Abstract
Using surgically resected tissue, we identified characteristic metabolites related to the diagnosis and malignant status of clear cell renal cell carcinoma (ccRCC). Specifically, we quantified these metabolites in urine samples to evaluate their potential as clinically useful noninvasive biomarkers of ccRCC. Between January 2016 and August 2018, we collected urine samples from 87 patients who had pathologically diagnosed ccRCC and from 60 controls who were patients with benign urological conditions. Metabolite concentrations in urine samples were investigated using liquid chromatography‐mass spectrometry with an internal standard and adjustment based on urinary creatinine levels. We analyzed the association between metabolite concentration and predictability of diagnosis and of malignant status by multiple logistic regression and receiver operating characteristic (ROC) curves to establish ccRCC predictive models. Of the 47 metabolites identified in our previous study, we quantified 33 metabolites in the urine samples. Multiple logistic regression analysis revealed 5 metabolites (l‐glutamic acid, lactate, d‐sedoheptulose 7‐phosphate, 2‐hydroxyglutarate, and myoinositol) for a diagnostic predictive model and 4 metabolites (l‐kynurenine, l‐glutamine, fructose 6‐phosphate, and butyrylcarnitine) for a predictive model for clinical stage III/IV. The sensitivity and specificity of the diagnostic predictive model were 93.1% and 95.0%, respectively, yielding an area under the ROC curve (AUC) of 0.966. The sensitivity and specificity of the predictive model for clinical stage were 88.5% and 75.4%, respectively, with an AUC of 0.837. In conclusion, quantitative analysis of urinary metabolites yielded predictive models for diagnosis and malignant status of ccRCC. Urinary metabolites have the potential to be clinically useful noninvasive biomarkers of ccRCC to improve patient outcomes.
Keywords: biomarker, metabolomics, predictive model, renal cell carcinoma, urinary metabolite
Quantitative analysis of urinary metabolites yielded predictive models for diagnosis and malignant status of ccRCC.

1. INTRODUCTION
In our previous studies, we investigated the metabolites of clear cell renal cell carcinoma (ccRCC) using tissue samples assessed with our global metabolomics (G‐Met) protocol. 1 , 2 Pathways associated with these metabolites (such as glutathione, tryptophan and glycolysis) were found to be associated with predictions for diagnosis and malignant status. We also reported that the glycoglycerolipid, carnitine, and tocopherol pathways had the potential for diagnosis and that the tricarboxylic acid (TCA) cycle, nucleotide sugar, and inositol pathways were associated with malignant status. 1 Therefore, we expected that these metabolites in urine samples could be useful noninvasive biomarkers for ccRCC.
Clinically useful biomarkers suitable for renal cell carcinoma (RCC) have not yet been identified. In recent years, biomarkers of various diseases have been sought by comprehensively analyzing metabolites using mass spectrometry. To date, cancer metabolism has been shown to utilize the Warburg effect, and intermediate metabolites have been shown to be associated with effects on cellular metabolism, cell proliferation, and immunosuppression. Accumulation of metabolites associated with cancer progression is expected to provide a biomarker for potential use in diagnosis and treatment. 3
Several studies have used metabolomics to evaluate urine biomarkers of RCC. 4 , 5 , 6 , 7 , 8 Falegan et al 5 reported alterations in the levels of multiple glycolytic and TCA cycle intermediate metabolites in RCC urine. In 2019, Liu et al 4 showed that a panel consisting of 9 metabolites distinguished patients with RCC from the healthy controls with area under the curve (AUC) values of 0.905 for the training dataset and 0.885 for the validation dataset. These previous reports suggested that models that combined multiple metabolites were likely to have better clinical utility for prediction than models that employed a single targeted metabolite.
However, most of the described measurement systems were semi‐quantitative and did not take urine dilution of each sample into account. In addition, many of these reports failed to examine the impact of matrix effects that might result from the presence of urinary protein, impurities, and volatilization of urine metabolites.
In the present study, we employed a quantitative measurement system for urine metabolites with correction using internal standards and urine creatinine, overcoming the problems of past measurement systems. This work permitted us to generate predictive models utilizing multiple urine metabolites; these models are expected to be clinically beneficial for diagnosis and evaluation of the malignant status of ccRCC.
2. MATERIALS AND METHODS
2.1. Design and study population
This study was approved by the institutional review board at Tohoku University Hospital, and study participants provided written informed consent. Patients who underwent radical nephrectomy or partial nephrectomy for clinical diagnosis of RCC at our institution between January 2016 and August 2018, and who were diagnosed pathologically as having ccRCC, were enrolled in this study (ccRCC group). None of these patients had received any neoadjuvant therapy, such as molecularly targeted therapy, before surgery. The control urine samples were obtained from 60 patients who were being treated for benign urological conditions (control group). The control group included patients with primary aldosteronism (N = 33), urinary calculi (N = 11), benign prostatic hyperplasia (N = 8), disorder of urination (N = 5), congenital urological disease (N = 2), renal simple cyst (N = 1), and renal angiomyolipoma (N = 1). All patients in the control group were evaluated for renal lesions by ultrasound examination or computerized tomography and were confirmed to have normal urinalyzes.
For both groups, patients who had a history of another malignant tumor within the preceding 5 y and those undergoing dialysis were excluded from this study. We predicted that urinary metabolites fluctuate after dietary intake. To exclude the effects of diet on urinary metabolites, urine samples were collected in the early morning or before a meal. Postoperative urine samples were collected about 1 wk (range: 6‐8 d) after surgery. The collected urine samples were immediately centrifuged at 1450 g for 10 min at 4°C, and the resulting supernatants were stored at −80°C until time of analysis. Clinical and pathologic tumor node‐metastasis (TNM) staging was performed according to the 7th edition of the American Joint Committee on Cancer (AJCC) staging system. 9 Estimated glomerular filtration rate (eGFR) was calculated using the Japanese Society of Nephrology equation: eGFR (mL/min/1.73 m2) = 194 × Cre − 1.094 × age − 0.28, 10 where Cre was the serum creatinine concentration (in units of mg/dL) and age was in years.
2.2. Chemicals and reagents
Forty‐seven metabolites were evaluated in the present study; these metabolites had been shown previously to be elevated in ccRCC tissue. 1 Standard substance used for the calibration curves and the isotope‐labeled internal standards for quantitative measurement are listed in Appendix S1.
2.3. LC‐MS/MS conditions
All liquid chromatography coupled with tandem mass spectrometry (LC‐MS/MS) analyses were performed using a LC‐MS‐8050 triple‐quadrupole mass spectrometer coupled with a Nexera X2 UHPLC system (Shimadzu) and Lab Solutions software (Shimadzu). For analyzing urinary metabolites, analyses were separated into 4 groups (designated Groups 1‐4) and analyzed with optimized methods to improve the measurement sensitivity for each metabolite. 11
The LC method for each compound and the optimized MS/MS conditions are summarized in Table S1. For all 4 analytical groups, the column oven temperature was set at 40°C and electrospray ionization mode was chosen as the ion source probe. The conditions of the ion source probe were set as follows: probe voltage, 4000 V; desolvation line temperature, 100°C; block heater temperature, 150°C; interface temperature, 400°C; nebulizing gas flow, 2 L/min; drying gas, 3 L/min; and heating gas flow, 17 L/min. Column and mobile phases were selected to optimize the sensitivity for each metabolite; those conditions, along with the preparation of the calibration standards and internal standards for each group, are listed in Appendix S2 and Table S2.
2.4. Sample preparation for LC‐MS/MS
At the time of analysis, 25 µL of each urine sample was combined with an equal volume of a given internal standard working solution 1, 2, or 3, along with 200 µL of acetonitrile, and the mixture was vortexed for 5 s and centrifuged at 15 000 g at 4°C for 5 min. Aliquots (120 µL each) of the supernatant were then transferred to separate 1.5‐mL microcentrifuge tubes (Fisher Scientific, Cat. No. 509‐GRD‐Q) and evaporated under reduced pressure for 1 h.
Separate aliquots from a given sample were reconstituted with 20 µL of either water (for Group 1) or a 75:25 (v:v) mixture of 20 µL acetonitrile: water (for Group 2), and 20 μL of each solution then was injected into the relevant analytical system.
For sample preparation for analytical Group 3, a 200‐µL aliquot of human urine was combined with 25 μL internal standard and 1 mL acetonitrile was added. The sample was mixed by vortexing for 5 s and then centrifuged at 15 000 g at 4°C for 5 min. The resulting supernatant was transferred to a new microcentrifuge tube and evaporated under reduced pressure for 1 h. Each aliquot was reconstituted with 50 µL of water, and 10 μL of the solution then was injected into the analytical system. 11
For sample preparation for analytical Group 4, a 50‐µL aliquot of human urine was combined with 175 μL pure water in a microcentrifuge tube, and 50 μL IS and 225 μL acetonitrile then were added. The sample was mixed by vortexing for 5 s and then centrifuged at 15 000 g at 4°C for 5 min. The supernatant was transferred to a new microcentrifuge tube, and 1 μL then was injected into the analytical system.
2.5. Urine creatinine assay
Urine creatinine concentrations were measured using an enzymatic protocol using the CRE‐CL commercial kit (Serotec Co., Ltd.) for all urine samples. Absorbances were read on an Infinite 200 Pro microplate reader (Tecan). Measured urine metabolite concentrations (in µmol/L) were adjusted for the urine creatinine concentration (in mmol/L) in the respective sample.
2.6. Statistical analysis
Numerical data regarding, age, body mass index (BMI), eGFR and the urine test results are presented as the mean and standard deviation (SD) and were compared using Student t tests. Categorical data concerning sex were compared using the Pearson chi‐squared test. Ratios of the level of a particular metabolite in the ccRCC patients to that seen in the control group are shown as fold change values. The stepwise method was used to select variables for multivariate analysis. 12 The multicollinearity of the selected variables was assessed by calculating variance inflation factor (VIF) values. Multivariate analysis was performed using multiple logistic regression analysis. Receiver operating characteristic (ROC) curve analysis was used to evaluate the diagnostic performance of the resulting regression model based on its AUC, sensitivity, and specificity values. A two‐tailed paired t test was performed on specimens from 20 patients from whom urine samples were obtained before and after surgery. All analyses were performed using JMP® Pro14 software (SAS Institute, Inc). Differences between groups were considered significant for values of P < .05.
3. RESULTS
3.1. Patient characteristics
Mean age of the 87 ccRCC patients (71 men, 16 women) was 63.3 y (range, 28‐85 y). Radical nephrectomy was performed in 54 cases and partial nephrectomy in 33 cases. Clinically diagnosed chronic kidney disease (CKD) with stage 3 or higher (eGFR, <60 mL/min/1.73 m2) was observed in 28 patients (32.6%) at the time of RCC diagnosis. Clinicopathological characteristics are presented in Table 1. The control group was age‐ and sex‐matched with the ccRCC group; the 2 groups did not differ significantly in Charlson comorbidity index (CCI), eGFR, or BMI, but the Karnofsky performance scale (PS) of the control group was significantly higher than that of the ccRCC group (P = .0121).
TABLE 1.
Clinicopathological characteristics of 87 patients with ccRCC and 60 patients comprising the benign control group
| Clear cell RCC | Control | P value | |
|---|---|---|---|
| No. pts | 87 | 60 | |
| No. Sex (%) | .334 | ||
| Male | 71 (81.6%) | 45(75.0%) | |
| Female | 16 (18.4%) | 15 (25.0%) | |
| Mean ± SD age | 63.3 ± 11.2 | 63.4 ± 9.1 | .97 |
| No. Karnofsky PS (%) | .0121 | ||
| 100 | 63 (72.4%) | 54 (90%) | |
| 90 | 16 (18.4%) | 6 (10%) | |
| ≤80 | 8 (9.2%) | 0 | |
| No. Charlson comorbidity index (%) | .671 | ||
| 0 | 50 (57.5%) | 38 (63.3%) | |
| 1 | 23 (26.4%) | 10 (16.7%) | |
| 2 | 8 (9.2%) | 10 (16.7%) | |
| ≥3 | 6 (6.9%) | 2 (3.3%) | |
| No. eGFR (%) | .209 | ||
| >60 | 59 (67.8%) | 34 (56.7%) | |
| 15‐60 | 26 (29.8%) | 21 (34.0%) | |
| <15 | 2 (2.3%) | 0 | |
| Unknown | 0 | 5 (8.3%) | |
| No. BMI (%) | .219 | ||
| <18.5 | 6 (6.9%) | 2 (3.3%) | |
| 18.5‐25 | 41 (47.1%) | 37 (61.7%) | |
| >25 | 40 (46.0%) | 21 (35.0%) | |
| No. Type of surgery | |||
| Radical nephrectomy | 54 (62.1%) | ||
| Partial nephrectomy | 33 (37.9%) | ||
| No. Clinical T stage (%) | |||
| T1 | 56 (64.4%) | ||
| T2 | 13 (14.9%) | ||
| T3 | 15 (17.2%) | ||
| T4 | 3 (3.4%) | ||
| No. Clinical N stage (%) | |||
| N0 | 82 (94.3%) | ||
| N1 | 2 (2.3%) | ||
| N2 | 3 (3.4%) | ||
| No. Clinical M stage (%) | |||
| M0 | 76 (87.4%) | ||
| M1 | 11 (12.6%) | ||
| No. Clinical stage (%) | |||
| I | 52 (59.8%) | ||
| II | 10 (11.5%) | ||
| III | 8 (9.2%) | ||
| IV | 17 (19.5%) | ||
| No. Pathological T stage (%) | |||
| T1 | 51 (58.6%) | ||
| T2 | 3 (3.4%) | ||
| T3 | 32 (36.8%) | ||
| No. Fuhrman grade (%) | |||
| Low (G1, G2) | 58 (66.7%) | ||
| High (G3, G4) | 8 (9.2%) | ||
| Unkown | 21 (24.1%) | ||
| No. Coagulation necrosis (%) | |||
| Presence | 24 (27.6%) | ||
| Absence | 44 (50.6%) | ||
| Unkown | 19 (21.8%) | ||
Abbreviation: ccRCC, clear cell renal cell carcinoma.
3.2. Comparison of urinary metabolite concentrations between the ccRCC and control groups
Of 47 targeted metabolites that were to be measured, 33 metabolites were quantitatively measurable in the urine samples (Table S3). Concentration levels of 14 of 33 metabolites were significantly higher in the ccRCC group than in the control group (P < .05, Table 2). A simple linear regression analysis of the 14 metabolites was performed, and AUC, sensitivity, and specificity values were evaluated for each of these 14 metabolites (Table 3). d‐Sedoheptulose 7‐phosphate showed AUC values of >0.8.
TABLE 2.
Urinary metabolites whose concentrations were significantly higher in the ccRCC group than in the control group
| Metabolite | ccRCC | Control | Fold change (ccRCC/control) | P value | ||
|---|---|---|---|---|---|---|
| Mean [µmol/L/u‐Cre (mmol/L)] | SD | Mean [µmol/L/u‐Cre (mmol/L)] | SD | |||
| GSH | 0.829 | 1.423 | 0.383 | 0.513 | 2.16 | .022 |
| Lactate | 75.868 | 216.226 | 26.818 | 25.148 | 2.83 | .083 |
| 2‐Hydroxyglutarate | 357.708 | 282.563 | 179.212 | 116.500 | 2.00 | <.0001 |
| Succinic acid | 0.148 | 0.222 | 0.050 | 0.069 | 2.95 | .001 |
| Phosphorylcholine | 0.509 | 0.446 | 0.345 | 0.316 | 1.47 | .015 |
| l‐Glutamine | 1268.288 | 2287.576 | 612.262 | 321.841 | 2.07 | .029 |
| d‐Fructose 6‐phosphate | 0.402 | 0.594 | 0.207 | 0.317 | 1.94 | .022 |
| Myoinositol | 23.728 | 35.035 | 6.183 | 5.706 | 3.84 | <.0001 |
| α‐d‐Glucose 1‐phosphate | 0.423 | 0.656 | 0.167 | 0.256 | 2.53 | .005 |
| d‐Sedoheptulose 7‐phosphate | 30.579 | 53.053 | 4.001 | 18.699 | 7.64 | <.0001 |
| l‐Tryptophan | 0.618 | 0.754 | 0.337 | 0.285 | 1.83 | .008 |
| l‐Kynurenine | 0.100 | 0.090 | 0.042 | 0.026 | 2.38 | <.0001 |
| Picolinic acid | 0.034 | 0.029 | 0.017 | 0.019 | 2.08 | <.0001 |
| Nicotinic acid | 0.005 | 0.005 | 0.003 | 0.004 | 1.63 | .014 |
Abbreviations: ccRCC, clear cell renal cell carcinoma; GSH, glutathione reduced form; u‐Cre, urinary creatinine.
TABLE 3.
A simple linear regression analysis of 14 metabolites with elevated concentrations in the ccRCC group
| Metabolite | Mean [µmol/L/u‐Cre (mmol/L)] | SD | AUC | Sensitivity (%) | Specificity (%) | Cut off [µmol/L/u‐Cre (mmol/L)] |
|---|---|---|---|---|---|---|
| d‐Sedoheptulose 7‐phosphate | 30.579 | 53.05 | 0.848 | 74.71 | 81.7 | 2.2454 |
| Lactate | 75.868 | 216.23 | 0.781 | 77.01 | 75.0 | 27.991 |
| Myoinositol | 23.728 | 35.04 | 0.780 | 66.7 | 81.7 | 8.78 |
| l‐Kynurenine | 0.100 | 0.09 | 0.740 | 64.4 | 80.0 | 0.056 |
| Picolinic acid | 0.034 | 0.03 | 0.720 | 85.1 | 53.3 | 0.011 |
| Succinic acid | 0.148 | 0.22 | 0.715 | 52.9 | 83.3 | 0.10 |
| 2‐Hydroxyglutarate | 357.708 | 282.56 | 0.690 | 46.0 | 95.0 | 324.08 |
| Nicotinic acid | 0.005 | 0.005 | 0.680 | 88.5 | 45.0 | 0.001 |
| GSH | 0.829 | 1.42 | 0.680 | 85.06 | 51.6 | 0.122 |
| l‐Tryptophan | 0.618 | 0.75 | 0.660 | 51.7 | 80.0 | 0.472 |
| Phosphorylcholine | 0.509 | 0.45 | 0.650 | 75.9 | 50.0 | 0.23 |
| α‐d‐Glucose 1‐phosphate | 0.423 | 0.66 | 0.640 | 57.5 | 44.4 | 0.17 |
| d‐Fructose 6‐phosphate | 0.402 | 0.59 | 0.638 | 64.4 | 61.7 | 0.16 |
| l‐Glutamine | 1268.288 | 2287.6 | 0.454 | 40.2 | 93.3 | 1076.49 |
Abbreviations: ccRCC, clear cell renal cell carcinoma; GSH, glutathione reduced form; u‐Cre, urinary creatinine.
3.3. Metabolite stratification of early diagnosis and clinicopathological factors
For the ccRCC group, we tested for correlation between the 33 metabolites for early diagnosis (control vs cT1a) and clinicopathological factors by analyzing the following parameters: clinical T stage (cT1 vs cT2‐4), pathological T stage (pT1 vs pT3), clinical M stage (cM0 vs cM1), Fuhrman grade (low vs high), and coagulation necrosis (absence vs presence). The concentrations of 6 metabolites (l‐tryptophan, l‐kynurenine, l‐glutamine, myoinositol, d‐sedoheptulose 7‐phosphate, and glutathione) were significantly increased in cT1a compared with the respective concentrations in the control group (P < .05 for each metabolite); therefore, these metabolites were inferred to be useful predictors for early diagnosis. Furthermore, l‐kynurenine concentrations were higher in cT2‐4 than in cT1 (P = .018). Butyrylcarnitine levels were higher in pT3 than in pT1 (P = .008). l‐Kynurenine levels were higher in cM1 than in cM0 (P = .009). The concentrations of 3 metabolites (l‐kynurenine, butyrylcarnitine, and 3‐methoxybenzopropanic acid) correlated with high Fuhrman grade (P < .05, respectively). Glutathione and lactate concentrations correlated with the presence of coagulation necrosis (P < .05 for each metabolite).
3.4. Construction of ccRCC predictive model
Multiple logistic regression analysis involving 33 metabolites was subjected to a stepwise variable selection method, and the results were used to construct a ccRCC diagnostic model. Five metabolites (lactate, 2‐hydroxyglutarate, l‐glutamic acid, myoinositol, and d‐sedoheptulose 7‐phosphate) were selected as variables for the diagnostic model (Figure 1A). The AUC, sensitivity, and specificity values of this model were 0.966, 93.1%, and 95.0%, respectively.
FIGURE 1.

Clear cell renal cell carcinoma (ccRCC) predictive model for diagnosis and clinical stage III/IV constructed by multiple logistic analysis. A, The black line on the graph is the receiver operating characteristics (ROC) curve for cancer diagnosis. The area under the curve (AUC), sensitivity, and specificity values obtained by multiple logistic regression analysis were 0.966, 93.1%, and 95.0%, respectively. The coefficients, P values, and variance inflation factor (VIF) values for each variable are shown in the table below the graph. The prediction formula is shown below the table. B, The black line is the ROC curve for clinical stage III/IV on the graph. The AUC, sensitivity, and specificity values obtained by multiple logistic regression analysis were 0.837, 88.5%, and 75.4%, respectively. The coefficients, P values, and VIF values for each variable are shown in the table below the graph. The predictive formula is shown below the table
Using the same process, another predictive model was generated to predict clinical stage III/IV. Four metabolites (l‐kynurenine, l‐glutamine, fructose 6‐phosphate, and butyrylcarnitine) were selected as variables for the predictive model of clinical stage III/IV. The AUC, sensitivity, and specificity values of this predictive model were 0.837, 88.5%, and 75.4%, respectively (Figure 1B).
3.5. Comparison of urinary metabolite concentrations before and after surgery for ccRCC
To confirm whether the 5 metabolites selected in the predictive diagnostic model for ccRCC before surgery indeed were decreased after surgery, we compared urine metabolite concentrations between samples collected before and after surgery in each of 20 patients (patient characteristics are presented in Table S4). The changed concentration levels of 5 selected metabolites were presented in Table S5. Four of the 5 selected urine metabolites exhibited significant decreases in concentration postoperatively; the sole exception was myoinositol (Figure 2).
FIGURE 2.
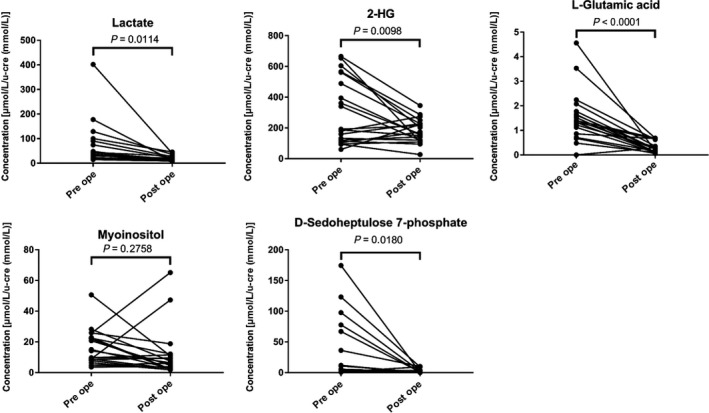
Changes in the concentrations of urinary metabolites before and after surgery in urine samples from 20 patients with clear cell renal cell carcinoma (ccRCC). A two‐tailed paired t test was performed on samples from 20 patients from whom urine samples were obtained both before and after surgery (range: 6‐8 d) for ccRCC. Each graph shows the concentration changes in the 5 metabolites that were selected as variables for the diagnostic model. Significant (P < .05 each) postoperative decreases in metabolite concentrations were observed for 4 metabolites; the exception was myoinositol
4. DISCUSSION
Our study of available urinary biomarkers for ccRCC indicated that d‐sedoheptulose 7‐phosphate, lactate, 2‐hydroxyglutarate, l‐glutamic acid, and myoinositol could be incorporated into a diagnostic model. We also found that l‐kynurenine, l‐glutamine, fructose 6‐phosphate, and butyrylcarnitine could be incorporated into a predictive model of clinical stage III/IV. In a previous G‐Met study using tissue samples, we broadly identified multiple metabolites that predicted the diagnosis and malignant status of ccRCC; that work also described the development of a system for quantitative measurement of these metabolites using LC‐MS. 1 Consequently, we were able to establish a precise ccRCC predictive model using internal standards and normalizing values based on urine creatinine; this model was constructed by combining measurements for multiple metabolites.
Five metabolites (including d‐sedoheptulose 7‐phosphate, lactate, 2‐hydroxyglutarate, l‐glutamic acid, and myoinositol) were selected for our predictive diagnostic model. Four metabolites (including l‐kynurenine, l‐glutamine, fructose 6‐phosphate, and butyrylcarnitine) were selected for our predictive model of clinical stage III/IV. These selected metabolites represent intermediates of the glycolysis, TCA cycle, tryptophan, and carnitine pathways, as indicated in the metabolomics map provided in Figure 3. d‐Sedoheptulose 7‐phosphate is produced via the fructose 6‐phosphate and pentose phosphate pathways. 13 Accumulation of d‐sedoheptulose 7‐phosphate appears to reflect the upregulation of glycolysis, but the role of this metabolite in cancer metabolism has not been reported. In previous RCC urine metabolite studies, lactate levels were found to be significantly elevated in patients with RCC compared with those in healthy controls, suggesting that RCC shifts metabolic flux from the TCA cycle to active glycolysis and the use of glucose for lactate fermentation. 6 , 14 The elevation of 2‐hydroxyglutarate levels, which also was noted in a previous RCC tissue study, may reflect the reduction of α‐ketoglutarate to 2‐hydroxyglutarate. 15 Glutamine is an alternative source of energy for living cells and is converted (via α‐ketoglutarate) to glutamic acid as part of the TCA cycle, thereby contributing to energy and biomass production. 16 Myoinositol is generated as part of the inositol pathway, which is involved in the control of a wide range of cellular processes; elevation of myoinositol concentrations also was reported in a previous RCC urine metabolomics study. 5 , 17 In the present study, the concentrations of these 4 metabolites were significantly decreased after surgery, and alteration of the levels of these molecules was inferred to reflect therapeutic effect. However, in 2 cases, myoinositol concentrations increased postoperatively. Although neither of these cases (which presented as pT1) has exhibited recurrence during the subsequent observation period, we propose that both patients should be followed closely, given our model's suggestion that myoinositol is a predictive factor related to malignant potential. Alterations in the levels of these 5 metabolites also have been reported in other cancers; their usefulness in ccRCC‐specific prediction models will require future validation studies including various other cancers.
FIGURE 3.
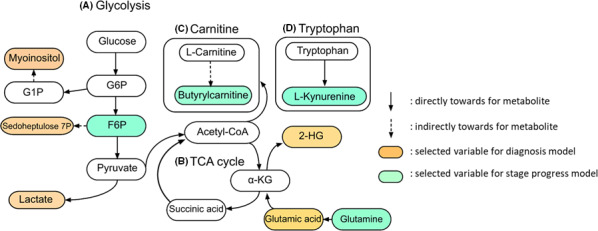
Positional relationships of the selected metabolites used for the diagnostic model and for the predictive model of clinical stage III/IV are shown on the metabolic pathways. The diagnostic model included 5 metabolites: myoinositol, d‐sedoheptulose 7‐phosphate, and lactate are derived from glycolysis; glutamic acid is incorporated via the tricarboxylic acid (TCA) cycle; and 2‐HG is an intermediate metabolite of the TCA cycle. For the predictive model of clinical stage III/IV, F6P belongs to glycolysis; glutamine is incorporated via the TCA cycle; l‐kynurenine is metabolized from tryptophan; and butyrylcarnitine is metabolized from the carnitine pathway. G1P, glucose 1‐phosphate; G6P, glucose 6‐phosphate; F6P, fructose 6‐phosphate; 2‐HG, 2‐hydroxyglutarate
In the present study, predictive models of clinical stage III/IV indicated elevated levels of l‐kynurenine and butyrylcarnitine in patients with RCC. These metabolites belong to the tryptophan and carnitine pathways, respectively. The expression of kynurenine in RCC tissue previously has been shown to indicate poor prognostic outcome. 18 Carnitine is known to derive from increased glycolysis, and has been shown (in a previous urine metabolomics study) to accumulate to increased levels in high‐grade RCC. 7 Fructose 6‐phosphate levels reflect increased glycolysis; metabolic processing of this compound may require increased energy at higher clinical stages.
Metabolomics has been evaluated in several diseases to identify new biomarkers and therapeutic targets in drug discovery. 19 , 20 , 21 Metabolomics also increasingly is being used in cancer research for prediction of disease progression and recurrence. Previously reported candidate protein markers have included carbonic anhydrase IX (CA‐IX), tumor necrosis factor receptor‐associated factor‐1 (TRAF‐1), serum amyloid A (SAA), kidney injury molecule‐1 (KIM‐1), aquaporin‐1 (AQP‐1), and perilipin‐2 (PLIN2); however, these markers were defined primarily in the context of advanced RCC. 22 , 23 , 24 , 25 , 26 Other candidate protein markers, including von Hippel‐Lindau (VHL), BRCA1‐associated protein 1 (BAP1), PBRM1, and nuclear factor (NF), have been detected from circulating tumor DNA (ctDNA). 27 However, it remains unclear whether these markers, which change dynamically with therapeutic effects, will be useful as biomarkers that can provide stable quantitative measurements in clinical practice.
Urine samples are ideal for biomarker research because such specimens can be collected noninvasively; additionally, given that renal epithelial tumors are in contact with the urinary space, urine is well suited for a metabolomics approach. 28 , 29 However, there could be some disadvantages for metabolomics analyses of urine samples. Indeed, major inconsistencies in the outcomes of metabolomics in urine have been observed among various studies; these inconsistencies presumably reflect differences in sample collection, handling, and manipulation, as well as in the analytical platforms used. 28
The advantages of our quantitative measurement system include the use of an internal standard and correction for dilution based on urine creatinine concentrations. Although there have been several previous reports of urinary metabolites as potential biomarkers in RCC, most of those measurement systems employed semi‐quantitative analysis without the use of internal standards and did not take the urinary matrix effect into account. 5 , 6 , 8 , 30 , 32 The matrix effect is a phenomenon whereby metabolite levels fluctuate as a result of the influence of urinary protein, impurities, and volatilization; the use of internal standards is expected to correct for the effect of the matrix on metabolite concentrations. 11
Urine creatinine has been used previously as a standard substance to correct for urine dilution. 33 , 34 Yoric et al 33 reported on the standardization of quantitative measurement of urinary metabolites via UHPLC‐RTOF‐MS; measurement of the levels of urinary creatinine and osmolality allowed those researchers to correct for variations in the levels of metabolites and was essential for analyzing urinary metabolites in cases with CKD. In our study, participants with CKD stage 3 or higher represented 32.1% of the ccRCC group and 35.0% of the control group. Thus, correction on the basis of urine creatinine concentration is considered crucial for analysis of urine metabolites in a real‐world study.
The use of a single metabolite as a biomarker for ccRCC was not expected to suffice, given the observation (in our previous study and in other reports on urinary metabolites) 4 , 5 that the heterogeneity of ccRCC is thought to be affected by multiple metabolic pathways. Therefore, to improve the predictive ability for diagnosis and malignant status, we combined key urinary metabolites to construct a predictive model using a stepwise method to perform multiple logistic regression analysis.
In the context of the constructed predictive model, the multicollinearity of the metabolites could not be confirmed because their VIF values were small. Therefore, the increased concentrations of these metabolites in the urine of patients with ccRCC may represent independent events. Thus, our predictive models for diagnosis of ccRCC via urine samples are expected to represent a new modality for ccRCC screening, permitting noninvasive sampling. Our predictive model of clinical stage III/IV RCC may improve clinical practice by enabling decision‐making for pharmaceutical treatment.
This study has several limitations. A stepwise feature selection in multiple logistic regression analysis can lead to overly optimistic figures of merit. Our result has limited evaluation due to the lack of validation by another cohort. Notably, there may have been sample collection bias. Urine samples in the control group represented patients with benign disorders or endocrine diseases; the effect of these diseases on urinary metabolites remains unclear. Additionally, the measurement procedure was conducted retrospectively. Furthermore, our predictive models were not specific to ccRCC. Future validation studies in other cancers may improve the usability of our models.
In conclusion, we used our system for accurate quantitative measurement of urinary metabolites to establish predictive models for diagnosis and malignant status of ccRCC; the resulting model exhibited high sensitivity and specificity. Further validation studies will be needed to overcome the limitations of our procedure. Nonetheless, our predictive models incorporating the levels of urinary metabolites may identify clinically useful noninvasive biomarkers that lead to improved outcomes for ccRCC patients.
DISCLOSURE
The authors declare no conflicts of interest.
Supporting information
Table S1
Table S2
Table S3
Table S4
Table S5
Appendix S2
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI (Grant Nos. JP17K07210 and JP18K16683).
Sato T, Kawasaki Y, Maekawa M, et al. Accurate quantification of urinary metabolites for predictive models manifest clinicopathology of renal cell carcinoma. Cancer Sci. 2020;111:2570–2578. 10.1111/cas.14440
REFERENCES
- 1. Sato T, Kawasaki Y, Maekawa M, et al. Value of global metabolomics in association with diagnosis and clinicopathological factors of renal cell carcinoma. Int J Cancer. 2019;145(2):484‐493. [DOI] [PubMed] [Google Scholar]
- 2. Saigusa D, Okamura Y, Motoike IN, et al. Establishment of protocols for global metabolomics by LC‐MS for biomarker discovery. PLoS ONE. 2016;11:e0160555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Danhier P, Banski P, Payen VL, et al. Cancer metabolism in space and time: beyond the Warburg effect. Biochim Biophys Acta. 2017;1858:556‐572. [DOI] [PubMed] [Google Scholar]
- 4. Liu X, Zhang M, Liu X, et al. Urine metabolomics for renal cell carcinoma (RCC) prediction: tryptophan metabolism as an important pathway in RCC. Front Oncol. 2019;9:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falegan OS, Ball MW, Shaykhutdinov RA, et al. Urine and serum metabolomics analyses may distinguish between stages of renal cell carcinoma. Metabolites. 2017;7(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ragone R, Sallustio F, Piccinonna S, et al. Renal cell carcinoma: a study through NMR‐based metabolomics combined with transcriptomics. Diseases (Basel, Switzerland). 2016;4(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ganti S, Taylor SL, Kim K, et al. Urinary acylcarnitines are altered in human kidney cancer. Int J Cancer. 2012;130:2791‐2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim K, Taylor SL, Ganti S, Guo L, Osier MV, Weiss RH. Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. OMICS. 2011;15:293‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471‐1474. [DOI] [PubMed] [Google Scholar]
- 10. Japan nephrology s . Special issue: clinical practice guidebook for diagnosis and treatment of chronic kidney disease 2012. Nihon Jinzo Gakkai shi. 2012;54:1034‐1191. [PubMed] [Google Scholar]
- 11. Abe K, Suzuki H, Maekawa M, Shimada M, Yamaguchi H, Mano N. Matrix effect‐corrected liquid chromatography/tandem mass‐spectrometric method for determining acylcarnitines in human urine. Clin Chim Acta; Int J Clin Chem. 2017;468:187‐194. [DOI] [PubMed] [Google Scholar]
- 12. Nishiumi S, Kobayashi T, Kawana S, et al. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple‐quadrupole mass spectrometry. Oncotarget. 2017;8:17115‐17126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stincone A, Prigione A, Cramer T, et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc. 2015;90:927‐963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wettersten HI, Hakimi AA, Morin D, et al. Grade‐dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. 2015;75:2541‐2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hakimi AA, Reznik E, Lee CH, et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu W, Pelicano H, Huang P. Cancer metabolism: is glutamine sweeter than glucose? Cancer Cell. 2010;18:199‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Serkova NJ, Gamito EJ, Jones RH, et al. The metabolites citrate, myo‐inositol, and spermine are potential age‐independent markers of prostate cancer in human expressed prostatic secretions. Prostate. 2008;68:620‐628. [DOI] [PubMed] [Google Scholar]
- 18. Lucarelli G, Rutigliano M, Ferro M, et al. Activation of the kynurenine pathway predicts poor outcome in patients with clear cell renal cell carcinoma. Urol Oncol. 2017;35(7):461.e15‐461.e27. [DOI] [PubMed] [Google Scholar]
- 19. Tumas J, Kvederaviciute K, Petrulionis M, et al. Metabolomics in pancreatic cancer biomarkers research. Med Oncol (Northwood, London, England). 2016;33:133. [DOI] [PubMed] [Google Scholar]
- 20. Farshidfar F, Weljie AM, Kopciuk KA, et al. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br J Cancer. 2016;115:848‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Budczies J, Denkert C. Tissue‐based metabolomics to analyze the breast cancer metabolome. Rec Results Cancer Res Fortschritte der Krebsforschung Progres dans les recherches sur le cancer. 2016;207:157‐175. [DOI] [PubMed] [Google Scholar]
- 22. Rajandram R, Yap NY, Pailoor J, et al. Tumour necrosis factor receptor‐associated factor‐1 (TRAF‐1) expression is increased in renal cell carcinoma patient serum but decreased in cancer tissue compared with normal: potential biomarker significance. Pathology. 2014;46:518‐522. [DOI] [PubMed] [Google Scholar]
- 23. Morrissey JJ, Mobley J, Song J, et al. Urinary concentrations of aquaporin‐1 and perilipin‐2 in patients with renal cell carcinoma correlate with tumor size and stage but not grade. Urology. 2014;83(1):256.e9‐256.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takacova M, Bartosova M, Skvarkova L, et al. Carbonic anhydrase IX is a clinically significant tissue and serum biomarker associated with renal cell carcinoma. Oncol Lett. 2013;5:191‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fischer K, Theil G, Hoda R, Fornara P. Serum amyloid A: a biomarker for renal cancer. Anticancer Res. 2012;32:1801‐1804. [PubMed] [Google Scholar]
- 26. Morrissey JJ, London AN, Lambert MC, Kharasch ED. Sensitivity and specificity of urinary neutrophil gelatinase‐associated lipocalin and kidney injury molecule‐1 for the diagnosis of renal cell carcinoma. Am J Nephrol. 2011;34:391‐398. [DOI] [PubMed] [Google Scholar]
- 27. Ball MW, Gorin MA, Guner G, et al. Circulating tumor DNA as a marker of therapeutic response in patients with renal cell carcinoma: a pilot study. Clin Genitourin Cancer. 2016;14:e515‐e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodrigues D, Monteiro M, Jeronimo C, et al. Renal cell carcinoma: a critical analysis of metabolomic biomarkers emerging from current model systems. Transl Res. 2017;180:1‐11. [DOI] [PubMed] [Google Scholar]
- 29. Weiss RH, Kim K. Metabolomics in the study of kidney diseases. Nat Rev Nephrol. 2011;8:22‐33. [DOI] [PubMed] [Google Scholar]
- 30. Niziol J, Bonifay V, Ossolinski K, et al. Metabolomic study of human tissue and urine in clear cell renal carcinoma by LC‐HRMS and PLS‐DA. Anal Bioanal Chem. 2018;410:3859‐3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Monteiro M, Moreira N, Pinto J, et al. GC‐MS metabolomics‐based approach for the identification of a potential VOC‐biomarker panel in the urine of renal cell carcinoma patients. J Cell Mol Med. 2017;21:2092‐2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim K, Aronov P, Zakharkin SO, et al. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics. 2009;8:558‐570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gagnebin Y, Tonoli D, Lescuyer P, et al. Metabolomic analysis of urine samples by UHPLC‐QTOF‐MS: impact of normalization strategies. Anal Chim Acta. 2017;955:27‐35. [DOI] [PubMed] [Google Scholar]
- 34. Miller RC, Brindle E, Holman DJ, et al. Comparison of specific gravity and creatinine for normalizing urinary reproductive hormone concentrations. Clin Chem. 2004;50:924‐932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3
Table S4
Table S5
Appendix S2


