Abstract
Lung adenocarcinoma with micropapillary pattern (MPP) has an aggressive malignant behavior. Limited resection should be avoided because of its high recurrence rate. If adenocarcinoma with MPP is diagnosed preoperatively, the selection of proper treatment is possible. To explore a preoperative biomarker for diagnosing MPP, we undertook RNA sequencing analysis of 25 clinical samples as the training set, including 6 MPP, 16 other adenocarcinoma subtypes, and 3 normal lung tissues. Unsupervised hierarchical clustering analysis suggested a presence of subgroup with MPP showing different gene expression phenotype. We extracted differentially expressed genes with high expression levels in MPP samples, and chose VSIG1, CXCL14, and BAMBI as candidate biomarkers for MPP. Reverse transcription‐quantitative PCR analysis confirmed a significantly higher expression of VSIG1 (P = .03) and CXCL14 (P = .02) in MPP than others. In a validation set of 4 MPP and 4 non‐MPP samples, CXCL14 expression was validated to be significantly higher in MPP than in non‐MPP (P = .04). Comparing a total of 10 MPP and 20 non‐MPP samples, the area under the curve of CXCL14 to distinguish MPP from others was 0.89. The threshold value was 0.0116, corresponding to sensitivity 80% and specificity 90%. In immunostaining of CXCL14, the staining score was significantly higher in MPP cases than others, where not only the MPP component but also other components showed heterogeneous staining in adenocarcinoma tissues with MPP. Moreover, a higher staining score of CXCL14 was significantly associated with poorer prognosis in all patients (P = .01) or within cases in stage I‐III (P = .01). In summary, we identified CXCL14 as a possible diagnostic biomarker of MPP.
Keywords: biomarker, CXCL14, lung adenocarcinoma, micropapillary pattern, RNA‐seq
In order to diagnose lung adenocarcinoma with micropapillary pattern (MPP) prior to the surgery, and to select appropriate treatment, eg lobectomy or supplemental treatment in addition to the surgery, biomarkers to predict the existence of MPP are desired. We undertook a transcriptome analysis in each subtype of lung adenocarcinoma by RNA sequencing of a training sample set, and a validation analysis of expression for mRNA level by RT‐qPCR and for protein level by immunohistochemistry in training sample and validation sets. We identified CXCL14 as a possible diagnostic biomarker of adenocarcinoma with MPP, which should be useful in preoperative decisions of suitable treatment against this cancer subtype and improvement of its prognosis.
Abbreviations
- AUC
area under the curve
- COPD
chronic obstructive pulmonary disease
- CXCL14
C‐X‐C motif chemokine ligand 14
- EGFR
epidermal growth factor receptor
- FFPE
formalin‐fixed paraffin‐embedded
- FPKM
fragments per kilobase of exon per million mapped fragments
- GO
gene ontology
- MPP
micropapillary pattern
- RNA‐seq
RNA sequencing
- ROC
receiver operating characteristic
- RT‐qPCR
reverse transcription‐quantitative PCR
- SCLC
small cell lung cancer
- SqCC
squamous cell carcinoma
1. INTRODUCTION
In 2011, an international multidisciplinary panel of lung cancer experts presented a novel classification system of lung adenocarcinoma by the predominant histologic subtype presenting more than 5% of tumor. The basis for the classification of lung adenocarcinomas was accepted in the recently published 2015 WHO classification of lung tumors, 4th edition, 1 where the predominant histologic subtype of adenocarcinoma and its correlation with oncological prognosis were also mentioned.
Histologic subtypes of adenocarcinoma differ in prognosis, and in particular, adenocarcinoma subtypes with MPP and solid pattern have been reported to show a poorer prognosis than other subtypes. 2 , 3 , 4 , 5 , 6 In our recent publication, MPP not only showed worse overall survival, but also its coexistence with free tumor clusters resulted in a further negative impact on the postoperative prognosis. 7 Adenocarcinoma with MPP with free tumor clusters was observed in approximately half of the cases in our previous cohort, and showed higher incidence of nodal metastasis, so that the survival curve was nearly equivalent to that in T3 cases of MPP‐negative adenocarcinoma.
Biomarkers to predict the existence of MPP are therefore expected to be developed, in order to diagnose lung adenocarcinoma with MPP prior to surgery and select appropriate treatment, eg lobectomy or supplemental treatment in addition to surgery, However, there is difficulty in the diagnosis using biopsied samples or frozen tissue samples obtained during surgery. 8 , 9 , 10 , 11 , 12
We undertook a transcriptome analysis in each subtype of lung adenocarcinoma by RNA‐seq of a training set of lung adenocarcinoma samples and explored the useful biomarker to diagnose the existence of MPP within adenocarcinoma. Through validation of the expression by RT‐qPCR and immunohistochemistry in the training set as well as the validation set of lung adenocarcinoma samples, we identified CXCL14 as a preoperative diagnostic biomarker of adenocarcinoma with MPP.
2. MATERIALS AND METHODS
2.1. Clinical samples
From 52 patients of lung adenocarcinoma who underwent lung surgery at Chiba University Hospital between 2009 and 2018, we collected 30 frozen primary lung adenocarcinoma samples, 3 frozen normal lung tissues, and 52 FFPE tissues of lung adenocarcinoma, with written informed consent. Twenty‐two frozen primary lung adenocarcinoma samples and 3 normal lung samples were used as training samples for RNA‐seq as well as RT‐qPCR analysis. The other 8 frozen samples were used for the RT‐qPCR analysis to validate the results of the training set. For immunohistochemistry, 52 FFPE samples were analyzed, including the 30 samples undergoing RT‐qPCR analysis. Clinicopathological data of these patients, including age, gender, pathological stage of tumors, mutation of EGFR, smoking history, presence/absence of COPD, recurrence of lung cancer, and overall survival, were extracted from our clinical record and used for correlation analyses between these factors and marker expression. EGFR mutation was analyzed by the peptide nucleic acid‐locked nucleic acid PCR clamp method. In addition to adenocarcinoma, FFPE samples of 15 SCLC and 15 SqCC tissues were also collected, and used for immunohistochemistry. The study was approved by the institutional review board at Chiba University (No. 1027).
2.2. RNA preparation and RNA‐seq analysis
All tissue samples were microscopically confirmed by 2 independent pathologists to confirm tumor cell content higher than 50%, and dissection was carried out if it was necessary to enrich tumor cells. RNA was extracted using an RNeasy mini kit (Qiagen). Libraries for RNA‐seq were prepared using the TruSeq Stranded mRNA Sample Prep Kit (Illumina), following the manufacturer’s protocol. Deep sequencing was carried out on the Illumina HiSeq 1500 platform using the TruSeq Rapid SBS Kit (Illumina) in 50‐base single‐end mode according to the manufacturer’s protocol. Gene expression levels were shown by FPKM. The RNA‐seq analysis was undertaken for 3 normal lung tissues and 22 adenocarcinoma samples including 3 MPP predominant, 8 papillary predominant (including 3 cases with 20%, 30%, and 30% of MPP fraction), 5 lepidic predominant, 3 acinar predominant, and 3 solid predominant cases. Among the 22 adenocarcinoma cases, 3 MPP dominant cases and 3 papillary dominant cases with MPP fraction (20% or higher) were regarded as MPP cases, and the remaining 16 adenocarcinoma subtypes were regarded as non‐MPP cases.
2.3. Clustering analysis
Unsupervised 2‐way hierarchical clustering was carried out to assess the presence of gene expression phenotypes in lung adenocarcinoma. Genes (n = 2153) showing significantly different gene expression levels between 22 adenocarcinoma and 3 normal lung samples (P < .05, t‐test) were extracted, and used for clustering analysis.
2.4. Extraction of highly expressed genes
The following criteria were used to extract candidate marker genes using RNA‐seq data of lung adenocarcinoma with and without MPP. We found 161 genes that met the following criteria: (i) average value of FPKM in adenocarcinoma with MPP is greater than 10; (ii) average FPKM value in adenocarcinoma with MPP is greater than 2‐fold compared with the average value of FPKM in non‐MPP adenocarcinoma; (iii) average FPKM value in normal lung tissue is greater than 10; and (iv) top 2 z‐score values in adenocarcinoma with MPP are greater than 0. We similarly found 58 genes that met the following criteria: (i) average FPKM value in non‐MPP adenocarcinoma is greater than 10; (ii) average FPKM value in non‐MPP adenocarcinoma is greater than 2‐fold compared with the average FPKM value in adenocarcinoma with MPP; (iii) average FPKM value in normal lung tissue is less than 10; and (iv) top 6 z‐score values in non‐MPP adenocarcinoma are greater than 0.
2.5. Gene ontology analysis
Gene annotation enrichment analysis was undertaken based on GO (biologic process, cellular component, and molecular function) using the Functional Annotation tool at DAVID Bioinformatics Resources (https://david.ncifcrf.gov/).
2.6. Reverse transcription‐qPCR analysis
Total RNA was treated with DNase I (Ambion), and cDNA was synthesized from 10 μg total RNA using a Superscript II kit (Life Technologies). Quantitative PCR was carried out using SYBR Green PCR Core Reagents (PE Biosystems) and an iCycler Thermal Cycler (Bio‐Rad Laboratories). The number of molecules of a specific gene in a sample was measured by comparing its amplification with the amplification of standard samples that contained 101‐1010 copies of the gene. 13 The quantity of mRNA of each gene was normalized with that of ACTB. The RT‐qPCR primers are shown in Table S1.
2.7. Immunohistochemistry
To evaluate the protein expression in cancer tissue, FFPE tissue samples were analyzed by immunohistochemistry, as previously described. 14 Rabbit anti‐CXCL14 Ab (ab46010; Abcam) was used to evaluate expression of CXCL14, which was scored by assigning a percentage of positive cells and an intensity score. Immunohistochemical staining intensity of 0, 1, 2, or 3, indicated absent, weak, moderate, or strong expression, respectively. The percentages of positive cells were scored as follows: 0, no staining; 1, 1%‐5%; 2, 5%‐25%; 3, 25%‐50%; 4, 50%‐75%; and 5, 75%‐100%. The intensity score was multiplied by the percentage score to obtain the staining score.
2.8. Statistical analysis
Z‐score was calculated by R and the heatmap was drawn in Java TreeView (http://jtreeview.sourceforge.net/). Unsupervised 2‐way hierarchical clustering was undertaken using Cluster 3.0 software (http://bonsai.hgc.jp/~mdehoon/software/cluster/). Fisher’s exact test, the χ2 test, and Wilcoxon’s signed rank test were used for analysis of clinical characteristics. The relative expression by RT‐qPCR and the staining score in immunohistochemistry were analyzed by Wilcoxon’s signed rank test with P < .05 considered statistically significant. Analyses of the AUC of the ROC curve were carried out using JMP Pro 13.0.0 (SAS Institute). Overall survival was undertaken using Kaplan‐Meier survival curves and the log‐rank statistic and Cox proportional hazard regression analyses were applied to calculate risk ratios and 95% confidence intervals for each risk factor using JMP Pro 13.0.0 software.
3. RESULTS
3.1. Clinical characteristics
The composition of all adenocarcinoma subtypes and present/absent EGFR mutation are shown in Table S2, and clinicopathologic features are summarized in Table 1. There was no significant difference in age, gender, pathological stage, smoking index, or COPD between MPP and non‐MPP adenocarcinoma, while death and recurrence after surgery were more frequently observed in adenocarcinoma with MPP than in non‐MPP adenocarcinoma. EGFR mutations were found in 40.3% of adenocarcinomas; there was no significant difference in frequencies of EGFR mutations between MPP and non‐MPP adenocarcinoma, or among any predominant subtypes of adenocarcinoma, when analyzing a total of 52 adenocarcinoma samples (Table 1).
Table 1.
Clinicopathologic features of adenocarcinoma with and without micropapillary pattern (MPP)
| RNA sequencing (n = 22) | RT‐qPCR (n = 30) | Immunohistochemistry (n = 52) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MPP | Non‐MPP | P value | MPP | Non‐MPP | P value | MPP | Non‐MPP | P value | |
| No. of samples | 6 | 16 | — | 10 | 20 | 21 | 31 | ||
| Age (y) | 68.2 | 67.5 | .6 | 70.7 | 66.1 | .1 | 69.0 | 67.8 | .5 |
| Gender | |||||||||
| Male | 3 | 8 | .6 | 6 | 11 | .3 | 11 | 18 | .3 |
| Female | 3 | 8 | 4 | 9 | 10 | 13 | |||
| Stage, n (%) | |||||||||
| I | 2 (33) | 9 (56) | .2 | 4 (40) | 12 (60) | .5 | 9 (43) | 18 (58) | .3 |
| II | 0 (0) | 3 (19) | 0 (0) | 3 (15) | 2 (10) | 6 (19) | |||
| III | 4 (67) | 4 (25) | 5 (50) | 5 (25) | 8 (38) | 6 (19) | |||
| IV | 0 (0) | 0 (0) | 1 (10) | 0 (0) | 2 (10) | 1 (3) | |||
| EGFR mutation (+), n (%) | 0 (0) | 11 (69) | .01 | 1 (10) | 11 (55) | .006 | 6 (29) | 15 (48) | .2 |
| Smoking (pack‐y) | 39.2 | 24.9 | .2 | 31.0 | 24.8 | .5 | 20.7 | 28.5 | .1 |
| COPD, n (%) | 3 (50) | 2 (13) | .1 | 4 (40) | 4 (20) | .2 | 5 (24) | 11 (36) | .2 |
| Recurrence after surgery, n (%) | 5 (83) | 3 (19) | .01 | 7 (70) | 5 (25) | .02 | 15 (71) | 8 (26) | .001 |
| Mortality during follow‐up, n (%) | 4 (67) | 3 (19) | .02 | 5 (50) | 3 (15) | .02 | 6 (29%) | 4 (13%) | .08 |
Clinicopathologic factors were compared between MPP and non‐MPP cases, in the cohort assessed by RNA sequencing (n = 22), RT‐quantitative (q)‐PCR (n = 30), and immunohistochemistry (n = 52).
COPD, chronic obstructive pulmonary disease; EGFR, epidermal growth factor receptor.
3.2. Transcriptome analysis by RNA‐seq
Using RNA‐seq data of 22 lung adenocarcinoma and 3 normal lung tissue samples, we undertook unsupervised 2‐way hierarchical clustering to assess the presence of any gene expression phenotypes corresponding to predominant patterns (Figure S1). Three normal samples formed a distinct cluster, and 22 cancer samples were classified into 3 clusters. All the 3 micropapillary predominant cases were included in a cluster of 4 samples (P < .05), and the remaining sample was a papillary predominant case also containing MPP fraction. As 4 of 6 MPP cases showed similar gene expression phenotype, we hypothesized that there might be marker genes with differential expression patterns to predict MPP cases.
We then extracted 161 genes showing high expression and 58 genes showing low expression in 6 adenocarcinoma samples with MPP, which contained an MPP component of at least 20% (Figure 1A). Gene ontology analysis of the 2 groups showed significant enrichment of GO terms such as immune response, inflammatory response, and chemokine activity in the low expression group (Figure 1B), and GO terms such as alpha‐amylase activity, extracellular exosome, and extracellular space in the high expression group (Figure 1C). Genes related with extracellular space included CXCL14, and 2 other genes, BAMBI and VSIG1, were selected as the candidate marker genes for adenocarcinoma with MPP.
FIGURE 1.
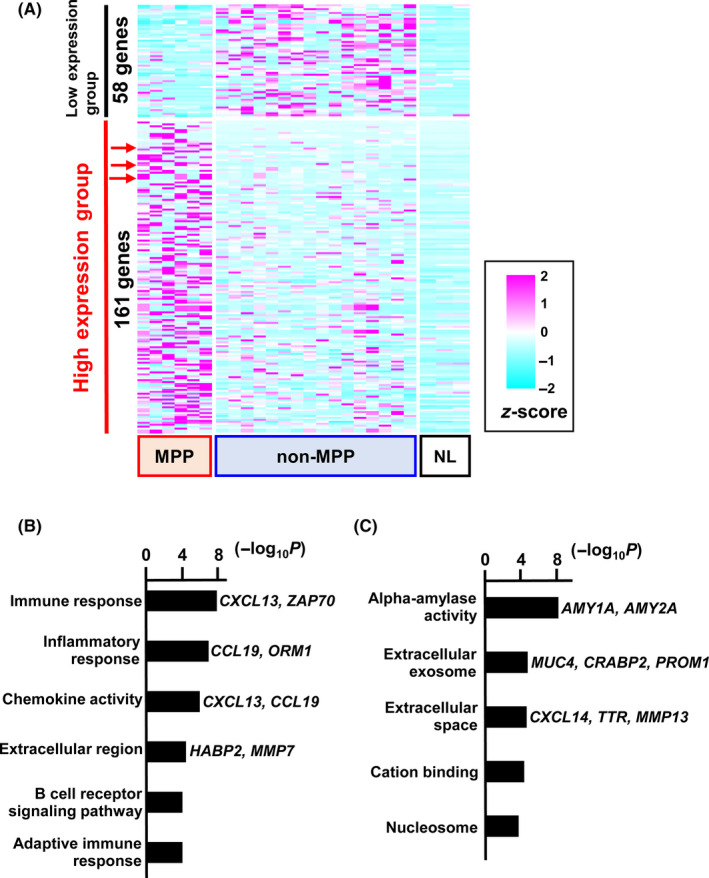
Differentially expressed genes. RNA sequencing was carried out for transcriptome analysis of lung adenocarcinoma with and without micropapillary pattern (MPP and non‐MPP, respectively), and normal lung tissues (NL). A, Heatmap showing differentially expressed genes. Of them, 161 genes were highly expressed specifically in MPP, but not in non‐MPP or NL (high expression group), and 58 genes were highly expressed specifically in non‐MPP, but not in MPP or NL (low expression group). Red arrows indicate the 3 candidate marker genes, VSIG1, CXCL14, and BAMBI. B, Gene ontology (GO) analysis of the low expression group of genes in MPP (high in non‐MPP). GO terms, eg immune response, inflammatory response, and chemokine activity, were significantly enriched. C, GO analysis of high expression genes in MPP (low in non‐MPP). GO terms, eg alpha‐amylase activity, extracellular exosome, and extracellular space, were significantly enriched
3.3. Validation of RNA‐seq results by RT‐qPCR
To quantitatively validate the results of the RNA‐seq analysis, RT‐qPCR was initially carried out using the training set, including 6 MPP and 16 non‐MPP samples. Although there was no marked difference in expression level of BAMBI, we found a significantly higher expression level of CXCL14 (P = .02) and VSIG1 (P = .03) in adenocarcinoma with MPP than non‐MPP (Figure 2A). Then, we additionally validated expression levels of the 2 genes using the validation set of 4 MPP and 4 non‐MPP samples and confirmed significantly higher expression of CXCL14 in adenocarcinoma with MPP than non‐MPP (P = .04) (Figure 2B). Using a total of 30 samples, including 10 MPP and 20 non‐MPP samples, we further compared CXCL14 expression level between adenocarcinoma with MPP and without MPP (Figure 2C), resulting in significant difference (P = .0006). When the AUC and ROC curves were evaluated using the relative expression level of CXCL14, the AUC was calculated to be 0.89. The CXCL14 threshold value, which offered the best sensitivity‐specificity relationship, was calculated to be 0.0116, with sensitivity 80% and specificity 90% (Figure 2D).
FIGURE 2.
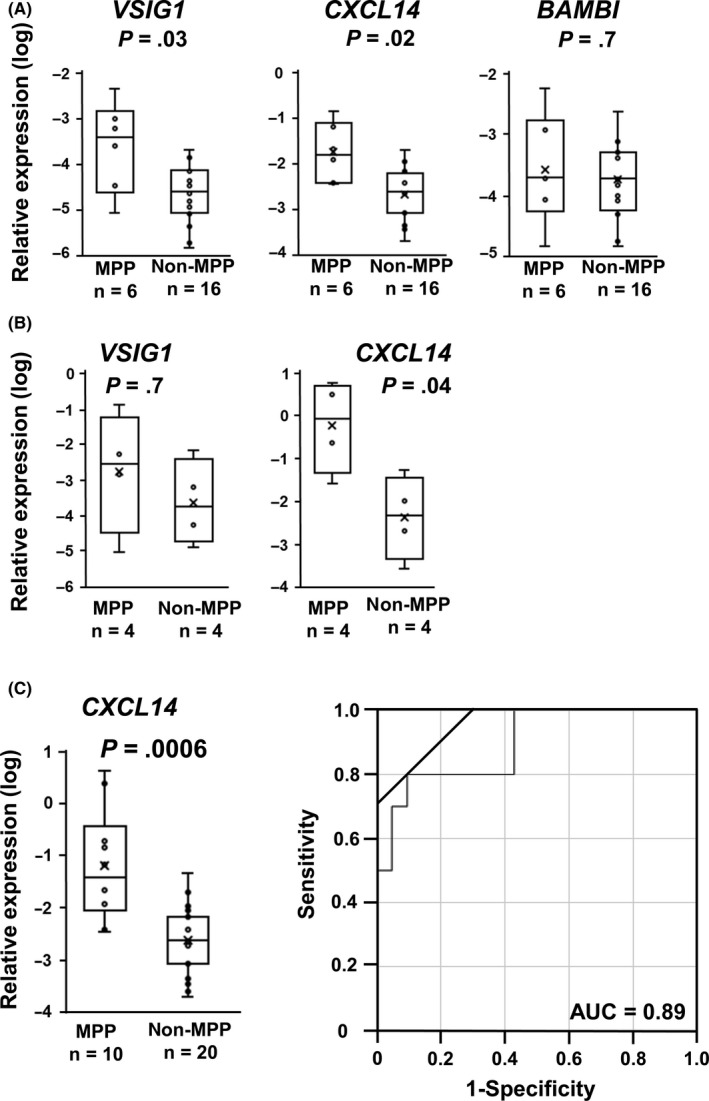
Reverse transcription‐quantitative (q)‐PCR analysis of candidate marker genes. Expression levels of VSIG1, CXCL14, and BAMBI were quantitatively analyzed by RT‐qPCR, and normalized by that of ACTB. A, RT‐qPCR analysis of the training set. Expression levels of VSIG1, CXCL14, and BAMBI, analyzed by RNA‐seq of the training set including 6 micropapillary pattern (MPP) and 16 non‐MPP samples, were measured by RT‐qPCR for validation. Significantly high expression of VSIG1 (P = .03) and CXCL14 (P = .02) in MPP was confirmed. B, RT‐qPCR analysis of the validation set. Expression levels of VSIG1 and CXCL14 were measured by RT‐qPCR using an additional sample set. Significantly high expression of CXCL14 in MPP was detected (P = .04). C, Relative expression of CXCL14 in a total of 30 samples, including 10 MPP and 20 non‐MPP samples. Significantly high expression of CXCL14 in MPP was detected (P = .0006). D, Receiver operating characteristic curve analysis of CXCL14 expression. The area under the curve (AUC) was 0.89. y‐axis, sensitivity; x‐axis, 1 − specificity
3.4. Immunostaining for CXCL14
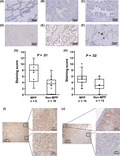
Protein expression of CXCL14 was analyzed by immunohistochemistry. We first analyzed FFPE tissue samples of the training set of 22 samples, including 6 MPP and 16 non‐MPP; frozen samples underwent RNA‐seq analysis. The staining scores of CXCL14 and the ratio of CXCL14 positive cells are summarized in Table S3. Expression of CXCL14 was detected in the cytoplasm of the tumor cells, confirming its expression in the tumor cells themselves (Figure 3A‐E), and CXCL14 staining was also clearly detected in the free tumor clusters in adenocarcinoma with MPP (Figure 3F). The staining score was significantly higher in MPP than non‐MPP samples (P = .01, Figure 3G). An additional 30 FFPE tissue samples, including 15 MPP and 15 non‐MPP, were analyzed as a validation set. The staining score was also significantly higher in MPP than non‐MPP samples (P = .02, Figure 3H), while the score was not significantly different amongst subtypes of non‐MPP adenocarcinoma (data not shown).
FIGURE 3.
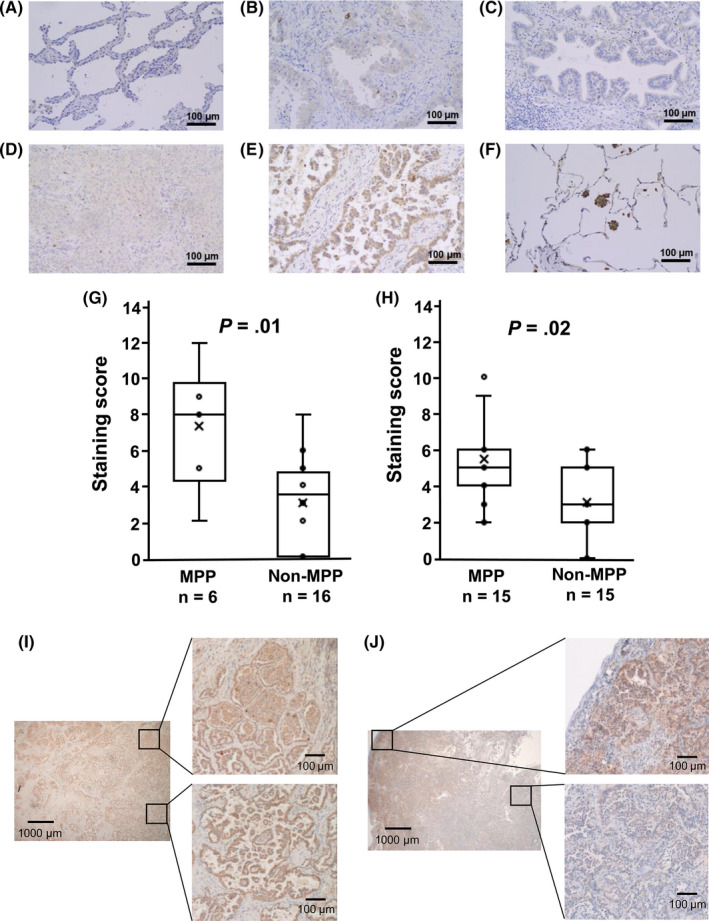
Immunohistochemistry of CXCL14. Expression of CXCL14 protein in lung adenocarcinoma specimens was analyzed by immunohistochemistry and shown (A‐F; magnification, ×100). A, Lepidic adenocarcinoma. B, Acinar adenocarcinoma. C, Papillary adenocarcinoma. D, Solid adenocarcinoma. E, Adenocarcinoma with micropapillary pattern (MPP). F, Free tumor cluster in adenocarcinoma with MPP. High expression of CXCL14 was confirmed in the cytoplasm of tumor cells in adenocarcinoma with MPP and its free tumor cluster. G, Staining score for the training set. Staining scores were calculated for the training set including 6 MPP and 16 non‐MPP samples and were shown to be significantly higher in MPP (P = .01). H, Staining score for the validation set. Immunohistochemistry of CXCL14 was carried out in 30 additional samples, including 15 MPP and 15 non‐MPP samples, confirming significantly higher staining score in MPP (P = .02). I, CXCL14 expression in components of other subtypes. Adenocarcinoma with MPP containing MPP and solid pattern components (left) was representatively shown to express CXCL14 at high level in both solid (right upper) and MPP (right lower) components. J, Heterogeneous CXCL14 expression. Adenocarcinoma with MPP containing MPP, acinar, and papillary pattern components (left) was representatively shown to express CXCL14 at high level in MPP and papillary pattern components (right upper), but at lower level in papillary and acinar pattern components (right lower)
Interestingly, the expression of CXCL14 was observed not only in the MPP component; homogeneous or scattered expression of CXCL14 was also observed in other components in adenocarcinoma with MPP (Figure 3I,J). In 21 MPP cases, CXCL14‐positive cells were observed at 78.3% in the MPP component and also observed at 61.9% in components of other subtypes (Table S3).
Immunostaining of CXCL14 protein was also undertaken for lung cancer other than adenocarcinoma: SCLC and SqCC. The staining scores in SCLC and SqCC were high, similar to MPP cases, and significantly higher than that of non‐MPP adenocarcinoma (P = .02 and P = .005, respectively) (Figure 4A). The high staining score did not correlate with smoking; the smoking index in MPP patients was significantly lower than SCLC and SqCC patients (P = .02 and P = .002, respectively) (Figure 4B).
FIGURE 4.
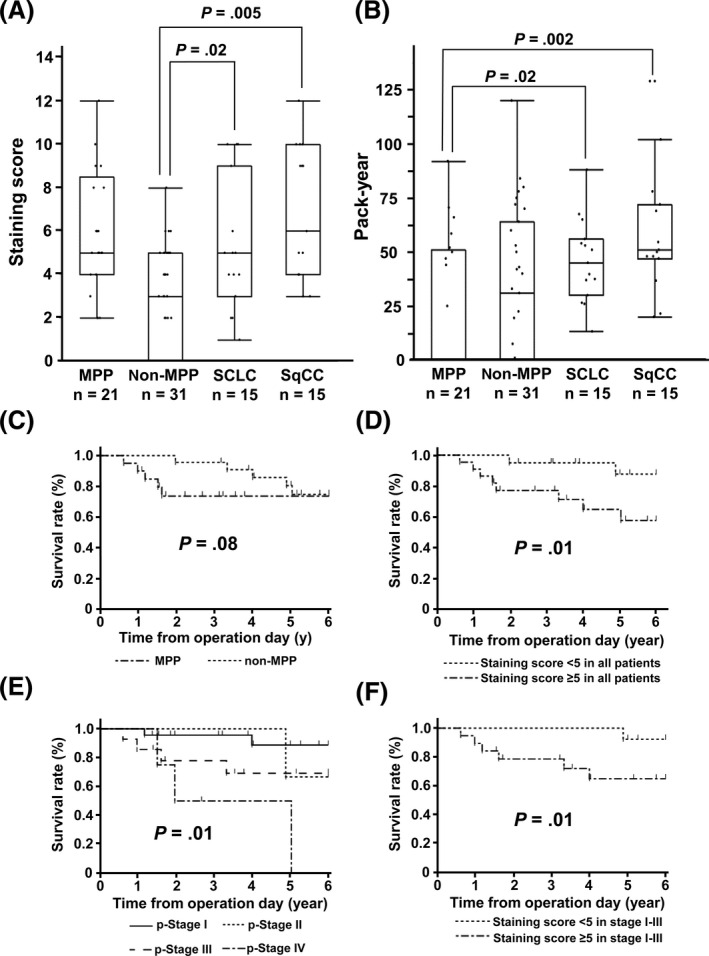
Association of CXCL14 expression with clinicopathologic factors. A, Small cell lung cancer (SCLC) and squamous cell cancer of the lung (SqCC) showed significantly higher expression of CXCL14 than non‐micropapillary pattern (MPP) adenocarcinoma (P = .02 and P = .005, respectively), which are at levels similar to adenocarcinoma with MPP. B, Smoking index. Adenocarcinoma with MPP showed significantly lower smoking index (pack‐year) than SCLC and SqCC (P = .02 and P = .002). C‐F, Kaplan‐Meier survival curve analyses of lung adenocarcinoma. Although a significant difference was not detected in overall survival between 21 MPP and 31 non‐MPP cases (P = .08) (C), high expression of CXCL14 (staining score ≥ 5) significantly correlated with worse prognosis (P = .01) (D). Overall survival was significantly different among pathological stages (I, II, III, and IV), and cases at stage IV showed worst prognosis (E). Even in analysis of stage I‐III cases only, high expression of CXCL14 (staining score ≥ 5) significantly correlated with worse prognosis (P = .01) (F)
3.5. Survival analysis
Correlation with overall survival was analyzed (Figure 4C‐F). Although MPP generally showed poorer prognosis than non‐MPP cases (Table 1), significant difference was not observed in the comparison of overall survival between MPP and non‐MPP using a total of 52 samples (P = .08, log‐rank test) (Figure 4C). Cases with higher staining score of CXCL14 (5 or higher) showed significantly worse overall survival than those with lower staining (less than 5) (Figure 4D). While prognosis was significantly different among pathological stages (P = .01), patients at stage IV showed the worst prognosis (Figure 4E). When analyzing patients at stage I‐III only, higher staining score of CXCL14 (5 or higher) significantly correlated with worse overall survival (Figure 4F).
Finally, correlation between overall survival and clinicopathologic factors was analyzed by the Cox proportional hazard model (Table 2). Only CXCL14 staining score was found to be significant in univariate analysis (P = .02) and in multivariate analysis (P = .02).
Table 2.
Univariate and multivariate analyses of clinicopathologic factors and prognosis in patients with adenocarcinoma
| Clinicopathologic factor | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| P value | Hazard ratio | 95% CI | P value | |||
| Sex | Male | 28 | .10 | 3.28 | 0.88‐15.59 | .08 |
| Female | 24 | |||||
| Age (y) | ≥65 | 33 | .90 | |||
| <65 | 19 | |||||
| Smoking index (pack‐y) | ≥20 | 26 | .80 | |||
| <20 | 26 | |||||
| MPP | MPP | 21 | .20 | 1.60 | 0.41‐6.21 | .50 |
| Non‐MPP | 31 | |||||
| CXCL14 staining score | ≥5 | 25 | .02 | 4.74 | 1.12‐32.29 | .03 |
| <5 | 27 | |||||
Univariate analysis was undertaken for sex, age, smoking index, presence of micropapillary pattern (MPP) component, and C‐X‐C motif chemokine ligand 14 (CXCL14) staining score, using Cox hazard regression model. Multivariate analysis was undertaken for sex, MPP, and CXCL14 staining score, using Cox hazard regression model.
CI, confidence interval.
4. DISCUSSION
The presence of MPP is known to be a factor associated with poor prognosis in lung adenocarcinoma. While the preoperative detection of MPP is difficult, limited resection including wide wedge resection and segmentectomy to adenocarcinoma with MPP reportedly resulted in a worse prognosis than lobectomy. 15 Approximately 50% of adenocarcinoma with MPP reportedly showed small tumor clusters, spreading in air space up to 2.5 cm from the main tumor. 7 , 16 Residual free tumor clusters in the resection margin could lead to tumor recurrence, thus it has been considered necessary to search for biomarkers enabling preoperative diagnosis of adenocarcinoma with MPP. In the present study, we undertook RNA‐seq screening in lung adenocarcinoma and identified that high expression of CXCL14 could be a biomarker for adenocarcinoma with MPP and poorer prognosis.
There have been abundant studies to analyze gene expression in lung cancer, and some articles pointed out their correlation with subgroups of adenocarcinoma. Bhattacharjee et al 17 classified 186 lung tumor samples into 4 distinct subgroups by hierarchical clustering analysis, and identified a poorer prognosis in the subgroup with neuroendocrine gene expression, in contrast to a relatively good prognosis in the subgroup containing bronchioloalveolar carcinoma. Garber et al 18 classified lung adenocarcinoma into 3 subgroups and reported that less differentiated adenocarcinoma showed a poorer prognosis. In terms of the correlation between molecular features and adenocarcinoma subtypes, The Cancer Genome Atlas reported that adenocarcinoma subtypes and prognostic diversity did not necessarily match the molecular features defined by transcriptional profiles, CpG island methylation, or oncogene mutations. 19 Conversely, Choi et al 20 undertook hierarchical clustering analysis using the mutation profile, and classified the acinar and the papillary subtypes of adenocarcinoma in 1 cluster, and the micropapillary and the solid subtypes in a different cluster. Another report showed that the solid and the lepidic subtypes showed a different gene expression profile from other subtypes. 21 Our hierarchical clustering analysis in this study showed that all the 3 MPP dominant cases and 1 papillary dominant case with MPP component formed a clearly distinct cluster (Figure S1), suggesting a presence of molecular marker genes to distinguish adenocarcinoma subtype with MPP component. We extracted genes differentially expressed at higher levels in adenocarcinoma with MPP, which showed significant enrichment of GO terms, eg alpha‐amylase activity, extracellular exosome, and extracellular space. A gene related with extracellular space, CXCL14, was validated to show significantly higher expression in adenocarcinoma with MPP and was thus identified as a possible biomarker of this subtype.
CXCL14, also called BRAK, was first identified to be expressed in the breast and kidney, and shown to be ubiquitously expressed in almost all cell types, especially epithelial cells, but not in many tumor cell lines in vitro. 22 , 23 In colorectal, breast, papillary thyroid cancer, and osteosarcoma, high expression of CXCL14 reportedly associated with metastasis and poor prognosis. 24 , 25 , 26 , 27 Shaykhiev et al 28 reported that clinical lung adenocarcinoma with high expression of CXCL14 had a poor prognosis. In this study, high staining score of CXCL14 significantly correlated with worse overall survival in lung adenocarcinoma (P = .01) (Figure 4D). Pathological stages also significantly correlated with worse prognosis (P = .01) (Figure 4E), and significant association was confirmed between high staining score of CXCL14 and advanced pathological stage (P < .05) (Table S4). We therefore excluded pathological stage from clinicopathologic factors in the univariate and multivariate analyses (Table 2), where only CXCL14 staining score was considered as a predictor of prognosis. In the analysis of stage I‐III patients who were eligible for surgery, high staining score of CXCL14 significantly correlated with poorer prognosis (Figure 4F). Although further study should be undertaken to validate this result using more lung adenocarcinoma samples, it is suggested that CXCL14 expression might be used to predict cases with poor prognosis.
Reportedly, CXCL14 bound to glycoproteins harboring heparan sulfate proteoglycans and sialic acids, leading to the proliferation and migration of cancer cells. 29 In contrast, CXCL14 expression also reportedly suppressed tumor growth by CXCL14‐mediated antitumor CD8+ T cell response in human papillomavirus‐related head and neck cancer, 23 , 30 , 31 and CXCL14 expression was a good prognostic factor contributing to decreased metastasis. 32 , 33 These reciprocal features of CXCL14 expression in cancer, tumorigenic or antitumorigenic, might depend on cancer cell types; the present study a showed significant correlation between high CXCL14 expression and lung adenocarcinoma with MPP. Although MPP was reportedly recognized as a poor prognostic factor 2 , 3 , 4 , 5 , 6 and generally tended to show poorer prognosis in our cohort (Table 1), significant difference was not observed in the Kaplan‐Meier analysis of overall survival (P = .08). This might be influenced by the relatively small sample size in our study (n = 52), whereas more than 120 cases were analyzed in the reported studies.
Cigarette smoke extract reportedly induced CXCL14 expression in the differentiated airway epithelium, and epidermal growth factor also mediated the upregulation of CXCL14. 28 Both SCLC and SqCC, which are known to be related to smoking in cancer development, showed high smoking index and high CXCL14 protein expression (Figure 4A,B). The MPP cases that showed lower smoking index, however, showed higher staining score of CXCL14, suggesting that high CXCL14 expression in adenocarcinoma with MPP could be independent of smoking.
Takiguchi et al 34 established clones of bone‐seeking lung cancer cells, which showed high expression of CXCL14, and enhanced cancer cell tropism to the bone and anchorage‐independent growth and/or recruitment of bone marrow cells. CXCL14 was also reported as a potent inhibitor of in vivo angiogenesis, stimulated by multiple angiogenic factors, eg interleukin‐8, fibroblast growth factor‐2, and vascular endothelial growth factor, to block endothelial cell chemotaxis in vitro. 35 Micropapillary pattern is characterized by a pattern showing micropapillary structures without a fibrovascular core; CXCL14 expression in lung adenocarcinoma cells and the surrounding environments might influence each other, which might contribute to formation of characteristic MPP features. Further studies should be investigated to explore the mechanism of developing MPP features and poor prognosis.
The limitation of this study might be the small number of frozen tissue samples examined by RT‐qPCR, as small as 10 for adenocarcinoma with MPP and 20 for non‐MPP. We evaluated the CXCL14 protein expression by immunohistochemistry using more FFPE tissue samples, including 21 MPP and 31 non‐MPP samples; however, more frozen tissue samples should be additionally analyzed by RT‐qPCR in order to evaluate the usefulness of CXCL14 as a pre‐ or intraoperatively assessed biomarker. Another limitation is that we have not evaluated preoperative biopsy specimens in this study. However, immunohistochemistry indicated that CXCL14 is highly expressed in adenocarcinoma with MPP, not only in the limited areas such as the free tumor clusters and MPP components, but also in other components homogenously or heterogeneously (Figure 3I,J). These scattered expression patterns of CXCL14 in other components could lead to advantage in analysis of biopsy specimens, because biopsy specimens of adenocarcinoma with MPP could show high expression of CXCL14 regardless of the content of the MPP lesions. Further studies to evaluate the usefulness of CXCL14 as a preoperative diagnostic marker should be carried out, by assessing CXCL14 expression and the content of MPP component in preoperative biopsy specimens.
In summary, we undertook a transcriptome analysis to search for biomarkers for lung adenocarcinoma with MPP and identified CXCL14 as a possible diagnostic biomarker of adenocarcinoma with MPP.
DISCLOSURE
All authors have no conflict of interest to disclose.
Supporting information
Supplementary Material
ACKNOWLEDGEMENT
We thank Eriko Ikeda and Haruka Maruyama for technical assistance. This work was supported by the Japan Agency for Medical Research and Development (AMED) (Practical Research for Innovative Cancer Control #19ck0106263h0003), Japan Society for the Promotion of Science (JSPS) (KAKENHI #17K16605), and a grant from Global and Prominent Research, Chiba University (2018‐Y9).
Sata Y, Nakajima T, Fukuyo M, et al. High expression of CXCL14 is a biomarker of lung adenocarcinoma with micropapillary pattern. Cancer Sci. 2020;111:2588–2597. 10.1111/cas.14456
REFERENCES
- 1. Travis WD, Brambilla E, Burke A. WHO classification of the tumours of the lung, pleura, thymus and heart (4th ed.). Lyon, France: IARC Press; 2015. [DOI] [PubMed] [Google Scholar]
- 2. Kawakami T, Nabeshima K, Makimoto Y, et al. Micropapillary pattern and grade of stromal invasion in pT1 adenocarcinoma of the lung: usefulness as prognostic factors. Mod Pathol. 2007;205:514‐521. [DOI] [PubMed] [Google Scholar]
- 3. Warth A, Muley T, Meister M, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage‐independent predictor of survival. J Clin Oncol. 2012;3013:1438‐1446. [DOI] [PubMed] [Google Scholar]
- 4. Zhang J, Wu J, Tan Q, Zhu L, Gao W. Why do pathological stage IA lung adenocarcinomas vary from prognosis?: a clinicopathologic study of 176 patients with pathological stage IA lung adenocarcinoma based on the IASLC/ATS/ERS classification. J Thorac Oncol. 2013;89:1196‐1202. [DOI] [PubMed] [Google Scholar]
- 5. Hung J‐J, Yeh Y‐C, Jeng W‐J, et al. Predictive value of the international association for the study of lung cancer/American Thoracic Society/European Respiratory Society classification of lung adenocarcinoma in tumor recurrence and patient survival. J Clin Oncol. 2014;3222:2357‐2364. [DOI] [PubMed] [Google Scholar]
- 6. Dai C, Xie H, Kadeer X, et al. Relationship of lymph node micrometastasis and micropapillary component and their joint influence on prognosis of patients with stage I lung adenocarcinoma. Am J Surg Pathol. 2017;419:1212‐1220. [DOI] [PubMed] [Google Scholar]
- 7. Morimoto J, Nakajima T, Suzuki H, et al. Impact of free tumor clusters on prognosis after resection of pulmonary adenocarcinoma. J Thorac Cardiovasc Surg. 2016;1521:64‐72.e1. [DOI] [PubMed] [Google Scholar]
- 8. Rodriguez EF, Monaco SE, Dacic S. Cytologic subtyping of lung adenocarcinoma by using the proposed International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society (IASLC/ATS/ERS) adenocarcinoma classification. Cancer Cytopathol. 2013;12111:629‐637. [DOI] [PubMed] [Google Scholar]
- 9. Trejo Bittar HE, Incharoen P, Althouse AD, Dacic S. Accuracy of the IASLC/ATS/ERS histological subtyping of stage I lung adenocarcinoma on intraoperative frozen sections. Mod Pathol. 2015;288:1058‐1063. [DOI] [PubMed] [Google Scholar]
- 10. Yeh YC, Nitadori J, Kadota K, et al. Using frozen section to identify histological patterns in stage I lung adenocarcinoma of </= 3 cm: accuracy and interobserver agreement. Histopathology. 2015;667:922‐938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matsuzawa R, Kirita K, Kuwata T, et al. Factors influencing the concordance of histological subtype diagnosis from biopsy and resected specimens of lung adenocarcinoma. Lung Cancer. 2016;94:1‐6. [DOI] [PubMed] [Google Scholar]
- 12. Huang K‐Y, Ko P‐Z, Yao C‐W, et al. Inaccuracy of lung adenocarcinoma subtyping using preoperative biopsy specimens. J Thorac Cardiovasc Surg. 2017;1541:332‐339.e1. [DOI] [PubMed] [Google Scholar]
- 13. Kaneda A, Kaminishi M, Yanagihara K, Sugimura T, Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res. 2002;6222:6645‐6650. [PubMed] [Google Scholar]
- 14. Nakagawa T, Matsusaka K, Misawa K, et al. Frequent promoter hypermethylation associated with human papillomavirus infection in pharyngeal cancer. Cancer Lett. 2017;407:21‐31. [DOI] [PubMed] [Google Scholar]
- 15. Nitadori J‐I, Bograd AJ, Kadota K, et al. Impact of micropapillary histologic subtype in selecting limited resection vs lobectomy for lung adenocarcinoma of 2cm or smaller. J Natl Cancer Inst. 2013;10516:1212‐1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Warth A, Muley T, Kossakowski CA, et al. Prognostic impact of intra‐alveolar tumor spread in pulmonary adenocarcinoma. Am J Surg Pathol. 2015;396:793‐801. [DOI] [PubMed] [Google Scholar]
- 17. Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;9824:13790‐13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;9824:13784‐13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Collisson EA, Campbell JD, Brooks AN, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi S, Kim HR, Sung CO, et al. Genomic alterations in the RB pathway indicate prognostic outcomes of early‐stage lung adenocarcinoma. Clin Cancer Res. 2015;2111:2613‐2623. [DOI] [PubMed] [Google Scholar]
- 21. Zabeck H, Dienemann H, Hoffmann H, et al. Molecular signatures in IASLC/ATS/ERS classified growth patterns of lung adenocarcinoma. PLoS One. 2018;1310:e0206132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hromas R, Broxmeyer HE, Kim C, et al. Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem Biophys Res Commun. 1999;2553:703‐706. [DOI] [PubMed] [Google Scholar]
- 23. Frederick MJ, Henderson Y, Xu X, et al. In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue. Am J Pathol. 2000;1566:1937‐1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oler G, Camacho CP, Hojaij FC, Michaluart P Jr, Riggins GJ, Cerutti JM. Gene expression profiling of papillary thyroid carcinoma identifies transcripts correlated with BRAF mutational status and lymph node metastasis. Clin Cancer Res. 2008;1415:4735‐4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pelicano H, Lu W, Zhou Y, et al. Mitochondrial dysfunction and reactive oxygen species imbalance promote breast cancer cell motility through a CXCL14‐mediated mechanism. Cancer Res. 2009;696:2375‐2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeng J, Yang X, Cheng L, et al. Chemokine CXCL14 is associated with prognosis in patients with colorectal carcinoma after curative resection. J Transl Med. 2013;11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang W, Yang C, Peng J, Qian Y, Wang Z. The expression of HSPD1, SCUBE3, CXCL14 and its relations with the prognosis in osteosarcoma. Cell Biochem Biophys. 2015;733:763‐768. [DOI] [PubMed] [Google Scholar]
- 28. Shaykhiev R, Sackrowitz R, Fukui T, et al. Smoking‐induced CXCL14 expression in the human airway epithelium links chronic obstructive pulmonary disease to lung cancer. Am J Respir Cell Mol Biol. 2013;493:418‐425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park CR, You D‐J, Kim D‐K, et al. CXCL14 enhances proliferation and migration of NCI‐H460 human lung cancer cells overexpressing the glycoproteins containing heparan sulfate or sialic acid. J Cell Biochem. 2013;1145:1084‐1096. [DOI] [PubMed] [Google Scholar]
- 30. Cicchini L, Westrich JA, Xu T, et al. Suppression of antitumor immune responses by human papillomavirus through epigenetic downregulation of CXCL14. MBio. 2016;7:e00270‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Westrich JA, Vermeer DW, Silva A, et al. CXCL14 suppresses human papillomavirus‐associated head and neck cancer through antigen‐specific CD8(+) T‐cell responses by upregulating MHC‐I expression. Oncogene. 2019;3846:7166‐7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hata R‐I, Izukuri K, Kato Y, et al. Suppressed rate of carcinogenesis and decreases in tumour volume and lung metastasis in CXCL14/BRAK transgenic mice. Sci Rep. 2015;5:9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu J, Wang D, Zhang C, et al. Identification of liver metastasis‐associated genes in human colon carcinoma by mRNA profiling. Chin J Cancer Res. 2018;306:633‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takiguchi S, Korenaga N, Inoue K, et al. Involvement of CXCL14 in osteolytic bone metastasis from lung cancer. Int J Oncol. 2014;444:1316‐1324. [DOI] [PubMed] [Google Scholar]
- 35. Shellenberger TD, Wang M, Gujrati M, et al. BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Res. 2004;6422:8262‐8270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


