Abstract
This study aimed to explore the phylogenetic and molecular characteristics of feline astrovirus. A total of 33 fecal samples of domestic cats with or without diarrhea were collected from the Anhui province, and two positive samples were detected. The complete genome and ORF2 of the two strains were sequenced and phylogenetically analyzed. AH-1-2020 and AH-2-2020 displayed 83.4% homology, and their homologies with other reference strains were 75.3%–83.4% and 83.4%–95.0%, respectively. Phylogenetic tree analysis revealed that all strains could be classified into three different clusters; therefore, the mean amino acid genetic distances (p-dist) among the three clusters were estimated. The results suggested that the two strains and other FeAstV strains were grouped into three genotypes, with AH-1-2020 belonging to a novel genotype. High similarity was observed (65.9%–66.5% nucleotide identity and 63.8%–64.8% amino acid identity) in ORF2 between porcine astrovirus type 1 and AH-1-2020. Furthermore, inter-specific recombination between porcine astrovirus type 1 and FeAstV was observed. We, therefore, inferred that inter-specific transmission may exist between pigs and cats; however, further studies are required to verify this. This is the first report on the genetic characterization and phylogenetic analysis of FeAstVs in the Anhui province and would further the current understanding of the genetic diversity and epidemiology of FeAstVs.
Keywords: FeAstV, Phylogenetic analysis, Epidemiology, Anhui province
Introduction
Astroviruses (AstVs) are small non-enveloped single-stranded positive-sense RNA viruses with a genome size of 6.2–7.7 kb, containing a 5′ untranslated region (5′ UTR), three overlapping open reading frames (ORFs) (ORF1a, ORF1b, and ORF2), a 3′ UTR, and a poly(A) tail (Lau et al. 2013). ORF1a and ORF1b, which are the more conserved regions, encode non-structural proteins including 3C-type serine protease, a viral genome-linked protein (VPg), RNA-dependent RNA polymerase (RdRp), and several uncharacterized proteins. ORF2 encodes a viral capsid precursor protein and determines the immunogenicity of the virus (Yi et al. 2018). AstVs are divided into two genera, Mamastrovirus and Avastrovirus, which contain 19 and 3 species, respectively (Mihalov-Kovacs et al. 2017). Generally, AstVs are considered enteroviruses that cause gastroenteritis in susceptible animals. Furthermore, Mamastrovirus is associated with diseases of the central nervous system, respiratory system, and circulatory system in humans, minks, pigs, cattle, dogs, and sheep (Blomstrom et al. 2010; Quan et al. 2010; Li et al. 2013; Padmanabhan and Hause 2016; Collinet et al. 2020). Avastrovirus causes gout in poultry, which may be potentially lethal (Panigrahi et al. 2019; Yu et al. 2020).
Feline astrovirus (FeAstV) was first reported in 1981 from the feces of a cat with diarrhea (Hoshino et al. 1981). Since then, numerous countries including Italy, United States, and Korea have reported FeAstV infections in cats (Moschidou et al. 2011; Cho et al. 2014; Lawler et al. 2018). FeAstV was first reported in Hong Kong, China, in 2013 (Lau et al. 2013) and later reported in northeast China in 2018 (Yi et al. 2018).
Thus far, few studies have isolated FeAstVs from infected cats in China. To better understand the genetic characteristics of FeAstV, in this study, the complete genome sequence of two FeAstV strains isolated from the Anhui province was amplified. The genetic diversity of the two viral strains was analyzed from their complete genome sequence and ORF2 sequence.
Materials and methods
Sample collection
Twenty-one fecal samples from domestic cats with diarrhea and 12 samples from healthy domestic cats were collected from animal hospitals in different areas of the Anhui province from June to October, 2019, in sterile centrifuge tubes using rectal swabs, and stored at − 80 °C until use.
RNA extraction and reverse transcription PCR (RT-PCR)-based detection
The samples were dissolved in 10% phosphate-buffered saline and mixed in an oscillating manner. The mixture was centrifuged at 10,000 × g for 10 min, and the supernatant was collected. Total RNA was extracted from feces, using the TIANamp Virus DNA/RNA Kit (TIANGEN, Beijing, China) in accordance with the manufacturer’s instructions. Reverse transcription was performed using the FastQuant cDNA Kit (TIANGEN) in accordance with the manufacturer’s instructions, and the cDNA was stored at − 20 °C until use.
FeAstVs were detected using conventional RT-PCR, as previously described (Zhang et al. 2019). Reactions comprised 12.5 μL of 2X Premix Taq® Version 2.0 (TaKaRa, Dalian, China), 1 μL of cDNA templates, 0.4-μM forward and reverse primers, and an appropriate volume of ddH2O to obtain a total volume of 25 μL. The cycling conditions for cDNA amplification were as follows: 94 °C for 5 min, followed by 40 cycles at 94 °C for 45 s, 51 °C for 45 s, and 72 °C for 30 s, and final extension at 72 °C for 10 min. The PCR products were visualized through agarose gel electrophoresis.
FeAstV complete genome amplification
A previously reported primer was used to amplify full-length ORF2 (Yi et al. 2018), and the specific primers used to amplify other genes were designed from other sequences in GenBank (Accession number: MK671306.1) using the Primer Premier 5 software (DNASTAR, Madison, WI, USA) (Table 1). PCR products were visualized through agarose gel electrophoresis and then purified using a DNA purification kit (TIANGEN) in accordance with the manufacturer’s instructions. After ligation of the PCR products into pMD19-T vector (TaKaRa), recombinant plasmids were sent to Sangon company (Shanghai, China) for sequencing. Complete genomes of the FeAstV strains obtained in this study were splined together with ORF1a, ORF1b, and ORF2, using SeqMan software (DNASTAR, Madison, WI, USA).
Table 1.
Primers and conditions used for the detection and complete sequence of FeAstV in this study
| Primer name | Nucleotide sequence (5′–3′) | Amplicon (bp) | Reaction condition |
|---|---|---|---|
| Detection | |||
| FeAstV-F | GCGGATTGGGCATGGTTTAGA | 645 | 94 °C for 5 min, followed by 40 cycles of 94 °C for 45 s, 51 °C for 45 s, and 72 °C for 30 s and a final extension at 72 °C for 10 min |
| FeAstV-R | ACCCCTCGTTTGGATCGTTACCT | ||
| ORF1a | |||
| F1a-1-F | TGGGCCAATTGGTCGAAG | 1167 | 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 48 °C for 30 s, and 72 °C for 1 min 15 s and a final extension at 72 °C for 10 min |
| F1a-1-R | CAAGCGTAGGGTGCGGAT | ||
| F1a-2-F | CTTGTCCCTTTGGGTTATGAATCT | 1046 | 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 51 °C for 30 s, and 72 °C for 1 min 10 s and a final extension at 72 °C for 10 min |
| F1a-2-R | ATTCCCTGTTGATTTCGTCTCTTAG | ||
| F1a-3-F | TGATTTCCACCCTCATAAGCCAC | 1081 | 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 52 °C for 30 s, and 72 °C for 1 min 10 s and a final extension at 72 °C for 10 min |
| F1a-3-R | AGATGACCAATGAGCGGAAAGTTAT | ||
| ORF1b | |||
| F1b-1-F | TGGGAGTCCTACGATTTTGAGT | 686 | 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 49 °C for 30 s, and 72 °C for 45 s and a final extension at 72 °C for 10 min |
| F1b-1-R | AGGGTAGGCAGGTGTTGAG | ||
| F1b-2-F | ATGATTCAAAGAAACCACGGGACG | 1422 | 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 53 °C for 30 s, and 72 °C for 1 min 30 s and a final extension at 72 °C for 10 min |
| F1b-2-R | GACCTCCACAGTCACTTCCTTACC | ||
| ORF2 | |||
| F2-F | ATGGCTAGCAAGYCTGGYAAAGAAG | 2438 | 94 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 65 °C for 1 min and 72 °C for 2.5 min and a final extension at 72 °C for 10 min |
| F2-R | GCGTGGCCTCGGCTCTCAA | ||
| poly(A) tail | |||
| Fpo-F | CAAACAAGCACGCAAAGC | 182 | 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 45 °C for 30 s, and 72 °C for 15 s and a final extension at 72 °C for 10 min |
| Fpo-R | TTTTTTTTTTTTTTTTGCTTCTGAT | ||
| 5′-UTR | |||
| FU-F | TGGGCCAACTGGTTGAAGAGGT | 284 | 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 53 °C for 30 s, and 72 °C for 20 s and a final extension at 72 °C for 10 min |
| FU-R | CAGTCGGAGTCATCATCAGGGTTG | ||
Sequence alignment and phylogenetic analyses
Sequence identity was analyzed by aligning nucleotide and amino acid sequences via the Clustal W method, and the amino acid distance was estimated using the p-distance model with 1000 bootstrap replicates, using the MegAlign 6.0 program (DNASTAR). Phylogenetic trees were constructed using the Maximum Likelihood method using the p-distance model with 1000 bootstrap replicates and default parameters in the MEGA X software (DNASTAR). The best substitution model was selected through ModelFinder in accordance with the Akaike Information Criterion score. All referenced FeAstV strains were obtained from GenBank (https://www.ncbi.nlm.nih.gov/) (Table 2). Recombination events were identified using the Recombination Detection Program v.4.39 (RDP 4.39) and the SimPlot software v.3.5.1.
Table 2.
General information and GenBank accession numbers of CanineCV strains used in this study
| Strains | GenBank Accession no. | District of origin | Nucleotides | Gene | Host |
|---|---|---|---|---|---|
| 17CC0308 | MK671306.1 | China | 6796 | Complete | Cat |
| 17HRB0511 | MK671307.1 | China | 6802 | Complete | Cat |
| 17JL0318 | MK671308.1 | China | 6802 | Complete | Cat |
| 17SP0801 | MK671309.1 | China | 6802 | Complete | Cat |
| 18CC0502 | MK671310.1 | China | 6802 | Complete | Cat |
| 18JL0310 | MK671311.1 | China | 6796 | Complete | Cat |
| 18JL0705 | MK671312.1 | China | 6796 | Complete | Cat |
| 18SY08703 | MK671313.1 | China | 6796 | Complete | Cat |
| 1637F | NC022249.1 | HongKong,China | 6795 | Complete | Cat |
| 1637F | KF499111.1 | HongKong,China | 6795 | Complete | Cat |
| FAstV-D1 | KM017741.1 | USA | 6598 | Complete | Cat |
| FAstV-D1 | NC024701.1 | USA | 6598 | Complete | Cat |
| FAstV-D2 | KM017742.1 | USA | 6813 | Complete | Cat |
| FAstV-D3 | KM017743.1 | USA | 6748 | Complete | Cat |
| Viseu | KF374704.1 | USA | 6780 | Complete | Cat |
| CS-2018 | MN148428.1 | China | 6820 | Complete | Tiger |
| Feline astrovirus | AF056197.1 | UK | 2511 | ORF2 | Cat |
| 17CC0311 | MH253878.1 | China | 2445 | ORF2 | Cat |
| 17CC0308 | MH253877.1 | China | 2445 | ORF2 | Cat |
| 17SY1204 | MH253876.1 | China | 2451 | ORF2 | Cat |
| 17JL0318 | MH253875.1 | China | 2451 | ORF2 | Cat |
| 17JL0312 | MH253874.1 | China | 2451 | ORF2 | Cat |
| 17JL0310 | MH253873.1 | China | 2451 | ORF2 | Cat |
| 17JL0303 | MH253872.1 | China | 2451 | ORF2 | Cat |
| 17HRB0905 | MH253871.1 | China | 2451 | ORF2 | Cat |
| 17HRB0511 | MH253870.1 | China | 2451 | ORF2 | Cat |
| 17HRB0505 | MH253869.1 | China | 2451 | ORF2 | Cat |
| 17CC1106 | MH253868.1 | China | 2451 | ORF2 | Cat |
| 17CC0806 | MH253867.1 | China | 2451 | ORF2 | Cat |
| 17CC0714 | MH253866.1 | China | 2451 | ORF2 | Cat |
| 17CC0704 | MH253865.1 | China | 2451 | ORF2 | Cat |
| 17CC0502 | MH253864.1 | China | 2448 | ORF2 | Cat |
| 16SY0715 | MH253862.1 | China | 2451 | ORF2 | Cat |
| 16SY0705 | MH253861.1 | China | 2451 | ORF2 | Cat |
| 16JZ0612 | MH253860.1 | China | 2451 | ORF2 | Cat |
| 16CC1104 | MH253859.1 | China | 2451 | ORF2 | Cat |
| 18SY0803 | MK671281.1 | China | 2445 | ORF2 | Cat |
| 18SY0302 | MK671280.1 | China | 2445 | ORF2 | Cat |
| 18SP0501 | MK671279.1 | China | 2454 | ORF2 | Cat |
| 18QQHE0707 | MK671278.1 | China | 2451 | ORF2 | Cat |
| 18JZ0701 | MK671277.1 | China | 2445 | ORF2 | Cat |
| 18JL0705 | MK671276.1 | China | 2445 | ORF2 | Cat |
| 18JL0310 | MK671275.1 | China | 2445 | ORF2 | Cat |
| 18HRB0701 | MK671274.1 | China | 2451 | ORF2 | Cat |
| 18DD0102 | MK671273.1 | China | 2445 | ORF2 | Cat |
| 18CC0903 | MK671272.1 | China | 2445 | ORF2 | Cat |
| 18CC0714 | MK671271.1 | China | 2451 | ORF2 | Cat |
| 18CC0512 | MK671270.1 | China | 2454 | ORF2 | Cat |
| 18CC0502 | MK671269.1 | China | 2451 | ORF2 | Cat |
| 18BC0502 | MK671268.1 | China | 2454 | ORF2 | Cat |
| 17SY0306 | MK671267.1 | China | 2451 | ORF2 | Cat |
| 17SP0801 | MK671266.1 | China | 2451 | ORF2 | Cat |
| 17JL0312 | MK671265.1 | China | 2451 | ORF2 | Cat |
| 17DD0902 | MK671264.1 | China | 2445 | ORF2 | Cat |
| 17BC0701 | MK671263.1 | China | 2445 | ORF2 | Cat |
| 16SY1102 | MK671262.1 | China | 2445 | ORF2 | Cat |
| 16LY0702 | MK671261.1 | China | 2451 | ORF2 | Cat |
| 16JZ1202 | MK671260.1 | China | 2451 | ORF2 | Cat |
| PAstV-GX1 | KF787112.2 | China | 6706 | Complete | Pig |
| Pig-wt/ESP/B333/2017 | MK962341.1 | Spain | 6460 | Complete | Pig |
| Pig-wt/ESP/B377/2017 | MK962342.1 | Spain | 6459 | Complete | Pig |
| PoAstV12-3/Canada/2006 | HM756258.1 | Canada | 2909 | ORF2 | Pig |
| Porcine astrovirus | Y15938.2 | Norway | 2459 | ORF2 | Pig |
| Mamastrovirus 3 | AB037272.1 | Japan | 2730 | ORF2 | Pig |
| HAstV 2 strain Oxford | MK059950.1 | USA | 6800 | Complete | Human |
| HAstV 3 strain Oxford | MK059951.1 | USA | 6815 | Complete | Human |
| HAstV 6 strain Oxford | MK059954.1 | USA | 6745 | Complete | Human |
| HAstV 7 strain Oxford | MK059955.1 | USA | 6786 | Complete | Human |
| HAstV 6 isolate Katano | HM237363.1 | Japan | 6772 | Complete | Human |
| HAstV 1 isolate JZ | KF211475.1 | China | 6771 | Complete | Human |
| HAstV 1 isolate Shanghai | FJ792842.1 | China | 2364 | ORF2 | Human |
| ITA/2005/PA124/type1d | JX087965.1 | Italy | 2977 | ORF2 | Human |
| HAstV 3 WH1859 | DQ630763.4 | China | 2478 | ORF2 | Human |
| O-33/87-25/Ehime1987 | AB025812.1 | Japan | 2329 | ORF2 | Human |
| CHN198 | AB037274.1 | Japan | 2750 | ORF2 | Human |
| BAstV-GX7/CHN/2014 | NC_024297.1 | USA | 6233 | Complete | Cattle |
| BAstV-GX27/CHN/2014 | KJ620980.1 | China | 6233 | Complete | Cattle |
| LVMS681 | MN200262.1 | Uruguay | 2243 | ORF2 | Cattle |
| LVMS2704 | MN200263.1 | Uruguay | 2238 | ORF2 | Cattle |
| Gillingham/2012/UK | NC026814.1 | USA | 6617 | Complete | Dog |
| CHN/2017/44 | MF973500.1 | China | 6626 | Complete | Dog |
| CHN/2017/58 | MF973501.1 | China | 6596 | Complete | Dog |
| CHN/2017/3 | MF973495.1 | China | 2526 | ORF2 | Dog |
| CHN/2017/S15 | MF973498.1 | China | 2505 | ORF2 | Dog |
| CHN/2017/Y6 | MF973499.1 | China | 2517 | ORF2 | Dog |
| Fox 5 | KC692365.1 | Netherlands | 6456 | Complete | Fox |
| Mink astrovirus | NC004579.1 | USA | 6610 | Complete/ORF2 | Mink |
| Mink astrovirus | AY179509.1 | Sweden | 6610 | Complete | Mink |
| SMS-AstV | GU985458.1 | Sweden | 2328 | ORF2 | Mink |
Results
Detection of FeAstV in the fecal samples
Among the 33 fecal samples obtained (21 from domestic cats with diarrhea and 12 from healthy cats), 2 (6.06%) samples from cats with diarrhea were positive for FeAstV infection as evidenced by a conventional RT-PCR analysis.
Characteristics of the FeAstV genomes
Complete genome sequences of the two FeAstV strains were successfully amplified and designated as AH-1-2020 (6797nt, encoding 2224aa) and AH-2-2020 (6800nt, encoding 2226aa) and deposited in GenBank (accession numbers: MN977118 and MN977119, respectively). The homology between the two strains was 83.4%. The nucleotide identity of complete sequence of strains AH-1-2020 and AH-2-2020 compared to that of other FeAstV strains was 75.3%–83.4% and 83.4%–95.0%, respectively. Moreover, the nucleotide identity of ORF2 was 57.1%–59.4% (AH-1-2020) and 59.4%–75.4% (AH-2-2020), and the amino acid identity was 48.9%–54.3% (AH-1-2020) and 71.6%–95.9% (AH-2-2020), indicating low homology in ORF2 of these two strains with that of other FeAstV strains, especially ORF2 of strain AH-1-2020.
Phylogenetic analyses of FeAstV
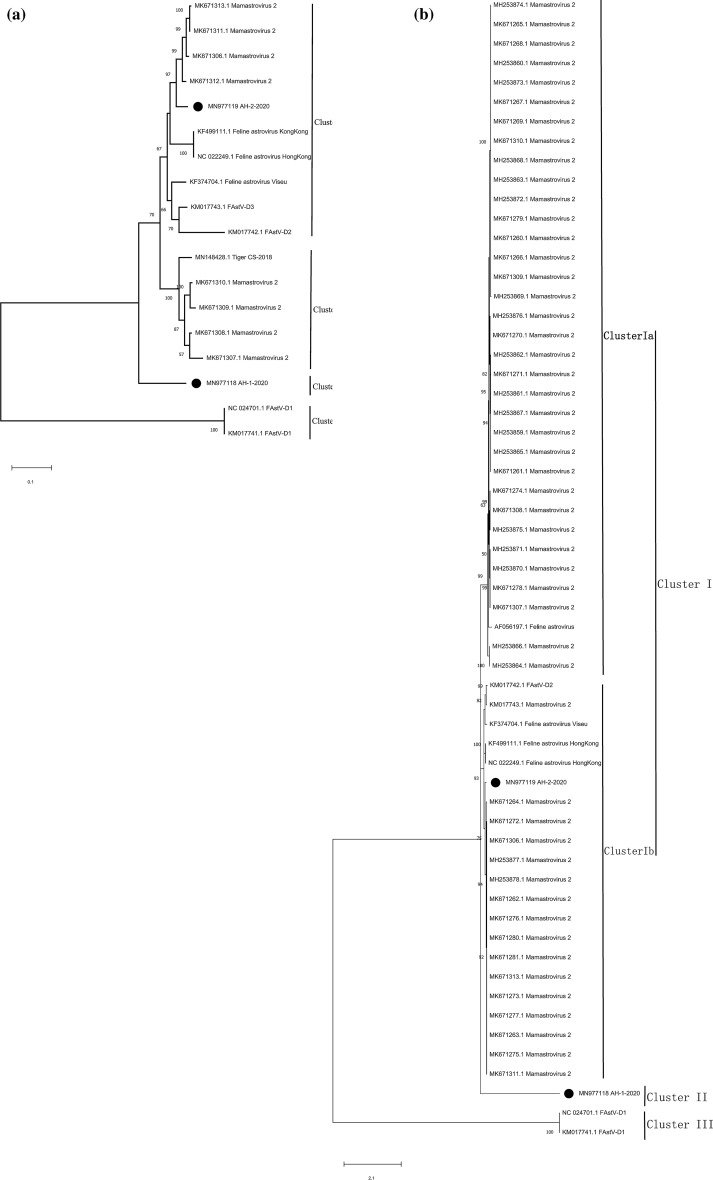
Phylogenetic analysis of the two strains and relevant FeAstV strains from mainland China, the United States, and Hong Kong based on complete genome sequences (Fig. 1a) that can represent the general characteristics of the genetic information revealed that they formed three clusters (Cluster I, Cluster II, and Cluster III). Cluster I contained two different clades (Cluster Ia and Cluster Ib). The FeAstV AH-2-2020 strain belonged to cluster Ia, which was closely related (94.6%–95.0%) to isolates from northeast China (GenBank accession numbers: MK671313.1, MK671312.1, MK671311.1, and MK671306.1) and shared a common evolutionary ancestor with isolates from Hong Kong (GenBank accession numbers: KF499111.1 and NC022249.1) and the United States (GenBank accession numbers: KM017742.1, KM017743.1, and KF374704.1). FeAstV AH-1-2020 strain and two strains from the United States (GenBank accession numbers: NC027701.1, KM017741.1) formed two unique branches, Cluster II and Cluster III, respectively. AH-1-2020 displayed low homology (75.3%–83.4%) with other FeAstVs strains. Furthermore, the phylogenetic tree based on ORF2 (Fig. 1b) revealed that ORF2 of the isolates was divided into three groups in a pattern similar to that of the phylogenetic tree based on complete genome sequences. The homology between ORF2 of the two strains isolated herein and other FeAstVs strains was very low (57.1%–59.4%).
Fig. 1.
Phylogenetic trees based on the complete genome sequences and ORF2 of feline astroviruses (FeAstVs). a Complete gene sequences (n = 16); b ORF2 (n = 59). Strains from this study are marked with black spots. Phylogenetic analysis based on complete genome sequences revealed that all strains formed three clusters (Cluster I, Cluster II, and Cluster III). Cluster I contained two different clades (Cluster Ia and Cluster Ib). AH-1-2020 and AH-2-2020 belong to Cluster I and Cluster II, respectively. The tree was constructed using the Maximum Likelihood method with 1000 bootstrap replicates in the MEGA X software. General Time Reversible (GTR) was selected as the substitution model through ModelFinder analysis
Mean amino acid genetic distances (p-dist) between the three clusters was estimated in accordance with the latest classification of AstV (To et al. 2017). The p-dist between Cluster I and Cluster II, Cluster I and Cluster III, and Cluster II and Cluster III was 0.489 ± 0.017, 0.771 ± 0.015, and 0.774 ± 0.015, respectively. The p-dist within Cluster I and Cluster II was 0.189 ± 0.007 and 0.000 ± 0.000, respectively. We concluded that these three clusters belonged to three different genotypes, among which the cluster comprising AH-1-2020 represented a novel genotype.
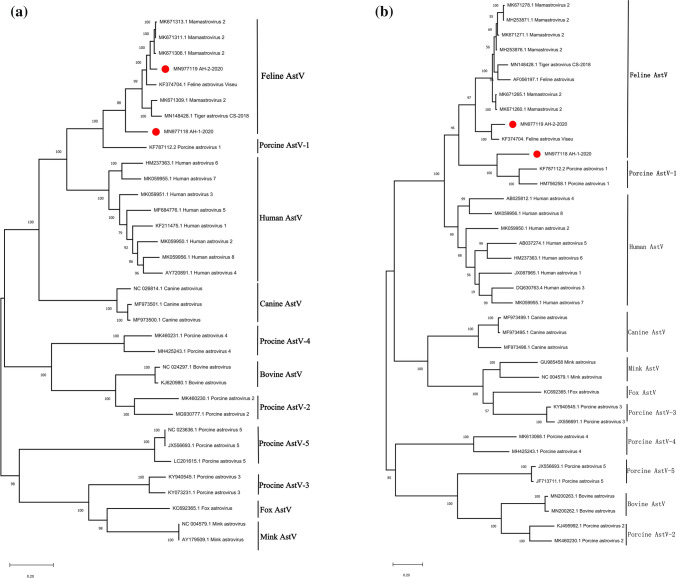
To analyze the relationships between the two FeAstV isolates of this study and other AstVs species, phylogenetic analyses based on the complete genome sequence and ORF2 were performed (Fig. 2). FeAstV isolates shared a common ancestor with porcine AstVs type 1 (PoAstV-1). As expected, AH-2-2020 formed a branch with FeAstVs. Interestingly, AH-1-2020 displayed high identity (73.1%) with PoAstV-1 strain (GenBank accession number: KF787112.2) isolated from Guangxi, China. ORF2 region of AH-1-2020 displayed 65.9%–66.5% nucleotide identity and 63.8%–64.8% amino acid identity with that of PoAstV-1 strain (GenBank accession numbers: KF787112.2, HM756258.1), which was higher than that with other FeAstV strains. These findings strongly suggest the possibility of inter-species viral transmission between cats and pigs.
Fig. 2.
Phylogenetic tree based on the complete genome sequence and ORF2 gene of feline astroviruses (FeAstVs) and other AstV strains. a Complete genome sequences (n = 34); b ORF2 (n = 37). Strains from this study are marked with red spots. The phylogenetic analyses based on the complete genome sequence indicate that FeAstV isolates share a common ancestor with PoAstV-1, and that AH-1-2020 displays a close relationship with PoAstV-1. The tree was constructed using the Maximum Likelihood method with 1000 bootstrap replicates in the MEGA X software. General Tim Reversible (GTR) was selected as the substitution model through ModelFinder analysis
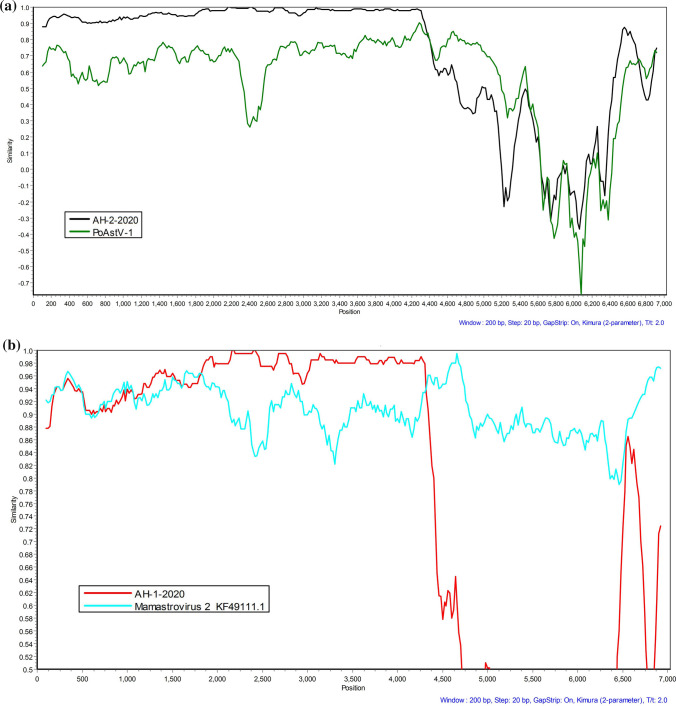
Analysis of recombination events using the RDP software and SimPlot revealed that AH-1-2020 and AH-2-2020 were both recombinant viruses. One recombination event was detected in AH-1-2020 (Fig. 3a) and AH-2-2020 (Fig. 3b). The major and minor parents of PoAstV-1 (GenBank accession number: KF787112.2) were AH-1-2020 and AH-2-2020, respectively. Similarity plots (Fig. 3a) show that the similarity between PoAstV-1 and AH-1-2020 in ORF1a, ORF1b and 3′ UTR region was lower than that between AH-2-2020 and AH-1-2020. Conversely, in the 5′ end of the ORF2 region, the similarity between PoastV-1 and AH-1-2020 was higher than that between AH-2-2020 and AH-1-2020. The result showed that the recombination region is concentrated at ORF1a, ORF1b, and the end of the 3′ UTR region. The parents of AH-2-2020 were all FeAstV strains.
Fig. 3.
Similarity plots of complete genomes. a AH-2020 (black line), PoAstV-1 (green line), and AH-1-2020 as query sequence; b AH-1-2020 (red line), Mamastrovirus 2 (blue line), and AH-2-2020 as query sequence. The AH-1-2020 recombination was primarily observed in the ORF1a and ORF1b regions, while that of AH-2-2020 was primarily observed in the ORF1a region. The SimPlot computer program was used to analyze similarity plots using a window size of 200 nucleotides that was moved along in 20-nt steps
Discussion
AstVs are prominent pathogens causing enteric diseases in both humans and animals (Karlsson et al. 2015). FeAstVs are speculated to be associated with gastroenteritis, and their infection in domestic cats has been reported in many countries. The genetic characteristics of FeAstV remain largely unknown, especially in eastern China. Herein, complete genomes of two FeAstV strains were successfully analyzed, providing molecular evidence of FeAstV circulation in the Anhui province. Herein, a FeAstV infection was identified in 6.06% (2/33) of the cat fecal samples analyzed. However, due to the limited sample size, we were unable to fully indicate the prevalence of FeAstV in the Anhui province. Hence, more extensive samples need to be collected to determine the positive rate of FeAstV in the Anhui province. Previous studies have reported that experimental infection with a FeAstV strain in specific pathogen-free (SPF) kittens induces enteritis and viral excretion for several days (Moschidou et al. 2011). Herein, although both positive samples were obtained from cats with diarrhea, the pathogenesis of FeAstVs requires further study.
In the 9th ICTV report, the Astroviridae Study Group updated the taxonomy based not only on the host range, but also on the genetic differences (mean amino acid genetic distances (p-dist) between and within genotypes ranging between 0.368–0.781 and 0–0.318, respectively) in the complete ORF2 sequence (https://talk.ictvonline.org/ictv/proposals/2010.018a-cV.A.v4.Mamastrovirus.pdf) (To et al. 2017). According to the latest proposed criteria, human astroviruses, PoAstV, and rodent astroviruses are divided into eight, five, and four genotypes, respectively (To et al. 2017; Jacobsen et al. 2018; Qin et al. 2019). Shushuai et al. divided the FeAstVs into two genotypes; however, our results are inconsistent with those of Shushuai et al. (Yi et al. 2018). This is the first study to have grouped FeAstVs into three genotypes with the discovery of the novel AH-1-2020 strain. Hence, it can be inferred that FeAstV genotypes are complex in China or worldwide, and their genetic diversity might have long been underestimated owing to limited sequence information.
Interestingly, this study reports a novel FeAstV isolate (AH-1-2020) sharing a common ancestor with PoAstV-1. ORF2 analyses revealed that AH-1-2020 shares greater similarity with PoAstV-1 than with other FeAstVs. ORF2 encodes the capsid protein and is the basis for AstV classification (Koci 2005). These results strongly suggest the possibility of inter-species transmission of AstVs between cats and pigs. Although AstVs are speculated to have stringent host specificity, some evidences from mammals and birds indicate that, in some cases, the host-species barrier may be breakable and that heterologous viruses may adapt stably to new hosts (Mihalov-Kovacs et al. 2017).
Recombination is common among numerous RNA viruses and in AstVs (Wohlgemuth et al. 2019). Recombination events were detected in bovine AstVs (Hirashima et al. 2018), canine AstVs (Li et al. 2018), porcine AstVs (Ito et al. 2017), and dromedary camel AstVs (Woo et al. 2015). Furthermore, inter-species recombination events in AstVs were increasingly noted. For instance, a recombination event was reported to have occurred between bovine AstVs and a roe deer astrovirus (Guan et al. 2018). Recombination requires co-infection of the same cell; therefore, inter-species recombination results from two AstVs infecting the same cell (Wohlgemuth et al. 2019). Herein, a potential recombination event might have occurred in FeAstV and PoAstV, further suggesting the possibility of inter-species transmission, consistent with the results of the phylogenetic tree analysis.
Such inter-species transmission increases the genetic diversity of FeAstVs and poses challenges for the prevention of their infections. Considering the relatively low prevalence of novel FeAstV among cats, the likelihood of cross-species transmission from cats may be minor, and hence, further studies are required to determine whether FeAstV can cause severe infections in pigs and domestic cats.
Conclusions
Herein, 2 of 33 fecal samples from domestic cats with or without diarrhea were positive for FeAstV, indicating that FeAstV may have been propagating among domestic cats in the Anhui province. Based on the phylogenetic analyses and p-dist estimation, three FeAstV genotypes were proposed and a novel genotype was discovered herein. Moreover, the inter-species transmission between pigs and cats warrants further investigation. This study furthers the current understanding of the genetic diversity and epidemiology of FeAstV in the Anhui province.
Author contributions
XG, YC, JS, ZF and YZ were involved in performing experiments. YC, YZ, KY and BC were involved in data analysis, experimental design. XG, YW, SJ and YL wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Ningbo Health Branding Subject Fund (No. ppxk2018-10), key research project of national science and technology (No. 2016YFD0501003), State Key Laboratory of Genetically Engineered Veterinary Vaccines (No. AGVSKL-ZD-202010) and the Science and Technology Promoting Agriculture Innovation Project of Shanghai (No. 2019 No.3-3).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical standards
All experiments were compliant with the ethical standards of Anhui Agricultural University.
Footnotes
Yong Wang and Xu Guo contributed equally to this work and considered to be the co-first authors.
Contributor Information
Shudong Jiang, Email: jshudong@163.com.
Yongdong Li, Email: liyd0551@126.com.
References
- Blomstrom AL, Widen F, Hammer AS, Belak S, Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol. 2010;48(12):4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YY, Lim SI, Kim YK, Song JY, Lee JB, An DJ. Molecular characterisation and phylogenetic analysis of feline astrovirus in Korean cats. J Feline Med Surg. 2014;16(8):679–683. doi: 10.1177/1098612X13511812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinet A, Garcia G, Wellehan J, Childress A, Carrera-Justiz S. Investigation of astrovirus and bornavirus in the cerebrospinal fluid of dogs clinically diagnosed with meningoencephalitis of unknown etiology. J Vet Intern Med. 2020;34(1):232–236. doi: 10.1111/jvim.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan TP, Teng JLL, Yeong KY, You ZQ, Liu H, Wong SSY, Lau SKP, Woo PCY. Metagenomic analysis of Sichuan takin fecal sample viromes reveals novel enterovirus and astrovirus. Virology. 2018;521:77–91. doi: 10.1016/j.virol.2018.05.027. [DOI] [PubMed] [Google Scholar]
- Hirashima Y, Okada D, Shibata S, Yoshida S, Fujisono S, Omatsu T, Mizutani T, Nagai M. Whole genome analysis of a novel neurotropic bovine astrovirus detected in a Japanese black steer with non-suppurative encephalomyelitis in Japan. Arch Virol. 2018;163(10):2805–2810. doi: 10.1007/s00705-018-3898-3. [DOI] [PubMed] [Google Scholar]
- Hoshino Y, Zimmer JF, Moise NS, Scott FW. Detection of astroviruses in feces of a cat with diarrhea. Brief report. Arch Virol. 1981;70(4):373–376. doi: 10.1007/BF01320252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Kuroda M, Masuda T, Akagami M, Haga K, Tsuchiaka S, Kishimoto M, Naoi Y, Sano K, Omatsu T, Katayama Y, Oba M, Aoki H, Ichimaru T, Mukono I, Ouchi Y, Yamasato H, Shirai J, Katayama K, Mizutani T, Nagai M. Whole genome analysis of porcine astroviruses detected in Japanese pigs reveals genetic diversity and possible intra-genotypic recombination. Infect Genet Evol. 2017;50:38–48. doi: 10.1016/j.meegid.2017.02.008. [DOI] [PubMed] [Google Scholar]
- Jacobsen S, Hohne M, Marques AM, Beslmuller K, Bock CT, Niendorf S. Co-circulation of classic and novel astrovirus strains in patients with acute gastroenteritis in Germany. J Infect. 2018;76(5):457–464. doi: 10.1016/j.jinf.2018.02.006. [DOI] [PubMed] [Google Scholar]
- Karlsson EA, Small CT, Freiden P, Feeroz MM, Matsen FAT, San S, Hasan MK, Wang D, Jones-Engel L, Schultz-Cherry S. Non-human primates harbor diverse mammalian and avian astroviruses including those associated with human infections. PLoS Pathog. 2015;11(11):e1005225. doi: 10.1371/journal.ppat.1005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koci MD. Immunity and resistance to astrovirus infection. Viral Immunol. 2005;18(1):11–16. doi: 10.1089/vim.2005.18.11. [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Yip CC, Bai R, Wu Y, Tse H, Yuen KY. Complete genome sequence of a novel feline astrovirus from a domestic cat in Hong Kong. Genome Announc. 2013;1(5):e00708-13. doi: 10.1128/genomeA.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler PE, Cook KA, Williams HG, Archer LL, Schaedel KE, Isaza NM, Wellehan JFX., Jr Determination of the diversity of astroviruses in feces from cats in Florida. J Vet Diagn Investig. 2018;30(2):275–279. doi: 10.1177/1040638717747322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Diab S, McGraw S, Barr B, Traslavina R, Higgins R, Talbot T, Blanchard P, Rimoldi G, Fahsbender E, Page B, Phan TG, Wang C, Deng X, Pesavento P, Delwart E. Divergent astrovirus associated with neurologic disease in cattle. Emerg Infect Dis. 2013;19(9):1385–1392. doi: 10.3201/eid1909.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yan N, Ji C, Wang M, Zhang B, Yue H, Tang C. Prevalence and genome characteristics of canine astrovirus in southwest China. J Gen Virol. 2018;99(7):880–889. doi: 10.1099/jgv.0.001077. [DOI] [PubMed] [Google Scholar]
- Mihalov-Kovacs E, Martella V, Lanave G, Bodnar L, Feher E, Marton S, Kemenesi G, Jakab F, Banyai K. Genome analysis of canine astroviruses reveals genetic heterogeneity and suggests possible inter-species transmission. Virus Res. 2017;232:162–170. doi: 10.1016/j.virusres.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschidou P, Martella V, Lorusso E, Desario C, Pinto P, Losurdo M, Catella C, Parisi A, Banyai K, Buonavoglia C. Mixed infection by Feline astrovirus and Feline panleukopenia virus in a domestic cat with gastroenteritis and panleukopenia. J Vet Diagn Investig. 2011;23(3):581–584. doi: 10.1177/1040638711404149. [DOI] [PubMed] [Google Scholar]
- Padmanabhan A, Hause BM. Detection and characterization of a novel genotype of porcine astrovirus 4 from nasal swabs from pigs with acute respiratory disease. Arch Virol. 2016;161(9):2575–2579. doi: 10.1007/s00705-016-2937-1. [DOI] [PubMed] [Google Scholar]
- Panigrahi S, Jindal N, Kumar P, Barua S, Kumar N, Riyesh T, Chander Y. Molecular characterization of chicken astroviruses in gout-affected commercial broiler chickens in Haryana, India. VirusDisease. 2019;30(4):551–561. doi: 10.1007/s13337-019-00554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Fang Q, Li X, Li F, Liu H, Wei Z, Ouyang K, Chen Y, Huang W. Molecular epidemiology and viremia of porcine astrovirus in pigs from Guangxi province of China. BMC Vet Res. 2019;15(1):471. doi: 10.1186/s12917-019-2217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis. 2010;16(6):918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KKW, Chan WM, Li KSM, Lam CSF, Chen Z, Tse H, Lau SKP, Woo PCY, Yuen KY. High prevalence of four novel astrovirus genotype species identified from rodents in China. J Gen Virol. 2017;98(5):1004–1015. doi: 10.1099/jgv.0.000766. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth N, Honce R, Schultz-Cherry S. Astrovirus evolution and emergence. Infect Genet Evol. 2019;69:30–37. doi: 10.1016/j.meegid.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, Lau SKP, Teng JLL, Tsang AKL, Joseph S, Xie J, Jose S, Fan RYY, Wernery U, Yuen KY. A novel astrovirus from dromedaries in the Middle East. J Gen Virol. 2015;96(9):2697–2707. doi: 10.1099/jgv.0.000233. [DOI] [PubMed] [Google Scholar]
- Yi S, Niu J, Wang H, Dong G, Guo Y, Dong H, Wang K, Hu G. Molecular characterization of feline astrovirus in domestic cats from Northeast China. PLoS One. 2018;13(10):e0205441. doi: 10.1371/journal.pone.0205441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhang D, Yang K, Bai C, Li Y, Li J, Jiang S, Wang Y. A simple and rapid diagnostic method to detect new goose astrovirus using reverse-transcription loop-mediated isothermal amplification. 3 Biotech. 2020;10(1):20. doi: 10.1007/s13205-019-2006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Niu J, Yi S, Dong G, Yu D, Guo Y, Huang H, Hu G. Development and application of a multiplex PCR method for the simultaneous detection and differentiation of feline panleukopenia virus, feline bocavirus, and feline astrovirus. Arch Virol. 2019;164(11):2761–2768. doi: 10.1007/s00705-019-04394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]