mattioli et al. 2009)).
In the developing axon growth cone, localization has been shown to be involved in growth cone pathfinding, allowing the axon to twist and turn as it navigates chemical gradients and forms immature synapses (Eom et al. 2003). In the mature neuron, dynamic RNA localization occurs in response to neural stimulation of specific dendrites, with RNAs localizing to their base and new protein synthesis occurring in the specifically stimulated spines (Yoon et al. 2016). Therefore, not only does the timing of translation play a key role when a dendrite is stimulated but also the proper RNA location at a specific synapse is also critical.
Mechanisms of RNA localization
It is now appreciated that RNA localization, facilitated by RBPs, is a highly conserved mechanism to spatially confine protein synthesis, amplify local protein concentration, or even direct integration into macromolecular complexes (reviewed in (Buxbaum et al. 2015; Glisovic et al. 2008)). Compartment-specific targeting of mRNA involves recognition of a short nucleotide sequence known as a cis-acting localization element by an RBP called the trans-acting factor. Many well-characterized RBPs bind sequence specifically while others have been shown to recognize specific stem loop structure or nonspecifically bind to single or double-stranded RNA. The direct association between RBPs and cis-acting localization elements leads to formation of a ribonucleoprotein (RNP) complex that travels along the cytoskeleton with the help of motor proteins (reviewed in (Eliscovich et al. 2013)). Identification of canonical trans-acting factors have been based on their i) ability to interact specifically with their target mRNAs, and ii) where loss of function or loss of expression results in mislocalization of the mRNA. Disruptions in the formation of transport RNPs can have significant consequences, especially in neurons where localization is necessary for synthesis of proteins and consequently synaptic plasticity.
Several models have been developed to understand how RNAs can become localized within a cell (Buxbaum et al. 2015). RNA molecules can passively diffuse until they reach an anchoring point (diffusion and entrapment). Different examples of anchors include cytoskeletal proteins or RBPs (Beach et al. 1999; Farina et al. 2003). RNAs can also be locally protected from degradation or conversely selectively degraded, leading to accumulation of RNA in areas that with lower decay rates (Tadros et al. 2007; Zaessinger et al. 2006). Additionally, RNAs can be localized by motor proteins, often through their interaction with an RBP which serves as an adapter (Long et al. 2000; Song et al. 2015). While each mechanism of localization can act alone, they can also be combined to perform biological functions. By combining directed transport with local entrapment it has been shown that ß-actin mRNA can rapidly localized to a stimulated dendritic spine where new proteins are then synthesized (Yoon et al. 2016). Key to both simple and complex modes of RNA localization are RNA binding proteins.
ZBP1: the RBP that controls ß-actin mRNA localization
Since the initial observation of polarized RNA within cells, ß-actin mRNA localization has provided a model system for understanding the mechanisms of RNA localization within eukaryotic cells (Fig 1, (Buxbaum et al. 2015; Das et al. 2019; Eliscovich and Singer 2017; Eliscovich et al. 2013). Biochemical methodologies, structural analysis and imaging based approaches have analyzed how a cis-acting element and a trans-acting factors act together to ensure the cytoplasmic fate of the mRNA once it is transcribed in the nucleus.
Figure 1. Timeline of ZBP1 and β-actin mRNA discoveries.
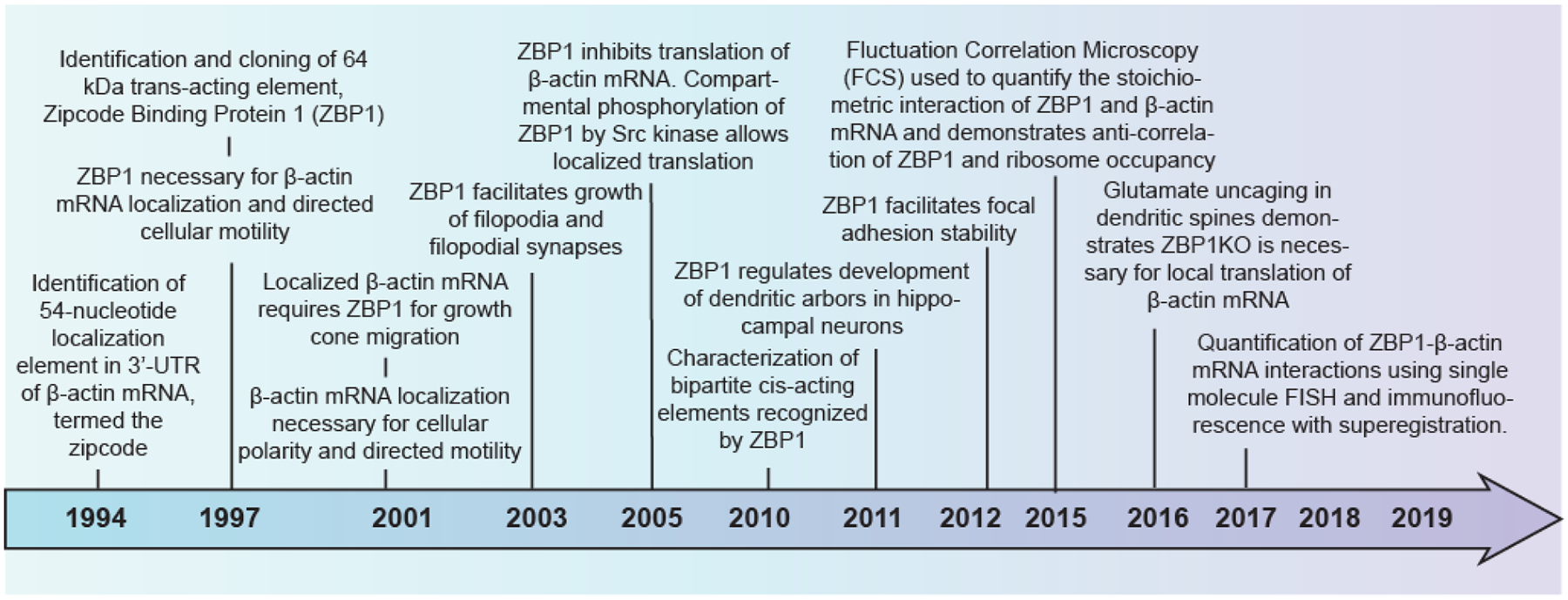
Over the past two decades, significant work has revealed the zipcode binding protein-1 (ZBP1) is involved in localization of mRNA. Presented here are a summary of important findings that has led to our current understanding of how ZBP1 acts to regulate β-actin mRNA.
Cytoplasmic ß-actin mRNA localization was initially observed using in situ hybridization in chicken embryonic skeletal myoblasts and fibroblasts (Lawrence and Singer 1986). Later reporter plasmids expressing different regions of the 3’-UTR of ß-actin mRNA demonstrated that a 54-nucleotide cis-acting “zipcode” element was responsible for the localization of ß-actin mRNA to the cellular periphery (Kislauskis et al. 1994). This 54-nucleotide zipcode sequence is conserved from chicken to mouse and human ß-actin 3’-UTRs. Zipcode Binding Protein 1 (ZBP1; also called IGF2BP1), the key RBP that binds the 54-nucleotide zipcode in the nucleus and regulates localization and translational repression of ß-actin mRNA in the cytoplasm (Farina et al. 2003; Hüttelmaier et al. 2005; Oleynikov and Singer 2003; Wu et al. 2015; Pan et al. 2007), was identified by UV cross linking and affinity purification (Ross et al. 1997). Subsequently, a number of RBPs homologous to ZBP1 were also discovered in human, mouse, flies and frogs (Yisraeli 2005). A timeline highlighting ZBP’s role as a critical RBP for β-actin mRNA localization in different cell types has been illustrated in Figure 1.
RNA reporter assays (Kislauskis et al. 1994), biochemical (Farina et al. 2003) and structural characterization of the 54-nucleotide zipcode led to the identification of a minimal 28-nucleotide consensus bipartite element that is specifically recognized by ZBP1 (Chao et al. 2010; Patel et al. 2012). The structural studies also showed that ZBP1KH34 (third and fourth hnRNP K-homology domains) specifically binds the bipartite ß-actin 3’-UTR element, with ZBP1KH4 and ZBP1KH3 recognizing ß-actin 5’-CGGAC-3’ and 5’-(C/A)CA(C/U)-3’ sequences, respectively (Chao et al. 2010; Patel et al. 2012). For each consensus sequence to bind to the two protein binding sites on opposite ends of ZBP1KH34, the RNA must loop around the protein. The distortion sequesters the stop codon of ß-actin mRNA and likely contributes to translational repression (Fig 2, Fig 3A–D, (Chao et al. 2010; Wu et al. 2015)).
Figure 2. Summary of the molecular interaction of ZBP1 with β-actin mRNA which determines cellular fate.
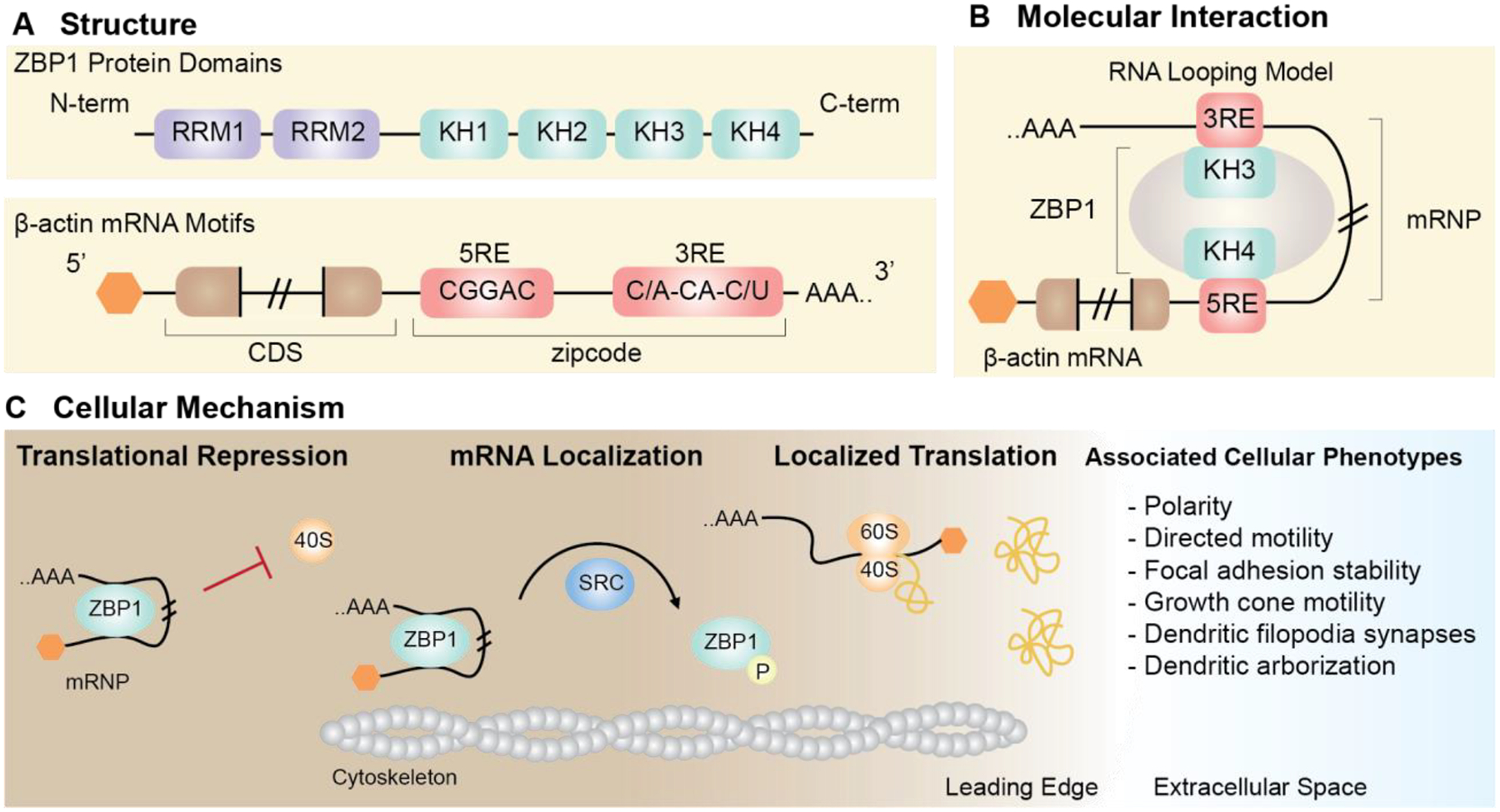
(A) The interaction of ZBP1 with cognate mRNA, β-actin is dependent on structural motifs. ZBP1 contains two RNA recognition motifs (RRM) (purple) and four hnRNP-K homology (KH) (green) domains. β-actin mRNA contains a bipartite localization element in the 3’-UTR, named the zipcode (red). (B) Interaction between the zipcode and ZBP1 has been proposed to occur by RNA looping, thereby forming the ribonucleoprotein complex (mRNP). (C) Formation of the mRNP complex has molecular and cellular consequences. The ZBP1-β-actin mRNP prevents ribosomal subunits (eg. 40S) from bindings β-actin mRNA, thus resulting in translational repression. Active transport of the ZBP1-β-actin mRNP along cytoskeletal filaments (motors not depicted) allows localization of β-actin mRNA. Disassembly of the ZBP1-β-actin mRNP is facilitated by Src-depedent phosphorylation of ZBP1 at the leading edge. Release of β-actin mRNA ultimately allows for ribosomal subunit assembly and local translation to occur. Localized translation of β-actin mRNA at the leading edge is associated with cellular phenotypes including polarity, directed motility, and focal adhesion stability in fibroblasts, and growth cone motility, dendritic filopodia synapses, and dendritic arborization in neurons.
Figure 3. Exploring RBP-RNA interactions by studying ZBP1-β-actin mRNA.
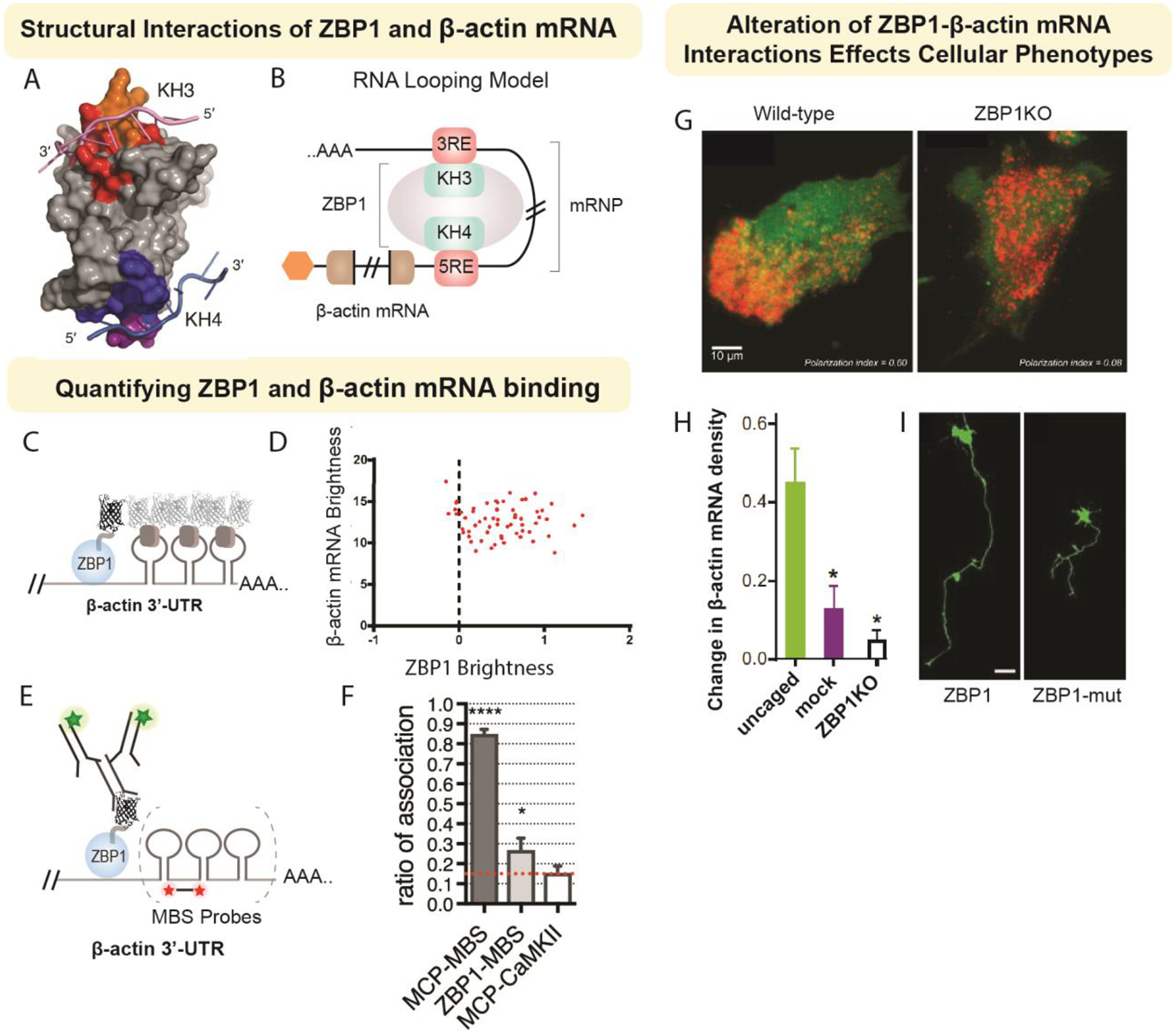
Here, we highlight studies that have contributed to our understanding of ZBP1-β-actin mRNA structural interactions (A-B), stoichiometry of binding (C-F), and how alterations of their interactions lead to respective changes in cellular phenotype (G-I). ZBP1 has two main domains (KH34) that are reported to interact with the zipcode element in the 3’-UTR of -β-actin (A; Modified from Patel, et al. 2012), which is proposed to form an RNA loop when bound by ZBP1 (B). The stoichiometric interaction of ZBP1 with β-actin mRNA has revealed a 1:1 binding. Two parallel imaging approaches have been utilized to confirm the stoichiometric associations of the ZBP1-β-actin complex: fluctuation correlation microscopy (FCS) (C-D) and fluorescence in situ hybridization and immunofluorescence (FISH-IF) (E-F). Using the bacteriophage MS2 system to label β-actin mRNA with stem loops (MBS), capsid proteins are labeled with fluorescent proteins. Simultaneous expression of ZBP1 with a fluorescent tag (C). Using live tracking of single particles in FCS videos, the fluorescent intensity of the both particles is used to evaluate stoichiometry (D; Modified from Wu, et al. 2015). Alternatively, fixed images from FISH-IF can be analyzed to determine stoichiometric association. Using a similar approach, ZBP1 is expressed with a fluorescent tag. To increase the signal from IF, staining is performed against thee fluorescent tag. FISH probes are used against the stem loop sequence (MBS) (E) and spatial association of signals are used to determine stoichiometry (F; Modified from Eliscovich, et al. 2017). Alteration of the ZBP1-β-actin mRNA complex leads to a variety of cellular defects. In fibroblasts, deletion of ZBP1 leads to loss of polarization. In neurons, ZBP1 is important for localization of β-actin mRNA in dendrites. The localization of β-actin mRNA occurs in response to glutamate stimulation (photoactivable uncaging of glutamate). Loss of ZBP1 results in a reduction of -β-actin mRNA in response to glutamate release (H; Yoon et al. 2016). Neurons also show alterations in outgrowth in response to mutations of the ZBP1 phosphorylation site Huttelmaier et al. 2005).
ZBP1 controls local ß-actin translation
Both the zipcode sequence within the 3’-UTR of ß-actin and KH34 domains of ZBP1 are necessary for the formation of the ZBP1-ß-actin mRNA complex (Fig. 2A, 2B). Regulation of ß-actin mRNA fate is dependent on these associations.
In the cytoplasm, formation of the ZBP1-ß-actin mRNA complex sterically inhibits the large ribosomal subunit from binding the ß-actin mRNA, thereby preventing translation (Hüttelmaier et al. 2005; Wu et al. 2015). Motorized movement of this complex along cytoskeletal filaments leads to peripheral localization (eg., leading edge in fibroblasts, dendrites or axonal cone in neurons) and accounts for the nonrandom distribution of ß-actin mRNA observed in migrating or stimulated cells (Bassell et al. 1998; Mukherjee et al. 2019). Src kinase-dependent phosphorylation of Tyr-396 in ZBP1 leads to disassembly of the ZBP1-ß-actin mRNA complex at the periphery. Subsequent release of ß-actin mRNA permits binding of ribosomal subunits and leads to localized translation (Fig 2C, (Hüttelmaier et al. 2005)). Thus, ZBP1 can both localize and control the translation of ß-actin mRNA, suggesting that RBPs can perform complex roles within the cells.
A similar pattern of events occurs in yeast when ASH1 mRNA localizes to the bud tip in a She2 dependent manner (Long et al. 2000). Motor dependent localization along cytoskeletal filaments and phosphorylation dependent translation initiation in both yeast and mammalian cells suggests there exists common mechanisms by which RBPs can localize RNAs and regulate their translation.
While ZBP1’s regulation upon ß-actin mRNA is well characterized, it is likely that multiple RBPs bind to and regulate a given RNA during its entire life cycle. ZBP1 binding to the ß-actin 3’-UTR is in coordination with other proteins, such as ZBP2 (human hnRNP protein K-homology splicing regulator protein, KHSRP). Binding of ZBP2/KHSRP in the nucleus facilitates nuclear ZBP1 association and further cytoplasmic localization in fibroblasts (Farina et al. 2003; Gu et al. 2002). Therefore, the binding of an RBP to its target can be facilitated or hindered by co-factors. Perturbations of mRNA-RBP interactions leads to functional consequences in multiple cell types. For example, in fibroblasts, absence of the mRNA zipcode or ZBP1 results in loss of polarity, focal adhesions, and random mRNA movement (Shestakova et al. 2001; Katz et al. 2012). Similarly in neurons, loss of either the zipcode or ZBP1 affects synaptic formation and dendritic branching (Perycz et al. 2011). The neuronal observations are consistent with ß-actin protein being an important cytoskeletal component involved in growth cone migration and facilitating synapse formation. Loss of the zipcode or ZBP1 in forebrain neurons is associated with decreased growth cone migration (Welshhans and Bassell 2011; Zhang et al. 2001). Hippocampal neurons, in contrast, have notable changes to dendritic arborization, with a severe reduction in branching and filopodia formation (Fig 3I, (Eom et al. 2003; Perycz et al. 2011)). Taken together, these studies demonstrate the importance of these conserved and coordinated molecular events underlying localized translation. However, the role of ZBP1 extends beyond translation by functioning as an adapter between the RNA and cytoskeletal motors and, thus, trafficking RNAs to their final destinations (Song et al. 2015, 11).
RNA transport in dendrites
The dendritic arbor represents a complex maze for mRNA transport and trafficking. Real-time imaging and tracking of mRNAs allows understanding of the basic rules for transport behavior. Endogenous and reporter mRNAs containing zipcodes in their 3′-UTRs exhibit similar velocities during directed motion, consistent with the cis-acting element and the trans-acting factor(s) being the critical determinants of RNP transport. With neuronal stimulation, the ß-actin mRNP can become unmasked and release a translatable pool of ß-actin mRNA in distal dendrites (Buxbaum et al. 2014). To determine whether mRNP localization and unmasking could be controlled, glutamate uncaging delivered neurotransmitter to a subset of dendritic spines. Upon localized stimulation, endogenous β-actin mRNAs localized to the stimulated synapses, resulting in an accumulation of mRNAs over two hours. This local capture of ß-actin mRNPs was blocked in neurons from the ZBP1 knockout mice, suggesting the role of ZBP1 as the targeting protein (Fig 3H, (Yoon et al. 2016)).
When the mRNP particles are in motion, they move processively (0.5–2.0 μm/s) or in a series of short distances (few microns) intervened by short pauses (< 10 s), or remain corralled (diffusion within a small volume of space). The directed movement is indicative of motor-driven transport along microtubule tracks, with instantaneous velocities ranging from 0.5 to 5 μm/s (Yoon et al. 2016; Das et al. 2018). Dendritic mRNPs can move in either direction, or switch directions—depending on the combined force of the bound motors and the orientation of the microtubules. Collectively, all dendritic mRNAs exhibit bidirectional motion, but with a slight bias toward the anterograde which allows them to be delivered to the distal dendrite to participate in local translation when needed.
mRNA localization in dendrites supports a ‘local entrapment’ model as a general mechanism of how local activity can captures mRNAs that are cruising along the dendrites. This represents a “sushi belt” where ß-actin mRNA(s) patrol through multiple synapses like a circling conveyor belt until they are captured by the recently activated synapses and anchored to the base of the spine (Doyle and Kiebler 2011). To determine the fate of the localized mRNP, ß-actin translation reporters showed that after an mRNA localizes to an activated spine, it persists there for hours undergoing multiple rounds of stimulation and translation to generate a pool of new ß-actin proteins which can be incorporated into the expanding dendritic spine structure. Future studies are required to validate whether this “sushi belt” model of mRNA transport is applicable to other dendritically localized mRNAs along the entire length of the dendrite.
To determine the conservation of ZBP1 dependent local translation for transcripts other than ß-actin, transcriptome wide profiling of ZBP1 interacting transcripts can subsequently be evaluated for physiologically relevant interactions.
Technological perspectives on RBPs – genome and proteome wide approaches
Given the complex nature of protein RNA interactions, it is now appreciated that few transcripts act with a single RBP and few RBPs act upon a single transcript. To determine the breadth of interactions development of genome wide approaches has been essential. While each technique focuses on one aspect of RNA regulation (binding to a target, downstream effects on stability, translation regulation) the integration of multiple approaches provides the opportunity to uncover the breadth and depth of RBP based regulation (Lapointe et al. 2018). Here we highlight a few techniques that have been used to study ZBP1 and related RBPs and hypothesize a few possible themes that may result from their integration.
To determine the targets of an RBP, the most notable approaches are RIP (RNA immunoprecipitation) and its successor CLIP (UV-Crosslinking Immunoprecipitation) (reviewed in (Wheeler et al. 2018)). Antibodies are used to isolate the RBP of interest and next generation sequencing is used to identify the fragments that are isolated. However, a number of limitations exist with this approach, the foremost being related to the indirect isolation of RBPs with antibodies. Varying degrees of antibody specificity and affinity are used, making the comparison of CLIP studies across groups, cell lines and experiments challenging. Recently an effort lead by the ENCODE project has characterized commercial antibodies and optimized the CLIP protocol (Van Nostrand et al. 2016). With the optimized protocol, hundreds of RBPs have been studied in HepG2 and K562 cells (Van Nostrand et al. 2016). However, the process of UV crosslinking, antibody based immunoprecipitation as well as most of the downstream processing steps have known associated biases. Complementary approaches to CLIP have been necessary to validate the putative targets that come from this approach.
One such promising in vitro approach performs highly multiplexed measurements of protein ON and OFF rates with thousands of randomly synthesized RNA targets. Named RNA arrays on High Throughput Sequences (RNA-HiTS), this approach utilizes the microscope, microfluidics and flow cells of DNA sequencers. Thousands of RNA sequences are synthesized during sequencing, individual cluster locations are annotated and then interrogated by flowing in fluorescent RNA binding proteins (reviewed in (Denny and Greenleaf 2019)). By determining the landscape of binding affinities in vitro, rules by which RNA binding proteins recognize the sequence and structures of their targets can be interrogated. This allows for refinement of CLIP and other in vivo data, motifs found by RNA-HiTS can be used to identify both false positives and false negative such as putative targets that may not be highly expressed in the cell type used.
Two antibody independent approaches to study RBPs were recently developed, RNA tagging in yeast and TRIBE (Targets or RNA binding proteins Identified By Editing) in Drosophila. Both approaches utilize enzymes fused to a RBP of interest to deposit covalent marks on RNA targets. RNA tagging utilizes yeast genetics to endogenously fuse poly U polymerase (C. elegans PUP-2 which lacks an RNA binding domain) to the RBP of interest. ‘U’ tailed RNAs are then isolated by poly A selection, after which paired end sequencing libraries are generated. Poly U tail length for RNA targets in yeast correlated with both affinity for the RBP as well as cellular function (Lapointe et al. 2015). TRIBE fuses Drosophila adenosine deaminase (ADAR) to an RBP of interest. After FACS sorting of cells expressing the ADAR-RBP fusion protein, standard RNA sequencing library preparation is then performed. Edited RNAs are then identified by the heterogeneity of nucleotides at individual positions along a transcript (McMahon et al. 2016; Rahman et al. 2018; Xu et al. 2018).
While neither RNA tagging nor TRIBE provide the nucleotide level resolution of CLIP, they may select for longer lived RNA-protein interactions and provide methods complementary to CLIP. The recent adaptation and application of TRIBE in mammalian cells (Biswas et al. 2019b) allowed for the profiling of mammalian RBPs such as ZBP1 and its family members (Biswas et al., in preparation). Data comparing mammalian TRIBE to CLIP suggests that TRIBE can avoid the biases and false positives associated with antibody immunoprecipitation and allow for all RBP binding sites to be discovered within a cell ((Biswas et al. 2019b), Biswas et al., in preparation).
To determine the proteins associated with a specific transcript, biochemical techniques such as complimentary biotinylated oligonucleotide based isolation of RNA followed by mass spectrometry have classically been used (Ross et al. 1997). Recent advances in proximity-based labeling techniques using the biotin ligases have emerged as a powerful complementary approach to map RNA-protein interactions. One of the most studied biotin ligase is BirA, a bifunctional protein expressed in E. coli, that mediates biotinylation of a specific lysine residue of a subunit of Acetyl-CoA carboxylase. Using a mutated BirA, which causes promiscuous biotinylation, proximity-dependent biotin identification (BioID) was developed which labels any proximal proteins within a radius of approximately 10 nm (Kim and Roux 2016).
The ß-actin mRNA associated proteome was identified by performing BioID upon ß-actin mRNA in MEFs (Mukherjee et al. 2019). Besides ZBP1, this approach identified novel regulators of ß-actin mRNA localization including FUBP1. Additionally, this approach determined which factors were constitutively loaded upon the RNA and which exchanged during the process of serum starvation or stimulation where localization to the leading edge is decreased or increased respectively. BioID has been extended to map large-scale RNP interactome by analyzing the biotinylation profile of 119 proteins associated at different stages of the mRNA life cycle (Youn et al. 2018).
One caveat to BioID is that it requires biotinylation over 16–24 hours and therefore captures multiple interactions occurring over a long period of time. For more rapid labeling, TurboID (Branon et al. 2018) and ascorbate peroxidase (APEX) (Lam et al. 2015) and RNA-protein interaction detection (RaPID) (Ramanathan et al. 2018) are the techniques of choice, which can capture transient interactions in situ, in different subcellular compartments. An engineered ascorbate peroxidase (APEX), which oxidizes biotin-phenol generates short-lived radicals that can covalently react with tyrosine and other electron-rich amino acids as well as the amino group on guanosine (Kim and Roux 2016). The optimization of the enzyme tags and the ability to biotinylate both RNA and proteins have extended the capability of APEX to map RNA-proteins interactions in cells with subcellular precision (Kaewsapsak et al. 2017; Fazal et al. 2019). More recently, proximity biotinylation by APEX has been used to profile the RNAs associated with different organelles and stress granules (Fazal et al. 2019; Padrón et al. 2019). Future studies may allow for the profiling of RNA granule components using APEX-based strategies.
Imaging based approaches to study mRNA-protein interactions in situ
The study of an mRNA and its binding protein(s) by ensemble biochemical approaches (Mili and Steitz 2004) lacks spatial information. Given the wealth of RBP-RNA interactions now defined by the aforementioned techniques, follow up experiments are required to determine in situ RNA protein interactions as well as subcellular RNA localization. Imaging based approaches therefore can provide spatial information currently inaccessible by other methods and this is particularly critical for the understanding of RNA localization.
High precision imaging, namely “super registration” is capable of determining whether two molecules are physically interacting in situ or simply in proximity by random chance using standard wide-field microscopy (Fig 3E, 3F, (Eliscovich et al. 2017)). This imaging technique corrects the chromatic aberration uniquely intrinsic to individual commercial microscope objectives so that two molecules can be super-imposed within 10 nm precision. By precisely localizing the mRNA and protein using single molecule FISH to detect mRNA and immunofluorescence (IF) against a RBP, a significant fraction of proteins biochemically defined to bind ß-actin mRNA were then shown to not interact with the RNA in situ. Therefore, super registration can complement information from biochemical interactions and visualize physical contacts in situ, with high precision in both neurons and cell lines (Eliscovich et al. 2017; Mukherjee et al. 2019).
Direct RNA-protein interactions within living cells can be interrogated with multi-color fluorescence correlation spectroscopy. By exciting a femtomolar volume within the cell, the association of different molecules can be correlated over time at a specific point in space. This approach can be used alongside super registration microscopy to determine live cell associations of RNA (labeled with an MBS array) with fluorescently tagged RBPs (Fig 3C, 3D, (Wu et al. 2015)). Future developments may combine this approach with super resolution STED imaging (Lanzanò et al. 2017) to allow for interactions to be measured within a dense environment, for instance in an individual dendritic spine.
While single molecule imaging often interrogates individual genes or proteins, recent advances in highly multiplexed RNA FISH can now localize thousands of RNA species inside the cell. By using iterative RNA FISH and multiplex barcoding of the FISH probes, techniques such as SeqFISH (Eng et al. 2019) and MERFISH (Xia et al. 2019) allow transcriptome wide localization of RNAs within both cells and tissues. These approaches have found novel transcripts localized to cellular protrusions, expanding our understanding of RNA localization (Eng et al. 2019). Future applications of these techniques may follow the entire breadth of ZBP1 targets (as defined by CLIP or TRIBE) in rapidly changing environments such as the migrating fibroblast, developing growth cone or stimulated synapse. While both approaches have been combined with single color immunofluorescence, recent multi-colored fluorescence imaging of barcoded antibodies has allowed for spatial profiling of over a dozen different proteins (Guo et al. 2019). Combined barcoding of RNA and protein may allow for future co-localization of the two molecules at a massively parallel scale within cells.
Instead of using oligonucleotide barcoding, other approaches have performed direct sequencing of RNAs in situ. Approaches such as in situ transcriptome accessibility sequencing, have begun to correlate in situ sequencing truncation events with the location of RBP binding sites. Using this approach, it was discovered that Drosophila ZBP1 (dIMP1) binding sites were found within the Act5C mRNA more localized to the periphery of retinal cells (Fürth et al. 2019). As evidenced by the above technologies, further development of high throughput protein and RNA imaging will allow for RNA protein interactions to be directly imaged in situ at a massively parallel scale.
Conclusions - Biological perspectives on RBPs
As evidenced above, work on β-actin-ZBP1 interactions has developed models for how, when and where RNAs become localized inside a cell. Future technical developments are required to allow the dimensions of RNA tracking to be expanded to include the entire life of an RNA as well as the entirety of its protein interactions. Tracking of RNA fate from transcription to decay is possible with the MS2 system (Tutucci et al. 2018) and high throughput methods allow a plethora of RBP interactions to be profiled. Missing from the current studies are in vivo dynamics of RNA-protein interactions, how are regulatory factors exchanged during a RNA’s lifetime? Genome wide studies like CLIP show that many RNA binding proteins interact with the same transcript, often at the same site. How does this multitude of interactions direct the fate of the transcript? Is there functional redundancy amongst RBP family members or RBPs from different families (Biswas et al. 2019a; Conway et al. 2016)? How do the targets of an RNA binding protein change as cellular identity changes during differentiation or reprogramming? Studies have yet to clearly define how changes in RBP expression or stoichiometry affect the process of finding RNA targets. These open questions will require high throughput, time resolved, non-destructive measurements of RNA-protein interactions, ideally within single cells.
Acknowledgements
The authors would like to thank members of the Singer lab past and present for their helpful discussions and contributions to the work presented in this perspective. JB was supported with funding from an MSTP Training Grant T32GM007288 and predoctoral fellowship F30CA214009. CE, YJY and RHS were supported by NIH grant NS083085. YY was also supported by MH120496. RHS was also supported by U01DA047729.
References
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. 1998. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci Off J Soc Neurosci 18: 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Salmon ED, Bloom K. 1999. Localization and anchoring of mRNA in budding yeast. Curr Biol CB 9: 569–578. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. 1998. Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445. [DOI] [PubMed] [Google Scholar]
- Biswas J, Patel VL, Bhaskar V, Chao JA, Singer RH, Eliscovich C. 2019a. The structural basis for RNA selectivity by the IMP family of RNA-binding proteins. Nat Commun 10: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas J, Rahman R, Gupta V, Rosbash M, Singer RH. 2019b. MS2-TRIBE evaluates protein-RNA interactions and nuclear organization of transcription by RNA editing. bioRxiv 829606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. 2018. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36: 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH. 2015. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16: 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Wu B, Singer RH. 2014. Single β-Actin mRNA Detection in Neurons Reveals a Mechanism for Regulating Its Translatability. Science 343: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. 2010. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev 24: 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway AE, Van Nostrand EL, Pratt GA, Aigner S, Wilbert ML, Sundararaman B, Freese P, Lambert NJ, Sathe S, Liang TY, et al. 2016. Enhanced CLIP Uncovers IMP Protein-RNA Targets in Human Pluripotent Stem Cells Important for Cell Adhesion and Survival. Cell Rep 15: 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. 2009. Translational control of long-lasting synaptic plasticity and memory. Neuron 61: 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Moon HC, Singer RH, Park HY. 2018. A transgenic mouse for imaging activity-dependent dynamics of endogenous Arc mRNA in live neurons. Sci Adv 4: eaar3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Singer RH, Yoon YJ. 2019. The travels of mRNAs in neurons: do they know where they are going? Curr Opin Neurobiol 57: 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny SK, Greenleaf WJ. 2019. Linking RNA Sequence, Structure, and Function on Massively Parallel High-Throughput Sequencers. Cold Spring Harb Perspect Biol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M, Kiebler MA. 2011. Mechanisms of dendritic mRNA transport and its role in synaptic tagging: Mechanisms of dendritic mRNA transport. EMBO J 30: 3540–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliscovich C, Buxbaum AR, Katz ZB, Singer RH. 2013. mRNA on the move: the road to its biological destiny. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliscovich C, Shenoy SM, Singer RH. 2017. Imaging mRNA and protein interactions within neurons. Proc Natl Acad Sci 114: E1875–E1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliscovich C, Singer RH. 2017. RNP transport in cell biology: the long and winding road. Curr Opin Cell Biol 45: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C-HL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan G-C, et al. 2019. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568: 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom T, Antar LN, Singer RH, Bassell GJ. 2003. Localization of a β-Actin Messenger Ribonucleoprotein Complex with Zipcode-Binding Protein Modulates the Density of Dendritic Filopodia and Filopodial Synapses. J Neurosci 23: 10433–10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Hüttelmaier S, Musunuru K, Darnell R, Singer RH. 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, Ting AY. 2019. Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell 178: 473–490.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fogarty K, Lifshitz LM, Carrington W, Singer RH. 2003. Visualization of single molecules of mRNA in situ. Methods Enzymol 361: 245–304. [DOI] [PubMed] [Google Scholar]
- Fürth D, Hatini V, Lee JH. 2019. In Situ Transcriptome Accessibility Sequencing (INSTA-seq). bioRxiv 722819. [Google Scholar]
- Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, Gerst JE. 2011. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA 17: 1551–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Delaveau T, Goussard S, Jacq C. 2010. Mitochondrial presequence and open reading frame mediate asymmetric localization of messenger RNA. EMBO Rep 11: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. 2002. A predominantly nuclear protein affecting cytoplasmic localization of β-actin mRNA in fibroblasts and neurons. J Cell Biol 156: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S-M, Veneziano R, Gordonov S, Li L, Danielson E, de Arce, Park D, Kulesa AB, Wamhoff E-C, Blainey PC, et al. 2019. Multiplexed and high-throughput neuronal fluorescence imaging with diffusible probes. Nat Commun 10: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC. 2010. Actin in dendritic spines: connecting dynamics to function. J Cell Biol 189: 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. 2005. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515. [DOI] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS. 2014. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346: 1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR, Tomlinson CR, Brodeur RD. 1983. Localization of actin messenger RNA during early ascidian development. Dev Biol 99: 408–417. [DOI] [PubMed] [Google Scholar]
- Kaewsapsak P, Shechner DM, Mallard W, Rinn JL, Ting AY. 2017. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking ed. E.R. Gavis. eLife 6: e29224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ZB, Wells AL, Park HY, Wu B, Shenoy SM, Singer RH. 2012. β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev 26: 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Roux KJ. 2016. Filling the Void: Proximity-Based Labeling of Proteins in Living Cells. Trends Cell Biol 26: 804–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. 1994. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol 127: 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY. 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12: 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzanò L, Scipioni L, Di Bona M, Bianchini P, Bizzarri R, Cardarelli F, Diaspro A, Vicidomini G. 2017. Measurement of nanoscale three-dimensional diffusion in the interior of living cells by STED-FCS. Nat Commun 8: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe CP, Stefely JA, Jochem A, Hutchins PD, Wilson GM, Kwiecien NW, Coon JJ, Wickens M, Pagliarini DJ. 2018. Multi-omics Reveal Specific Targets of the RNA-Binding Protein Puf3p and Its Orchestration of Mitochondrial Biogenesis. Cell Syst 6: 125–135.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe CP, Wilinski D, Saunders HAJ, Wickens M. 2015. Protein-RNA networks revealed through covalent RNA marks. Nat Methods 12: 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. 1986. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45: 407–415. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. 1985. Quantitative analysis of in situ hybridization methods for the detection of actin gene expression. Nucleic Acids Res 13: 1777–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. 2007. Global Analysis of mRNA Localization Reveals a Prominent Role in Organizing Cellular Architecture and Function. Cell 131: 174–187. [DOI] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Pezo RC, Singer RH. 2002. Single-Cell Gene Expression Profiling. Science 297: 836–840. [DOI] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. 2000. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J 19: 6592–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. 2009. mRNA Localization: Gene Expression in the Spatial Dimension. Cell 136: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AC, Rahman R, Jin H, Shen JL, Fieldsend A, Luo W, Rosbash M. 2016. TRIBE: Hijacking an RNA-Editing Enzyme to Identify Cell-Specific Targets of RNA-Binding Proteins. Cell 165: 742–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA N Y N 10: 1692–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee J, Hermesh O, Eliscovich C, Nalpas N, Franz-Wachtel M, Maček B, Jansen R-P. 2019. β-Actin mRNA interactome mapping by proximity biotinylation. Proc Natl Acad Sci 116: 12863–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. 2003. Real-Time Visualization of ZBP1 Association with β-Actin mRNA during Transcription and Localization. Curr Biol 13: 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrón A, Iwasaki S, Ingolia NT. 2019. Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Mol Cell 75: 875–887.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Hüttelmaier S, Singer RH, Gu W. 2007. ZBP2 Facilitates Binding of ZBP1 to β-Actin mRNA during Transcription. Mol Cell Biol 27: 8340–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, et al. 2012. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev 26: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perycz M, Urbanska AS, Krawczyk PS, Parobczak K, Jaworski J. 2011. Zipcode Binding Protein 1 Regulates the Development of Dendritic Arbors in Hippocampal Neurons. J Neurosci 31: 5271–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman R, Xu W, Jin H, Rosbash M. 2018. Identification of RNA-binding protein targets with HyperTRIBE. Nat Protoc 13: 1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M, Majzoub K, Rao DS, Neela PH, Zarnegar BJ, Mondal S, Roth JG, Gai H, Kovalski JR, Siprashvili Z, et al. 2018. RNA–protein interaction detection in living cells. Nat Methods 15: 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebagliati MR, Weeks DL, Harvey RP, Melton DA. 1985. Identification and cloning of localized maternal RNAs from xenopus eggs. Cell 42: 769–777. [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. 1997. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova EA, Singer RH, Condeelis J. 2001. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci 98: 7045–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer RH, Ward DC. 1982. Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc Natl Acad Sci U S A 79: 7331–7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Zheng Y, Wang Y, Katz Z, Liu X, Chen S, Singer RH, Gu W. 2015. Specific interaction of KIF11 with ZBP1 regulates the transport of β-actin mRNA and cell motility. J Cell Sci 128: 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 12: 143–155. [DOI] [PubMed] [Google Scholar]
- Tutucci E, Vera M, Biswas J, Garcia J, Parker R, Singer RH. 2018. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods 15: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M, Biswas J, Senecal A, Singer RH, Park HY. 2016. Single-Cell and Single-Molecule Analysis of Gene Expression Regulation. Annu Rev Genet 50: 267–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshhans K, Bassell GJ. 2011. Netrin-1-induced local β-actin synthesis and growth cone guidance requires zipcode binding protein 1. J Neurosci Off J Soc Neurosci 31: 9800–9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Nostrand ELV, Yeo GW. 2018. Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. Wiley Interdiscip Rev RNA 9: e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. 2014. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346: 748–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Buxbaum AR, Katz ZB, Yoon YJ, Singer RH. 2015. Quantifying Protein-mRNA Interactions in Single Live Cells. Cell 162: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Fan J, Emanuel G, Hao J, Zhuang X. 2019. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc Natl Acad Sci 116: 19490–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Rahman R, Rosbash M. 2018. Mechanistic implications of enhanced editing by a HyperTRIBE RNA-binding protein. RNA N Y N 24: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli JK. 2005. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell 97: 87–96. [DOI] [PubMed] [Google Scholar]
- Yoon YJ, Wu B, Buxbaum AR, Das S, Tsai A, English BP, Grimm JB, Lavis LD, Singer RH. 2016. Glutamate-induced RNA localization and translation in neurons. Proc Natl Acad Sci 113: E6877–E6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J-Y, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. 2018. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol Cell 69: 517–532.e11. [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M. 2006. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Dev Camb Engl 133: 4573–4583. [DOI] [PubMed] [Google Scholar]
- Zappulo A, van den Bruck D, Mattioli CC, Franke V, Imami K, McShane E, Moreno-Estelles M, Calviello L, Filipchyk A, Peguero-Sanchez E, et al. 2017. RNA localization is a key determinant of neurite-enriched proteome. Nat Commun 8: 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. 2001. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron 31: 261–275. [DOI] [PubMed] [Google Scholar]


