Abstract
We performed a genome‐wide association study to investigate the association between single nucleotide polymorphisms and anthracycline‐induced cardiotoxicity (ACT) in patients diagnosed with early breast cancer. From January 2000 to December 2015, 8490 patients underwent breast surgery at the National Cancer Center in Korea. Patients who received doxorubicin (cumulative dose 240 mg/m2‐300 mg/m2) with or without trastuzumab as a neoadjuvant/adjuvant therapy were included in our cohort. Sixty‐seven patients in our cohort were diagnosed with ACT. Clinical data, including age, body weight, height, cancer stage, trastuzumab treatment, comorbidities, and concomitant medications, were collected retrospectively. Patients were classified as having either persistent or transient ACT based on their clinical course. In total, 346 946 single nucleotide polymorphisms in 42 cases and 215 controls were tested in this study. Body mass index (BMI) ≥25 kg/m2 [odds ratio (OR) = 2.45, 95% confidence interval (CI), 1.23‐4.88, P = .011] and trastuzumab use (OR = 2.40, 95% CI, 1.11‐5.17, P = .026) were identified as significant risk factors. We found 7 genetic variants for ACT including rs17530621 (SHISA3, P = 3.10E−06), rs11894115 (MPP4, P = 4.71E−06), rs58328254 (RPL7, P = 6.09E−06), and rs117299725 (PRUNE2, P = 8.53E−06), although none of these variants reached the Bonferroni‐corrected significance level when adjusted for BMI and trastuzumab use ( = α1.44E−07 based on 0.05/346 946). rs117299725 was a common variant when only the persistent ACT group was analyzed separately. It is meaningful that our study analyzed comprehensively the influence of genetic variation on ACT, along with some clinical factors in Asian breast cancer patients who received anthracycline with or without trastuzumab. Further research will be needed on candidate genetic variants found in this study.
Keywords: anthracycline, breast cancer, cardiotoxicity, case‐control study, genome‐wide association study
In this study, we addressed potent genetic variants related with anthracycline induced cardiotoxicity in Korean early breast cancer patients adjusting various clinical factors with a long follow‐up period. The results of our study showed that in anthracycline treatments with a cumulative dose not exceeding 300 mg/m2, clinical factors were more associated with cardiotoxicity than genetic factors. Several possible genetic variants found in this study may require further study.

1. INTRODUCTION
Anthracycline has long been used as the primary treatment for breast cancer. Despite its potential benefits, the number of doses of anthracycline needs to be limited due to its adverse effects on the heart, including congestive heart failure. 1 , 2 Doxorubicin is an anthracycline with an overall incidence of cardiotoxicity in up to 3.3% of patients who received a cumulative dose of at least 450 mg/m2. 1 Clinical factors associated with the development of cardiotoxicity include the cumulative dose of doxorubicin, age, and existing heart disease. 1 , 3 The concomitant use of other chemotherapeutic agents, such as trastuzumab and bevacizumab, with anthracyclines also increases the cardiotoxicity risk. 4 , 5
Anthracycline‐induced cardiotoxicity (ACT) involves irreversible cardiomyocyte death due to reactive oxygen species produced by doxorubicin metabolism within the cardiomyocytes. 6 , 7 Many studies have been conducted to identify genetic risk factors that predict the development of cardiotoxicity. 8 , 9 , 10 , 11 , 12 , 13 Results from these studies have not been consistent, however, due to the heterogeneity of the study populations and differences in the definitions of cardiotoxicity.
Anthracyclines have been included as neoadjuvant or adjuvant treatments with or without other chemotherapeutic agents in the treatment of breast cancer. Under such treatment regimens, the cumulative dose of doxorubicin usually does not exceed 300 mg/m2, nevertheless cardiotoxicity occurred in 1.5%‐2.0% of patients who took part in a large clinical trial. 4 , 14 Given the severity of cardiotoxicity, there is a need to develop reliable predictive biomarkers to identify high‐risk patients.
In this study, we conducted a genome‐wide association study (GWAS) of patients with early breast cancer to assess single nucleotide polymorphisms (SNPs) across the whole genome. The goal of this GWAS was to identify genetic variants and clinical factors associated with risk of ACT in this population.
2. MATERIALS AND METHODS
2.1. Study population
From January 2000 to December 2015, 8490 patients underwent breast surgery at the National Cancer Center in Korea. Of these patients, 3910 patients received doxorubicin as a neoadjuvant or adjuvant treatment, and 67 were diagnosed with ACT. In this study, cardiotoxicity was defined as a >10% reduction in the left ventricular ejection fraction (LVEF) from baseline and LVEF < 50% on multi‐gated blood pool scan (MUGA), or <55% on echocardiography. 15 Among patients in the case group, those who completely recovered from ACT during the follow‐up period were classified into the transient ACT group and those who needed medication and regular monitoring were classified into the persistent ACT group. We analyzed separately genetic differences between the persistent ACT group and the control group.
For the control group, we selected 317 age‐matched patients with breast cancer who underwent breast surgery before or after doxorubicin‐containing chemotherapy as neoadjuvant/adjuvant treatment. The cardiac function of the control group was normal before and after chemotherapy as confirmed by MUGA or echocardiography. For all patients, clinical data, including age, body weight, height, cancer stage, cumulative dose of doxorubicin, radiation therapy, trastuzumab treatment, comorbidities, and concomitant medications, were collected retrospectively. This study was approved by the Institutional Review Board of the National Cancer Center in Korea (IRB no. NCC2017‐0161) and all patients provided written informed consent before participation.
2.2. Genotyping
Peripheral blood samples were drawn into vacutainer tubes and genomic DNA was extracted from whole blood using the QIAamp DNA Mini Kit (Qiagen, CA) in accordance with the manufacturer's instructions. DNA samples from both the case and control groups were genotyped using the Korea Biobank Array platform (KoreanChip). 16 The KoreanChip is comprised of 833 535 markers, including more than 247 000 rare frequency or functional variants based on the sequencing data of 2500 Koreans. 16
2.3. Quality control for samples
Samples were subjected to quality control (QC). Among the 384 samples, 10 samples were excluded due to multidimensional scaling and 30 samples due to a low call rate and excessive heterozygosity rate. An additional 87 samples were further excluded for the following reasons: male patient (n = 1), stage IV cancer (n = 2), anthracycline cumulative dose < 240 mg/m2 (n = 7), no chemotherapy (n = 6), and anthracycline cumulative dose > 300 mg/m2 (n = 71). The final association test was performed with 257 samples (Figure 1A).
FIGURE 1.
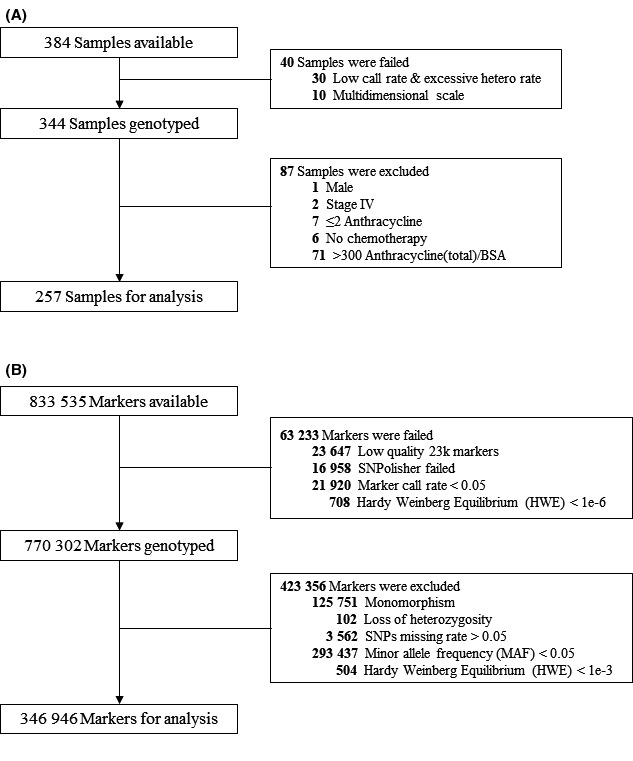
Consort diagram of quality control (QC) for (A) each samples and (B) SNPs
2.4. Quality control for markers
SNP QC steps were performed on 833 535 available markers. SNPolisher software designed by Affymetrix was used to select markers for better QC. 17 In total, 63 233 markers were removed due to low quality (n = 23 647), SNPolisher failure (n = 16 958), marker call rate < 0.05 (n = 21 920), and Hardy‐Weinberg equilibrium P‐value ≤ 1E−6 (n = 708). Additional SNP QC steps were performed on the final 257 samples included in the analysis. Among the 770 302 remaining markers, SNPs exhibiting monomorphism (n = 125 751) or loss of heterozygosity (n = 102) were first filtered out. Then SNPs with a missing rate > 5% (n = 3562), minor allele frequency (MAF) ≤5% (n = 293 437), or Hardy‐Weinberg equilibrium P‐value < .001 (n = 504) were further excluded. The final marker set included 346 946 SNPs across 257 patients. The P‐value threshold for significance was = α1.44E−07 based on 0.05/346 946 (Figure 1B).
2.5. Statistical analysis
Before the study, statistical power was calculated based on a maximum sample size of 60 cases and 240 controls. This sample size (1:4 ratio of cases to controls) was expected to achieve 87.4% and 67.3% power at significance levels of 10−2 and 10−3, respectively, with an assumed odds ratio (OR) of 3.0 in the additive model under a high‐risk allele frequency of 0.1 and disease prevalence of 1.7%. The distribution of clinical characteristics between cases and controls was presented as means ± SD for continuous variables and frequency (percentage) for categorical variables. Differences in clinical variables were tested using the Student t test for continuous variables and Pearson chi‐square test or Fisher exact test for categorical variables. To test the association between ACT and SNPs, we used several genetic models, including the additive, dominant, and recessive models. Univariate and multivariate logistic regression models were applied to perform the association tests with adjustment for significant clinical factors in the multivariate model. Significance of the association of SNPs was examined using the minimum of the P‐value among the additive and dominant genetic models. 18 The effective number of SNPs was calculated with respect to the linkage disequilibrium (LD) correlation structure. 19 The LD between genetic markers were calculated using the joint probability and the product of the individual allele probabilities. We did this using the LD() function of the ‘genetics’ packages in R software. Regional plots were created using LocusZoom (http://locuszoom.sph.umich.edu/locuszoom/). Expression quantitative trait loci (eQTL) analysis was performed using the Genotype‐Tissue Expression (GTEx) database 20 (http://gtexportal.org/). To compare the overall survival between patients in the case and control groups, the difference was tested using the log‐rank test and plotted as Kaplan‐Meier curves.
3. RESULTS
3.1. Patient characteristics
Forty‐two cases and 215 controls were included in this study after excluding samples for QC. Across all patients, the median follow‐up duration was 8.43 y (interquartile range: 5.15‐10.42 y; maximum: 15.45 y). The median onset time for ACT was 220 d (range: 55‐2472 d) after completion of chemotherapy. Most clinical variables were not significantly different between the case and control groups, however body mass index (BMI) >25 kg/m2 (45.2% vs 25.6%, P = .010), higher weight (61.6 vs 57.5 kg, P = .002), and a history of hyperlipidemia (16.7% vs 6.1%, P = .028) were more frequently observed in the case group. In addition, the number of patients with HER2 amplification (83.3% vs 60.5%, P = .005) and trastuzumab use (76.2% vs 57.7%, P = .025) was significantly higher in the case group (Table 1). Most patients with HER2‐amplified tumors received trastuzumab treatment concurrently with adjuvant chemotherapy, therefore trastuzumab use but not HER2 positivity was included as a factor in the analysis, to account for any interaction between the 2 factors. In the multivariate logistic regression model, BMI ≥ 25 kg/m2 (OR = 2.45, 95% CI, 1.23‐4.88, P = .011) and trastuzumab use (OR = 2.40, 95% CI, 1.11‐5.17, P = .026) remained as significant factors associated with ACT (Table 2). These 2 factors were included as covariates in the adjusted model.
TABLE 1.
Distribution of clinical characteristics between case and control
| Characteristics | Total | Case | Control | P‐value | |
|---|---|---|---|---|---|
| (n = 257) | (n = 42) | (n = 215) | |||
| Age at diagnosis (mean ± SD) | 49.3 ± 9.5 | 49.8 ± 8.8 | 49.2 ± 9.7 | .720 | |
| Height (cm) | 157.2 ± 5.0 | 158.1 ± 5.7 | 157.0 ± 4.9 | .221 | |
| Weight (kg) | 58.2 ± 7.9 | 61.6 ± 9.2 | 57.5 ± 7.5 | .002 | |
| Body mass index (BMI) | <25 | 183 | 23 (54.8) | 160 (74.4) | .010 |
| ≥25 | 74 | 19 (45.2) | 55 (25.6) | ||
| Hypertension | No | 214 | 34 (81.0) | 180 (83.7) | .660 |
| Yes | 43 | 8 (19.0) | 35 (16.3) | ||
| Hyperlipidemia | No | 237 | 35 (83.3) | 202 (94.0) | .028 |
| Yes | 20 | 7 (16.7) | 13 (6.1) | ||
| Diabetes | No | 247 | 41 (97.6) | 206 (95.8) | >.999 |
| Yes | 10 | 1 (2.4) | 9 (4.2) | ||
| Other heart disease | No | 252 | 40 (95.2) | 212 (98.6) | .189 |
| Yes | 5 | 2 (4.8) | 3 (1.4) | ||
| Taxol | No | 85 | 11 (26.2) | 74 (34.4) | .300 |
| Yes | 172 | 31 (73.8) | 141 (65.6) | ||
| ER | Negative | 89 | 16 (38.1) | 73 (34.0) | .606 |
| Positive | 168 | 26 (61.9) | 142 (66.0) | ||
| PR | Negative | 104 | 17 (40.5) | 87 (40.5) | .999 |
| Positive | 153 | 25 (59.5) | 128 (59.5) | ||
| HER2 | Negative | 92 | 7 (16.7) | 85 (39.5) | .005 |
| Positive | 165 | 35 (83.3) | 130 (60.5) | ||
| Radiotherapy | No | 21 | 3 (7.1) | 18 (8.4) | >.999 |
| Yes | 236 | 39 (92.9) | 197 (91.6) | ||
| Endocrine therapy | No | 86 | 15 (35.7) | 71 (33.0) | .735 |
| Yes | 171 | 27 (64.3) | 144 (67.0) | ||
| Trastuzumab use | No | 101 | 10 (23.8) | 91 (42.3) | .025 |
| Yes | 156 | 32 (76.2) | 124 (57.7) | ||
| Death | Survival | 231 | 39 (92.9) | 192 (89.3) | |
| Death from breast cancer | 22 | 3 (7.1) | 19 (8.8) | ||
| Death from other cause | 4 | 0 (0.0) | 4 (1.9) | ||
| Progress of ACT | Persistent ACT | 30 | 30 (71.4) | ||
| Transient ACT | 12 | 12 (28.6) |
Abbreviations: ACT: anthracycline‐induced cardiotoxicity; HER2, human epidermal growth factor receptor 2.
TABLE 2.
Univariable and multivariable logistic regression model for clinical variables
| Characteristics | Univariate model | Multivariate model | |||
|---|---|---|---|---|---|
| OR (95% CI) | P‐value | OR (95% CI) | P‐value | ||
| Body mass index (BMI) | <25 | 1 (ref) | 1 (ref) | ||
| ≥25 | 2.40 (1.22‐4.75) | .012 | 2.45 (1.23‐4.88) | .011 | |
| Trastuzumab use | No | 1 (ref) | 1 (ref) | ||
| Yes | 2.35 (1.10‐5.02) | .028 | 2.40 (1.11‐5.17) | .026 | |
Abbreviations: CI, confidence interval; OR, odds ratio.
Thirty patients (71.4%) in the case group were classified into the persistent ACT group. In contrast, 12 patients in the case group completely recovered from ACT within a median of 410 d (range: 125‐850 d) and were classified into the transient ACT group. A further analysis of the genes within the persistent ACT group was conducted.
3.2. Genetic association study
A quantile‐quantile (Q‐Q) plot showed the distribution of the observed P‐value from association testing of 346 946 SNPs and all case and control samples distributed into the same cluster in the principal component analysis (PCA) plot (Figure S1A,B). A Manhattan plot was used to display the 346 946 SNPs that passed QC (Figure 2). None of the SNPs reached the Bonferroni‐corrected significance level (α = 1.44E−07 from 0.05/346 946). We therefore identified 86 SNPs with a minimum P‐value < 1.0E−04 in the crude or adjusted model (Table S1). Because several SNPs selected on the same gene were in LD, we calculated the effective number of 86 SNPs based on LD correlation structure. The effective number was 68.9 due to duplicated genes among the 86 SNPs. We identified 7 SNPs with a minimum P‐value < 1.0E−05 in the adjusted model, corresponding to an effective number of 6.9 (Table 3).
FIGURE 2.
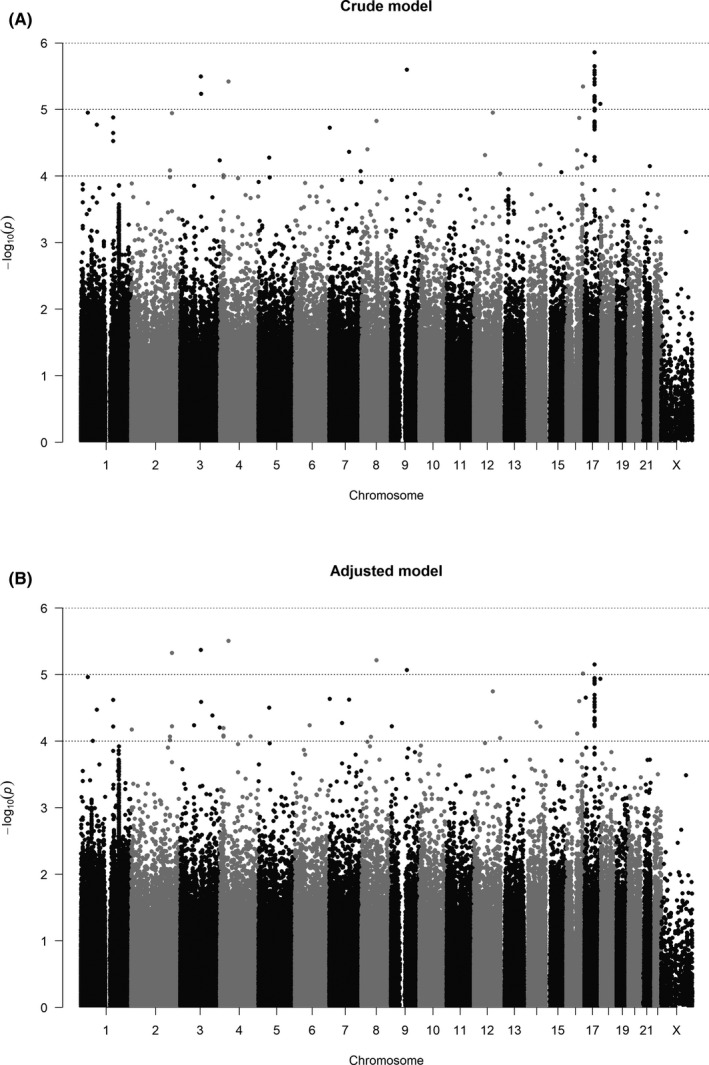
Manhattan plots for association test of 346 946 SNPs on (A) crude and (B) adjusted model
TABLE 3.
Frequency and P‐values using crude and adjusted logistic regression model for candidate SNPs (P‐value < 1.E−05)
| Gene name | RS ID | CHR | Physical. position | Genotype | Frequency | Crude model | Adjusted model a | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Case | OR1 (95% CI) | OR2 (95% CI) | P‐value | OR1 (95% CI) | OR2 (95% CI) | P‐value | |||||
| BEND4/SHISA3/ATP8A1 | rs17530621 | 4 | 42 227 497 | G/G | 170 | 18 | 1 | 1 | 3.81.E−06 | 1 | 1 | 3.10.E−06 |
| G/T | 42 | 23 | 4.24 (2.23‐8.06) | 5.15 (2.57‐10.33) | 4.53 (2.33‐8.84) | 5.66 (2.73‐11.72) | ||||||
| T/T | 2 | 1 | 17.97 (4.97‐64.93) | 5.15 (2.57‐10.33) | 20.56 (5.41‐78.09) | 5.66 (2.73‐11.72) | ||||||
| MIR548AB | rs6804462 | 3 | 103 342 570 | G/G | 198 | 27 | 1 | 1 | 3.18.E−06 | 1 | 1 | 4.26.E−06 |
| G/C | 15 | 14 | 5.60 (2.59‐12.11) | 6.87 (3.06‐15.47) | 6.24 (2.86‐13.61) | 7.33 (3.13‐17.16) | ||||||
| C/C | 1 | 1 | 31.35 (6.70‐146.56) | 6.87 (3.06‐15.47) | 38.91 (8.17‐185.28) | 7.33 (3.13‐17.16) | ||||||
| MPP4 | rs11894115 | 2 | 202 512 449 | C/C | 76 | 2 | 1 | 1 | 1.13.E−05 | 1 | 1 | 4.71.E−06 |
| C/T | 102 | 22 | 3.39 (1.97‐5.84) | 10.78 (2.53‐45.92) | 3.96 (2.20‐7.15) | 11.57 (2.69‐49.79) | ||||||
| T/T | 32 | 16 | 11.49 (3.86‐34.16) | 10.78 (2.53‐45.92) | 15.72 (4.83‐51.13) | 11.57 (2.69‐49.79) | ||||||
| RPL7/RDH10 | rs58328254 | 8 | 74 203 722 | G/G | 135 | 12 | 1 | 1 | 1.49.E−05 | 1 | 1 | 6.09.E−06 |
| G/A | 70 | 23 | 3.48 (1.98‐6.11) | 4.29 (2.07‐8.90) | 3.98 (2.19‐7.23) | 4.90 (2.30‐10.47) | ||||||
| A/A | 6 | 6 | 12.09 (3.91‐37.37) | 4.29 (2.07‐8.90) | 15.81 (4.78‐52.28) | 4.90 (2.30‐10.47) | ||||||
| CA10/C17orf112 | rs2113374 | 17 | 50 508 719 | C/C | 186 | 23 | 1 | 1 | 1.38.E−06 | 1 | 1 | 7.06.E−06 |
| C/T | 24 | 18 | 5.29 (2.62‐10.69) | 6.15 (2.94‐12.85) | 5.07 (2.48‐10.36) | 5.73 (2.68‐12.28) | ||||||
| T/T | 1 | 1 | 27.96 (6.85‐114.23) | 6.15 (2.94‐12.85) | 25.71 (6.15‐107.42) | 5.73 (2.68‐12.28) | ||||||
| PRUNE2 | rs117299725 | 9 | 79 423 611 | G/G | 191 | 24 | 1 | 1 | 2.53.E−06 | 1 | 1 | 8.53.E−06 |
| G/A | 24 | 18 | 5.97 (2.84‐12.56) | 5.97 (2.84‐12.56) | 5.69 (2.65‐12.24) | 5.69 (2.65‐12.24) | ||||||
| A/A | 0 | 0 | 35.63 (8.04‐157.79) | 5.97 (2.84‐12.56) | 32.41 (7.01‐149.92) | 5.69 (2.65‐12.24) | ||||||
| CDH13 | rs147631684 | 16 | 83 632 820 | G/G | 202 | 28 | 1 | 1 | 4.50.E−06 | 1 | 1 | 9.61.E−06 |
| G/A | 11 | 13 | 6.21 (2.64‐14.60) | 7.82 (3.25‐18.82) | 6.20 (2.51‐15.34) | 7.97 (3.18‐19.97) | ||||||
| A/A | 1 | 0 | 38.57 (6.98‐213.15) | 7.82 (3.25‐18.82) | 38.50 (6.30‐235.26) | 7.97 (3.18‐19.97) | ||||||
RS ID, reference SNP cluster ID; CHR, chromosome; OR1, odds ratio (OR) and 95% confidence interval (CI) in additive model; OR2, odds ratio (OR) and 95% confidence interval (CI) in dominant model.
Adjusted for body mass index and trastuzumab use.
All these 7 SNPs, except rs11894115, were found in non‐exonic regions (Table 3, Figures 3 and S2). rs17530621 on chromosome 4 had the smallest observed P‐value of 3.10E−06 and was located in an intergenic region between BEND4 and SHISA3; this region also included the 5′‐end of ATP8A1 (Figure 3A). Although the allele frequency of rs17530621 was known to be 0.60 for the reference allele (T) and 0.40 for an alternative allele (G) in European populations, the frequency was 0.14 for T and 0.86 for G in our study population. These frequencies were similar to those reported in the dbSNP database (https://www.ncbi.nlm.nih.gov/snp) for Asian populations, with T of 0.12 and G of 0.88. rs11894115 on chromosome 2 was a missense variant in the exonic region of the MPP4 gene with a P‐value of 4.71E−06. rs58328254 on chromosome 8 and rs117299725 on chromosome 9 were located in the introns of RPL7 and PRUNE2 genes, respectively and showed strong associations, with P‐values of 6.09E−06 and 8.53E−06, respectively. rs2113374 and rs147631684 were placed in the intron region of the long intervening non‐coding RNA (lincRNA) of C17orf112 and CDH13 genes, respectively. rs6804462 was located near MIR548AB. In all results, denoting the homozygote of the reference allele as A/A, heterozygous as A/B, and homozygote of the alternative (risk, minor) allele as B/B, the odds of having the cardiotoxicity significantly increased as the number of alternative alleles increased (Table 3).
FIGURE 3.
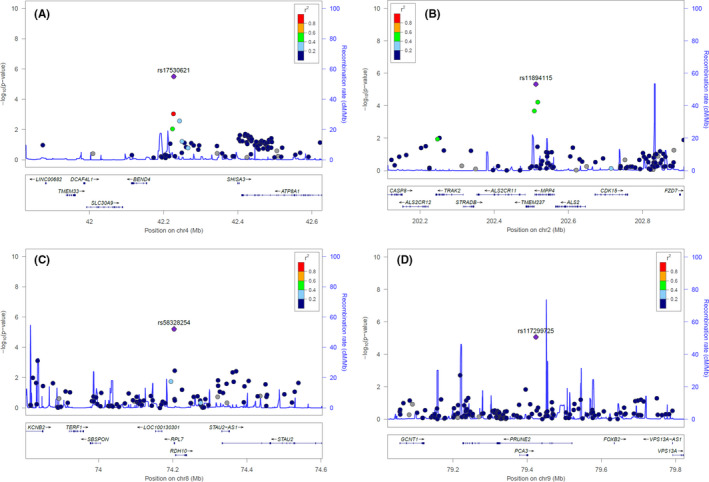
Regional association plots for (A) rs17530621, (B) rs11894115, (C) rs58328254, and (D) rs117299725
Only among the persistent ACT group, we identified 62 SNPs with a minimum P‐value < 1.0E−04 in the crude or adjusted model, corresponding to an effective number of 53.4 (Table S2). In the adjusted model, rs4336659, which had the smallest P‐value, was located near the GLIS3 and SLC1A1 genes (P = 3.03E−06). The WWOX gene, which had a P‐value of 7.24E−05 in the crude model of the entire study population data set (Table S1), had a higher association in the persistent ACT group (P = 7.28E−06, Table S2). The PRUNE2 gene was a common gene that was identified both in the entire study population data set and in the persistent ACT group. (Table S2, Figure S3).
3.3. Expression analysis of the candidate SNPs
To identify the association between the genetic variants and expression of related genes, we conducted expression quantitative trait loci (eQTL) analysis using the Genotype‐Tissue Expression (GTEx) database. 20 As a result, 2 SNPs showed association with the expression of nearby genes (P < .05). rs17530621 was associated with the expression of the ATP8A1 gene in the tibial artery (P = 3.8E−03) and in the left ventricle of the heart (P = 2.0E−02) with normalized effect sizes (NES) of 0.084 and 0.113, respectively. rs58328254 was associated with expression of the RDH10‐AS1 (antisense 1) gene in the tibial artery (P = 2.7E−12) and aorta artery (P = 1.0E−06) with NESs of −0.393 and −0.310, respectively (Table S3).
3.4. Overall survival
During the follow‐up period, 26 (10.1%) patients died. The most common cause of death was breast cancer recurrence, which occurred in 3 (7.1%) patients in the case group and 19 (8.8%) patients in the control group (Table 1). There was no death from ACT in the case group. In addition, there was no significant difference in overall survival (P = .464) between the 2 groups (Figure S4).
4. DISCUSSION
Cardiotoxicity is a major side effect of anthracycline treatment, with a higher incidence in patients who received a cumulative dose of more than 450 mg/m2, as well as those with cardiovascular comorbidities or those who receive combination treatment with other chemotherapeutic agents such as trastuzumab. 3 , 5 Cardiotoxicity is a serious lifelong problem for both children and adults and is associated with high morbidity and mortality. The extent and frequency of cardiotoxicity varies significantly across patients. Susceptible genetic factors, as well as various clinical risk factors, have been studied to identify high‐risk patients before anthracycline administration. Previous studies have revealed that genes related to the generation of excess reactive oxygen species (encoded by POR, NCF4, RAC2, and CYBA), intracellular accumulation of toxic metabolites (encoded by CBR3, SLC8A3 and UGT1A6), and interaction of anthracycline with topoisomerase‐2β (encoded by RARG) were significant in the development of ACT. 21 , 22 In particular, the RARG variant (rs2229774) was found in one discovery cohort (280 European population) and 2 validation cohorts (96 European and 80 non‐European) that collectively included more than 450 childhood cancer patients. 11 RARG is known to bind the topoisomerase IIβ promoter and in a mouse model was expressed in cardiomyocytes after injury. 23 , 24 However, the RARG variant was not found in a GWAS of more than 7800 breast cancer patients from 3 large randomized clinical trials who had received anthracycline treatment. 13 This discrepancy in findings may be due to differences in study populations, methodologies, and concurrent treatments used across studies. In addition, the impact of age and other environmental factors such as weight and other comorbidities may be more significant in adult patient populations compared with cohorts involving children.
To date, most studies have been conducted on Western populations and only a few large‐scale clinical studies involving Asian patient cohorts have been conducted. The current study only included Korean women who had been diagnosed with early breast cancer. Every patient in our cohort received 4‐6 cycles of doxorubicin sequentially with or without trastuzumab as a neoadjuvant/adjuvant treatment. Based on our results, higher BMI and trastuzumab use were important clinical factors that were strongly associated with ACT; these findings were consistent with the results of prior studies. 25 , 26 , 27 In our GWAS analysis, we found that 7 susceptible variants, including rs17530621, rs6804462, rs11894115, rs58328254, rs2113374, rs117299725, and rs147731684, were associated with ACT across the entire study population (minimum P‐value < 1.0E−05). However, the SNPs discovered in this study did not reach statistical significance when adjusting for clinical factors, higher BMI, and trastuzumab use.
Our expression analysis found only 2 SNPs (rs17530621 and rs58328254) that were associated with expression of nearby genes, because most variants, except rs118941115, were located in non‐coding regions. Their biological significance in relation to ACT development was also unclear, however several previous studies have suggested that the genes we identified may be linked functionally to cardiotoxicity. In a model of heart failure, SHISA3, which was associated with rs17530621 and primarily expressed in the vascular cells of the fetal heart, was involved in pathological tissue remodeling via reduction of KLF15 and WNT signaling activation. 28 PRUNE2 is reported to be highly expressed in patients with idiopathic dilated heart failure, as well as in patients with ischemic heart failure and also is functionally related to myocardial muscle tension. 29 Genetic polymorphisms of CDH13 are associated with plasma adiponectin levels and are involved in the development of cardiometabolic disease. 30 , 31 These data suggest that it is necessary to further study the relationship between these genes and development of ACT.
In this study, SNPs in RARG, SLC8A3, UGT1A6, NCF4, and RAC2 did not emerge as significant, despite reports in previous studies that these genes were associated with susceptibility to ACT. 11 , 19 , 20 There may be several reasons for these discrepancies, including the relatively small size of our control group, differences in ethnicity, and differences in combined chemotherapeutic regimens. For example, the MAF of the RARG variant rs2229774 was 6.7% in Europeans, while this value was significantly lower (0.7%) in East Asians. The MAF of rs17530621 found in our study also showed a significant difference between European and Asian populations. Such differences may have caused the lack of identification of results between studies. One advantage of our study, however, is that we were able to adjust for possible clinical risk factors in patients with long‐term follow‐up. Our study, therefore, enabled us to infer that the effect of genetic variations in the development of ACT may be diluted by the effects of various clinical risk factors.
In conclusion, we found that clinical factors such as higher BMI and trastuzumab use were important factors in the development of ACT among patients with early breast cancer. Our GWAS identified candidate SNPs that were associated with the development of ACT in a Korean female population. Although none of these SNPs reached a significance threshold, these candidate SNPs warrant further study.
DISCLOSURE
The authors have no conflicts of interest.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Tables S1‐S3
ACKNOWLEDGMENTS
We thank all the patients who participated in this study. This work was supported by NCCK grant number 1940520‐1.
Park B, Sim SH, Lee KS, Kim HJ, Park IH. Genome‐wide association study of genetic variants related to anthracycline‐induced cardiotoxicity in early breast cancer. Cancer Sci. 2020;111:2579–2587. 10.1111/cas.14446
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Volkova M, Russell R. Anthracycline cardiotoxicity: prevalence, pathogenesis and treatment. Curr Cardiol Rev. 2011;7:214‐220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kremer L, van der Pal H, Offringa M, van Dalen EC, Voûte PA. Frequency and risk factors of subclinical cardiotoxicity after anthracycline therapy in children: a systematic review. Ann Oncol. 2002;13:819‐829. [DOI] [PubMed] [Google Scholar]
- 3. Singal PK, Iliskovic N. Doxorubicin‐induced cardiomyopathy. N Engl J Med. 1998;339:900‐905. [DOI] [PubMed] [Google Scholar]
- 4. Cameron D, Brown J, Dent R, et al. Adjuvant bevacizumab‐containing therapy in triple‐negative breast cancer (BEATRICE): primary results of a randomised, phase 3 trial. Lancet Oncol. 2013;14:933‐942. [DOI] [PubMed] [Google Scholar]
- 5. Gianni L, Salvatorelli E, Minotti G. Anthracycline cardiotoxicity in breast cancer patients: synergism with trastuzumab and taxanes. Cardiovasc Toxicol. 2007;7:67‐71. [DOI] [PubMed] [Google Scholar]
- 6. Kotamraju S, Konorev EA, Joseph J, et al. Doxorubicin‐induced apoptosis in endothelial cells and cardiomyocytes is ameliorated by nitrone spin traps and ebselen. Role of reactive oxygen and nitrogen species. J Biol Chem. 2000;275:33585‐33592. [DOI] [PubMed] [Google Scholar]
- 7. Dowd NP, Scully M, Adderley SR, Cunningham AJ, Fitzgerald DJ. Inhibition of cyclooxygenase‐2 aggravates doxorubicin‐mediated cardiac injury in vivo. J Clin Invest. 2001;108:585‐590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanco JG, Leisenring WM, Gonzalez‐Covarrubias VM, et al. Genetic polymorphisms in the carbonyl reductase 3 gene CBR3 and the NAD(P)H:quinone oxidoreductase 1 gene NQO1 in patients who developed anthracycline‐related congestive heart failure after childhood cancer. Cancer. 2008;112:2789‐2795. [DOI] [PubMed] [Google Scholar]
- 9. Wang X, Liu W, Sun C‐L, et al. Hyaluronan synthase 3 variant and anthracycline‐related cardiomyopathy: a report from the children's oncology group. J Clin Oncol. 2014;32:647‐653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Armenian SH, Ding Y, Mills G, et al. Genetic susceptibility to anthracycline‐related congestive heart failure in survivors of haematopoietic cell transplantation. Br J Haematol. 2013;163:205‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aminkeng F, Bhavsar AP, Visscher H, et al. A coding variant in RARG confers susceptibility to anthracycline‐induced cardiotoxicity in childhood cancer. Nat Genet. 2015;47:1079‐1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Visscher H, Ross CJD, Rassekh SR, et al. Pharmacogenomic prediction of anthracycline‐induced cardiotoxicity in children. J Clin Oncol. 2012;30:1422‐1428. [DOI] [PubMed] [Google Scholar]
- 13. Schneider BP, Shen F, Gardner L, et al. Genome‐wide association study for anthracycline‐induced congestive heart failure. Clin Cancer Res. 2017;23:43‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sparano JA, Wang M, Martino S, et al. Weekly paclitaxel in the adjuvant treatment of breast cancer. N Engl J Med. 2008;358:1663‐1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zamorano JL, Lancellotti P, Rodriguez Muñoz D, et al. 2016 ESC Position Paper on cancer treatments and cardiovascular toxicity developed under the auspices of the ESC Committee for Practice Guidelines: The Task Force for cancer treatments and cardiovascular toxicity of the European Society of Cardiology (ESC). Eur Heart J. 2016;37:2768‐2801. [DOI] [PubMed] [Google Scholar]
- 16. Moon S, Kim YJ, Han S, et al. The Korea Biobank Array: design and identification of coding variants associated with blood biochemical traits. Sci Rep. 2019;9:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. SNPolisher User Guide (Version 1.5.2.). Affymetrix Inc; 2015. http://tools.thermofisher.com/content/sfs/manuals/SNPolisher_User_Guide.pdf. Accessed December 20, 2018. [Google Scholar]
- 18. Joo J, Kwak M, Chen Z, et al. Efficiency robust statistics for genetic linkage and association studies under genetic model uncertainty. Stat Med. 2010;29:158‐180. [DOI] [PubMed] [Google Scholar]
- 19. Cheverud JM. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87:52‐58. [DOI] [PubMed] [Google Scholar]
- 20. GTEx Consortium . The Genotype‐Tissue Expression (GTEx) project. Nat Genet. 2013;45:580‐585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang VY, Wang JJ. Pharmacogenetics of chemotherapy‐induced cardiotoxicity. Curr Oncol Rep. 2018;20:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Linschoten M, Teske AJ, Cramer MJ, et al. Chemotherapy‐related cardiac dysfunction: a systematic review of genetic variants modulating individual risk. Circ Genom Precis Med. 2018;11:e001753. [DOI] [PubMed] [Google Scholar]
- 23. Delacroix L, Moutier E, Altobelli G, et al. Cell‐specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010;30:231‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bilbija D, Haugen F, Sagave J, et al. Retinoic acid signalling is activated in the postischemic heart and may influence remodelling. PLoS ONE. 2012;7:e44740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez EA, Suman VJ, Davidson NE, et al. Cardiac safety analysis of doxorubicin and cyclophosphamide followed by paclitaxel with or without trastuzumab in the North Central Cancer Treatment Group N9831 adjuvant breast cancer trial. J Clin Oncol. 2008;26:1231‐1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Slamon D, Eiermann W, Robert N, et al. Adjuvant trastuzumab in HER2‐positive breast cancer. N Engl J Med. 2011;365:1273‐1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guenancia C, Lefebvre A, Cardinale D, et al. Obesity as a risk factor for anthracyclines and trastuzumab cardiotoxicity in breast cancer: a systematic review and meta‐analysis. J Clin Oncol. 2016;34:3157‐3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noack C, Iyer LM, Liaw NY, et al. KLF15‐Wnt‐dependent cardiac reprogramming up‐regulates SHISA3 in the mammalian heart. J Am Coll Cardiol. 2019;74:1804‐1819. [DOI] [PubMed] [Google Scholar]
- 29. Song Y, Ahn J, Suh Y, et al. Identification of novel tissue‐specific genes by analysis of microarray databases: a human and mouse model. PLoS ONE. 2013;8:e64483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chung C‐M, Lin T‐H, Chen J‐W, et al. A genome‐wide association study reveals a quantitative trait locus of adiponectin on CDH13 that predicts cardiometabolic outcomes. Diabetes. 2011;60:2417‐2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morisaki H, Yamanaka I, Iwai N, et al. CDH13 gene coding T‐cadherin influences variations in plasma adiponectin levels in the Japanese population. Hum Mutat. 2012;33:402‐410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Tables S1‐S3
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


