Abstract
With advances in computing, agent-based models (ABMs) have become a feasible and appealing tool to study biological systems. ABMs are seeing increased incorporation into both the biology and mathematics classrooms as powerful modeling tools to study processes involving substantial amounts of stochasticity, nonlinear interactions, and/or heterogeneous spatial structures. Here we present a brief synopsis of the agent-based modeling approach with an emphasis on its use to simulate biological systems, and provide a discussion of its role and limitations in both the biology and mathematics classrooms.
Keywords: Agent-based models (ABMs), Simulation, Pedagogy, Education
Introduction
Agent-based models (ABMs) are computational structures in which system-level (macro) behavior is generated by the (micro) behavior of individual agents, which may be persons, cells, molecules or any other discrete quantities. Typical ABMs contain three elements: agents, an environment, and rules governing each agent’s behavior and its local interactions with other agents and with the environment. Decades of advancement in computer power has made agent-based modeling a feasible and appealing tool to study a variety of complex and dynamic systems, especially within the life sciences. As the use of ABMs in research has grown, so too has the inclusion of ABMs in life science and mathematical modeling courses as a means of exploring and predicting how individual-level behavior and interactions among individuals lead to system-level observable patterns. ABMs are now one of the many types of models students studying the life sciences or applied mathematics should encounter in their undergraduate education.
Prior to the introduction of ABMs into biological and applied mathematics curricula, the clear model format of choice was the ordinary differential equation (ODE), or maybe a pair of them; occasionally, discrete difference equations and/or matrix equations would also be introduced. Exponential growth and decay were ready examples, paving the way for extensions of the exponential growth process toward a carrying capacity in the form of the logistic growth process (Voit 2020). This logistic process was easily generalized to two populations, which were at first independent, but then allowed to interact. Depending on these interactions, the result was a pair of two populations competing for the same resource or a simple predator–prey model in the format of a two-variable Lotka–Volterra system.
Although ODEs and any other types of “Diff-E-Qs” are a priori dreaded by almost all but mathematicians and physicists, the concept of an ODE, if adequately explained, becomes quite intuitive. For instance, one may ease a novice into the world of ODEs by considering changes in the water level W of a lake over time (Ayalew 2019). Whereas these dynamics are difficult to formulate as an explicit function W(t), newcomers readily understand that changes in the water level depend on influxes from tributaries, rain, and other sources on the supply side, and on effluxes, evaporation and water utilization on the side of reducing the amount of water. Just putting these components into an equation leads directly to a differential equation of the system (Weisstein 2011). On the left side, one finds the change over time as dW/dt, and this change is driven, on the right-hand side, by a sum of augmenting and diminishing processes.
There is hardly a limit to what can be achieved with ODEs in biology, with the very important exception of processes that have genuine spatial features. And while it is not difficult to ease a biology undergraduate into ordinary differential equations, the same is not necessarily true for partial differential equations (PDEs). However, spatial phenomena in biology seldom occur in homogeneous conditions. As examples, consider the formation of tumors with angiogenesis and necrosis; the local patterns of cell-to-cell signaling that governs the embryonic development; the spread of the red fire ant (Solenopsis invicta) from Mobile, AL, its alleged port of entry into the USA, all along the Gulf and East Coasts; or the population size and dynamics of the Santa Cruz island fox (Urocyon littoralis santacruzae) being driven by territory size which in turn depends on local vegetation (Scott 2019). Until relatively recently, the conundrum of space was often dealt with in the final chapter of mathematical modeling in biology. A sea change came with the development of ABMs, which are natural formats for both stochasticity and spatial phenomena. By their nature, these models are computationally expensive, which initially prevented their use in most classrooms. However, this situation has obviously changed. As the Bio2010 Report (2003) stated: “Computer use is a fact of life of all modern life scientists. Exposure during the early years of their undergraduate careers will help life science students use current computer methods and learn how to exploit emerging computer technologies as they arise.”
Classroom use of ABMs has thus become not just logistically feasible, but also very appealing for demonstrating spatial dynamics in a wide range of biological systems (Kottonau 2011; Triulzi & Pyka 2011; Shiflet 2013; Pinder 2013). Supporting this appeal is a repertoire of software tools, such as SimBio and NetLogo (see Sect. 3), that contain predefined examples and require minimal computer coding skills for model analysis. Here, we present a brief synopsis and history of this modeling approach with emphasis on life science applications (Sect. 2), describe some of the software tools most frequently used in the classroom (Sect. 3), and then focus on some of its roles and limitations in the classroom (Sect. 4).
Background, Rationale, and Pitfalls of ABMs
The Origins of Agent-Based Modeling. The true origins of any method or procedure are seldom identifiable in an unambiguous manner. In the case of agent-based modeling, one could think of Craig Reynolds’ 1987 seminal article on the formation of bird flocks (with the agents denoted as boids, short for “bird-oid object”), which he was able to represent with just three rules of behavior: (1) avoid collisions with nearby birds; (2) attempt to match the velocity of nearby birds; and (3) attempt to stay close to nearby birds in the flock (Reynolds 1987; Gooding 2019). The result of simulations with this simple ABM was very realistic-looking flocking behavior. Particularly intriguing in this study was the fact that there was no leader or a global organizing principle. Instead, the virtual birds were truly individual agents that self-organized locally, thereby generating a globally coherent flight pattern.
While Reynolds’ work was a milestone, key concepts leading to modern ABMs can be found much earlier. One notable contributor of ideas was Nobel laureate Enrico Fermi, who used mechanical addition machines to generate probabilities for stochastic models with which he solved otherwise unwieldy problems (Gooding 2019). This procedure was an early form of the method of a Monte Carlo simulation, which was later independently developed and published by Stanislav Ulam, like Fermi a member of the Manhattan Project (Metropolis & Ulam 1949; Metropolis 1987). Another very important contribution to the budding development of ABMs was the Turing machine (Turing 1936), which is a mathematical model of computation that uses a set of rules to manipulate symbols in discrete cells on an infinite tape. Much closer to ABMs were ideas of Ulam, who was fascinated by the “automatic” emergence of patterns in two-dimensional games with very simple rules (Ulam 1950; Metropolis 1987). Together with the new concept of game theory (von Neumann & Morgenstern 1944), all these ideas were developed into the concept of cellular automata, which are direct predecessors of ABMs (Ulam 1950; Ulametal 1947; von Neumann & Morgenstern 1944). A very appealing implementation of a cellular automaton was John Conway’s famous Game of Life (Gardner 1970).
The social sciences adopted computational methods in the 1960s for microanalytic simulations or, simply, micro-simulations (Gilbert & Troitzsch 2005; Gooding 2019). In contrast to today’s ABMs, which use simple rules to recreate observed or unknown patterns, the agents in the original microsimulations acted according to empirical data (Bae 2016). The advantage of this strategy is that model analysis can reveal systemic behaviors under different realistic scenarios (Gooding 2019). A seminal paper in this context described generic segregation processes (Schelling 1971). Other agent-based modeling work in sociology and economics was gleaned from the biological ABM work of Smith (1982), who formulated Darwin’s ideas of evolution as a computer simulation. This idea inspired Nelson & Winter to apply similar concepts and implementations to market studies, where firms were modeled like animals that followed specific routines. In particular, the firms were in competition, and the market weeded out bad routines while rewarding the fittest. Nelson & Winter’s influential book An Evolutionary Theory of Economic Change (Nelson & Winter 1982) strongly proposed the use of computer simulations, which today would fall within the scope of agent-based modeling. Their ideas led to a school of thought called evolutionary economics (Hanappi 2017). An early and particularly influential paper in this context tried to shed light on the stock market (Palmer 1994).
Initially, ABMs of artificial life simulated simple homogeneous agents that acted like huge colonies of ants that could just move and eat in the pursuit of food. Somewhat more sophisticated, economic simulations used as the main agent homo economicus, a consistently rational human pursuing the optimization of some economic goal or utility with exclusive self-interest (Persky 1995). Based on these humble beginnings, sophistication in computing soon permitted heterogeneous agents and much more complicated landscapes than before. The successes in economics were so tantalizing that simulation studies eventually reached the most prestigious journals of economics and the social sciences (Axtell 1996; Epstein & Axtell 1996; Geanakoplos 2012; Hanappi 2017). Modern ABMs in economics are capable of capturing much of the complexity of macroeconomic systems (e.g., Caiani (2016)).
Following directly the principles of cellular automata, Kauffman studied large grids with elements that changed features in a binary fashion (Kauffman 1993). For instance, a white agent could turn black, and this shift occurred according to Boolean rules that usually involved some or all neighboring grid points. Kauffman was able to demonstrate the emergence of complex patterns, such as oscillations and percolation. Starting in the 1980, Wolfram performed systematic studies of cellular automata, which led to his influential 2002 book A New Kind of Science (Wolfram 2002) that assigns cellular automata a wide range of applications in a variety of fields.
Biological Applications of Agent-Based Modeling. ABMs have been constructed to study a wide range of biological phenomenon. Numerous reviews and research efforts using ABMs have focused on specific biomedical systems. Issues of gene expression were modeled by Thomas (2019). Morphogenetic and developmental processes were discussed in Grant (2006), Robertson (2007), Thorne (2007), Tang (2011), and Glen (2019). Models of tissue mechanics and microvasculature are captured in Bailey (2007) and van Liedekerke (2015). Inflammation, wound healing and immune responses were addressed in An (2004), An (2009), Chavali (2008), and Castiglione & Celada (2015). Other authors (Wang 2015; Segovia 2004; Cisse 2013) used ABMs to model cancer growth, tuberculosis, and schistosomiasis, respectively. Lardon and colleagues (Lardon 2011) used ABMs to analyze biofilm dynamics. Butler and colleagues described the use of ABMs in pharmacology (Butler 2015). ABMs studying multicellular systems provide a unique capability to examine interactions and feedback loops across the different hierarchies of the system (Hellweger 2016).
Reviews of ABMs in the context of ecology, environmental management, and land use include (Bousquet 2004; Matthews 2007; Grimm & Railsback 2005; Caplat 2008; DeAngelis & Diaz 2019). In some applications, interventions or treatments were addressed and therefore required the adaptation of agents to changing scenarios (Berry 2002).
ABMs have also been used to simulate epidemics with analyses examining the impact of implemented or potential intervention measures (e.g., quarantining/physical distancing, mask wearing, and vaccination) (Mniszewski 2013; Perez & Dragicevic 2009; Tracy 2018). Visual representations of epidemiological ABMs have even been used by news outlets during the COVID-19 pandemic to help explain to the public how various intervention methods change the shape of an epidemic (i.e., “flatten the curve) or the basic reproduction number () of an epidemic; see, for example, Fox (2020) and Stevens (2020).
Rationale & Pitfalls of ABMs. The two most frequent goals of an ABM analysis are (1) the elucidation and explanation of emergent behaviors of a complex system and (2) the inference of rules that govern the actions of the agents and lead to these emerging system behaviors. This type of inference is based on large numbers of simulations, i.e., replicate experiments with the ABM using the same assumptions and parameters, and different experiments over which assumptions or parameter values are systematically changed. Simulations of ABMs essentially always yield different outcomes, because movements, actions and interactions of agents with each other or with the environment are stochastic events. The inference of rules from simulation results is an abductive process (Voit 2019) that is challenging, because one can easily demonstrate that different rule sets may lead to the emergence of the same systemic behaviors, and because even numerous simulations seldom cover the entire repertoire of a system’s possible responses. In fact, Hanappi (2017) warned: “assumptions on microagents that play the role of axioms from which the aggregate patterns are derived need not—and indeed never should—be the end of ABM research.”
Arguably the greatest appeal of ABMs, and at the same time a treacherous pitfall, is their enormous flexibility, which is attributatble to the fact that any number of rules can be imposed on the agents, and that the environment may be very simple but can also be exceedingly complicated. For instance, the environment may exhibit gradients or even different individually programmable patches (Barth 2012; Gooding 2019) including importing geographic information systems (GIS) data to define the characteristics of each patch (see Scott (2019) for an example with a detailed explanation of how GIS data are incorporated into an ABM). In particular, the repeated addition of new elements to a model can quickly increase the complexity of the model, thereby possibly distracting from the core drivers of the system’s behavior, obscuring the importance of each of the rules the agents must follow, and generally making interpretations of results more difficult. Related to this option of adding elements is the critique that ABMs can be ‘tuned’ by researchers to create outcomes that support the researcher’s narrative (Gooding 2019). To counteract increases in complexity, some authors have begun to develop methods of model reduction that retain the core features of a model, but eliminate unnecessary details (Zou 2012). Another critique of ABMs is that simulations are not readily reproducible, because the rules can be complicated and stochasticity rarely repeats the same model trajectories. A strategy to increase the reproducibility of ABMs was the establishment of protocols for the standardized design and analysis of ABMs (Lorek & Sonnenschein 1999; Grimm 2006, 2010; Heard 2015).
Support of ABMs from the Scientific Community. Several introductory tutorials describe the features of ABMs, including Bonabeau (2002), Matthews (2007), Macal (2010), Heath (2010), Niazi & Hussain (2011), Heard (2015), and Weimer (2016). Generic reviews of ABMs include Gu & Blackmore (2015), Gooding (2019), Heath (2009), Grimm (2010), and Hanappi (2017). In a slight extension of ABMs, Lattilä (2010) and Cisse (2013) described hybrid simulations involving ABMs and dynamic systems, and Heard (2015) discussed statistical methods of ABM analysis. These introductory tutorials and reviews, however, are typically not designed for undergraduates with limited mathematical or computational modeling experience. The scientific community has also worked hard on facilitating the use of ABMs by offering software like NetLogo, Swarm, RePast, and Mason. Summaries and evaluations of some of the currently pertinent software are available in Berryman (2008) and Abar (2017), and NetLogo is described more fully in Sect. 3.
Software Tools for the Classroom
A variety of software tools can be used to construct, simulate, and analyze ABMs. When ABMs are taught or used in biology or mathematics courses, software should be chosen to align with pedagogical objectives. Note that the pedagogy of ABMs in life science and mathematical modeling courses is discussed in more detail in Sect. 4. In this section, we highlight some of the most used software packages in an educational setting.
EcoBeaker & SimBio. EcoBeaker was developed by Eli Meir and first released in 1996 as software that ran simulated experiments designed to explore ecological principles. In 1998, SimBio (Meir 1998) was founded (then called BeakerWare) for the release of the second version of EcoBeaker and has since grown to include simulated experiments in evolution, cell biology, genetics, and neurobiology. Many of the simulated experiments in the SimBio Virtual Labs software are agent-based simulations. In a SimBio Virtual Lab, the user interacts with a graphical interface portraying the agents (individuals in the experiment) and sometimes their environment, which can be manipulated in various ways for different experimental designs. The graphical interface also contains relevant graphs which are updated as the simulated experiment runs. EcoBeaker and SimBio Virtual Labs are examples of software where the focus is on experimental design and the use of simulation to understand biological concepts. The user never interfaces with the code and does not need to understand the underlying algorithms which produce the simulation. SimBio Virtual Labs are used in many biology classrooms across the United States. The software is priced per student (currently $6/lab/student or $49/student for unlimited labs).
NetLogo. The agent-based modeling environment NetLogo (Wilensky 1999) was developed by Uri Wilensky and first released in 1999. NetLogo is free and continues to be improved and updated (the software is currently on version 6). Additionally, a simplified version of NetLogo can be run through a Web browser at http://netlogoweb.org/. NetLogo is a programming platform allowing the implementation of any ABM a user might design. As such, its user interface includes a tab where ABM code is written in the NetLogo programming language, and a tab for the user to view a visualization of the ABM and user specified outputs as the ABM is simulated. Textbooks by Railsback & Grimm (2012) and Wilensky & Rand (2015) provide introductions to the NetLogo prgramming languages as well as providing a thorough overview of the algorithmic structures of ABMs.
Since its initial development, NetLogo has built up an extensive model library of ABMs. Additionally, over the years, faculty at various institutions have developed ABM modules through NetLogo that allow students to explore a variety of biological phenomenon. For example, the Virtual Biology Lab (Jones 2016) has created 20 different virtual laboratory modules using NetLogo through a web browser for exploring topics in ecology, evolution, and cell biology. The Virtual Biology Labs are similar in scope to the EcoBeaker and SimBio labs. Another example is Infections On NeTWorks (IONTW) which provides an ABM framework and teaching modules for examining aspects of disease dynamics on various network structures (Just 2015a, b, c).
Pedagogy of ABMs in Life Science & Math Modeling Courses
One of the responses to the Bio2010 Report (2003) has been a push to create biocalculus courses or to insert more biological application examples and projects within traditional calculus courses. Indeed, studies have shown that including applications from the life sciences in classic math courses like calculus leads to students gaining equivalent or better conceptual knowledge than from similar courses without life science applications (Comar 2008; Eaton & Highlander 2017). However, many mathematics and biology educators have pointed out that the subset of mathematics applicable to biology extends well beyond calculus, and undergraduates (especially those majoring in biology) should be exposed to a variety of mathematical models and methods of analysis across biology and mathematics courses (Bressoud 2004; Gross 2004; Robeva & Laubenbacher 2009).
The only prerequisites for analyzing the simulations of an ABM are a basic understanding of the underlying biology and, in some instances, knowledge of how to perform basic statistical calculations or generate graphical representations of results. Many pre-built ABMs in SimBio Virtual Labs and NetLogo generate relevant graphs and/or produce spreadsheets or text files of the relevant data. Thus, a student can utilize ABMs in life science courses without having to learn how to implement (by writing code) an ABM.
The prerequisites for learning how to implement ABMs are not as extensive as for other forms of mathematical models. The most essential prerequisite is some exposure to the fundamentals of computer programming such as understanding loop structures and conditional statements, implementing stochastic processes, and understanding how the order of executed operations within a program impacts the program’s output. Agent-based modeling software like NetLogo (Wilensky 1999) keeps model implementation and analysis relatively simple by providing built-in model visualization tools and automatically randomizing the order in which agents execute programmed operations.
Using ABMs in Life Science Courses
Due to their appealing visual representation, ABMs can easily be used in the classroom to demonstrate biological processes ranging from chemical reactions to interactions among species, as if the model were simply an animation. However, using ABMs in this way is much like using a cell phone to hammer nails (q.v., Theobald (2004)): It may work in the desired fashion, but represents an utter waste of the tool’s real potential. Adding just one more step, the collection of model-generated data, transforms a passive learning experience into an active one. Students can be asked to calculate means and variances, graph relationships between variables, discuss the sample size needed for reliable results, and generate quantitative predictions under different hypotheses, all in the context of a specific biological question. This can be done even in a large classroom with only a single, instructor-controlled computer, bridging the gap between lecture and lab.
If students have access to individual computers, much more is possible. Either individually or in small groups, students can use ABMs to collect and analyze their own data. Free file-sharing resources such as GoogleDocs make it easy to pool data across many groups, thereby crowd-sourcing problems that would be too large for any one group to handle on their own. In smaller classes and lab sections, individuals or groups can be assigned to model different scenarios (e.g., the interaction effects between different parameters), prompting discussions of the most appropriate parameter values and model settings. Such models can even be extended into miniature research projects. For example, in a unit on community ecology, students might be assigned a question about two interacting species, then use online resources to find relevant information and parameter estimates, design and conduct a series of model runs, analyze their data using simple statistical techniques, and present their findings to the class.
Although ABMs can be used to simulate almost any biological process, meaningful exploration of a model typically requires a substantial commitment of class time and instructor engagement. As a result, except in modeling courses, it is seldom practical to incorporate ABMs into every lesson plan. In our experience, their educational value is highest for studying processes involving (a) substantial amounts of stochasticity, (b) nonlinear interactions, and/or (c) a defined spatial structure.
Using ABMs in Math Modeling Courses
The inclusion of ABMs in math courses generally comes in two different modes: (1) ABMs are taught as one modeling technique in a course covering multiple modeling techniques, and (2) the construction and analysis of ABMs are taught in a course where ABMs are the only type of modeling being used. However, due to the minimal prerequisites for learning agent-based modeling, both types of courses can potentially be offered with one or no prerequisite courses. Bodine (2018) provides an example of a course where ABMs are taught as one type of discrete-time modeling technique in an undergraduate course designed for biology, mathematics, biomathematics, and environmental science majors that has no prerequisites beyond high-school algebra. An example of a course where ABMs are the only type of modeling being used is given by Bodine (2019); this course has only a single prerequisite: either a prior math modeling course which introduces basic computer programming or an introduction to computer science course.
The use of biological applications in teaching mathematical modeling (including modeling with ABMs) is often viewed as having a lower entry point with less new vocabulary overhead than other types of applications (e.g., those from physics, chemistry, or economics). In particular, models of population dynamics, even those involving interactions between multiple subpopulations or different species, usually do not require any new vocabulary for most students, which allows for a more immediate focus on mechanisms of population change and impact of interactions between individuals within the populations.
Challenges & Best Practices for Courses Using/Teaching ABMs
Video Game vs. Scientific Process. In our experience, students sometimes react to an ABM’s many controls and visual output by treating the model as a video game, clicking buttons at random to see what entertaining patterns they can create. Other students prefer to complete the exercise as quickly as possible by blindly following the prescribed instructions. Neither of these approaches substantially engages students in thinking about the question(s) underlying the model, different strategies for collecting and analyzing data, or the model’s limitations.
To foster these higher-order cognitive skills, use of the ABM should be explicitly framed as an example of the scientific process. This approach begins with a set of initial observations and a specific biological question. For example, what management practices would be most effective in controlling the invasive brown tree snake? After familiarizing themselves with the biological system, students propose hypotheses about the factors contributing to the snake’s success on Guam, suggest management strategies, and set model parameters that reflect their chosen strategy. Finally, they run the model multiple times to collect data that allow them to measure their strategy’s effectiveness. Under this pedagogical approach, ABMs become a vehicle for designing and conducting a miniature research project, enabling experiments that would not otherwise be practical due to cost, logistics, or ethical considerations. The modeling exercise can also reinforce lessons on how scientific knowledge is constructed and tested (e.g., the three P’s of science, namely, problem posing, problem solving, and peer persuasion (Watkins 1992)).
As part of this exercise, students should engage in the process of deciding how best to collect, analyze, and present their data. For example, as part of the brown tree snake project, students might be asked to explore the practical steps that other Pacific islands could take to prevent invasions or eradicate invaders. One group of students decides to focus on two different control measures: cargo checks of all inbound flights and deployment of poison baits around airstrips. Following an overview of different statistical approaches, the students determine that a multiple regression analysis would best allow them to address their question. Allowed only a limited ‘budget’ of 30 model runs, students settle on a factorial design using three treatment levels of cargo checks, three levels of baiting, and three replicates of each combination. The students set up a spreadsheet to record the data from each model run, graph their data in a scatter plot, and use software such as Microsoft Excel’s Analysis ToolPak to conduct their analysis.
A Model as a Caricature of the Real World. Students at early stages of their academic career often envision science as a collection of factual information and fixed procedures. Students with this mindset may dismiss as useless any model that does not incorporate every detail of a particular biological system. By contrast, scientists recognize that models, whether mathematical, physical, or conceptual, are deliberate simplifications that attempt to capture certain properties of a biological system while ignoring others (Dahlquist 2017). For example, the standard epidemiological SIR model (diagrammed in Fig. 1a) divides a population in three subpopulations (susceptible, infectious, and removed) while ignoring any potential heterogeneity within each subpopulation (e.g., age, treatment status, groups at higher risk for transmission).
Fig. 1.
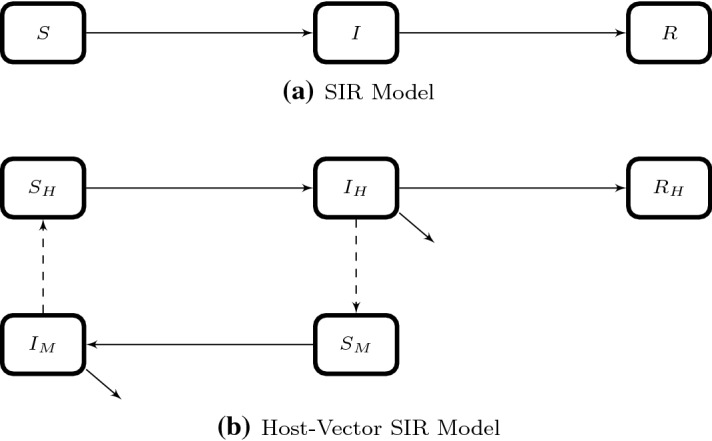
Diagram of compartmental models of disease dynamics where S, I, and R, susceptible, infectious, and recovered, respectively, while the subscripts H and M represent humans and mosquitoes, respectively
Students will need to engage in activities that frame the ABM as a hypothesis about the organization and function of a specific biological system (Weisstein 2011). After a description (and possible exploration) of the basic model, students can work in groups to suggest additional processes and variables that seem relevant to understanding the system. They can then choose one or two of the factors that they consider most important to addressing the question being asked. Finally, they should consider how to modify the model to incorporate the chosen features. For example, a standard epidemiological model divides the host population into susceptible, infectious, and removed subpopulations, and studies the movement of individuals among these subpopulations (Fig. 1a). A group of students decides to modify this model to track a malarial epidemic. After discussing mortality rates, prevention and treatment options, and genetic and age-related variation in host susceptibility, the students decide to focus on incorporating vector transmission into their model. Through guided discussion with the instructor, they realize that transmission now occurs in two directions: from infected vectors to susceptible hosts and from infected hosts to uninfected vectors. They therefore develop a schematic model (Fig. 1b) that depicts these revised rules for each agent in the ABM. Even if the students do not actually build the corresponding computational model, this exercise in extending a model to reflect specific biological assumptions helps students understand the iterative process by which models are developed and the utility of even simple models to clarify key features of the system’s behavior.
Algorithms vs. Equations. The concept of an equation is introduced fairly early in mathematics education. In the United States, children can encounter simple algebraic equations in elementary school (Common Core 2019) and then continue to see increasingly complex equations in math classes through college. Because of this long exposure to equations, the use of functions and systems of equations to model systems in the natural world feels “natural” or logical to students when they first encounter differential equation models or matrix models. ABMs, on the other hand, can seem confusing to students because they lack the ability to be expressed as an equation or set of equations. An ABM is constructed as an algorithm describing when and how each agent interacts with their local environment (which may include other agents). Often these interactions are governed by stochastic processes, and thus “decisions” by agents are made through the generation of random numbers. When first introducing students to ABMs, it can be helpful to teach students how to read and construct computer program flowcharts and to create a visual representation of what is occurring within an algorithm or portion of an algorithm (see Bodine (2019) for example assignments that utilize program flowchart construction). In life science classes where ABMs are being analyzed but not constructed and implemented, a flow diagram can be a useful tool for conveying the order processes occur in the model. Class discussions can question whether the order of processes make biological sense, and whether there are alternatives. In math modeling classes, the construction of flowcharts, even for simple ABMs, can help students elucidate where decision points are within the code, and what procedures are repeated through loop structures. The construction of flowcharts as students progress to more complicated ABMs can help students rectify the order of events in their implemented algorithm against the order in which events should be occurring biologically. Whether students are working with ABMs in life science or math modeling classes, it is helpful for them to learn how to read and understand flow diagrams as they are often included in research publications that use agent-based modeling.
Describing an ABM. Much to the alarm of many math students beginning to develop ABMs, the formal description of an ABM requires more than just writing the computer code. The standard method for describing ABMs in scientific publications, referred to as the Overview, Design Concepts, and Details (ODD) Protocol (Grimm 2006, 2010; Railsback & Grimm 2012), often requires more than one page to fully describe even a simple ABM. This can make ABMs seem overwhelming to students as they first begin to explore ABMs. In courses which teach the implementation and description of ABMs, instructors should take care not to introduce the computer code implementation simultaneous to the model description via the ODD protocol. Note that the introductory text on agent-based modeling by Railsback & Grimm (2012) does not introduce the concept of the ODD protocol until Chapter 3, which comes after the introduction and implementation (in NetLogo) of a simple ABM in Chapter 2. In the course materials by Bodine (2019), the concept of the ODD protocol is introduced prior to Project 2, but the students are not required to write their own ODD Protocol description until the final project, once they have seen ODD descriptions for multiple models.
Model Implementation vs. the Modeling Cycle. Courses that aim to teach methods in mathematical modeling often start with a discussion of the modeling cycle, which is typically presented as a flow diagram showing the loop of taking a real world question, representing it as a mathematical model, analyzing the model to address the question, and then using the results to ask the next question or refine the original real world question. Figure 2 shows an example of a modeling cycle diagram. In courses where the mathematical models are encapsulated in one or a small handful of equations, the time spent on representing the real world as a mathematical model (the green box in Fig. 2) is relatively short. The construction of ABMs, however, can be a fairly lengthy process, as ABMs are designed to simulate interactions between individuals and the local environment. When students are in the middle of constructing their first few ABMs, they often lose sight of where they are in the modeling cycle because the model implementation becomes a cycle of its own; a cycle of writing bits of code, testing the code to see if it runs and produces reasonable results, and repeating this process to slowly add all the components needed for the full algorithm of the ABM. As students are first learning agent-based modeling, they need to be reminded often to pull back and view where they are in the modeling cycle; to see the flock for the boids, as it were.
Fig. 2.
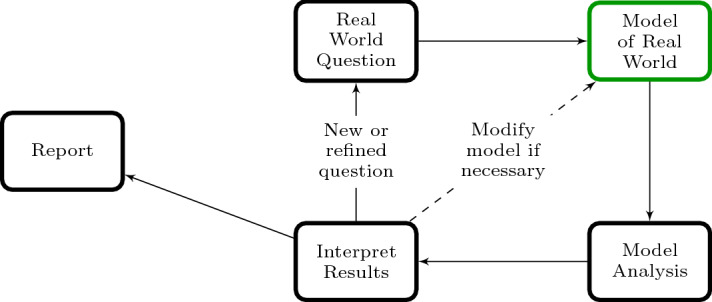
Diagram of the modeling cycle. The time spent constructing and numerically implementing the model of the real world (green node) can take more time if the model is an ABM than compared to other types of mathematical models (Color figure online)
Model Validation. Within the modeling cycle, there is a smaller cycle of model validation (see dashed line in Fig. 2). In a course where students are first introduced to the classic Lotka–Volterra predator-prey model, the students are usually first introduced to a predator-prey data set (like the 200-year data set of Canadian lynx and snowshoe hare pelts purchased by the Hudson Bay Company (MacLulich 1937; Elton & Nicholson 1942)), which shows the oscillating population densities of the predator and prey populations. When the students then simulate the Lotka–Volterra model for various parameter sets, they find that they are able to produce the same oscillating behavior of the predator and prey populations. This is a form of model validation, and a model which did not display this distinctive trend seen in the data would be considered invalid for that data set. A similar process must occur for validating ABMs against observed biological patterns. However, in order to engage in this validation process for ABMs, students must first understand how decisions of individual agents and interactions between neighboring agents can lead to system-level observable patterns, a phenomenon referred to as “emergence” or the “emergent properties” of an ABM. The classic ABM example for easily identifying an emergent property is a flocking example (a stock example of flocking exists in the NetLogo models library, and is explored in Chapter 8 of the ABM textbook by Railsback & Grimm (2012)).
The concept of an emergent property can take a little while for students to fully understand. By definition, it is an observable outcome that the system was not specifically programmed to produced. In particular, it is not a summation of individual-level characteristics, and is typically not easily predicted from the behaviors and characteristics of the agents. For example, a variation of the Reynolds (1987) flocking model is included in the NetLogo Library and is explored in Railsback & Grimm (2012, Chapter 8). In the model, each agent moves based on three rules:
Separate: Maintain a minimum distance from nearby agents
Align: Move in the same direction as nearby agents
Cohere: Move closer to nearby agents
where all agents move at the same speed and different schemes for determining who is a nearby agent can be used. Additionally, there are model parameters for the minimum distance to be maintained between agents, and the degree to which an agent can turn left or right in a single time step in order to align, cohere, and separate. It is not immediately evident from this set of rules that individual agents might be able to form flocks (or swarm together), and indeed that system-level behavior does not emerge for all parameter sets. However, certain parameter sets do lead to the agents forming one or more flocks that move together through the model landscape.
Students learning agent-based modeling will likely need multiple examples of ABMs with emergent properties in order to understand the concept enough to identify emergent properties on their own. A few other examples to consider from the NetLogo library are:
The emergence of synchronized flashing in a population of fireflies (Fireflies Model).
The tipping point forest density at which a forest fire burns the majority of the forest (Fire Model).
The emergence of population oscillations in linked predator and prey populations (Wolf Sheep Predation Model).
Observations: Algorithms for Pattern Recognition. One of the most exciting moments for students when they first begin running simulations of an ABM is seeing system-level patterns emerge before their eyes. One of the challenges for students (and agent-based modelers, in general) is to develop algorithms to identify and/or quantify the patterns we can easily identify by sight. For example, in the flocking ABM discussed above, an observer watching the locations of individual agents change at each time step can easily see the formation of flocks. If however, the observer wanted to systematically explore the parameter space of the flocking ABM and determine the regions of the parameter space under which flocking occurs (a process which might involve running hundreds or thousands of simulations), it would be a tedious and time-consuming task to physically watch each simulation and record whether the agents formed a flock or not. Instead, the observer must choose the criteria that indicate the formation of a flock, and then determine a measure or an algorithm for determining whether a flock (or multiple flocks) have formed. In a course designed to teach the construction and analysis of ABMs, this is a point where students should be encouraged to be both creative and methodical about developing such measures and algorithms. The development of these observational measures and algorithms also provides a great opportunity for collaboration between students. It is especially helpful if students with a diversity of academic backgrounds can be brought together to brainstorm ideas; for instance, mixing students with various levels of exposure in mathematics, computer science, and biology can be very beneficial.
Conclusions
Rapid advances in computing power over the past decades have made agent-based modeling a feasible and appealing tool to study biological systems. In undergraduate mathematical biology education, there are multiple modes by which ABMs are utilized and taught in the classroom. In biology classrooms, ABMs can be used to engage students in hypothesis testing and in the experimental design and data collection processes of otherwise infeasible experiments, and to enable students to utilize models as a part of the scientific process. All of this can be done without students having to learn a programming language. By contrast, students who have had some exposure to computer programming can learn the construction, implementation, and analysis of agent-based models in a math or computer science modeling class. Biological applications are ideal systems for first attempts at agent-based models as they typically do not necessitate learning extensive new vocabulary and theory to understand the basic components that need to be included in the model. Throughout this article, we endeavored to articulate the benefits and challenges of including ABMs in undergraduate life science and math modeling courses.
Consistent with the Bio2010 Report (2003), we recommend that undergraduate biology and life science curricula be structured to ensure that all students have some exposure to mathematical modeling. We additionally recommend that this includes agent-based modeling. While not every student necessarily needs to take a course exclusively focused on agent-based modeling, every undergraduate biology student should have the opportunity to utilize an ABM to perform experiments and to collect and analyze data. As we educate the next-generation of life scientists, let us empower them with the ability to utilize ABMs to simulate and better understand our complex and dynamic world.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Erin N. Bodine, Email: bodinee@rhodes.edu
Robert M. Panoff, Email: rpanoff@shodor.org
Eberhard O. Voit, Email: eberhard.voit@bme.gatech.edu
Anton E. Weisstein, Email: weisstae@truman.edu
References
- Abar S, Theodoropoulos GK, Lemarinier P, O’Hare GMP. Agent based modelling and simulation tools: A review of the state-of-art software. Comp Sci Rev. 2017;24:13–33. doi: 10.1016/j.cosrev.2017.03.001. [DOI] [Google Scholar]
- An In silico experiments of existing and hypotheticalcytokine-directed clinical trials using agent-based modeling. Crit Care Med. 2004;32(10):2050–2060. doi: 10.1097/01.CCM.0000139707.13729.7D. [DOI] [PubMed] [Google Scholar]
- An G, Mi Q, Dutta-Moscato J, Vodovotz Y. Agent-based models in translational systems biology. WIREs Syst Biol Med. 2009;1(2):159–171. doi: 10.1002/wsbm.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell R, Axelrod R, Epstein JM, Cohen MD. Aligning simulation models: a case study and results. Comput Math Organ Theory. 1996;1(2):123–141. doi: 10.1007/BF01299065. [DOI] [Google Scholar]
- Ayalew M, Hylton D, Sistrunk J, Melton J, Johnson K, Voit EO (2020) Integration of biology, mathematics and computing in the classroom through the creation and repeated use of transdisciplinary modules. PRIMUS, In Revision [DOI] [PMC free article] [PubMed]
- Bae JW, Paik E, Kim K, Singh K, Sajjad M. Combining microsimulation and agent-based model for micro-level population dynamics. Procedia Comput Sci. 2016;80:507–517. doi: 10.1016/j.procs.2016.05.331. [DOI] [Google Scholar]
- Bailey AM, Thorne BC, Peirce SM. Multi-cell agent-based simulation of the microvasculature to study the dynamics of circulating inflammatory cell trafficking. Ann Biomed Eng. 2007;35:916–936. doi: 10.1007/s10439-007-9266-1. [DOI] [PubMed] [Google Scholar]
- Barth R, Meyer M, Spitzner J. Typical pitfalls of simulation modeling-lessons learned from armed forces and business. J Artif Soc S. 2012;15(2):5. doi: 10.18564/jasss.1935. [DOI] [Google Scholar]
- Berry BJL, Kiel LD, Elliott E. Adaptive agents, intelligence, and emergent human organization: capturing complexity through agent-based modeling. Proc Natl Acad Sci USA. 2002;99:7187–7188. doi: 10.1073/pnas.092078899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryman M (2008) Review of software platforms for agent based models. Land Operations Division DSTO, Defence Science and Technology Organisation, PO Box 1500, Edinburgh South Australia 5111, Australia
- Bonabeau E. Agent-based modeling: Methods and techniques for simulating human systems. Proc Natl Acad Sci USA. 2002;99(Suppl 3):7280–7287. doi: 10.1073/pnas.082080899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet F, Le Page C. Multi-agent simulations and ecosystem management: a review. Ecol Model. 2004;176(3):313–332. doi: 10.1016/j.ecolmodel.2004.01.011. [DOI] [Google Scholar]
- Bressoud DM. The changing face of calculus: first- and second- semester calculus as college courses. MAA Focus. 2004;11:14–16. [Google Scholar]
- Bodine EN, Lenhart S, Gross LJ. Mathematics for the life sciences. Princeton: Princeton University Press; 2014. [Google Scholar]
- Bodine EN. Discrete math modeling with biological applications (course materials) QUBES Edu Resour. 2018 doi: 10.25334/Q42T54. [DOI] [Google Scholar]
- Bodine EN. Agent-based modeling course materials. QUBES Edu Resour. 2019 doi: 10.25334/Q4VF0K. [DOI] [Google Scholar]
- Butler J, Cosgrove J, Alden K, Read M, Kumar V, Cucurull-Sanchez L, Timmis J, Coles M. Agent-based modeling in systems pharmacology. CPT Pharm Syst Pharmacol. 2015;4(11):615–629. doi: 10.1002/psp4.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiani A, Godin A, Caverzasi E, Gallegati M, Kinsella S, Stiglitz JE. Agent based-stock flow consistent macroeconomics: towards a benchmark model. J Econ Dyn Control. 2016;69:375–408. doi: 10.1016/j.jedc.2016.06.001. [DOI] [Google Scholar]
- Caplat P, Anand M, Bauch C. Symmetric competition causes population oscillations in an individual-based model of forest dynamics. Ecol Model. 2008;211(3–4):491–500. doi: 10.1016/j.ecolmodel.2007.10.002. [DOI] [Google Scholar]
- Castiglione F, Celada F. Immune system modelling and simulation. Boca Raton: CRC Press; 2015. p. 274. [Google Scholar]
- Chavali AK, Gianchandani EP, Tung KS, Lawrence MB, Peirce SM, Papin JA. Characterizing emergent properties of immunological systems with multicellular rule-based computational modeling. Trends Immunol. 2008;29(12):589–599. doi: 10.1016/j.it.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Cisse PA, Dembele JM, Lo M, Cambier C. Assessing the spatial impact on an agent-based modeling of epidemic control: case of schistosomiasis. Complex Sci. 2013;126:58–69. doi: 10.1007/978-3-319-03473-7_6. [DOI] [Google Scholar]
- Comar TD. The integration of biology into calculus courses. PRIMUS. 2008;18(1):49–70. [Google Scholar]
- Common Core State Standards Initiatives (2019) http://www.corestandards.org/Math/. Accessed 08 Sep 2019
- Dahlquist KM, Aikens ML, Dauer JT, Donovan SS, Eaton CD, Highlander HC, Jenkins KP, Jungck JR, LaMar MD, Ledder G, Mayes RL, Schugart RC. An invitation to modeling: building a community with shared explicit practices. PeerJ Preprints. 2017;5:e3215ve3215v1. doi: 10.7287/peerj.preprints.3215v1. [DOI] [Google Scholar]
- DeAngelis D, Diaz SG. Decision-making in agent-based modeling: a current review and future prospectus. Front Ecol Evol. 2019;6:237. doi: 10.3389/fevo.2018.00237. [DOI] [Google Scholar]
- Eaton CD, Highlander HC. The case for biocalculus: design, retention, and student performance. CBE–Life Sci Educ. 2017;16(2):ar25. doi: 10.1187/cbe.15-04-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton C, Nicholson M. The ten-year cycle in numbers of the lynx in Canada. J Animal Ecol. 1942;11(2):215–244. [Google Scholar]
- Epstein J, Axtell R. Growing artificial societies: social science from the bottom up. Cambridge: MIT Press; 1996. [Google Scholar]
- Fox J, Shin Y, Emamdjomeh A (2020) How epidemics like covid-19 end (and how to end them faster). Washington Post, Feb 19. https://www.washingtonpost.com/graphics/2020/health/coronavirus-how-epidemics-spread-and-end/
- Gardner M. Mathematical Games-The fantastic combinations of John Conway’s new solitaire game “life”. Sci Am. 1970;223(4):120–123. [Google Scholar]
- Geanakoplos J, Axtell R, Farmer DJ, Howitt P, Conlee B, Goldstein J, Hendrey M, Palmer NM, Yang C-Y. Getting at systemic risk via an agent-based model of the housing market. Am Econ Rev. 2012;102(3):53–58. doi: 10.1257/aer.102.3.53. [DOI] [Google Scholar]
- Gilbert N, Troitzsch K. Simulation for the social scientist. Oxford: Oxford University Press; 2005. [Google Scholar]
- Glen CM, Kemp ML, Voit EO. Review: agent-based modeling of morphogenetic systems—advantages and challenges. PLoS Comp Biol. 2019;15(3):e1006577. doi: 10.1371/journal.pcbi.1006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding T (2019) Agent-based model history and development. In: Economics for a fairer society–going back to basics using agent-based models. Palgrave Pivot, Cham, pp 25–36. 10.1007/978-3-030-17020-2
- Graner F, Glazier J. Simulation of biological cell sorting using a two-dimensional extended Potts model. Phys Rev Lett. 1992;69(13):2013–2017. doi: 10.1103/PhysRevLett.69.2013. [DOI] [PubMed] [Google Scholar]
- Grant MR, Mostov KE, Tlsty TD, Hunt CA. Simulating properties of in vitro epithelial cell morphogenesis. PLoS Comput Biol. 2006;2(10):e129. doi: 10.1371/journal.pcbi.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm V, Berger U, Bastiansen F, Eliassen S, Ginot V, Giske J, Goss-Custard J, Grand T, Heinz S, Huse G, Huth A, Jepsen JU, Jørgensen C, Mooij WM, Müller B, Pe’er G, Piou C, Railsback SF, Robbins AM, Robbins MM, Rossmanith E, Rüger N, Strand E, Souissi S, Stillman RA, Vabø R, Visser U, DeAngelis DL. A standard protocol for describing individual-based and agent-based models. Ecol Model. 2006;198(1–2):115–126. doi: 10.1016/j.ecolmodel.2006.04.023. [DOI] [Google Scholar]
- Grimm V, Berger U, DeAngelis DL, Polhill JG, Giske J, Railsback SF. The ODD protocol: a review and update. Ecol Model. 2010;221(23):2760–2768. doi: 10.1016/j.ecolmodel.2010.08.019. [DOI] [Google Scholar]
- Grimm V, Railsback SF. Individual-based modeling and ecology. Princeton: Princeton University Press; 2005. p. 485. [Google Scholar]
- Gross LJ. Interdisciplinarity and the undergraduate biology curriculum: finding a balance. Cell Biol Educ. 2004;3(2):85–87. doi: 10.1187/cbe.04-03-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Blackmore KL. A systematic review of agent-based modelling and simulation applications in the higher education domain. High Educ Res Dev. 2015;34(5):883–898. doi: 10.1080/07294360.2015.1011088. [DOI] [Google Scholar]
- Hanappi G. Agent-based modelling. History, essence, future. PSL Quart Rev. 2017;70(283):449–472. doi: 10.13133/2037-3643\_70.283\_3. [DOI] [Google Scholar]
- Heath BL (2010) The history, philosophy, and practive of agent-based modeling and the development of the conceptual model for simulation diagram. Doctoral Dissertation, Wright State University, Dayton, OH
- Heath B, Hill R, Ciarallo F. A survey of agent-based modeling practices (January 1998 to July 2008) J Artif Soc Sci. 2009;12(4):9. [Google Scholar]
- Heard D, Dent G, Schifeling T, Banks D. Agent-based models and microsimulation. Annu Rev Stat Appl. 2015;2:259–272. doi: 10.1146/annurev-statistics-010814-020218. [DOI] [Google Scholar]
- Hellweger FL, Clegg RJ, Clark JR, Plugge CM, Kreft J-U. Advancing microbial sciences by individual-based modelling. Nat Rev Microbiol. 2016;14:461–471. doi: 10.1038/nrmicro.2016.62. [DOI] [PubMed] [Google Scholar]
- Hirashima T, Rens EG, Merks RMH. Cellular Potts modeling of complex multicellular behaviors in tissue morphogenesis. Dev Growth Differ. 2017;59(5):329–339. doi: 10.1111/dgd.12358. [DOI] [PubMed] [Google Scholar]
- Jones, TC (2016) Virtual Biology Lab: an Inquiry-Based Learning Environment. http://virtualbiologylab.org/
- Just W, Callendar H, LaMar D (2015) IONTW https://qubeshub.org/iontw
- Just W, Callender H, LaMar MD, Toporikova N (2015) Transmission of Infectious Diseases: Data, Models, and Simulations. In: Algebraic and Discrete Mathematica Methods for Modern Biology. Academic Press, New York, pp 193–215. 10.1016/B978-0-12-801213-0.00008-3
- Just W, Callender H, LaMar MD (2015) Disease Transmission dynamics on networks: network structure versus disease dynamics. In: Algebraic and discrete mathematica methods for modern biology. Academic Press, New York, pp 193–215. 10.1016/B978-0-12-801213-0.00009-599
- Kauffman SA. The origins of order: self organization and selection in evolution. New York: Oxford University Press; 1993. [Google Scholar]
- Kottonau J. An interactive computer model for improved student understanding of random particle motion and osmosis. J Chem Educ. 2011;88(6):772–775. doi: 10.1021/ed100088h. [DOI] [Google Scholar]
- Lardon LA, Merkey BV, Martins S, Dötsch A, Picioreanu C, Kreft JU, Smets BF. iDynoMiCS: next-generation individual-based modelling of biofilms. Environ Microbiol. 2011;13(9):2416–2434. doi: 10.1111/j.1462-2920.2011.02414.x. [DOI] [PubMed] [Google Scholar]
- Lattilä L, Hilletofth P, Bishan L. Hybrid simulation models–when, why, how? Expert Syst Appl. 2010;37:7969–7975. [Google Scholar]
- Lorek H, Sonnenschein M. Modelling and simulation software to support individual-oriented ecological modelling. Ecol Model. 1999;115:199–216. [Google Scholar]
- Macal CM, North MJ. Tutorial on agent-based modelling and simulation. J Simulat. 2010;4(3):151–162. [Google Scholar]
- MacLulich DA. Fluctuations in the numbers of the varying hare (Lepus americanus) Toronto: University of Toronto Press; 1937. [Google Scholar]
- Matthews RB, Gilbert NG, Roach A, Polhill JG, Gotts NM. Agent-based land-use models: a review of applications. Landscape Ecol. 2007;22(10):1447–1459. [Google Scholar]
- Meir E (1998) SimBio Virtual Labs. https://simbio.com/. SimBio, Missoula, MT
- Metropolis N. The beginning of the Monte Carlo method. Los Alamos Sci Spec Issue. 1987;15:125–130. [Google Scholar]
- Metropolis N, Ulam S. The Monte Carlo method. J Am Stat Assoc. 1949;44:335–341. doi: 10.1080/01621459.1949.10483310. [DOI] [PubMed] [Google Scholar]
- Mniszewski SM, Del Valle SY (2013) EpiSimS: large-scale agent-based modeling of the spread of disease. Technical report, Los Alamos National Laboratory (LANL)
- National Research Council . BIO2010: transforming undergraduate education for future research biologists. Washington: The National Academies Press; 2003. [PubMed] [Google Scholar]
- Nelson R, Winter S. An evolutionary theory of economic change. Cambridge: The Belknap Press of Harvard University Press; 1982. [Google Scholar]
- Niazi M, Hussain A. Agent-based computing from multi-agent systems to agent-based models: a visual survey. Scientometrics. 2011;88:497. doi: 10.1007/s11192-011-0468-9. [DOI] [Google Scholar]
- Palmer RG, Arthur WB, Holland JH, LeBaron B, Tayler P. Artificial economic life: a simple model of a stockmarket. Physica D. 1994;75(1–3):264–274. doi: 10.1016/0167-2789(94)90287-9. [DOI] [Google Scholar]
- Persky J. Retrospectives: the ethology of homo economicus. J Econ Perspect. 1995;9(2):221–231. [Google Scholar]
- Perez L, Dragicevic S. An agent-based approach for modeling dynamics of contagious disease spread. Int J Health Geogr. 2009;8:50. doi: 10.1186/1476-072X-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder JP. An active learning exercise for introducing agent-based modeling. Decision Sci J Innovative Educ. 2013;11(3):221–232. doi: 10.1111/dsji.12010. [DOI] [Google Scholar]
- Railsback SF, Grimm V. Agent-based and individual-based modeling: a practical introduction. Princeton: Princeton University Press; 2012. [Google Scholar]
- Reynolds C. Flocks, herds and schools: a distributed behaviour model. Comput Gr. 1987;21(4):25–34. [Google Scholar]
- Robertson SH, Smith CK, Langhans AL, McLinden SE, Oberhardt MA, Jakab KR, Dzamba B, DeSimone DW, Papin JA, Peirce SM. Multiscale computational analysis of Xenopus laevis morphogenesis reveals key insights of systems-level behavior. BMC Syst Biol. 2007;1:46. doi: 10.1186/1752-0509-1-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robeva R, Laubenbacher R. Mathematical biology education: beyond calculus. Science. 2009;325:542–543. doi: 10.1126/science.1176016. [DOI] [PubMed] [Google Scholar]
- Segovia-Juarez JL, Ganguli S, Kirschner D. Identifying control mechanisms of granuloma formation during M. tuberculosis infection using an agent-based model. J Theor Biol. 2004;231:357–376. doi: 10.1016/j.jtbi.2004.06.031. [DOI] [PubMed] [Google Scholar]
- Schelling TC. Dynamic models of segregation. J Math Sociol. 1971;1(2):143–186. [Google Scholar]
- Scott SM, Middleton CE, Bodine EN. An agent-based model of the spatial distribution and density of the Santa Cruz Island Fox. Integrated population biology and modeling Part B. Oxford: North-Holland; 2019. pp. 3–32. [Google Scholar]
- Shiflet AB, Shiflet GW, Sanders WE., Jr Undergraduate module on computational modeling: introducing modeling the cane toad invasion. Procedia Comput Sci. 2013;18:1429–1435. [Google Scholar]
- Smith JM. Evolution and the theory of games. Cambridge: Cambridge University Press; 1982. [Google Scholar]
- Stevens H (2020) Why outbreaks like coronavirus spread exponentially, and how to “flatten the curve”. Washington Post, Mar 14. https://www.washingtonpost.com/graphics/2020/world/corona-simulator/
- Tang J, Enderling H, Becker-Weimann S, Pham C, Polyzos A, Chen C, Costes S. Phenotypic transition maps of 3D breast acini obtained by imaging-guided agent-based modeling. Integr Biol. 2011;3(4):408–421. doi: 10.1039/c0ib00092b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald D (2004) 29+ Evidences for Macroevolution, Part 2: Past History. http://www.talkorigins.org/faqs/comdesc/section2.html
- Thomas P. Intrinsic and extrinsic noise of gene expression in lineage trees. Sci Rep-UK. 2019;9(1):474. doi: 10.1038/s41598-018-35927-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne BC, Bailey AM, DeSimone DW, Peirce SM. Agent-based modeling of multicell morphogenic processes during development. Birth Defects Res C Embryo Today. 2007;81:344–353. doi: 10.1002/bdrc.20106. [DOI] [PubMed] [Google Scholar]
- Tracy M, Cerdá M, Keyes KM. Agent-based modeling in public health: current applications and future directions. Ann Rev Pub Health. 2018;39:77–94. doi: 10.1146/annurev-publhealth-040617-014317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triulzi G, Pyka A. Learning by modeling: insights from an agent-based model of university-industry relationships. Cybernet Syst. 2011;42(7):484–501. [Google Scholar]
- Turing AM. Intelligent machinery a heretical theory. Philos Math. 1936;4(3):256. [Google Scholar]
- Ulam S, Richtrnyer RD, von Neumann J (1947) Statistical methods in neutron diffusion. Alamos Scientific Laboratory Report LAMS-55 1
- Ulam S. Random processes and transformations. Proc Int Congr Math. 1950;2:264–275. [Google Scholar]
- van Liedekerke P, Palm MM, Jagiella N, Drasdo D. Simulating tissue mechanics with agent-based models: concepts, perspectives and some novel results. Comp Part Mech. 2015;2:401–444. [Google Scholar]
- Voit EO. Perspective: dimensions of the scientific method. PLoS Comp Biol. 2019;15(9):e1007279. doi: 10.1371/journal.pcbi.1007279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit EO. Very short introduction: systems biology. Oxford: Oxford University Press; 2020. [Google Scholar]
- von Neumann J, Morgenstern O. Theory of games and economic behaviour. Princeton: Princeton University Press; 1944. [Google Scholar]
- Voss-Böhme A. Multi-scale modeling in morphogenesis: a critical analysis of the cellular potts model. PLoS One. 2012;7(9):e42852. doi: 10.1371/journal.pone.0042852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins BT. Teaching students to think like scientists: software enables experiments to be conducted that would be impossible in a laboratory. Washington: Chronicle of Higher Education; 1992. [Google Scholar]
- Wang Z, Butner JD, Kerketta R, Cristini V, Deisboeck TS. Simulating cancer growth with multiscale agent-based modeling. Semin Cancer Biol. 2015;30:70–78. doi: 10.1016/j.semcancer.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisstein AE. Building mathematical models and biological insights in an introductory biology course. Math Model Nat Phenom. 2011;6(6):198–214. doi: 10.1051/mmnp/20116610. [DOI] [Google Scholar]
- Wilensky U (1999) NetLogo. http://ccl.northwestern.edu/netlogo/. Center for Connected Learning and Computer-Based Modeling, Northwestern University, Evanston, IL
- Wilensky U, Rand W. An introduction to agent-based modeling: modeling natural, social, and engineers complex systems with NetLogo. Cambridge: MIT Press; 2015. [Google Scholar]
- Weimer CW, Miller JO, Hill RR (2016) Agent-based modeling: an introduction and primer. In: Proceedings IEEE 2016 winter simulation conference, Washington, DC, December 11–14, 2016. 10.1109/WSC.2016.7822080
- Wolfram S. A new kind of science. Champaign: Wolfram Media; 2002. [Google Scholar]
- Zou Y, Fonoberov VA, Fonoberova M, Mezic I, Kevrekidis IG. Model reduction for agent-based social simulation: coarse-graining a civil violence model. Phys Rev E. 2012;85(6):066106. doi: 10.1103/PhysRevE.85.066106. [DOI] [PubMed] [Google Scholar]


