Abstract
Cyanobacteria are prolific producers of natural products and genome mining has shown that many orphan biosynthetic gene clusters can be found in sequenced cyanobacterial genomes. New tools and methodologies are required to investigate these biosynthetic gene clusters and here we present the use of Anabaena sp. strain PCC 7120 as a host for combinatorial biosynthesis of natural products using the indolactam natural products (lyngbyatoxin A, pendolmycin, and teleocidin B-4) as a test case. We were able to successfully produce all three compounds using codon optimized genes from Actinobacteria. We also introduce a new plasmid backbone based on the native Anabaena 7120 plasmid pCC7120ζ and show that production of teleocidin B-4 can be accomplished using a two-plasmid system, which can be introduced by co-conjugation.
Keywords: cyanobacteria, indolactam, secondary metabolites, heterologous expression
Graphical Abstract
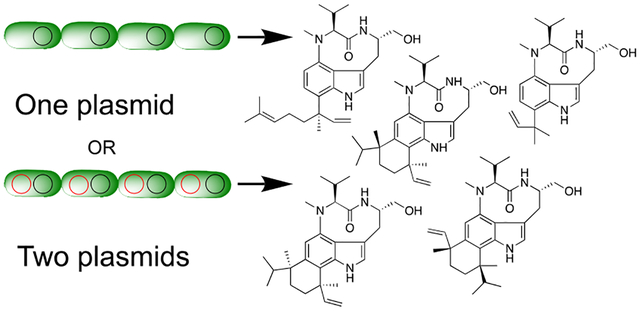
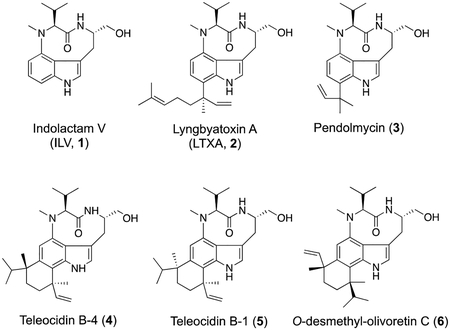
Cyanobacteria are prevalent producers of natural products with diverse structures and interesting bioactivities. Over 1,100 secondary metabolites have been identified from cyanobacteria, and the majority of those are produced by only four genera: Hapalosiphon, Lyngbya (now known as Moorea), Microcystis, and Nostoc.1 Natural products of known or predicted cyanobacterial origin account for roughly 20% of marine-inspired small molecules currently in clinical trials.2 The compounds produced by cyanobacteria can be quite complex, often tailored with rare and unique modifications.3 The ability to produce such an array of compounds hints at the likely untapped biosynthetic capacity of the cyanobacteria. One group of compounds based on the indolactam-V (ILV, 1) core has been the focus of our recent research.
To date, at least three gene clusters have been shown to produce ILV as a biosynthetic intermediate: the ltxA-C genes from the filamentous marine cyanobacterium Moorea producens (formerly Lyngbya majuscula) yield lyngbyatoxin A (2),4–5 a protein kinase C activator;6 the mpnB-D genes from the marine actinomycete Marinactinospora thermotolerans SCSIO 00652 yield pendolmycin (3),7 a compound shown to inhibit phosphatidylinositol turnover;8 and the tleA-D genes of Streptomyces blastmyceticus NBRC 12747 yield teleocidin B-4, teleocidin B-1, and O-desmethylolivoretin C (4–6),9 which are protein kinase C activators.10 The first gene of each of these clusters encodes a two module non-ribosomal peptide synthetase (NRPS; LtxA, MpnB, and TleA, respectively) that forms N-methyl-L-valyl-L-tryptophanol (NMVT, 7). Following the reductive release of 7, a cytochrome P450 (LtxB, MpnC, and TleB, respectively) cyclizes 7 into 1. Pendolmycin and lyngbyatoxin A are formed by reverse prenylation of 1 by MpnD or LtxC/TleC in the presence of dimethylallyl diphosphate or geranyl diphosphate, respectively. Finally, 4, 5, and 6 are formed by the methylation and cyclization of the prenyl group of lyngbyatoxin A onto the indole ring by TleD (Figure 1). The presence of multiple tailoring enzymes that modify the common intermediate (1) has led to a natural combinatorial library of compounds across genera.
Figure 1.
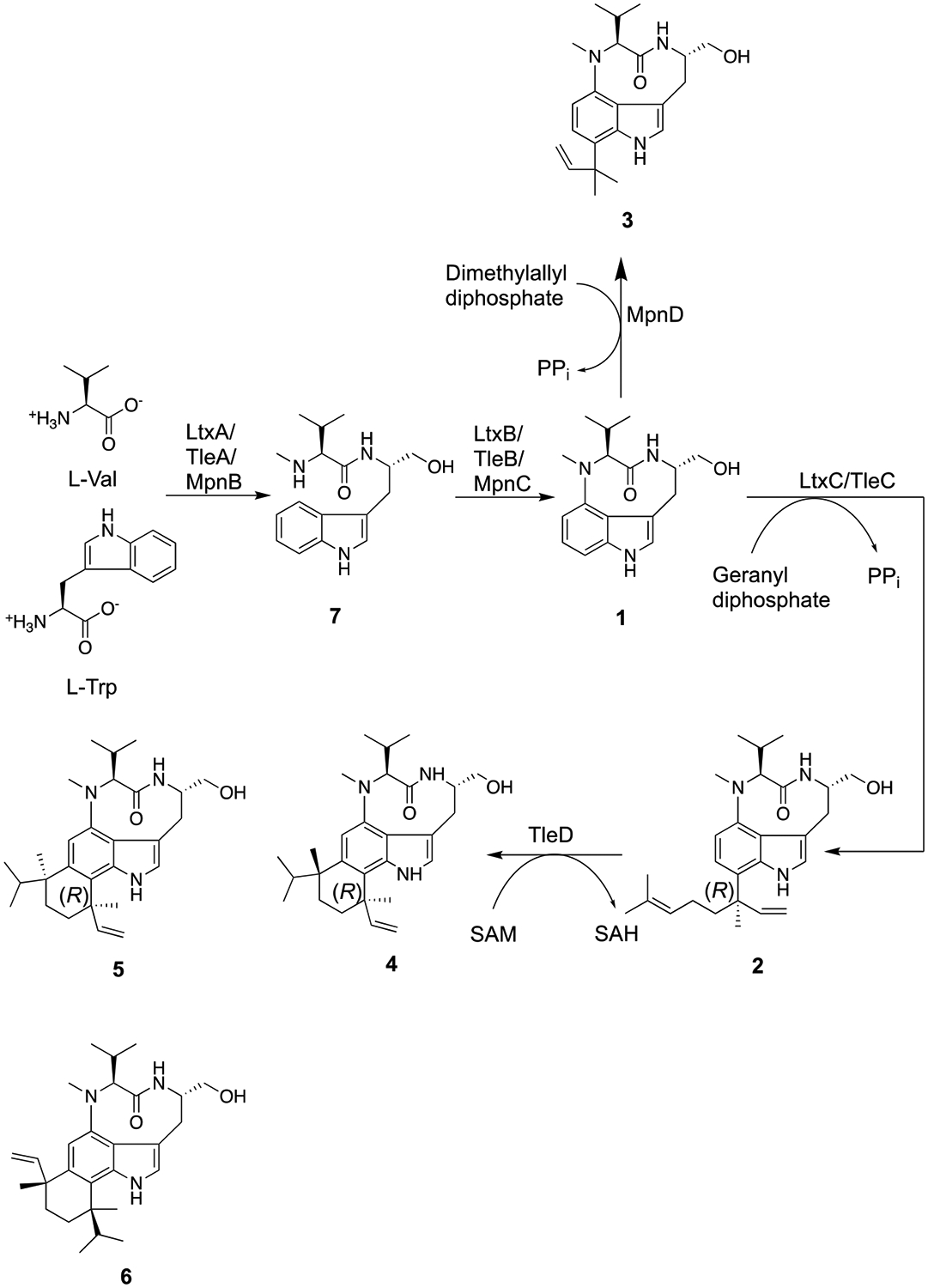
Biosynthetic pathway for the formation of indolactam natural products.
The production of desirable cyanobacterial compounds is often hampered by slow growth of the producing strain, low compound yields, recalcitrance to laboratory culture or genetic manipulation, and the spontaneous cessation of compound production. To overcome these hurdles, Anabaena sp. strain PCC 7120 (herein Anabaena 7120) was assessed as a general heterologous expression host for cyanobacterial natural products.11 Introduction of the ltxA-C genes into Anabaena 7120 on a replicative plasmid resulted in lyngbyatoxin A production with no biosynthetic intermediates (1 or 7) being observed. Exchange of the native ltxA promoter region with the glnA promoter from Anabaena 7120, coupled with a preliminary optimization of growth conditions, resulted in a 13-fold increase in lyngbyatoxin A yield up to 3.2 mg/L. Promoter regions from four other cyanobacterial natural products gene clusters were recognized and expressed by the native Anabaena 7120 transcriptional machinery. Taken together, these results indicate that Anabaena 7120 is a viable candidate for a general cyanobacterial heterologous expression host.11
Here we describe our efforts to expand the toolbox for heterologous expression of natural products in Anabaena 7120 using the indolactam family as a proof of concept. To expand the functional capacity of Anabaena 7120 as a heterologous host, we used different combinations of cyanobacterial (ltxA-C) and codon-optimized non-cyanobacterial genes (mpnD, tleC, and tleD) to produce 2, 3, and 4 in good yields. Here we describe compound production in both BG-11(Nit) and BG-11(NH4) media and utilize fructose to increase the resulting dry cell mass. We also capitalize on the introduction of the glnA promoter to increase compound titers as previously described.11 A new replicative origin of replication, based on the endogenous plasmid pCC7120ζ, was defined and validated for plasmid selection and maintenance in Anabaena 7120. Assessment of the introduction of one or two plasmids simultaneously into Anabaena 7120 demonstrated that, though possible, the efficiency drops with complexity of the conjugation mixture.
Results and Discussion
Heterologous Production of Pendolmycin and Teleocidin B-4 in Anabaena 7120 Using Codon-Optimized Genes from Cyanobacterial and Non-Cyanobacterial Genera in Mixed Gene Clusters.
To extend the utility of Anabaena 7120 as a heterologous host, we sought to determine whether derivations of 1 could be accomplished using tailoring genes of non-cyanobacterial origin. Given the increased yields we previously observed using solid media over liquid media, our experiments were performed primarily using BG-11(Nit) and BG-11(NH4) solid media.11
Three tailoring genes were chosen for study: tleC and tleD from S. blastmyceticus NBRC 12747,9 which encode proteins responsible for the formation of 2 from 1 and the conversion of 2 to 4, 5, and 6 respectively; and mpnD from M. thermotolerans SCSIO 00652,7 which encodes a prenyltransferase that catalyzes the production of 3 from 1. As tleC, tleD, and mpnD are derived from Actinobacteria with high G+C content genomes, the coding regions of tleC, tleD, and mpnD were codon optimized for Anabaena 7120 and commercially synthesized for the following experiments.
The tleC gene encodes a prenyltransferase that has been shown to transfer a geranyl group to 1 to create 2.9, 12 To verify the function of TleC in Anabaena 7120, ltxC was replaced with codon-optimized tleC to retain the gene order if these are transcribed as an operon and use the native promoters if they are not, creating pPJAV642, which was introduced into Anabaena 7120. The resulting strain was cultured on BG-11(Nit) plates, which contain sodium nitrate as the nitrogen source. The cells were collected, lyophilized, and extracted as previously described,11 and the extract was analyzed by HPLC-MS/MS and HPLC-HRMS as described in the Experimental Section. A peak was present only for the extract from the culture harboring ltxAB-tleC (pPJAV642, Figure 2) with a HRESIMS (m/z 438.3112; calcd 438.3115, 0.7 ppm error), and MS/MS fragmentation (Figure S1) pattern consistent with 2.11, 13 To confirm the produced compound was 2 and not the epimer at the C-18 position we pursued isolation of the compound. To facilitate the isolation we exchanged the native promoter with the strong, constitutive PglnA promoter to create pPJAV647.11 Introduction of pPJAV647 to Anabaena 7120 was followed by cultivation in 20 L of liquid BG11(Nit) media supplemented with 50 mM fructose. Addition of fructose was previously shown to increase the growth of Anabaena 7120 under mixotrophic conditions.14 Here we utilized the addition of 50 mM fructose to BG-11(Nit) to increase the yield of dried cell mass that we obtained, which increases the total yield of compound. This allowed sufficient compound to be isolated for characterization via 1H NMR spectroscopy. Comparison to literature data validated that this compound was 2 (Figure S2). These results show that TleC is functional in Anabaena 7120 and can convert 1 to 2 as previously described.9
Figure 2. Production of lyngbyatoxin A (2) and teleocidin B-4 (4).
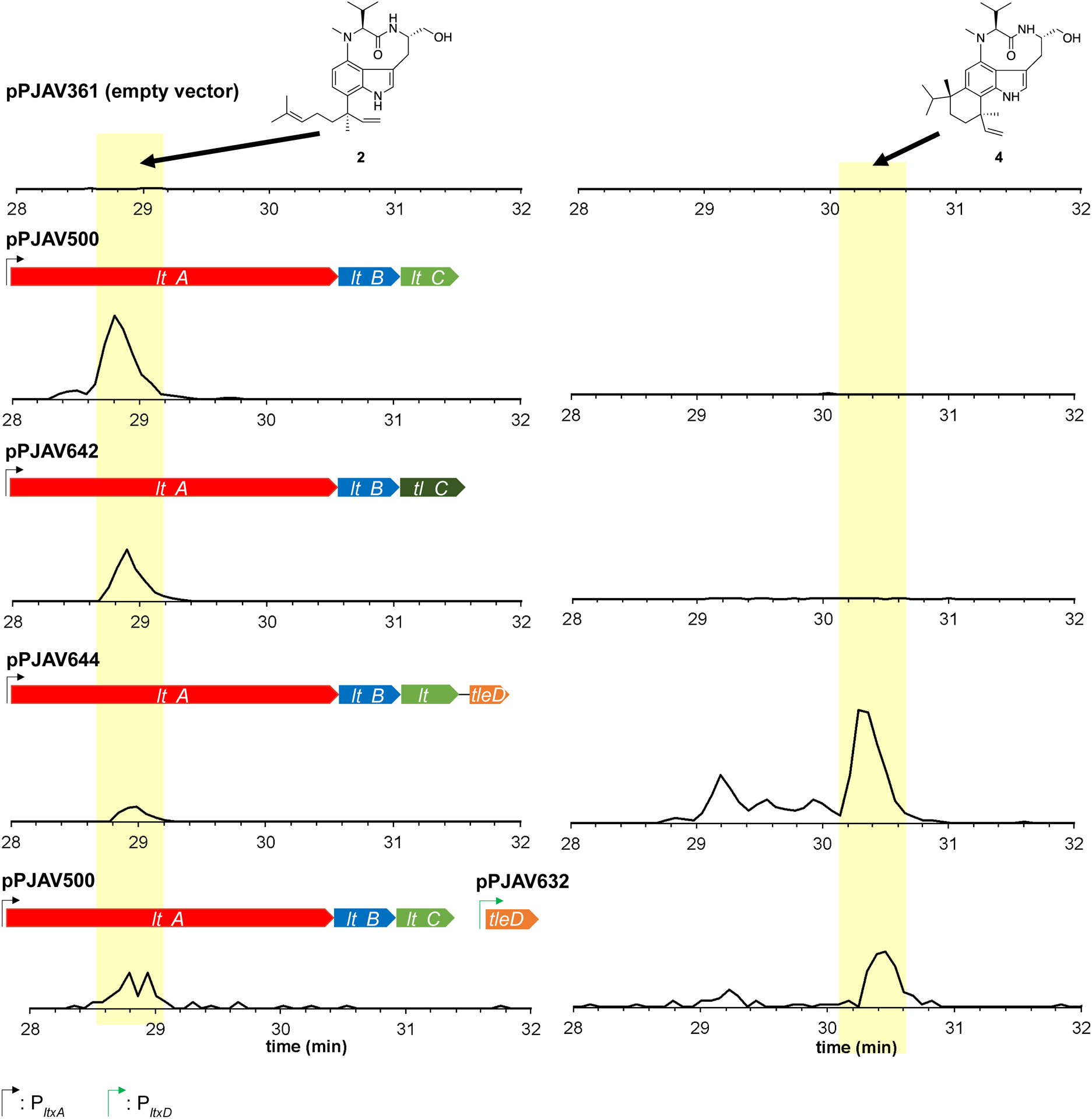
Zoomed LCMS MRM chromatogram showing the constructs used and production of 2 (438.3 → 410.3), left column; and constructs used and production of 4 (452.3 → 424.3), right column. Traces are from a single replicate of culture grown on BG-11(Nit) media containing 1.5% agar (1 plate = approximately 40 mL) and are representative of all samples.
The tleD gene encodes a protein with C-methyltransferase and terpene cyclase activities that converts 2 to 4 as the major product, while 5 and 6 are produced in lower yield.9 To verify the function of TleD in Anabaena 7120, tleD was cloned into the position of ltxD in ltxABCD to create pPJAV644, such that tleD was expressed at the native level of the ltxD gene. Following plasmid introduction, extraction, and analysis (see Experimental Section), four peaks were present between 29.2–30.4 min for the extract from the culture carrying ltxABC-tleD (pPJAV644, Figure 2). Each of these peaks displayed a UV absorption spectrum (Figure S3), high-resolution protonated molecules of 452.3247, 452.3263, 452.3277, and 452.3268 ([M+H]+ calcd 452.3272, Figure S4), and MS/MS fragmentation patterns consistent with teleocidin B-like compounds (Figure S5). To definitively identify the structure of the compounds produced we placed ltxABC-tleD under the control of PglnA to create pPJAV657 (Table S8). Roughly 1.1 mg of the major compound was isolated from Anabaena 7120 containing pPJAV657 (PglnA-ltxABC-tleD) cultivated in 20 L of BG11(Nit) medium supplemented with 50 mM fructose and analyzed by 1H NMR, which confirmed the major product to be 4 (Figure S6, Table S1) in agreement with previous work.9 The other three compounds were produced at low levels and co-eluted with contaminating compounds despite the use of multiple HPLC columns and conditions, which prevented definitive structure characterization.
Under our standard culture conditions, the PltxA-ltxABC-tleD construct produced the four compounds in a 1.6 ± 0.3:1.1 ± 0.1:1.0 ± 0.0:6.0 ± 0.6 ratio as determined through peak area integration in the MRM chromatogram (Figure 2, Figure S7), and inclusion of fructose in the growth medium resulted in a similar distribution of metabolites (2.1 ± 0.5:1.1 ± 0.2:1.1 ± 0.2:6.3 ± 1.1). The production of four compounds here is in contrast to the observation of three compounds by Abe and co-workers.9 However, we note that it is possible that a fourth compound in the in vitro reaction co-eluted with unreacted 2 and was therefore unobservable.9 Given the previous work, two of the other teleocidin B-like peaks are presumed to be O-desmethylolivoretin C and teleocidin B-1. Though the structure of the fourth compound is currently unknown, we postulate that it could be either 9 or 10 (Figure S8), possibly arising from deprotonation of the carbocation intermediate as proposed by Abe and co-workers.9 Experiments to identify the fourth compound are underway in our lab. Production levels of the teleocidin B-like compounds were consistent with those from our previous data on lyngbyatoxin A,11 and the addition of fructose had variable effects on compound production (Figure 3, Table S2–S3). While fructose did occasionally reduce the yield per dried cell mass, as calculated in ng/mg of dried cell mass (Figure 3, Table S2–S3), it almost universally increased the mass of the cells harvested (Table S4), which often increased total compound yield.
Figure 3. Total “Teleocidin B” Production in Anabaena 7120.
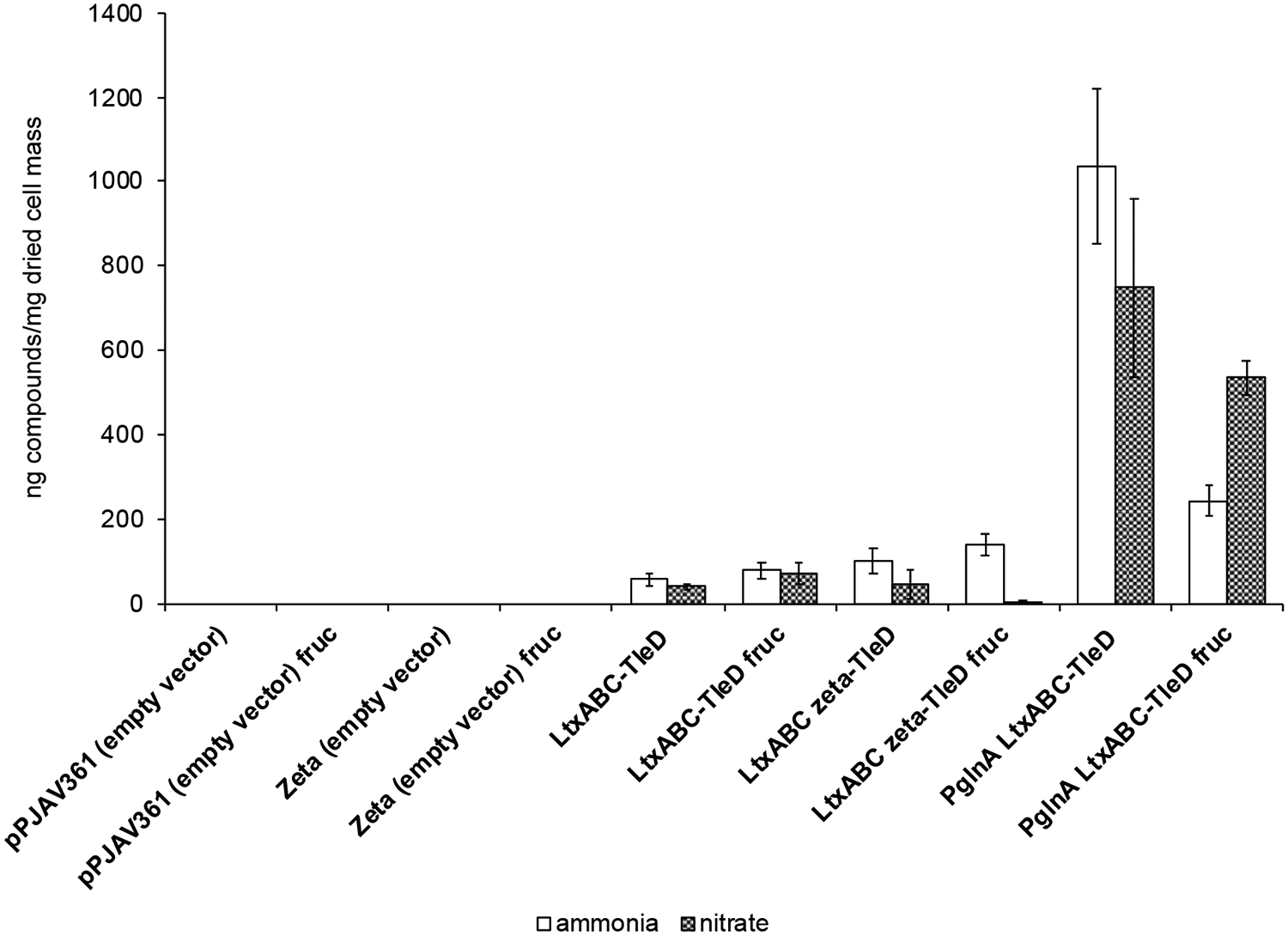
This is the summed total amount of all four compounds displaying the MRM transition m/z 452.3 → 424.3. Production of the individual compounds can be seen in Table S2. Values are given as the average ng/mg of dried cell mass ± standard deviation from three replicates. Each replicate culture was grown on media containing 1.5% agar (1 plate = approximately 40 mL). afruc, denotes media containing 50 mM fructose.
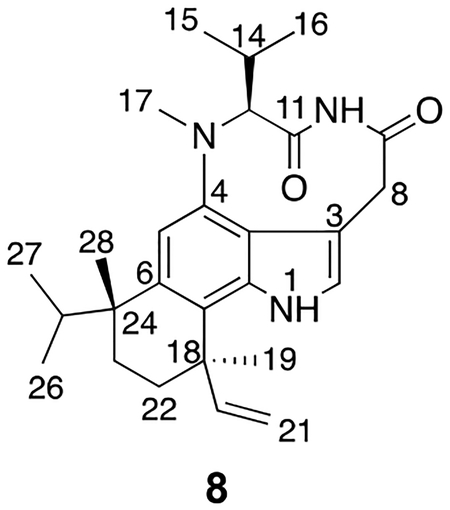
Multiple metabolites with an m/z of 436 (16 amu less than teleocidin B-4) were found in minor quantities in strains producing teleocidin B-4 (Figure S9), and one of the compounds was targeted for isolation and structure determination. HRESIMS produced a protonated molecule at m/z 436.2956 which corresponds to the molecular formula C27H38N3O2+ (calcd 436.2956, 0.46 ppm error, Figure S10). Examination of the 1H, COSY, HSQC, HMBC, and NOESY NMR data established the structure as 8, a presumed oxidative degradation product of teleocidin B-4 (4), the major compound isolated above. In this structure, the hydroxymethylene group has been lost and replaced by a carbonyl group to generate an imide through an oxidative degradation (8, Figure S11–S17, Table S5). Conversion of 4 to 8 results in a single conformer being observed in the 1H NMR spectrum, which is in contrast to the mixture of two conformers found in the other members of the indolactam natural products. Given the extremely low quantities in the extract compared to teleocidin B-4 and related compounds (Figure S9), we propose that these are non-enzymatic degradation products that accumulate during purification and the initial oxidation could be catalyzed by contaminating chlorophyll, which elutes at a similar time. Given that the oxidation occurs at C-9 of teleocidin B-4, we propose that the configurations of the remaining centers are unchanged. Because the XIC of m/z 435.5–436.5 showed four compounds with a similar distribution to the teleocidin B family of compounds, we propose that all four compounds can undergo this oxidative degradation process; however, we only characterized one because the others co-elute with contaminating compounds.
The mpnD gene encodes a prenyltransferase that, unlike TleC or LtxC, transfers a dimethylallyl group to the C-7 position of ILV to complete the biosynthesis of pendolmycin.7 Like the verification of tleC above, mpnD was cloned into the position of ltxC to create pPAJV643 (ltxAB-mpnD). After introduction, culture, and extraction, and LCMS analysis, we noted the presence of a peak only for the extract from the culture containing ltxAB-mpnD and not for plasmids containing ltxC or tleC (Figure 4). This peak had a shorter retention time (23.6 min) than both 2 or 4 and displayed a protonated molecule of 370.2474 Da, consistent with the molecular formula of pendolmycin (calcd 370.2489, 4.0 ppm error, Figure S18). Additionally, the MS/MS fragmentation pattern showed a loss of 28 Da, which was identical to the loss observed in the MS/MS fragmentation of 2 and 4 (Figure S1, S5, S19). Isolation of this compound from a 20 L culture of Anabaena 7120 containing pPJAV659 (PglnA-ltxAB-mpnD) in BG11(Nit) media supplemented with 50 mM fructose, followed by 1H NMR analysis, confirmed it to be pendolmycin (Figure S20, Table S6). Pendolmycin production levels were comparable with our previous data on lyngbyatoxin A production,11 and the addition of fructose had variable effects on the production of pendolmycin, as mentioned above (Figure 5).
Figure 4. Production of pendolmycin.
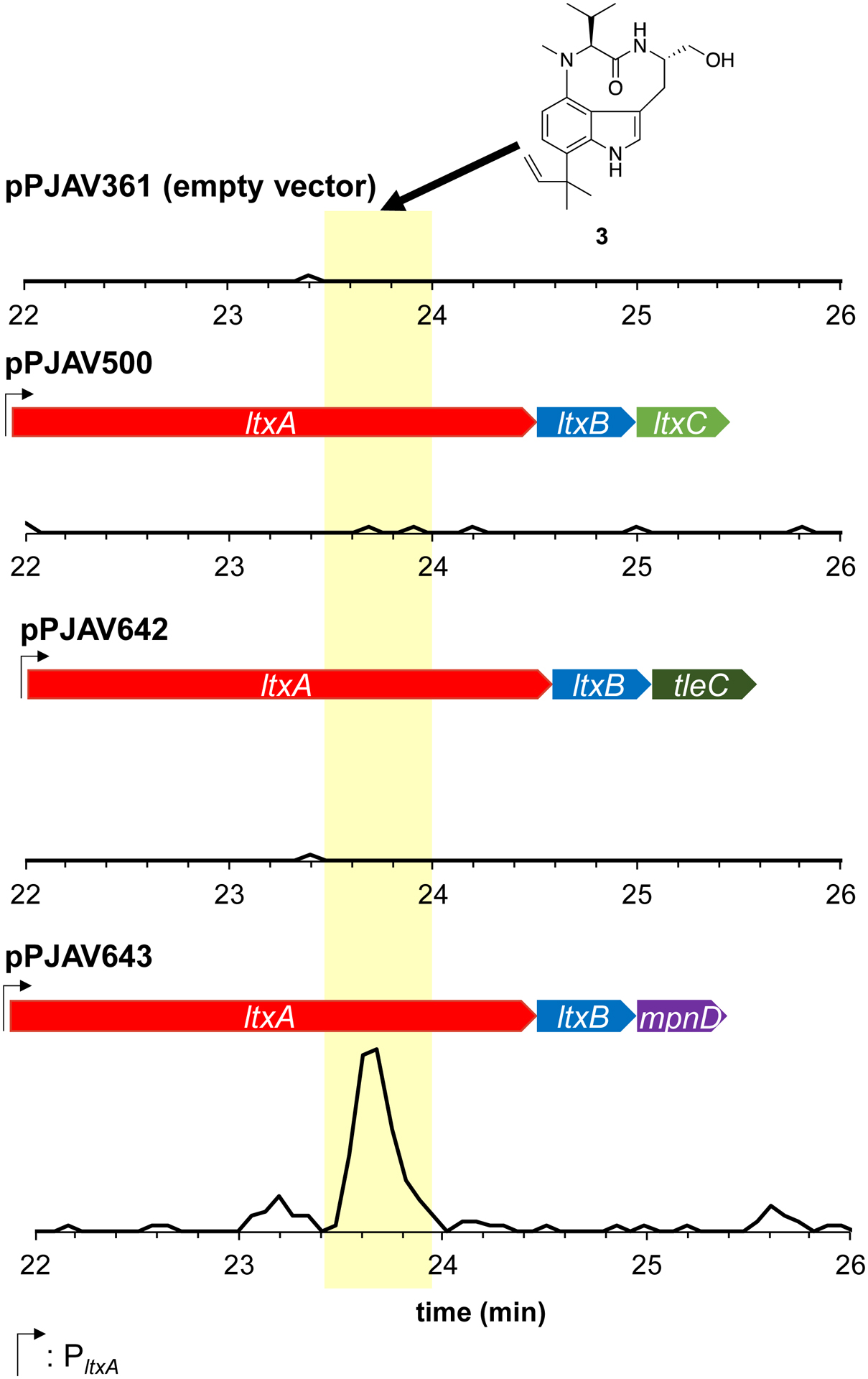
Zoomed LCMS MRM chromatogram showing the constructs used and production of 3 (370.3 → 342.3). Y-axis are all scaled identically. Traces are from a single replicate of culture grown on BG-11(Nit) media containing 1.5% agar (1 plate = approximately 40 mL) and are representative of all samples.
Figure 5. Pendolmycin Production in Anabaena 7120.
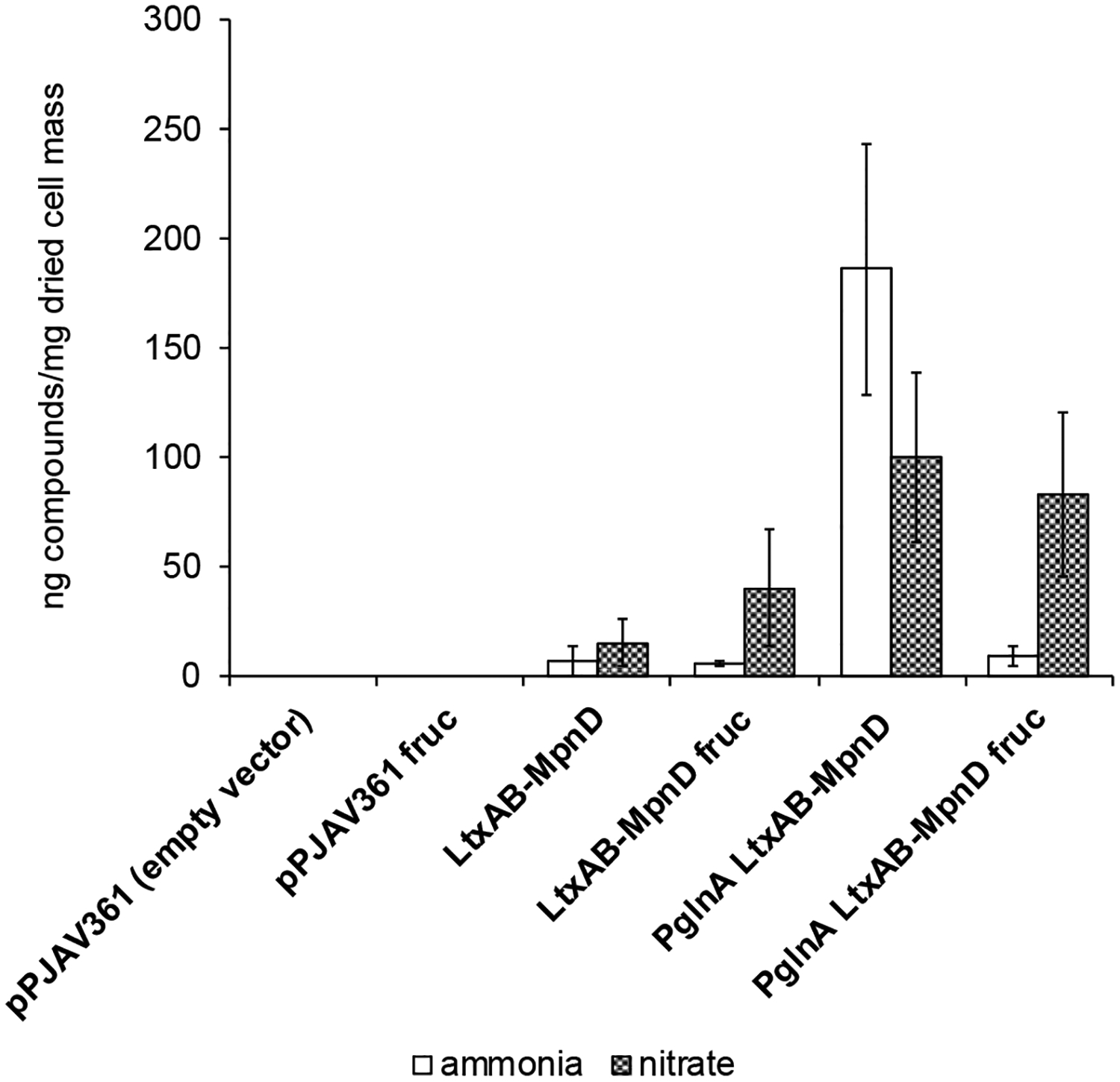
Values are given as the average ng/mg of dried cell mass ± standard deviation from three replicates. Each replicate culture was grown on media containing 1.5% agar (1 plate = approximately 40 mL). afruc, denotes media containing 50 mM fructose.
These results show that the codon-optimized tleC, tleD, and mpnD all produce active proteins with the expected activity when heterologously expressed in Anabaena 7120.9, 15 This demonstrates that the native levels of transcriptional and translational regulation exerted when these genes occupy the same positions as ltxC (tleC, mpnD) and ltxD (tleD) in the ltx gene cluster are sufficient to produce proteins that efficiently catalyzed the desired reactions; each protein completely converted its substrate to the expected product(s) because 1 was not observed in extracts of ltxABC-tleD or ltxAB-mpnD. We did note the presence of trace amounts of 2 in Anabaena 7120 containing pPJAV644. We also note that no other compounds were identified by LC-HRESIMS analysis other than the expected products (e.g. pendolmycin in extracts of Anabaena 7120/pPJAV642) despite the fact that purified TleC and MpnD were found to accept C5-C25 prenyl groups in vitro.15
Creation and Validation of a New Plasmid Vector for Use in Anabaena 7120.
In the above section, we used traditional synthetic biology to stitch together gene clusters, which is a labor-intensive process, and so we endeavored to create a new vector (with orthogonal antibiotic resistance) and co-transformation protocols for synthetic biology efforts in Anabaena 7120 that could be used to quickly screen potential natural product tailoring genes from diverse sources. To date, only the pDU1, pANS, and RSF1010 origins of replication (oriV), derived from Nostoc sp. strain PCC 7524,16 Synechococcus elongatus strain PCC 7942,17 and Salmonella typhimurium,18 respectively, have been shown to facilitate plasmid replication in Anabaena 7120.19–20 To provide another oriV for use in Anabaena 7120, we sought to define a region harboring the oriV from the endogenous Anabaena 7120 plasmid pCC7120ζ.21 Plasmid pCC7120ζ is 5.58 kb in size and has a copy number of roughly six plasmids per chromosome;22 Anabaena 7120 is thought to carry 10 to 20 copies of its chromosome per cell.23 This low copy number is advantageous when carrying larger inserts as lower copy numbers typically correspond with greater insert stability.
We amplified pCC7120ζ using PCR such that every quarter, half, three‐quarter, and linearized whole-plasmid combination was prepared and cloned onto the pBR322 oriV and Sp/Smr cassette from pPJAV361 for replication in E. coli (Figure S15B). Of the nine amplification products generated, only three of these products produced positive clones: the half segments of PCC7120ζ derived from amplification with the primers zeta-SmaI-3F and zeta-SmaI-1R and zeta-SmaI-1F and zeta-SmaI-3R, respectively, and the three-quarter segment derived from the primers zeta-SmaI-2F and zeta-SmaI-1R, which were used to create pPJAV504, pPJAV505, and pPJAV506, respectively (Tables S7 and S8). Anabaena 7120 colonies transformed with plasmid pPJAV504 grew on selection following conjugation (carrying pCC7120ζ DNA nucleotides 4533–1703, relative to the annotated starting position, e. g. the region between primers 3F and 1R, (Figure S21A)). Two independent isolates of pPJAV504, named pPJAV579 and pPJAV580, were re-isolated from Anabaena 7120 using a DNA methylation-deficient strain of E. coli. Sequencing of pPJAV579 and pPJAV580 showed that both had acquired an IS10-type transposon insertion in the pCC7120ζ portion of the plasmids (Figure S21B).24 When pPJAV579 and pPJAV580 were re-conjugated into Anabaena 7120, pPJAV579 displayed a far higher conjugation efficiency so the remaining studies were conducted with this isolate.
To determine which fragment of pCC7120ζ DNA flanking the transposon insertion was required for replication, as well as whether the transposon was involved in this process, the pCC7120ζ DNA fragments and the proximal IS10 insertion sequence were individually deleted from pPJAV579 and the ability to replicate in Anabaena 7120 was assessed. Only introduction of pPJAV606 (harboring the segment of pCC7120ζ proximal to primer zeta-SmaI-1R, Figure S21C), and not pPJAV607 (harboring the segment of pCC7120ζ proximal to primer zeta-SmaI-3F, Figure S21D, Table S8), resulted in Anabaena 7120 colonies following antibiotic selection harboring a plasmid that could be re-isolated. This indicates that the DNA necessary for replication of pCC7120ζ is contained with nucleotides 5427–1703 of pCC7120ζ (relative to the annotated starting condition). Additionally, the ability of the transposon to transpose out of the plasmid was not involved in plasmid replication because pPJAV606, which harbored the transposon with an insertion sequence removed to prohibit transposition, yielded colonies upon introduction in Anabaena 7120. To increase the utility of this finding, a kanamycin/neomycin resistant version of pPJAV606 was also generated (pPJAV626). It was also possible to create a version of pPJAV626 harboring an ~42 kb DNA fragment from Moorea producens (pPJAV655) that was introduced and maintained in Anabaena 7120 (Figure S22).4 This indicates that the pCC7120ζ oriV can support the replication of plasmids much larger than the native pCC7120ζ plasmid (5.58 kb). After nearly two years of repeated cultivation following the initial introduction and continued selection of pPJAV504 and, later, pPJAV606, we were unable to cure native pCC7120ζ. The inability to cure pCC7120ζ indicates that it may contain an essential factor for Anabaena 7120 growth or a portion of it aids in the replication or maintenance of pPJAV504 and pPJAV606. In previous work, a copB homolog (asl9502), shown to be involved in plasmid replication in other systems,25 was required for plasmid maintenance in Anabaena 7120.22 It is possible that curing pCC7120ζ did not occur because replication of pPJAV504 and pPJAV606 required the continued presence of asl9502 and curing of pCC7120ζ would have resulted in colony death on continued antibiotic selection.
Determination of Plasmid Copy Numbers in Anabaena 7120.
Anabaena 7120 is thought to maintain 10–20 copies of its chromosome per cell.23 Previous studies have shown that pDU1-based vectors can be maintained at up to 1800 copies per chromosome while roughly six copies of pCC7120ζ per chromosome have been observed.25–26 To determine the influence of relevant growth conditions and cargo of plasmids on plasmid copy number, qPCR was conducted and the copy number was assessed relative to the chromosomal concentration. The growth conditions tested were liquid and solid medium supplemented with ammonia or nitrate and liquid diazotrophic growth. In general, the copy numbers observed for pDU1- and pCC7120ζ-based plasmids used in this study are similar to those previously published (Table 3).16–20 Though copy numbers were commonly higher in liquid medium than from cultures on solid medium, they were not significantly different (t-test, p = 0.19). In our previous work, strengths of promoters tested with plasmid-borne transcriptional fusions were highest during growth in liquid cultures.11 It is very likely that the increased levels of expression previously recorded were due to the comparably higher copy numbers of pPJAV361-based plasmids during liquid growth. Copy numbers recorded in this work were statistically higher during growth on media supplemented with ammonia (t-test, p > 0.0001), possibly because of the ease of assimilation into metabolic processes. The copy number of the native pCC7120ζ plasmid remained fairly constant within each growth condition irrespective of the presence of pCC7120ζ-based plasmids pPJAV626 or pPJAV632. In most cases, the copy number of pPJAV500 was higher than pPJAV361, which is consistent with previous work indicating that the cargo of the plasmid can influence the copy number of pDU1-derived plasmids in Anabaena 7120.27 The copy numbers of pCC7120ζ-based plasmids with either neomycin or spectinomycin/streptomycin resistance were not statistically different (t-test, p = 0.98), which suggests that the resistance mechanism does not exert much control over copy number. In most growth conditions, the presence of a pDU1-based plasmid resulted in an increased copy number of the pCC7120ζ-based plasmids (pPJAV626 or pPJAV632), though not to a statistically different level (t-test, p = 0.13). In contrast, the pDU1-based plasmid copy number was not generally altered by the presence of a pCC7120ζ-based plasmid (t-test, p = 0.31). This suggests that the pCC7120ζ oriV controls replication in Anabaena 7120 because these two construct types contain the same E. coli oriV. Together, these results indicate that pDU1- and pCC7120ζ-based plasmids can be maintained individually or together in Anabaena 7120 and that their relative copy numbers are modulated by growth condition, plasmid cargo, and, in some cases, the presence of additional plasmids.
Simultaneous Addition of Multiple Plasmids Via Conjugation.
Plasmids are generally introduced into Anabaena 7120 via conjugation from E. coli host strains28 containing a self-mobilizable plasmid such as pRK24 or pRK2013.29 To increase the efficiency of plasmid maintenance following the conjugal event, an Anabaena 7120 DNA methylase on plasmid pRL528 is included to methylate mobilizable plasmids so they are not digested by the native Anabaena 7120 restriction systems.28 This can proceed as a bi- or triparental mating. In a biparental mating, the E. coli donor strain UC585 (or similar conjugal strain) harbors three plasmids: pRK24, pRL528, and the mobilizable plasmid to be introduced, which contains a compatible oriT.30 In a triparental mating, two E. coli strains are utilized: one carrying pRK2013 and pRL528 (e.g. strain JCM113) and a second strain with the oriT-containing mobilizable plasmid to be introduced into Anabaena 7120. Using biparental, triparental, or quadriparental mating strategies, it is theoretically possible to introduce up to two plasmids into Anabaena 7120 simultaneously. As a triparental mating, both mobilizable plasmids are introduced separately into E. coli strain UC585, and a quadriparental mating proceeds when E. coli strain JCM113 is mixed with two E. coli strains each harboring only a single mobilizable plasmid for transfer. We introduced pPJAV361 (pDU1-based empty vector, Spectinomycin/Streptomycin resistant (Sp/Smr)), pPJAV500 (harboring ltxABC, Sp/Smr),11 pPJAV626 (pCC7120ζ-based empty vector, Neomycin resistant (Neor)), and pPJAV632 (harboring PltxD-tleD, Neor) into Anabaena 7120 singly or in combinations of two plasmids with different selectable markers (Neor or Sp/Smr), and utilized both triparental and quadriparental mating strategies to demonstrate the feasibility of simultaneous addition of two plasmids to Anabaena 7120. We successfully selected for the introduction of two plasmids using both triparental mating and quadriparental mating with varying efficiencies (Table S7).
This is the first report of simultaneous addition of multiple plasmids into Anabaena 7120 and indicates that pDU1 and pCC7120ζ are in different plasmid incompatibility groups. This work also opens up the possibility of cloning multiple biosynthetic gene clusters (BGCs) into a single Anabaena 7120 strain or splitting a BGC into two vectors to facilitate conjugation and maintenance of said BGC.
The Use of Multiple Plasmids for Heterologous Compound Production.
Following the assessment of a new oriV for plasmid maintenance in Anabaena 7120 and ascertaining that multiple plasmids can be conjugated into Anabaena 7120 simultaneously, we set out to determine if both plasmids (pPJAV500 and pPJAV632) were functional in tandem. Anabaena 7120 strains carrying either pPJAV361 (empty vector) and pPJAV626 (empty vector), or pPJAV500 (PltxA-ltxABC) and pPJAV632 (PltxD-tleD) were cultured, harvested, extracted, and assessed for the production of 2 and/or 4. While the negative control did not display peaks corresponding to either compound, the two-plasmid combination resulted in the production of the teleocidin B family of compounds in every condition tested (Figure 2, Table S2–S3). We observed incomplete conversion of 2 to the teleocidin B family from the pPJAV500/pPJAV632 dual-plasmid system. During the media optimization experiments noted above, we noted that the addition of fructose (50 mM) increased both LTXA yield and cell mass obtained, greatly increasing the overall yield. We tested the effect of fructose addition on compound production in Anabaena 7120 containing pPJAV500/pPJAV632. Cultivation on BG-11(Nit) supplemented with 50 mM fructose resulted in no 2 being observed as well as an increased yield of the teleocidin B family (Figure 3, Table S2–S3). This observation and the experiments modulating plasmid copy number described above suggest that incomplete conversion of intermediates, formed by the action of proteins encoded on one plasmid, by accessory proteins encoded by the second plasmid may be overcome by media optimization, promoter exchange, or shuffling genes between plasmids.
In the S. blastmyceticus NBRC 12747 genome, the tleD gene is located distally from tleABC.9 It is entirely possible that the tailoring genes associated with other natural product gene clusters are spread throughout the producers’ genomes. Using this two-plasmid system, it would be possible to clone the main gene cluster into one plasmid and then create a cosmid library using the other plasmid to screen for tailoring genes of interest. This strategy could also be employed to test a library of random mutants of a single gene to identify amino acids required for function.
Conclusions/Summary
In this study, we showed that Anabaena 7120 is a viable host for combinatorial biosynthesis of natural products and introduced a new plasmid backbone to enable these investigations. Using the indolactam natural products as a test case and codon optimized genes from Actinobacteria, we were able to produce pendolmycin and teleocidin B-4 in acceptable yields in Anabaena 7120. We were also able to isolate and structurally characterize a previously unknown oxidative degradation product of the indolactam natural product family. We believe that the co-conjugation protocols and new plasmid backbone described in this paper will be useful in studying cyanobacterial natural products and in combinatorial biosynthesis studies in the future.
Experimental Section
General Experimental Procedures.
All UV-vis spectroscopy was performed using a BioSpectrometer Kinetic (Eppendorf). NMR spectra were obtained on a Bruker DPX-500 MHz instrument with a 5mm TXI triple resonance (HCN) probe or a Bruker Avance III-800 MHz instrument equipped with a 4-channel 5 mm TCI cryoprobe using TopSpin version 3.5pl7. Data was processed using TopSpin version 4.0 (Bruker). Quantitation of metabolites was obtained using a Shimadzu Prominence HPLC (consisting of a degasser, two LC-10AD HPLC pumps, an autosampler, a photodiode array, and system controller) upstream of a 3200 QTrap mass spectrometer (AbSciex) operated using the Analyst software package. Data was analyzed offline using Peakview version 2.2 software. High-resolution mass spectrometry was performed using an Agilent 6230 time-of-flight mass spectrometer downstream of an Agilent 1260 Infinity HPLC system consisting of a degasser, quaternary pump, autosampler, and diode array detector. The instrument was operated using MassHunter software and data was processed offline using MassHunter Qualitative software.
LCMS grade H2O and MeOH were purchased from MilliporeSigma, while all restriction enzymes, polynucleotide kinase, Escherichia coli NEB10β and DH5αMCR cells, and T4 DNA ligase were purchased from New England Biolabs. The QIAquick PCR purification kit, QIAquick gel extraction kit, and QIAprep Spin Miniprep Kit were purchased from Qiagen and were used according to the manufacturer’s instructions. LCMS grade H2O, MeCN, and formic acid were purchased from Fisher Chemicals. All other chemicals were purchased from Sigma-Aldrich and used without further purification unless otherwise specified. Sanger sequencing was performed with Big Dye Terminator chemistry at the Center for Genome Resources and Biocomputing (Oregon State University). Primestar GXL was purchased from CloneTech and KOD Hot Start DNA polymerase was purchased from EMD Millipore and both were used according to the manufacturer’s instructions. Oligonucleotides were purchased from IDT Technologies with standard desalting and used without further purification.
Bacterial Strains and Growth Conditions.
The strains used in this study are listed in Table S8. E. coli was routinely cultured in Lysogeny Broth (LB), Miller supplemented with spectinomycin (100 μg/mL), kanamycin (50 μg/mL), ampicillin (100 μg/mL), or chloramphenicol (30 μg/mL) for plasmid selection as previously described and solidified with 1.5% agar for plate culture.11 Anabaena PCC 7120 was routinely grown on BG-11 medium with nitrate (BG-11(Nit)) or ammonia (BG-11(NH4)) as the nitrogen source and supplemented with streptomycin and spectinomycin (2.5 μg/mL each) or neomycin (90 μg/mL) for plasmid selection as previously described.11, 31 Plasmids were introduced into Anabaena 7120 by conjugation from E. coli as previously described.28, 32 Bacteria were handled using aseptic technique and all molecular biology protocols were performed according to standard procedures unless otherwise stated.33
Plasmid Construction.
The plasmids used in this study are listed in Table S8. The oligonucleotides used in this study are listed in Table S9. The preparation of inserts and vectors for ligase-mediated cloning and λ/Red recombination using pKD46 in E. coli strain BW25113 were conducted as previously described.11, 34 Plasmids pUC57-tleC, pUC57-tleD, and pUC57-mpnD, which contain codon-optimized genes encoding TleC and TleD from Streptomyces blastmyceticus NBRC 12747 and MpnD from Marinactinospora thermotolerans SCSIO 00652, respectively, were purchased from GenScript, and individually cloned into the EcoRV site of pUC57 to create pUC57-tleC, pUC57-tleD, and pUC57-mpnD.
Construction of Plasmids pPJAV504, pPJAV505, and pPJAV506.
Plasmids pPJAV504, pPJAV505, and pPJAV506 are mobilizable shuttle vectors based on pPJAV36111 with fragments of the endogenous Anabaena 7120 Zeta plasmid individually replacing the pDU1 oriV.16, 21 Fragments 2, 3, and 6 of the Zeta plasmid were amplified by PCR from Anabaena 7120 chromosomal DNA with the primer pairs zeta-SmaI-3F and zeta-SmaI-1R, zeta-SmaI-4F and zeta-SmaI-2R, zeta-SmaI-2F and zeta-SmaI-1R, respectively (Figure S16A, Table S9), and all digested with SmaI. A fragment from pPJAV361 containing the pBR322 oriV, oriT, and the Spr/Smr Ω interposon was amplified by PCR from pPJAV361 with the primers pAM504-pBR-F and pAM504-pBR-R and each fragment of Zeta was individually cloned with the product to create pPJAV504, pPJAV505, and pPJAV506.
Construction of Plasmids pPJAV579 and pPJAV580.
Plasmids pPJAV579 and pPJAV580 are mobilizable shuttle vectors based on pPJAV361 harboring a transposon insertion within the fragment of Zeta DNA (pCC7120ζ). Following introduction and selection of pPJAV504 in Anabaena 7120, positive colonies were isolated, genomic DNA was extracted, and transformed into E. coli strain DH5αMCR. Plasmids were extracted from the spectinomycin resistant E. coli colonies arising from transformation with DNA extracted from two independent Anabaena 7120 isolates. PCR analysis showed that the Zeta fragment amplified from each isolate was larger than analogous fragment amplified from pPJAV504. The region was sequenced with the walking primers 505-BamHI, pAM504Ecoliup-R, Zeta2-1-walk1-F, Zeta2-1-walk1-R, and zeta-SmaI-4R. Both isolates harbor a transposon insertion within the fragment of Zeta DNA to create pPJAV579 and pPJAV580.
Construction of Plasmids pPJAV606 and pPJAV607.
Plasmids pPJAV606 and pPJAV607 are mobilizable shuttle vectors based on pPJAV361 harboring half of the fragment of Zeta DNA on either side of the transposon insertion present in pPJAV579. pPJAV579 was linearized by PCR with the primer pairs Tnp-int-F and pAM504-pBR-R and Tnp-int-R and pAM504-pBR-F, respectively, and phosphorylated with polynucleotide kinase). The products were self-ligated removing the up- and downstream portions of Zeta DNA and the transposon insertion sequence relative to the orientation of the transposon in pPJAV579 to create pPJAV606 and pPJAV607.
Construction of Plasmid pPJAV626.
Plasmid pPJAV626 is a mobilizable shuttle vector based on pPJAV50435 harboring the fragment of Zeta DNA allowing pPJAV606 to replicate in Anabaena 7120. A fragment containing Zeta and transposon DNA was amplified by PCR from pPJAV606 with the primers Zeta-SmaI-3F and pAM504-Ecoliup-R, which was phosphorylated with polynucleotide kinase. This product was digested with SmaI and cloned onto the portion of pAM504 required for kanamycin resistance, conjugation, and replication in E. coli, amplified by PCR from pAM504 with the primers pAM504-pBR-F and pAM504-pBR-R, to create pPJAV626.
Construction of Plasmid pPJAV631.
Plasmid pPJAV631 is a source of the ltxA promoter. The ltxA promoter was amplified by PCR from the fosmid fos-DE3–864 with the primers PlxtA-XhoI-F and Pltx-R. The product was cloned into the EcoRV site of pBlueScript SK+ (Stratagene) and screened for directionality by PCR such that the promoter reads toward the SmaI site to create pPJAV631.
Construction of Plasmid pPJAV632.
Plasmid pPJAV632 is a mobilizable shuttle vector based on pPJAV626 carrying PltxD-tleD. The ltxD promoter region was amplified by PCR from the fosmid fos-DE3–86 with the primers PltxD-tleC-red-F and PltxD-R and the coding region of tleD was amplified from pUC57-tleD with the primers tleD-PltxD-OEX-F and tleD-R. The products were fused by overlap extension,36 cloned into the SmaI site of pPJAV626, and screened for directionality by PCR to read away from the Zeta DNA involved in replication to create pPJAV632.
Construction of Plasmid pPJAV642.
Plasmid pPJAV642 is a mobilizable shuttle vector based on pPJAV361 carrying PltxA-ltxAB-tleC. The coding region of tleC was amplified by PCR from pUC57-tleC with the primers tleC-ltxB-red-F and tleC-R and cloned into the SmaI site of pPJAV631. A fragment containing PltxA-tleC was amplified by PCR from the previous construct with the primers PltxA-XhoI-F and tleC-R, cloned into the SmaI site of pPJAV361, and checked for directionality such it read away from the spectinomycin/streptomycin resistance cassette. The resulting plasmid was linearized by PCR with the primers Pltx-R and tleC-ltxB-red-F and electroporated into competent E. coli as described above for lambda red recombination to recombineer ltxA-C. Resulting transformants were screened by PCR for the presence of ltxA-C to create pPJAV642.
Construction of Plasmid pPJAV643.
Plasmid pPJAV643 is a mobilizable shuttle vector based on pPJAV361 carrying PltxA-ltxAB-mpnD. The coding region of mpnD was amplified by PCR from pUC57-mpnD with the primers mpnD-ltxB-red-F and mpnD-R and cloned into the SmaI site of pPJAV631. A fragment containing PltxA-mpnD was amplified by PCR from the previous construct with the primers PltxA-XhoI-F and mpnD-R, cloned into the SmaI site of pPJAV361, and checked for directionality such it read away from the spectinomycin/streptomycin resistance cassette. The resulting plasmid was linearized by PCR with the primers Pltx-R and mpnD-ltxB-red-F and electroporated into competent E. coli as described above for lambda red recombination to recombineer ltxA-C. Resulting transformants were screened by PCR for the presence of ltxA-C to create pPJAV643.
Construction of Plasmid pPJAV644.
Plasmid pPJAV644 is a mobilizable shuttle vector based on pPJAV361 carrying PltxA-ltxABC-tleD. The coding region of tleD was amplified by PCR from pUC57-tleD with the primers tleD-PltxD-OEX-F and tleD-R and PltxD was amplified by PCR from fos-DE3–86 with the primers PltxD-tleC-red-F and PltxD-R. These products were fused by overlap extension and cloned into the SmaI site of pPJAV631. A fragment containing PltxA-PltxD-tleD was amplified by PCR from the previous construct with the primers PltxA-XhoI-F and tleD-R, cloned into the SmaI site of pPJAV361, and checked for directionality such it read away from the spectinomycin/streptomycin resistance cassette. The resulting plasmid was linearized by PCR with the primers Pltx-R and PltxD-tleC-red-F and electroporated into competent E. coli as described above for lambda red recombination to recombineer ltxA-C. Resulting transformants were screened by PCR for the presence of ltxA-C to create pPJAV644.
Construction of Plasmid pPJAV647.
Plasmid pPJAV647 is a mobilizable shuttle vector based on pPJAV361 carrying PglnA-ltxAB-tleC. A fragment harboring PglnA-ltxA was amplified by PCR from pPJAV50311 with the primers PglnA-XhoI-F and ltxA-int-SmaI-R. The product was cloned as an XhoI-NdeI fragment into the same sites of pPJAV642 to create pPJAV647.
Construction of Plasmid pPJAV650, pPJAV657, and pPJAV659.
Plasmids pPJAV650, pPJAV657, and pPJAV659 are mobilizable shuttle vectors based on pPJAV361 carrying PglnA-ltxAB-mpnD-tleD, PglnA-ltxABC-tleD, and PglnA-ltxAB-mpnD, respectively. A fragment harboring PglnA-ltxA was amplified by PCR from pPJAV503 with the primers PglnA-XhoI-F and ltxA-int-SmaI-R, digest with XhoI, and cloned into the XhoI-EcoRV sites of pBlueScript SK+. Fragments containing mpnD-tleD, ltxC-tleD, tleC-tleD, and mpnD were amplified by PCR from pPJAV645, pPJAV644, pPJAV646, and pPJAV643, respectively, with the forward primer ltxB-int-NdeI-F and the corresponding reverse primer tleD-R or mpnD-R. The products were digested with NdeI and individually cloned into the NdeI-SmaI sites downstream of PglnA-ltxA in pBlueScript SK+. Fragments containing PglnA-ltxA-mpnD-tleD, PglnA-ltxA-ltxC-tleD, and PglnA-ltxA-mpnD were excised from pBlueScript SK+ via XhoI-SacI digestion and cloned into the SalI-SacI sites of pPJAV361. Each of the resulting constructs was linearized by NdeI digestion and electroporated into competent E. coli as described above for Lambda Red recombination to recombineer ltxAB. Resulting transformants were screened by PCR for the presence of ltxAB, ltxC, tleC, tleD, and mpnD as appropriate to create pPJAV650, pPJAV657, and pPJAV659, respectively.
Construction of Plasmid pPJAV653.
Plasmid pPJAV653 is a mobilizable shuttle vector based on pAM50437 carrying the coding region of hetR as well as an Ω interposon conferring resistance to spectinomycin and streptomycin. The coding region of hetR was amplified by PCR from Anabaena 7120 chromosomal DNA with the primers HetR-NdeI-F and HetR-R and the product was cloned into the SmaI site of pAM504. This construct was digested with EcoRI, blunt-ended, and the Ω interposon from pDW938 was cloned in as a blunt-ended HindIII fragment to create pPJAV653.
Construction of Plasmid pPJAV655.
Plasmid pPJAV655 is a mobilizable shuttle vector based on pPJAV626 carrying the ~40 kb M. producens DNA fragment from fos-DE3–86. Regions up- and downstream of the M. producens insert were amplified by PCR from fos-DE3–86 with the primer pairs fos-up-F and fos-up-BamHI-R and fos-dn-BamHI-F and fos-dn-EcoRI-R, respectively. The up- and downstream products were digested with BamHI and cloned into the SmaI site of pPJAV626. The resulting plasmid was linearized by PCR with the primers fos-dn-BamHI-F and fos-up-BamHI-R and electroporated into competent E. coli as described above for Lambda Red recombination to recombineer the M. producens DNA fragment from fos-DE3–86. Resulting transformants were screened by PCR for the presence of ltxA-D to create pPJAV655.
Quantification of Conjugation Efficiency.
Anabaena 7120 was cultured in 100 mL of BG-11 (Nit) until mid-log phase (OD750 of 0.6 – 0.9), cells were pelleted using centrifugation at 2,000 × g for 3 min, all but 10 mL of the supernatant was decanted, and then the cells were transferred to a glass culture tube. The culture was disrupted by sonication with a Branson 3510 sonicator until the average filament length was roughly 10 cells long as determined by visual assessment. E. coli strains NEB10β (New England Biolabs) and UC585,30 individually harboring pPJAV361, pPJAV500, pPJAV606, or pPJAV636, and JCM113 (HB101 with pRL528 and pRK2013 for conjugation, a kind gift of J. C. Meeks) were grown overnight in 2 mL liquid cultures of LB containing the appropriate antibiotic, and were used to inoculate fresh 2 mL LB cultures in the morning (1:100 dilution). Once these new cultures grew to an OD600 of 0.7–0.9, they were pelleted by centrifugation at 7,000 × g and washed twice with BG-11(Nit). For conjugation, 200 μL of sonicated Anabaena 7120 (about 5.6 × 108 CFU) was mixed with 50 μL of each washed E. coli strain (about 2.1 × 108 CFU) and allowed to dry onto the surface of BG-11(Nit) plates supplemented with 5% LB broth as previously described.32 Every bi-, tri-, and quadraparental mating was mixed in triplicate to create all possible combinations that would result in the introduction of one or two plasmids. After 2 days of growth on the conjugation plates, each mix was individually resuspended in 1 mL of BG-11(Nit) and used to create five ten-fold serial dilutions from which 300 uL was plated onto BG-11(Nit) supplemented with the appropriate antibiotics. Colonies were counted 3 weeks later and used to calculate the combined efficiency of conjugation, plate transfer, and selection as compared to the possible growth of the unconjugated sonicated Anabaena 7120 culture. The data was expressed in this manner because it more accurately represents the number of colonies expected following completion of this protocol rather than solely an approximation of conjugal efficiency.
Production, Purification, and LC-MS/MS Quantification of Lyngbyatoxin A, Pendolmycin A, and Teleocidin B.
Assays for the production of heterologously expressed compounds were prepared, carried out, and compounds were extracted as previously described.11 Assay conditions were identical to those previously published.11 LC-MS/MS analyses were conducted as previously described with minor modifications. Briefly, a Shimadzu Prominence HPLC consisting of a degasser, two LC-10AD HPLC pumps, an autosampler, a photodiode array, and system controller, upstream of a 3200 QTrap mass spectrometer (AbSciex) was used for separation and quantitation. Separation was achieved using a Luna C18(2) column (2.0 Å~ 150 mm, 3 μm, Phenomenex) with a flow rate of 0.2 mL/min with line A containing H2O + 0.1% (v/v) formic acid and line B containing MeCN + 0.1% (v/v) formic acid, which operated under the following program. The column was pre-equilibrated in 95% A/5% B, and upon injection, this composition was held for 1 min. The composition of mobile phase was then changed to 0% A/100% B over 29 min utilizing a linear gradient. This composition was held for 5 min, followed by changing to 95% A/5% B over 3 min. The column was equilibrated in 95% A/5% B for 5 min prior to the next injection. Under these chromatographic conditions, pendolmycin eluted at 23.6 min, lyngbyatoxin A eluted at 29 min, and the four compounds with protonated molecules at m/z 452.3 eluted from 29.2–30.4 min. MS/MS analysis was done in MRM mode for pendolmycin (Q1, 370.3; Q2, 242.3; 40 ms), lyngbyatoxin A (Q1, 438.3; Q2, 410.2; 40 ms), and m/z 452.3 compounds (Q1, 452.3; Q2, 424.2; 40 ms). The instrument was operated with Analyst 1.5.1, build 5218, and data analysis was performed with PeakView, ver. 2.1.0.11041 (AbSciex). Known concentrations of purified pendolmycin, lyngbyatoxin A, and teleocidin B-4 were run on the same program and standard curves were created for the low range from the MRM programs described above and for the high range by integrating the area under the UV curve at 300 ± 2 nm.
DNA Extraction and Plasmid Copy Number Determination by qPCR.
Anabaena 7120 strains harboring pPJAV361, pPJAV500, pPJAV606, pPJAV626, and/or pPJAV636 were grown on plates or in 30 mL liquid cultures in identical conditions to those used for heterologous expression, as well as in liquid BG-11(NH4) or liquid BG-11(Nit) 48 h after the removal of combined nitrogen, to represent many physiologically relevant growth conditions. After the required culture duration, cells were scraped from plates or pelleted by centrifugation at 2,000 × g for 3 min, washed with TES (10 mM Tris HCl, pH 7.5; 25 mM EDTA, pH 8.0, 500 mM NaCl) to remove exopolysaccharides as previously described,39 and stored at −80 °C until processing. The pellets were treated with lysozyme as previously described40 and DNA was purified by phenol-chloroform extraction as described.26, 41 DNA concentrations were measured using a BioSpectrometer kinetic (Eppendorf) and all samples were diluted to 100 pg/uL for use. All qPCRs were performed in MicroAmp Fast Optical 96 well reaction plates (Thermo Fisher Scientific) covered with MicroAmp Optical Adhesive film (Thermo Fisher Scientific). qPCR reactions were assembled manually and contained 10 pmol of each primer, 100 pg of template DNA, and 10 μL of iTaq Universal SYBR Green Supermix (Bio-Rad) in 20 μL reactions. The primers AMO-645 and AMO-646 were utilized to amplify a portion of the hetR coding region to assesses the concentration of Anabaena 7120 chromosomes as previously described.22 Portions of the alr9504, nptII, and aadAI genes were amplified from purified plasmid DNA with the primer sets Zeta-qPCR-F and Zeta-qPCR-R, AMO-679 and AMO-680, and aadA1-F and aadA1-R, respectively, as previously described.22 The reaction profile was 3 min at 95 °C followed by 40 cycles of 30 s at 95°C, 30 s at 56 °C, and 30 s at 72 °C. An annealing temperature of 56°C was used because this temperature elicited similar amplification levels from all primer sets. Negative controls (no template DNA) were included and a melting curve analysis was performed in all cases. qPCRs were performed with two biological replicates and technical triplicate of each DNA sample in a StepOnePlus Real-Time PCR System (Applied Biosystems). Standard curves were performed with eight ten-fold serial dilutions for each primer set in technical triplicate, using purified PCR products, derived from either pPJAV653 or chromosomal DNA, as templates. The relative quantities of each sample were calculated using the ΔΔCt method, considering each primer’s specific efficiency calculated from the standard curves. All values are expressed as the plasmid copy number per chromosome as previously described.22
Supplementary Material
Table 1. Copy Number of pDU1- and Zeta-based Plasmids in Anabaena 7120 During Culture in Liquid or Solid BG-11 Medium Supplemented with Nitrate or Ammonia as Nitrogen Sources or Grown Diazotrophically.
Experiments not conducted are denoted as N/A.
| Liquid BG-11 | |||
|---|---|---|---|
| Plasmids | pDU1 | Zeta-Sp/Km | Native Zeta |
| pPJAV606 | N/A | 4.9 ± 0.01 | 12.9 ± 0.6 |
| pPJAV626 | N/A | 2.1 ± 0.2 | 1.9 ± 0.1 |
| pPJAV632 | N/A | 4.2 ± 0.1 | 12.2 ± 0.2 |
| pPJAV361 | 2.9 ± 0.1 | N/A | 10.7 ± 0.3 |
| pPJAV500 | 15.9 ± 0.3 | N/A | 12.2 ± 0.9 |
| pPJAV361 + pPJAV626 | 15.5 ± 0.0 | 1.4 ± 0.1 | 11.8 ± 0.5 |
| pPJAV361 + pPJAV632 | 3.2 ± 0.2 | 3.6 ± 0.1 | 9.9 ± 0.1 |
| pPJAV500 + pPJAV626 | 8.1 ± 0.1 | 8.9 ± 0.2 | 9.9 ± 0.2 |
| pPJAV500 + pPJAV632 | 4.4 ± 2.1 | 4.9 ± 2.5 | 10.4 ± 0.2 |
| Liquid BG-11(NH4) | |||
| Plasmids | pDU1 | Zeta-Sp/Km | Native Zeta |
| pPJAV606 | N/A | 6.1 ± 0.8 | 15.6 ± 2.1 |
| pPJAV626 | N/A | 1.7 ± 0.1 | 1.5 ± 0.1 |
| pPJAV632 | N/A | 5.0 ± 1.0 | 17.3 ± 5.3 |
| pPJAV361 | 182.2 ± 8.7 | N/A | 17.4 ± 5.3 |
| pPJAV500 | 414.5 ± 64.1 | N/A | 25.4 ± 5.4 |
| pPJAV361 + pPJAV626 | 296.4 ± 84.3 | 2.8 ± 0.7 | 30.3 ± 5.1 |
| pPJAV361 + pPJAV632 | 93.9 ± 16.6 | 92.2 ± 15.5 | 13.9 ± 1.4 |
| pPJAV500 + pPJAV626 | 237.7 ± 5.2 | 230.5 ± 6.4 | 19.0 ± 3.4 |
| pPJAV500 + pPJAV632 | 199.0 ± 44.7 | 206.9 ± 34.8 | 20.1 ± 1.2 |
| Liquid BG-11 Diazotrophy | |||
| Plasmids | pDU1 | Zeta-Sp/Km | Native Zeta |
| pPJAV606 | N/A | 5.9 ± 0.7 | 1.5 ± 0.01 |
| pPJAV626 | N/A | 2.4 ± 0.2 | 2.2 ± 0.02 |
| pPJAV632 | N/A | 5.2 ± 0.4 | 2.2 ± 0.2 |
| pPJAV361 | 3.1 ± 0.1 | N/A | 1.7 ± 0.02 |
| pPJAV500 | 17.9 ± 1.4 | N/A | 1.9 ± 0.01 |
| pPJAV361 + pPJAV626 | 6.7 ± 0.1 | 1.3 ± 0.04 | 1.9 ± 0.2 |
| pPJAV361 + pPJAV632 | 13.0 ± 1.0 | 3.8 ± 0.4 | 2.0 ± 0.1 |
| pPJAV500 + pPJAV626 | 2.9 ± 0.1 | 9.0 ± 2.3 | 1.5 ± 0.3 |
| pPJAV500 + pPJAV632 | 7.4 ± 1.9 | 7.5 ± 1.1 | 1.4 ± 0.1 |
| BG-11 Plates | |||
| Plasmids | pDU1 | Zeta-Sp/Km | Native Zeta |
| pPJAV606 | N/A | 7.5 ± 1.5 | 1.9 ± 0.1 |
| pPJAV626 | N/A | 4.2 ± 0.2 | 1.9 ± 0.05 |
| pPJAV632 | N/A | 4.7 ± 0.6 | 1.4 ± 0.1 |
| pPJAV361 | 2.8 ± 0.1 | N/A | 1.5 ± 0.01 |
| pPJAV500 | 12.9 ± 0.3 | N/A | 1.4 ± 0.05 |
| pPJAV361 + pPJAV626 | 2.7 ± 0.2 | 13.8 ± 0.04 | 1.7 ± 0.1 |
| pPJAV361 + pPJAV632 | 1.8 ± 0.5 | 23.3 ± 1.7 | 1.3 ± 0.1 |
| pPJAV500 + pPJAV626 | 1.9 ± 0.02 | 24.6 ± 1.9 | 1.1 ± 0.01 |
| pPJAV500 + pPJAV632 | 2.2 ± 0.02 | 29.7 ± 1.0 | 1.2 ± 0.01 |
| BG-11(NH4) Plates | |||
| Plasmids | pDU1 | Zeta-Sp/Km | Native Zeta |
| pPJAV606 | N/A | 10.4 ± 5.3 | 1.3 ± 1.2 |
| pPJAV626 | N/A | 1.1 ± 0.3 | 1.1 ± 1.0 |
| pPJAV632 | N/A | 38.0 ± 0.1 | 1.9 ± 0.04 |
| pPJAV361 | 99.5 ± 33.0 | N/A | 2.3 ± 0.2 |
| pPJAV500 | 244.8 ± 12.8 | N/A | 5.6 ± 0.6 |
| pPJAV361 + pPJAV626 | 203.4 ± 39.5 | 30.8 ± 1.2 | 3.5 ± 0.2 |
| pPJAV361 + pPJAV632 | 493.7 ± 149.5 | 438.1 ± 56.4 | 1.5 ± 0.1 |
| pPJAV500 + pPJAV626 | 692.7 ± 17.3 | 744.1 ± 2.9 | 1.7 ± 0.1 |
| pPJAV500 + pPJAV632 | 763.7 ± 28.6 | 828.1 ± 77.0 | 1.8 ± 0.01 |
Acknowledgements.
We thank Dr. Neil K. Garg (UCLA) for synthetic standards of lyngbyatoxin A and pendolmycin. We thank Drs. William Gerwick and Lena Gerwick (UCSD) for fos-DE-96 containing the lyngbyatoxin gene cluster. We acknowledge the support of the Oregon State University NMR Facility and the SOU Biotechnology Research Center.
Funding sources: This work was supported by the College of Pharmacy, Oregon State University, a New Investigator Grant from the Medical Research Foundation of Oregon (grant #1415), and an AREA award grant from the National Institutes of Health (1R15GM117541) to BP and startup funds from Southern Oregon University to PV. The Oregon State University NMR Facility was partially funded by the National Institutes of Health, HEI Grant 1S10OD018518, and by the M. J. Murdock Charitable Trust grant #2014162.
Abbreviations
- BG-11(NH4)
BG-11 media with ammonium chloride as the nitrogen source
- BG-11(Nit)
BG-11 media with sodium nitrate as the nitrogen source
- BGC
biosynthetic gene cluster
- COSY
correlation spectroscopy
- DNA
deoxyribonucleic acid
- HMBC
heteronuclear multiple bond correlation
- HPLC
high pressure liquid chromatography
- HRESIMS
high resolution electrospray ionization mass spectrometry
- HSQC
heteronuclear single quantum coherence quantum correlation
- ILV
indolactam V
- MRM
multiple reaction monitoring
- MS/MS
tandem mass spectrometry
- Neor
neomycin resistant
- NMR
nuclear magnetic resonance
- NMVT
N-methyl-L-valyl-L-tryptophanol
- NOESY
Nuclear Overhauser spectroscopy
- NRPS
non-ribosmal peptide synthetase
- ori
origin of replication
- oriT
origin of transfer
- Smr
spectinomycin resistant
- Spr
spectinomycin resistant
- XIC
extracted ion chromatogram
Footnotes
Supporting Information: Supplemental figures (S1–S23), tables (S1–S9) and supplemental discussion are supplied as Supporting Information.
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Dittmann E; Gugger M; Sivonen K; Fewer DP, Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol 2015, 23 (10), 642–652. [DOI] [PubMed] [Google Scholar]
- 2.Gerwick William H.; Moore Bradley S., Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol 2012, 19 (1), 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleigrewe K; Gerwick L; Sherman DH; Gerwick WH, Unique marine derived cyanobacterial biosynthetic genes for chemical diversity. Nat. Prod. Rep 2016, 33 (2), 348–364. [DOI] [PubMed] [Google Scholar]
- 4.Edwards DJ; Gerwick WH, Lyngbyatoxin biosynthesis: Sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J. Am. Chem. Soc 2004, 126 (37), 11432–11433. [DOI] [PubMed] [Google Scholar]
- 5.Cardellina JH 2nd; Marner FJ; Moore RE, Seaweed dermatitis: structure of lyngbyatoxin A. Science 1979, 204 (4389), 193–195. [DOI] [PubMed] [Google Scholar]
- 6.Basu A; Kozikowski AP; Lazo JS, Structural requirements of lyngbyatoxin A for activation and downregulation of protein kinase C. Biochemistry 1992, 31 (15), 3824–3830. [DOI] [PubMed] [Google Scholar]
- 7.Ma J; Zuo D; Song Y; Wang B; Huang H; Yao Y; Li W; Zhang S; Zhang C; Ju J, Characterization of a single gene cluster responsible for methylpendolmycin and pendolmycin biosynthesis in the deep sea bacterium Marinactinospora thermotolerans. ChemBioChem 2012, 13 (4), 547–552. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita T; Imoto M; Isshiki K; Sawa T; Naganawa H; Kurasawa S; Zhu B-Q; Umezawa K, Isolation of a new indole alkaloid, pendolmycin, from Nocardiopsis. J. Nat. Prod 1988, 51 (6), 1184–1187. [Google Scholar]
- 9.Awakawa T; Zhang L; Wakimoto T; Hoshino S; Mori T; Ito T; Ishikawa J; Tanner ME; Abe I, A methyltransferase initiates terpene cyclization in teleocidin B biosynthesis. J. Am. Chem. Soc 2014, 136 (28), 9910–9913. [DOI] [PubMed] [Google Scholar]
- 10.Hitotsuyanagi Y; Fujiki H; Suganuma M; Aimi N; Sakai S; Endo Y; Shudo K; Sugimura T, Isolation and structure elucidation of teleocidin B-1, B-2, B-3, and B-4. Chem Pharm Bull (Tokyo) 1984, 32 (10), 4233–4236. [DOI] [PubMed] [Google Scholar]
- 11.Videau P; Wells KN; Singh AJ; Gerwick WH; Philmus B, Assessment of Anabaena sp. strain PCC 7120 as a heterologous expression host for cyanobacterial natural products: Production of lyngbyatoxin A. ACS Synth. Biol 2016, 5 (9), 978–988. [DOI] [PubMed] [Google Scholar]
- 12.Jones AC; Ottilie S; Eustáquio AS; Edwards DJ; Gerwick L; Moore BS; Gerwick WH, Evaluation of Streptomyces coelicolor A3(2) as a heterologous expression host for the cyanobacterial protein kinase C activator lyngbyatoxin A. FEBS J 2012, 279 (7), 1243–1251. [DOI] [PubMed] [Google Scholar]
- 13.Ongley SE; Bian X; Zhang Y; Chau R; Gerwick WH; Müller R; Neilan BA, High-titer heterologous production in E. coli of lyngbyatoxin, a protein kinase C activator from an uncultured marine cyanobacterium. ACS Chem. Biol 2013, 8 (9), 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stebegg R; Wurzinger B; Mikulic M; Schmetterer G, Chemoheterotrophic growth of the cyanobacterium Anabaena sp. strain PCC 7120 dependent on a functional cytochrome c oxidase. J. Bacteriol 2012, 194 (17), 4601–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori T; Zhang L; Awakawa T; Hoshino S; Okada M; Morita H; Abe I, Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases. Nat. Commun 2016, 7 (EARLY EDITION). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolk CP; Vonshak A; Kehoe P; Elhai J, Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc. Natl. Acad. Sci. U. S. A 1984, 81 (5), 1561–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y; Taton A; Go M; London RE; Pieper LM; Golden SS; Golden JW, Self-replicating shuttle vectors based on pANS, a small endogenous plasmid of the unicellular cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 2016, 162 (12), 2029–2041. [DOI] [PubMed] [Google Scholar]
- 18.Anderson ES; Lewis MJ, Characterization of a transfer factor associated with drug resistance in Salmonella typhimurium. Nature 1965, 208 (5013), 843–849. [DOI] [PubMed] [Google Scholar]
- 19.Wolk CP; Fan Q; Zhou R; Huang G; Lechno-Yossef S; Kuritz T; Wojciuch E, Paired cloning vectors for complementation of mutations in the cyanobacterium Anabaena sp. strain PCC 7120. Arch. Microbiol 2007, 188 (6), 551–563. [DOI] [PubMed] [Google Scholar]
- 20.Fan Q; Huang G; Lechno-Yossef S; Wolk CP; Kaneko T; Tabata S, Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol. Microbiol 2005, 58 (1), 227–243. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko T; Nakamura Y; Wolk CP; Kuritz T; Sasamoto S; Watanabe A; Iriguchi M; Ishikawa A; Kawashima K; Kimura T; Kishida Y; Kohara M; Matsumoto M; Matsuno A; Muraki A; Nakazaki N; Shimpo S; Sugimoto M; Takazawa M; Yamada M; Yasuda M; Tabata S, Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 2001, 8 (5), 205–213. [DOI] [PubMed] [Google Scholar]
- 22.Lee MH; Scherer M; Rigali S; Golden JW, PlmA, a new member of the GntR family, has plasmid maintenance functions in Anabaena sp. strain PCC 7120. J. Bacteriol 2003, 185 (15), 4315–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang JD; Haselkorn R, A vector for analysis of promoters in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol 1991, 173 (8), 2729–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalmers RM; Kleckner N, IS10/Tn10 transposition efficiently accommodates diverse transposon end configurations. EMBO J 1996, 15 (18), 5112–5122. [PMC free article] [PubMed] [Google Scholar]
- 25.del Solar G; Hernández-Arriaga AM; Gomis-Rüth FX; Coll M; Espinosa M, A genetically economical family of plasmid-encoded transcriptional repressors involved in control of plasmid copy number. J. Bacteriol 2002, 184 (18), 4943–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bancroft I; Wolk CP, Characterization of an insertion sequence (IS891) of novel structure from the cyanobacterium Anabaena sp. strain M-131. J. Bacteriol 1989, 171 (11), 5949–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y; Huang X-Z; Wang L; Risoul V; Zhang C-C; Chen W-L, Phenotypic variation caused by variation in the relative copy number of pDU1-based plasmids expressing the GAF domain of Pkn41 or Pkn42 in Anabaena sp. PCC 7120. Res. Microbiol 2013, 164 (2), 127–135. [DOI] [PubMed] [Google Scholar]
- 28.Elhai J; Wolk CP, [83] Conjugal transfer of DNA to cyanobacteria In Methods Enzymol, Academic Press: 1988; Vol. Volume 167, pp 747–754. [DOI] [PubMed] [Google Scholar]
- 29.Figurski DH; Helinski DR, Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A 1979, 76 (4), 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang J; Scappino L; Haselkorn R, The patB gene product, required for growth of the cyanobacterium Anabaena sp. strain PCC 7120 under nitrogen-limiting conditions, contains ferredoxin and helix-turn-helix domains. J. Bacteriol 1993, 175 (6), 1697–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitschke J; Vioque A; Haas F; Hess WR; Muro-Pastor AM, Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (50), 20130–20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elhai J; Vepritskiy A; Muro-Pastor AM; Flores E; Wolk CP, Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol 1997, 179 (6), 1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J; Russell D, Molecular Cloning: A Laboratory Manual Cold Spring Harbor Labs Publishing: Cold Spring Harbor, N.Y, 2001; p 2344. [Google Scholar]
- 34.Datsenko KA; Wanner BL, One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A 2000, 97 (12), 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei TF; Ramasubramanian TS; Golden JW, Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol 1994, 176 (15), 4473–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higuchi R; Krummel B; Saiki R, A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 1988, 16 (15), 7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei TF; Ramasubramanian TS; Pu F; Golden JW, Anabaena sp. strain PCC 7120 bifA gene encoding a sequence-specific DNA-binding protein cloned by in vivo transcriptional interference selection. J. Bacteriol 1993, 175 (13), 4025–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golden JW; Wiest DR, Genome rearrangement and nitrogen fixation in Anabaena blocked by inactivation of xisA gene. Science 1988, 242 (4884), 1421–1423. [DOI] [PubMed] [Google Scholar]
- 39.Smith RJ; Carr NG, An investigation of RNA synthesis in Anacystis nidulans during exponential growth using techniques of RNA-DNA hybridization. J. Gen. Microbiol 1977, 98 (2), 559–567. [Google Scholar]
- 40.Craig IW; Leach CK; Carr NG, Studies with deoxyribonucleic acid from blue-green algae. Arch. Microbiol 1969, 65 (3), 218–227. [DOI] [PubMed] [Google Scholar]
- 41.Pigott GH; Midgley JE, Characterization of rapidly labelled ribonucleic acid in Escherichia coli by deoxyribonucleic acid-ribonucleic acid hybridization. Biochem. J 1968, 110 (2), 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


