Abstract
The F-box protein 22 (FBXO22), one of F-box proteins, has been identified to be critically involved in carcinogenesis. FBXO22 promotes proliferation in breast cancer and lung cancer, but suppresses migration and metastasis. FBXO22 exerts oncogenetic functions via promoting the ubiquitination and degradation of its substrates, including KDM4A, KDM4B, methylated p53, p21, KLF4, LKB1, Snail, CD147, Bach1, PTEN, and HDM2. FBXO22 is also regulated by several regulatory factors such as p53, miR-155, SNHG14, and circ_0006282. In this review, we summarize the regulatory factors and downstream targets of FBXO22 in cancers, discuss its functions in tumorigenesis, and further highlight the alteration of FBXO22 expression in a variety of human malignancies. Finally, we provide novel insights for future perspectives on targeting FBXO22 as a promising strategy for cancer therapy.
Subject terms: Ubiquitylation, Ubiquitin ligases
Facts
FBXO22 targets multiple substrates for ubiquitination and degradation.
FBXO22 is critically involved in tumorigenesis and tumor progression.
FBXO22 might be a therapeutic target for cancer treatment.
Open questions
Which targets of FBXO22 are pivotal for cancer development and malignant progression?
Do E3 liagses regulate the protein levels of FBXO22?
How the inhibitors of FBXO22 could be developed and discovered for cancer therapy?
Introduction
Post-translational modification (PTM) is one of the critical pathways in regulation of cellular events such as proliferation, apoptotic death, cell cycle, mitosis, motility, and innate immunity1–3. PTMs include, but are not limited to, ubiquitination, phosphorylation, acetylation, methylation, succinylation, and sumoylation4–6. Among these PTMs, ubiquitination is one of the most studied and is mediated by ubiquitin proteasome system (UPS) to trigger protein degradation. In general, three enzymes are involved in UPS-induced protein degradation, including ubiquitin activating E1 enzyme, ubiquitin conjugating E2 enzyme, and ubiquitin E3 ligase7. The target protein is labeled by ubiquitins and subsequently degraded by the 26S proteasome complex, leading to reduction of substrate protein. E3 ligases recognize and recruit the target protein for ubiquitination, thus they were extensively characterized8. Among E3 ligases, Cullin-RING E3 ligase (CRL) complex is one of the largest families, including CRL1–3, 4A, 4B, 5, 7, and 99–11. CRL1, also known as SKP1-cullin 1-F-box protein (SCF) E3 ligase complex, contains cullin-1 acting as the scaffold protein, RBX1 for recruiting ubiquitin-loaded E2, SKP1 working as an adaptor protein to connect F-box protein, and F-box protein for selecting substrates for degradation12. A total of 69 F-box proteins encoding by human genome, are divided into three subclasses according to their variable domains: FBXW proteins with WD40 repeat domains, FBXL proteins with leucine-rich repeat domains, and FBXO proteins with other domains like kelch repeats or proline-rich domains13,14.
F-box proteins have been validated to play a pivotal role in carcinogenesis and progression13,15. They are involved in various physiological and pathological processes which include proliferation, apoptosis, metastasis, cancer stem cell generation, epithelial–mesenchymal transition (EMT), drug resistance in human cancers16–18. It is important to notice that F-box proteins, including FBXO22, have oncogenic or tumor suppressive role in cancer development and progression. In the following sections, we will describe the regulatory factors and downstream targets of FBXO22 in a variety of human cancers, and the alteration of its expression levels in human cancer tissues.
Upstream regulators of FBXO22
The p53 protein as a traditional tumor suppressor has been identified to be mutated in a variety of human malignancies19. Mutant p53 proteins lose anticancer function due to impaired cellular homeostasis and damaged genome stability, leading to enhancement of survival, invasion, and metastasis19,20. Evidence has demonstrated that p53-mediated tumor suppressive activity is in part through regulation of downstream targets and multiple signaling pathways21. One study validated that wild-type p53 increases the transcription of FBXO22 via binding to DNA and promotion of histone acetylation at FBXO22 promoter22. Specifically, p53 overexpression elevated FBXO22 mRNA level by real-time RT-PCR analysis. Data from chromatin immunoprecipitation (ChIP) on chip analysis demonstrated that FBXO22 is a direct p53 target22. Since FBXO22 might target numerous substrates for degradation or inactivation, p53 exerts its tumor suppressive activity partly via induction of FBXO22 expression. Another study reported that miR-155 could target FBXO22 in anterior uveitis23. It has been well known that miRNAs are small, non-coding RNAs that regulate hundreds of target genes at the post-transcriptional level, so that miRNAs govern multiple biological functions, such as differentiation, proliferation, stemness and oncogenesis24. Through TargetScan online computational algorithm and validation by a luciferase reporter gene assay, FBXO22 is identified as a target of miR-15523. Several studies have revealed that miR-155 plays an essentical role in oncogenesis and progression. For example, miR-155 promoted cell growth and invasion via regulation of epidermal growth factor receptor (EGFR) and nuclear factor-kappa B (NF-κB) in salivary adenoid cystic carcinoma25. One group reported that miR-155 was highly expressed in sera of hepatocellular carcinoma (HCC) patients, which is associated with blood telomerase level26. In addition, miR-155 inhibited proliferation, migration, invasion and triggered cell cycle arrest and apoptosis via regulating the expression of collagen triple repeat containing 1 (CTHRC1) in human melanoma27. Moreover, exosome-mediated miR-155 delivery led to cisplatin resistance of oral squamous cell carcinoma (OSCC) cells via induction of EMT28. However, it is unclear whether miR-155 targets FBXO22 in human cancer cells, which is required to further determine.
Small nucleolus RNA host gene 14 (SNHG14), one long noncoding RNA, was reported to act as a competing endogenous RNA (ceRNA) to decoy miR-433-3p and subsequently increase FBXO22 expression in osteosarcoma cells29. Downregulation of FBXO22 or SNHG14 inhibited proliferation, motility of osteosarcoma cells, but stimulated apoptosis29. Token together, SNHG14 enhanced osteosarcoma progression through modulation of miR-433-3p/FBXO22 pathway. Recently, circular RNA (circRNA) circ_0006282 was revealed to facilitate tumor progression via sponging miR-155 to increase FBXO22 expression in gastric cancer30. Specifically, circ_0006282 functions as a ceRNA to spong miR-155 and cause the upregulation of its target, FBXO22, resulting in enhancement of proliferation and metastasis of gastric cancer cells30 (Fig. 1).
Fig. 1. The regulation of FBXO22 and its downstream substrates.
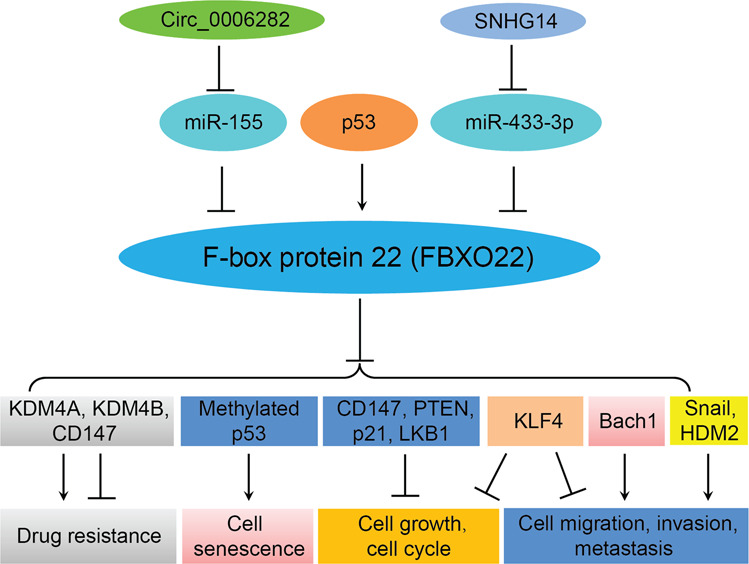
FBXO22 targets several substrates for ubiquitination and degradation, and is regulated by multiple regulatory factors. FBXO22 regulates proliferation, cell cycle, senescence, migration, invasion, metastasis, and drug resistance in human cancers.
Downstream targets of FBXO22
Accumulated evidence has demonstrated that FBXO22 targets several substrates for uiquitination and degradation (Table 1 and Fig. 1). The histone lysine demethylase 4 (KDM4) subfamily includes four proteins, KDM4A, KDM4B, KDM4C and KDM4D, which control chromatin structure and gene expression31. KDM4A, also termed as JMJD2A and JHDM3A, can demethylate histone H3 lysine 9 (H3K9) and 36 (H3K36) and H1.4K26, leading to regulation of genome replication and stability31. KDM4A is identified as a substrate of FBXO2232. Therefore, FBXO22 is potentially involved in development, differentiation and cancer via controlling KDM4A stability, leading to regulation of H3K9 and H3K36 methylation, which are important factors to maintain normal cellular function32. Similalry, KDM4B degradation is mediated by FBXO22 in breast cancer cells, resulting in regulation of selective estrogen receptor modulators (SERMs) activity, leading to modulation of tamoxifen resistance in ER-positive breast cancer cells33. FBXO22 is required for cell growth inhibition induced by tamoxifen, and FBXO22-induced KDM4B degradation is necessary for the antagonistic function of SERMs in breast cancer33. One study showed that FBXO22-KDM4A act as an E3 ubqiquitin ligase to govern methylated p53 stability via its degradation, leading to regulation of senescence34. In line with this, Fbxo22−/− mice exhibited an increase of p53 expression level, and mouse embryonic fibroblast (MEFs) from Fbxo22−/− mice had increased methylated p53, suggesting that FBXO22 might regulate the amount of methylated p5334.
Table 1.
FBXO22 targets substrates for degradation in human diseases.
| Substrates | Cell lines | Functions | Refs |
|---|---|---|---|
| KDM4A | HeLa, 293T, 293T-Rex | Regulation of cell cycle, involves in development, differentiation, cancer | 32 |
| KDM4B | MCF7, T47D | Tomaxifen resistance | 33 |
| Methylated p53 | HeLa, U2OS, MCF7, 293T, HCA2, MEFs, HCT116, RPE-1 | Regulating senescence | 34 |
| p21 | HL-7702, HepG2, Huh7, Hep3B, Bel-7402, HLF, LM3, 293T | Promotes proliferation and tumor growth | 35 |
| KLF4 | HepG2, Huh7, Hep3B | Promotes proliferation and invasion | 38 |
| LKB1 | H322, H446, H460, H661, H1299, BT549 | Promotes cell growth | 43 |
| CD147 | 293T, A549, SMMC-7721, Huh-7 | Inhibits cisplatin resistance | 45 |
| Bach1 | A549, H2009, 293T, KP, KPK. | Inhibits migration and metastasis | 47 |
| PTEN | 293T, HeLa, SW620, SW480, LS174T | Promotes tumor growth | 48 |
| Snail | MDA-MB-231, Hs578T, MCF-7, ZR-75-1, T47D | Inhibits migration, invasion, and metastasis; promotes proliferation | 39 |
| HDM2 | HeLa, MDA-MB-231, BT-549, 4T1 | Inhibits invasion and metastasis | 42 |
FBXO22 elevated proliferation of HCC cells and enhanced tumor growth in mice. Knockdown of FBXO22 in HLF and HepG2 cells led to suppression of proliferation and inhibition of colony formation, whilest overexpression of FBXO22 in LM3 and Hep3B cells promoted cell viability and colony formation35. Moreover, results from both subcutaneous and orthotopic mouse models showed that downregulation of FBXO22 slowed down the tumor growth in vivo35. Mechansitically, FBXO22 interacted with p21 and subsequently ubiquitinated p21 via its F-box domain for degradation. Strikingly, FBXO22 accelerated cell growth partly and modulated cell cycle progression via downregulation of p21. Consistently, FBXO22 expression was negatively associated with p21 level in HCC tumor samples35.
Kruppel-like factor 4 (KLF4) has tumour suppressive functions in a variety of human malignancies36. KLF4 is often downregulated in tumor specimens and associated with poor survival in cancer patients37. It has been shown that FBXO22 interacts with and destabilizes KLF4 via ubiquitination in HCC cells, leading to promotion of proliferation and invasion38. A negative correlation between FBXO22 and KLF4 was observed in HCC tumor samples38. Interestingly, FBXO22 was reported to exhibit a different function in breast tumorigenesis and metastasis39. Overexpression of FBXO22 elevated proliferation and facilitated colony formation in vitro and in vivo, but inhibited EMT, cell motility and invasion as well as metastasis in breast cancer39. Moreover, FBXO22 targets Snail, a key factor to trigger EMT process, for ubiquitination and degradation in a glycogen synthase kinase 3β (GSK-3β) phosphorylation-dependent manner. It is worth noting that Snail/Slug and ZEB-1/SIP1 families not only control EMT process, but also inhibit cell cycle progression by repression of cyclin D40,41. This study indicated that FBXO22 may act as an upstream regulator and play a dual role in mammary cancer by inducing Snail degradation: promotion of proliferation and suppression of metastasis39. Human homolog of mouse double minute 2 (HDM2) is often highly expressed in various types of cancers. The stability of HDM2 oncoprotein is regulated by FBXO22 by ubiquitin-dependent degradation in breast cancer cells42. FBXO22 targets HDM2 for ubquitination and degradation, leading to inhibitory effects on invasion and metastasis in breast cancer42. Consistently, FBXO22 level is negatively associated with HDM2 expression in patients with breast cancer42.
Liver kinase B1 (LKB1), a serine/threonine kinase, has been identified to involve in oncogenesis in various types of human cancers. LKB1 expression level is regulated by FBXO22 via proteasome-mediated degradation in non-small cell lung cancer (NSCLC) cells43. FBXO22 interacts with and triggers LKB1 for K63-mediated ubiquitination, leading to inhibition of LKB1 activity and subsequent inactivation of AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signaling pathways43. FBXO22-mediated inactivation of LKB1 causes promotion of cell growth via modulation of AMPK and mTOR pathways in NSCLC cells43. CD147 as a transmembrane glycoprotein is often overexpressed in human malignancies and is involved in chemoresistance in cancer cells44. FBXO22 could ubiquitinate CD147 and result in its degradation, leading to enhancement of cisplatin sensitivity in lung cancer cells45. The transcription factor BTB and CNC homology 1 (Bach1) plays a regulatory role in cell cycle, senescence, angiogenesis, immunity, and carcinogenesis and metastasis. Bach1 expression is linked to recurrence of breast cancer patients, and Bach1 promotes migration and invasion in colon and prostate cancer cells46. Recently, FBXO22 was identified to mediate the Bach1 degradation and inhibit migration in lung cancer cells47. More recently, in agreement with the oncogenic role of FBXO22, phosphatase and tensin homolog on chromosome 10 (PTEN), a bona fide tumor suppressor, is validated as a direct substrate of FBXO2248. FBXO22 ubiquitinates and degrades nuclear PTEN via proteasome-mediated degradation in colorectal cancer, leading to tumor development48 (Table 1 and Fig. 1).
Multiple studies have dissected that FBXO22 could regulate the expression of several downstream targets, such as hypoxia-inducible factor (HIF1α), vascular endothelial growth factor A (VEGFA), tissue inhibitor of matrix metalloproteinase-1 (TIMP-1) and metalloproteinase-9 (MMP-9) in human cancer cells49,50. FBXO22 downregulation in melanoma cells suppressed migration, invasion and angiogenesis, and decreased the formation of blood vessels in nude mice49. Moreover, FBXO22 promoted the motility of melanoma cells and angiogenesis through upregulation of HIF1α and VEGFA49. In RCC cells, FBXO22 has no any effect on proliferation, but FBXO22 restricted RCC cell motility and reversed EMT via an increase of the activity of TIMP-1 and an decrease of MMP-9 expression as well as a reduction of VEGF secretion50. In line with this in vitro result, in vivo study showed that FBXO22 inhibited RCC metastasis. Altogether, FBXO22 mainly reduced migration, invasion and metastasis in RCC through suppression of MMP-9 and VEGF pathways50. These paradoxical findings suggest that the role of FBXO22 in metastasis is in a context dependent manner. Thus, future investigations should be directed to elucidate the molecular mechanisms how FBXO22 regulates VEGF pathway.
Functions of FBXO22 in tumorigenesis
FBXO22 performs its functions via targeting its substrates by proteasome-mediated degradation in human malignancies, and exhibits its functions in controlling proliferation, cell cycle, apoptosis, migration, invasion, and metastasis. Numerous studies have been conducted to determine the role of FBXO22 in carcinogenesis by in vitro and in vivo experiments. The expression level of FBXO22 in various types of cancers has also been determined. To clarify the physiological function of FBXO22, the Fbxo22 knockout mice have been established using the CRISPR-Cas9 approach. Two Fbxo22−/− mice were viable, but had smaller size with the body weight reduced by 50% at six months of age, as compared to Fbxo22+/+ or Fbxo22+/− mice34. Although most Fbxo22−/− mice died within two days after birth, the genotype distribution of the offsprings from intercrossing Fbxo22+/− mice is consistent with the Mendelian-based ratio of 1:2:1, indicating that Fbxo22 is dispensable for early embryonic development34. Given that most Fbxo22−/− mice died within a couple of days after birth, it is of high demanding in the field to generate conditional knockout mouse model (Fbxo22fl/fl) to investigate the role of Fbxo22 in tumorigenesis. Specifically, Fbxo22 is deleted in various organs in combination with other established genetically modified mouse tumor models such as tumor suppressor inactivation (Ptenfl/fl, p53fl/fl, or Lkb1fl/fl), particularly PTEN, p53, and LKB1 acting as the substrates of FBXO22, or oncogene activation (e.g. KRasG12D). Fbxo22 deletion promoting or blocking tumorigenesis in these mouse models will elucidate the physiological role of FBXO22 in tumorigenesis in a given organ. In the following paragraphs, we will summarize the alteration of FBXO22 levels and its association with carcinogenesis and progression.
FBXO22 expression in human tumor tissues
One study measured the expression of FBXO22 on a tissue microarray with 110 pairs of HCC specimens by immunohistochemistry (IHC) approach and indicated that FBXO22 is highly expressed in tumors, compared to adjacent non-tumor tissues35. Another study also observed the overexpression of FBXO22 in HCC tumor tissures38. Notably, FBXO22 expression level is correlated with serum AFP, tumor size, and vascular invasion. Furthermore, high expression of FBXO22 is associated with poor prognosis in patients with HCC35. High FBXO22 expression is also observed in melanoma tissues, compared with normal skin tissues49. Additionally, FBXO22 mRNA level is increased in lung squamous cell carcinoma and lung adenocarcinoma according to the data from the cancer genome atlas (TCGA) database43. Moreover, IHC staining result indicated that higher expresson of FBXO22 existed in lung adenocarcinoma tissues than adjacent normal tissues. The result of Western blotting analysis confirmed the increased FBXO22 expression in lung cancer tissues as well43. In support of the oncogenic role of FBXO22 in lung cancer, the high expression of FBO22 is correlated with poor overall survival in lung cancer patients43.
On the other hand, FBXO22 also exhibits tumor-suppressing characteristics. Lower expression of FBXO22 is associated with worse prognosis in estrogen receptor (ER)-positive and human epidermal growth factor receptor type 2 (HER2)-negative breast cancer patients33. Although one group found that FBXO22 expression is increased in primary breast tumor specimens39, FBXO22 expression is correlated with favorable clinical outcomes in patients with breast cancer39. Similarly, low expression of FBXO22 is associated with poor survival in patients with breast cancer42. Intriguingly, FBXO22 is downregulated in pregnancy-associated breast cancer via analysis of NCBI-GEO datasets51. IHC analysis in renal cell carcinoma (RCC) tissues found that FBXO22 expression levels were decreased in RCC specimens compared with those in normal renal tissues50. Lower expression of FBXO22 in RCC patients is associated with tumor size, TNM stage, and poor survival50.
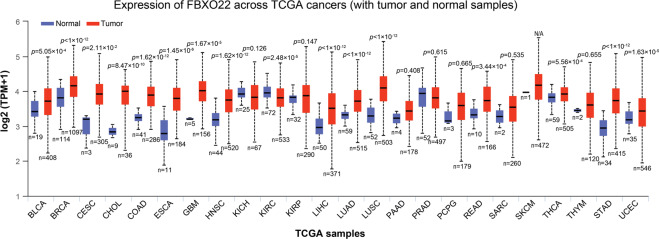
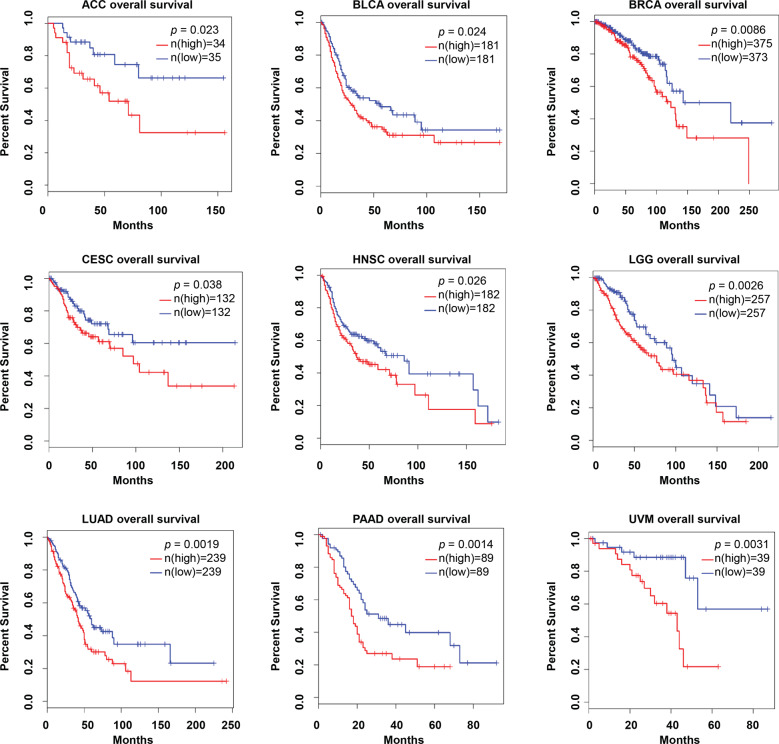
To better clarify the expression pattern and the correlation with patient survival of FBXO22 in human cancers, we examined FBXO22 mRNA expression using the data obtained from the TCGA database. The mRNA levels of FBXO22 were significantly increased in various types of human tumor tissues compared to that in normal tissues. The long list includes bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangio carcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), rectum adenocarcinoma (READ), thyroid carcinoma (THCA), stomach adenocarcinoma (STAD), and uterine corpus endometrial carcinoma (UCEC) (Fig. 2). Moreover, high levels of FBXO22 were significantly associated with short overall survival. The long list includes adrenocortical carcinoma (ACC), BLCA, BRCA, CESC, HNSC, brain lower grade glioma (LGG), LUAD, pancreatic adenocarcinoma (PAAD), and uveal melanoma (UVM) (Fig. 3). These results imply FBXO22 containing oncogenic characteristics.
Fig. 2. The expression of FBXO22 in human tumor and normal tissues.
The height of bar represents the mRNA expression of FBXO22 in TCGA cancer types. TPM transcripts per million. BLCA bladder urothelial carcinoma, BRCA breast invasive carcinoma, CESC cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL cholangio carcinoma, COAD colon adenocarcinoma, ESCA esophageal carcinoma, GBM glioblastoma multiforme, HNSC head and neck squamous cell carcinoma, KICH kidney Chromophobe, KIRC kidney renal clear cell carcinoma, KIRP kidney renal papillary cell carcinoma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, PAAD pancreatic adenocarcinoma, PRAD prostate adenocarcinoma, PCPG pheochromocytoma and paraganglioma, READ rectum adenocarcinoma, SARC sarcoma, SKCM skin cutaneous melanoma, THCA thyroid carcinoma, THYM thymoma, STAD stomach adenocarcinoma, UCEC uterine corpus endometrial carcinoma.
Fig. 3. FBXO22 expression is associated with poor survival in a variety of human cancers.
Blue line: low FBXO22; red line: High FBXO22. The cutoff-high (%) and cutoff-low (%) for ACC, BLCA, BRCA, CESC, HNSC, LGG, LUAD, PAAD, and UVM are 55/45, 55/45, 65/35, 55/45, 65/35, 50/50, 50/50, 50/50, and 50/50, respectively. ACC adrenocortical carcinoma, BLCA bladder urothelial carcinoma, BRCA breast invasive carcinoma, CESC cervical squamous cell carcinoma and endocervical adenocarcinoma, HNSC head and neck squamous cell carcinoma, LGG brain lower grade glioma, LUAD lung adenocarcinoma, PAAD pancreatic adenocarcinoma, UVM uveal melanoma.
Conclusion and future perspectives
In conclusion, FBXO22 is critically involved in oncogenesis through degradation of multiple substrates (Table 1 and Fig. 1). FBXO22 exerts its tumor promoting role in HCC, lung cancer, breast cancer, but inhibits migration and metastasis in lung cancer and breast cancer, indicating FBXO22 either acting as a tumor suppressor or acting as an oncogene. Thus, there are many fundamental questions that should be addressed to fully understand the role of FBXO22 in tumorigenesis. For example, what are functions of FBXO22 in other types of human cancers other than HCC, lung cancer, breast cancer? What are unknown substrates of FBXO22 that are pivotal in carcinogenesis? What are new regulatory factors to control the expression of FBXO22 in human cancer cells? To answer these questions, it is required to use the FBXO22 knockout or knockin mice to further validate the in vitro data. How can we discover the FBXO22 inhibitors for FBXO22 suppression? A complementary chemical and genomic screening approach might be a novel strategy for achieving FBXO22 inhibitors for treating cancer patients. Without a doubt, more investigations are essential to determine the underlying molecular mechanism of FBXO22-mediated tumorigenesis.
Acknowledgements
This work was supported by the National Key R&D Program of China (2016YFA0501800 to Y.Z.), the National Natural Science Foundation of China (81672728, 81972591, and 81721091 to Y.Z.).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Emre Sayan
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liu J, Qian C, Cao X. Post-translational modification control of innate immunity. Immunity. 2016;45:15–30. doi: 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 2.Vu LD, Gevaert K, De Smet I. Protein language: post-translational modifications talking to each other. Trends Plant Sci. 2018;23:1068–1080. doi: 10.1016/j.tplants.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Hirano A, Fu YH, Ptacek LJ. The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 2016;23:1053–1060. doi: 10.1038/nsmb.3326. [DOI] [PubMed] [Google Scholar]
- 4.Murn J, Shi Y. The winding path of protein methylation research: milestones and new frontiers. Nat. Rev. Mol. Cell Biol. 2017;18:517–527. doi: 10.1038/nrm.2017.35. [DOI] [PubMed] [Google Scholar]
- 5.Han ZJ, Feng YH, Gu BH, Li YM, Chen H. The post-translational modification, SUMOylation, and cancer (Review) Int J. Oncol. 2018;52:1081–1094. doi: 10.3892/ijo.2018.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat. Rev. Cancer. 2017;17:184–197. doi: 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]
- 7.Rape M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018;19:59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 8.Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen HC, Wang W, Xiong Y. Cullin-RING E3 ubiquitin ligases: bridges to destruction. Subcell. Biochem. 2017;83:323–347. doi: 10.1007/978-3-319-46503-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Sun Y. Cullin-RING ligases as attractive anti-cancer targets. Curr. Pharm. Des. 2013;19:3215–3225. doi: 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui D, Xiong X, Zhao Y. Cullin-RING ligases in regulation of autophagy. Cell Div. 2016;11:8. doi: 10.1186/s13008-016-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 2014;13:889–903. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat. Rev. Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin J, et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tekcham DS, et al. F-box proteins and cancer: an update from functional and regulatory mechanism to therapeutic clinical prospects. Theranostics. 2020;10:4150–4167. doi: 10.7150/thno.42735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan L, et al. Emerging roles of F-box proteins in cancer drug resistance. Drug Resist Updat. 2019;49:100673. doi: 10.1016/j.drup.2019.100673. [DOI] [PubMed] [Google Scholar]
- 17.Song Y, Lin M, Liu Y, Wang ZW, Zhu X. Emerging role of F-box proteins in the regulation of epithelial-mesenchymal transition and stem cells in human cancers. Stem Cell Res. Ther. 2019;10:124. doi: 10.1186/s13287-019-1222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen KM, Busino L. The biology of F-box proteins: the SCF family of E3 Ubiquitin ligases. Adv. Exp. Med. Biol. 2020;1217:111–122. doi: 10.1007/978-981-15-1025-0_8. [DOI] [PubMed] [Google Scholar]
- 19.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26:199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Q, Su Z, Gu W, Rustgi AK. Mutant p53 on the Path to Metastasis. Trends Cancer. 2020;6:62–73. doi: 10.1016/j.trecan.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitolli, C. et al. p53-mediated tumor suppression: DNA-damage response and alternative mechanisms. Cancers (Basel), 11, 1983 (2019). [DOI] [PMC free article] [PubMed]
- 22.Vrba L, Junk DJ, Novak P, Futscher BW. p53 induces distinct epigenetic states at its direct target promoters. BMC Genomics. 2008;9:486. doi: 10.1186/1471-2164-9-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Rourke M, Trenkmann M, Connolly M, Fearon U, Murphy CC. Novel gene targets for miRNA146a and miRNA155 in anterior uveitis. Br. J. Ophthalmol. 2019;103:279–285. doi: 10.1136/bjophthalmol-2018-312885. [DOI] [PubMed] [Google Scholar]
- 24.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 25.Liu L, Hu Y, Fu J, Yang X, Zhang Z. MicroRNA155 in the growth and invasion of salivary adenoid cystic carcinoma. J. Oral. Pathol. Med. 2013;42:140–147. doi: 10.1111/j.1600-0714.2012.01189.x. [DOI] [PubMed] [Google Scholar]
- 26.Ezzat WM, et al. Relationship between serum microRNA155 and telomerase expression in hepatocellular carcinoma. Arch. Med Res. 2016;47:349–355. doi: 10.1016/j.arcmed.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, et al. Overexpression of CTHRC1 in human melanoma promotes tumorigenesis targeted by miRNA155. Int J. Clin. Exp. Pathol. 2017;10:8199–8210. [PMC free article] [PubMed] [Google Scholar]
- 28.Kirave P, et al. Exosome mediated miR-155 delivery confers cisplatin chemoresistance in oral cancer cells via epithelial-mesenchymal transition. Oncotarget. 2020;11:1157–1171. doi: 10.18632/oncotarget.27531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou XK, Mao JS. Long noncoding RNA SNHG14 promotes osteosarcoma progression via miR-433-3p/FBXO22 axis. Biochem. Biophys. Res. Commun. 2020;523:766–772. doi: 10.1016/j.bbrc.2020.01.016. [DOI] [PubMed] [Google Scholar]
- 30.He Y, et al. Circular RNA circ_0006282 contributes to the progression of gastric cancer by sponging miR-155 to upregulate the expression of FBXO22. Onco Targets Ther. 2020;13:1001–1010. doi: 10.2147/OTT.S228216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DH, et al. Advances in histone demethylase KDM4 as cancer therapeutic targets. Faseb J. 2020;34:3461–3484. doi: 10.1096/fj.201902584R. [DOI] [PubMed] [Google Scholar]
- 32.Tan MK, Lim HJ, Harper JW. SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol. Cell Biol. 2011;31:3687–3699. doi: 10.1128/MCB.05746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johmura Y, et al. Fbxo22-mediated KDM4B degradation determines selective estrogen receptor modulator activity in breast cancer. J. Clin. Invest. 2018;128:5603–5619. doi: 10.1172/JCI121679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johmura Y, et al. SCF(Fbxo22)-KDM4A targets methylated p53 for degradation and regulates senescence. Nat. Commun. 2016;7:10574. doi: 10.1038/ncomms10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, et al. FBXO22 promotes the development of hepatocellular carcinoma by regulating the ubiquitination and degradation of p21. J. Exp. Clin. Cancer Res. 2019;38:101. doi: 10.1186/s13046-019-1058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghaleb AM, Yang VW. Kruppel-like factor 4 (KLF4): What we currently know. Gene. 2017;611:27–37. doi: 10.1016/j.gene.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu M, Hao B, Zhan Y, Luo G. Kruppel-like factor 4 expression in solid tumor prognosis: a meta-analysis. Clin. Chim. Acta. 2018;485:50–59. doi: 10.1016/j.cca.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 38.Tian X, et al. F-box protein FBXO22 mediates polyubiquitination and degradation of KLF4 to promote hepatocellular carcinoma progression. Oncotarget. 2015;6:22767–22775. doi: 10.18632/oncotarget.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun R, et al. FBXO22 possesses both protumorigenic and antimetastatic roles in breast cancer progression. Cancer Res. 2018;78:5274–5286. doi: 10.1158/0008-5472.CAN-17-3647. [DOI] [PubMed] [Google Scholar]
- 40.Vega S, et al. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mejlvang J, et al. Direct repression of cyclin D1 by SIP1 attenuates cell cycle progression in cells undergoing an epithelial mesenchymal transition. Mol. Biol. Cell. 2007;18:4615–4624. doi: 10.1091/mbc.E07-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai J, et al. SCF(FBXO22) targets HDM2 for degradation and modulates breast cancer cell invasion and metastasis. Proc. Natl Acad. Sci. USA. 2019;116:11754–11763. doi: 10.1073/pnas.1820990116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu XN, et al. FBXO22 mediates polyubiquitination and inactivation of LKB1 to promote lung cancer cell growth. Cell Death Dis. 2019;10:486. doi: 10.1038/s41419-019-1732-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grass GD, Dai L, Qin Z, Parsons C, Toole BP. CD147: regulator of hyaluronan signaling in invasiveness and chemoresistance. Adv. Cancer Res. 2014;123:351–373. doi: 10.1016/B978-0-12-800092-2.00013-7. [DOI] [PubMed] [Google Scholar]
- 45.Wu, B. et al. F-Box protein FBXO22 mediates polyubiquitination and degradation of CD147 to reverse cisplatin resistance of tumor cells. Int. J. Mol. Sci.18, 212 (2017). [DOI] [PMC free article] [PubMed]
- 46.Zhang, X. et al. Bach1: function, regulation, and involvement in disease. Oxid Med. Cell Longev. 2018, 1347969 (2018). [DOI] [PMC free article] [PubMed]
- 47.Lignitto L, et al. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell. 2019;178:316–329 e318. doi: 10.1016/j.cell.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ge MK, et al. FBXO22 degrades nuclear PTEN to promote tumorigenesis. Nat. Commun. 2020;11:1720. doi: 10.1038/s41467-020-15578-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Y, et al. Knockdown of FBXO22 inhibits melanoma cell migration, invasion and angiogenesis via the HIF-1alpha/VEGF pathway. Invest N. Drugs. 2020;38:20–28. doi: 10.1007/s10637-019-00761-z. [DOI] [PubMed] [Google Scholar]
- 50.Guo F, et al. FBXO22 suppresses metastasis in human renal cell carcinoma via inhibiting MMP-9-mediated migration and invasion and VEGF-mediated angiogenesis. Int J. Biol. Sci. 2019;15:647–656. doi: 10.7150/ijbs.31293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, J. et al. Identification of core genes and clinical roles in pregnancy-associated breast cancer based on integrated analysis of different microarray profile datasets. Biosci. Rep. 39, BSR20190019 (2019). [DOI] [PMC free article] [PubMed]