Abstract
A pneumonia of unknown causes, which was detected in Wuhan, China, and spread rapidly throughout the world, was declared as Coronavirus disease 2019 (COVID-19). Thousands of people have lost their lives to this disease. Its negative effects on public health are ongoing. In this study, an intelligence computer-aided model that can automatically detect positive COVID-19 cases is proposed to support daily clinical applications. The proposed model is based on the convolution neural network (CNN) architecture and can automatically reveal discriminative features on chest X-ray images through its convolution with rich filter families, abstraction, and weight-sharing characteristics. Contrary to the generally used transfer learning approach, the proposed deep CNN model was trained from scratch. Instead of the pre-trained CNNs, a novel serial network consisting of five convolution layers was designed. This CNN model was utilized as a deep feature extractor. The extracted deep discriminative features were used to feed the machine learning algorithms, which were k-nearest neighbor, support vector machine (SVM), and decision tree. The hyperparameters of the machine learning models were optimized using the Bayesian optimization algorithm. The experiments were conducted on a public COVID-19 radiology database. The database was divided into two parts as training and test sets with 70% and 30% rates, respectively. As a result, the most efficient results were ensured by the SVM classifier with an accuracy of 98.97%, a sensitivity of 89.39%, a specificity of 99.75%, and an F-score of 96.72%. Consequently, a cheap, fast, and reliable intelligence tool has been provided for COVID-19 infection detection. The developed model can be used to assist field specialists, physicians, and radiologists in the decision-making process. Thanks to the proposed tool, the misdiagnosis rates can be reduced, and the proposed model can be used as a retrospective evaluation tool to validate positive COVID-19 infection cases.
Keywords: COVID-19, Medical decision support system, Deep learning, Deep feature extraction, Machine learning
Highlights
-
•
In this study, COVID-19 detection system is proposed to support daily clinical applications.
-
•
The proposed model is based on the CNN and can automatically reveal discriminative features on chest X-ray images.
-
•
Instead of the pre-trained CNNs, a novel serial network consisting of five convolution layers was designed.
1. Introduction
COVID-19, a new type of Coronavirus, has created a very critical chaotic situation, negatively affecting a large number of deaths and people’s lives worldwide. It first appeared in Wuhan, China, in December 2019. It has spread to approximately 200 countries worldwide. In many countries, rulers and governments have taken new measures and created new lifestyles to combat COVID-19. Today’s science and technology have made an extremely valuable contribution to the implementation of these new policies of states in this unknown and unpredictable process. As an example of technological developments, robots, and drones have been used to transport food and medicines to hospitals [1], [2].
While many researchers in the medical field develop vaccines to prevent the virus, many medicines and medical practices are being developed to heal infected patients and prevent them from passing on to others [3].
On the other hand, artificial intelligence and computer scientists have proposed and implemented real-life hybrid systems based on X-ray images and computed tomography (CT) to detect COVID-19. This artificial intelligence (AI) applications have been successfully applied in many areas [4]. The studies carried out in the literature and the studies carried out to give a more detailed description are given in the form of a table.
Some studies and diagnostic methods regarding COVID-19 in the literature are briefly summarized below. In Y. Pathak et al. study [5], they used Chest Computed Tomography (CT) images and Deep Transfer Learning (DTL) method to detect COVID-19 and obtained a high diagnostic accuracy. Mesut Toğaçar et al. proposed a novel hybrid method called the Fuzzy Color technique + deep learning models (MobileNetV2, SqueezeNet) with a Social Mimic optimization method to classify the COVID-19 cases and achieved high success rate in their work [6]. In the Ali Abbasian Ardakani et al. work [7], they used the deep learning models including AlexNet, VGG-16, VGG-19, SqueezeNet, GoogLeNet, MobileNet-V2, ResNet-18, ResNet-50, ResNet-101, and Xception to diagnose the COVID-19 and compared them with each other with respect to the obtained classification accuracy. Ferhat Ucar et al. proposed a novel method called Deep Bayes-SqueezeNet based COVIDiagnosis-Net to classify the COVID-19 cases as the COVID-19 or normal (healthy) [8]. As for other work of Tulin Ozturk et al. [9], they suggested a new method called the DarkCovidNet model for diagnosing the COVID-19 cases. Table 1 presents the conducted works regarding COVID-19 detection and diagnosis in the literature.
Table 1.
The conducted works regarding the COVID-19 detection and diagnosis in the literature.
| The authors and year of the conducted work in the literature | The used method | The used dataset and images |
|---|---|---|
| Y. Pathak et al. (2020) [5] | Deep Transfer Learning (DTL) | Chest Computed Tomography (CT) images |
| Mesut Toğaçar et al. (2020) [6] | Fuzzy Color technique + deep learning models (MobileNetV2, SqueezeNet) with Social Mimic optimization method | Chest X-ray images |
| Ali Abbasian Ardakani et al. (2020) [7] | Deep learning models including AlexNet, VGG-16, VGG-19, SqueezeNet, GoogleNet, MobileNet-V2, ResNet-18, ResNet-50, ResNet-101, and Xception | Chest Computed Tomography (CT) images |
| Ferhat Ucar et al. (2020) [8] | Deep Bayes-SqueezeNet based COVIDiagnosis-Net | Chest X-ray images |
| Tulin Ozturk et al. (2020) [9] | DarkCovidNet model | Chest X-ray images |
| Shreshth Tuli et al. (2020) [10] | Machine Learning and Cloud Computing | The outbreak dataset of COVID-19 Coronavirus |
| Turker Tuncer et al. (2020) [11] | Automated Residual Exemplar Local Binary Pattern and iterative ReliefF based corona detection method | Lung X-ray images |
| H. Kang et al. (2020) [12] | Structured Latent Multi-View Representation Learning | Chest computed tomography (CT) images |
| X. Wang et al. (2020) [13] | Weakly supervised deep learning framework | Chest computed tomography (CT) images |
| Yujin Oh et al. (2020) [14] | Deep learning model with Limited Training Data Sets | Chest X-ray images |
| Abdul Waheed et al. (2020) [15] | Auxiliary Classifier Generative Adversarial Network (ACGAN) based model called CovidGAN | Chest X-ray images |
In this study, we propose an intelligence diagnosis COVID-19 infection diagnosis model based on the convolutional neural networks (CNNs) and machine learning techniques. The proposed model ensures an end-to-end learning schema that can directly learn discriminative features from the input chest CT X-ray images and eliminate handcrafted feature engine. Contributions of the proposed model can be listed as follows:
-
(1)
CNNs with rich filter family, convolution, abstraction, and weight sharing have ensured an effective deep feature extraction engine.
-
(2)
The deep features extracted from deep layers of CNNs have been applied as the input to machine learning models to further improve COVID-19 infection detection.
-
(3)
As a result, a cheap, fast, and reliable intelligence tool has been provided for COVID-19 infection detection.
-
(4)
The developed model can be used to assist the field specialists, physicians, and radiologists in the decision-making process.
-
(5)
Thanks to this study, the misdiagnosis rates can be reduced, and the proposed model can be used as a retrospective evaluation tool.
The rest of this study is organized as follows: the dataset and the related methods are presented in Section 2. The results are reported in Section 3. A discussion is presented in Section 4, and lastly, concluding remarks are given in Section 5.
2. Material and methods
2.1. COVID-19 radiology database
Not only the structures of the samples in a database but also the distribution of the recordings among the classes have a great impact on the model to be developed. The morphological features, color, shape, and texture-based features directly affect the achievements of the intelligence computer-aided models [16]. Besides, it is important to ensure an equal number of samples, which cover all situations or cases for each class to produce a consistent and robust model.
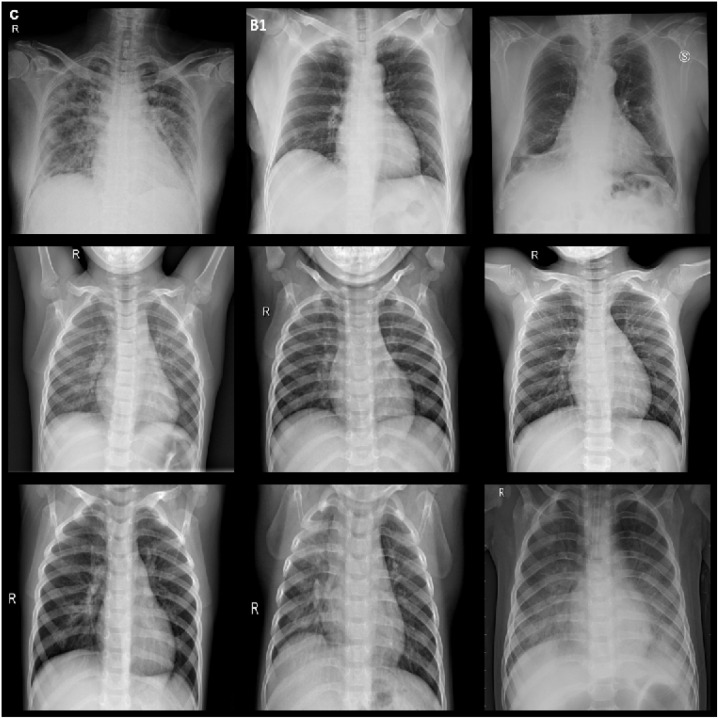
Recently, many studies have pointed out that chest CT images can be a vital evaluation means for diagnosing COVID-19 infection [6], [7], [8], [9]. Several specific patterns, including bilateral, peripheral and basal predominant ground-glass opacity (GGO), multifocal patchy consolidation, crazy-paving pattern with a peripheral distribution observed on chest CT images have been adopted as the findings of COVID-19 infection [17], [18], [19]. A subsample of the recordings belonging to COVID-19, normal and viral Pneumonia classes is shown in Fig. 1.
Fig. 1.
The samples correspond to the COVID-19, normal and viral pneumonia from the COVID-19 radiology database.
An open-access database that covers the posterior-to-anterior chest X-ray images was used in this study [20]. In fact, the COVID-19 Radiology database was generated by collecting the samples from four different resources. In other words, the samples collected from the Italian Society of Medical and Interventional Radiology (SIRM) COVID-19 Database [21], Novel Corona Virus 2019 Dataset [22], COVID-19 positive chest X-ray images from different articles and lastly chest X-ray [23] pneumonia images were combined. Totally 2905 images are presented with three classes in this database, as shown in Table 2.
Table 2.
The distribution of the samples between the classes.
| Class | # of samples |
|---|---|
| COVID-19 | 219 |
| Normal | 1341 |
| Viral Pneumonia | 1345 |
| Total | 2905 |
2.2. Proposed CNN model and training algorithm
2.2.1. CNN layers
CNNs are architectures consisting of a large number of sequenced layers. Layers that perform different functions are used in these architectures to reveal the distinctive features of the data applied as input [24]. In general, the tasks of these layers can be summarized as follows:
-
(1)Convolution layer: This layer is the main building block of CNN architectures, and it is used to reveal the discriminative features of the input data. This layer applies some filter families to the data so as to reveal low and high-level features in the data [25]. After the convolution process, the size of the input data changes. These charges vary depending on the stride and padding. The outputs of the convolution layers are called activation maps and defined as follows:
The convolution process is defined as in Eq. (1). Herein, the previous layers are shown with , the learnable kernels are and the bias term is . matches the input map section.(1) -
(2)Non-linearity layer: The convolution layer is ordinarily followed by the nonlinearity layer. This layer gives the system a non-linearity feature and called the activation layer. Since the neural network acts as a single perceptron, the outputs of the neural network can be calculated using linear combinations, so activation maps are used [26]. To this aim, the most commonly used activation function is Rectifier (ReLU), and it is defined as follows:
(2) -
(3)
Pooling (Down-sampling) layer: This layer is often added between consecutive convolutional layers to reduce the number of the computational nodes. Average pooling, maximum pooling, and L2-norm pooling are used frequently.
-
(4)
Flatting layer: This layer collects the data in a single vector and prepares the data for the neural network.
-
(5)
Fully-connected layers: This layer is used to transfer the activations that are obtained by passing the data throughout the network for the next unit. Fully connected layers are located at the end of the architecture to ensure the connections between all activations and computational nodes in these layers [27], [28], [29], [30]. These layers are exploited when the CNNs are used as the feature extractors.
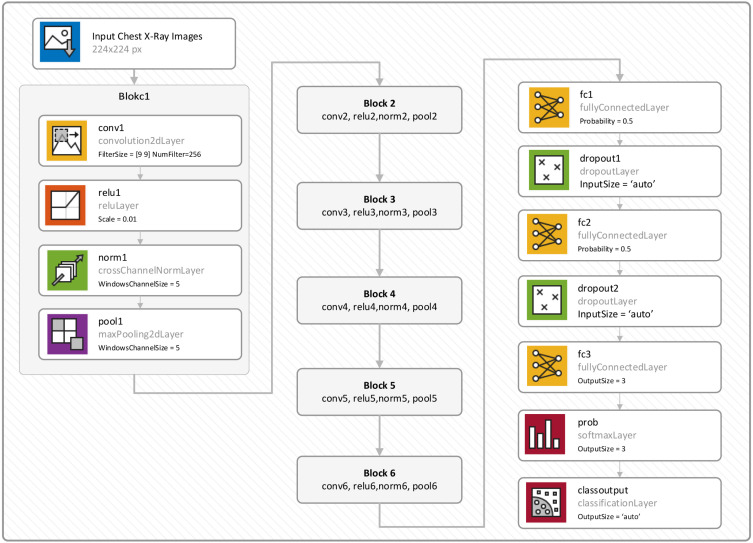
In this study, a new CNN model that consists of five basic blocks is proposed for COVID-19 infection detection, as shown in Fig. 2. In each block, convolution, ReLU, normalization, and pooling layers are used. At the end of the proposed model, three fully connected layers and the softmax layer are also used. The details of the proposed model are given in Table 3.
Fig. 2.
The block diagram of the proposed CNN model.
Table 3.
The details of the proposed CNN model.
| Name | Type | Activations | Learnables | |
|---|---|---|---|---|
| 1 | ChestXrayCT | Image Input | 227 × 227 × 3 | – |
| 2 | conv1 | Convolution | 74 × 74 × 128 | Weights 9 × 9 × 3 × 128 Bias 1 × 1x128 |
| 3 | relu1 | ReLU | 74 × 74 × 128 | – |
| 4 | norm1 | Cross channel normalization | 74 × 74 × 128 | – |
| 5 | pool1 | Max Pooling | 37 × 37 × 128 | – |
| 6 | conv2 | Convolution | 19 × 19 × 256 | Weights 3 × 3 × 128 × 256 Bias 1 × 1 × 256 |
| 7 | relu2 | ReLU | 19 × 19 × 256 | – |
| 8 | norm2 | Cross channel normalization | 19 × 19 × 256 | – |
| 9 | pool2 | Max Pooling | 10 × 10 × 256 | – |
| 10 | conv3 | Convolution | 5 × 5 × 256 | Weights 3 × 3 × 256 × 256 Bias 1 × 1 × 256 |
| 11 | relu3 | ReLU | 5 × 5 × 256 | – |
| 12 | norm3 | Cross channel normalization | 5 × 5 × 256 | – |
| 13 | pool3 | Max Pooling | 3 × 3 × 256 | – |
| 14 | conv4 | Convolution | 2 × 2 × 512 | Weights 3 × 3 × 256 × 512 Bias 1 × 1 × 512 |
| 15 | relu4 | ReLU | 2 × 2 × 512 | – |
| 16 | norm4 | Cross channel normalization | 2 × 2 × 512 | – |
| 17 | pool4 | Max Pooling | 1 × 1 × 512 | – |
| 18 | conv5 | Convolution | 1 × 1 × 512 | Weights 3 × 3 × 512 × 512 Bias 1 × 1 × 512 |
| 19 | relu5 | ReLU | 1 × 1 × 512 | – |
| 20 | norm5 | Cross channel normalization | 1 × 1 × 512 | – |
| 21 | pool5 | Max Pooling | 1 × 1 × 512 | – |
| 22 | fc1 | Fully connected | 1 × 1 × 1024 | Weights 1024 × 512 Bias 1024 × 1 |
| 23 | drop1 | Dropout | 1 × 1 × 1024 | – |
| 24 | fc2 | Fully connected | 1 × 1 × 1024 | Weights 1024 × 1024 Bias 3 × 1 |
| 25 | relu6 | ReLU | 1 × 1 × 1024 | – |
| 26 | drop2 | Dropout | 1 × 1 × 1024 | – |
| 27 | fc3 | Fully connected | 1 × 1 × 3 | Weights 3 × 1024 Bias 3 × 1 |
| 28 | Softmax | Softmax | 1 × 1 × 3 | – |
| 29 | classoutput | Classification output | – | – |
2.2.2. Training of the proposed CNN model
Once the model design has been carried out, the developed COVID-19 infection diagnosis model needs to be trained. In this step, hyperparameters such as initial learning rate, mini-batch size, the maximum number of iterations, the number of images to be processed in each iteration should be determined. In addition, an optimization algorithm must be selected for backpropagation and updating of model weights. For the proposed model training, the steps described in Algorithm 1 are followed, and the proposed CNN model is trained from scratch.
Herein, the training and test sets are shown with and , respectively. The learning rate that is one of the most important hyperparameters and determines how rapidly a modal adopts to the problem is shown with . The total number of iterations is denoted with . As for , it points to the number of samples processed in each iteration. The values of , and hyperparameters were determined by trial and error in the experiments [31].
ADAM optimization algorithm was used as a solver. The epoch was set to 64. The number of recordings per epoch was 21, and the maximum iteration was 1344. The initial learning was adjusted to 0.0001. By the way, the learning rate was reduced gradually by 0.1 for every 16 epochs.
2.2.3. Data augmentation approach
Offline or online data augmentation techniques can be used to realize a more efficient training for the computational models [24]. However, it is essential to be aware that the data augmentation techniques should not be used on the test set because of the overfitting problem.
In the experiment, the whole data set was divided into two parts as the training and test sets with 70% and 30% rates, respectively. The distribution of the samples over the classes is imbalanced. To overcome this issue, the data augmentation approach has been used. To this aim, we focused on only the COVID-19 class since the number of samples in this class was lower compared to other classes, as shown in Table 4.
Table 4.
The distribution of the samples between the classes after the data augmentation approach in the training set.
| Class | Training set |
Test set | |
|---|---|---|---|
| Before augmentation | After augmentation | (Frozen) | |
| # of samples | # of samples | # of samples | |
| COVID-19 | a | 918 | 66 |
| Normal | 939 | 939 | 402 |
| Viral pneumonia | 941 | 941 | 404 |
| Total | 2033 | 2798 | 872 |
The data augmentation approach was applied to only COVID-19 class in the training set.
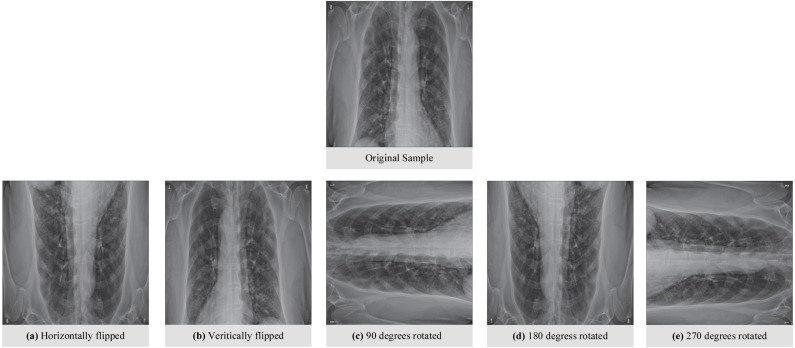
Each original sample was represented with five additional samples derived from the original sample, as shown in Fig. 3. The rotate and flip data augmentation approaches were used in this process. As a result, each sample in the training set was represented with a total of six images. The number of COVID-19 samples in the training set was increased from 153 to 918 after the data augmentation process. In this manner, the distribution of the samples for each class was almost equal.
Fig. 3.
An original and augmented sample from the training set.
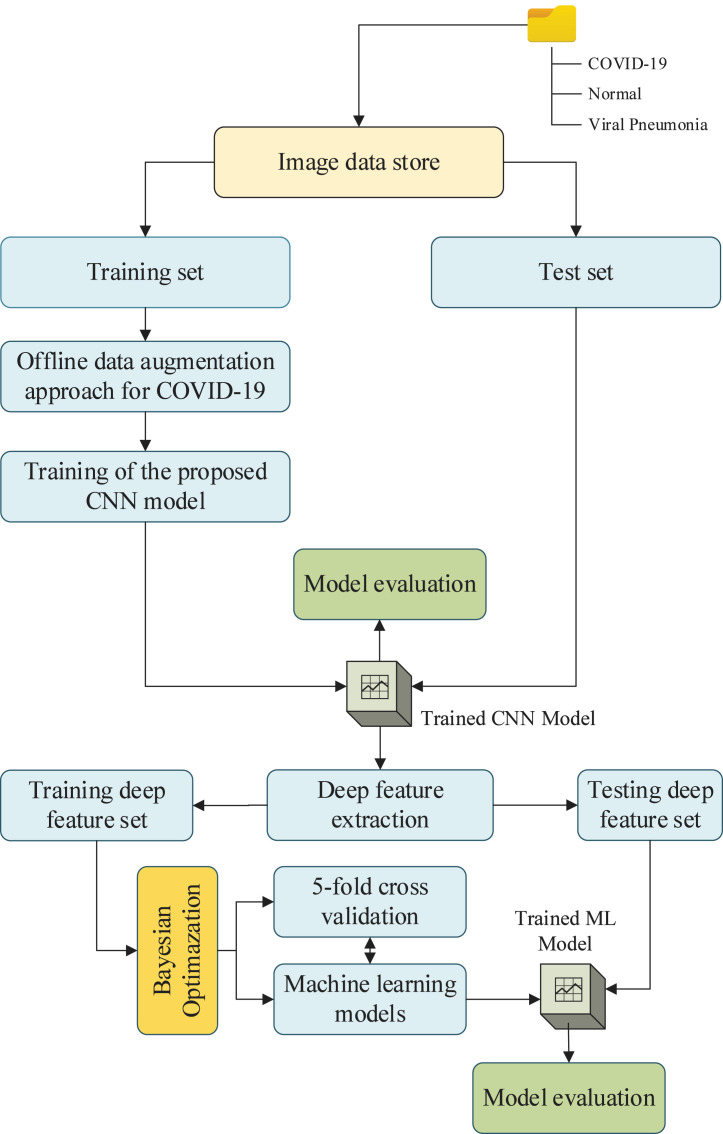
The overall block diagram of the proposed model is given in Fig. 4. The whole dataset is divided into two sets as training and test sets with 70% and 30% rates, respectively. Only the number of samples in the COVID-19 class is increased by using the offline data augmentation approach, and then the proposed CNN model is trained and tested. Then, the deep features extracted from the proposed CNN model is considered. A combination of deep feature extraction and machine learning techniques are utilized to achieve a consistent and robust diagnosis model for COVID-19 infection diagnosis.
Fig. 4.
The overall block diagram of the proposed model.
2.3. Machine learning techniques
Three different classification algorithms have been used to detect COVID-19 infection detection in this study. These classification algorithms are different in structure and have high performance. Each classifier algorithm was trained and tested using the 70%–30% training and testing data partition. The used classifier algorithms were explained in the following subsections.
2.3.1. Support vector machine
Support vector machines (SVM) is a consulting machine learning algorithm that can be used to solve both classification and regression problems. Although, it is most often used in the solution of classification problems. In the SVM algorithm, each pair of data can be represented as a point in n-dimensional space with each property value in a special coordinate plane. Then, to solve a two-class classification problem, a hyper-plane is found, and the classification is performed. In the SVM classifier, it is easy to have a linear hyper-plane between the two classes. The gap between linear equations and classes on the hyper-plane needs to be optimized.
In our SVM model, the RBF (Radial Basis Function) kernel function has been used in the classification of the datasets. The Radial basis function kernel called the RBF kernel, or Gaussian kernel is a kernel that is in the form of a radial basis function. We chose the RBF kernel because it gives the highest performance with respect to the classification performance. The parameters in the RBF kernel function have been optimized by using the Bayesian optimization method in our study. The used kernel function is given in Eq. (3) as follows:
| (3) |
Where is the distance between data points of and . For more information about the multi-class-SVM classifier, the readers can refer to [32], [33], [34].
2.3.2. Decision tree
The decision tree classifier is used to solve simple and mostly classification problems. Applies the correct way to solve the classification problem. The decision tree classifier has a structure consisting of roots, leaves, and branches descending from top to bottom. The most used decision tree classification algorithms are ID3, C4.5, and C5. In our applications, we have used the C4.5 decision tree classifier. For more information about the decision tree classifier, the readers can refer to [35], [36], [37], [38], [39], [40].
2.3.3. k-nearest neighbor
The -NN (-nearest neighbor) algorithm is one of the simplest and most widely used classification algorithms. NN is a non-parametric, lazy learning algorithm. Unlike eager learning, if we try to understand the concept of lazy, lazy learning does not have a training phase. It does not learn the training data; instead, it “memorizes” the training data set. When we want to make an estimate, it looks for the nearest neighbors in the whole dataset [41].
In the study of the algorithm, a k value is determined. The meaning of this value is the number of elements to be looked at. When a value arrives, the distance between the incoming value is calculated by taking the nearest element. The Euclidean function is generally used in distance calculation. As an alternative to the Euclidean function, City Block, Minkowski, and Chebyshev functions can also be used [42]. After the distance is calculated, it is sorted, and the incoming value is assigned to the appropriate class. The parameters in the NN classifier have been optimized by using the Bayesian optimization method in our study.
2.4. Model evaluation
To evaluate the proposed model, we have used the confusion matrix, and some commonly used performance metrics such as accuracy (Acc), specificity (Sp), sensitivity (Se), and F-score derived from this matrix. It consists of four indices that are true positive (TP), true negative (TN), false positive (FP) and false-negative (FN) and the mentioned performance metrics are calculated using these indices as described follows:
| (4) |
| (5) |
| (6) |
| (7) |
Herein, TP and TN represent the number of correctly predicted positive and negative samples, whereas FP and FP correspond to the number of incorrectly predicted positive and negative samples.
Besides, the area under curve (AUC) of receiving operating characteristic (ROC) has been taken into account to evaluate the model performance. ROC is a 2D graph that is drawn the true positive rate (TPR) against the false-negative rate (FNR). This curve indicates a trade-off between Se and Sp, and it is useful to understand the overall achievement of the models [31].
3. Results
The experiments were carried out on a workstation with Intel® Xeon® Gold 6132 CPU @2.60 GHz and NVIDIA Quadro P6000 GPU. The simulation environment was MATLAB (2019a).
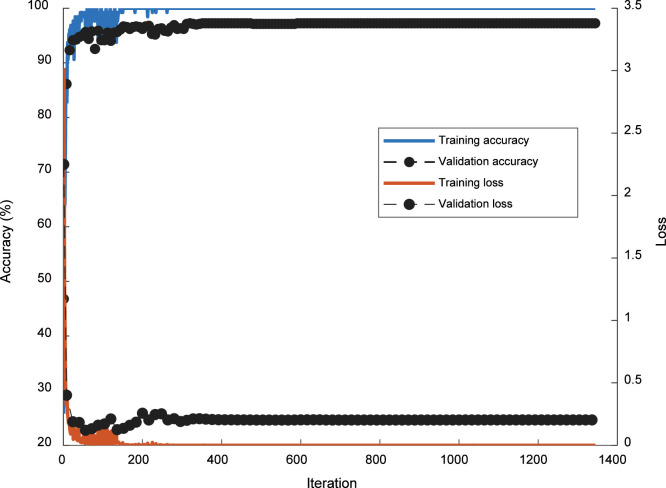
The training of the proposed CNN model was realized in 64 epoch, and the mini-batch size was 128. 21 samples were processed per epoch, and the training of the model was completed in a total of 1344 iteration. The time elapsed for the training of the model was 85.15 min. The initial learning rate was 0.0001. We employed a learning rate schedule approach in the training of the model. In this scope, the learning rate was gradually decreased. The learning drop factor was set to 0.1, and the learning drop period was adjusted to 16. ADAM optimization method was used as a solver. The training and validation graphs with the loss of the proposed CNN model are given in Fig. 5. In Fig. 5, lefty-axis shows the training and validation accuracies of the proposed CNN model, while the right -axis shows loss values. As a result, the final training accuracy and training loss were obtained as 100% and 0, respectively.
Fig. 5.
The training and validation graphs with the loss of the proposed CNN model.
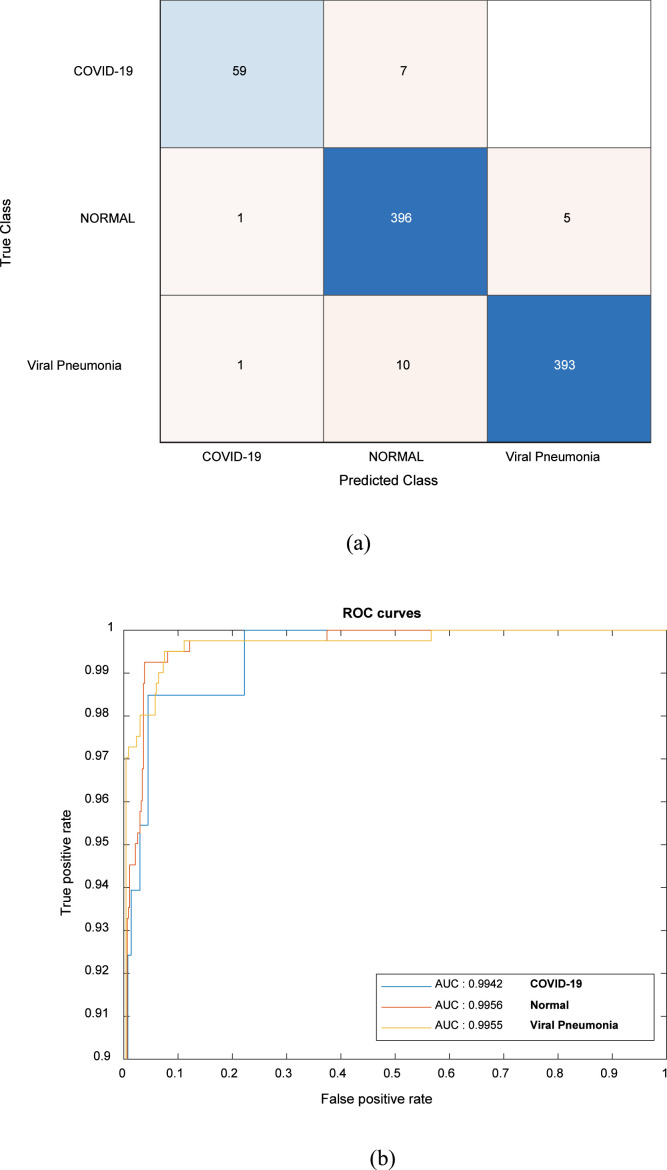
As for a prediction, the confusion matrix is given in Fig. 6(a). As mentioned before, the test set was separated and frozen at the starting of the experiment. The number of samples belonging to the COVID-19 class in the test set was 66. 59 of these samples were identified correctly by the proposed CNN model. The rates of the classification achievements for normal and viral pneumonia cases were rather satisfactory. The final validation accuracy and final validation loss were 97.25% and 0.2032, respectively. The Se, Sp, and F-score were achieved as 94.61%, 98.29%, and 95.75%, respectively. The ROC curves of the proposed CNN model are also presented in Fig. 6(b). The AUCs were obtained as 0.9942, 0.9956, 0.9955 for COVID-19, normal, and viral pneumonia cases, respectively. As a result, an efficient CNN model ensured for diagnosis of COVID-19 infection.
Fig. 6.
(a) Confusion matrix of the proposed CNN model. (b) ROC curves of the proposed CNN model.
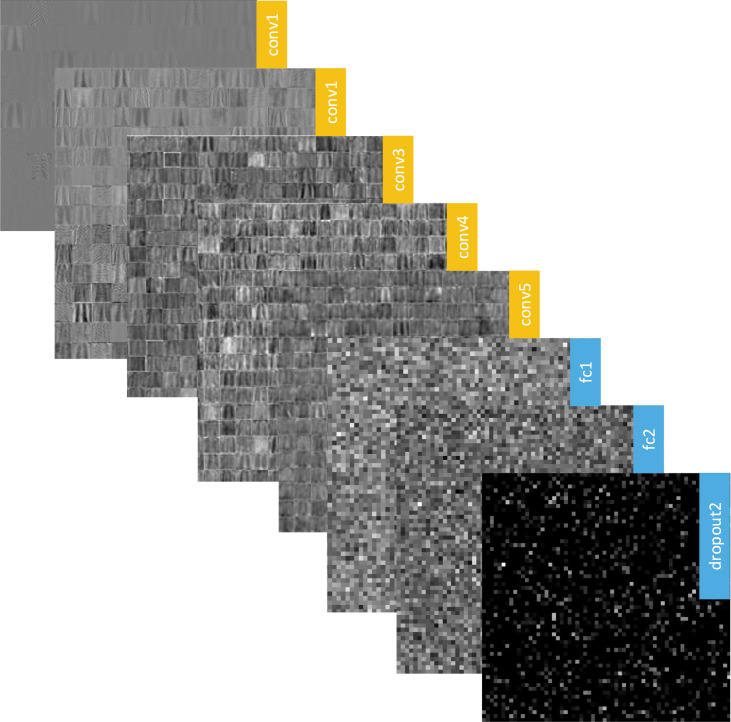
In the second step of the experiment, we focused on the activation maps in the proposed CNN architecture. These activation maps with different levels keep the discriminative features of the input data and finally collected in the fully connected layers. The activations may help us to understand what the model has learned. A visual representation of the activation maps is given in Fig. 7.
Fig. 7.
A visual representation of some activation maps in the proposed CNN models.
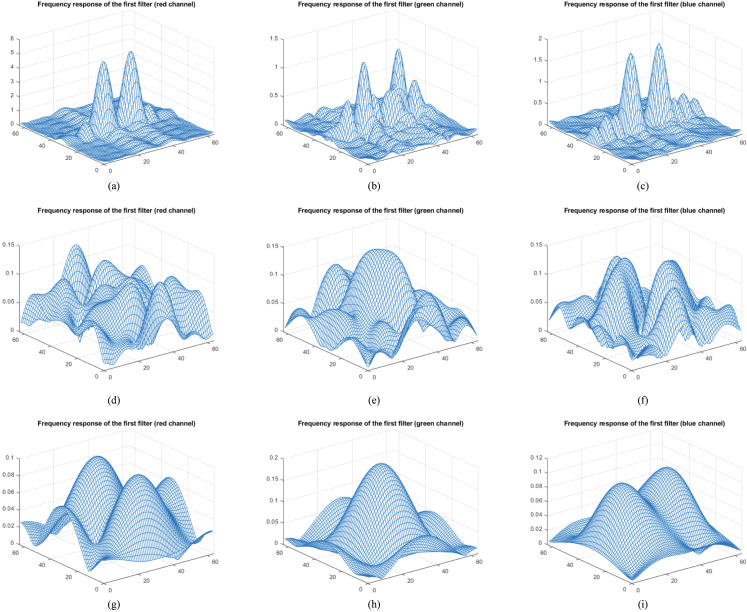
Depending on the progress of the input data throughout the model, the significant changes realize in the activation maps, and the abstraction in the training process can be observed via activations. Our main purpose in this step was to separate the activations that have the best discrimination capacity compared to others with relatively weak representation power. The rich filter families are used in the convolution layers, and numerous forms of the input data are processed in the CNNs. The basic features such as color and edges can be learned in the first convolution layers, while more complicated features can be revealed in deeper convolution layers. Besides, the discriminative capacity of the activations may vary depending on the structures of the problems. For this reason, determining the most efficient activations is a rather difficult task. To visualize this challenge, the frequency responses on the RGB channels of the first filters in the first three convolution layers in the proposed CNN model are illustrated in Fig. 8.
Fig. 8.
The frequency responses of the weights of the convolution layers on RGB channels.
Since all activations were collected from fully connected layers, the deep features were extracted from fc1 and fc2 layers. These two different deep feature sets were applied individually as the input to machine learning models. As a result, the activations were used effectively for COVID-19 infection detection.
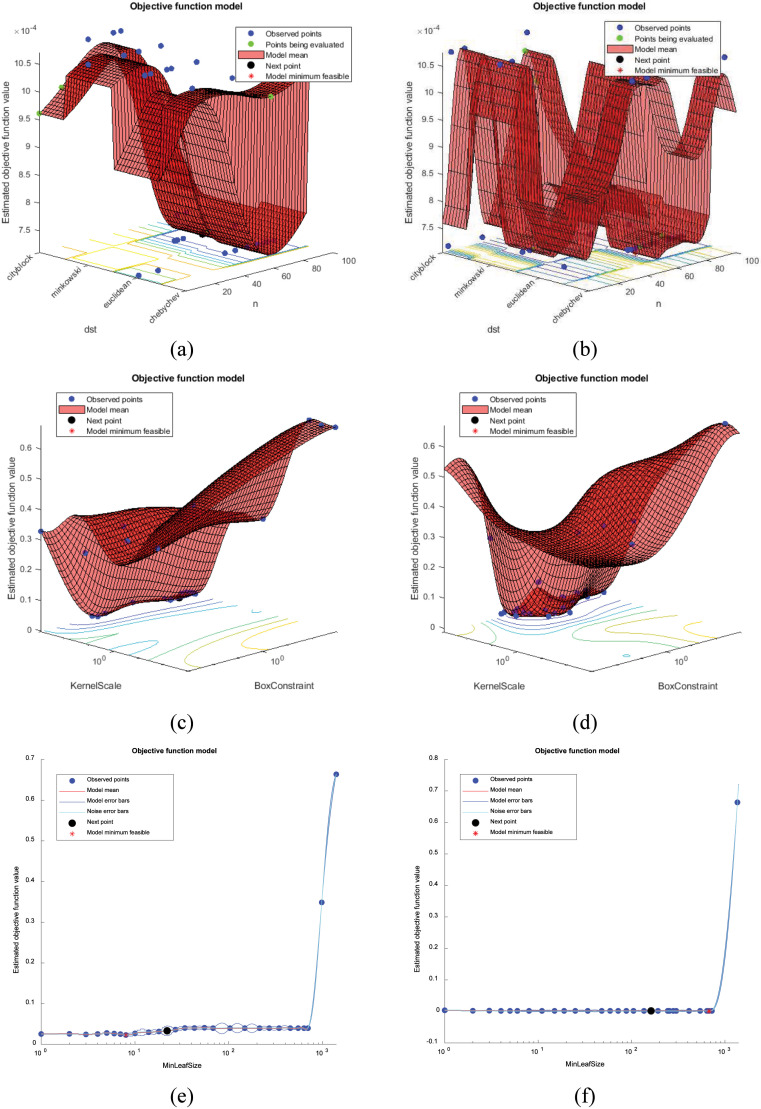
The hyperparameters of the machine learning models were optimized using the Bayesian optimization algorithm. For NN classifier, four distance functions that were City Block, Minkowski, Euclidean, and Chebyshev were evaluated with different values in the range of and . A result, the best points were obtained when was 82 and distance function was for fc1 deep feature set whereas was 65 and distance function was for fc2 deep feature set as shown in Fig. 9(a) and (b). The NN produced the best results on fc2 deep feature set with 1024 deep features. The Acc, Se, Sp, F-score were obtained as 95.76%, 92.29%, 97.43%, and 93.97%, respectively. Besides, the kNN model fed with fc1 feature was also yielded promising results with an Acc of 95.07%, Se of 90.11%, Sp of 96.97%, and F-score of 92.61%.
Fig. 9.
Bayesian optimization results. (a) NN fed with fc1. (b) NN fed with fc2. (c) SVM fed with fc1. (d) SVM fed with fc2. (e) DT fed with fc1. (f) DT fed with fc2.
As for the SVM classifier, the kernel was adjusted Radial Basis Function (RBF). The optimum kernel scale and box constraint were searched in the range of and . As a result, the most efficient results were observed when the box constraint was 815.17, and the kernel scale was 999.64 for fc1 feature set, as shown in Fig. 9(c). The Acc, Se, Sp and F-score were 98.62%, 89.39%, 99.38% and 90.77%, respectively. The model achievement was evaluated on fc2 deep feature set, the best observed feasible points were 0.4569 for box constraint and 635.62 for kernel scale, as shown in Fig. 9(d). The model ensured satisfactory results with an Acc of 98.97%, Se of 89.39%, Sp of 99.75%, and F-score of 96.72%, respectively.
DT algorithm was optimized as in NN and SVM classifier. To this aim, the determination of the minimum leaf size was realized by the Bayesian optimization algorithm and set to 6 for fc1 deep feature, as shown in Fig. 9(e). The Acc, Se, Sp, and F-score were 93.35%, 90.55%, 96.29%, and 90.06%, respectively. In addition, the best estimated feasible point considering the DT algorithm was 675 for fc2 deep feature set, as shown in Fig. 9(f). The Acc was 96.10%, Se was 93.81%, Sp was 97.70% and F-score was 94.56%.
All scores of the classifiers are reported in Table 5, considering the two different deep feature sets. The SVM classifier was superior to NN and DT machine learning algorithms. It was seen that the SVM model ensured an improvement in the automated COVID-19 infection detection task. Unlike it was observed that the classification achievement was lightly decreased when the classification task was realized by NN and DT.
Table 5.
The performance metrics of the machine learning models.
| Classifier | Feature set | Acc (%) | Se (%) | Sp (%) | F-score (%) |
|---|---|---|---|---|---|
| NN | fc1 with 1024 deep features | 95.07 | 90.11 | 96.97 | 92.61 |
| SVM | 98.62 | 89.39 | 99.38 | 90.77 | |
| DT | 92.09 | 87.53 | 95.54 | 87.69 | |
| NN | fc2 with 1024 features | 95.76 | 92.29 | 97.43 | 93.97 |
| SVM | 98.97 | 89.39 | 99.75 | 96.72 | |
| DT | 96.10 | 93.81 | 97.70 | 94.56 | |
| Proposed CNN | – | 97.14 | 94.61 | 98.29 | 95.75 |
4. Discussion
In this section, we evaluate the superior aspects as well as the limitations of the proposed model by taking into account the state-of-art models. However, it is important to be aware of a one-to-one comparison is not feasible due to differences in datasets, methods, and various simulation environments.
There are many people affected by COVID-19 disease. However, large-scale datasets labeled by field experts are still not available. So, computational works on automatic COVID-19 infection detection have been conducted on the combined datasets. The samples in these datasets were collected from different resources, as inferred from Table 6.
Table 6.
Comparison of the state-of-art models.
| Methods | Dataset | # of classes | Acc (%) | Se (%) | Sp (%) |
|---|---|---|---|---|---|
| DarkCovidNet [9] | Public | 3 | 87.02 | 92.18 | 89.96 |
| COVIDiagnosis-Net [8] | Public | 3 | 98.26 | 99.13 | – |
| The pretrained CNNs [43] | Public | 3 | 93.48 | 92.85 | 98.75 |
| COVID-Net [44] | Public | 3 | 92.64 | 91.37 | 95.76 |
| Deep features, ResNet-50, SVM [45] | Public | 2 | 95.38 | – | – |
| Deep CNNs [46] | Public | 2 | 90.00 | 100 | 80.00 |
| Deep CNN, ResNet-50 [47] | Public | 2 | 98.00 | – | – |
| DRE-Net, deep CNN [48] | Private dataset | 2 | 86.00 | 96.00 | – |
| Deep CNN, Inception, transfer learning [49] | Private dataset | 2 | 89.50 | 87.00 | 88.00 |
| nCOVnet, transfer learning, deep CNN [31] | Public | 2 | 88.10 | 97.62 | 89.13 |
| A novel CNN model, training from scratch strategy, deep feature extraction, SVM | Public | 3 | 98.97 | 89.39 | 99.75 |
Recently, it is seen that the scientific community has focused on chest X-ray images in order to contribute to the clinical evaluation of COVID-19 cases that have increased day by day. Many computational models based on CNN architecture have been proposed. The greatest advantage of these models is that they provide an end-to-end learning scheme by isolating handcrafted feature engine. To this aim, the transfer learning approach has been generally adopted to train the CNNs. Some of the computational studies have been focused on the deep features provided by the pre-trained models [45]. In this aspect, our study offers a novel CNN model that was trained from scratch, not a transfer learning approach. Also, instead of using pre-trained CNNs, fully-connected layers in the proposed architecture were considered, examined, and used for the COVID-19 infection detection task. Our study contains the innovative components in this respect. Besides, the proposed model works according to the end-to-end learning principle, and a handcrafted feature extraction engine is not applied. As a result, an efficient, fast, reliable model was developed, and promising results were achieved.
It should not be forgotten that the proposed model is evaluated at the COVID-19 Radiology Database scale. Considering the number of positive COVID-19 cases worldwide, it can be argued that the database is not large enough. However, we think that there is nothing to worry about this issue. Because the performances of the CNN networks increase depending on the scale of the number of samples used in the training process, in such a case, it is only necessary to consider the calculation time and hardware resources. Another important issue is that when the positive COVID-19 cases are detected using X-ray images, the infection may have already significantly advanced. In other words, X-ray images may be a very significant means to confirm positive COVID-19 cases, but may not be clinically relevant for early diagnosis.
5. Conclusion
General public health, global economy, and our routine life continue with new norms with the effect of COVID-19. The number of people affected by this infection is still increasing significantly. In this study, an automated COVID-19 diagnostic system has been proposed to contribute to clinical trials. The proposed model is based on the CNN architecture, and it is trained from scratch, as opposed to the transfer learning approach. Thanks to its convolution with rich filter families, abstraction, and weight sharing features, it automatically provides highly efficient deep, distinctive features. Thus, the handcrafted feature extraction engine is not performed. As a result, positive COVID-19 cases can be detected easily and with high sensitivity via the proposed tool using chest X-ray images. As a result of this study, a cheap, fast, and reliable diagnostic tool was obtained. The model provided an accuracy of 98.97%, a sensitivity of 89.39%, the specificity of 99.75%, and F-score of 95.75%. When it is evaluated clinically, the developed model can support the decision-making processes of field specialists, physiologists, and radiologists. With this model, the misdiagnosis rate can be reduced, and positive COVID-19 cases can be detected quickly without having to wait for days.
CRediT authorship contribution statement
Majid Nour: Literature analysis, Interpretation of results, Preparation of the manuscript. Zafer Cömert: Literature analysis, Interpretation of results, Preparation of the manuscript. Kemal Polat: Literature analysis, Interpretation of results, Preparation of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, Saudi Arabia , under grant No. (GCV19-4-1441) The authors, therefore, gratefully acknowledge DSR technical and financial support.
References
- 1.Ceylan Z. Estimation of COVID-19 prevalence in Italy, Spain, and France. Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Awadhi A.M., Alsaifi K., Al-Awadhi A., Alhammadi S. Death and contagious infectious diseases: Impact of the COVID-19 virus on stock market returns. J. Behav. Exp. Financ. 2020;27 doi: 10.1016/j.jbef.2020.100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W. 2020. AI-Assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. MedRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S., Shukla P.K. Deep transfer learning based classification model for COVID-19 disease. IRBM. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ucar F., Korkmaz D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U. Rajendra P.K. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuli S., Tuli S., Tuli R., Gill S.S. Predicting the growth and trend of COVID-19 pandemic using machine learning and cloud computing. Internet of Things. 2020;11 doi: 10.1016/j.iot.2020.100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuncer T., Dogan S., Ozyurt F. An automated residual exemplar local binary pattern and iterative relieff based corona detection method using lung X-ray image. Chemom. Intell. Lab. Syst. 2020;203 doi: 10.1016/j.chemolab.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang H., Xia L., Yan F., Wan Z., Shi F., Yuan H. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE. Trans. Med. Imaging. 2020 doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- 13.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;1 doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 14.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;1 doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 15.Waheed A., Goyal M., Gupta D., Khanna A., Al-Turjman F., Pinheiro P.R. CovidGAN: Data augmentation using auxiliary classifier GAN for improved Covid-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toğaçar M., Ergen B., Cömert Z. Application of breast cancer diagnosis based on a combination of convolutional neural networks, ridge regression and linear discriminant analysis using invasive breast cancer images processed with autoencoders. Med. Hypotheses. 2020;135 doi: 10.1016/j.mehy.2019.109503. [DOI] [PubMed] [Google Scholar]
- 17.J.P. Kanne, B.P. Little, J.H. Chung, B.M. Elicker, L.H. Ketai, Essentials for radiologists on COVID-19: An update—Radiology scientific expert panel, Radiology 200527. 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed]
- 18.Chang T.-H., Wu J.-L., Chang L.-Y. Clinical characteristics and diagnostic challenges of pediatric COVID-19: A systematic review and meta-analysis. J. Formos. Med. Assoc. 2020 doi: 10.1016/j.jfma.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. Prokop, W. van Everdingen, T. van Rees Vellinga, J. van Ufford, L. Stöger, L. Beenen, et al. CO-RADS – A categorical CT assessment scheme for patients with suspected COVID-19: definition and evaluation, Radiology 201473. 10.1148/radiol.2020201473. [DOI] [PMC free article] [PubMed]
- 20.Muhammad E.H.C., Tawsifur R., Amith K., Rashid M., Muhammad Abdul K., Zaid Bin M. 2020. COVID-19 radiology database. Can AI help screen viral COVID-19 pneumonia? pp. 1–14. https://arxiv.org/abs/2003.13145. [Google Scholar]
- 21.Radiology IS of M and I. Italian society of medical and interventional radiology. https://www.sirm.org/category/senza-categoria/covid-19/.
- 22.Joseph Paul C., Paul M., Lan D. 2020. COVID-19 image data collection; pp. 1–4. https://arxiv.org/pdf/2003.11597.pdf. [Google Scholar]
- 23.Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Başaran E., Cömert Z., Çelik Y. Convolutional neural network approach for automatic tympanic membrane detection and classification. Biomed. Signal Process. Control. 2020;56 doi: 10.1016/j.bspc.2019.101734. [DOI] [Google Scholar]
- 25.Toğaçar M., Ergen B., Cömert Z. BrainMRNet: Brain tumor detection using magnetic resonance images with a novel convolutional neural network model. Med. Hypotheses. 2020;134 doi: 10.1016/j.mehy.2019.109531. [DOI] [PubMed] [Google Scholar]
- 26.Toğaçar M., Ergen B., Cömert Z. Waste classification using autoencoder network with integrated feature selection method in convolutional neural network models. Measurement. 2019 doi: 10.1016/j.measurement.2019.107459. [DOI] [Google Scholar]
- 27.Budak Ü., Cömert Z., Rashid Z.N., Şengür A., Çıbuk M. Computer-aided diagnosis system combining FCN and bi-LSTM model for efficient breast cancer detection from histopathological images. Appl. Soft Comput. 2019;85 doi: 10.1016/j.asoc.2019.105765. [DOI] [Google Scholar]
- 28.Ullo S.L., Khare S.K., Bajaj V., Sinha G.R. Hybrid computerized method for environmental sound classification. IEEE Access. 2020;8:124055–124065. doi: 10.1109/ACCESS.2020.3006082. [DOI] [Google Scholar]
- 29.Khare S., Bajaj V. Time-frequency representation and convolutional neural network based emotion recognition. IEEE Trans. Neural Netw. Learn. Syst. 2020 doi: 10.1109/TNNLS.2020.3008938. in press. [DOI] [PubMed] [Google Scholar]
- 30.Bajaj V., Taran S., Tanyildizi E., Sengur A. Robust approach based on convolutional neural networks for identification of focal EEG signals. IEEE Sensors Lett. 2019;3(5) doi: 10.1109/LSENS.2019.2909119. 1–4. [DOI] [Google Scholar]
- 31.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos Solitons Fractals. 2020 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cortes C., Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–297. doi: 10.1007/BF00994018. [DOI] [Google Scholar]
- 33.Nayak J., Naik B., Behera H.S. A comprehensive survey on support vector machine in data mining tasks: Applications & challenges. Int. J. Database Theory Appl. 2015;8:169–186. doi: 10.14257/ijdta.2015.8.1.18. [DOI] [Google Scholar]
- 34.Awad M., Khanna R. In: Support Vector Machines for Classification BT - Efficient Learning Machines: Theories, Concepts, and Applications for Engineers and System Designers. Awad M., Khanna R., editors. Apress; Berkeley, CA: 2015. pp. 39–66. [DOI] [Google Scholar]
- 35.Safavian S.R., Landgrebe D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991;21:660–674. [Google Scholar]
- 36.Rokach L., Maimon O. Decision Trees. Springer US; Boston, MA: 2005. pp. 165–192. (Data Min. Knowl. Discov. Handb.). [DOI] [Google Scholar]
- 37.Aha D.W., Kibler D., Albert M.K. Instance-based learning algorithms. Mach. Learn. 1991;6:37–66. doi: 10.1007/BF00153759. [DOI] [Google Scholar]
- 38.Arican Murat, Polat Kemal. Binary particle swarm optimization (BPSO) based channel selection in the EEG signals and its application to speller systems. J. Artif. Intell. Syst. 2020;2:27–37. doi: 10.33969/AIS.2020.21003. [DOI] [Google Scholar]
- 39.Ozdemir Akin., Polat Kemal. Deep learning applications for hyperspectral imaging: A systematic review. J. Inst. Electron. Comput. 2020;2:39–56. doi: 10.33969/JIEC.2020.21004. [DOI] [Google Scholar]
- 40.Demir F., Bajaj V., Ince M.C. Surface EMG signals and deep transfer learning-based physical action classification. Neural Comput. Appl. 2019;31:8455–8462. doi: 10.1007/s00521-019-04553-7. [DOI] [Google Scholar]
- 41.Wang H., Duntsch I. Brock Univ; 2003. Nearest Neighbours Without K: A Classification Formalism Based on Probability; pp. 1–10. [Google Scholar]
- 42.Suguna N., Thanushkodi K. An improved k-nearest neighbor classification using genetic algorithm. IJCSI Int. J. Comput. Sci. 2010;7:7–10. [Google Scholar]
- 43.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020 doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Wong A. 2020. COVID-NEt: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sethy P.K., Behera S.K. 2020. Detection of coronavirus disease (COVID-19) based on deep features. Preprints . [DOI] [Google Scholar]
- 46.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. [Google Scholar]
- 47.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional. Neural Netw. 2020 doi: 10.1007/s10044-021-00984-y. arXiv preprint https://arxiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z. 2020. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. MedRxiv . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J. 2020. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19) MedRxiv . [DOI] [PMC free article] [PubMed] [Google Scholar]