Abstract
Carbapenem-resistant Klebsiella pneumoniae (CRKP) poses a significant clinical problem given the lack of therapeutic options available. Alternative antibacterial agents, such as bacteriophages, can be used as a valuable tool to treat the infections caused by these highly resistant bacteria. In this study, we isolated 54 phages from medical and domestic sewage wastewater between July and September 2019 and determined their host ranges against 54 clinical CRKP isolates, collected from a tertiary hospital in eastern China. The 54 CRKP isolates were from 7 sequence types (STs) and belonged to 9 capsular K locus types, harboring blaKPC–2 (n = 49), blaNDM–1 (n = 5), and blaIMP–4 (n = 3). Among them, the epidemic KPC-2-producing ST11 strains were most predominant (88.9%). The 54 phages showed different host ranges from 7 to 52 CRKP isolates. The total host ranges of three phages can potentially cover all 54 CRKP isolates. Among the 54 phages, phage P545, classified as a member of Myoviridaes, order Caudovirales, had a relatively wide host range (96.3%), a short latent period of 20 min, and a medium burst size of 82 PFU/cell and was stably maintained at different pH values (4–10) and temperatures (up to 60°C). P545 showed the ability to inhibit biofilm formation and to degrade the mature biofilms. Taken together, the results of our study showed that the newly isolated phage P545 had a relatively wide host range, excellent properties, and antibacterial activity as well as antibiofilm activity against a clinical CRKP ST11 isolate, providing a promising candidate for future phage therapy applications.
Keywords: bacteriophages, carbapenem-resistant, host range, Klebsiella pneumoniae, sewage
Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) has spread globally and become a significant clinical threat because of the lack of therapeutic options available. The carbapenem-resistant genes were mostly plasmid borne, thereby facilitating the horizontal transfer of resistance between different species and strains. Worrisome is that CRKP strains have recently developed resistance to some last-line antibiotics, such as polymyxin and tigecycline (Zhang et al., 2018). Although some novel β-lactam/β-lactamase inhibitor combinations, e.g., cefatazidime/avibactam, meropenem/vaborbactam, and imipenem/relebactam, were approved for the treatment of CRKP infections, they were not effective against all carbapenemase producers. Consequently, antibiotic therapeutic choices against CRKP remain limited, while other measures, such as phage therapy, have been considered as alternative measures to prevent and control CRKP infections (Ciacci et al., 2018; Anand et al., 2019).
Bacteriophages, also known as phages, are viruses that can invade and replicate in bacterial cells, and for the case of lytic phages, they are able to disrupt bacterial metabolism and cause the bacteria to lyse. Phages were discovered and clinically implemented against bacterial infections approximately 100 years ago, even before the discovery of antibiotics (Kwan et al., 2005). However, phage therapy was gradually ignored along with the wide application of antibiotics (Criscuolo et al., 2017; Lazaro-Perona et al., 2018). The progressive reduction of the effectiveness of antibiotics has recently generated renewed interest in revisiting phage therapy.
During clinical practice, phages can be used systematically through oral or injection administration, or locally on wounds or burns, as well as in the hospital environment and on medical devices. Phage therapy has been used against some Enterobacteriaceae pathogens, and among them, Escherichia coli phages (Chen et al., 2017; Cieplak et al., 2018) and Salmonella phages (Huang et al., 2018) were the most studied and applied. K. pneumoniae phages also had some applications, such as in burns (Chadha et al., 2017) and diabetes-related foot ulcers (Morozova et al., 2018). However, studies on phages against clinical CRKP strains were still limited. In the study reported here, we isolated 54 phages from domestic and hospital sewage wastewater and examined their activities against 54 CRKP strains collected from a tertiary hospital in eastern China, within one of the CRKP endemic regions.
Materials and Methods
Bacteria and Growth Conditions
A total of 54 unique (one isolate per patient) CRKP isolates, collected from blood (n = 7), abdominal cavity (n = 5), respiratory tract (n = 35), urinary tract (n = 4), and other body sites (n = 3), from a tertiary hospital in eastern China between 2016 and 2018 were used as host bacteria to determine the host ranges of phages. Details of the bacterial isolates used in this study are listed in Table 1. All isolates were routinely cultured in Luria–Bertani (LB) broth at 37°C on an orbital shaker at 180 rpm. Phosphate-buffered saline (0.1 M Na2HPO4, 0.15 M NaCl2, pH 7.2) was used for dilution and wash of K. pneumoniae cultures.
TABLE 1.
Molecular characteristics of the 54 K. pneumoniae clinical isolates.
| Sequence types (STs) | Number (%) | K locus (KL) types (n,%) | Carbapenemases (n,%) | ESBLs and AmpC (n,%) | Sources (n,%) |
| ST11 | 48 (88.9) | KL64 (24, 50.0) KL47 (23, 47.9) KL25 (1, 2.1) | KPC-2 (48, 100.0) NDM-1 (2, 4.2) | CTX-M-65 (43, 89.6) CTX-M-14 (1, 2.1) SHV-12 (16, 33.3) DHA-1 (10, 20.8) | Blood (7, 13.0) Abdominal cavity (5, 9.3) Respiratory tract (30, 55.6) Urinary tract (4, 7.4) Others (2, 3.7) |
| ST147 | 1 (1.9) | KL107 (1, 100.0) | KPC-2 (1, 100.0) | − | Respiratory tract (1, 1.9) |
| ST230 | 1 (1.9) | KL136 (1, 100.0) | IMP-4 (1, 100.0) | CTX-M-3 (1, 100.0) | Respiratory tract (1, 1.9) |
| ST3155 | 1 (1.9) | KL139 (1, 100.0) | IMP-4 (1, 100.0) | − | Others (1, 1.9) |
| ST3369 | 1 (1.9) | KL116 (1, 100.0) | NDM-1 (1, 100.0) | − | Respiratory tract (1, 1.9) |
| ST367 | 1 (1.9) | KL1 (1, 100.0) | IMP-4 (1, 100.0) NDM-1 (1, 100.0) | CTX-M-55 (1, 100.0) | Respiratory tract (1, 1.9) |
| ST656 | 1 (1.9) | KL14 (1, 100.0) | NDM-1 (1, 100.0) | CTX-M-3 (1, 100.0) | Respiratory tract (1) |
−, negative.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was conducted using the standard broth microdilution method and followed the Clinical & Laboratory Standards Institute (CLSI) guidelines. Meropenem and imipenem were tested from 0.25 to 32 μg/mL. The 2018 CLSI breakpoints were used to interpret the susceptibility results. This experiment was performed in three biological replicates. Escherichia coli ATCC®25922 was used as a negative control.
Genetic Background of CRKP Strains
Multilocus sequence typing (MLST) was used to investigate the sequence types (STs) of the total 54 CRKP isolates. In brief, polymerase chain reaction (PCR) detection followed by Sanger sequencing was used to examine seven conserved housekeeping genes including gapA, infB, mdh, pgi, phoE, rpoB, and tonB (Diancourt et al., 2005). Allelic profiles and sequence types were determined based on the Klebsiella locus/sequence definitions database1. Capsular types were determined by wzi sequencing (Brisse et al., 2013) and the K locus types (KLs) were inferred using the same Klebsiella locus/sequence definitions database as described above. In addition, ST11 KL64 and KL47 capsular types were examined by a multiplex PCR scheme described previously (Yu et al., 2018).
Detection of Carbapenemases, Extended-Spectrum Beta-Lactamases, and AmpC Genes
Polymerase chain reaction was performed to investigate the presence of carbapenemase-encoding genes, including blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA–48-like. In addition, we examined the extended-spectrum beta-lactamases (ESBLs) (CTX-M, SHV, and TEM) and AmpC cephalosporinases (CMY, ACT, and DHA) using PCR followed by Sanger sequencing. Oligonucleotide primers used for screening the aforementioned genes were described in previous reports (Mammeri et al., 2010; Bokaeian et al., 2015; Candan and Aksoz, 2015).
Isolation of Phages
Phages were isolated from the filtered (0.22-μm pore size) sewage, including medical sewage from three sewage treatment stations of local hospitals (site A, B, C) and domestic sewage from a local sewage treatment plant (site D) (Supplementary Table S1).
The purification, counting, and propagation of phages were performed using the double-layer agar plate method (Adams, 1959). SM buffer (50 mM Tris−HCl [pH 7.5], 100 mM NaCl, 10 mM MgSO4, and 0.01% gelatin) was used for the dilution of the phages. Phage suspensions were centrifuged at 5000 × g, 4°C for 10 min, and then the supernatants were filtered through 0.22-μm filters (Merck Millipore, Germany) and stored at either 4°C or at -80°C in glycerol (3:1 [v/v]) (Gu et al., 2011). All the sterile operations were carried out in class II biosecurity cabinets with institutional safety approval.
Host Range Determination of Phages
The host ranges of the phages were determined against 54 CRKP strains using spot testing (Chen et al., 2017). Briefly, 10 μL of the purified phage suspensions (∼109 PFU/mL) were spotted onto freshly seeded lawns of the isolates and left to dry before incubation for 6 h at 37°C (Kutter, 2009). Host bacteria were determined by the observation of lytic spots. The host-range frequencies were analyzed using the GraphPad Prism 5 software package. Host ranges of 54 phages were illustrated by HemI software (Deng et al., 2014).
Examination of Bacteriophage Morphology by Electron Microscopy
We selected three phages−P545, P539, and P507−for morphological analysis by transmission electron microscopy (TEM), as they had different host ranges and plaque morphology. The filtrates of phages P545, P539, and P507 were applied to a copper grid, respectively, before negative staining with phosphotungstic acid (PTA, 2% w/v). Electron micrographs were observed using an H_7650 transmission electron microscope (Hitachi, Tokyo, Japan). Bacteriophage morphology and taxonomy were confirmed following the guidelines from the International Committee on Taxonomy of Viruses2.
Optimal Multiplicity of Infection
A relatively wide host-range phage, P545, identified from the current study was further selected to determine its basic characteristics, including optimal multiplicity of infection, one-step growth curve, and thermo and pH stability.
Multiplicity of infection (MOI) refers to the ratio of phages to host bacteria during the processes for infection (Ji et al., 2019). K. pneumoniae ST11 strain KP4 was used as the host strain and was grown to logarithmic phase at a final concentration of approximately 2 × 108 CFU/mL (OD600 ∼0.4). Phage P545 suspensions were then added at different MOIs (phage/bacteria = 10, 1, 0.1, 0.01, 0.001, 0.0001, 0.00001, and 0.000001), and the mixtures were incubated at 37°C for 10 h. The phage titers were then determined immediately by plaque assay after serial dilution (Gong et al., 2016). All assays were conducted in triplicate. The MOI that generated the highest phage titer within 10 h was considered as the optimal MOI.
One-Step Growth Curve of Phage P545
Bacteriophage latent time and burst size were determined by one-step growth curve as previously described (Pajunen et al., 2000). Phage P545 was added to the exponential-phase KP4 culture (∼2 × 108 CFU/mL) at an MOI of 0.1, followed by adsorption at 37°C for 10 min. The precipitated KP4 strains were then resuspended in 10 mL of fresh LB medium after discarding unabsorbed phages by centrifugation at 5000 × g for 10 min at 4°C, followed by incubation at 37°C with shaking at 180 rpm. Samples were taken out at 5-min intervals for 120 min, and a 1% final concentration of chloroform was added to each subsample to release the intracellular phages for determining phage titers using the double-layer agar plate method. All assays were conducted in triplicate.
Thermo and pH Stability
Thermo and pH stability tests of phage P545 were conducted following a previously described protocol with minor modifications (Laemmli, 1970). In brief, 100 μL of phage was suspended in 900 μL of SM buffer (pH 7.5) and kept at different temperature (i.e., 4, 25, 37, 50, 60, 70, and 80°C) for 1 h. Phage survival rates were determined by the double-layer agar plate method described above. All assays were conducted in triplicate.
To determine pH stability of P545, 100 μL of phage suspension was added to 900 μL of SM buffer with a gradient of pH (range from 1 to 12), followed by incubation at 37°C for 1 h. SM buffer with a gradient of pH (pH 1–12) was adjusted by HCl and NaOH solution, and measured by a pH meter. Similarly, all the samples were diluted and tested immediately by the double-layer agar plate method. All assays were conducted in triplicate.
DNA Extraction and Genome Sequencing
The phage P545 suspensions were concentrated and purified for genomic DNA extraction as previously described (Sambrook and Russell, 2006). The phage genome was sequenced using the Illumina HiSeq system (Illumina, San Diego, CA, United States). Sequencing reads were de novo assembled using Spades 3.11.1 (Bankevich et al., 2012). The whole-genome sequence of phage P545 was deposited in the GenBank database under accession number MN781108. Annotation of the phage genome was conducted by the RAST server (Aziz et al., 2008), followed by manual curation. Phages closely related to P545 were examined using online BLASTn against the NCBI database3. Genome alignment of the phages was illustrated using EasyFig.
Bacteriophage Potency Against Planktonic Cells and Biofilm
To assess the inhibition effectiveness of phage P545 against KP4 planktonic cells, phages were added to the KP4 cultures (OD 0.1, ∼5 × 107CFU/mL) at MOIs of 0.1, 0.01, and 0.001, followed by incubation at 37°C with shaking at 180 rpm. Bacterial cultures with no phages was used as an untreated group. The OD600 values of the cultures were determined at 5-min intervals for 70 min. The inhibition effectiveness of P545 were analyzed by the growth kinetics of host bacteria.
To assess the inhibition effectiveness of phage P545 in biofilm formation, 100 μL of exponential-phase bacteria (OD 0.4, ∼2 × 108CFU/mL), 20 μL of phages (at MOIs of 0.1, 0.01, and 0.001), and 80 μL of fresh tryptic soy broth (TSB) medium (Merck, Darmstadt, Germany) were added into each well of 96-well flat-bottomed polystyrene microtiter plates (Sigma-Aldrich, St. Louis, MO, United States) as phage-treated groups. The untreated group consisted of 100 μL of bacteria mixed with 100 μL of fresh TSB medium, while 200 μL of fresh TSB medium was used as the negative control group. All plates were incubated for 24 h at 37°C without shaking. The biofilms were stained with 1% crystal violet as described previously (Wu Y. et al., 2019). Optical density values at 595nm (OD595) were measured using an ELISA microplate reader (Biotek, Winooski, VT, United States). Each experiment was performed in triplicate.
The degradation activity of P545 against the established mature biofilms of KP4 was evaluated using previously described protocols with minor modification (Cerca et al., 2005). Following the incubation of logarithmic-phase bacteria (see above) for 24 h at 37°C in 96-well plates, 100 μL of phage P545 at MOIs of 0.1, 0.01, and 0.001 (phage-treated groups), or 100 μL of fresh medium (untreated group) was added to each well. The negative control group was the same as above. Biofilm degradation was evaluated by the crystal violet staining method described above. Each experiment was performed in triplicate.
Statistical Analysis
All statistical analyses in this study were carried out using the GraphPad Prism 5 software package. Mean differences of negative control, untreated, and phage-treated groups of planktonic or biofilm assay were analyzed by Student’s t-test. Differences at P ≤ 0.05 were considered significant.
Results
Molecular Characteristics of the Clinical Isolates
Multilocus sequence typing showed that 54 K. pneumoniae belonged to seven different STs, with ST11 being the most common (48/54, 88.9%). Among 48 ST11 isolates, 24 isolates belonged to capsular K locus type KL64, 23 isolates belonged to KL47, and 1 isolate belonged to KL25. The other 6 isolates belonged to ST147, ST230, ST3155, ST3369, ST367, and ST656, and were assigned as capsular K locus type KL10, KL136, KL139, KL116, KL1, and KL30, respectively. All 54 K. pneumoniae isolates were carbapenemase producers, harboring blaKPC–2 (49/54, 90.7%), blaNDM–1 (5/54, 9.3%), and blaIMP–4 (3/54, 5.6%). Among them, three isolates coharbored two carbapenemase genes, with two strains harboring both blaKPC–2 and blaNDM–1, and the other one harboring both blaNDM–1 and blaIMP–4. Meanwhile, 48 isolates (88.9%) were identified to carry ESBLs and/or AmpC-encoding genes. Among them, 47 isolates were found to carry blaCTX–M, including blaCTX–M–65 (n = 43), blaCTX–M–3 (n = 2), blaCTX–M–14 (n = 1), and blaCTX–M–55 (n = 1), while 16 isolates carried blaSHV–12 and 10 isolates contained blaDHA–1. The results are summarized in Table 1.
The result of antibiotic susceptibility testing showed 54 (100%) K. pneumoniae were all carbapenem resistant (Table 2). Fifty (92.6%) isolates (46 KPC-2, 1 IMP-4, and 2 NDM-1 producers and 1 KPC-2 and NDM-1 producer) exhibited minimum inhibitory concentrations (MICs) of 32 or 16 mg/L for meropenem. Fifty (92.6%) isolates (47 KPC-2, 1 NDM-1, and 2 KPC-2 and NDM-1 producers) exhibited MICsof 32 or 16 mg/L for imipenem, while the other 4 (7.4%) isolates (2 IMP-4, 1 NDM-1, and 1 IMP-4 and NDM-1 producer) exhibited MICs of 4 or 8 mg/L for imipenem (Table 2).
TABLE 2.
Minimal inhibitory concentration of meropenem and imipenem against Klebsiella pneumoniae.
| Antimicrobial | |||||
| Strain no. | Carbapenemase | MIC (mg/L) | susceptibility | ||
| MEM | IPM | MEM | IPM | ||
| KP1 | KPC-2 | 32 | 32 | R | R |
| KP2 | KPC-2 | 16 | 16 | R | R |
| KP3 | NDM-1 | 16 | 8 | R | I |
| KP4 | KPC-2 | 16 | 16 | R | R |
| KP5 | KPC-2 | 16 | 16 | R | R |
| KP6 | KPC-2 | 16 | 16 | R | R |
| KP7 | KPC-2 | 32 | 32 | R | R |
| KP8 | KPC-2 | 16 | 16 | R | R |
| KP9 | KPC-2 | 16 | 16 | R | R |
| KP10 | KPC-2 | 16 | 16 | R | R |
| KP11 | KPC-2 | 16 | 16 | R | R |
| KP12 | KPC-2 | 16 | 16 | R | R |
| KP13 | KPC-2 | 16 | 16 | R | R |
| KP14 | KPC-2 | 16 | 16 | R | R |
| KP15 | KPC-2 | 16 | 16 | R | R |
| KP16 | KPC-2 | 16 | 16 | R | R |
| KP17 | KPC-2 | 16 | 16 | R | R |
| KP18 | KPC-2 | 16 | 16 | R | R |
| KP19 | KPC-2 | 16 | 16 | R | R |
| KP20 | KPC-2 | 16 | 16 | R | R |
| KP21 | KPC-2 | 16 | 16 | R | R |
| KP22 | KPC-2 | 16 | 16 | R | R |
| KP23 | KPC-2 | 32 | 32 | R | R |
| KP24 | KPC-2 | 16 | 16 | R | R |
| KP25 | KPC-2 | 16 | 16 | R | R |
| KP26 | KPC-2 | 16 | 16 | R | R |
| KP27 | KPC-2 | 16 | 16 | R | R |
| KP28 | KPC-2 | 16 | 16 | R | R |
| KP29 | IMP-4 | 16 | 8 | R | I |
| KP30 | KPC-2 | 16 | 16 | R | R |
| KP31 | KPC-2 | 16 | 16 | R | R |
| KP32 | KPC-2 | 16 | 16 | R | R |
| KP34 | KPC-2 | 16 | 32 | R | R |
| KP35 | KPC-2 | 16 | 16 | R | R |
| KP36 | NDM-1 | 16 | 16 | R | R |
| KP37 | KPC-2 | 16 | 16 | R | R |
| KP38 | KPC-2 | 16 | 16 | R | R |
| KP39 | KPC-2 | 32 | 32 | R | R |
| KP40 | KPC-2 | 16 | 16 | R | R |
| KP41 | KPC-2 | 16 | 16 | R | R |
| KP42 | KPC-2 | 16 | 16 | R | R |
| KP43 | KPC-2 | 16 | 16 | R | R |
| KP44 | KPC-2 | NA | 16 | NA | R |
| KP45 | KPC-2 | 16 | 16 | R | R |
| KP46 | KPC-2 | 16 | 16 | R | R |
| KP47 | KPC-2 | 16 | 16 | R | R |
| KP48 | KPC-2 | 16 | 16 | R | R |
| KP49 | KPC-2 | 16 | 16 | R | R |
| KP50 | KPC-2 | 16 | 16 | R | R |
| KP51 | KPC-2 | 16 | 16 | R | R |
| KP55 | IMP-4; NDM-1 | NA | 8 | NA | I |
| KP56 | IMP-4 | 1 | 4 | S | I |
| KP57 | KPC-2; NDM-1 | 16 | 16 | R | R |
| KP58 | KPC-2; NDM-1 | NA | 16 | NA | R |
I, intermediate; IPM, imipenem; MEM, meropenem; R, resistant; S, susceptible. NA, not available. I, intermediate; IPM, imipenem; MEM, meropenem; S, susceptible. NA, not available.
Phage Isolation
A total of 54 phages were isolated from two types of sewage water samples (medical sewage and domestic sewage) between July and September 2019. Among the 54 phages, 27, 14, and 8 phages were isolated from three medical sewage samples from site C, B, and A respectively, while five phages were isolated from a domestic sewage sample from site D (Supplementary Table S1).
Host Ranges of 54 Phages
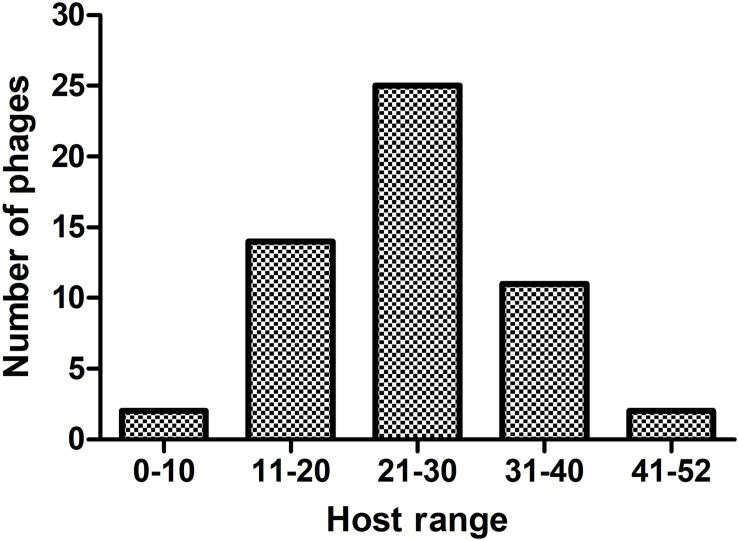
The 54 phages can lyse on average 25 CRKP strains, and each phage can lyse at least one ST11 strain. The host-range frequency analysis of phages showed that 2 (3.7%), 11 (20.4%), 25 (46.3%), 14 (25.9%), and 2 (3.7%) phages lysed >41, 31–40, 21–30, 11–20, and <10 CRKP strains, respectively (Figure 1).
FIGURE 1.
The host-range frequency distribution of phages. The x-axis represents five host-range groups based on the number of CRKP host strains. The y-axis represents the number of phages.
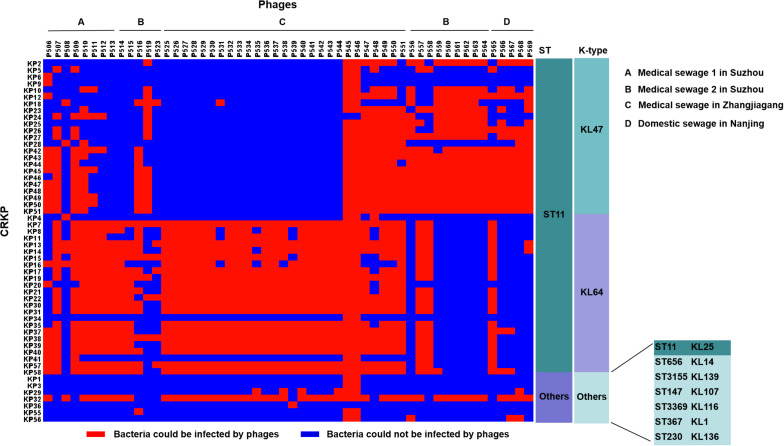
Among these 54 phages, phages P545 and P546, both isolated from a sewage wastewater sample from a local hospital (site C), formed clear lysis spots on the lawns of 52 out of 54 (96.3%) CRKP strains, including both ST11 (KL47, KL64, and KL25) and non-ST11 strains, indicating that P545/P546 had a relatively wide host range (Figure 2). Two CRKP strains−KP24 (ST11, KL47) and KP36 (ST230, KL136) −were insensitive to both phage P545 and P546. By contrast, phage P523 and P508 had narrow host ranges and could form clear lysis spots on the lawns of only eight and seven strains, respectively.
FIGURE 2.
The host ranges of 54 phages. The host range was determined using 54 clinical CRKP isolates. Blue squares represent the CRKP strains (left) that cannot be infected by the corresponding phages (top). Red squares represent the CRKP strains that could be infected by corresponding phages. (A–D) represent different sources of sewage. (A–C) were from hospital and (D) was from domestic sewage wastewater.
The host ranges of some phages appear to be correlated with the capsular types of host bacteria. Fourteen phages (25.9%) showed an ability to lyse both ST11 KL47-type and KL64-type strains, including phages P506, P507, P509–P512, P547–P551, P557, and P558 isolated from medical sewage, and one phage P565 isolated from domestic sewage. By contrast, eight phages lysed mainly ST11 KL47 strains, including P556, P559–P564 from medical sewage, and P568 from domestic sewage (Figure 2). Twenty-three phages lysed mainly KL64-type CRKP strains, for example, P513–P515 and P525–P544, isolated mostly from medical sewage (Figure 2).
Morphology Analysis
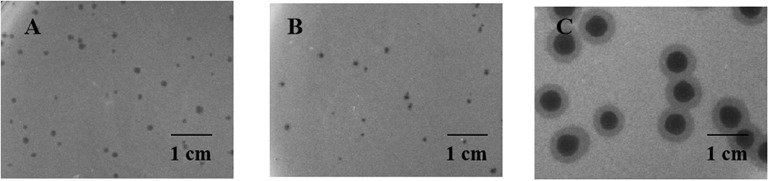
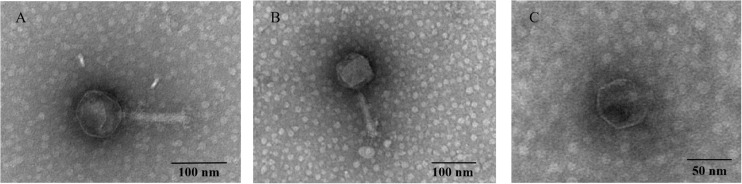
Two relatively wide host-range phages, P545 and P539, and a high lytic activity phage, P507, were selected for plaque morphology analysis. Both P545 and P539 formed small clear and round plaques with a diameter of approximately 0.1 cm. P507 formed large clear and round plaques with a diameter of approximately 0.5 cm surrounded by 0.15-cm enlarged haloes, indicating that P507 most likely encodes depolymerases with polysaccharide-degrading activity (Figure 3). In addition, phages P560, P569, and P551 also formed big clear and round plaques surrounded by enlarged haloes (data not shown). TEM confirmed that P545 and P539 belonged to T4-like phages, a member of Myoviridaes, order Caudovirales. P507 belonged to T7-like phage, a member of Podoviridaes, order Caudovirales. The diameter of capsid of P507 was estimated at 50 ± 0.5 nm (Figure 4).
FIGURE 3.
Plaque morphology of P545 (A), P539 (B), and P507 (C). Both P545 (A) and P539 (B) formed small clear and round plaques with a diameter of approximate 0.1 cm on double layer agar plate. P507 (C) formed big clear and round plaques with a diameter of approximately 0.5 cm surrounded by 0.15-cm enlarged haloes.
FIGURE 4.
Transmission electron micrograph of phage P545 (A), P539 (B), and P507 (C). The phage filtrate was applied to a copper grid before negative staining with phosphotungstic acid (PTA, 2% w/v). Electron micrographs were observed using an H_7650 (Hitachi, Tokyo, Japan) TEM.
Optimal MOI and One-Step Growth Curve
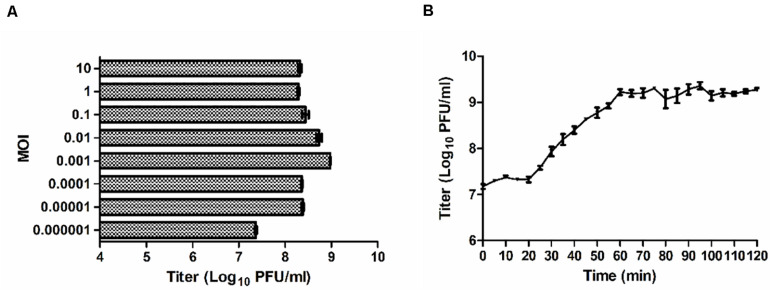
When the MOI was 0.01 or 0.001, the titers of P545 reached maximum values after propagation with a titer of approximately 109 PFU/mL (Figure 5A). Considering the input–output ratio (or growth multiple) of phages, the low MOI (0.001) was considered as the optimal MOI. The one-step growth curve experiment indicated that the latent time period of phage P545 was approximately 20 min, and the burst size was about 82 phages/cell (Figure 5B).
FIGURE 5.
Growth characteristics of P545. (A) The phage titers under different MOI (phage/bacteria = 10, 1, 0.1, 0.01, 0.001, 0.0001, 0.00001, 0.000001) are indicated on the y-axis. (B) One-step growth curve test of P545 was carried out at MOI = 0.1. The results are shown as mean ± SEM from three biological replicates.
Thermo and pH Stability of the Phages
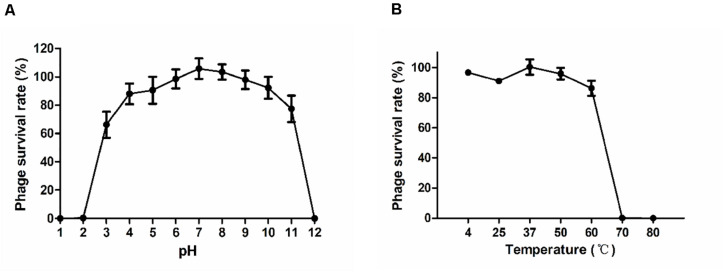
The stability of phage P545 at various pH values and temperatures was examined. The survival rates of P545 were stable at pH 4 to pH 10 (phage survival rates >80%). However, the survival rates decreased significantly at the acidic (pH < 3) or alkaline (pH > 11) conditions (Figure 6A). After incubation for1 h, high survival rates were maintained between 4°C and 37°C (approaching 100.0%). When the temperature reached 50°C to 60°C, the survival rates declined to 95.9% and 86.2%, respectively. When the temperature reached 70°C and 80°C, nearly no phages survived (Figure 6B).
FIGURE 6.
Stability tests of P545. (A) pH stability: phage particles were incubated under different pH conditions for 1 h. (B) Thermal stability: phage particles were incubated at various temperatures for 1 h. Phage survival rate = (titer after incubation)/(initial titer). SEM are shown as vertical lines.
Genome Analysis of P545
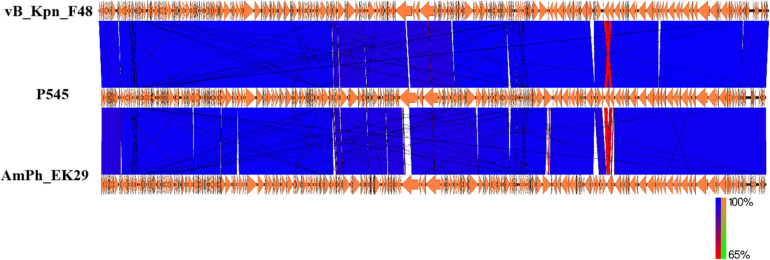
Next-generation sequencing analysis showed that P545 was a linear dsDNA molecule of 169,725 bp and a GC content of 40.8%. No repeated terminal sequences were detected at the 5′- and 3′-ends. P545 contained a total of 287 putative coding regions (CDSs) and 7 tRNAs. No lysogeny or virulence associated genes were identified in the genome of P545, showing its lytic nature and the potential for therapeutic application. Two putative tail lysozymes (gene loci: 7096–7590 and 157173–158912) were identified based on the RAST predication. Online BLASTn analysis of P545 genome sequence revealed that it is mostly close to Klebsiella phage vB_Kpn_F48 (Accession no. MG746602) isolated from a hospital sewage wastewater in Italy (Ciacci et al., 2018), with 99% query coverage and 99% nucleotide identity, and to Klebsiella phage AmPh_EK29 (MN434092), isolated from sewage and wastewater in Australia, with 96% query coverage and 98% nucleotide identity (Figure 7).
FIGURE 7.
Comparative genome analysis of K. pneumoniae P545 bacteriophage. Schematics were produced using EasyFig. The genome sequence of the P545 was compared to those of two most closely related phages, vB_Kpn_F48 (MG746602) and AmPh_EK29 (MN434092) published in NCBI.
Antibacterial and Antibiofilm Activity of Bacteriophage P545
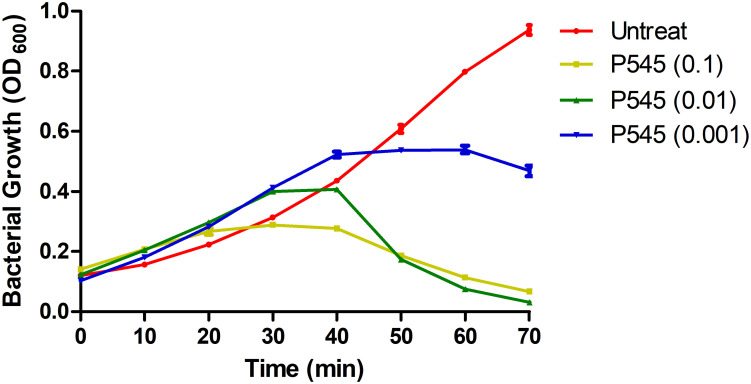
To evaluate the inhibitory activity of phage P545 against planktonic bacteria, P545 suspensions of different concentrations (MOI = 0.1, 0.01, and 0.001) were incubated with CRKP strain KP4 for 70 min. The antibacterial assay showed that the OD600 values of three phage-treated groups (MOI = 0.1, 0.01, and 0.001) were significantly lower than those of the untreated group after 50, 60, and 70 min of incubation (Figure 8). The results indicated that phage P545 can effectively inhibit bacterial growth.
FIGURE 8.
In vitro activity of phage P545 at 37°C. Panels show the OD600 values of bacterial cultures. The yellow line represents the OD600 value of CRKP KP4 treated with phage P545 at MOI 0.1; the green line represents that of KP4 treated with phage P545 at MOI 0.01; the blue line represents that of KP4 treated with phage P545 at MOI 0.001; the red line represents that of untreated KP4.
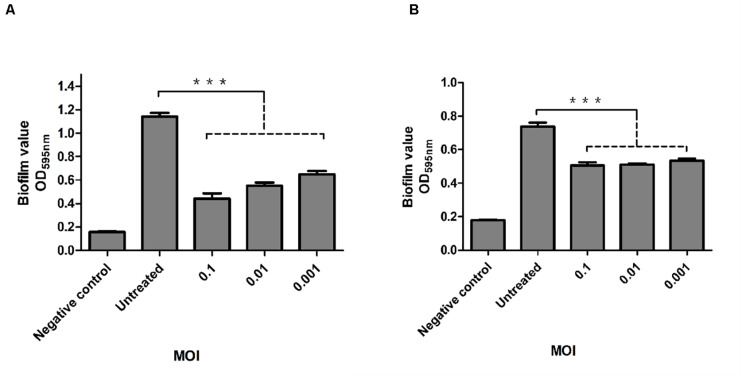
The inhibitory effect of phage P545 on biofilm formation in the KP4 strain was analyzed using 96-well plates. To evaluate the ability of phage P545 to inhibit biofilm formation, phage P545 suspensions were incubated with CRKP strain KP4 at different MOIs (0.1, 0.01, and 0.001) for 24 h. The results of crystal violet staining assay showed that the OD595 values of three different phage-treated groups (MOI 0.1, 0.01, and 0.001) decreased 0.70, 0.59, and 0.49, respectively, in comparison to that of the untreated group (Figure 9A, P < 0.001). The ability of P545 to degrade biofilms was also assessed using 24-h-old mature biofilms. The OD595 values of the total biomass that remained attached in the phage-treated groups decreased 0.24 (MOI = 0.1), 0.23 (MOI = 0.01), and 0.21 (MOI = 0.001) compared with that of the untreated group (Figure 9B, P < 0.001). These results suggested that P545 can inhibit biofilm formation and degrade the formed mature biofilm.
FIGURE 9.
Antibacterial and antibiofilm activity of phage P545. (A) P545 inhibited the formation of biofilms. P545 was incubated with CRKP strain KP4 at MOIs of 0.1, 0.01, and 0.001 in 96-well plates for 24 h. (B) P545 disrupted the mature biofilm of KP4. Biofilm formation of KP4 was induced in 96-well plates for 24 h, and then the biofilm was treated with P545 at MOIs of 0.1, 0.01, and 0.001 for 24 h. Fresh TSB medium was used as a negative control. The OD595 values of the residual biofilms were measured after crystal violet staining. All the data are shown as the mean ± SEM. ***indicates p < 0.001.
Discussion
Carbapenem-resistant Klebsiella pneumoniae strains have emerged as significant multidrug-resistant bacterial pathogens that pose a serious threat to global public health. The international spreads of CRKP strains were associated with a few high-risk clonal strains, including ST258, ST11, ST14, ST147, and ST307 etc. Among them, ST258 predominates in North America and Europe, whereas ST11 is the primary CRKP clone in East Asia, especially China (Chen et al., 2014; Zhang et al., 2019). In China, ST11 strains accounted for >70% CRKP strains (Liu et al., 2018). CRKP ST11 strains were primarily associated with the blaKPC–2 gene in China, but other carbapenemase genes, such as blaNDM, blaOXA–48–like, blaVIM, and blaIMP, were frequently identified in ST11 strains elsewhere (Chen et al., 2018; Liu et al., 2018; Wang et al., 2018). For example, in this study, two ST11 strains were also found to carry both blaNDM–1 and blaKPC–2. In addition, some CRKP ST11 strains have evolved to be extensively drug resistant (Dorp et al., 2019; Wu X. et al., 2019). ccordingly, combating the infection caused by epidemic CRKP ST11 strains has already become a priority in controlling CRKP strains in China and some other countries.
Bacteriophages are being considered as a potential therapeutic approach against highly drug-resistant pathogens, including CRKP strains (Wu X. et al., 2019). In this study, we isolated 54 phages from domestic and hospital sewage wastewater. Our results suggested that sewage, especially medical sewage, may be a better phage source against clinical CRKP strains, since phages can be efficiently isolated (Peng et al., 2020). Clinically, phages with relatively wide host ranges could be ideal candidates for phage cocktail in phage therapy. Based on the host-range analysis, three selected phages (i.e., P545, P539, and P507) can potentially cover all 54 clinical CRKP isolates, collected from various clinical sources and harboring different carbapenemase genes.
Our results also indicated that the phage lytic activity was independent of the carbapenemase genotypes. Importantly, all 54 phages showed lytic activity against some epidemic ST11 strains. The host ranges of certain phages appeared to be narrow and were specific to certain K. pneumoniae strains. This result was similar to the results of other studies that some phages were specific to certain capsular serotypes. For example, bacteriophage NTUH-K2044-K1-1 isolated from Taiwan exhibited specificity for capsular type K1 (Lin et al., 2014). Klebsiella bacteriophage K5-2 exhibited specificity for capsular types K5, K30, and K69, and bacteriophage K5-4 exhibited specificity for capsular types K8 and K5 (Hsieh et al., 2017). Three bacteriophages−KpV41, KpV475, and KpV71−had specific lytic activity against mainly K1 stains (Solovieva et al., 2018). Moreover, two phages, KpV74, and KpV763, had specific lytic activity against K2 strains, and the phage KpV767 had specific lytic activity against K57 strains (Solovieva et al., 2018).
Interestingly, we identified a phage, P545, with a relatively wide host range, that can lyse 52 of 54 (96.3%) CRKP strains from our collection. These 52 CRKP strains included 24 ST11 KL64, 22 ST11 KL47, 1 ST11 KL25, 1 ST367 KL1, 1 ST147 KL10, 1 ST656 KL14, 1 ST3155 KL139, and 1 ST230 KL136. The molecular mechanism underlying the relatively wide host range of phage P545 remains unclear. TEM of phage P545 revealed that P545 belonged to T4-like lytic phage, a member of Myoviridaes, order Caudovirales. BLASTn analysis showed that P545 was highly similar to Klebsiella phage vB_Kpn_F48 (MG746602) from Italy and AmPh_EK29 (MN434092) from Australia. However, both vB_Kpn_F48 and AmPh_EK29 appear to be narrow host-range phages. Phage vB_Kpn_F48 had a lytic activity specific to K. pneumoniae strains belonging to ST101 (K17) and its closely related strains (Ciacci et al., 2018). Similarly, AmPh_EK29 was specifically active against K. pneumoniae ST258 clade 1 strains (KL106). Our phage P545 had a lytic activity against ST11 strains (KL47 and KL64) and six other STs. Further studies will be performed to examine whether P545 can actively lyse other CRKP strains, especially ST258 and ST101 strains. Our genome alignment results showed that P545 harbors >2600 and >4600 core single nucleotide polymorphisms and some insertion or deletion versus vB_Kpn_F48 and AmPh_EK29, respectively. For T4-like phages, the tail fiber protein gene in general determined the specificity and host range. BLASTn analysis revealed the tail fiber protein gene sequence of P545 had only 91% query coverage and 89.4% nucleotide identity (genome level: 99% query coverage and 99% nucleotide identity) with that of Klebsiella phage vB_Kpn_F48. We suspected the genetic variation in tail fiber protein gene of P545, in comparison to vB_Kpn_F48 and AmPh_EK29, may contribute to the relatively wide host range observed in P545, which deserves further study.
Phage P545 had a short latent period of 20 min and a medium burst size of 82 PFU/cell and was stable at pH values (4–10) and temperatures (up to 60°C). When the MOI was 0.01 or 0.001, the titers of P545 reached the maximum value (no significant difference). Considering the input–output ratio (or growth multiple) of phage, the lower MOI (0.001) was considered the optimal MOI.
In this study, phage P545 inhibited bacterial growth within 70 min. The result of crystal violet staining assay suggested that phage P545 can not only inhibit biofilm formation but also degrade the formed mature biofilms. Many bacteriophages produced specific depolymerases, which are able to destroy the capsular polysaccharides, thereby allowing adsorption of phages to the outer membrane receptors (Pires et al., 2016) and penetration of phage DNA into the bacterial cells (Hughes et al., 1998). These phage-derived capsule depolymerases can be potentially used as therapeutic agents against capsulated bacteria, such as CRKP strains. However, the plaque morphology analysis suggested that phage P545 was a non-depolymerase-producing phage, because no clear enlarged haloes were observed (Figure 4). In fact, besides depolymerases, phage-encoded lysozymes (endolysins, lysins, etc.) can also digest bacterial cell wall material (Chan and Abedon, 2015; Ajuebor et al., 2016). Two tail lysozyme genes were found in the genome of P545 based on RAST predication. Therefore, we suspected that the mechanism of biofilm formation inhibition and biofilm degradation may be associated with phage-encoded tail lysozymes of phage P545. The results suggested that P545 could be an attractive candidate as an anti-CRKP agent. Moreover, in this study, P507 formed large clear and round plaques surrounded by enlarged haloes, suggesting that it most likely encodes depolymerases with polysaccharide-degrading activity (Pan et al., 2017; Wu X. et al., 2019).
One limitation of current study was that we determined the phage activities against CRKP isolates collected from only a single medical center. Nevertheless, these CRKP isolates were collected from various clinical sources and harbored a wide battery of carbapenemase genes, including the most predominant STs and capsular types of strains spreading in China. In addition, another limitation is that only one of the 54 phages (P545) was fully characterized. Further work will be conducted to characterize other phages isolated in the current study, such as P507, and to test the activities of these phages against CRKP isolates from other genetic backgrounds. Today, the sources and rapid availability of phages against clinically isolated pathogens remains the most significant challenge for phage therapy. Our study described that the 54 phages can be successfully obtained within a short period of time (1.5 months) from local sewage, especially from medical wastewater. Medical sewage in the surrounding area could serve as a rich naturally occurring phage source against clinically important pathogens. Natural phage, in combination with engineered phages and phage-encoded enzymes, may provide a more effective solution for combating epidemic CRKP infections.
Data Availability Statement
The datasets generated for this study can be found in the annotated whole-genome sequence of phage P545 was deposited in the GenBank database under accession number: MN781108.
Ethics Statement
This study was approved by the Institutional Review Board (IRB) of The Second Affiliated Hospital of Soochow University. The clinical isolates were retrospectively collected, and patient data were not included in this study, therefore informed consent was waived by the Institutional Review Board (IRB) of The Second Affiliated Hospital of Soochow University.
Author Contributions
ML, WZ, and HD conceived the study. ML, MG, LoC, PL, YX, and WZ performed the experiment and computational analysis. ML, LiC, HG, and WZ wrote the article. All authors read, revised, and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by grants from the National Key Research and Development Program of China (2018YFC1602500), the National Natural Science Foundation of China (81572032), the National Natural Science Foundation of China (U1803109), the Six Talent Peaks Project in Jiangsu Province (2016-WSN-112), the Key Research and Development Project of Jiangsu Provincial Science and Technology Department (BE2017654), the Gusu Key Health Talent of Suzhou, Jiangsu Youth Medical Talents Program (QN-867), and the Science and Technology Program of Suzhou (SZS201715).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01554/full#supplementary-material
References
- Adams M. (1959). Bacteriophage. New York, NY: Interscience Publishers. [Google Scholar]
- Ajuebor J., McAuliffe O., O’Mahony J., Ross R. P., Hill C., Coffey A. (2016). Bacteriophage endolysins and their applications. Sci. Prog. 99 183–199. 10.3184/003685016X14627913637705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand T., Virmani N., Kumar S., Kumar Mohanty A., Pavulraj S., Ch Bera B., et al. (2019). Phage Therapy for treatment of virulent Klebsiella pneumoniae infection in mouse model. J. Glob. Antimicrob. Resist. 21 34–41. 10.1016/j.jgar.2019.09.018 [DOI] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokaeian M., Shahraki Zahedani S., Soltanian Bajgiran M., Ansari Moghaddam A. (2015). Frequency of PER, VEB, SHV, TEM and CTX-M genes in resistant strains of Pseudomonas aeruginosa producing extended spectrum beta-lactamases. Jundishapur J. Microbiol. 8:e13783. 10.5812/jjm.13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse S., Passet V., Haugaard A. B., Babosan A., Kassis-Chikhani N., Struve C., et al. (2013). wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 51 4073–4078. 10.1128/JCM.01924-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candan E. D., Aksoz N. (2015). Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim. Pol. 62 867–874. 10.18388/abp.2015_1148 [DOI] [PubMed] [Google Scholar]
- Cerca N., Martins S., Pier G. B., Oliveira R., Azeredo J. (2005). The relationship between inhibition of bacterial adhesion to a solid surface by sub-MICs of antibiotics and subsequent development of a biofilm. Res. Microbiol. 156 650–655. 10.1016/j.resmic.2005.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadha P., Katare O. P., Chhibber S. (2017). Liposome loaded phage cocktail: enhanced therapeutic potential in resolving Klebsiella pneumoniae mediated burn wound infections. Burns 43 1532–1543. 10.1016/j.burns.2017.03.029 [DOI] [PubMed] [Google Scholar]
- Chan B., Abedon S. (2015). Bacteriophages and their enzymes in biofilm control. Curr. Pharm. Des. 21 85–99. 10.2174/1381612820666140905112311 [DOI] [PubMed] [Google Scholar]
- Chen C. M., Guo M. K., Ke S. C., Lin Y. P., Li C. R., Vy Nguyen H. T., et al. (2018). Emergence and nosocomial spread of ST11 carbapenem-resistant Klebsiella pneumoniae co-producing OXA-48 and KPC-2 in a regional hospital in Taiwan. J. Med. Microbiol. 67 957–964. 10.1099/jmm.0.000771 [DOI] [PubMed] [Google Scholar]
- Chen L., Mathema B., Chavda K. D., DeLeo F. R., Bonomo R. A., Kreiswirth B. N. (2014). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22 686–696. 10.1016/j.tim.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Zhang L., Abdelgader S. A., Yu L., Xu J., Yao H., et al. (2017). Alterations in gp37 expand the host range of a T4-like phage. Appl. Environ. Microbiol. 83:e01576-17. 10.1128/AEM.01576-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciacci N., D’Andrea M. M., Marmo P., Dematte E., Amisano F., Di Pilato V., et al. (2018). Characterization of vB_Kpn_F48, a newly discovered lytic bacteriophage for Klebsiella pneumoniae of sequence type 101. Viruses 10:482. 10.3390/v10090482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieplak T., Soffer N., Sulakvelidze A., Nielsen D. S. (2018). A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes 9 391–399. 10.1080/19490976.2018.1447291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo E., Spadini S., Lamanna J., Ferro M., Burioni R. (2017). Bacteriophages and their immunological applications against infectious threats. J. Immunol. Res. 2017:3780697. 10.1155/2017/3780697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Wang Y., Liu Z., Cheng H., Xue Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. 10.1371/journal.pone.0111988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diancourt L., Passet V., Verhoef J., Grimont P. A., Brisse S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43 4178–4182. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorp L. V., Wang Q., Shaw L. P., Acman M., Brynildsrud O. B., Eldholm V., et al. (2019). Rapid phenotypic evolution in multidrug-resistant Klebsiella pneumoniae hospital outbreak strains. Microb. Genom. 5:e000263. 10.1099/mgen.0.000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Cheng M., Li X., Jiang H., Yu C., Kahaer N., et al. (2016). Characterization of Enterococcus faecium bacteriophage IME-EFm5 and its endolysin LysEFm5. Virology 492 11–20. 10.1016/j.virol.2016.02.006 [DOI] [PubMed] [Google Scholar]
- Gu J., Xu W., Lei L., Huang J., Feng X., Sun C., et al. (2011). LysGH15, a novel bacteriophage lysin, protects a murine bacteremia model efficiently against lethal methicillin-resistant Staphylococcus aureus infection. J. Clin. Microbiol. 49 111–117. 10.1128/JCM.01144-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P. F., Lin H. H., Lin T. L., Chen Y. Y., Wang J. T. (2017). Two T7-like bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: isolation and functional characterization. Sci. Rep. 7:4624. 10.1038/s41598-017-04644-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Shi J., Ma W., Li Z., Wang J., Li J., et al. (2018). Isolation, characterization, and application of a novel specific Salmonella bacteriophage in different food matrices. Food Res. Int. 111 631–641. 10.1016/j.foodres.2018.05.071 [DOI] [PubMed] [Google Scholar]
- Hughes K. A., Sutherland I. W., Jones M. V. (1998). Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology 144(Pt 11), 3039–3047. 10.1099/00221287-144-11-3039 [DOI] [PubMed] [Google Scholar]
- Ji Y., Cheng M., Zhai S., Xi H., Cai R., Wang Z., et al. (2019). Preventive effect of the phage VB-SavM-JYL01 on rabbit necrotizing pneumonia caused by Staphylococcus aureus. Vet. Microbiol. 229 72–80. 10.1016/j.vetmic.2018.12.021 [DOI] [PubMed] [Google Scholar]
- Kutter E. (2009). Phage host range and efficiency of plating. Methods Mol. Biol. 501 141–149. 10.1007/978-1-60327-164-6_14 [DOI] [PubMed] [Google Scholar]
- Kwan T., Liu J., DuBow M., Gros P., Pelletier J. (2005). The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 102 5174–5179. 10.1073/pnas.0501140102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lazaro-Perona F., Sotillo A., Troyano-Hernaez P., Gomez-Gil R., de la Vega-Bueno A., Mingorance J. (2018). Genomic path to pandrug resistance in a clinical isolate of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 52 713–718. 10.1016/j.ijantimicag.2018.08.012 [DOI] [PubMed] [Google Scholar]
- Lin T. L., Hsieh P. F., Huang Y. T., Lee W. C., Tsai Y. T., Su P. A., et al. (2014). Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J. Infect. Dis. 210 1734–1744. 10.1093/infdis/jiu332 [DOI] [PubMed] [Google Scholar]
- Liu L., Feng Y., Tang G., Lin J., Huang W., Qiao F., et al. (2018). Carbapenem-resistant isolates of the Klebsiella pneumoniae complex in western china: the common ST11 and the surprising hospital-specific types. Clin. Infect. Dis. 67(Suppl._2), S263–S265. 10.1093/cid/ciy662 [DOI] [PubMed] [Google Scholar]
- Mammeri H., Guillon H., Eb F., Nordmann P. (2010). Phenotypic and biochemical comparison of the carbapenem-hydrolyzing activities of five plasmid-borne AmpC beta-lactamases. Antimicrob. Agents Chemother. 54 4556–4560. 10.1128/AAC.01762-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova V. V., Kozlova Y. N., Ganichev D. A., Tikunova N. V. (2018). Bacteriophage treatment of infected diabetic foot ulcers. Methods Mol. Biol. 1693 151–158. 10.1007/978-1-4939-7395-8_13 [DOI] [PubMed] [Google Scholar]
- Pajunen M., Kiljunen S., Skurnik M. (2000). Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182 5114–5120. 10.1128/jb.182.18.5114-5120.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y. J., Lin T. L., Chen C. C., Tsai Y. T., Cheng Y. H., Chen Y. Y., et al. (2017). Klebsiella phage PhiK64-1 encodes multiple depolymerases for multiple host capsular types. J. Virol. 91:e02457-16. 10.1128/JVI.02457-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Fang M., Liu X., Zhang C., Liu Y., Yuan Y. (2020). Isolation and characterization of a novel phage for controlling multidrug-resistant Klebsiella pneumoniae. Microorganisms 8:542. 10.3390/microorganisms8040542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. P., Oliveira H., Melo L. D., Sillankorva S., Azeredo J. (2016). Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 100 2141–2151. 10.1007/s00253-015-7247-0 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2006). Precipitation of bacteriophage lambda particles from large-scale lysates. CSH Protoc. 2006:pdb.prot3966. 10.1101/pdb.prot3966 [DOI] [PubMed] [Google Scholar]
- Solovieva E. V., Myakinina V. P., Kislichkina A. A., Krasilnikova V. M., Verevkin V. V., Mochalov V. V., et al. (2018). Comparative genome analysis of novel Podoviruses lytic for hypermucoviscous Klebsiella pneumoniae of K1, K2, and K57 capsular types. Virus Res. 243 10–18. 10.1016/j.virusres.2017.09.026 [DOI] [PubMed] [Google Scholar]
- Wang Q., Wang X., Wang J., Ouyang P., Jin C., Wang R., et al. (2018). Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012-2016). Clin. Infect. Dis. 67(Suppl._2), S196–S205. 10.1093/cid/ciy660 [DOI] [PubMed] [Google Scholar]
- Wu X., Han H., Chen C., Zheng B. (2019). Genomic characterisation of a colistin-resistant Klebsiella pneumoniae ST11 strain co-producing KPC-2, FloR, CTX-M-55, SHV-12, FosA and RmtB causing a lethal infection. J. Glob. Antimicrob. Resist. 19 78–80. 10.1016/j.jgar.2019.08.023 [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang R., Xu M., Liu Y., Zhu X., Qiu J., et al. (2019). A novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front. Microbiol. 10:2768. 10.3389/fmicb.2019.02768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Lv J., Niu S., Du H., Tang Y. W., Pitout J. D. D., et al. (2018). Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (Sequence Type 258 [ST258] and ST11) and hypervirulent (ST23, ST65, ST86, and ST375) strains. J. Clin. Microbiol. 56:e00731-18. 10.1128/JCM.00731-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Dong N., Huang Y., Zhou H., Xie M., Chan E. W., et al. (2018). Evolution of tigecycline- and colistin-resistant CRKP (carbapenem-resistant Klebsiella pneumoniae) in vivo and its persistence in the GI tract. Emerg. Microbes Infect. 7:127. 10.1038/s41426-018-0129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang W., Yu H., Wang M., Zhang H., Lv J., et al. (2019). New delhi metallo-beta-lactamase 5-producing Klebsiella pneumoniae sequence Type 258, Southwest China, 2017. Emerg. Infect Dis. 25 1209–1213. 10.3201/eid2506.181939 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the annotated whole-genome sequence of phage P545 was deposited in the GenBank database under accession number: MN781108.