Abstract
Methylmalonic acidemia (MMA)-affected patients may have developmental, hematological, neurological, metabolic, ophthalmological, and dermatological clinically abnormal findings. This study aimed to identify mutations in 13 Chinese MMA cases. We provided genetic counseling, treatment, and prenatal diagnosis for the families with MMA. Liquid chromatography-tandem mass spectrometry (LC–MS/MS) was performed and the results were confirmed by gas chromatography and mass spectrometry (GC/MS). Variant screening in probands was performed by targeted next-generation sequencing. Identified variants were confirmed by Sanger sequencing. Of these 13 MMA cases, seven were isolated MMA, and among them, six were caused by variants in MMUT and one was caused by a variant in MCEE. The other six cases were MMA with homocystinuria, which was caused by variants in MMACHC. We found six novel variants in three MMA-causing genes as follows: c.2008G>A, c.301_302insTA, c.984delC, and c.319A>T of MMUT; c.445T>C of MMACHC; and c.296T>C of MCEE. We provided prenatal diagnosis for two families with MMA at their next pregnancy, and one family had a healthy newborn. In conclusion, our findings expand the spectrum of genotypes in MMA. Effective genetic counseling is required to allow awareness of the patients’ families that MMA disease is treatable and a good prognosis can be obtained.
Subject terms: Diseases, Medical research, Molecular medicine
Introduction
Methylmalonic acidemia is a group of inborn errors of metabolism causing multisystem disease. MMA has two subtypes of isolated MMA and MMA with homocystinuria. Two genetic defects of deficiency of methylmalonyl CoA mutase and defects in the synthesis of the coenzyme adenosylcobalamin cause isolated MMA1. The worldwide estimated incidence of MMA is approximately 1/48,000–1/250,0002. The incidence of MMA is different among the different MMA subtypes and regions of China. The total incidence of MMA in China is approximately 1: 28,000 for MMA at birth3, but in Shandong Province, the incidence of cobalamin (cbl) C type is approximately 1: 3,920 live births4. According to the age of onset, MMA can be divided into two forms as follows. In the early-onset form, patients with MMA present with symptoms within the first year. The late-onset form is rarer than the early-onset form and is easily misdiagnosed or missed. In the late-onset form, patients present with a milder clinical phenotype with acute or slowly progressive nervous system symptoms and behavioral disorders at any time from childhood to adulthood5. The clinical manifestations of patients with MMA are complex and varied, range in severity, and can manifest as an acute or chronic course. Severe cases of MMA and death occur in the neonatal period, and mild disease can occur in adulthood.
MMA is a genetically heterogeneous disease. Isolated MMA can be caused by mutations in MMUT (OMIM, 609058), which result in deficiency of methylmalonyl CoA mutase, and this form is unresponsive to vitamin B12 therapy. Mutations in MCEE lead to deficiency of methylmalonyl-CoA epimerase, which can result in mild isolated MMA6. Other forms of isolated MMA are found in a subset of patients with defects in synthesizing the coenzyme adenosylcobalamin and are classified into three groups: cblA (OMIM, 251100), which is caused by mutations in MMAA (OMIM, 607481); cblB (OMIM, 251110), which is caused by mutations in MMAB (OMIM, 607568)7; and cb1D variant 2, which is caused by mutations in MMADHC (OMIM, 611935). The finding of biallelic pathogenic variants in one of the five genes (MMUT, MMAA, MMAB, MCEE, and MMADHC) associated with isolated MMA with confirmation of carrier status in the parents can establish the diagnosis of MMA. Approximately 97% of isolated MMA cases were caused by mutations in MMUT, MMAA, and MMAB, and mutations in MCEE and MMADHC only account for approximately 1–3%6,8. MMA with homocystinuria can be found in the following complementation groups: cblC (OMIM, 277400), which is caused by mutations in MMACHC (OMIM, 609831); cblD (OMIM, 277410), which is caused by mutations in MMADHC (OMIM, 611935); and cblF (OMIM, 277380), which is caused by mutations in LMBRD1 (OMIM, 612625)9.
In our study, we recruited 13 unrelated patients with MMA and studied the clinical, biochemical, and hereditary pathogenesis of these patients. We performed accurate genetic metabolic disease screening, molecular diagnosis, and genetic counseling for these families. We provided invasive prenatal diagnosis for two families and followed up their pregnancy results.
Patients and methods
Patients
A total of 13 unrelated patients with MMA (seven with isolated MMA and six with MMA with homocystinuria) and their families were recruited. The patients were from Gansu Provincial Maternal and Child Health Care Hospital during November 2016 to May 2019. The age of the probands ranged from 1 day to 5 years old. All patients were from non-consanguineous families. This study was performed according to the tenets of the Declaration of Helsinki. This study was approved by the Ethics Committee of Gansu Provincial Maternal and Child Health Care Hospital (No. 4 of the hospital ethics review, 2016). Written informed consent was obtained from all participants in this study, and written informed consent of patients younger than 18 years old was obtained from their parents.
Methods
Genetic metabolic disease screening
We collected dried blood spots (in whole blood) of the probands. Blood amino acids, free carnitine, and acylcarnitines were measured by using liquid chromatography-tandem mass spectrometry (LC–MS/MS) (TQD; Waters, USA). The reference levels of propionylcarnitine (C3) and C3/acetylcarnitine (C2) in the blood were 0.3–4.95 μmol/L and 0.05–0.27 μmol/L, respectively. Concentrations of organic acids in urine were measured by gas chromatography and mass spectrometry (GC/MS) (GCMS-QP 2010; Shimadzu Corporation, Japan) in these patients. The normal value of urine methylmalonic acid ranges from 0.0–5.34 μmol/L.
Treatment and follow up
Metabolic therapy, including a high-calorie diet with special formula supplementation (no isoleucine, threonine, methionine, and valine), stopping protein intake and intramuscular cobalamin were provided to most patients. Treatment for the patients was then adjusted according to their response to vitamin B12 and other individual conditions7,10–12.
Two families had an invasive prenatal diagnosis during 19–20 weeks of pregnancy. We obtained 15 ml of amniotic fluid and then used Sanger sequencing to detect the same mutations as those found in the probands.
Genomic DNA preparation
A total of 2–3 ml of blood samples were collected from the probands and their parents. Genomic DNA was extracted using the Tiangen Biotech DNA extraction kit (Beijing, China).
Targeted next generation sequencing and Sanger Sequencing
Mutation screening for the probands was performed by targeted gene capturing and sequencing of 175 related genes by MyGenostics Corporation (MyGenostics GenCap Enrichment Technologies, Beijing, China).The list of genes is shown in Table S1. Candidate variants were confirmed in the parents in each family by Sanger sequencing. Primers for polymerase chain reaction were designed by Primer3 Input (v.0.4.0, https://bioinfo.ut.ee/primer3-0.4.0/) (Table S2). Amplification conditions for Sanger sequencing are shown in Table S2. DNA sequencing was performed using an ABI 3500DX Genetic Analyzer (Applied Biosystems, USA).
Bioinformatics analysis
The variants were described according to the nomenclature recommended by the Human Genomic Variation Society. Novel variants were checked in the Human Gene Variant Database (HGMD) (https://www.hgmd.cf.ac.uk/) and ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/). We used PolyPhen2 (https://genetics.bwh.harvard.edu/pph2) and PROVEAN (https://provean.jcvi.org/index.php) tools to predict the possible functional role of novel variants. InterVar software (https://wintervar.wglab.org/) was used to evaluate the pathogenicity of the novel variants with reference to the standards and guidelines of the American College of Medical Genetics and Genomics (ACMG)13. This process has been described previously14,15.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Gansu Provincial Maternal and Child Health Care Hospital (No. 4 of hospital ethics review, 2016). Written informed consents have been obtained from all participants the study procedure started.
Result
Clinical features, biochemical indicators and variant analysis
Our 13 MMA cases from 13 unrelated families were recruited from the genetic counseling clinic and the neonatal intensive care unit of Gansu Provincial Maternal and Child Health Care Hospital. Two of the patients had the late-onset form (> 1 year old) and 11 had the early-onset form (< 1 year old). The clinical features and biochemical indicators are shown in Table 1. Seven of the cases were isolated MMA and six were MMA with homocystinuria.
Table 1.
Novel mutations of MUT, MMACHC and MCEE.
| Gene | Nucleotide change | Amino acid Change | PolyPhen2 result (Score) | PROVEN result (Score) | Pathogenicity | Evidence |
|---|---|---|---|---|---|---|
| MUT | c.2008G>A | p.G670S | PD (1.000) | D (− 5.850) | LP | PM1 PM2 PM5 PP3 |
| c.301_302insTA | p.T101Ifs*80 | NA | NA | P | PVS1, PS2 and PM2 | |
| c.984delC | p.W329Gfs*4 | NA | NA | P | PVS1, PS2 and PM2 | |
| c.319A>T | p.I107F | PD (1.000) | D (− 3.950) | LP | PM1 PM2 PP3 PP4 | |
| MMACHC | c.445T>C | p.C149R | PD (0.884) | D (− 7.990) | P | PM1 PM2 PP3 PP4 |
| MCEE | c.296T>C | p.L99P | PD (1.000) | D (− 6.950) | LP | PM1 PM2 PP3 PP4 |
PD Probably Damaging, D Deleterious, P Pathogenic, LP Likely pathogenic.PolyPhen2 result: The score is closer to 1, the damaging will be more strong; Proven Result: Variants with a score equal to or below − 2.5 are considered “deleterious”, Variants with a score above − 2.5 are considered “neutral”. The type of evidence refers to. ACMG/AMP 2015 guideline (https://wintervar.wglab.org/).
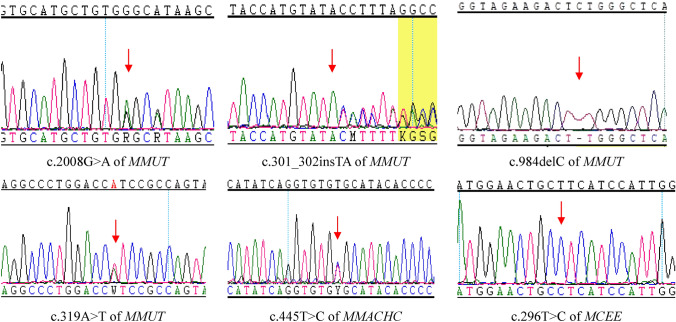
Variants in MMUT: MMA was caused by a homozygous variant in one patient and MMA was caused by compound heterozygous variants in MMUT in five patients. Eleven different variants of MMUT were identified in these six patients (Table 2). Eight were missense variants and three were frame-shift variants. Seven of the variants were previously reported11,16–20 and four variants, c.2008G>A (p.G670S), c.301_302insTA(p.T101Ifs*80), c.984delC (p.W329Gfs*4), and c.319A>T(p.I107F), were not been previously reported in the HGMD and ClinVar databases (Table 2, Fig. 1).
Table 2.
Results of DQ/IQ tests of the 5 patients.
| Case No | Age at test | DQ | Full-scale IQ | ||||
|---|---|---|---|---|---|---|---|
| Gross motor | Fine motor | Language | Adaptability | Personal-social | |||
| 2 | 2Ys/5Ys | 75 | 69 | 68 | 63 | 64 | 62 |
| 7 | 8Mons | 72 | 63 | 78 | 62 | 59 | |
| 8 | 5Ys/7Ys | / | / | / | / | / | 54/48 |
| 9 | 3Ys | 78 | 75 | 72 | 77 | 69 | |
| 13 | 10Mons/2Ys | 68/76 | 95/61 | 95/61 | 95/60 | 95/61 | |
Mon month, D day, Y Year.
Figure 1.
Novel variants of MMUT, MMACHC, MCEE.
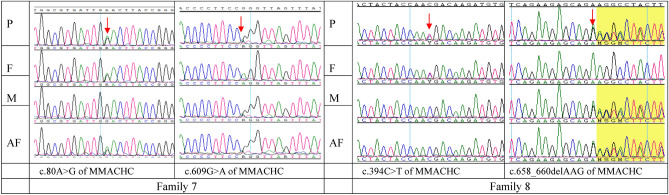
Variants in MMACHC: MMA was caused by a homozygous variant in two patients and MMA was caused by a compound heterozygous variant in MMACHC in four patients. Six types of variants in MMACHC were detected in these six patients (Table 2). Two missense variants, two frame-shift variants, and two nonsense variants were identified. Among them, c.445T>C (p.C149R) was not been previously reported in the HGMD and ClinVar databases (Table 2, Fig. 1). Two families with MMA caused by variants in MMACHC chose prenatal diagnosis through amniotic fluid puncture at their next pregnancy. The fetus from family 7 had the same compound heterozygous variant detected as that in the proband, and the fetus from family 8 only had a maternal heterozygous variant and there was no paternal variant(Fig. 2).
Figure 2.
Prenatal diagnosis of MMA families. P proband, F father, M mother, AF amniotic fluid.
Variants in MCEE: One patient with MMA had the homozygous variant c.296T>C(p.L99P) in MCEE (Table 2, Fig. 1). This variant has not been previously reported.
Bioinformatics analysis: We used PolyPhen2 and PROVEAN tools to predict the possible functional role of these six novel variants. According to the ACMG guidelines and InterVar software, these six novel variants were categorized as “likely pathogenic” (Table 2).
Follow up
Because of severe metabolic acidosis, eight families (numbers 1, 3, 4, 5, 6, 10, 11, and 12) abandoned treatment for the probands, and these patients eventually died. Family 7 provided treatment for the patient. However, the patient had a poor vitamin B12 response, and the patient died of metabolic acidosis at 11 months because of poor treatment. Patients 2 and 13 received oral L-carnitine, vitamin B12, and folate. Patients 8 and 9 received oral L-carnitine, betaine, vitamin B12, and folate (Table 3). One month later, the levels of serum C3 and C3/C2, urine MMA, and serum homocysteine (HCY) were slightly decreased. Six months later, serum C3, C3/C2, and HCY levels, and urine MMA levels of the patients were in the normal range (Table 3).
Table 3.
Clinical characteristics, diagnosis, genotype information, treatment and prognosis of MMA patients in this study.
| Case no | Age of onset | Curr-ent age | Sex | Blood C3 0–1–6 month (normal: 0.3–4.95 μmol/L) | Blood C3/C2 0–1–6 month (normal: 0.05–0.27) | Urinary MMA 0–1–6 month (normal: 0.0–5.34 μmol/L) | HCY 0–1–6 month (normal: 0.0–15.0 μmol/L) | Treatment (L-carnitine, betaine, B12 and folate) | Methylcobal-amin (B12) response | Prognosis | Gene | Genotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 14 Ds | / | F | 10.2 | 0.79 | 391.1 | 7.5 | No | / | Dead | MUT | c.2008G>A /c.103C>T |
| 2 | 2 Ys | 5Y | F | 12.11/2.98/2.19 | 0.7/0.23/0.09 | 242.15/31.87/4.98 | 6.3 | Yes | Effective | DD | MUT | c.301_302insTA /c.613G>A |
| 3 | 6 Ds | / | M | 10.59 | 0.78 | 403.59 | 4.3 | No | / | Dead | MUT | c.984delC/ c.984delC |
| 4 | 1 D | / | M | 11.28 | 1.38 | 512.3 | 3.5 | No | / | Dead | MUT | c.1106G>A /c.323G>A |
| 5 | 2 Ds | / | M | 11.35 | 1.63 | 1,117.47 | 5.3 | No | / | Dead | MUT | c.914 T>C/ c.494A>G |
| 6 | 1 D | / | M | 13.2 | 0.45 | 300.46 | 2.2 | No | / | Dead | MUT | c.1038_1040delTCT/ c.319A>T |
| 7 | 8 Mons | / | F | 14.83 | 0.69 | 334.5 | 63.9 | No | Ineffective | Dead | MMACHC | c.80A>G /c.609G>A |
| 8 | 5 Ys | 7Y | F | 8.87/3.35/1.27 | 1.77/0.19/0.14 | 149.86/36.09/3.25 | 65.4/23.2/8.9 | Yes | Effective | DD | MMACHC | c.394C>T /c.658_660delAAG |
| 9 | 56 Ds | 3Y | M | 8.98/3.48/2.15 | 0.84/0.21/0.18 | 358.2/6.02/2.98 | 62.1/52.5/6.8 | Yes | Effective | DD | MMACHC | c.609G>A/ c.609G>A |
| 10 | 15 Ds | / | M | 10.32 | 1.01 | 431.2 | 47.8 | No | / | Dead | MMACHC | c.609G>A/ c.609G>A |
| 11 | 3 Mons | / | M | 11.64 | 0.62 | 175.85 | 41.1 | No | / | Dead | MMACHC | c.445 T>C/ c.609G>A |
| 12 | 40 Ds | / | M | 9.94 | 0.89 | 331.2 | 54.3 | No | / | Dead | MMACHC | c.80A>G /c.567dupT |
| 13 | 7 Mons | 3Y | F | 14.38/6.42/3.27 | 0.8/0.31/0.16 | 522/81.74/3.63 | 4.9 | Yes | Effective | DD | MCEE | c.296 T>C/ c.296 T>C |
Mon: month; D: day; Y: Year; M: male; F: female; DD: development delay.
The developmental quotient (DQ) was evaluated using the Revised Gesell Developmental Evaluation for children (< 4 years). This evaluation provides a developmental profile in five domains, including adaptive, gross motor, fine motor, language, and personal-social domains9. If the DQ of a child in any specific domain was ≤ 759, development was below average. The intelligence quotient (IQ) was assessed by the Wechsler Intelligence Scale for Children (> 4 years). Low intelligence was defined as an IQ score < 809. We evaluated the DQ for four patients (patient numbers 2, 7, 9, 13) and the IQ for patient 8 (Table 2). The patients from families 2 and 8 were treated too late, which resulted in mental retardation and motor dysfunction. Patient 9 had a good vitamin B12 response. However, because of a lack of timely treatment, he still had a certain degree of neurodevelopmental delay. He is currently 3 years old and his language is also slightly delayed. Patient 13 achieved better treatment results, but unfortunately, because of the COVID-19 pandemic, continuous treatment was interrupted, and she is currently in a serious condition.
Families 7 and 8 had prenatal diagnosis during pregnancy. The same compound heterozygous variants as those in the proband were found in the fetus of family 7, and this family stopped the pregnancy (Fig. 2). The fetus of family 8 only carried the maternal heterozygous variant (Fig. 2), and this family chose to continue the pregnancy with successful delivery.
Discussion
In this study, we investigated the genetic etiology for 13 unrelated patients with MMA by using molecular testing. Six patients had MMA caused by homozygous/compound heterozygous variants in MMUT, 11 types of variants were found, and four of these variants were novel variants. Six patients had MMA caused by variants in MMACHC, six different variants were found, and c.445T>C was a novel variant. One patient had MMA caused by a novel homozygous variant, c.296T>C, in MCEE.
In our study, we found the six novel variants c.2008G>A, c.301_302insTA, c.984delC, and c.319A>T in MMUT, c.445T>C in MMACHC, and c.296T>C in MCEE. By using PolyPhen2 and PROVEAN tools and InterVar software, and according to the ACMG/AMP guidelines, these six novel variants were categorized as “likely pathogenic” (Table 2). With regard to the c.2008G>A(p.G670S) variant of MMUT, the same amino acid position variant p.G670R was reported in a 7-day-old female neonate21, but p.G670S has not been reported previously.
In our study, all of the six patients with MMA with homocystinuria had variants in MMACHC. The frequency of the variants in MMACHC was associated with the genetic background of the patients. In our patients, c.609G>A was the most frequent variant (50%, 6/12), which is consistent with reports in the northern Chinese population20. However, the frequency of this variant is not high in the European population22. Lerner-Ellis et al.22 reported that the variant c.271dupA accounted for 40% of all disease alleles in the European population, but we did not find this variant in our patients. Liu et al.20 showed that the frequency of c.271dupA was low (1/140). The c.609G>A, c.658_660delAAG, c.482G>A, c.394C>T, and c.80A>G mutations are the most common mutations in Chinese people, and in European people, the most common mutations are c.271dupA, c.331C>T, and c.394C>T. The variant c.394C>T is common in both European and Chinese people. Lerner-Ellis et al22 reported that the variant c.394C>T was associated with late-onset MMA. However, in Liu et al.’s study20, eight patients carried the variant c.394C>T, and only two patients had late-onset MMA. Therefore, we consider that the variant c.394C>T is also associated with early-onset MMA.
In the seven patients with isolated MMA, six were caused by variants in MMUT and one was caused by a homozygous variant in MCEE. Only approximately 21 patients with isolated MMA have been reported to have MMA caused by variants in MCEE23–29. To the best of our knowledge, we report the first patient with MMA caused by a variant in MCEE in Chinese people, and this was caused by the novel homozygous variant c.296T>C. We did not perform functional analysis of this novel variant at the cellular and animal levels. However, we used function prediction software to access the pathogenicity of this variant, and it was categorized as a “likely pathogenic” variant.
The girl with MCEE revisited the hospital at 7 months old because of a positive LC–MS/MS result, and she had genetic diagnosis and treatment at 10 months old. During the re-examination, LC–MS/MS and GCMS indicated MMA. A comprehensive evaluation of our patient was normal. The gross motor DQ was 68, the fine motor DQ was 95, the language DQ was 95, the adaptability DQ was 95, and the personal-social DQ was 95. Oral L-carnitine and folate, and intramuscular injection of methylcobalamin led to a reduction in C3 and C3/C2 levels. Unfortunately, because of the COVID-19 pandemic, the patient’s parents did not insist on continuous treatment, and currently, the patient is in a serious condition. Currently, the patient is 2 years old. A comprehensive evaluation of this patient showed retardation, the gross motor DQ was 76, the fine motor DQ was 61, the language DQ was 61, the adaptability DQ was 60, and the personal-social DQ was 61. Abily-Donval et al.28 summarized the treatment prognosis of eight patients with MCEE23–27. After suitable treatments, growth and psychomotor development of four patients were satisfactory, and their development was normal. The other four patients also had suitable treatment, but they still had problems in psychomotor, language, and nervous system development. Because our patient discontinued treatment, unfortunately, we do not have sufficient experience for treating patients with MCEE.
All of the patients with MMA were from the same Province, Gansu, which is located in the northwest of China. This region is economically insular with limited population mobility. MMA was caused by a homozygous variant in MMUT in one patient, by homozygous variants in MMACHC in two patients, and by a homozygous variant in MCEE in one patient. Although these patients were born from non-consanguineous families, we suspect that these homozygous cases might be due to a founder mutation in the local population.
In recent years, an increasing number of provinces in China have carried out newborn screening for genetic metabolic disorders by using MS/MS and GC/MS. With the development of molecular diagnostic technology and a decrease in cost of tests, currently, next-generation sequencing is a more effective method of providing accurate molecular diagnosis for some rare diseases30. Therefore, we can provide accurate and rapid diagnosis for patients who are affected by genetic metabolic disorders. We can also provide genetic counseling for the families and prenatal diagnosis for their next child. In our study, we provided prenatal diagnosis for two families with MMA caused by variants in MMACHC. The fetus from family 7 carried the same variants as those in the proband, and this family terminated the pregnancy. Intrauterine therapy is used to treat fetuses with MMA. Trefz et al.31 reported a patient with MMA in whom prenatal maternal treatment from week 15 of pregnancy prevented disease manifestation in a girl who is currently 11 years old with a normal IQ. However, we did not offer prenatal B12 therapy to the mother of family 7, and the pregnant woman chose to terminate the pregnancy. The fetus from family 8 only carried the maternal heterozygous variant, and this child was successfully delivered. Newborn screening at 72 h and biochemical examinations after 1 month were normal.
In our 13 patients with MMA, nine (69.2%) of them were detected by newborn screening through MS/MS and GC/MS. Four (30.8%) of these patients were diagnosed on the basis of clinical symptoms and biochemical examinations, including elevated serum C3 and C3/C2 levels and urine MMA levels, and MMA was confirmed by molecular testing. Unfortunately, only five patients received treatment. While four patients are currently still alive, the chance of recovery of these patients is not good.
Conclusion
We studied 13 patients with MMA from the northwest of China, and by using a molecular test, we identified their genetic etiology. We found six novel variants in three MMA-causing genes, MMUT, MMACHC, and MCEE. We provided genetic counseling for these families with MMA, and two families had prenatal diagnosis during their next pregnancy. Our findings extend the mutation spectrum of isolated MMA in the Chinese population. Newborn screening and next-generation sequencing can provide an accurate diagnosis for genetic metabolic diseases at an early age. However, more effective genetic counseling, and timely treatment and follow-up are required to make the patients’ families aware that some genetic metabolic diseases are treatable and can achieve a good prognosis.
Supplementary information
Acknowledgements
We would like to thank all the participants in this study. We thank Ellen Knapp, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript. This work was supported by theGansu Natural Science Foundation (Grant: No.18JR3RA036; 1606RJZA151). The National Key Research and Development Program of China (Grant No.: 2016YFC1000307); Non-profit Central Research Institute Fund of National Research Institute For Family Planing (Grant No.:2019GJZ07); the National Population and Reproductive Health Science Data Center (Grant No.:2005DKA32408), People’s Republic of China.
Author contributions
C.Z. and X.W. designed the study and write the manuscript. C.Z., S.H., Q.Z., B.Z., F.L., X.F., X.C., L.Z., P.M. and C.x.C. collected the data and performed the molecular test. Z.C. and X.M. reviewed the manuscript and Made lots of suggestions. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Data availability
All data analysed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zongfu Cao, Email: zongfu_cao@163.com.
Xu Ma, Email: maxubioinfo@163.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-69565-z.
References
- 1.Zwickler T, et al. Metabolic decompensation in methylmalonic aciduria: Which biochemical parameters are discriminative? J. Inherit. Metab. Dis. 2012;35(5):797–806. doi: 10.1007/s10545-011-9426-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhou W, et al. Newborn screening for methylmalonic acidemia in a Chinese population: Molecular genetic confirmation and genotype phenotype correlations. Front. Genet. 2019;9(1):726–735. doi: 10.3389/fgene.2018.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, et al. Clinical, biochemical, and molecular analysis of combined methylmalonic acidemia and hyperhomocysteinemia (cblC type) in China. J. Inherit. Metab. Dis. 2010;33(Suppl 3):S435–S442. doi: 10.1007/s10545-010-9217-0. [DOI] [PubMed] [Google Scholar]
- 4.Han B, et al. Clinical presentation, gene analysis and outcomes in young patients with early-treated combined methylmalonic acidemia and homocysteinemia (cblC type) in Shandong province, China. Brain Dev. 2016;38(5):491–497. doi: 10.1016/j.braindev.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Martinelli D, et al. Cobalamin C defect: Natural history, pathophysiology, and treatment. J. Inherit. Metab. Dis. 2011;34(1):127–135. doi: 10.1007/s10545-010-9161-z. [DOI] [PubMed] [Google Scholar]
- 6.Keyfi F, et al. Methylmalonic acidemia diagnosis by laboratory methods. Rep. Biochem. Mol. Biol. 2016;5(1):1–14. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou XY, et al. Methylmalonic acidemia: Current status and research priorities. Intractable Rare Dis. Res. 2018;7(2):73–78. doi: 10.5582/irdr.2018.01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shafaat M, et al. Autozygosity mapping of methylmalonic acidemia associated genes by short tandem repeat markers facilitates the identification of five novel mutations in an Iranian patient cohort. Metab. Brain Dis. 2018;33(5):1689–1697. doi: 10.1007/s11011-018-0277-4. [DOI] [PubMed] [Google Scholar]
- 9.Ledley FD. Perspectives on methylmalonic acidemia resulting from molecular cloning of methylmalonyl CoA mutase. BioEssays. 1990;12(7):335–409. doi: 10.1002/bies.950120706. [DOI] [PubMed] [Google Scholar]
- 10.Baumgartner MR, et al. Proposed guidelines for the diagnosis and management of methylmalonic and propionic acidemia. Orphanet. J. Rare Dis. 2014;9(9):130. doi: 10.1186/s13023-014-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han B, et al. Clinical presentation, molecular analysis and follow-up of patients with mut methylmalonic acidemia in Shandong province, China. Pediatr. Neonatol. 2020;61(2):148–154. doi: 10.1016/j.pedneo.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Kang L, et al. A study on a cohort of 301 Chinese patients with isolated methylmalonic acidemia. J. Inherit. Metab. Dis. 2020;43(3):409–423. doi: 10.1002/jimd.12183. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Wang K. InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet. 2017;100(2):267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang J, et al. Mutation screening of crystallin genes in Chinese families with congenital cataracts. Mol. Vis. 2019;25(8):427–437. [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Z, et al. Novel mutations in HSF4 cause congenital cataracts in Chinese families. BMC Med. Genet. 2018;19(1):150. doi: 10.1186/s12881-018-0636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu MY, et al. Mutation profile of the MUT gene in Chinese methylmalonic aciduria patients. JIMD Rep. 2012;6(1):55–64. doi: 10.1007/8904_2011_117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janata J, et al. Expression and kinetic characterization of methylmalonyl-CoA mutase from patients with the mut- phenotype: Evidence for naturally occurring interallelic complementation. Hum. Mol. Genet. 1997;6(9):1457–1464. doi: 10.1093/hmg/6.9.1457. [DOI] [PubMed] [Google Scholar]
- 18.Acquaviva C, et al. N219Y, a new frequent mutation among mut(degree) forms of methylmalonic acidemia in Caucasian patients. Eur. J. Hum. Genet. 2001;9(8):577–582. doi: 10.1038/sj.ejhg.5200675. [DOI] [PubMed] [Google Scholar]
- 19.Worgan LC, et al. Spectrum of mutations in mut methylmalonic acidemia and identification of a common Hispanic mutation and haplotype. Hum. Mutat. 2006;27(1):31–43. doi: 10.1002/humu.20258. [DOI] [PubMed] [Google Scholar]
- 20.Liu MY, et al. Mutation spectrum of MMACHC in Chinese patients with combined methylmalonic aciduria and homocystinuria. J. Hum. Genet. 2010;55(9):621–626. doi: 10.1038/jhg.2010.81. [DOI] [PubMed] [Google Scholar]
- 21.O'Shea CJ, et al. Neurocognitive phenotype of isolated methylmalonic acidemia. Pediatrics. 2012;129(6):e1541–e1551. doi: 10.1542/peds.2011-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerner-Ellis JP, et al. Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat. Genet. 2006;38(1):93–100. doi: 10.1038/ng1683. [DOI] [PubMed] [Google Scholar]
- 23.Bikker H, et al. A homozygous nonsense mutation in the methylmalonyl-CoA epimerase gene (MCEE) results in mild methylmalonic aciduria. Hum. Mutat. 2006;27(7):640–643. doi: 10.1002/humu.20373. [DOI] [PubMed] [Google Scholar]
- 24.Dobson CM, et al. Homozygous nonsense mutation in the MCEE gene and siRNA suppression of methylmalonyl-CoA epimerase expression: A novel cause of mild methylmalonic aciduria. Mol. Genet. Metab. 2006;88(4):327–333. doi: 10.1016/j.ymgme.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Gradinger AB, et al. Atypical methylmalonic aciduria: Frequency of mutations in the methylmalonyl CoA epimerase gene (MCEE) Hum. Mutat. 2007;28(10):1045. doi: 10.1002/humu.9507. [DOI] [PubMed] [Google Scholar]
- 26.Mazzuca M, et al. Combined sepiapterin reductase and methylmalonyl-CoA epimerase deficiency in a second patient: Cerebrospinal fluid polyunsaturated fatty acid level and follow-up under L-DOPA, 5-HTP and BH4 trials. JIMD Rep. 2015;22(3):47–55. doi: 10.1007/8904_2015_410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waters PJ, et al. Methylmalonyl-coA epimerase deficiency: A new case, with an acute metabolic presentation and an intronic splicing mutation in the MCEE gene. Mol. Genet. Metab. Rep. 2016;9(9):19–24. doi: 10.1016/j.ymgmr.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abily-Donval L, et al. Methylmalonyl-CoA epimerase deficiency mimicking propionic aciduria. Int. J. Mol. Sci. 2017;18(11):2294. doi: 10.3390/ijms18112294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heuberger K, et al. Genetic, structural, and functional analysis of pathogenic variations causing methylmalonyl-CoA epimerase deficiency. Biochim. Biophys. Acta Mol. Basis. Dis. 2019;1865(6):1265–1272. doi: 10.1016/j.bbadis.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C, et al. A novel LOXHD1 variant in a Chinese couple with hearing loss. J. Int. Med. Res. 2019;47(12):6082–6090. doi: 10.1177/0300060519884197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trefz FK, Scheible D, Frauendienst-Egger G, et al. Successful intrauterine treatment of a patient with cobalamin C defect. Mol. Genet. Metab. Rep. 2016;6:55–59. doi: 10.1016/j.ymgmr.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analysed during this study are included in this published article.